Abstract
Any pathogen population sufficiently large is expected to harbor spontaneous drug-resistant mutants, often responsible for disease relapse after antibiotic therapy. It is seldom appreciated, however, that while larger populations harbor more mutants, the abundance distribution of these mutants is expected to be markedly uneven. This is because a larger population size allows early mutants to expand for longer, exacerbating their predominance in the final mutant subpopulation. Here, we investigate the extent to which this reduction in evenness can constrain the genetic diversity of spontaneous drug resistance in bacteria. Combining theory and experiments, we show that even small variations in growth rate between resistant mutants and the wild type result in orders-of-magnitude differences in genetic diversity. Indeed, only a slight fitness advantage for the mutant is enough to keep diversity low and independent of population size. These results have important clinical implications. Genetic diversity at antibiotic resistance loci can determine a population’s capacity to cope with future challenges (i.e., second-line therapy). We thus revealed an unanticipated way in which the fitness effects of antibiotic resistance can affect the evolvability of pathogens surviving a drug-induced bottleneck. This insight will assist in the fight against multidrug-resistant microbes, as well as contribute to theories aimed at predicting cancer evolution.
Keywords: genetic diversity, clonal heterogeneity, antibiotic resistance cost, population size, mutation
NATURAL populations often experience stressful conditions that can drastically reduce their numbers. Well-documented examples include exposure to novel predators (Schoener et al. 2001), competition for essential resources (Petren and Case 1996), or changes in the physicochemical properties of the habitat (Harvey and Jackson 1995). Declining populations can avoid extinction through different mechanisms, depending on several genetic, demographical, and ecological factors whose relative contributions have long been debated (Gomulkiewicz and Holt 1995; Orr and Unckless 2008; Carlson et al. 2014). When the stress is too sudden and severe, adaptation through mutation or migration may not be possible; thus, a population’s survival chances would rely on its genetic variation. This scenario is especially relevant in the context of antimicrobial and antitumoral chemotherapy, where a substantial number of treatments fail due to the existence of resistant cells at the time of the first drug administration (Komarova and Wodarz 2005; García 2009).
The probability that a bacterial population contains a subpopulation of resistant cells is a classical question in genetics, dating back to the seminal work of Luria and Delbrück (1943). They showed that the final number of resistant cells in a growing population is highly fluctuating, as expected under the hypothesis that mutations are spontaneous and not caused by the selective agent. This fluctuation stems from the fact that each mutation can give rise to not just one cell, but also to a clone of cells whose size depends on the timing of the mutational event. Apart from settling a controversy over one of the basic tenets of modern evolutionary theory, the Luria–Delbrück experiment became established as the standard method to estimate mutation rates (Foster 1999). For these reasons, a variety of alternate formulations, extensions, and analyses of their basic model have been proposed over the decades (Zheng 1999, 2015; Foster 2006).
While the determinants of the size of a resistant subpopulation are relatively well understood (Lipsitch and Levin 1997), less attention has been paid to the determinants of genetic diversity. As a consequence of rebounding from a fraction of the original population, the surviving population after a drug-induced bottleneck is expected to display low genetic diversity. Recovery from this state will take a variable amount of time, depending on the rate of introduction of new alleles. This opens a window of time during which the new population may be less able to cope with a second stressful challenge (Willi et al. 2006). Studying this issue is thus clinically relevant because genetic diversity will influence the immediate evolution of resistant populations, including response to second-line therapy, evasion of the immune response, or host recolonization (Lipsitch and Levin 1997; Woolhouse et al. 2001; Tenaillon et al. 2010). Indeed, in the related field of cancer evolution, consideration of genetic diversity within premalignant lesions and tumors is becoming increasingly recognized as key to improving diagnosis, prognosis, and the choice of optimal treatment strategies (Maley et al. 2006; Park et al. 2010; Saunders et al. 2012).
At first glance, it seems trivial that genetic diversity is mainly determined by population size. Certainly, in an asexually growing population, the total number of mutant clones is roughly equal to the product of population size and mutation rate. However, diversity has long been recognized to depend not only on the number but also on the relative abundance of classes, a concept originally introduced into the ecological debate by Simpson (1949). In his classic article, Simpson asked whether a population with five equally abundant species should be considered as diverse as a population with the same five species, one of which comprises 95% of the individuals. Simpson reasoned that the probability of sampling two individuals of the same species is obviously greater in the latter case, leading him to establish the distinction between richness (in our context, the number of clones in a population) and evenness (the distribution of individuals across clones). For a given a number of clones, the maximum diversity is attained when all of them are equally abundant (Pielou 1966).
The present work was prompted by the realization that, in a bacterial population, the growth of clones appearing at different times will generate a very unequal distribution of clone sizes (see Figure 1). This is because an increase in population size allows early mutants to expand for longer, exacerbating the differences among the sizes of the first and last clones to appear. As a result, although larger populations will exhibit greater richness, they will also exhibit lower evenness. A natural question then arises: To what extent can the inequality introduced by clonal growth reduce genetic diversity in expanding populations?
Figure 1.
Clonal growth renders mutant populations increasingly unequal. Large enough bacterial populations produce new mutants each generation. The number of mutants, however, grows due to both new mutations and the expansion of preexistent ones. Typical outcomes of this process are illustrated. Histograms represent clone size distributions under different conditions (clones are ordered according to decreasing size). Pie charts provide a visual indication of the probability of randomly sampling two mutants from the same clone. (A–D) Four successive generations of an idealized population in which mutations occur in a deterministic way, strictly proportional to population size. In the fourth generation (D), the oldest clone is eight times more abundant than any of the eight clones created last: the clone size distribution became richer, but less even. The effect is exacerbated when mutations occur stochastically, since earlier-than-average mutations are allowed to expand for longer (E, the first mutant occurred two generations earlier than in A–D). Evenness can also be reduced if mutants grow faster than their wild-type counterpart (F, mutants grow four times faster than in A–D). In this work, we sought to understand how all these factors determine genetic diversity in expanding bacterial populations.
