Abstract
In small populations, genetic linkage between a polymorphic neutral locus and loci subject to selection, either against partially recessive mutations or in favor of heterozygotes, may result in an apparent selective advantage to heterozygotes at the neutral locus (associative overdominance) and a retardation of the rate of loss of variability by genetic drift at this locus. In large populations, selection against deleterious mutations has previously been shown to reduce variability at linked neutral loci (background selection). We describe analytical, numerical, and simulation studies that shed light on the conditions under which retardation vs. acceleration of loss of variability occurs at a neutral locus linked to a locus under selection. We consider a finite, randomly mating population initiated from an infinite population in equilibrium at a locus under selection. With mutation and selection, retardation occurs only when S, the product of twice the effective population size and the selection coefficient, is of order 1. With S >> 1, background selection always causes an acceleration of loss of variability. Apparent heterozygote advantage at the neutral locus is, however, always observed when mutations are partially recessive, even if there is an accelerated rate of loss of variability. With heterozygote advantage at the selected locus, loss of variability is nearly always retarded. The results shed light on experiments on the loss of variability at marker loci in laboratory populations and on the results of computer simulations of the effects of multiple selected loci on neutral variability.
Keywords: associative overdominance, background selection, deleterious mutations, heterozygote advantage, neutral variability
THERE has recently been much interest in the effects of selection at one locus on patterns of evolution and variation at linked neutral or nearly neutral loci, with mounting evidence for such effects from surveys of genome-wide patterns of molecular variability and evolution (Cutter and Payseur 2013; Neher 2013; Charlesworth and Campos 2014). Attention has especially been directed at the possibility of enhanced neutral variability at nucleotide sites that are closely linked to sites under long-term balancing selection (Charlesworth 2006; Gao et al. 2015) and at the reduction in variability caused by the hitchhiking effects of directional selection, involving either positive selection (selective sweeps) (Maynard Smith and Haigh 1974) or negative selection (background selection) (Charlesworth et al. 1993). There seems little doubt that such linkage effects play an important role in shaping patterns of variability across the genome (Cutter and Payseur 2013; Charlesworth and Campos 2014).
Recent studies have, however, made little or no reference to classical work on associative overdominance (AOD), dating back to >40 years ago. Following a proposal by Frydenberg (1963), who coined the term, it was shown by Sved (1968, 1971, 1972) and by Ohta and Kimura (1970; Ohta 1971, 1973) that linkage disequilibrium (LD) between a polymorphic neutral locus and a locus subject either to selection in favor of heterozygotes or to selection against recessive or partially recessive mutant deleterious alleles could result in apparent heterozygote advantage at the neutral locus. This is because such LD, generated by genetic drift in a randomly mating finite population, leads to an association between the homozygosities of the two loci (Haldane 1949). If homozygosity at the selected locus results in reduced fitness, homozygotes at the neutral locus will appear to be at a selective disadvantage. A heuristic treatment of this effect is given by Charlesworth and Charlesworth (2010, pp. 396–397 and 403–404).
This effect of LD in a randomly mating population should be distinguished from the effect of identity disequilibrium (ID) in a population with a mixture of random mating and matings between close relatives (Haldane 1949; Cockerham and Weir 1968), as exemplified by species that reproduce by a mixture of outcrossing and self-fertilization. Here, AOD can arise because of variation among individuals in their inbreeding coefficients, which causes correlations among loci in their levels of homozygosity even if they are unlinked (Haldane 1949; Cockerham and Weir 1968). Theoretical models show that this type of AOD does not require either finite population size or LD (Ohta and Cockerham 1974; Charlesworth 1991).
Associations between heterozygosities at putatively neutral molecular marker loci and higher values of fitness components have frequently been found in natural populations, leading to a debate as to whether AOD due to LD with random mating or to ID caused by variation in inbreeding levels is the main cause of these associations (David 1998; Hansson and Westerberg 2002). Current evidence appears to favor the latter model (Szulkin et al. 2010; Hoffman et al. 2014), especially because the LD model seems to require a relatively small effective population size to produce substantial effects, unless linkage is very tight (Ohta 1971, 1973).
