Abstract
Sex is generally determined by sex chromosomes in vertebrates, and sex chromosomes exhibit the most rapidly-evolving traits. Sex chromosome evolution has been revealed previously in numerous cases, but the association between sex chromosome origin and the reproduction mode transition from unisexual to sexual reproduction remains unclear. Here, we have isolated a male-specific sequence via analysis of amplified fragment length polymorphism from polyploid gibel carp (Carassius gibelio), a species that not only has the ability to reproduce unisexually but also contains males in wild populations. Subsequently, we have found through FISH analysis that males have several extra microchromosomes with repetitive sequences and transposable elements when compared to females. Moreover, we produced sex-reversed physiological females with a male-specific marker by using estradiol hormone treatment, and two gynogenetic families were established from them. In addition, the male incidence rates of two gynogenetic families were revealed to be closely associated with the extra microchromosome number of the sex-reversed physiological females. These results suggest that the extra microchromosomes in males might resemble a common feature of sex chromosomes and might play a significant role in male determination during the evolutionary trajectory of the reproduction mode transition from unisexual to sexual reproduction in the polyploid fish.
Keywords: microchromosome, sex determination, sex chromosome, polyploid, gibel carp, genetics of sex
SEX is a common phenomenon in nature and also one of the most important topics in life sciences, especially in evolutionary biology and genetics (Graves 2008). Most vertebrates are gonochoristic and reproduce by sexual reproduction. Different sex-determination systems, such as male heterogametic XX/XY sex chromosomes and female heterogametic ZZ/ZW sex chromosomes, as well as their numerous variants, have been revealed (Bachtrog et al. 2014; Mei and Gui 2015). Along with the rapid development of genomics and molecular genetic techniques, labile sex-determination systems and rapid sex chromosome turnovers have been noticed in both animals and plants recently (Bachtrog et al. 2014; Chen et al. 2014; Cortez et al. 2014; Graves 2014; Wei and Barbash 2015). Although how neo-sex chromosomes evolved (Roberts et al. 2009; Cortez et al. 2014; Vicoso and Bachtrog 2015) and how unisexual and sexual reproduction modes transformed (Jokela et al. 2009; Zhang et al. 2015) have been revealed, the association between sex chromosome origin and reproduction mode transition from unisexual to sexual reproduction remains unclear in vertebrates.
The gibel carp, Carassius gibelio, has a wide geographic distribution in the Eurasian continent (Hanfling et al. 2005; Li and Gui 2008; Gui and Zhou 2010; Jakovlic and Gui 2011; Jiang et al. 2013) and diverse gynogenetic strains have been identified using biological traits and genetic markers (Zhou et al. 2000a; Yang and Gui 2004; Guo and Gui 2008). As a hexaploid with >150 chromosomes (Zhou and Gui 2002), the gibel carp had two rounds of polyploidy origins in its evolutionary history, as revealed recently (Li et al. 2014b). In contrast to other unisexual vertebrates, the gibel carp has dual reproduction modes consisting of unisexual gynogenesis and sexual reproduction (Zhou et al. 2000b; Gui and Zhou 2010; Wang et al. 2011; Zhang et al. 2015); and a rare but significant proportion of males have been observed in many natural habitats, such as northeast Asia (Jiang et al. 2013), Russia (Abramenko et al. 2004), Greece (Liasko et al. 2010), and Croatia (Jakovlic and Gui 2011). Thus, the gibel carp is a great model system to investigate the origin of sex chromosomes and the transition between different reproduction modes (Gui and Zhou 2010; Liu et al. 2015).
In this study, we have isolated a male-specific genetic marker and identified several extra microchromosomes in males, which were revealed to be closely related to male determination of the gibel carp. In addition, these extra microchromosomes in males, which resemble a common feature of sex chromosomes, might play an important role in the origin of sex chromosomes during the transition from unisexual to sexual reproduction in gibel carp. These findings provide clear evidence for understanding the association between sex chromosome origin and sexual reproduction evolution.
Materials and Methods
Experimental fish source
Gibel carp (C. gibelio) and common carp (Cyprinus carpio) were collected from Guanqiao Experimental Station of the Institute of Hydrobiology, Chinese Academy of Sciences, which is located in the Wuchang District, Wuhan, China. The experimental protocols were approved by the animal care and use committee of the Institute of Hydrobiology, Chinese Academy of Sciences.
Various gynogenetic strains of gibel carp collected from wild populations were named in the style of A, B, C, D, etc., according to their biological characters or genetic markers (Zhu and Jiang 1993; Yang et al. 2001; Zhou and Gui 2002). Also some artificial strains, such as strain DA and strain A+, which were also main strains in aquaculture, were generated by sexual reproduction and maintained through successive gynogenesis (Zhou et al. 2000b; Zhou and Gui 2002; Wang et al. 2011; Gui and Zhu 2012). In this study, artificial propagation of a maternal fish from strain DA mating with a paternal fish from strain A (DA♀ × A♂) was performed, as this mating could simulate hybrid-similar reproduction (Zhang et al. 2015). Additionally, this mating could exclude almost all influences of the paternal parent and also generate males in the offspring (Zhang et al. 2015). Thus, the nearly-identical genetic background in the offspring makes it easier to isolate sex-specific genetic markers.
Artificial propagation and fish culture
During the propagation season of the gibel carp, the selected maternal fish were induced into spawning by intraperitoneal injection with a mixture of acetone-dried carp pituitary, human chorionic gonadotropin, and luteinizing hormone releasing hormone as described previously (Sun et al. 2010). About 8–10 hr after injection, the experimental fish began to ovulate and the maternal eggs released during ovulation were inseminated with paternal sperm from common carp or gibel carp. The embryos were incubated in culture dishes and the fry were reared at 20° in water boxes equipped with an inflator pump. After hatching in the incubators, they were fed with fairy shrimp for 30 days and then maintained in an outdoor tank (5 m × 4 m × 1.5 m) with normal feed. The phenotypic sex of the offspring was distinguished based on whether they ovulated and released eggs or produced sperm in the propagation season of the following year.
Genomic DNA extraction and analysis of amplified fragment length polymorphism
Genomic DNA was extracted from a small piece of fin for each sample using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI). The amplified fragment length polymorphism (AFLP) procedure was performed as described previously (Vos et al. 1995; Wang et al. 2009) with five main procedures: (1) digestion of DNA with endonucleases EcoRI and MseI and subsequent ligation of adaptors, (2) amplification of the resulting fragments using preselective primers with one selective nucleotide, (3) amplification of the diluted products using selective primers with three selective nucleotides, (4) electrophoretic separation of the fragments in 6% denatured polyacrylamide gels, and (5) visualization of the fragments via silver staining. The sequences of the adapters and the primers are summarized in Supplemental Material, Table S1.