In this article, we present a simple mathematical model that describes the change in genetic diversity (in the resistant subpopulation) as a function of clonal growth and population size. The analytical solutions show that when mutants grow at a rate equal to or slower than that of the parental strain, genetic diversity is directly proportional to the population size. However, when mutants grow faster, the loss in evenness outweighs the contribution of de novo mutations in increasing genetic diversity. Interestingly, in such cases the correlation between diversity and population size disappears in a few generations. This means that no matter how large a population grows, diversity will remain constantly low. These predictions were extended using a more realistic stochastic simulation model and confirmed by manipulating size and the mutant’s growth rate in experimental populations of the bacterium Pseudomonas aeruginosa.
Materials and Methods
Computer simulations
To produce the simulation results presented here, two basic algorithms were employed. The first one produces distributions of clone sizes by simulating the growth of an asexual population and is described as follows. At the start of each simulation, a matrix where the population data are to be stored is allocated according to the expected number of clones (∼Ntμ). Simulations begin with a single wild-type cell. Each generation, the wild-type population doubles its size and mutations are calculated using a Poisson-distributed pseudorandom number generator (the function rpois in R, where the mutation rate per cell per division acts as the parameter of the Poisson distribution). This number of mutations is subtracted from the wild-type population size, and an equivalent number of clones are initialized. Slots in the population matrix are assigned based on the order of occurrence. Clones produce exactly r offspring per generation, and back mutations and cell death are neglected. All mutants are detected at the end of the run with 100% efficiency.
The second algorithm utilizes the resulting clone size distributions to simulate the random sampling, without reposition, of two mutants per population. Briefly, the population matrix (excluding the wild type) is normalized so that each clone occupies a range proportional to its size within the interval [0, 1]. The first mutant is chosen by drawing a uniformly distributed pseudorandom number (the function runif in R). After recording the clone to which it belongs, the size of its clone is adjusted and the population matrix is renormalized. The second mutant is chosen following the same procedure as above.
To compare the experimental results with those predicted by the simulation model, we ran the programs using the parameter values estimated from the experiments (see next section). In all regimes, the mutation rate was set to 1.19 × 10−7. When mimicking the no-antibiotic regimes, the mutants grew at the same rate as the parental type. The regimes with antibiotic were simulated by adjusting the mutant’s growth rate (r) to 5.4 offspring per generation. Simulations ended when the population size reached the following values: 3.4 × 107 (25 generations) and 2.7 × 1011 (38 generations) for small and large size, respectively, in the no-antibiotic regimes and 1.7 × 107 (24 generations) and 1.4 × 1011 (37 generations) for small and large size, respectively, in the regimes with antibiotic. For each of the four regimes, we computed the expected frequency of cases, from 12 experiments, where two randomly sampled mutants belong to the same clone. These frequencies were obtained for 1000 replicates. All programming was carried out using the R statistical programming language (R Development Core Team 2013) and basic codes are freely available at https://github.com/ACouce/Genetics2016.
Experimental system
All experiments were conducted using P. aeruginosa strain PA14, kindly provided by Frederick M. Ausubel (Rahme et al. 1995). Strains are available upon request. Populations were initiated with ∼103 cells from overnight cultures. Incubation at 37° with vigorous shaking was carried out in Erlenmeyer flasks (50 ml) with 10 ml of lysogeny broth (LB) medium. Incubation times varied between the small-population and the large-population regimes and were optimized to yield ∼107 and ∼1011 cells, respectively. Population sizes were estimated by plating appropriate aliquots onto LB agar. These values were (mean ± SD of 12 experiments) as follows: Nfinal = 2.8 × 107 ± 7.2 × 106 for the small-population no-antibiotic regime (6 hr of incubation), Nfinal = 2.3 × 1011 ± 3.7 × 1010 for the large-population no-antibiotic regime (16 hr of incubation), Nfinal = 1.6 × 107 ± 4.2 × 106 for the small-population with fosfomycin regime (9.5 hr of incubation), and Nfinal = 1.1 × 1011 ± 3.4 × 1010 for the large-population with fosfomycin regime (17 hr of incubation). Resistant mutants were selected by plating onto LB agar supplemented with 128 mg/liter fosfomycin. Loss of GlpT transporter activity is the only mechanism that provides resistance at this concentration in this bacterium, and it is known to be cost-free under standard laboratory conditions (Castañeda-García et al. 2009). However, we determined that in the presence of 8 mg/liter of fosfomycin (1/2 × Minimal Inhibitory Concentration (MIC) for the wild type, estimated by the microdilution method), knocking out glpT confers a growth rate 2.7 ± 0.4 times greater than the wild type’s, which, in terms of our model, is equal to r = 5.4 ± 0.8 offspring per generation (values represent mean ± SD). Growth rates were estimated in triplicate as the maximum slope of the logarithm of the optical density vs. time (Supplemental Material, Figure S1).
To calculate the mutation rate to fosfomycin resistance, a fluctuation test with 12 independent cultures was conducted. The Ma-Sandri-Sarkar Maximum Likelihood (MSS-ML) method (Sarkar et al. 1992), implemented in a custom-made program (Couce and Blázquez 2011), was used to yield an estimate of μ = 1.19 × 10−7 and a 95% confidence interval of (1.85 × 10−7–0.65 × 10−7). To characterize the genetic diversity within the glpT locus, two independent colonies per population were picked at random by proximity to arbitrary points. Their glpT locus was then PCR amplified and subjected to Sanger sequencing. Both amplification and sequencing were performed with oligonucleotides 5′-ACG AAG GCG GCG AGT ATT GC-3′ and 5′-CCT GTC GAG CCT GCA TGT GTA TG-3′. Sequence curation and alignment were performed with the freely available Ridom TraceEdit (www.ridom.de/traceedit) and MAFFT v6 (mafft.cbrc.jp/alignment/software) programs.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results and Discussion
Genetic diversity is highly sensitive to the fitness effect of resistance
Here we consider an idealized bacterial population growing exponentially by binary fission from a single, drug-sensitive cell. The population grows unrestricted and accumulates drug-resistant cells during a number of generations before the sudden occurrence of a lethal selection with antibiotic. We confine our analysis to the genetic diversity of drug-resistant mutations in the drug-resistant subpopulation. To this end, we introduce a haploid, one-locus, infinite-alleles model (see Figure 2). We make the simplifying assumption that every mutational event gives rise to a unique resistance allele and that all alleles are equivalent in terms of fitness. For further simplicity, both reproduction and mutation are treated deterministically, back mutation and cell death are neglected, and generations are assumed to be discrete. Later on, we will use computer simulation to examine the consequences of relaxing some of these strong assumptions.
