Abstract
Copy-number alterations are widespread in animal and plant genomes, but their immediate impact on gene expression is still unclear. In animals, copy-number alterations usually exhibit dosage effects, except for sex chromosomes which tend to be dosage compensated. In plants, genes within small duplications (<100 kb) often exhibit dosage-dependent expression, whereas large duplications (>50 Mb) are more often dosage compensated. However, little or nothing is known about expression in moderately-sized (1–50 Mb) segmental duplications, and about the response of small RNAs to dosage change. Here, we compared maize (Zea mays) plants with two, three, and four doses of a 14.6-Mb segment of chromosome 1 that contains ∼300 genes. Plants containing the duplicated segment exhibit dosage-dependent effects on ear length and flowering time. Transcriptome analyses using GeneChip and RNA-sequencing methods indicate that most expressed genes and unique small RNAs within the duplicated segments exhibit dosage-dependent transcript levels. We conclude that dosage effect is the predominant regulatory response for both genes and unique small RNA transcripts in the segmental dosage series we tested. To our knowledge this is the first analysis of small RNA expression in plant gene dosage variants. Because segmental duplications comprise a significant proportion of eukaryotic genomes, these findings provide important new insight into the regulation of genes and small RNAs in response to dosage changes.
Keywords: dosage effect, copy-number alterations, phenotypic variations, small RNAs, maize
DNA copy-number alterations, whether involving chromosome segments or entire chromosomes, can have dramatic phenotypic impacts. Aneuploidy, one kind of DNA copy-number alteration (Birchler 2013), results from changes of chromosome number, i.e., gain or loss of one or more entire chromosome(s). In most animals aneuploidy is detrimental and may cause severe genetic disorders. In humans, the gain of a single extra copy of chromosome 21 (trisomy 21) causes Down syndrome; most other aneuploidies result in severe developmental disorders and do not survive to term. For instance, most spontaneous abortions and developmental abnormalities in humans are caused by aneuploidy (Hassold and Hunt 2001). In addition, many human cancer cells are highly aneuploid, although the mutual causality of aneuploidy and tumorigenesis is still unclear (Weaver and Cleveland 2007). However, aneuploidy in plants is typically far less detrimental. Trisomics for each of the chromosomes have been recovered in several plant species, and even monosomics for all of the 10 maize chromosomes have been recovered and are viable (Weber 1994).
In contrast to the severe effects of aneuploidy, other copy-number alterations such as segmental aneuploidy and copy-number variation (CNV) often have milder phenotypic effects, which may facilitate their retention and possible accumulation in a population. CNV, involving segments of 1 kb to 1 Mb in size (Birchler 2013), is widespread across many species including mammals (Iafrate et al. 2004; Sebat et al. 2004; Redon et al. 2006) and major crop plants (Springer et al. 2009; Lai et al. 2010; Swanson-Wagner et al. 2010; Saintenac et al. 2011; P. Yu et al. 2011; Zheng et al. 2011; Jiao et al. 2012; McHale et al. 2012). For example, >10% of the human genome is composed of CNVs and segmental duplications with sizes ranging from a few kb to several Mb (Iafrate et al. 2004; Sebat et al. 2004; Redon et al. 2006; Stankiewicz and Lupski 2010). In rice, tandem-arrayed genes account for up to 20% of the duplicated genes (Rizzon et al. 2006).
In addition to the effects of CNV and segmental duplications on creating genomic diversity, CNV and segmental duplications in eukaryotes can dramatically affect organismal phenotype. For example, gain or loss of one copy of human 1q21.1, a genomic region associated with developmental abnormalities (O’Donovan et al. 2008), confers a higher risk of mental disorders (Stefansson et al. 2008, 2009). In addition, microdeletion or microduplication of human 16p11.2 has an association with autism (Weiss et al. 2008). CNV can be advantageous as well. For instance, changes in copy number of an amylase gene (AMY2B) greatly facilitated the adaptation of dogs to starch-rich diets associated with domestication (Axelsson et al. 2013), and increased copy number of the human AMY1 gene may aid the digestion of starchy foods (Perry et al. 2007). In plants, several important traits are directly affected by CNV (Xiao et al. 2008; Cook et al. 2012; Diaz et al. 2012; Li et al. 2012; Maron et al. 2013). For example, a recent study in rice showed that a tandem duplication that increases copy number of the GL7 locus has a significant effect on grain length (Wang et al. 2015). However, despite the fact that CNV and segmental duplications are pervasive and often have major effects, the question of precisely how copy-number alterations affect gene expression and contribute to phenotypic diversity remains largely unanswered.
Dosage compensation and dosage sensitivity are two major responses of gene expression to changes in DNA dosage (Guo et al. 1996; Gupta et al. 2006; Birchler 2010; Birchler and Veitia 2012). Several recent studies in humans and plants showed that the change in the expression of genes with altered copy number is a causative factor for the phenotypic impact of CNV (Golzio et al. 2012; Li et al. 2012). Specifically, elevated expression [messenger RNA (mRNA) and/or protein levels] of genes located within the trisomy or segmental trisomy region is largely responsible for the aneuploid syndromes (Kahlem et al. 2004; Vacik et al. 2005; Huettel et al. 2008; Williams et al. 2008; Pavelka et al. 2010; Stingele et al. 2012). These results suggest that genes are expressed in proportion to their dosage. However, expression of most genes located on the X chromosome of humans, Caenorhabditis elegans, or Drosophila is typically dosage compensated, though different mechanisms are involved in maintaining gene-expression balance between males and females in these species. In addition to sex-chromosomal genes, the expression of autosomal genes in Drosophila could also be dosage compensated (Devlin et al. 1982; Devlin et al. 1988; Birchler et al. 1990; Stenberg et al. 2009; Sun et al. 2013a). In maize, the alcohol dehydrogenase (Adh1) gene showed a dosage effect in small segmental aneuploids (Birchler 1981); when tested in larger segmental aneuploids, however, the same Adh1 gene as well as other linked genes exhibited equivalent levels of expression when compared to diploids (Birchler 1979; Guo and Birchler 1994). These results suggest that, in general, the size of the duplicated sequence may affect the regulation of the included genes (Birchler 1981; Birchler and Newton 1981). In addition, genes with altered copy numbers could exert trans effects on the expression of genes whose dosage is constant (Birchler 1979; Rabinow et al. 1991; Guo and Birchler 1994; Xie and Birchler 2012). Dosage effects and trans effects have also been observed in dosage series of variable lengths (Guo and Birchler 1997) and thus the particular response of any region likely depends on the types of genes varied. Maize is one of the most diverse plant species (Buckler et al. 2006), and variation in maize CNVs is prevalent not only among modern inbred lines (Springer et al. 2009; Lai et al. 2010; Jiao et al. 2012), but also between domesticated maize and its wild progenitor, teosinte (Swanson-Wagner et al. 2010; Chia et al. 2012; Hufford et al. 2012). However, less is known about the association between phenotypic divergence and CNV. Besides affecting expression of protein-coding genes, CNV could potentially affect expression of noncoding RNAs which have been recently recognized as having important roles in regulating gene expression and maintaining genomic stability. How small RNAs respond to copy-number alterations is an interesting and as yet unexplored question.
We have shown previously that alternative transposition involving two Ac/Ds transposon termini can induce genomic rearrangements, including inversion, deletion, duplication, and translocation (Zhang and Peterson 1999; Zhang and Peterson 2004; Zhang and Peterson 2005; C. Yu et al. 2010, 2011). We isolated a number of such duplications and deletions, and conducted phenotypic and transcriptional analysis on one case (p1-ww714) with a tandem inverted duplication on chromosome 1S. The region duplicated in p1-ww714 is 14.6 Mb in size and is predicted to contain ∼300 gene models in the maize filtered gene set (ZmB73_5b_FGS). Using this segmental duplication in combination with a normal chromosome 1, we produced sibling plants containing two, three, and four copies of the affected region. We then conducted phenotypic and transcriptional studies on this segmental dosage series. We implemented both high-throughput RNA sequencing [RNA-seq and small RNA-seq (sRNA-seq)] and GeneChip [new Affymetrix Maize whole transcriptome (WT) 100K array] approaches to study the relationship between CNV and transcript accumulation. Our study provides valuable insights into disclosing CNV effects in maize and understanding how gene expression responds to copy-number change.
Materials and Methods
Phenotypic characterization of CNV plants
CNV plants (p1-ww714/p1-ww714, p1-ww714/B73, and B73/B73) were grown in the field season of summer 2012 in Iowa State and in the winter season of 2012 in Chile with three and two replications, respectively. Each genotype was randomly assigned to a four-row plot in every replication. Individual plant heights were measured after flowering as the distance from the surface of the soil to the top of the tassel. Flowering date was recorded for each plot when more than half of its plants were shedding pollen. Ear length was measured as the distance from the bottom to top of the mature cob. A linear mixed-effects model that included fixed effects for genotypes and random effects for replications was used to test for phenotypic differences among genotypes.
Plant growth, tissue collection, and RNA extraction
For mRNA sequencing, sibling seeds of three different genotypes (p1-ww714/p1-ww714, p1-ww714/B73, and B73/B73) were sown in SB 300 Universal Soil and grown in a PGW-40 growth chamber as described by Swanson-Wagner et al. (2006); 15 hr of light and 9 hr of dark; 25° at day time and 20° at night time. Ten days after sowing, individual plants were genotyped by genomic PCR. At 14 days, aboveground tissues of six random plants per genotype per replication were harvested and pooled for preparation of total RNA. Nine pooled maize tissues (three genotypes × three replications per genotype) were ground in liquid nitrogen and total RNA was extracted using RNeasy Plant Mini Kit (QIAGEN, Valencia, CA) as per manufacturer’s instructions. Total RNA was further purified by performing on-column DNase digestion (QIAGEN), and RNA quality and intensity was evaluated by a NanoDrop 1000 spectrophotometer and Agilent 2100 Bioanalyzer.