To detect genomic differences between the female and male offspring of DA♀ × A♂, bulked segregant analysis (BSA) through AFLP was carried out as described previously (Pan et al. 2015). Nine female and nine male individuals were used to make three respective pools, and a total of 256 EcoRI/MseI primer combinations were screened in the six pools (male pools one, two, and three; and female pools four, five, and six). Subsequently, a primer combination that produced the sex-specific fragment in bulk segregant analysis was further analyzed in individual screening using 15 female offspring, 15 male offspring, maternal fish from strain DA, and paternal fish from strain A. Finally, the sex-specific AFLP fragment was cloned and sequenced.
Six randomly chosen EcoRI/MseI primer combinations, including E4/M10, E9/M13, E9/M16, E11/M16, E15/M12, and E13/M9, were used in AFLP analysis to evaluate the genetic diversity in the offspring of DA♀ × A♂.
Genomic walking and PCR amplification
Genomic walking was conducted using the Genome Walker Universal Kit (Clontech) as described previously (Dan et al. 2013), and the genomic DNA of a male in the offspring of DA♀ × A♂ was used as a template. Positive PCR products were then purified by the QIAEX II Gel Extraction Kit (QIAGEN, Valencia, CA) and cloned into the PMD19-T vector (Takara) for sequencing. Sequence assembly was performed using DNAMAN software. Repetitive sequences were detected through the Basic Local Alignment Search Tool by comparing the male-specific sequence with itself, and the structure of a transposable element was detected via LTR_FINDER (Xu and Wang 2007). The male-specific sequence characterized amplified region (SCAR) marker was obtained by placing the primer pair Cg-MSM-F and Cg-MSM-R in 25 μl of Taq DNA Polymerase (Fermentas) reaction mix. PCR cycling conditions were as follows: 94° for 4 min; 35 cycles of 94° for 30 sec, 58° for 30 sec, and 72° for 30 sec; 72° for 10 min; and endless 4°. The sequence of Cg-MSM-F was 5′-GCCACACTCACTTCTGTCTACA-3′. The sequence of Cg-MSM-R was 5′-ACTGCCATCTAACTCAGCCC-3′. FISH probe 1 was amplified by the primer pair Cg-FISH-F1 and Cg-FISH-R1 in 50 μl of LA Taq (Takara) reaction mix. PCR cycling conditions were as follows: 94° for 4 min; 35 cycles of 98° for 10 sec, and 68° for 8 min; 72° for 10 min; and endless 4°. The sequence of Cg-FISH-F1 was 5′-TCAAAATTAGTGGGCACGGTTTGTTAAACAC-3′. The sequence of Cg-FISH-R1 was 5′-TAAAAACAAACAAATTGAGCCACTCACATAG-3′. FISH probe 2 was amplified by the primer pair Cg-FISH-F2 (5′-TAGTATTGGAGGCTCAAGGCAGGATCAGTG-3′) and Cg-FISH-R2 (5′-ATCTAGTTAAACTGCCATCTAACTCAGCCC-3′) in 50 μl of LA Taq (Takara) reaction mix. PCR cycling conditions were as follows: 94° for 4 min; 35 cycles of 98° for 10 sec, 65° for 5 min, and 72° for 10 min; and endless 4°. The positive PCR products were then purified using the QIAEX II Gel Extraction Kit (QIAGEN) for subsequent FISH analyses.
Chromosome preparation and FISH
Chromosome preparations were obtained through the method of head-kidney cell-phytohemagglutinin culture in vivo as described previously (Zhu et al. 2006). FISH analysis was performed according to the method described previously (Yi et al. 2003), with minor modifications. In brief, purified fragments were labeled with Biotin-16-dUTP by using the Biotin-Nick Translation Mix (Roche). The slide with chromosome metaphase spreads was first treated at 65° for 2 hr and then exposed to RNase A solution at a final concentration of 100 μg/ml in 2× SSC. The slide was denatured in 70% deionized formamide/2× SSC for 3 min at 72°; dehydrated in a precooled 70, 90, and 100% ethanol series for 5 min each; and lastly air-dried. A 60 μl hybridization mixture containing a 100 ng labeled probe, 50% deionized formamide, 20% dextran sulfate, 0.5 μg/μl sheared salmon sperm DNA, 0.1% SDS, and 2× SSC was denatured in boiling water for 10 min and then put in ice immediately. The denatured mixture was dropped on the slide and covered with a coverslip. Next, hybridization was carried out in a moist chamber at 37° for ∼20 hr. The slide was washed for 10 min in each of the following: 2× SSC, 2× SSC with 50% formamide, and 0.1× SSC with 0.1% Tween 20. It was then washed three times for 5 min each in 1× PBS at room temperature. The probe with Biotin was first stained with ExtraAvidin-Cy3 antibody (Sigma Chemical, St. Louis, MO) and the signal was amplified by orderly adding monoclonal Biotin-conjugated anti-avidin antibody (Sigma Chemical) and ExtraAvidin-Cy3 antibody. Lastly, the slides were stained with 0.25 μg/ml DAPI in 1× PBS solution, and then the images were observed and acquired under confocal microscopy (NOL-LSM 710; Carl Zeiss, Thornwood, NY) as described (Li et al. 2014b). The number of microchromosomes stained by the FISH probe was relatively dispersed because of the high chromosome numbers of gibel carp (Zhou and Gui 2002). Thus, the number of microchromosomes in each tested fish was obtained according to the most frequent number by counting 100 metaphases.
DAPI staining of nuclear behavior in the fertilized eggs
The ovulated eggs of the female from strain DA in the gibel carp were inseminated by sperm from strain A or from another species, red common carp. The fertilized eggs were digested by 0.25% trypsin to remove their shells and then incubated at 23° for cytological observation. The fertilized eggs of different developmental stages were fixed with 4% paraformaldehyde in PBS at 4° overnight. After washing with PBS three times, the nuclei were stained using DAPI, and the images were acquired under confocal microscopy as described (Wang et al. 2013; Li et al. 2014a).
Artificial sex reversal
A female was mated with a male in strain DA to produce male offspring through sexual reproduction (Gui and Zhou 2010; Zhang et al. 2015). The fish fry were fed with fairy shrimp that had been placed in estradiol (Sigma Chemical) at a final concentration of 20 μg/liter for 0.5 hr. The duration of treatment lasted for 40 days from the hatching date, and then the treated fry were maintained in an outdoor tank (5 m × 4 m × 1.5 m) with normal feed. After treatment with estradiol, some genotypic males were transformed into phenotypic females. The phenotypic sex of these fish was distinguished based on whether they ovulated and released eggs or produced sperm in the propagation season of the following year, and the genotypic sex was detected by the male-specific SCAR marker. Thus, the mature sex-reversed physiological females with the male-specific marker (genetic male but phenotypic female) were selected to establish gynogenetic families with male offspring.