Figure 2.
A simple model of the accumulation of mutants in an asexual population. Here we consider a mutant subpopulation emerging within a much larger wild-type population (not shown). For simplicity, mutation and growth are treated deterministically: each generation the wild-type population doubles its size and produces twice as many new mutants, whereas preexistent mutants produce exactly r offspring (indicated with an arrow in the diagram). These assumptions will be relaxed later in the computer simulation models. Generation count starts at 0, when the wild-type population reaches a size of N0 = 1/μ individuals, where μ is the per-generation mutation rate. This period is referred to in the literature as the Luria–Delbrück period. General formulas for the size of clones from generation t’ at time t (st(t’)), number of mutants (mt), and number of clones (ct) are shown. Note how the clonal distribution becomes increasingly uneven provided that clones undergo some growth (r > 1).
The process starts at time t = 0 when the wild-type, drug-sensitive population reaches a size of exactly N0 = 1/μ, where μ denotes the mutation rate per generation at which new mutant cells are produced [that is, we consider only the so-called Luria–Delbrück period (Rosche and Foster 2000; Refsland and Livingston 2005; Pope et al. 2008)]. Since N0μ = 1, the first resistant mutant, and therefore the first clone, appears when t = 0. To clarify nomenclature, we define “clone” as the set of genetically identical cells derived from a single mutational event. Each generation the wild-type population doubles its size, giving rise to Ntμ = 2tN0μ = 2t new clones, whereas the preexistent clones increase their size by a factor of r, the mutant’s growth rate [interpreted here as the average number of offspring per generation, i.e., the Wrightian fitness (Wu et al. 2013)]. We consider values of the mutant’s growth rate larger or larger or smaller than r = 2 (that of the wild type) motivated by the attention that the fitness effect of resistance has received over the past decade (Andersson and Hughes 2010; Melnyk et al. 2015). While resistance mutations typically impair growth to a certain extent (Schrag et al. 1997; Reynolds 2000), some can be advantageous even in the absence of the drug (Luo et al. 2005; Marcusson et al. 2009; Vickers et al. 2009; Baker et al. 2013; Miskinyte and Gordo 2013; Rodríguez-Verdugo et al. 2013). In addition, there is a growing concern regarding the selection for resistance under the nonlethal antibiotic concentrations commonly found in many clinical and natural environments (Gullberg et al. 2011; Andersson and Hughes 2014; Larsson 2014; Johnning et al. 2015).
According to the assumptions stated above, the distribution of mutants among clones will follow a geometric sequence; that is, the clone arising in the first generation will be r times more abundant than those appearing in the second generation, which in turn will be r times more abundant than those of the third generation, and in general the clones of generation t will be rt’−t times more abundant than those of generation t’. The number of mutants (mt) in the tth generation is given by
| (1a) |
When r = 2, this expression simplifies to
| (1b) |
When r ≠ 2, however, we need to use the formula for the geometric series to arrive at the following solution:
| (1c) |
As a proxy for genetic diversity we use the probability Pt of sampling, without replacement, two mutants of the same clone. This probability is usually referred to in the ecological literature as Simpson’s index (Gregorius and Gillet 2008), and it is commonly applied to describe intratumor heterogeneity in the field of cancer evolution (Maley et al. 2006; Durrett et al. 2011; Iwasa and Michor 2011; Gatenby et al. 2014) [note that this metric is conceptually similar to the probability of identity-by-descent used in classical population genetics (Malécot and Blaringhem 1948)]. When t = 0, Simpson’s index is not defined, since there is only one mutant in the whole population. When t = 1, it can be calculated by applying Laplace’s rule as
| (2) |
We decided not to further simplify this expression, since it will be useful for inferring the general one. For the second generation, we can write
| (3) |
Hence the general formula can be written as
| (4) |
A glance at numerical solutions of this equation (Figure 3A) reveals a strong nonlinear dependence of genetic diversity on the mutant growth rate. In the following, we derive approximate analytical solutions when t → ∞ for the cases r = 2, r > 2, and r < 2.
Figure 3.
Genetic diversity as a function of total population size. Values represent the probability, Pt, of randomly sampling two mutants from the same clone against the number of generations, tLD, in the Luria–Delbrück period (i.e., after a population size of N = 1/μ is reached). (A and B) Results from the deterministic analytical model (A) and the stochastic computational model (B). Lines correspond, from top to bottom, to the following mutant growth rates: r = 5, r = 3, r = 2 (same as wild type), r = 1.6, and r = 1.2. In this work, we show analytically that diversity increases with size unless mutants grow faster than the wild type, in which case it converges to a constantly low value. Allowing for stochasticity in the mutational timing reduces diversity mainly due to the contribution of rare jackpot events, cases where the final mutant population is flooded by the members of an earlier-than-average clone. However, this effect is only quantitative: the threshold that marks the loss of correlation between diversity and population size remains the same (r > 2). (C) Equilibrium values of Simpson’s index calculated by the approximate solution represented by Equation 7 (open squares), the exact computer solution of expression (4) (shaded diamonds), or the stochastic computational model (circles with dark shading). In all cases, the expected value for r ≤ 2 is zero, which implies that slight differences in growth rate between resistant mutants and the wild type translate into orders-of-magnitude differences in genetic diversity (note the logarithmic scale).
As a general approach, when t → ∞ and r ≠ 2 we will proceed by taking into account only the behavior of the highest exponential term of the numerator of expression (4), which is expected to dominate its value under such circumstances. In the case of r > 2, the dominant term will be that with the highest power of r. This term is the first one in the summation (i = 0). Using Equation 1c, Pt can then be written as
| (5) |
Rearranging and simplifying, we obtain
| (6) |
Thus P∞ can be approximated as
| (7) |
The good agreement between this result and the exact computer solution of expression (4) can be observed in Figure 3C.
When t → ∞ and r < 2, the dominant term will be that with the highest power of 2. This corresponds to the last term of the numerator of expression (4). Recalling Equation 1c, Pt becomes
| (8a) |
which further simplifies to
| (8b) |
and therefore P∞ → 0.
When r = 2, we can no longer use the same approach as above. However, from Equation 1b, Pt can be written as
| (9a) |
Rearranging the numerator yields a summation that can be shown, using the formula from the geometric series (see Appendix), to converge exactly to 2; so, we have
| (9b) |
and therefore P∞ → 0.