For sRNA-seq, six plants from each of the four tested genotypes (p1-vv9D9A/p1-vv9D9A, id1/id1, id4/id4, and B73/B73) were grown in the growth chamber in the same conditions described above. Roots and seedlings were harvested at 14 days after sowing. Tissues from the same genotype were pooled together. For the p1-ww714/p1-ww714 and p1-ww714/B73 genotypes, tissue samples were taken from one of the replications used for mRNA sequencing. Total RNA was made from each pooled root and seedling tissue by the PureLink Plant RNA Reagent, followed by DNase treatment (DNase I; New England Biolabs, Beverly, MA).
Quantitative real-time PCR
Plants were grown in the same conditions as described above, with two replications, and were harvested at 14 days after sowing. Each replication included six plants that were later pooled to form a single sample. The methods and reagents used to prepare purified total RNA were the same as described for mRNA sequencing. Total RNA was reverse transcribed to complementary DNA (cDNA) using Omniscript reverse transcription kit (QIAGEN). PCR was performed using SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA), and analyzed on a Stratagene Mx4000 multiplex quantitative PCR system with three technical replicates.
GeneChip hybridization and statistical analysis of GeneChip data
Purified total RNA samples for three biological replicates of each genotype were sent to the Iowa State University GeneChip Facility for labeling and hybridization to new maize 100 format whole transcriptome arrays according to the instructions of GeneChip WT terminal labeling and hybridization user manual (http://www.plexdb.org/modules/PD_general/Maize100WT_description.php). Raw GeneChip data were normalized using the Robust Multi-array Average approach which is implemented in the PLEXdb pipeline (Dash et al. 2012). Differentially-expressed genes (DEGs) were identified by linear model analyses carried out with the R package limma (Smyth 2005). Each gene-specific linear model included effects for genotypes and replications. P-values from the genotype tests were converted to q-values (Nettleton 2006). Genes with a false discovery rate (FDR) of <0.05 were identified as differentially expressed.
mRNA and sRNA-seq and data processing of Illumina reads
cDNA sequencing libraries were constructed by the Iowa State University DNA Facility, and then sequenced on an Illumina HiSequation 2000 to generate 100-bp single-end reads, also at the Iowa State University DNA Facility. Random barcodes were used for each sample and three biological replicates of each genotype were performed and run on three independent lanes. Sequencing reads were trimmed by FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and then aligned to the maize B73 reference genome (RefGen_V2) by Tophat (Trapnell et al. 2009) using default parameters except a maximum intron size of 50,000 bp. Raw read count of each gene model (ZmB73_5b_FGS; http://ftp.maizesequence.org/release-5b/filtered-set/) was computed by HTSeq package (Anders et al. 2015) for uniquely-mapped reads. Only genes having at least 45 mapped reads were considered to be expressed in the sampled tissue. DEGs with FDR <0.05 were identified based on the raw read counts by using QuasiSeq package (Lund et al. 2012).
For sRNA-seq, libraries were constructed and sequenced at the Beijing Genome Institute. Small RNA reads were trimmed by FASTX toolkit and then mapped to maize B73 reference genome (RefGen_V2) by Bowtie (Langmead et al. 2009). The genome was subdivided into 100-kb windows across the 10 maize chromosomes, and the expression of small RNAs was represented by the number of mapped reads per 100-kb window.
Statistical tests of transcript regulation models
The experiment included three replications. Within each replication, one RNA sample was sequenced from each of the three genotypes. For each gene, we compared three generalized linear models, henceforth referred to as M0, M1, and M2 in increasing order of complexity. In each model, the log of the mean read count was assumed to be a linear function of gene-specific parameters plus a sample-specific normalization factor, determined for each sample by the log of the 0.75 quantile of sample-specific read counts (Bullard et al. 2010). The linear function for model M0 included an intercept and replication effects but no genotype effects. The linear function for model M1 included an intercept, replication effects, and a slope coefficient (β1) on the log of the number of copies of the duplicated region (two, three, or four depending on the genotype). The linear function for the most general model (M2) included an intercept as well as replication and unrestricted genotype effects.
Genes for which model M0 is adequate are not differentially expressed across genotypes. Genes that require a more complicated model (either M1 or M2) are differentially expressed across genotypes. When model M1 is adequate relative to model M2, and when the β1 in model M1 is not significantly different from 1, a gene’s expression pattern is consistent with a dosage effect that implies mean expression level is proportional to the number of copies of the duplicated region. To determine which scenario holds for each gene, we performed the following analyses.
Genes with an average of at least one uniquely-mapped read across samples were analyzed using the R package QuasiSeq (http://cran.r-project.org/web/packages/QuasiSeq). The negative binomial QLShrink method implemented in the QuasiSeq package and described by Lund et al. (2012) was used to compute a P-value for each gene and each model comparison (M0 vs. M1, M0 vs. M2, and M1 vs. M2). Using the P-values for each comparison, the approach of Nettleton et al. (2006) was used to estimate the number of genes with true null hypotheses among all genes tested. Using this estimate, q-values were computed from P-values according to the method of Storey (2002). To obtain approximate control of the FDR at 5%, the null hypothesis was rejected for all tests with q-values no larger than 0.05. To assess the plausibility of effects proportional to dosage of the duplicated region, ∼95% C.I.s were constructed for the β1 in model M1. Genes with intervals containing 1 are consistent with effects proportional to dosage.
Data availability
The datasets supporting the conclusions of this article are available in the following repositories. All MIAME-compliant GeneChip profiling data are available as accession ZM50 at the PLEXdb expression resource for plants and plant pathogens (www.plexdb.org). RNA-seq data are deposited as accession GSE71448 at NCBI-GEO and accession SRP061705 in the NCBI’s Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov); sRNA-seq data are deposited as accession GSE71959 at NCBI-GEO and accession SRP062285 in the SRA (http://www.ncbi.nlm.nih.gov).
Results
Identification of a 14.6-Mb tandem inverted duplication on maize chromosome 1
p1-ww714 is an allele of the maize p1 gene that encodes a Myb-like transcription factor, which regulates maize kernel pericarp and cob pigmentation (Grotewold et al. 1994). The p1-ww714 allele was isolated in a screen for inverted duplications derived from the progenitor allele p1-vv9D9A that contains insertions of Ac/fAc transposable elements (TEs). Previous research has shown that the paired Ac/fAc elements in p1-vv9D9A can undergo aberrant transposition reactions termed sister chromatid transposition (SCT) to generate inverted duplications that begin at the p1 gene and extend variable lengths proximally (Zhang and Peterson 1999). The structure of p1-ww714 was tested by PCR of genomic DNA using a series of primers diagnostic for the presence of inverted duplications (Supplemental Material, Figure S1A). These PCR tests confirm that the progenitor p1-vv9D9A structure was disrupted by the presence of an inverted duplication in p1-ww714 (Figure S1B). The SCT model predicts that the duplication endpoint will be adjacent to the 3′ end of the fAc element; therefore we used inverse PCR to isolate this junction sequence and used it in Basic Local Alignment Search Tool searches of the maize reference genome. The endpoint junction is located at position 62.7 Mb on maize chromosome 1, a distance of 14.6 Mb from the p1 locus at position 48.1 Mb. Taken together, these results indicate that p1-ww714 allele contains a ∼14.6-Mb inverted duplication in the short arm of maize chromosome 1 from 48.1 to 62.7 Mb, representing ∼5% of chromosome 1.
Phenotypic impact of the 14.6-Mb duplication
Initial observations found that plants that carry the p1-ww714 allele exhibit significant phenotypic differences compared to their progenitor. To better study the phenotypic effect of this copy-number alteration, the original p1-ww714 allele was backcrossed with the maize inbred B73 for five generations to achieve a near-B73 genetic background. In the succeeding generation, heterozygous p1-ww714/B73 plants were self-pollinated to generate sibling plants of the genotypes B73/B73, p1-ww714/B73, and p1-ww714/p1-ww714; which carry two, three, and four copies of the 14.6 Mb segment, respectively. The sibling plants of BC5F2 were used for transcriptome studies, whereas the subsequent BC5F3 plants were field-grown to maturity at Iowa State University in summer 2012 in a randomized complete-block design with three replications.