Data availability
The male-specific sequence in gibel carp (Cg-M-s) has been submitted to GenBank with accession number KT260068.
Results
Isolation of a male-specific DNA sequence in gibel carp
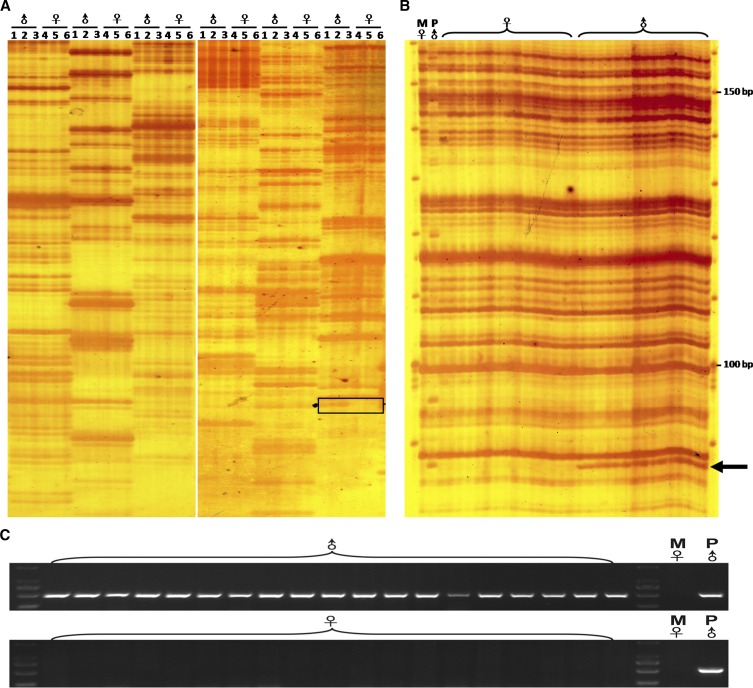
We used a female from strain DA mating with a male from strain A (DA♀ × A♂) in gibel carp, and 19 males and 71 females were observed in the offspring, for which the male ratio was 21.1%. To screen the genetic difference between females and males, we employed an AFLP-based screening approach (Pan et al. 2015) using 256 EcoRI/MseI primer combinations (Table S1). All the primer combinations produced almost identical AFLP patterns between pools of males (pools one, two, and three) and pools of females (pools four, five, and six), except the E15/M5 primer combination, which was able to be used to produce a male-specific AFLP band (Figure 1A). The male-specific fragment was present in 15 randomly-picked male individuals and the paternal fish from strain A, whereas it was absent in 15 randomly-picked female individuals and the maternal fish from strain DA (Figure 1B). Subsequently, the male-specific AFLP fragment was cloned, and its flanking sequence was obtained via genomic walking. A male-specific sequence with a total of 8335 bp was obtained, and a 515 bp male-specific SCAR marker was converted from the sequence to check its specificity for males (Figure 1C).
Figure 1.
Isolation of the male-specific genetic marker in gibel carp. (A) BSA of the male pools and female pools. Lines 1, 2, and 3 indicate male pools (♂), while lines 4, 5, and 6 indicate female pools (♀). Primer combinations from left to right are: E10/M16, E10/M15, E10/M14, E15/M7, E15/M6, and E15/M5. The black box shows the differentially-amplified fragment between females and males. (B) Individual screening of 15 females and 15 males using the primer combination of E15/M5. The arrow indicates the male-specific AFLP fragment. (C) PCR detection analysis of the male-specific SCAR marker in the 19 randomly-picked males and the 19 randomly-picked females in the offspring, as well as of the parental individuals. ♀, female; ♂, male; M♀, maternal fish from strain DA; P♂, paternal fish from strain A.
Extra microchromosomes in males are closely related to male determination
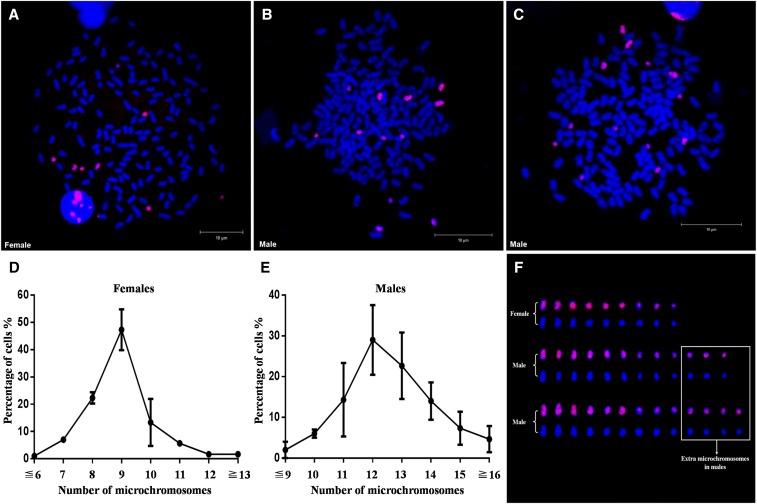
A 7687-bp fragment of a male-specific sequence (Figure 5A) was used as probe 1 to perform FISH detection on metaphases prepared from the offspring of DA♀ × A♂. Interestingly, some chromosomes were positively stained by the probe in both metaphases from the females and males, but the numbers of positively stained chromosomes were different between females and males. There were generally 9 positive chromosomes stained by the FISH probe in female metaphases (Figure 2A and Figure S1A), whereas there were ∼12 in the male metaphases (Figure 2, B and C, and Figure S1B). A total of three females (Figure 2D) and three males (Figure 2E) in the offspring were examined for statistical analysis, and the numbers of positive chromosomes were moderately variable between male individuals (Figure S1B). As shown in Figure 2, the red positive chromosomes stained by the FISH probe appear to be smaller than the rest of the chromosomes, no matter if they were from females or males; thus, they are called microchromosomes. Six microchromosomes show slightly stronger FISH signals than the others and exist identically in both females and males (Figure 2F). In comparison with female metaphases, the male metaphases have three or four extra microchromosomes with slightly weaker FISH signals (Figure 2F), which indicates that the extra microchromosomes are associated with male determination in gibel carp.