These results are amenable to a very intuitive interpretation. As discussed above, clonal diversity is controlled by two opposing forces: mutation and clonal growth. The former increases diversity by increasing the richness of the clonal distribution, whereas the latter reduces it by increasing its inequality. The magnitude of mutation scales with powers of 2, whereas the magnitude of clonal growth scales with powers of r. When r ≤ 2, mutation is more influential than clonal expansion, and thus diversity increases with each generation (i.e., the probability of sampling two siblings tends to zero). On the contrary, with values of r > 2, clonal growth is able to counterbalance the action of mutation, producing the equilibrium value shown in expression (7). In this respect, it is worth noting that these results are readily generalizable to any other biologically relevant values of the wild type’s growth rate, here arbitrarily set to 2 for simplicity.
The impact of “jackpot” events on genetic diversity
The analytical model helped us to understand how the dynamic balance between mutation and clonal growth determines genetic diversity in the resistant subpopulation. However, the assumption that mutation is deterministic, albeit convenient for tractability, is clearly unnatural and could introduce some bias in the model’s predictions. Specifically, by imposing that the first mutation emerges when the size of the wild-type population satisfies N0 = 1/μ, the model actually places an upper limit on the size of the most abundant clone. The model thus neglects the contribution of the rare but notorious jackpot events: populations filled with mutants due to the occurrence of the first mutation very early on in the growth of the culture. In such cases diversity is expected to be particularly low, because the first clone represents the vast majority of the final mutant population (thus increasing Pt). It is likely, then, that the model is overestimating genetic diversity. To study how this simplification affects the main analytical findings, we developed a more realistic computer simulation model that treats mutations as stochastic events (see Materials and Methods).
The simulation results indeed show that the inclusion of jackpot events has a significant impact on reducing genetic diversity (Figure 3B). In particular, compared with the analytical model, the probability of sampling two mutants from the same clone increases uniformly across the explored range of parameters. However, the threshold that marks the change in the dynamics of the system (r ≤ 2) does not seem to be affected by jackpot events, and therefore the qualitative behavior remains the same: diversity in the resistant subpopulation increases with size unless mutants grow faster than the wild type, in which case it converges to a constantly low value. This invariance of the threshold value could have been anticipated to some extent. When r = 2, the analytical model shows that, in each generation, the decrease in genetic diversity due to the growth of preexistent clones exceeds its increase due to novel mutations. This dynamic relationship between the two opposing forces is by no means affected by an early appearance of the first mutation, and hence the critical value of r that marks the transition between the two regimes remains unchanged.
The impact of other biologically relevant features on genetic diversity
Beyond the stochasticity of mutation, a number of extensions of our basic model are possible. At least three merit brief consideration here due to their relevance to the biology of antibiotic resistance. First, resistance mutations can display phenotypic lag, which stems from delays in the synthesis of functional products or the turnover of sensitive molecules (Newcombe 1948). If more than one generation elapses between the occurrence of a resistance mutation and its phenotypic manifestation, the clones generated last will not be available for sampling, thus diminishing genetic diversity. Figure 4 shows that this effect is important only in small populations. This is explained by the fact that, as long as r > 1, the fraction of resistant cells accounted for by last-generation clones decreases with population size. On a related note, this is the same reason why including cell death, which leads to the stochastic loss of small clones, reduces diversity (increases Pt) mainly at low population sizes (see Figure S2).
Figure 4.
The effect of phenotypic lag on genetic diversity. Values represent the probability, Pt, of randomly sampling two mutants from the same clone against the number of generations, tLD, in the Luria–Delbrück period (i.e., after a population size of N = 1/μ is reached). Lines correspond, from top to bottom, to the following mutant growth rates: r = 5, r = 3, r = 2 (same as wild type), r = 1.6, and r = 1.2. (A and B) Results from the stochastic computational model without phenotypic lag (A) and with a phenotypic lag of one generation (B). Phenotypic lag reduces diversity because it prevents last-generation clones from being sampled (thus increasing Pt). Nonetheless, this effect is relevant only at small population sizes, because last-generation clones account for a decreasing fraction of resistant cells in larger populations (as long as r > 1). C highlights this phenomenon by plotting the difference between the corresponding values from A and B. Note that, to aid visualization, all y-axes are zoomed in with respect to Figure 3.
Second, so far we have considered an infinite-alleles model. However, in most situations of interest the number of different resistance alleles is in the order of tens to hundreds (Garibyan et al. 2003; Nilsson et al. 2003; Schenk et al. 2012; Monti et al. 2013; Couce et al. 2015). Under such circumstances mutants belonging to independent clones can nevertheless exhibit the same genotype, hence diminishing the amount of genetic diversity that can be effectively observed [although this will not necessarily affect genetic diversity at linked sites (Pennings and Hermisson 2006)]. Figure 5, A–C, shows that the introduction of a finite number of alleles does not have an appreciable impact on the overall dynamics: it only sets a lower limit on the probability of sampling two identical mutants. The effect is thus largely confined to cases where diversity is expected to be the highest (i.e., costly mutations in large populations).
Figure 5.
The impact of finite alleles and variable fitness effects on genetic diversity. Values represent the probability, Pt, of randomly sampling two mutants from the same clone against the number of generations, tLD, in the Luria–Delbrück period (i.e., after a population size of N = 1/μ is reached). Lines correspond, from top to bottom, to the following mutant growth rates: r = 5, r = 3, r = 2 (same as wild type), r = 1.6, and r = 1.2. (A–C) Results from the stochastic computational model when the number of different alleles is limited to 300 (A), 100 (B), or 30 (C). Limiting the number of alleles effectively establishes an upper limit on the maximum observable diversity. The effect is thus generally important only for deleterious mutations at large population sizes. (D–F) Results from the stochastic computational model for different degrees of variability in the mutant’s growth rate. This variability was simulated by randomly drawing from a Gaussian distribution with mean r and the following values of standard deviation: σ = 0.1 (D), σ = 0.3 (E), and σ = 0.5 (F) (see Figure S4 for details on the shape of these distributions). Variability in the mutant’s growth rate reduces genetic diversity, especially at large population sizes. This is because, in larger populations, the mutant’s average growth rate becomes increasingly dominated by that of the fastest-growing mutants.