It has also been reported that the level of variability in both quantitative traits and marker loci in laboratory populations can sometimes decline less rapidly over time than is expected under the standard neutral model (e.g., Rumball et al. 1994; Latter 1998; Gilligan et al. 2005). In addition, laboratory populations and populations of domesticated animals and plants often have levels of quantitative trait variability that are surprisingly high, given their low effective population sizes (Johnson and Barton 2005; Hill 2010). Sved (1968) and Ohta (1971) proposed that AOD caused by randomly generated LD leads to a retardation in the loss of variability at neutral loci linked to loci under selection. Ohta suggested that this effect could be substantial when the effects of partially recessive mutations distributed over a whole chromosome are considered, provided that the population size is sufficiently small (Ohta 1971, 1973). Computer simulations of multilocus systems have indeed shown that a retardation of the rate of loss of neutral variability can occur in small populations (Latter 1998; Pamilo and Palsson 1998; Palsson and Pamilo 1999; Wang and Hill 1999; Wang et al. 1999). However, an accelerated rate of loss is observed when the level of dominance of deleterious mutations is sufficiently high and/or selection is sufficiently strong. This effect reflects background selection (BGS), whereby the effective population size experienced by a neutral locus is reduced by the presence of linked, deleterious mutations (Charlesworth et al. 1993).
AOD due to LD is an attractive potential explanation for the maintenance of unexpectedly high levels of variability in small populations. However, we lack a quantitative theory concerning the conditions under which AOD produces noticeable effects on levels of variability, other than for the cases of selfing and sib-mating lines studied by Wang and Hill (1999). It is also unclear when retardation vs. acceleration of the rate of loss of variability is likely to prevail in randomly mating populations, especially with multiple loci subject to mutation and selection. As a first step toward such a theory, we present some analytical, numerical, and simulation results on the simplest possible model: a neutral locus linked to another locus subject to selection. We also present simulation results for a neutral locus surrounded by a pair of selected loci. Selection can involve either partially recessive deleterious mutations or heterozygote advantage.
Our main focus is on a small, randomly mating population, founded from a large initial equilibrium population, mimicking experiments on the rates of loss of neutral variability in laboratory populations. We derive approximate expressions for the apparent selection coefficients against homozygotes at the neutral locus, as well for the rate of loss of neutral variability. We show that, contrary to what seems to have been widely assumed, these apparent selection coefficients have nothing to do with the rate of loss of variability, assuming that selection is sufficiently weak that second-order terms in the selection coefficients can be neglected. In particular, with selection against partially recessive deleterious mutations there is a wide range of parameter space in which BGS increases the rate of loss of variability, but there is always an apparent selective advantage to heterozygotes at the neutral locus. When selection is very weak, however, AOD always causes a retardation of the loss of neutral variability, unless mutations are close to being semidominant in their fitness effects.
Theory
Apparent selection coefficients against homozygotes at a neutral locus caused by a linked locus subject to mutation to deleterious alleles
We assume that the neutral locus is segregating for two alleles, A1 and A2, with frequencies x and y = 1 – x, respectively, in a given generation. The selected locus has wild-type and deleterious mutant alleles, B1 and B2, respectively, with frequencies p and q = 1 – p. The selection and dominance coefficients at this locus are h and s, such that the relative fitnesses of B1B1, B1B2, and B2B2 are 1, 1 – hs, and 1 – s. The mutation rates for B1 to B2 and B2 to B1 are u and v, respectively. Let the haplotype frequencies of A1B1, A1B2, A2B1, and A2B2 be y1, y2, y3, and y4. We have x = y1 + y2, q = y2 + y4. The frequency of recombination between the two loci is c.
With random mating, the apparent fitnesses (denoted by tildes) of the three genotypes A1A1, A1A2, and A2A2 at the neutral locus, conditional on these haplotype frequencies, are as follows (Sved 1968, 1972; Ohta 1971):
| (1a) |
| (1b) |
| (1c) |
The extent of apparent selection at a neutral locus, induced by LD in a randomly mating finite population, can be assessed by determining the expectations of these apparent fitnesses over the distribution of haplotype frequencies induced by drift, conditioning on segregation at the A locus (Ohta and Kimura 1970; Ohta 1971). In the present case, the starting point is assumed to be a population of infinite size, at equilibrium under mutation and selection at the B locus and with no LD between the two loci. It is thereafter maintained at a population size of N breeding individuals each generation. For simplicity, a Wright–Fisher population model is assumed, so that N is also the effective population size.