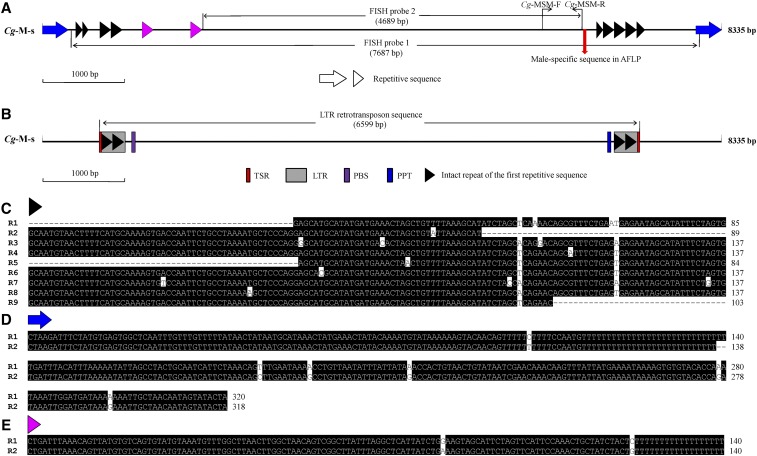
Figure 5.
Repetitive sequence analysis in the male-specific sequence (Cg-M-s). (A) Schematic diagram of repetitive sequences in the male-specific sequence (Cg-M-s). Arrows and arrowheads with the same color indicate repetitive sequences of similar sequences. The sites of primers including Cg-MSM-F and Cg-MSM-R are marked by black arrows. The FISH probes are marked by black line arrows. The red arrow shows the position of the male-specific sequence found in the AFLP analysis. (B) Schematic diagram of the predicted LTR retrotransposon sequence revealed by LTR_FINDER. Red boxes indicate TSR. Gray boxes indicate LTR. Purple boxes indicate primer binding sites. Blue boxes indicate the PPT. The positions and sequences of the above elements are given in Table S2. (C–E) The alignment of repetitive sequences is indicated by arrowheads or arrows with the same color. The similar repetitive sequences are numbered from left to the right as shown in (A). Cg, C. gibelio; R, repetitive sequence.
Figure 2.
Discovery of extra microchromosomes in males via FISH analysis. (A) FISH analysis in metaphase of female 1. (B) FISH analysis in metaphase of male 1. (C) FISH analysis in metaphase of male 3. (D) Statistical data of microchromosome number in females. (E) Statistical data of microchromosome number in males. The number of microchromosomes is shown on the x-axis, and the percentage of total cells is shown on the y-axis. Three females and three males in the offspring were used in statistical analysis, and a total of 100 metaphases were counted for each tested individual. (F) Morphological comparisons of microchromosomes in (A–C). Extra microchromosomes in males are indicated in the white box. The probe was labeled with Biotin, and red fluorescence was produced. All metaphase chromosomes were counterstained with DAPI and appeared blue.
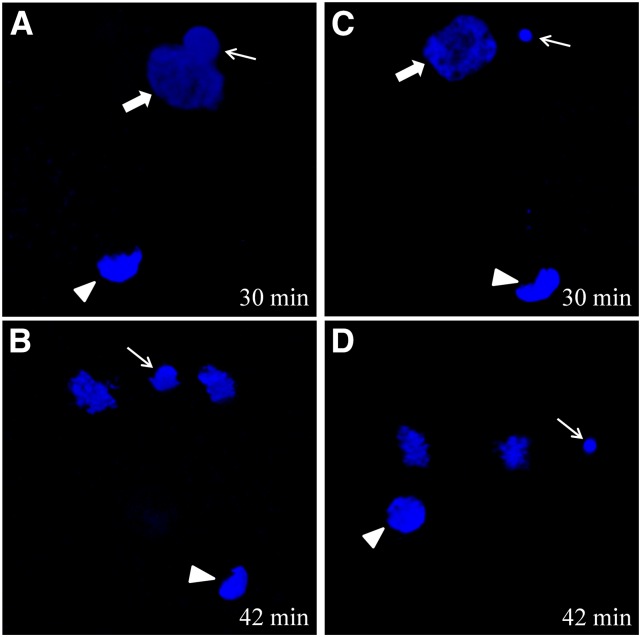
Moreover, we used DAPI staining to observe nuclear behavior in the fertilized eggs by different sperms. When the eggs of maternal fish from strain DA were fertilized by the sperm from strain A, several early nuclear events; including second polar extrusion, sperm nucleus swelling, and fusing with the female pronucleus (Figure 3A); were similar to the classical nuclear events of sexual reproduction in bisexual fishes. However, when the fertilized egg entered first mitosis, it underwent an unusual hybrid-similar development behavior in which the male pronucleus failed to integrate into the first zygotic mitosis and became the divorced chromatin (Figure 3B). When the eggs of maternal fish from strain DA were fertilized by the sperm from C. carpio, the typical process of unisexual gynogenesis, in which the sperm nucleus remains in a condensed status throughout the whole process, occurred (Figure 3, C and D). The significant nuclear difference also implies an association of hybrid-similar developmental behavior and extra microchromosome occurrence in strain DA eggs fertilized by strain A sperm.
Figure 3.
DAPI-stained nuclear behavior in two eggs with different types of development at 30 min and 42 min after fertilization. (A and B) Nuclear behavior in hybrid-similar development eggs of a DA-strain female inseminated by strain A sperm. (C and D) Nuclear behavior in gynogenetic eggs of a DA-strain female activated by C. carpio sperm. The thin arrow indicates the sperm nucleus or male pronucleus. The thick arrow indicates the female pronucleus. The arrowhead indicates the second polar body. The corresponding time (min) after fertilization is shown in the right corner.