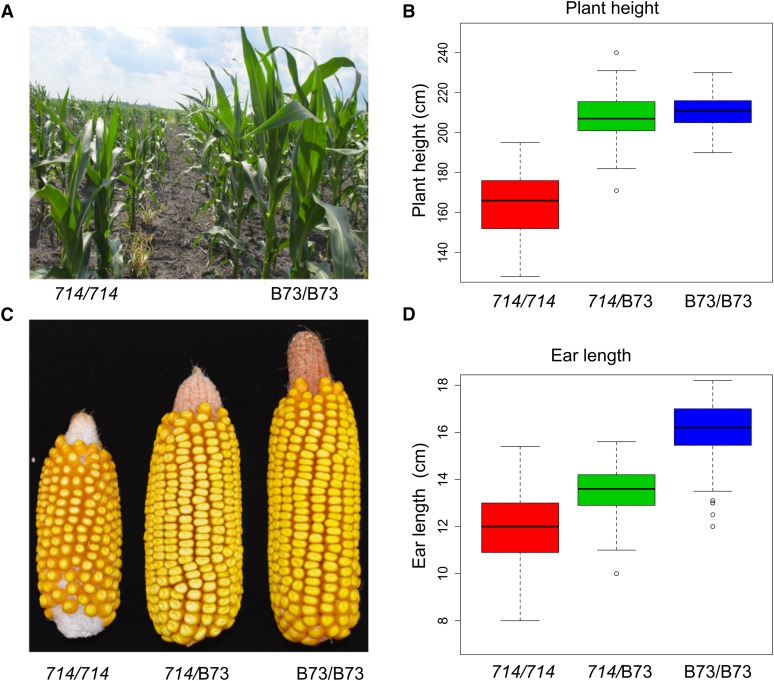
In plants field-grown in Iowa, p1-ww714/p1-ww714 and p1-ww714/B73 genotypes exhibited delayed flowering time compared to that of B73/B73. Homozygous p1-ww714/p1-ww714 plants flowered ∼10 days later than B73/B73, while p1-ww714/B73 heterozygotes flowered ∼6 days later than B73/B73 but 4 days earlier than p1-ww714/p1-ww714 (Table S1). Additional phenotypic impacts on plant height and ear length were observed: homozygous p1-ww714 plants were 46 and 43 cm shorter than homozygous B73 plants and heterozygous p1-ww714/B73 plants, respectively, with a P-value < 0.0001 (Figure 1; Table S1). However, the average heights of p1-ww714/B73 and B73/B73 plants were not significantly different. The average ear length of p1-ww714/p1-ww714 was 4.1 cm shorter than that of sibling B73/B73 (P < 0.0001) and 1.7 cm shorter than that of sibling p1-ww714/B73 (P = 0.0007), while ear length of p1-ww714/B73 was 2.4 cm (P = 0.0001) shorter than that of B73/B73 (Figure 1, C and D; Table S1).
Figure 1.
Effects of copy-number alterations on plant stature and ear size. In three BC5 F3 families, homozygous p1-ww714 plants have shorter stature, are delayed in development (A and B), and have shorter ears (C and D), than the standard maize inbred B73. (C and D) Plants heterozygous for the duplication allele (p1-ww714/B73) are intermediate in ear length.
Phenotypic measurements were repeated using sibling plants field-grown during 2012 winter nursery in Rancagua, Chile with two replications. Similarly, homozygous p1-ww714 plants were significantly shorter than B73/B73 and p1-ww714/B73 plants; while ears of p1-ww714/p1-ww714 were on average 3.9 cm (P = 0.0061) and 2.2 cm (P = 0.0159) shorter than that of sibling B73/B73 and p1-ww714/B73, respectively (Table S1). The average height of p1-ww714/B73 plants was not significantly different from that of B73/B73. In summary, homozygous p1-ww714/p1-ww714 plants exhibited delayed development, shorter plant stature, and shorter ear length. Whereas, heterozygous p1-ww714/B73 plants were intermediate for flowering time and ear length. Finally, ∼15% of homozygous p1-ww714 plants exhibited severe development defects, such as extreme dwarfing, absent or barren tassels, and other morphological aberrations. Because these aberrant plants did not appear to have a common mutant phenotype, we suggest that they may represent the heterogeneous products of genome instability, likely induced by the presence of the large segmental duplication. Whatever the cause of these exceptional plants, they were excluded from the phenotypic and molecular analyses since they were not representative of the majority of homozygous p1-ww714 plants.
Differential transcript abundance in duplication genotypes detected by GeneChip
The phenotypic differences observed among copy-number variants suggest that some differences in transcript levels might be induced by the altered copy number of the 14.6-Mb segment. To study the effects of CNV on gene expression, a new Affymetrix Maize 100K format WT array containing 103,262 probe sets was used to profile global gene expression in the copy-number variants. Nine collected RNA samples from sibling plants of the p1-ww714/p1-ww714, B73/B73, and p1-ww714/B73 genotypes were used to prepare probes for hybridization with the arrays. Hybridization data were processed using the PLEXdb pipeline (Dash et al. 2012). In a preliminary analysis a series of statistical tests were performed to identify DEGs between genotypes, using twofold changes as cutoff and controlling FDR at 5% level (see Materials and Methods). This analysis identified a total of 67 genes differentially expressed between the p1-ww714/p1-ww714 and B73/B73 plant seedling tissues. Eighteen (27%) of these DEGs are located in the 14.6-Mb duplicated region; this is a relatively high proportion considering that this duplication comprises <1% of the maize genome. Among these 18 DEGs, 15 are overexpressed and 3 are underexpressed. For the other 49 DEGs, 15 are located in nonduplicated regions of chromosome 1, and the remaining 34 are from other maize chromosomes. In contrast, comparisons of the GeneChip transcriptional data from heterozygous p1-ww714/B73 samples with either p1-ww714/p1-ww714 or B73/B73 homozygous plants did not identify any genes as differentially expressed.
Interestingly, most genes within the 14.6-Mb duplicated segment exhibited increased expression in the higher copy genotypes, as the log2 fold change distributions of each comparison are clearly centered above zero (Figure S2). Although transcript abundance ratios are somewhat smaller than their corresponding copy-number ratios, they are still positively and proportionally correlated with gene copy number. In contrast, we did not observe any similar regional shifts in transcript levels outside the duplicated segment, either on the same or other chromosomes. These GeneChip data suggest that many transcripts mapping within the duplicated chromosome region are differentially expressed in accordance with the gene dosage in each genotype.
Dosage-dependent expression of genes in duplicated region detected by RNA sequencing
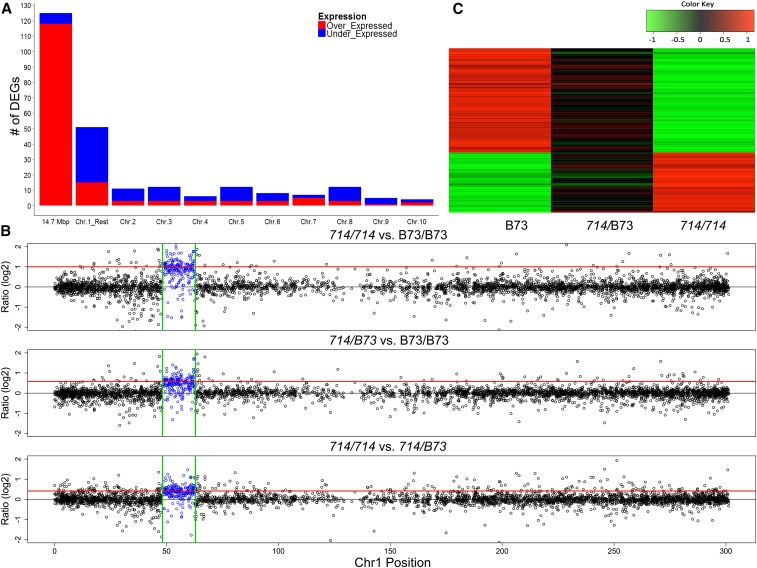
To further examine gene expression in the copy-number variant genotypes, we employed high-throughput RNA-seq of the same 9 RNA samples used for GeneChip. In total, we generated 149.7 million 100 bp raw reads; 76.2% of them were uniquely mapped to the B73 reference genome (RefGen_V2). The number of uniquely-mapped reads per sample ranged from 9.5 to 15.2 million (Table S2). The Pearson correlations of read counts within genotypes were >98%. A series of bioinformatics tools and statistical tests were then applied to analyze the data and identify DEGs (see Materials and Methods). Among 23,730 expressed genes, 253 were differentially expressed between p1-ww714/p1-ww714 and B73/B73 in the 14-day-old seedling tissues: 125 of 253 (49%) DEGs are located within the 14.6-Mb chromosomal duplicated region, 51 DEGs are within the remainder of chromosome 1, and the remaining 77 DEGs are distributed in the other nine chromosomes (Figure 2A). These results indicate that genes within the duplicated segment are clearly overrepresented among all DEGs detected.
Figure 2.
DEGs and dosage-dependent expression pattern revealed by RNA-seq. (A) The distribution of DEGs between homozygous p1-ww714 and B73 plants identified by RNA-seq. Bars indicate the numbers of overexpressed (red) and underexpressed (blue) DEGs identified in the 14.6-Mb duplicated region (first bar), the remainder of chromosome 1 (second bar), and other chromosomes. (B) Ratios of chromosome 1 gene expression among three genotypes as determined by RNA-seq. Log2 fold changes in transcript levels were plotted for pairwise comparisons among p1-ww714/p1-ww714, B73/B73, and p1-ww714/B73 sibling plants. Expression ratios are shown for all genes on chromosome 1. The gene copy-number ratios (log2) of p1-ww714/p1-ww714 vs. B73/B73, p1-ww714/B73 vs. B73/B73, and p1-ww714/p1-ww714 vs. p1-ww714/B73 are 1, 0.585 and 0.415, respectively; these values are indicated as the solid red lines. The x-axis indicates the position of each gene in Mb on maize chromosome 1; the y-axis indicates the transcript level ratios (log2) among the three tested genotypes. The segment duplicated in p1-ww714 (48.1–62.7 Mb) is indicated by the two green vertical lines; expression ratios of genes within this segment are indicated in blue. (C) Trans effects on unduplicated genes. Heat map of transcript ratios (log2) of 90 DEGs unlinked with the duplicated region: 33 genes overexpressed (red) and 57 genes underexpressed (green) in p1-ww714/p1-ww714 vs. B73/B73. The transcript ratios were compared among B73/B73, p1-ww714/B73, and p1-ww714/p1-ww714 sibling plants as determined from mRNA-seq data.