The expectations for a given generation t after the foundation of the population are most simply found by expressing the haplotype frequencies in terms of the products of the relevant allele frequencies and the coefficient of linkage disequilibrium, D = y1 y4 – y2 y3. The expectation of D, E{D}, remains at zero, since selection, mutation, and drift do not affect the direction of association between the two loci; this applies generally to E{Dqi}, where i is an arbitrary nonnegative integer (a proof is given in section S4 of Supplemental Material, File S1). This does not, however, imply that quantities involving the expectation of products of D and other functions of the allele frequencies at the two loci can be ignored, which complicates the analyses.
Simple algebra (Ohta 1971) yields expressions for the expected apparent selection coefficients against A1A1 and A2A2 homozygotes (neglecting second-order terms in s) over a set of replicate populations that are segregating at the A locus. For simplicity, we simply refer to these as the apparent selection coefficients and use tildes to distinguish them from the selection coefficients at the selected locus. We have
| (2a) |
| (2b) |
It has previously been assumed that the terms in D in Equations 2 can be neglected, so that the extent of apparent overdominance at the neutral locus is given by the terms in D2 alone. This allows approximations for the apparent selection coefficients to be derived; two different approaches have been used, as described by Sved (1968, 1972), Ohta and Kimura (1970), and Ohta (1971) and by Bierne et al. (2000), respectively. We also neglect the terms in D, which are numerically small. We use an approach that takes into account the initial allele frequency at the neutral locus in the founding generation, as well as the subsequent effects of drift in creating a probability distribution around this frequency.
The measures of apparent overdominance in Equations 2 can be conveniently be written as
| (3a) |
| (3b) |
where r2 = D2/[x(1 − x)pq] is the squared correlation coefficient in allelic state between the two loci (Hill and Robertson 1968). The difference between Equations 2 and 3 in the notation for the apparent selection coefficients is intended to emphasize the absence of terms in D from Equations 3.
To obtain useful approximate expressions for these apparent selection coefficients at an arbitrary time t, we assume that the probability distributions of x, q, and r are independent of each other, which is of course not exact (the effects of departures from independence are considered in the Results section on the apparent selection coefficients). We can then write
| (4a) |
| (4b) |
To first order in s, the quantity s(1 – 2h)E{pq} is equal to the expectation of the inbred load, B, i.e., the expected difference in fitness between the logarithms of the mean fitness of a randomly mating population and of a completely homozygous population with the same allele frequencies (Greenberg and Crow 1960). Because of the loss of variability due to drift, E{B} will in general be smaller than the inbred load for an infinite population, B* = p*q*s(1 – 2h), where q* is the equilibrium frequency of B2 under mutation and selection in an infinite population (Glémin et al. 2003). If v << u, as assumed here, q* is given by the following expression (Crow and Kimura 1970, p. 260):
| (5a) |
With hs >> u, this reduces to the familiar result of Haldane (1927):
| (5b) |
This yields the widely used formula for the inbred load in an infinite population:
| (6) |
Weakly selected loci may contribute substantially to the inbred load in the initial population, so that q* could be substantially greater than zero. The more general expressions for q* and B* described above are, therefore, used here.
An exact analytic treatment of how E{B} changes over time in a finite population is difficult. Since the expected change in allele frequency due to selection is a third-degree function of q when h ≠ 0.5, the third and higher moments of q enter into the expected change per generation in B under drift, mutation, and selection, so that closed expressions cannot be obtained without using a full solution to the relevant diffusion equation under selection (e.g., Balick et al. 2015). The simplest way to obtain tractable analytical results is to make the assumption that departures caused by drift from q* are sufficiently small that the change in allele frequency due to selection per generation, (Δsq), can be linearized around q* and hence equated to (Δsq)q* + (q – q*)(dΔsq/dq)q* (e.g., Charlesworth and Charlesworth 2010, p. 355). If the mutational contributions to the change in allele frequency are included, the net change in q is equal to (q – q*)(dΔsq/dq)q* – (u + v). The expected change in q in a finite population is then zero, and a recursion equation for the variance of q, Vq, can easily be obtained, yielding a simple expression for the approximate value of E{pq} in a given generation (see Appendix).