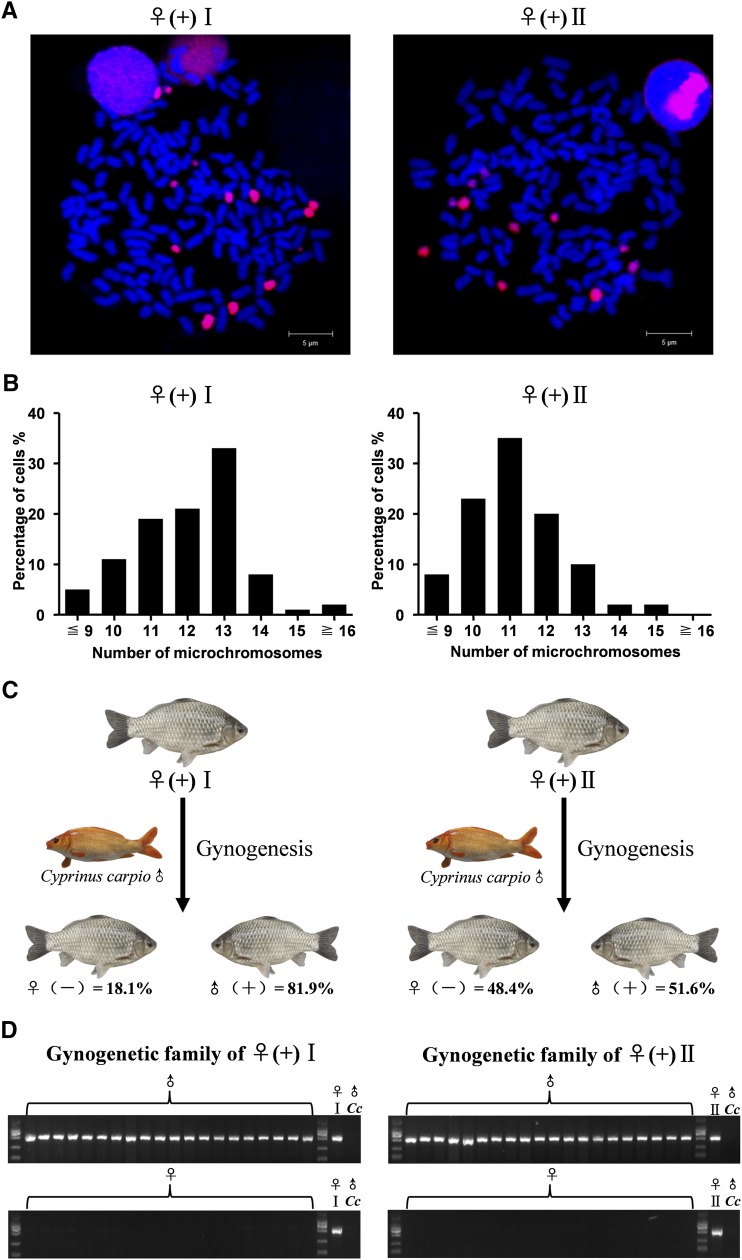
Male incidence is associated with extra microchromosome number of parents
In gibel carp, all-female offspring are easily produced by unisexual gynogenesis in response to sperm from another species, whereas males generally result from sexual reproduction in response to sperm from male gibel carp (Gui and Zhou 2010; Zhang et al. 2015). To reveal the association between microchromosomes and male incidence, we first produced sex-reversed physiological females with a male-specific marker from genetic males via estradiol treatment. Subsequently, these sex-reversed physiological females with the male-specific marker [♀(+)] were used as maternal parents, and the eggs of each maternal parent were inseminated by the sperm of common carp (C. carpio) to establish gynogenetic families (Figure S2). In comparison with common gynogenetic offspring, which consisted of all-female individuals (Gui and Zhou 2010; Zhang et al. 2015), these gynogenetic families produced a high incidence of male individuals (Figure S2).
The ratios of male occurrence were also found to be variable and related to the number of microchromosomes in these gynogenetic families. As shown in Figure 4, the first gynogenetic family, which comes from one sex-reversed physiological female [♀(+) I] with 13 microchromosomes (Figure 4, A and B, left panel), comprises ∼81.9% males (113 males in 138 individuals) (Figure 4C, left panel); while the second gynogenetic family, which is reproduced from the other sex-reversed physiological female [♀(+) II] with 11 microchromosomes (Figure 4, A and B, right panel), contains ∼51.6% males (47 males in 91 individuals) (Figure 4C, right panel). Additionally, all the examined males from the two gynogenetic families carry the male-specific marker, while the marker is absent in all the examined females (Figure 4D). Compared with normal genetic females with nine microchromosomes (Figure 2, A and D), the ♀(+) I should possess four extra microchromosomes, while the ♀(+)II has only two extra microchromosomes. The meaning of this is just that the more extra microchromosomes there are in the maternal parent, the higher the male incidence in the gynogenetic offspring. Therefore, these results indicate that male incidence is closely associated with the number of extra microchromosomes.
Figure 4.
Comparison of two gynogenetic families of sex-reversed physiological females ♀(+)I and ♀(+)II. (A) FISH analysis in metaphases of sex-reversed physiological females ♀(+)I (left panel) and ♀(+)II (right panel). (B) Histogram analysis of microchromosome number in ♀(+)I (left panel) and ♀(+)II (right panel). The number of microchromosomes is indicated on the x-axis, and the percentage of total cells is shown on the y-axis. (C) Sex ratios in the gynogenetic families of ♀(+)I (left panel) and ♀(+)II (right panel). (D) Detection of the male-specific SCAR marker in 20 randomly-picked females, 20 randomly-picked males, and parental individuals in each family. ♀, female; ♂, male; (+), with male-specific marker; (-), without male-specific marker; Cc, C. carpio.
Moreover, we also examined three females and three males in the offspring of the two gynogenetic families of ♀(+)I and ♀(+)II. In the offspring of the gynogenetic family of ♀(+)I, the tested females had 9 microchromosomes, whereas the tested males possessed 12–13 microchromosomes (Figure S3A). In the offspring of the gynogenetic family of ♀(+)II, the examined females also had 9 microchromosomes, while the examined males contained 11–12 microchromosomes (Figure S3B). Again, the data further validate the male determination role that the extra microchromosomes play in males.
Detection of repetitive sequences in male-specific sequence
Strong fluorescence appearance on almost whole microchromosomes in both males and females (Figure 2) implicates the existence of repetitive sequences in the isolated male-specific sequence. As a result, three kinds of repetitive sequences (Figure 5A) and a similar LTR retrotransposon sequence without reverse transcriptase and integrase domains (Lerat 2010) (Figure 5B) were detected in this male-specific sequence. The first kind of repetitive sequence is 137 bp in length; in it there are five intact repeats and four incomplete repeats in the male-specific 8335 sequence (Figure 5, A and C). The second kind of repetitive sequence is composed of 320 or 318 bp (Figure 5, A and D), and the third kind of repetitive sequence contains 140 bp (Figure 5, A and E). Besides its characterized repeats, such as target site repeats (TSR), LTR, primer binding sites, and the polypurine tract (PPT); significantly, the similar LTR retrotransposon sequence even includes two intact repeats of the first repetitive sequence in each of the two LTRs (Figure 5B and Table S2). Thus, these results indicate that that the microchromosomes contain numerous repetitive sequences and transposable elements, which display a common feature similar to neo-sex chromosomes (Kaiser and Bachtrog 2010; Zhou et al. 2013; Palacios-Gimenez et al. 2015). In addition, these extra microchromosomes in males, which are closely related to male determination, might evolve to sex chromosomes or provide primary material for the origin of sex chromosomes in gibel carp.