Among 304 genes in the duplicated segment, 212 genes were expressed in the seedling tissue; 125 of these 212 genes (∼60%) were differentially expressed. Among the 125 DEGs, the great majority (118/125) were overexpressed in p1-ww714/p1-ww714 compared with B73/B73 sibling plants, indicating a predominantly positive effect of copy-number change on gene expression. Whereas, among the 128 DEGs located outside the duplicated segment, fewer than a third (41/128) of them were overexpressed. Gene ontology analysis on genes within the duplicated segment did not identify any functional categories enriched among the DEGs. We also compared the transcript data of p1-ww714/B73 (three copies) with that of sibling homozygous B73 (two copies) and p1-ww714 (four copies) plants. Interestingly, only one gene was identified as differentially expressed in the comparison between p1-ww714/B73 and p1-ww714/p1-ww714, and only 10 DEGs were identified in the comparison between p1-ww714/B73 and B73/B73. Most likely our statistical analysis was not sufficiently sensitive to detect expression changes caused by single-copy differences.
As a complement to statistical identification of individual DEGs, comparisons of transcript expression ratios (Sun et al. 2013a,b) between genotypes can also elucidate copy-number effects, especially for those genes that exhibit smaller changes in dose-dependent expression. Consequently, we computed transcript abundance ratios among the three tested genotypes derived from RNA-seq data and plotted the log2 fold changes in expression with corresponding genome position. We found that the majority of maize chromosome 1 genes (Figure 2B) as well as genes located in other chromosomes (Figure S3) have similar expression levels across the three tested genotypes, because the log-expression ratio distributions are centered on or near zero. The striking exception is the duplicated segment located between 48.1 and 62.7 Mb on maize chromosome 1. The p1-ww714/p1-ww714, p1-ww714/B73, and B73/B73 genotypes carry four, three, and two copies of this 14.6-Mb segment, respectively. The log2 ratio of gene transcripts located in this region is clearly shifted above 0 for all three pairwise comparisons; whereas, the ratios of transcript levels for other regions of maize chromosome 1 are not similarly skewed (Figure 2B). Further analysis of this 14.6-Mb region revealed that >80% of genes have a log2 ratio value >0 in the genotype containing additional copy or copies of this 14.6 Mb-segment; i.e., most genes within the altered region show increased transcript number as copy number is increased. The distribution of the log2 fold change in the comparison between p1-ww714/p1-ww714 and B73/B73 is very closely centered near 1.0, which is the log ratio of copy number between homozygous p1-ww714 (four copies) and B73 (two copies). Similarly, the distributions of the log2 fold change obtained from the other two comparisons are centered close to their corresponding log2 ratios of gene copy numbers; specifically, the gene copy-number ratios (log2) of p1-ww714/p1-ww714 vs. B73/B73, p1-ww714/B73 vs. B73/B73, and p1-ww714/p1-ww714 vs. p1-ww714/B73 are 1.0, 0.585 and 0.415, respectively; and the median transcript abundance ratios (log2) are 0.91, 0.52 and 0.38, respectively. The direct correlation between gene-transcript levels and copy number suggests that most genes located within the duplicated region are expressed in proportion to their dosage.
Trans-acting dosage effect for genes unlinked with duplicated region
In addition to dosage effects of genes within the 14.6-Mb duplicated segment, trans-acting effects could alter the expression of unlinked target genes (Rabinow et al. 1991; Xie and Birchler 2012; Li et al. 2013). RNA-seq experiments comparing homozygous p1-ww714 and B73 identified 128 DEGs located outside the duplicated segment; 38 of these are in the short arm of chromosome 1, closely linked with the duplicated region, whereas the other 90 DEGs are in the long arm of chromosome 1 or in other chromosomes, segregating independently with the 14.6-Mb region. Among the 90 unlinked DEGs, 33 are overexpressed and 57 are underexpressed in p1-ww714/p1-ww714, suggesting the existence of both activating and repressing trans effects. To test whether trans-acting dosage effects (Guo and Birchler 1994; Cooper and Birchler 2001) are implicated, transcript levels of these 90 genes across three copy-number variants were further compared. We hypothesized that if the expression of these 90 genes is in fact subject to trans-acting dosage effects, we should observe proportional expression levels of these 90 genes; i.e., expression of these genes in the heterozygote p1-ww714/B73 should be intermediate between that of p1-ww714 and B73. Examination of the log2 fold changes in expression between p1-ww714/B73 and B73, and p1-ww714 and B73, shows that all 90 genes exhibit consistent increasing or decreasing expression as the copy number of the 14.6-Mb segment changes (Figure 2C). These results suggest that both positive and negative trans-acting dosage effects do in fact impact the expression of genes outside the duplicated region, although specific statistical analyses are required to test this conclusion (below).
Statistical tests of cis- and trans-dosage effects
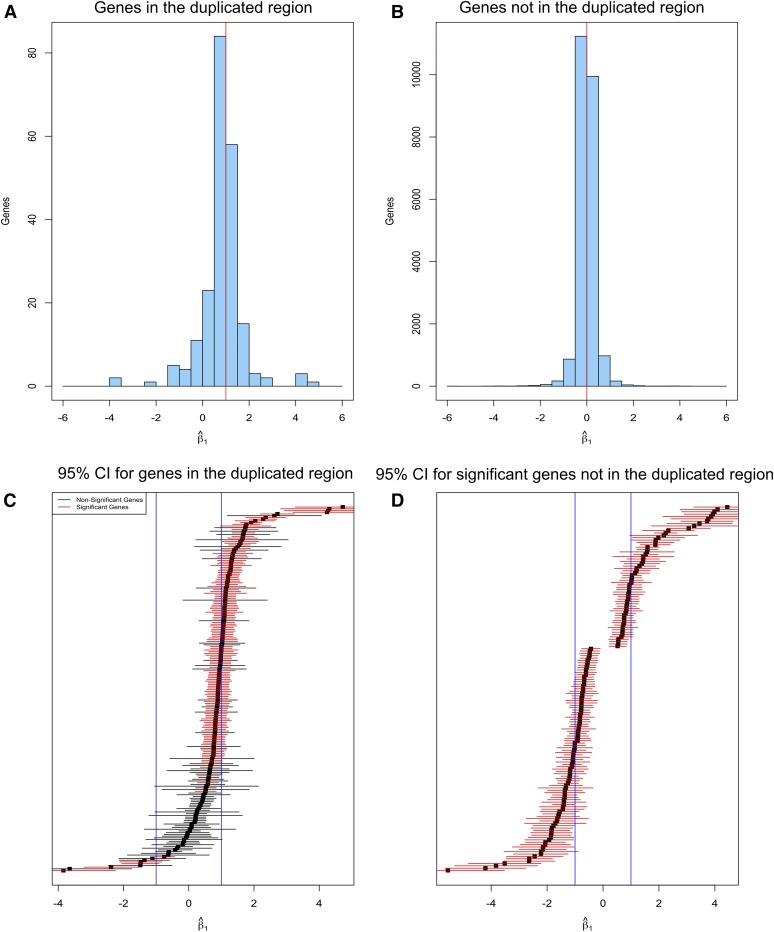
As described above, we observed dosage-dependent expression of most genes located in the duplicated region, as well as trans-acting dosage effects for some genes outside the duplicated region. We then developed and applied statistical approaches (see Materials and Methods) to rigorously test the hypothesis of dosage effect by virtue of transcript level of genes across all three genotypes. For each gene, we compared three generalized linear models (M0, M1, and M2) in increasing order of complexity. The log of the mean read count for each gene was assumed to be a linear function of gene-specific parameters plus a sample-specific normalization factor, determined for each sample by the log of the 0.75 quantile of sample-specific read counts (Bullard et al. 2010). We found no evidence that the full model (M2) for the means is needed. In fact, the log-linear model (M1) appears to be sufficient (see Materials and Methods). According to the M1 model, the dosage compensation hypothesis is represented by the equation β1 = 0. The nondosage compensation effect (dosage effect or others) is represented by the equation β1 ≠ 0, whereas the dosage effect is represented by β1 = 1. We found that 135 of 212 (63.7%) genes in the duplicated region have β1 significantly different from 0 when controlling the FDR at 5%, indicating that these genes do not exhibit dosage compensation. The distribution of the β1 estimate for each gene in the duplicated region is depicted as a histogram (see Figure 3A), where the center point is significantly different from 0 and in fact is close to 1. To further estimate the number of genes within the duplicated region that satisfy the dosage-effect hypothesis (β1 = 1), we constructed 95% C.I.s for the β1 value for each gene. We found that the β1 value of 108 out of 135 genes was not significantly different from the value 1 (Figure 3C), suggesting that expression levels of 50.9% (108/212) of genes in the duplicated region were consistent with the positive dosage-effect hypothesis. Expression levels of 3 of 135 genes exhibited β1 estimates not significantly differently from −1, consistent with regulation by a negative dosage effect. Expression levels of the remaining 11.3% (24/212) of duplicated genes could not be explained by either the dosage-compensation or dosage-effect model (Figure 3C).
Figure 3.