An approximate linear recursion equation for the expectation of r2 can be obtained in a similar way, following the approach of Sved (1971) (see Appendix). The quantity σ2d = E{D2}/E{xypq} is also frequently used as a descriptor of LD in finite populations, since it can be calculated using diffusion equations (Ohta 1971; Ohta and Kimura 1971), as discussed below. While a simple recursion for σ2d does not exist, a heuristic approach is to assume that it has a similar form to that for r2, replacing the equilibrium value of r2 given by Sved’s approach by the (smaller) equilibrium value of σ2d (see Appendix). We can then substitute the expected value of σ2d for the corresponding expectation of r2 in Equations 3. Both of these approaches ignore any effects of selection on LD.
We also need to obtain the expectations of x–1 and (1 – x)–1 for a given generation, conditioned on segregation at the neutral locus. This can be done by means of the diffusion equation solution for the probability distribution at a biallelic locus under pure drift (Kimura 1955), using the terms involving the first few eigenfunctions of the power series representation of the probability distribution, conditioning on an initial allele frequency x0 (see Appendix). The approximate expectations of x–1 and (1 – x)–1 can then easily be determined (see Appendix). As was shown by Fisher (1930), this distribution is asymptotically close to a uniform distribution, with some slight deviations in the terminal classes and with a mean allele frequency of 0.5 regardless of the value of x0. The asymptotic values of x–1 and (1 – x)–1 can then be calculated, which both approach the same value asymptotically (Equation A11). This implies that the apparent selection coefficients against each homozygote should approach the same asymptotic value, provided that any effects of mutation at the neutral locus can be ignored over the timescale under consideration.
Apparent selection coefficients induced by linkage to a locus with heterozygote advantage
The same machinery can be used when the selected locus is segregating for a pair of alleles where the B1B1 and B2B2 homozygotes have fitnesses 1 – s and 1 – t relative to a fitness of 1 for B1B2 . We have
| (7a) |
| (7b) |
| (7c) |
(Sved 1968; Ohta and Kimura 1970).
The approximate apparent selection coefficients are then given by
| (8a) |
| (8b) |
Using the same argument that led to Equations 4, these apparent selection coefficients can be further approximated by
| (9a) |
| (9b) |
The expectation of the inbred load B in a finite population is now equal to (s + t) E{pq}, so that Equations 4 and 9 both involve the same function of E{B}. An approximation for E{pq} can again be found by linearizing the recursion relation for q around the deterministic equilibrium, q* = s/(s + t). The coefficient κ that measures the rate of approach to equilibrium is now equal to –st/(s + t) (Charlesworth and Charlesworth 2010, p. 355). In this case, no mutation is allowed at the locus under selection; this is because we are considering a single nucleotide site instead of a whole gene sequence, and mutations to new variants are unlikely to occur over the timescale with which we are concerned.
The effect of selection at a linked locus on the amount of neutral variability
Lack of relevance of the apparent selection coefficients to the rate of loss of variability:
It was assumed in previous theoretical studies that the apparent selection coefficients obtained by ignoring terms in D in Equations 2 and 8 provide a measure of the extent to which the loss of variability at a neutral locus, caused by drift, is retarded by linkage to a selected locus (Sved 1968; Ohta and Kimura 1970; Ohta 1971). But this assumption is mistaken, as can be shown as follows.
This can be seen for the case of deleterious mutations at the B locus; a similar argument holds for the case of heterozygote advantage. The expected change in x can be obtained from Equations 2, yielding
This gives
| (10) |
In other words, the terms in D2 contribute nothing to the change in the frequency of allele B1 at the neutral locus, which is determined entirely by the expected product of D and the additive effect of B1 on fitness, a(q) = s[h + q(1 – 2h)], provided that terms in s2 are neglected. An equivalent result is used in models of the effects of selective sweeps on neutral allele frequencies at linked sites (Charlesworth and Charlesworth 2010, p. 410). It is an example of the Price equation (Price 1970), since Da is the covariance between fitness and the allelic state with respect to A1 at the neutral locus (Santiago and Caballero 1995).