Discussion
For fishes with unisexual reproduction modes of gynogenesis or hybridogenesis, the existence of rare males was noted in Poecilia formosa (Hubbs et al. 1959; Lamatsch et al. 2010), Squalius alburnoides (Sousa-Santos et al. 2007), Phoxinus eos-neogaeus (Goddard and Dawley 1990), Cobitis sp. (Vasil’ev et al. 2003), and Misgurnus anguillicaudatus (Itono et al. 2006). In comparison with the minor proportion of males in the above fishes, the gynogenetic gibel carp was observed to have variable male incidences in natural habitats (Gui and Zhou 2010), and an unusual high male incidence of up to 23% was even found in a northeast Asian river (Jiang et al. 2013). Even though the evolutionary implications and mechanisms had been discussed (Jiang et al. 2013), the genetic evidence for why and how the lineages were able to produce variable proportions of males was still undiscovered. In this study, we isolated a male-specific sequence from the polyploid gibel carp (Figure 1) and identified several extra microchromosomes in males (Figure 2). Furthermore, these microchromosomes, which resemble a common feature of sex chromosomes (Figure 5), were revealed to play an important role in male determination (Figure 2 and Figure 4) in gibel carp.
Microchromosomes were also reported in other fishes and most of these microchromosomes were considered to be supernumerary chromosomes, which were suggested to have an effect on the phenotype of host species, especially the sex (Schartl et al. 1995; Noleto et al. 2012). In Chilomycterus spinosus (Noleto et al. 2012) and one clone of P. formosa (Lamatsch et al. 2000; Lamatsch et al. 2010), they were revealed to be associated with male determination; whereas in Sphoeroides spengleri, they were found to be related to female determination (Noleto et al. 2012). Intriguingly, the extra microchromosomes in male gibel carp are dispensable for a normal life cycle, and their number and morphology varies between individuals (Figure 2 and Figure S1). Moreover, these microchromosomes can be lost or accumulated during their inheritance, as evidenced by the number of microchromosomes in some offspring being different from that of the maternal parents (Figure 4 and Figure S3). These microchromosomes are autonomously independent from the host genome, which makes the extra microchromosomes in males resemble supernumerary chromosomes (Camacho 2005; Martis et al. 2012; Bauerly et al. 2014; Houben et al. 2014). Recently, the supernumerary chromosomes that are restricted to females in a Lake Victoria cichlid species, Lithochromis rubripinnis, have been revealed to have a functional effect on female determination (Yoshida et al. 2010). In contrast with the shown effect in cichlid fish, these extra microchromosomes in males may play a role in male determination in gibel carp.
On the other hand, these extra microchromosomes might be able to exude from the male pronucleus and incorporate into the zygotic nucleus more easily than other chromosomes with a normal size during the hybrid-similar developmental process (Figure 3, A and B). Also, this hybrid-similar development (Figure 3, A and B) might be the stage of transition from unisexual gynogenesis (Figure 3, C and D) to sexual reproduction (Zhang et al. 2015), which might cause a nearly-identical genotype in the offspring individuals (Figure S4) except for a male-specific sequence (Figure 1). Thus, these data further indicate that extra microchromosomes in males are closely related to male determination in gibel carp, even though their number is variable in different male individuals.
As male requirement is a strong selective pressure for the unisexual polyploid gibel carp, these microchromosomes with a role in male determination may be selected and preserved. These microchromosomes in males indeed display a common feature similar to neo-sex chromosomes because they contain numerous repetitive sequences and transposable elements (Figure 5). In insects and fishes, the accumulation of repetitive sequences has been revealed to be the initial stage of neo-sex chromosome evolution (Zhou and Bachtrog 2012; Zhou et al. 2013; Palacios-Gimenez et al. 2015), and the expansion of repetitive sequence repeats can trigger heterochromatin formation, restrict recombination, and lead to subsequent chromosomal rearrangements (Kaiser and Bachtrog 2010; Zhou et al. 2013; Palacios-Gimenez et al. 2015). The current data in polyploid gibel carp indicate that the extra microchromosomes with repetitive sequences and transposable elements in males play a male determination role in gibel carp, and they might evolve to sex chromosomes or provide primary material for the origin of sex chromosomes during the reproduction mode transition from unisexual gynogenesis to sexual reproduction (Zhang et al. 2015).
However, it is still unclear how these extra microchromosomes are related to male sex determination in gibel carp. One explanation might be that the presence of extra microchromosomes creates a genetic imbalance between males and females and thereby leads to male development. Another explanation might be that some extra microchromosomes may contain a male-specific locus with a male determination role. The second explanation seems to be more likely than the first one, as the male-specific sequence has been identified (Figure 1) and it is always related to male occurrence in gibel carp (Figure 1 and Figure 4D). Unfortunately, we cannot yet identify the male-specific extra microchromosomes using the male-specific sequence (FISH probe 1). We even used the nonrepetitive part of the male-specific sequence to design another probe (FISH probe 2, Figure 5A) for FISH analysis, but all of the microchromosomes were still stained by red fluorescence (Figure S5), and the male-specific extra microchromosomes could not be distinguished. The reasons might derive from two characteristics. One might be that only a lower genetic differentiation exists between this male-specific sequence and the corresponding female-homologous sequence, as reported in other fish species (Dan et al., 2013; Pan et al., 2015). Another might be related to the LTR retrotransposon sequence and various repetitive sequences (Figure 5): perhaps it is the complicated retrotransposon and repetitive sequences and their accumulation (Estep et al. 2013) in the polyploid gibel carp, especially after two rounds of polyploidy (Li et al., 2014b), that leads to the difficulty in discrimination. Of course, our ongoing complete genome sequencing will be of benefit in solving these problems (Gui and Zhu 2012).
Acknowledgments
We thank Wei Liu for providing confocal services (Analytical and Testing Center, Institute of Hydrobiology, Chinese Academy of Sciences). This work was supported by the National Natural Science Foundation of China (31502148), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA08030201), the earmarked fund for Modern Agro-Industry Technology Research System (NYCYTX-49), the Special Fund for Agro-Scientific Research in the Public Interest (grant number 200903046), the Autonomous Project of the State Key Laboratory of Freshwater Ecology and Biotechnology (2011FBZ17), and the Autonomous Project of the Institute of Hydrobiology, Chinese Academy of Sciences (Y25A171).
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.185843/-/DC1.