Third, we explored the consequences of relaxing the assumption that all mutations are equivalent in terms of fitness. Figure 5, D–F, reveals that allowing for variability in the mutant’s growth rate (r) has the general effect of reducing diversity. This is because the mutant subpopulation quickly becomes dominated by the clones with the largest values of r, which increases the effective average growth rate of the mutants with respect to the wild type. As a logical consequence, the reducing effect becomes increasingly significant with larger population sizes. Taken together, these results suggest that the main prediction of our basic model (the high sensitivity of genetic diversity to the fitness effects of resistance) is expected to hold for many real-world scenarios.
Genetic diversity in experimental populations of spontaneous drug-resistant bacteria
Finally, we sought to experimentally validate the prediction that genetic diversity becomes independent of population size when mutants grow faster than the wild type. To this end we set out to empirically estimate, under different conditions, the frequency with which two mutants picked at random belong to the same clone. This was accomplished by characterizing fosfomycin-resistance mutations in large and small populations of the opportunistic pathogen P. aeruginosa.
Fosfomycin resistance in this organism is acquired exclusively through the inactivation of the glycerol-3-phosphate antiporter GlpT (Castañeda-García et al. 2009). Such inactivation can presumably arise from a high variety of mutations in the glpT gene; and so it is reasonable to assume that, within the same population, two mutants displaying the same mutation probably belong to the same clone. It is well known, however, that particular DNA sequences exhibit a greater-than-average propensity to mutate, leading to the concentration of mutations at certain positions called hotspots (Coulondre et al. 1978). Our experiments indeed revealed the presence of several mutational hotspots in the sequence of glpT, which are problematic because they increase the probability that two independent mutants exhibit the same mutation. The most prominent example is the motif GCCATC, repeated twice consecutively starting at base position 211 and whose expansions and contractions represent almost 11% (9/83) of the independent mutations that were initially observed. Other detected hotspots were A916 → C and A1006 → C, although their relative frequency was lower (5/83 and 4/83, respectively). The complications posed by these mutations were resolved by discarding them whenever they appeared in the two samples from the same population and conducting a new experiment.
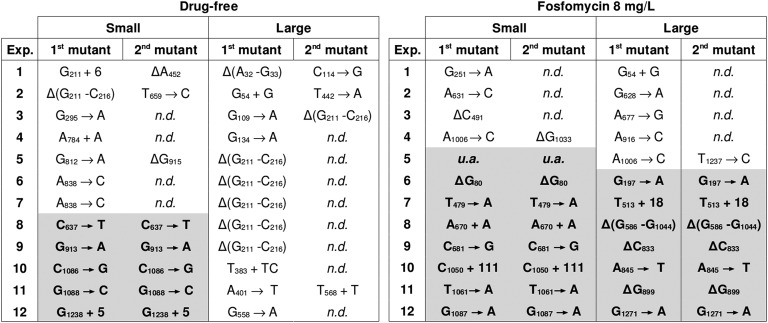
The experimental design comprised the manipulation of population size and the mutant’s relative growth rate (see Materials and Methods). The former was achieved simply through the adjustment of incubation times, and the latter was controlled by means of the presence or absence of sublethal concentrations of fosfomycin (Figure S2). Four experimental regimes were established, involving two population sizes (∼107 vs. ∼1011) and two mutant relative growth rates (r = 1 vs. r = 2.7). For each of the four parameter combinations, 12 replicate cultures were employed. Two fosfomycin-resistant colonies were selected at random from each culture, and their glpT genes were sequenced. The list of identified mutations is presented in Figure 6.
Figure 6.
Mutational spectrum of spontaneous fosfomycin resistance. Sequence data are shown from the 12 replicate populations propagated in each experimental regime. Two colonies per population were randomly selected and their glpT locus was sequenced. Shaded background indicates that the pair shared the same mutation, in which case they were considered to belong to the same clone (see text). Due to the gene’s length (1347 bp), the sequencing was performed from both ends, in two separate rounds. When, in the first run, only one member of a couple was found to carry a mutation, the mutants were assumed to be different and the second mutation was reported as not determined (“n.d.”). In one occasion (experiment 5, small population with antibiotic regime), we were unable to amplify (“u.a.”) the glpT locus in both mutants, which were thus assumed to share the same large deletion or insertion.
In qualitative terms, the experimental results are consistent with the theoretical predictions: genetic diversity increases with size in the absence of antibiotic, whereas it remains low regardless of population size in the presence of the drug. To check whether these results are also satisfactory in quantitative terms, we incorporated the experimental parameters into the simulation model and then computed the expected frequency of cases from 12 experiments where two randomly sampled mutants belong to the same clone (see Materials and Methods). The empirical data also showed a good quantitative agreement with the stochastic model predictions (Figure 7). Of note, this agreement was obtained despite the various sources of experimental error, including the uncertainty in the estimates of mutation rates, population sizes, and growth rates (see Materials and Methods) and the biases introduced by discarding or not detecting hotspot mutations.
Figure 7.
Empirical estimates of genetic diversity in drug-resistant bacterial populations. To validate the theoretical insights, we empirically estimated the probability of randomly sampling two mutants from the same clone. (A) Solid bars represent the experimental data (frequency of matches from 12 replicates), while open bars are the median of 1000 simulated experiments run with appropriate parameter values (error bars indicate interquartile range). Bars are arranged according to the four different experimental regimes as indicated on the lower x-axis. (A) left, results from the no-antibiotic regimes. The mutant’s growth rate is indistinguishable from that of the wild type, and thus diversity increases with population size. A, right, shows results from the regimes with sublethal concentrations of fosfomycin. Since mutants grow faster than the wild type, the loss in evenness outweighs the gain in richness and consequently diversity remains low regardless of population size. (B) Average composition of the replicate populations for each experimental setting (expressed as total colony-forming units). Open bars indicate total population size, while superimposed solid bars represent the resistant subpopulation. Note that, despite huge differences in size and composition (B), both small and large populations from the antibiotic regime exhibit the same level of genetic diversity (A).