Statistical test of cis- and trans-acting dosage effects. (A and B) Distributions of estimated values of β1 for levels of gene transcripts in p1-ww714/p1-ww714 vs. B73/B73. A log-linear model was applied to test the hypothesis that gene transcript level is directly proportional to gene dosage. The histograms show the numbers of genes (y-axis) with the indicated estimates of β1 (x-axis). The dosage-effect hypothesis predicts that β1 = 1 for genes within the duplicated segment (A), and β1 = 0 for genes not in the duplicated region (B). The predicted values of β1 are indicated by the red lines. (C and D) 95% C.I. for each estimated β1 value. (C) 95% C.I. of β1 value for each gene within the duplicated region. Red represents significantly DEGs, and black indicates non-DEGs. (D) 95% C.I. of β1 value for each significantly DEG located outside the duplicated region.
A similar statistical test was performed for the 23,518 genes located outside the 14.6-Mb duplicated region. In contrast, only 154 of the 23,518 (0.65%) genes had β1 estimates that are significantly different from 0 (FDR ≤ 0.05), suggesting that the expression of the majority of unduplicated genes is not altered by the 14.6-Mb duplication in chromosome 1. The distribution of the β1 estimate for each gene that is not in the duplicated region is clearly centered near 0 (Figure 3B). Among 113 of 154 genes that are unlinked with the duplicated region, results from the calculated 95% C.I.s show that 27% (30/113) of genes were positively regulated by trans-acting dosage effects, whereas 40% (46/113) of genes were negatively regulated (Figure 3D). The changes of RNA level of the remaining 37 genes cannot be explained solely by trans-acting dosage effects.
Parallel verification of DEGs in an independent duplication allele
In addition to p1-ww714, we also isolated a number of shorter inverted duplications generated by Ac/Ds-induced SCT. One such allele, p1-wwid1, carries a 3.3-Mb duplicated segment (Zhang and Peterson 1999), which is completely overlapped by the 14.6-Mb duplication in p1-ww714. The 3.3-Mb region (chromosome 1: 48.1–51.4 Mb) contains 42 annotated genes, 27 of which were identified as DEGs between homozygous p1-ww714 and B73. If expression of these 27 DEGs were in fact regulated by gene dosage, we would expect these genes should also be upregulated in the p1-wwid1 allele. We randomly chose 5 of the 27 total DEGs for testing, and 1 non-DEG located outside the duplicated region as a control. Homozygous p1-wwid1 and B73 plants were grown under the same conditions as for p1-ww714/p1-ww714, and transcript levels were determined by quantitative real-time PCR, using tissues from the same developmental stage as used previously for the p1-ww714 series. We found that all five DEGs were overexpressed in p1-wwid1/p1-wwid1 compared to B73/B73, while expression of the non-DEG control was not significantly different between the two genotypes (Figure S4). These results using an independent segmental duplication further support the conclusions discussed above.
Small RNA dosage effect
The additional copy of the 14.6-Mb fragment in p1-ww714 will not only increase the copy number of protein-coding genes, but will also double the dose of intergenic regions, transposons, and small RNA genes. To investigate the effects of dosage on small RNA expression, we performed sRNA-seq of p1-ww714/p1-ww714, p1-ww714/B73 and B73/B73 genotypes using the identical 14-day whole aboveground seedling tissues described above from six pooled sibling plants. Our sRNA-seq data included 31.3-million reads for p1-ww714/p1-ww714, 33.5-million reads for p1-ww714/B73, and 65.5-million reads for B73/B73. The size of small RNA reads varied from 18 to 27 nt, but the majority were within the range of 20 to 24 nt. Among them, 24 nt was the most abundant class, accounting for >50% of the total small RNAs. We selected reads that could be uniquely and perfectly mapped to the maize B73 reference genome because the origin of these uniquely-mapped reads is more tractable. For each genotype ∼30% of sRNA-seq reads mapped to a single site in the maize B73 reference genome (RefGen_V2). The genome was subdivided into 100-kb windows across the 10 maize chromosomes, and the total number of uniquely-mapped reads (18–27 nt) per 100-kb window was calculated. The levels of small RNAs in homozygous p1-ww714 and B73 are very similar in most genomic positions (Figure S5), although p1-ww714/p1-ww714 has some dispersed regions with apparently lower expression levels, possibly due to residual sequence polymorphisms (unreplaced background). However, within chromosome 1 where the duplication is located, a large (∼35 Mb) region shows substantial variation in small RNA levels between p1-ww714/p1-ww714 and B73/B73 (Figure 4A), p1-ww714/p1-ww714 and p1-ww714/B73, as well as p1-ww714/B73 and B73/B73 (Figure S6). We believe that many of these apparent differences in small RNA levels between p1-ww714 and B73 are actually caused by residual sequence polymorphisms in the p1-ww714 chromosome that reduce the efficiency of mapping the p1-ww714 reads to the B73 reference genome. Most likely this ∼35-Mb region was not replaced during backcrossing to B73 due to suppression of recombination by the inverted duplication.
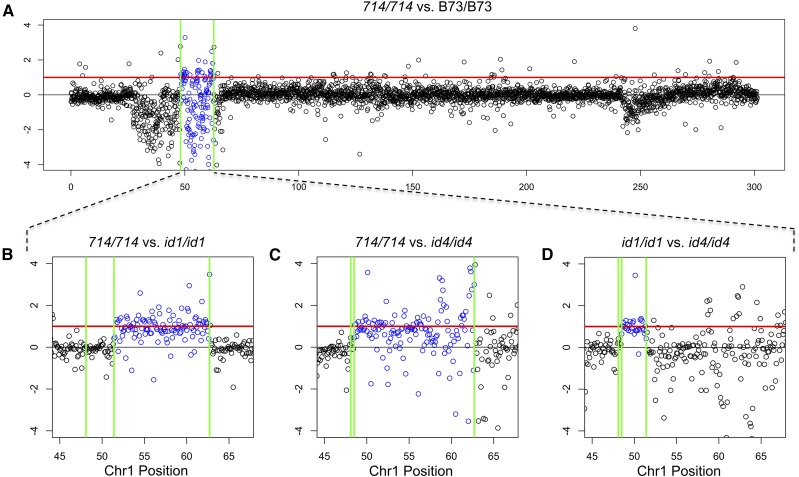
Figure 4.
Dosage-dependent expression of small RNAs. Log2 fold changes in small RNA levels per 100-kb window were plotted for comparisons between (A) p1-ww714/p1-ww714 and B73/B73, (B) p1-ww714/p1-ww714 and p1-wwid1/p1-wwid1, (C) p1-ww714/p1-ww714 and p1-wwid1/p1-wwid1 and (D) p1-wwid1/p1-wwid1 and p1-wwid4/p1-wwid4. The x-axis indicates the position of each 100-kb window in Mb on maize chromosome 1; the y-axis indicates the ratios (log2) of small RNA level between the tested genotypes. Vertical lines (green) indicate the duplication borders. The numbers within two vertical lines represent the size of duplications. The thicker red lines indicate the twofold copy-number ratio between genotypes. (A) Expression difference between p1-ww714/p1-ww714 and B73/B73. p1-ww714 carries a 14.6-Mb segmental duplication (48.1–62.7 Mb), indicated by the two green vertical lines; log expression ratios of genes within this segment are indicated in blue. (B) Expression difference between p1-ww714/p1-ww714 and p1-wwid1/p1-wwid1. p1-wwid1 has a 3.3-Mb segmental duplication which is overlapped by the 14.6-Mb duplication of p1-ww714. The 3.3-Mb overlapped duplication is represented by the first two green lines. (C) Expression difference between p1-ww714/p1-ww714 and p1-wwid4/p1-wwid4. p1-wwid4 has a 400-kb segmental duplication which is overlapped by the duplication in p1-ww714. The 400-kb overlapped duplication is represented by the first two green lines. (D) Expression difference between p1-wwid1/p1-wwid1 and p1-wwid4/p1-wwid4. p1-wwid4 has a 400-kb segmental duplication which is overlapped by the 3.3-Mb duplication of p1-wwid1. The 400-kb overlapped duplication is represented by the first two green lines.
To circumvent this mapping bias, we compared p1-ww714 with two other smaller duplication alleles (p1-wwid1 and p1-wwid4). Both p1-wwid1 (id1) and p1-wwid4 (id4) were also generated by SCT from p1-vv9D9A, the same progenitor of p1-ww714; however, id1 and id4 carry smaller duplications of ∼3.3 Mb and ∼400 kb, respectively, which are completely overlapped by the 14.6-Mb duplication in p1-ww714. These alleles were also backcrossed to B73 in parallel with p1-ww714. During the backcross program the p1 locus was selectively retained in each generation; hence the genome region near p1 has likely been retained in each of the three duplication stocks, so comparisons among these alleles will provide a better test of small RNA expression levels. Our results show that the 3.3-Mb duplicated region in id1 displays a twofold increase in small RNA transcripts relative to the single-copy progenitor p1-vv9D9A (Figure S7). Also, the results (Figure 4B) show a region of doubled small RNA expression in p1-ww714 when compared with id1. This region of overexpression extends from ∼52.4 to 62.7 Mb in chromosome 1, which is exactly the extent of the duplication in p1-ww714 that is not contained within id1. Whereas, the 3.3-Mb region (48.1–52.4 Mb) that is duplicated in both p1-ww714 and id1 shows equal levels of small RNAs. Similar pairwise comparisons were also made between homozygous p1-ww714 and id4, and id1 and id4 (Figure 4, C and D). A clear positive dosage effect of the 3.3-Mb region is now visible for both comparisons since both p1-ww714 and id1 carry an extra copy of this 3.3-Mb region with respect to id4.