Communicating editor: K. M. Nichols
Literature Cited
- Abramenko M. I., Nadtoka E. V., Makhotkin M. A., Kravchenko O. V., Poltavtseva T. G., 2004. Distribution and cytogenetic features of triploid male goldfish in Azov basin. Ontogenez 35: 375–386. [PubMed] [Google Scholar]
- Bachtrog D., Mank J. E., Peichel C. L., Kirkpatrick M., Otto S. P., et al. , 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12: e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerly E., Hughes S. E., Vietti D. R., Miller D. E., McDowell W., et al. , 2014. Discovery of supernumerary B chromosomes in Drosophila melanogaster. GENETICS 196: 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho, J. P. M., 2005 B chromosomes, pp. 224–286 in The Evolution of the Genome, edited by T. R. Gregory. Elsevier Academic Press, San Diego. [Google Scholar]
- Chen S., Zhang G., Shao C., Huang Q., Liu G., et al. , 2014. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46: 253–260. [DOI] [PubMed] [Google Scholar]
- Cortez D., Marin R., Toledo-Flores D., Froidevaux L., Liechti A., et al. , 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508: 488–493. [DOI] [PubMed] [Google Scholar]
- Dan C., Mei J., Wang D., Gui J. F., 2013. Genetic differentiation and efficient sex-specific marker development of a pair of Y- and X-linked markers in yellow catfish. Int. J. Biol. Sci. 9: 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep M. C., DeBarry J. D., Bennetzen J. L., 2013. The dynamics of LTR retrotransposon accumulation across 25 million years of panicoid grass evolution. Heredity 110: 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard K. A., Dawley R. M., 1990. Clonal inheritance of a diploid nuclear genome by a hybrid fresh-water minnow (Phoxinus eos-neogaeus, Pisces: Cyprinidae). Evolution 44: 1052–1065. [DOI] [PubMed] [Google Scholar]
- Graves J. A. M., 2008. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu. Rev. Genet. 42: 565–586. [DOI] [PubMed] [Google Scholar]
- Graves J. A. M., 2014. The epigenetic sole of sex and dosage compensation. Nat. Genet. 46: 215–217. [DOI] [PubMed] [Google Scholar]
- Gui J. F., Zhou L., 2010. Genetic basis and breeding application of clonal diversity and dual reproduction modes in polyploid Carassius auratus gibelio. Sci. China Life Sci. 53: 409–415. [DOI] [PubMed] [Google Scholar]
- Gui J. F., Zhu Z. Y., 2012. Molecular basis and genetic improvement of economically important traits in aquaculture animals. Chin. Sci. Bull. 57: 1751–1760. [Google Scholar]
- Guo W., Gui J. F., 2008. Microsatellite marker isolation and cultured strain identification in Carassius auratus gibelio. Aquacult. Int. 16: 497–510. [Google Scholar]
- Hanfling B., Bolton P., Harley M., Carvalho G. R., 2005. A molecular approach to detect hybridisation between crucian carp (Carassius carassius) and non-indigenous carp species (Carassius spp. and Cyprinus carpio). Freshw. Biol. 50: 403–417. [Google Scholar]
- Houben A., Banaei-Moghaddam A. M., Klemme S., Timmis J. N., 2014. Evolution and biology of supernumerary B chromosomes. Cell. Mol. Life Sci. 71: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbs C., Drewry G. E., Warburton B., 1959. Occurrence and morphology of a phenotypic male of a gynogenetic fish. Science 129: 1227–1229. [DOI] [PubMed] [Google Scholar]
- Itono M., Morishima K., Fujimoto T., Bando E., Yamaha E., et al. , 2006. Premeiotic endomitosis produces diploid eggs in the natural clone loach, Misgurnus anguillicaudatus (Teleostei: Cobitidae). J Exp Zool A Ecol Genet Physiol 305A: 513–523. [DOI] [PubMed] [Google Scholar]
- Jakovlic I., Gui J. F., 2011. Recent invasion and low level of divergence between diploid and triploid forms of Carassius auratus complex in Croatia. Genetica 139: 789–804. [DOI] [PubMed] [Google Scholar]
- Jiang F. F., Wang Z. W., Zhou L., Jiang L., Zhang X. J., et al. , 2013. High male incidence and evolutionary implications of triploid form in northeast Asia Carassius auratus complex. Mol. Phylogenet. Evol. 66: 350–359. [DOI] [PubMed] [Google Scholar]
- Jokela J., Dybdahl M. F., Lively C. M., 2009. The maintenance of sex, clonal dynamics, and host-parasite coevolution in a mixed population of sexual and asexual snails. Am. Nat. 174: S43–S53. [DOI] [PubMed] [Google Scholar]
- Kaiser V. B., Bachtrog D., 2010. Evolution of sex chromosomes in insects. Annu. Rev. Genet. 44: 91–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamatsch D. K., Nanda I., Epplen J. T., Schmid M., Schartl M., 2000. Unusual triploid males in a microchromosome-carrying clone of the Amazon molly, Poecilia formosa. Cytog. C. Gen. 91: 148–156. [DOI] [PubMed] [Google Scholar]
- Lamatsch D. K., Stock M., Fuchs R., Dobler M., Wacker R., et al. , 2010. Morphology, testes development and behaviour of unusual triploid males in microchromosome-carrying clones of Poecilia formosa. J. Fish Biol. 77: 1459–1487. [DOI] [PubMed] [Google Scholar]
- Lerat E., 2010. Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity 104: 520–533. [DOI] [PubMed] [Google Scholar]
- Li F. B., Gui J. F., 2008. Clonal diversity and genealogical relationships of gibel carp in four hatcheries. Anim. Genet. 39: 28–33. [DOI] [PubMed] [Google Scholar]
- Li X. Y., Li Z., Zhang X. J., Zhou L., Gui J. F., 2014a Expression characterization of testicular DMRT1 in both Sertoli cells and spermatogenic cells of polyploid gibel carp. Gene 548: 119–125. [DOI] [PubMed] [Google Scholar]
- Li X. Y., Zhang X. J., Li Z., Hong W., Liu W., et al. , 2014b Evolutionary history of two divergent Dmrt1 genes reveals two rounds of polyploidy origins in gibel carp. Mol. Phylogenet. Evol. 78: 96–104. [DOI] [PubMed] [Google Scholar]
- Liu W., Li S. Z., Li Z., Wang Y., Li X. Y., et al. , 2015. Complete depletion of primordial germ cells in an all-female fish leads to sex-biased gene expression alteration and sterile all-male occurrence. BMC Genomics 16: 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liasko R., Liousia V., Vrazeli P., Papiggioti O., Chortatou R., et al. , 2010. Biological traits of rare males in the population of Carassius gibelio (Actinopterygii: Cyprinidae) from Lake Pamvotis (north-west Greece). J. Fish Biol. 77: 570–584. [DOI] [PubMed] [Google Scholar]