Conclusions
The aim of this work was to gain insight into the determinants of genetic diversity of spontaneous drug resistance in bacteria. The topic of genetic diversity in asexuals has received renewed attention during the last decade, spurred by the observation of clonal interference in microbial experimental evolution (de Visser and Rozen 2006; Kosheleva and Desai 2013) and by the recognition of intratumor diversity as a predictor of cancer outcomes (Maley et al. 2006; Durrett et al. 2011). Here we focus on the relevant but still unexplored case of the diversity at antibiotic-resistance loci within a subpopulation of resistant cells. In particular, we asked how the balance between mutation and clonal growth shapes the abundance distribution of resistance alleles after a drug-induced bottleneck. We uncovered the existence of two different dynamical regimes separated by a critical value of the fitness effect of resistance (advantageous vs. neutral or deleterious). As a result, slight differences in growth rate between resistant mutants and the wild type translate into orders-of-magnitude differences in genetic diversity.
The existence of these two regimes is consistent with previous modeling of other biological scenarios. When mutations are neutral (r = 2), the well-known Ewens’ sampling formula from inferential population genetics implicitly predicts that diversity will increase unboundedly with population size (Ewens 1972). This result was shown to hold true for deleterious mutations (r < 2) under different sets of assumptions (Slatkin and Rannala 1997; Wakeley 2008). In turn, the existence of an upper limit to diversity when mutations are advantageous (r > 2) was predicted by Durrett et al. (2011) in their analysis of exponentially expanding tumor cell populations. Interestingly, this observation contrasts with that of Pennings and Hermisson (2006) in the context of soft selective sweeps, where diversity was found to follow Ewens’ sampling formula. The discrepancy presumably arises from their assumption of a constant population size. While richness increases with size both in constant-sized and exponentially expanding populations, evenness exhibits opposing behaviors. In the exponential case, an increase in population size makes early clones larger and late clones more numerous. This exacerbates the dominance of a few early clones over the final population census. In the constant-sized case, however, an increase in size is reflected only in an increase in the number of clones from all generations. As a consequence, dominance becomes spread among a greater number of early clones (see Figure S3) and diversity increases unboundedly with population size.
In practical terms, it is important to emphasize that the effects described in our work do not require unnatural parameter values. Clinical infections display a wide range of bacterial loads, with cases reporting cell densities as high as 109 colony-forming units per gram of sputum (Son et al. 2007) or per milliliter of pus (Hamilton et al. 2006). Resistance mutations typically impair growth due to either the disruption of physiological functions or the imposition of metabolic expenditures (Melnyk et al. 2015). Yet these same mutations will readily confer a growth advantage in the presence of sublethal drug concentrations (MacLean and Buckling 2009; Gullberg et al. 2011). Such conditions are not rare in clinical and agricultural settings, where antibiotic gradients occur naturally in wastewater or inside human and animal body compartments (Baquero and Negri 1997; Kümmerer 2004). In addition, recent reports showed that some resistance mutations can be advantageous in the absence of antibiotics (Luo et al. 2005; Marcusson et al. 2009; Vickers et al. 2009; Baker et al. 2013; Miskinyte and Gordo 2013; Rodríguez-Verdugo et al. 2013). Interestingly, some of these benefits were described to arise as a by-product of adaptation to common circumstances, such as thermal stress (Rodríguez-Verdugo et al. 2013), macrophage phagocytosis (Miskinyte and Gordo 2013), or growth impairment caused by previously acquired resistance mutations (Marcusson et al. 2009).
It is worth considering the relevance of genetic diversity to the subsequent evolution of bacterial populations after a drug-induced bottleneck. The evenness of a population will determine the probability that low-frequency mutants are lost following a random bottleneck, such as in the event of a subsequent intra- or interhost colonization. If the mutants, for example, vary in their capacity to confer resistance to second-line drugs (Marcusson et al. 2009) or tolerance to novel stressors (Rodríguez-Verdugo et al. 2013), this random loss could include the mutants best able to ensure the population’s survival in the future environment. Interestingly, recent insights have revealed that the accumulation of multiple-resistance mutations can be severely constrained by epistatic interactions (Trindade et al. 2009; Salverda et al. 2011). As a consequence, the availability of evolutionary trajectories will generally be reduced if a population is dominated by just one or a few clones. This suggests the intriguing possibility that costly resistance mutations may favor the exploration of multiple-resistance combinations, challenging the notion that clinical practice should give priority to antibiotics for which resistance comes at the highest possible cost (Andersson 2006; Martínez et al. 2007; Perron et al. 2007).
We finally note that our results are also applicable to the study of somatic evolution in cancer (de Bruin et al. 2013). Intratumoral diversity has emerged over the past years as a promising predictor for cancer initiation (Maley et al. 2006), progression (Park et al. 2010), and chemotherapy resistance (Saunders et al. 2012). Since diagnostic biopsies typically sample only a small portion of the lesion, inferences about total diversity rely heavily on the accuracy of available population genetic models (Beerenwinkel et al. 2015). The theoretical literature has largely focused on the dynamics of diversity during the successive sweeps of beneficial “driver” mutations, generally neglecting neutral and deleterious variation not linked to the drivers (Bozic et al. 2010; Durrett et al. 2011; Iwasa and Michor 2011). This nonadaptive variation can become important, however, in the event of a sudden lethal selection, such as in the case of chemoresistance, often associated with a fitness cost (Liang et al. 2008; Silva et al. 2012). We showed that small differences in growth rate between resistant mutants and the wild type lead to widely divergent expectations concerning the genetic diversity that survives a drug-induced bottleneck (Figure 4). This fact, therefore, needs to be taken into account to ensure the optimal choice of diversity-based biomarkers for risk stratification and prognosis (Merlo et al. 2010; Felip and Martinez 2012).
Acknowledgments
We thank F. Cantero for mathematical modeling advice and H. Kemble, J. Poyatos, J. Rodríguez-Beltrán, J. Rolff, O. Makarova, O. Tenaillon, D. Weinreich, J. Wakeley, and two anonymous referees for helpful comments on the manuscript. This work was supported by predoctoral fellowship FI05/00569 to A.C. and grants Red Española de Investigación de Patologías infecciosas (REIPI) RD12/0015/0012 and Fondo de Investigaciones Sanitarias (FIS) PI13/00063 to J.B. from Instituto de Salud Carlos III, Spain (www.isciii.es). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Appendix: Solution to a Geometric Series
Let S be the sum of the terms of a geometric series:
| (A1) |
If x ≠ 1, its value can be easily calculated recalling that
| (A2) |
and therefore
| (A3) |
Footnotes
These authors contributed equally to this work.
Communicating editor: D. M. Weinreich
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.185355/-/DC1.