We also performed sRNA-seq for some alleles (B73, p1-vv9D9A, id1, and id4) from 14-day-old root tissues. The comparisons of small RNA expression from the root tissue are very similar to those from the seedling tissue: compared with p1-vv9D9A and id4, the id1 allele contains a 3.3-Mb region with overexpressed small RNAs that is completely coincident with the extent of the duplicated segment (Figure S8). This indicates the consistency of dosage effect on small RNA transcripts between the leaf and root tissue.
Recent studies have shown that different size classes of small RNAs may have specific functional roles; e.g., 21-nt microRNAs regulate gene expression, while 24 nt small RNAs silence heterochromatic regions via RNA-directed DNA methylation (Axtell 2013; Bologna and Voinnet 2014). To determine if small RNAs of different sizes respond differently to dosage change, we examined the expression of small RNAs in size classes ranging from 20 to 24 nt. We found a clear trend of increased small RNA expression in all five size classes in the duplicated region, although the pattern is most pronounced in the 24 nt class. These results indicate a general positive dosage effect on small RNA expression similar to that observed for mRNA genes.
Discussion
Dosage effect and dosage compensation are two contrasting mechanisms of gene regulation in response to copy-number alterations. Aneuploids in human and mouse frequently exhibit increased expression of genes located in a trisomic region, suggesting that dosage effect is relatively more common in these species. Dosage effect is also reported for relatively small duplications in plants (Xiao et al. 2008; Cook et al. 2012; Wang et al. 2015). However, studies of large segmental duplications in maize suggest that dosage compensation is a commonly-found response of many genes (Birchler and Newton 1981; Guo and Birchler 1994; Makarevitch et al. 2008).
Here, we constructed a de novo series of copy-number variants in maize and compared levels of transcripts of protein-coding genes and small RNAs by RNA-seq and GeneChip. Results of RNA-seq showed that 58.7% of protein-coding genes (125/213) within the duplicated region exhibited significantly different expression levels when comparing genotypes containing two vs. four copies. However, the number of identified DEGs dropped dramatically when comparisons were made between genotypes that differ by a single copy. For example, only 1 gene was differentially expressed between p1-ww714/p1-ww714 and p1-ww714/B73 (four vs. three copies), and 10 genes were differentially expressed between p1-ww714/B73 and B73/B73 (three vs. two copies). Nevertheless, transcript levels of most genes within the altered region in p1-ww714/p1-ww714 and p1-ww714/B73 were elevated compared to B73, even though some of the increments were not large enough to be classified as statistically significant. To make the conclusion of dosage effect and dosage compensation more reliable, we developed and applied a statistical procedure to test dosage effect and dosage compensation by integrating transcript data of all three genotypes. The expression of ∼63.7% (135/212) of the duplicated genes were clearly not dosage compensated; 52.4% showed a proportional gene dosage effect, while the remaining 11.3% had transcript levels that do not fit the predictions of either the dosage compensation or dosage effect models.
It is important to note that our results may actually underestimate the proportion of dosage-effect genes, due to the effects of residual sequence polymorphisms between the p1-ww714 and B73 genotypes. Although the p1-ww714 stock was backcrossed five generations to B73, our analysis of RNA-seq data indicates that a ∼35-Mb region (chromosome 1: 28–63 Mb) retained a high frequency of SNPs in the p1-ww714 genotype. This region of unreplaced background includes the 14.6-Mb inverted duplication and a ∼20 Mb distal region, and is not surprising considering the well-documented suppression of recombination in inversion heterozygotes (Sturtevant and Beadle 1936). The ∼20-Mb distal region contains a significantly higher number of underexpressed genes than the genome-wide average (Figure S9), suggesting that the estimates of transcript levels within this region are reduced due to less-efficient mapping of the p1-ww714 transcripts to the B73 reference genome. This same negative bias would impact the detection of transcripts in the 14.6-Mb duplicated region, resulting in an underestimate of the number of dosage-effect expressed genes.
Studies of larger aneuploids in maize suggested that dosage compensation was a major gene-regulatory response to copy-number changes (Birchler 1979; Birchler and Newton 1981; Guo and Birchler 1994). These conclusions were based on analysis of selected genes within large segmental aneuploids, e.g., the long arm of maize chromosome 1, produced using maize B-A translocation chromosomes. Possibly, the larger size of duplication may have influenced the gene expression pattern, since the same Adh1 gene did exhibit dosage-dependent expression in a shorter duplication (Birchler 1981). More recently, Makarevitch and colleagues performed large scale RNA profiling of maize segmental aneuploid stocks using the Affymetrix maize 17K GeneChip and reported both dosage compensation and dosage-dependent expression (Makarevitch et al. 2008; Makarevitch and Harris 2010). They found ∼50% or more of genes present in three copies were dosage compensated when compared to the expression of the same genes present in two copies, suggesting that dosage compensation is more common than dosage-dependent expression. Because the stocks they used contained duplications of nearly the entire short arm of maize chromosome 5 (∼50 Mb), a region considerably larger than our ∼15-Mb duplication, it is possible that the results are impacted by the size of duplication as noted above for Adh1. On the other hand, the number of dosage-dependent genes identified in Makarevitch et al. (2008) might be underestimated. The comparison of transcript levels was made between two genotypes (duplication-deficient vs. normal) that differ by a single copy (three vs. two copies); hence dose-dependent changes in transcript abundance would be only 1.5-fold. Given this small difference, and considering the inherent high variability of gene expression, statistical methods may lack the power to reliably detect significant changes in expression between the tested genotypes. In our study, we used both GeneChip hybridizations and high-throughput sequencing to determine transcript levels in a segmental dosage series comprising three genotypes containing two, three, and four copies of the variant region. This approach provides greater statistical power to detect differences in gene expression. Interestingly, Makarevitch and colleagues found that qualitative changes in expression are common in their segmental aneuploidy material, and that some of them are directly responsible for some aspects of the aneuploid phenotype. In contrast, we did not detect any genes ectopically expressed in our experiments, despite the use of similar tissue and developmental stage in both studies. The distinction between smaller and larger aneuploids is likely a reflection of trans-acting genes that produce global effects across the genome including the aneuploid segment. Indeed, small aneuploids to the level of single genes can exhibit dosage effects of trans-regulators that modulate other genes in a dosage-sensitive manner (Rabinow et al. 1991; Birchler et al. 2001; Xie and Birchler 2012). Larger aneuploids have a higher probability of including such trans-acting dosage sensitive genes.
Issues of transcript normalization
The comparisons of transcript levels employed in our study are based on the commonly-held assumption that total RNA levels per cell are similar among the tested genotypes. However, several recent studies reported two- to threefold increased total RNA levels in certain tumor cells compared with normal cells (Lin et al. 2012; Nie et al. 2012). This observation calls into question the assumption of similar RNA expression levels, which could result in misinterpretation of GeneChip and/or RNA-seq results if not correctly controlled (Loven et al. 2012). In our study, we found that genes within the duplicated region exhibited significantly different transcript levels, while the expression of most genes located outside the duplication was unchanged. Because these expressional differences are largely restricted to genes within the duplicated segment, our results cannot be readily explained as an artifact of global transcriptional amplification or repression.
Another recent paper reported that in Drosophila triple X metafemale flies, the expression of most genes located in the autosomal chromosomes was downregulated by 1/3 due to increased X-chromosome number (Sun et al. 2013b). This observed inverse relationship is called inverse dosage effect; the combination of inverse and gene dosage effects are proposed to result in dosage compensation (Birchler et al. 2001; Birchler and Veitia 2012). In our study, we found that expression of most genes within the varied chromosome segment is upregulated, while the expression of genes outside the varied segment is largely unchanged. Considering the inverse dosage effect and the potential normalization issue raised by Birchler (2014), one possible explanation of our results is that the duplicated genes were mostly dosage compensated, while genes outside the duplication were downregulated in proportion to the change in dosage of the varied segment. In the present case the duplicated segment is 14.6 Mb, which represents only ∼0.6% of the total maize genome; it seems unlikely that a 0.6% change in genome size would result in a global ∼1/3 reduction in total RNA level per cell. Moreover, we reanalyzed our data based on the methods implemented by Sun et al. (2013b), and found that dosage effects were still evident when the duplicated segment and other chromosome regions were treated separately (Figure S10). Therefore, we conclude that dosage effect, not dosage compensation, is the prevalent response in the maize copy-number alteration stocks we tested.
Dosage effects on noncoding RNAs
Although the effects of copy-number alterations on mRNA and protein levels have been studied in several systems, much less is known about how copy-number alterations affect small RNA transcripts from noncoding and intergenic sequences such as TEs. For example, ∼45% of the human genome is composed of TE-derived sequences (Lander et al. 2001), whereas in maize the amount of TEs is as high as 85% (Schnable et al. 2009). Hence, large duplications will include a substantial portion of TE sequences, and proper regulation (silencing) of these extra TEs is likely critical to maintain genome stability. In plants, small RNA-guided methylation plays an important role in deactivating transposons (Law and Jacobsen 2010). We performed sRNA-seq of our dosage series and analyzed the levels of small RNA transcripts produced from consecutive 100-kb windows across the genome. When examining the levels of small RNA including reads that map at up to 100 different genome sites (multiply-mapped reads), there was little or no indication of overexpression of small RNA transcripts within the duplicated segment. This is not surprising considering the large number of other genomic loci outside the duplicated segment that can encode the bulk of multiply-mapped reads. However, analysis of the uniquely- and perfectly-mapped reads located within the duplicated segments indicates that the levels of many of these uniquely-mapped small RNA transcripts are increased in a dosage-dependent manner (Figure 4). Whether these results extend to multi-copy TEs and other highly-repetitive heterochromatic sequences remains unknown.