Since the change in the heterozygosity, 2x(1 – x), at the neutral locus caused by the change in allele frequency due to selection at the B locus is ∼2(1 – 2x)Δx, the accompanying change in the expected heterozygosity, H = 2E{xy}, is given by
| (11) |
Similar results apply to the case of heterozygote advantage. Here, a(q) = qt – ps = q(s+t) – s, so that
| (12) |
| (13) |
We cannot, therefore, use the apparent selection coefficients to assess the extent to which loss of variability at the neutral locus is affected by selection at a linked locus.
How to calculate the rate of loss of variability:
To solve this problem, we employ the linear diffusion operator method to obtain a closed system of recursion equations for the expectations of the quantities of interest (Ohta and Kimura 1971; Ohta 1971); this is closely related to the generation matrix approach of Hill and Robertson (1968) and Hill (1977). These methods provide recursion equations for the expectations of functions of the allele frequencies at the two loci and the linkage disequilibrium coefficient.
We have extended this method to include the effects of selection, mutation, and drift on the allele frequency q at the selected locus, reducing the dimensionality of the set of recursion equations by ignoring third- and higher-order moments of q around q*. The complexity of the relations between the different variables means that a closed system can be obtained only by use of a nine-dimensional vector, Y, of the expectations of functions of allele frequencies and D, with a corresponding 9 × 9 recursion matrix, R (see section S1 of File S1 for details).
The elements of Y are as follows: Y1 = E{xy}, Y2 = E{xyq}, Y3 = E{xyq2}, Y4 = E{D(x – y)}, Y5 =E{D(x – y)q}, Y6 = E{D(x – y)q2}, Y7 = E{D2}, Y8 = E{D2q}, and Y9 = E{D2q2}. Iteration of the R matrix provides an approximation for σ2d in an arbitrary generation (which can be used instead of E{r2} in Equations 3) as well as for H, since σ2d = Y7/(Y2 – Y3) and H = 2Y1.
Computer simulations
One neutral and one selected locus:
The theoretical predictions described above were compared with Monte Carlo simulation results for a population started in mutation–selection balance at the selected locus, which was in linkage equilibrium with a neutral locus. This was then run for a chosen number of generations at a fixed population size. In each generation, the new haplotype frequencies were calculated using the standard deterministic equations, and 2N uniform random numbers were generated to sample each new haplotype from the cumulative distribution of haplotype frequencies.
Since the effects of a single selected locus on a linked neutral locus are very small, it was necessary to run a large number of replicate simulations (usually 107) to obtain tight confidence intervals on the mean heterozygosity at the neutral locus.
Multiple loci with selection and mutation:
To simulate multiple loci subject to mutation and selection, we assumed additive fitness interactions across the relevant loci. For weak selection, this should yield very similar results to those of a multiplicative fitness model. The state of the population in a given generation is represented by the haplotype frequencies with respect to n loci subject to mutation and selection, following the same rules as in the single-locus case, with a neutral locus in the center of the chromosome. The recombination frequency between the loci under selection is c; the frequency of recombination between the neutral locus and each of the adjacent selected loci is c/2. Complete interference is assumed, so that only single crossovers are allowed, generating a frequency of recombination between the two terminal loci of (n – 1)c.
The frequencies of diploid genotypes at the start of a given generation are assumed to be given by random combinations of the frequencies of the haplotypes determined in the previous generation, thus tacitly assuming an infinite number of new zygotes. The frequencies of haplotypes after recombination were calculated deterministically, using an extension of the three-locus algorithm described by Crow and Kimura (1970, p. 51). The haplotype frequencies after mutation and selection were then calculated deterministically. The haplotype frequencies after drift were obtained by choosing 2N haplotypes from a pool with the new haplotype frequencies, in the same way as above.
FORTRAN programs for all the cases mentioned here are available on request from B. Charlesworth.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
A single selected locus with mutation and selection
To generalize the results described below as far as possible, we have used the principle from diffusion equation theory that the outcome of the evolutionary process is determined by the products of the effective population size, Ne, and the deterministic parameters, measuring time in units of 2Ne generations (Ewens 2004, p. 157). In the present case, where Ne = N, the simulation results for a given N and a set of mutation, selection, and recombination parameters can be applied to a system with a population size of KN and deterministic parameters that are a factor of K–1 times those used here. For this reason, the selection and mutation parameters in most of the material described below are scaled by a factor of 2N, thereby avoiding reference to a specific population size. We use S = 2Ns, C = 2Nc, U = 2Nu, and V = 2Nv to denote these scaled parameters.