Literature Cited
- Andersson D. I., 2006. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr. Opin. Microbiol. 9: 461–465. [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D., 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8: 260–271. [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D., 2014. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12: 465–478. [DOI] [PubMed] [Google Scholar]
- Baker S., Duy P. T., Nga T. V. T., Dung T. T. N., Phat V. V., et al. , 2013. Fitness benefits in fluoroquinolone-resistant Salmonella Typhi in the absence of antimicrobial pressure. eLife 2: e01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero F., Negri M. C., 1997. Selective compartments for resistant microorganisms in antibiotic gradients. BioEssays 19: 731–736. [DOI] [PubMed] [Google Scholar]
- Beerenwinkel N., Schwarz R. F., Gerstung M., Markowetz F., 2015. Cancer evolution: mathematical models and computational inference. Syst. Biol. 64: e1–e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic I., Antal T., Ohtsuki H., Carter H., Kim D., et al. , 2010. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. USA 107: 18545–18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson S. M., Cunningham C. J., Westley P. A. H., 2014. Evolutionary rescue in a changing world. Trends Ecol. Evol. 29: 521–530. [DOI] [PubMed] [Google Scholar]
- Castañeda-García A., Rodríguez-Rojas A., Guelfo J. R., Blázquez J., 2009. The glycerol-3-phosphate permease GlpT is the only fosfomycin transporter in Pseudomonas aeruginosa. J. Bacteriol. 191: 6968–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couce A., Blázquez J., 2011. Estimating mutation rates in low-replication experiments. Mutat. Res. Mol. Mech. Mutagen. 714: 26–32. [DOI] [PubMed] [Google Scholar]
- Couce A., Rodríguez-Rojas A., Blázquez J., 2015. Bypass of genetic constraints during mutator evolution to antibiotic resistance. Proc. R. Soc. Lond. B Biol. Sci. 282: 20142698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W., 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274: 775–780. [DOI] [PubMed] [Google Scholar]
- de Bruin E. C., Taylor T. B., Swanton C., 2013. Intra-tumor heterogeneity: lessons from microbial evolution and clinical implications. Genome Med. 5: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J. A. G. M., Rozen D. E., 2006. Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli. Genetics 172: 2093–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett R., Foo J., Leder K., Mayberry J., Michor F., 2011. Intratumor heterogeneity in evolutionary models of tumor progression. Genetics 188: 461–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens W. J., 1972. The sampling theory of selectively neutral alleles. Theor. Popul. Biol. 3: 87–112. [DOI] [PubMed] [Google Scholar]
- Felip E., Martinez P., 2012. Can sensitivity to cytotoxic chemotherapy be predicted by biomarkers? Ann. Oncol. 23: x189–x192. [DOI] [PubMed] [Google Scholar]
- Foster P. L., 1999. Sorting out mutation rates. Proc. Natl. Acad. Sci. USA 96: 7617–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 2006. Methods for determining spontaneous mutation rates. Methods Enzymol. 409: 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M. S., 2009. Early antibiotic treatment failure. Int. J. Antimicrob. Agents 34(Suppl. 3): S14–S19. [DOI] [PubMed] [Google Scholar]
- Garibyan L., Huang T., Kim M., Wolff E., Nguyen A., et al. , 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2: 593–608. [DOI] [PubMed] [Google Scholar]
- Gatenby R. A., Cunningham J. J., Brown J. S., 2014. Evolutionary triage governs fitness in driver and passenger mutations and suggests targeting never mutations. Nat. Commun. 5: 5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomulkiewicz R., Holt R. D., 1995. When does evolution by natural selection prevent extinction? Evolution 49: 201–207. [DOI] [PubMed] [Google Scholar]
- Gregorius H.-R., Gillet E. M., 2008. Generalized Simpson-diversity. Ecol. Modell. 211: 90–96. [Google Scholar]
- Gullberg E., Cao S., Berg O. G., Ilbäck C., Sandegren L., et al. , 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7: e1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A., Robinson C., Sutcliffe I. C., Slater J., Maskell D. J., et al. , 2006. Mutation of the maturase lipoprotein attenuates the virulence of Streptococcus equi to a greater extent than does loss of general lipoprotein lipidation. Infect. Immun. 74: 6907–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey H. H., Jackson D. A., 1995. Acid stress and extinction of a spring-spawning fish population. Water Air Soil Pollut. 85: 383–388. [Google Scholar]
- Iwasa Y., Michor F., 2011. Evolutionary dynamics of intratumor heterogeneity. PLoS One 6: e17866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnning A., Kristiansson E., Fick J., Weijdegård B., Larsson D. G. J., 2015. Resistance mutations in gyrA and parC are common in Escherichia communities of both fluoroquinolone-polluted and uncontaminated aquatic environments. Antimicrob. Resist. Chemother. 6: 1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova N. L., Wodarz D., 2005. Drug resistance in cancer: principles of emergence and prevention. Proc. Natl. Acad. Sci. USA 102: 9714–9719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosheleva K., Desai M. M., 2013. The dynamics of genetic draft in rapidly adapting populations. Genetics 195: 1007–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerer K., 2004. Resistance in the environment. J. Antimicrob. Chemother. 54: 311–320. [DOI] [PubMed] [Google Scholar]
- Larsson D. G. J., 2014. Antibiotics in the environment. Ups. J. Med. Sci. 119: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.-J., Finkel T., Shen D.-W., Yin J.-J., Aszalos A., et al. , 2008. SIRT1 contributes in part to cisplatin resistance in cancer cells by altering mitochondrial metabolism. Mol. Cancer Res. 6: 1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Levin B. R., 1997. The population dynamics of antimicrobial chemotherapy. Antimicrob. Agents Chemother. 41: 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N., Pereira S., Sahin O., Lin J., Huang S., et al. , 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. USA 102: 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M., 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean R. C., Buckling A., 2009. The distribution of fitness effects of beneficial mutations in Pseudomonas aeruginosa. PLoS Genet. 5: e1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malécot G., Blaringhem L., 1948. Les Mathématiques de l’Hérédité. Masson et Cie, Paris. [Google Scholar]
- Maley C. C., Galipeau P. C., Finley J. C., Wongsurawat V. J., Li X., et al. , 2006. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet. 38: 468–473. [DOI] [PubMed] [Google Scholar]
- Marcusson L. L., Frimodt-Møller N., Hughes D., 2009. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 5: e1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez J. L., Baquero F., Andersson D. I., 2007. Predicting antibiotic resistance. Nat. Rev. Microbiol. 5: 958–965. [DOI] [PubMed] [Google Scholar]