Long noncoding RNAs (lncRNAs) are recently disclosed to play key roles in regulation of gene expression (Wilusz et al. 2009). A previous study identified 1704 high-confidence lncRNAs in the maize B73 reference genome (Li et al. 2014). Among them, 10 lncRNAs transcribed from the 14.6-Mb duplicated region are expressed in the seedling tissue we studied. We found that 8 of 10 lncRNAs are significantly overexpressed in the p1-ww714 background compared to B73 genotype, suggesting that, similar to protein-coding genes, the majority of expressed lncRNAs in the duplicated region exhibit dosage-dependent expression.
Phenotypic differences are likely caused by both cis- and trans-acting dosage effects
The effects of aneuploidy on gene expression may be observed not only for genes with altered dosage, but also for genes unlinked to the varied chromosomal segment. This phenomenon, termed trans-acting dosage effect, is a mechanism to proportionally regulate genes outside the altered chromosomal segment (Birchler 1979; Birchler and Newton 1981; Guo and Birchler 1994). In our study, we provide further evidence through the identification of 30 genes upregulated by positive trans-acting dosage effect and 46 genes downregulated by negative trans-acting dosage effect. Considering that the 14.6-Mb duplication studied here comprises ∼0.6% of the total maize genome, these findings suggest that trans-acting dosage effects may be common in segmental aneuploid conditions. We propose that the cumulative effects of both cis- and trans-acting dosage effects likely play important roles in producing the pronounced phenotypic effects of the p1-ww714/p1-ww714 for the following reasons. First, we did not find any gene and/or QTL in the 14.6-Mb region that has significantly large effects on plant height (Peiffer et al. 2014) or flowering time (Buckler et al. 2009). Second, if a single gene were responsible for these phenotypes, id1 plants which carry a 3.3-Mb duplication would resemble either p1-ww714 or B73, depending on whether the responsible gene is located in the 3.3-Mb region or not. However, id1 plants exhibit plant height, ear size, and flowering time phenotypes that are intermediate between p1-ww714 and B73. Third, maize plants trisomic for chromosome 1 have shorter stature (reduced height, ear, and kernel sizes) and develop later than normal. Interestingly, in our segmental aneuploid series, p1-ww714/p1-ww714 and p1-ww714/B73 plants exhibit similar but less-severe phenotypes compared with maize chromosome 1 trisomics. Taken together, these observations indicate that the severity of phenotype is correlated with the extent of the affected chromosome region, suggesting that cumulative effects play a predominant role in determining phenotype in these copy-number alteration plants.
Alternative transposition of Ac/Ds elements as a tool to induce maize CNVs
Like many other eukaryotic genomes, the maize genome is large and complex, due to its high level of TEs and segmental duplications (Emrich et al. 2007). In addition, striking differences in genome content and organization have been reported among maize inbred lines, including significant levels of CNV and presence/absence variation. However, the functional impacts of segmental duplications in maize are still unclear. While most examples of extensive copy-number changes are deleterious, several recent studies in yeast revealed a beneficial role of aneuploidy in stress response (Torres et al. 2007; Pavelka et al. 2010; Sheltzer et al. 2012; Siegel and Amon 2012), although comparable aneuploidy in developing organisms provides no such advantage (Birchler 2013). By taking advantage of alternative transposition of Ac/Ds elements, we generated a 14.6-Mb segmental duplication in maize chromosome 1 with pronounced phenotypic and transcriptional effects. We recently showed that large tandem direct duplications can be generated at high frequency by another type of alternative transposition reaction termed reversed-ends transposition (Zhang et al. 2013; Zhang et al. 2014). Thus, alternative transposition may be a productive mechanism to induce segmental duplications for research purposes, as well as a new approach to generate agronomically-beneficial duplications for maize breeding programs.
Acknowledgments
We thank Lisa Coffey and Patrick Schnable for assistance in growing plants in their growth chambers, Dafang Wang for preparing RNA samples for sRNA-seq, and James Birchler for helpful comments on the manuscript. We thank the Iowa State University’s Genome Technologies Facility for providing the quantitative real-time PCR service. The author(s) declare(s) that they have no competing interests.
Author contributions: T.Z., J.Z., and T.P. conceived and designed the research; T.Z., J.Z., and D.F.W. performed the experiments; T.P., J.Z., D.N. R.W., and S.D. contributed new reagents/materials/analysis tools; T.Z., J.Z., A.L., S.D., R.W., D.N., and T.P. analyzed the data; and T.Z., A.L., D.N., and T.P. wrote the paper.
Footnotes
Communicating editor: J. A. Birchler
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.188235/-/DC1.
Literature Cited
- Anders S., Pyl P., Huber W., 2015. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E., Ratnakumar A., Arendt M.-L., Maqbool K., Webster M. T., et al. , 2013. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495: 360–364. [DOI] [PubMed] [Google Scholar]
- Axtell M., 2013. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Birchler J., 1979. A study of enzyme activities in a dosage series of the long arm of chromosome one in maize. Genetics 92: 1211–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J., 1981. The genetic basis of dosage compensation of alcohol dehydrogenase-1 in maize. Genetics 97: 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J., 2010. Reflections on studies of gene expression in aneuploids. Biochem. J. 426: 119–123. [DOI] [PubMed] [Google Scholar]
- Birchler J., 2013. Aneuploidy in plants and flies: the origin of studies of genomic imbalance. Semin. Cell Dev. Biol. 24: 315–319. [DOI] [PubMed] [Google Scholar]
- Birchler J., 2014. Facts and artifacts in studies of gene expression in aneuploids and sex chromosomes. Chromosoma 123: 459–469. [DOI] [PubMed] [Google Scholar]
- Birchler J., Newton K., 1981. Modulation of protein levels in chromosomal dosage series of maize: the biochemical basis of aneuploid syndromes. Genetics 99: 247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J., Veitia R. A., 2012. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. USA 109: 14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J., Hiebert J., Paigen K., 1990. Analysis of autosomal dosage compensation involving the alcohol dehydrogenase locus in Drosophila melanogaster. Genetics 124: 677–698. [PMC free article] [PubMed] [Google Scholar]
- Birchler J., Bhadra U., Bhadra M., Auger D., 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 234: 275–288. [DOI] [PubMed] [Google Scholar]
- Bologna N., Voinnet O., 2014. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65: 473–503. [DOI] [PubMed] [Google Scholar]
- Buckler E., Gaut B., McMullen M., 2006. Molecular and functional diversity of maize. Curr. Opin. Plant Biol. 9: 172–176. [DOI] [PubMed] [Google Scholar]
- Buckler E., Holland J., Bradbury P., Acharya C., Brown P., et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Bullard J., Purdom E., Hansen K., Dudoit S., 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia J., Song C., Bradbury P., Costich D., de Leon N., et al. , 2012. Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44: 803–807. [DOI] [PubMed] [Google Scholar]
- Cook D., Lee T., Guo X., Melito S., Wang K., et al. , 2012. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338: 1206–1209. [DOI] [PubMed] [Google Scholar]
- Cooper J., Birchler J., 2001. Developmental impact on trans-acting dosage effects in maize aneuploids. Genesis 31: 64–71. [DOI] [PubMed] [Google Scholar]
- Dash S., Van Hemert J., Hong L., Wise R., Dickerson J., 2012. PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res. 40: D1194–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin R., Holm D., Grigliatti T., 1982. Autosomal dosage compensation in Drosophila melanogaster strains trisomic for the left arm of chromosome 2. Proc. Natl. Acad. Sci. USA 79: 1200–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin R., Holm D., Grigliatti T., 1988. The influence of whole-arm trisomy on gene expression in Drosophila. Genetics 118: 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A., Zikhali M., Turner A., Isaac P., Laurie D., 2012. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS One 7: e33234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich S., Li L., Wen T., Yandeau-Nelson M., Fu Y., et al. , 2007. Nearly identical paralogs: implications for maize (Zea mays L.) genome evolution. Genetics 175: 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golzio C., Willer J., Talkowski M. E., Oh E. C., Taniguchi Y., et al. , 2012. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 485: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E., Drummond B., Bowen B., Peterson T., 1994. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553. [DOI] [PubMed] [Google Scholar]
- Guo M., Birchler J., 1994. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 266: 1999–2002. [DOI] [PubMed] [Google Scholar]
- Guo M., Birchler J., 1997. Dosage regulation of Zea mays homeobox (ZmHox) genes and their relationship with the dosage-sensitive regulatory factors of Shrunken 1 (Sh1) in maize. Dev. Genet. 20: 67–73. [Google Scholar]
- Guo M., Davis D., Birchler J., 1996. Dosage effects on gene expression in a maize ploidy series. Genetics 142: 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Parisi M., Sturgill D., Nuttall R., Doctolero M., et al. , 2006. Global analysis of X-chromosome dosage compensation. J. Biol. 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T., Hunt P., 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2: 280–291. [DOI] [PubMed] [Google Scholar]
- Huettel B., Kreil D., Matzke M., Matzke A., 2008. Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet. 4: e1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford M., Xu X., van Heerwaarden J., Pyhajarvi T., Chia J., et al. , 2012. Comparative population genomics of maize domestication and improvement. Nat. Genet. 44: 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A., Feuk L., Rivera M., Listewnik M., Donahoe P., et al. , 2004. Detection of large-scale variation in the human genome. Nat. Genet. 36: 949–951. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Zhao H., Ren L., Song W., Zeng B., et al. , 2012. Genome-wide genetic changes during modern breeding of maize. Nat. Genet. 44: 812–815. [DOI] [PubMed] [Google Scholar]