- Martis M. M., Klemme S., Banaei-Moghaddam A. M., Blattner F. R., Macas J., et al. , 2012. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl. Acad. Sci. USA 109: 13343–13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J., Gui J. F., 2015. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 58: 124–136. [DOI] [PubMed] [Google Scholar]
- Noleto R. B., Vicari M. R., Cestari M. M., Artoni R. F., 2012. Variable B chromosomes frequencies between males and females of two species of pufferfishes (Tetraodontiformes). Rev. Fish Biol. Fish. 22: 343–349. [Google Scholar]
- Palacios-Gimenez O. M., Marti D. A., Cabral-de-Mello D. C., 2015. Neo-sex chromosomes of Ronderosia bergi: insight into the evolution of sex chromosomes in grasshoppers. Chromosoma 124: 353–365. [DOI] [PubMed] [Google Scholar]
- Pan Z. J., Li X. Y., Zhou F. J., Xiang X. G., Gui J. F., 2015. Identification of sex-specific markers reveals male heterogametic sex determination in Pseudobagrus ussuriensis. Mar. Biotechnol. (NY) 17: 441–451. [DOI] [PubMed] [Google Scholar]
- Roberts R. B., Ser J. R., Kocher T. D., 2009. Sexual conflict resolved by invasion of a novel sex determiner in lake Malawi cichlid fishes. Science 326: 998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M., Nanda I., Schlupp I., Wilde B., Epplen J. T., et al. , 1995. Incorporation of subgenomic amounts of DNA as compensation for mutational load in a gynogenetic fish. Nature 373: 68–71. [Google Scholar]
- Sousa-Santos C., Collares-Pereira M. J., Almada V., 2007. Fertile triploid males—an uncommon case among hybrid vertebrates. J Exp Zool A Ecol Genet Physiol 307A: 220–225. [DOI] [PubMed] [Google Scholar]
- Sun M., Li Z., Gui J. F., 2010. Dynamic distribution of spindlin in nucleoli, nucleoplasm and spindle from primary oocytes to mature eggs and its critical function for oocyte-to-embryo transition in gibel carp. J Exp Zool A Ecol Genet Physiol 313A: 461–473. [DOI] [PubMed] [Google Scholar]
- Vasil’ev V. P., Akimova N. V., Emel’yanova N. G., Pavlov D. A., Vasil’eva E. D., 2003. Reproductive capacities in the polyploid males of spined loaches from the unisexual-bisexual complex, occurred in the Moscow river. Folia Biol. (Krakow) 51: 67–73. [PubMed] [Google Scholar]
- Vicoso B., Bachtrog D., 2015. Numerous transitions of sex chromosomes in diptera. PLoS Biol. 13: e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., Hogers R., Bleeker M., Reijans M., Vandelee T., et al. , 1995. AFLP: a new technique for DNA-fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Mao H. L., Chen H. X., Liu H. Q., Gui J. F., 2009. Isolation of Y- and X-linked SCAR markers in yellow catfish and application in the production of all-male populations. Anim. Genet. 40: 978–981. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhou L., Li Z., Gui J. F., 2013. Apolipoprotein C1 regulates epiboly during gastrulation in zebrafish. Sci. China Life Sci. 57: 975–984. [DOI] [PubMed] [Google Scholar]
- Wang Z. W., Zhu H. P., Wang D., Jiang F. F., Guo W., et al. , 2011. A novel nucleo-cytoplasmic hybrid clone formed via androgenesis in polyploid gibel carp. BMC Res. Notes 4: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K. H. C., Barbash D. A., 2015. Never settling down: frequent changes in sex chromosomes. PLoS Biol. 13: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wang H., 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35: 265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Gui J. F., 2004. Positive selection on multiple antique allelic lineages of transferrin in the polyploid Carassius auratus. Mol. Biol. Evol. 21: 1264–1277. [DOI] [PubMed] [Google Scholar]
- Yang L., Yang S. T., Wei X. H., Gui J. F., 2001. Genetic diversity among different clones of the gynogenetic silver crucian carp, Carassius auratus gibelio, revealed by transferrin and isozyme markers. Biochem. Genet. 39: 213–225. [DOI] [PubMed] [Google Scholar]
- Yi M. S., Li Y. Q., Liu J. D., Zhou L., Yu Q. X., et al. , 2003. Molecular cytogenetic detection of paternal chromosome fragments in allogynogenetic gibel carp, Carassius auratus gibelio Bloch. Chromosome Res. 11: 665–671. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Terai Y., Mizoiri S., Aibara M., Nishihara H., et al. , 2010. B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet. 7: e1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun M., Zhou L., Li Z., Liu Z., et al. , 2015. Meiosis completion and various sperm responses lead to unisexual and sexual reproduction modes in one clone of polyploid Carassius gibelio. Sci. Rep. 5: 10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Gui J. F., 2002. Karyotypic diversity in polyploid gibel carp, Carassius auratus gibelio Bloch. Genetica 115: 223–232. [DOI] [PubMed] [Google Scholar]
- Zhou L., Wang Y., Gui J. F., 2000a Analysis of genetic heterogeneity among five gynogenetic clones of silver crucian carp, Carassius auratus gibelio Bloch, based on detection of RAPD molecular markers. Cytogenet. Cell Genet. 88: 133–139. [DOI] [PubMed] [Google Scholar]
- Zhou L., Wang Y., Gui J. F., 2000b Genetic evidence for gonochoristic reproduction in gynogenetic silver crucian carp (Carassius auratus gibelio bloch) as revealed by RAPD assays. J. Mol. Evol. 51: 498–506. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Bachtrog D., 2012. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science 337: 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Ellison C. E., Kaiser V. B., Alekseyenko A. A., Gorchakov A. A., et al. , 2013. The epigenome of evolving Drosophila neo-sex chromosomes: dosage compensation and heterochromatin formation. PLoS Biol. 11: e1001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. P., Ma D. M., Gui J. F., 2006. Triploid origin of the gibel carp as revealed by 5S rDNA localization and chromosome painting. Chromosome Res. 14: 767–776. [DOI] [PubMed] [Google Scholar]
- Zhu L. F., Jiang Y. G., 1993. A comparative study of the biological characters of gynogenetic clones of silver crucian carp (Carassius auratus gibelio). Acta Hydrobiol. Sinica 17: 112–120. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The male-specific sequence in gibel carp (Cg-M-s) has been submitted to GenBank with accession number KT260068.