- Melnyk A. H., Wong A., Kassen R., 2015. The fitness costs of antibiotic resistance mutations. Evol. Appl. 8: 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo L. M. F., Shah N. A., Li X., Blount P. L., Vaughan T. L., et al. , 2010. A comprehensive survey of clonal diversity measures in Barrett’s esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev. Res. 3: 1388–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskinyte M., Gordo I., 2013. Increased survival of antibiotic-resistant Escherichia coli inside macrophages. Antimicrob. Agents Chemother. 57: 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti M. R., Morero N. R., Miguel V., Argaraña C. E., 2013. nfxB as a novel target for analysis of mutation spectra in Pseudomonas aeruginosa. PLoS One 8: e66236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcombe H. B., 1948. Delayed phenotypic expression of spontaneous mutations in Escherichia coli. Genetics 33: 447–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson A. I., Berg O. G., Aspevall O., Kahlmeter G., Andersson D. I., 2003. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob. Agents Chemother. 47: 2850–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Unckless R. L., 2008. Population extinction and the genetics of adaptation. Am. Nat. 172: 160–169. [DOI] [PubMed] [Google Scholar]
- Park S. Y., Gönen M., Kim H. J., Michor F., Polyak K., 2010. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J. Clin. Invest. 120: 636–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings P. S., Hermisson J., 2006. Soft sweeps II–molecular population genetics of adaptation from recurrent mutation or migration. Mol. Biol. Evol. 23: 1076–1084. [DOI] [PubMed] [Google Scholar]
- Perron G. G., Gonzalez A., Buckling A., 2007. Source–sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proc. Biol. Sci. 274: 2351–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petren K., Case T. J., 1996. An experimental demonstration of exploitation competition in an ongoing invasion. Ecology 77: 118–132. [Google Scholar]
- Pielou E. C., 1966. The measurement of diversity in different types of biological collections. J. Theor. Biol. 13: 131–144. [Google Scholar]
- Pope C. F., O’Sullivan D. M., McHugh T. D., Gillespie S. H., 2008. A practical guide to measuring mutation rates in antibiotic resistance. Antimicrob. Agents Chemother. 52: 1209–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme L. G., Stevens E. J., Wolfort S. F., Shao J., Tompkins R. G., et al. , 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268: 1899–1902. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2013 R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. Available at: http://www.R-project.org.
- Refsland E. W., Livingston D. M., 2005. Interactions among DNA ligase I, the flap endonuclease and proliferating cell nuclear antigen in the expansion and contraction of CAG repeat tracts in yeast. Genetics 171: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds M. G., 2000. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics 156: 1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Verdugo A., Gaut B. S., Tenaillon O., 2013. Evolution of Escherichia coli rifampicin resistance in an antibiotic-free environment during thermal stress. BMC Evol. Biol. 13: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W. A., Foster P. L., 2000. Determining mutation rates in bacterial populations. Methods 20: 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salverda M. L. M., Dellus E., Gorter F. A., Debets A. J. M., van der Oost J., et al. , 2011. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 7: e1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Ma W. T., Sandri G. H., 1992. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica 85: 173–179. [DOI] [PubMed] [Google Scholar]
- Saunders N. A., Simpson F., Thompson E. W., Hill M. M., Endo-Munoz L., et al. , 2012. Role of intratumoural heterogeneity in cancer drug resistance: molecular and clinical perspectives. EMBO Mol. Med. 4: 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk M. F., Szendro I. G., Krug J., de Visser J. A. G. M., 2012. Quantifying the adaptive potential of an antibiotic resistance enzyme. PLoS Genet. 8: e1002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoener T. W., Spiller D. A., Losos J. B., 2001. Predators increase the risk of catastrophic extinction of prey populations. Nature 412: 183–186. [DOI] [PubMed] [Google Scholar]
- Schrag S. J., Perrot V., Levin B. R., 1997. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc. Biol. Sci. 264: 1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. S., Kam Y., Khin Z. P., Minton S. E., Gillies R. J., et al. , 2012. Evolutionary approaches to prolong progression-free survival in breast cancer. Cancer Res. 72: 6362–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E. H., 1949. Measurement of diversity. Nature 163: 688. [Google Scholar]
- Slatkin M., Rannala B., 1997. The sampling distribution of disease-associated alleles. Genetics 147: 1855–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M. S., Matthews W. J., Kang Y., Nguyen D. T., Hoang T. T., 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75: 5313–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Skurnik D., Picard B., Denamur E., 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8: 207–217. [DOI] [PubMed] [Google Scholar]
- Trindade S., Sousa A., Xavier K. B., Dionisio F., Ferreira M. G., et al. , 2009. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 5: e1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers A. A., Potter N. J., Fishwick C. W. G., Chopra I., O’Neill A. J., 2009. Analysis of mutational resistance to trimethoprim in Staphylococcus aureus by genetic and structural modelling techniques. J. Antimicrob. Chemother. 63: 1112–1117. [DOI] [PubMed] [Google Scholar]
- Wakeley J., 2008. Conditional gene genealogies under strong purifying selection. Mol. Biol. Evol. 25: 2615–2626. [DOI] [PubMed] [Google Scholar]
- Willi Y., Buskirk J. V., Hoffmann A. A., 2006. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 37: 433–458. [Google Scholar]
- Woolhouse M. E. J., Taylor L. H., Haydon D. T., 2001. Population biology of multihost pathogens. Science 292: 1109–1112. [DOI] [PubMed] [Google Scholar]
- Wu B., Gokhale C. S., van Veelen M., Wang L., Traulsen A., 2013. Interpretations arising from Wrightian and Malthusian fitness under strong frequency dependent selection. Ecol. Evol. 3: 1276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., 1999. Progress of a half century in the study of the Luria-Delbrück distribution. Math. Biosci. 162: 1–32. [DOI] [PubMed] [Google Scholar]
- Zheng Q., 2015. A new practical guide to the Luria-Delbrück protocol. Mutat. Res. Mol. Mech. Mutagen. 781: 7–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.