- Kahlem P., Sultan M., Herwig R., Steinfath M., Balzereit D., et al. , 2004. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res. 14: 1258–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., Li R., Xu X., Jin W., Xu M., et al. , 2010. Genome-wide patterns of genetic variation among elite maize inbred lines. Nat. Genet. 42: 1027–1030. [DOI] [PubMed] [Google Scholar]
- Lander E., Linton L., Birren B., Nusbaum C., Zody M., et al. , 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J., Jacobsen S., 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xiao J., Wu J., Duan J., Liu Y., et al. , 2012. A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol. 196: 282–291. [DOI] [PubMed] [Google Scholar]
- Li L., Petsch K., Shimizu R., Liu S., Xu W., et al. , 2013. Mendelian and non-Mendelian regulation of gene expression in maize. PLoS Genet. 9: e1003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Eichten S., Shimizu R., Petsch K., Yeh C., et al. , 2014. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 15: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Loven J., Rahl P., Paranal R., Burge C., et al. , 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J., Orlando D., Sigova A., Lin C., Rahl P., et al. , 2012. Revisiting global gene expression analysis. Cell 151: 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund S., Nettleton D., McCarthy D., Smyth G., 2012. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat. Appl. Genet. Mol. Biol. 11: 8. [DOI] [PubMed] [Google Scholar]
- Makarevitch I., Harris C., 2010. Aneuploidy causes tissue-specific qualitative changes in global gene expression patterns in maize. Plant Physiol. 152: 927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I., Phillips R., Springer N., 2008. Profiling expression changes caused by a segmental aneuploid in maize. BMC Genomics 9: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron L., Guimaraes C., Kirst M., Albert P., Birchler J., et al. , 2013. Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl. Acad. Sci. USA 110: 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale L., Haun W., Xu W., Bhaskar P., Anderson J., et al. , 2012. Structural variants in the soybean genome localize to clusters of biotic stress-response genes. Plant Physiol. 159: 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton D., 2006. A discussion of statistical methods for design and analysis of Microarray experiments for plant scientists. Plant Cell 18: 2112–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton D., Hwang J., Caldo R., Wise R., 2006. Estimating the number of true null hypotheses from a histogram of p values. J. Agric. Biol. Environ. Stat. 11: 337–356. [Google Scholar]
- Nie Z., Hu G., Wei G., Cui K., Yamane A., et al. , 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan M., Kirov G., Owen M., 2008. Phenotypic variations on the theme of CNVs. Nat. Genet. 40: 1392–1393. [DOI] [PubMed] [Google Scholar]
- Pavelka N., Rancati G., Zhu J., Bradford W., Saraf A., et al. , 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer J., Romay M., Gore M., Flint-Garcia S., Zhang Z., et al. , 2014. The genetic architecture of maize height. Genetics 196: 1337–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G., Dominy N., Claw K., Lee A., Fiegler H., et al. , 2007. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39: 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinow L., Nguyen-Huynh A. T., Birchler J., 1991. A trans-acting regulatory gene that inversely affects the expression of the white, brown and scarlet loci in Drosophila. Genetics 129: 463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K., Feuk L., Perry G., et al. , 2006. Global variation in copy number in the human genome. Nature 444: 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C., Ponger L., Gaut B., 2006. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLOS Comput. Biol. 2: 989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac C., Jiang D., Akhunov E., 2011. Targeted analysis of nucleotide and copy number variation by exon capture in allotetraploid wheat genome. Genome Biol. 12: R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P., Ware D., Fulton R., Stein J., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., et al. , 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528. [DOI] [PubMed] [Google Scholar]
- Sheltzer J., Torres E., Dunham M., Amon A., 2012. Transcriptional consequences of aneuploidy. Proc. Natl. Acad. Sci. USA 109: 12644–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J., Amon A., 2012. New insights into the troubles of aneuploidy. Annu. Rev. Cell Dev. Biol. 28: 189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K., 2005. Limma: linear models for microarray data, pp. 397–420 in Bioinformatics and Computational Biology Solutions Using R and Bioconductor, edited by Gentleman R., Carey V., Dudoit S., Irizarry W. R. Springer-Verlag, New York. [Google Scholar]
- Springer N., Ying K., Fu Y., Ji T., Yeh C., et al. , 2009. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J. R., 2010. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61: 437–455. [DOI] [PubMed] [Google Scholar]
- Stefansson H., Rujescu D., Cichon S., Pietilainen O. P., Ingason A., et al. , 2008. Large recurrent microdeletions associated with schizophrenia. Nature 455: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H., Ophoff R. A., Steinberg S., Andreassen O. A., Cichon S., et al. , 2009. Common variants conferring risk of schizophrenia. Nature 460: 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg P., Lundberg L., Johansson A.-M., Ryden P., Svensson M., et al. , 2009. Buffering of segmental and chromosomal aneuplodies in Drosophila melanogaster. PLoS Genet. 5: e1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S., Stoehr G., Peplowska K., Cox J., Mann M., et al. , 2012. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 8: 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J., 2002. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol 64: 479–498. [Google Scholar]
- Sturtevant A. H., Beadle G. W., 1936. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics 21: 554–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Johnson A., Li J., Lambdin A., Cheng J., et al. , 2013a Differential effect of aneuploidy on the X chromosome and genes with sex-biased expression in Drosophila. Proc. Natl. Acad. Sci. USA 110: 16514–16519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Johnson A., Donohue R., Li J., Cheng J., et al. , 2013b Dosage compensation and inverse effects in triple X metafemales of Drosophila. Proc. Natl. Acad. Sci. USA 110: 7383–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner R., Jia Y., DeCook R., Borsuk L., Nettleton D., et al. , 2006. All possible modes of gene action are observed in a global comparison of gene expression in a maize F-1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. USA 103: 6805–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner R., Eichten S., Kumari S., Tiffin P., Stein J., et al. , 2010. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 20: 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E., Sokolsky T., Tucker C., Chan L., Boselli M., et al. , 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacik T., Ort M., Gregorova S., Strnad P., Blatny R., et al. , 2005. Segmental trisomy of chromosome 17: a mouse model of human aneuploidy syndromes. Proc. Natl. Acad. Sci. USA 102: 4500–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xiong G., Hu J., Jiang L., Yu H., et al. , 2015. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47: 944–948. [DOI] [PubMed] [Google Scholar]
- Weaver B., Cleveland D., 2006. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18: 658–667. [DOI] [PubMed] [Google Scholar]
- Weber D. F., 1994. Use of maize monosomics for gene localization and dosage studies, pp. 350–358 in The Maize Handbook, edited by Freeling M., Walbot V. Springer-Verlag, New York. [Google Scholar]
- Weiss L. A., Shen Y., Korn J. M., Arking D. E., Miller D. T., et al. , 2008. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 358: 667–675. [DOI] [PubMed] [Google Scholar]
- Williams B., Prabhu V., Hunter K., Glazier C., Whittaker C., et al. , 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J., Sunwoo H., Spector D., 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 23: 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Jiang N., Schaffner E., Stockinger E., van der Knaap E., 2008. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527–1530. [DOI] [PubMed] [Google Scholar]
- Xie W., Birchler J., 2012. Identification of inverse regulator-a (Inr-a) as synonymous with pre-mRNA cleavage complex II protein (Pcf11) in Drosophila. G3 (Bethesda) 2: 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Zhang J., Pulletikurti V., Weber D., Peterson T., 2010. Spatial configuration of transposable element Ac termini affects their ability to induce chromosomal breakage in maize. Plant Cell 22: 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Zhang J., Peterson T., 2011. Genome rearrangements in maize induced by alternative transposition of reversed Ac/Ds termini. Genetics 188: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Wang C., Xu Q., Feng Y., Yuan X., et al. , 2011. Detection of copy number variations in rice using array-based comparative genomic hybridization. BMC Genomics 12: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Peterson T., 1999. Genome rearrangements by nonlinear transposons in maize. Genetics 153: 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Peterson T., 2004. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics 167: 1929–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Peterson T., 2005. A segmental deletion series generated by sister-chromatid transposition of Ac transposable elements in maize. Genetics 171: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zuo T., Peterson T., 2013. Generation of tandem direct duplications by reversed-ends transposition of maize Ac Elements. PLoS Genet. 9: e1003691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zuo T., Wang D., Peterson T., 2014. Transposition-mediated DNA re-replication in maize. eLife 3: e03724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Guo X., He B., Sun L., Peng Y., et al. , 2011. Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biol. 12: R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are available in the following repositories. All MIAME-compliant GeneChip profiling data are available as accession ZM50 at the PLEXdb expression resource for plants and plant pathogens (www.plexdb.org). RNA-seq data are deposited as accession GSE71448 at NCBI-GEO and accession SRP061705 in the NCBI’s Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov); sRNA-seq data are deposited as accession GSE71959 at NCBI-GEO and accession SRP062285 in the SRA (http://www.ncbi.nlm.nih.gov).