An additional point to note is that the assumption that the initial state of the population has D = 0 implies that only the first three elements of the initial Y vector are nonzero. Without loss of generality, the elements of Y can be divided by the product of the initial frequencies of A1 and A2, x0y0, and the resulting vector, Y′, can then be iterated using the R matrix. The ratio of the first element of Y′ in generation t to (1 – 1/2N)t measures the ratio of the mean heterozygosity at the neutral locus to its value in the absence of selection at the B locus, Hrel. This means that the matrix predictions for Hrel and σ2d are independent of the initial allele frequencies at the neutral locus, since the initial value of Y′ is independent of these frequencies. If, however, D were nonzero in the initial generation, only the asymptotic behavior of the system would be independent of the initial allele frequencies.
Accuracy of the matrix approximation:
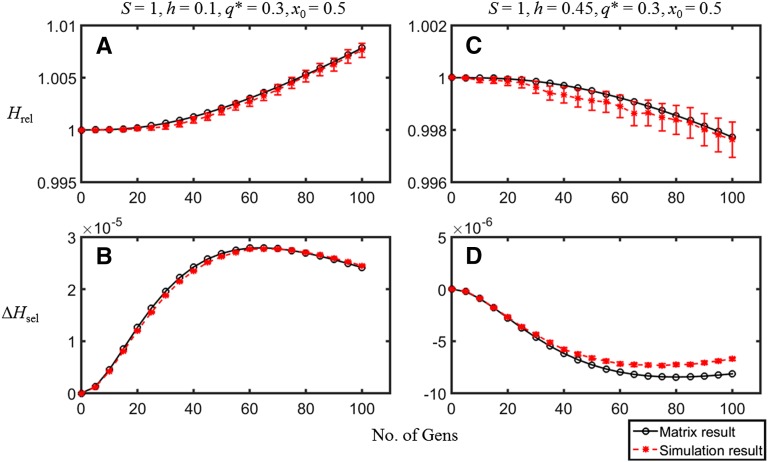
We first consider the accuracy of the 9 × 9 matrix approximation. Figure 1 shows the values of Hrel and 2ΔE{xy} = ΔHsel from Equation 11 over 2N generations, for two different dominance coefficients and C = 0.1. We are assuming that selection and mutation at the B locus relate to deleterious mutations affecting the gene as a whole rather than individual nucleotide sites, whereas the alleles at the A locus represent a pair of variants chosen by an experimenter as neutral markers and are not affected by mutation over the time course of the experiment. The selection and mutation parameters were chosen such that the equilibrium frequency of the deleterious mutation at the B locus was ∼0.3. This may seem to be very high, but is consistent with the typical frequency per gene of putatively deleterious nonsynonymous mutations in Drosophila populations (Haddrill et al. 2010), although the selection coefficient and mutation rates used in Figure 1 are probably much higher than is realistic.
Figure 1.
A comparison of the simulation results (red solid circles, with 95% confidence interval error bars) with the matrix approximation (black open circles) for the case of mutation and selection. The scaled selection coefficient and recombination rates are S = 1 and C = 0.1. The population size is N = 50; the initial frequency of the neutral allele A1 is x0 = 0.5; h = 0.1 for A and B and h = 0.45 for C and D. The mutation rates are such that the equilibrium frequency of the deleterious allele B2, q*, is ∼0.3, with v = 0.1u. A and C show the heterozygosity relative to neutral expectation at the A locus, Hrel; B and D show the change in heterozygosity per generation at the A locus due to selection at the B locus, ΔHsel. h = 0.1 and h = 0.45 are on opposite sides of the critical h value (∼0.37 at 2N generations), so that A and B show a retardation of loss of neutral variability, whereas C and D show an acceleration.
Agreement between the simulation and matrix results is remarkably good for this parameter set. A value of h = 0.1 is associated with Hrel > 1 in later generations (retardation of loss of neutral variability), whereas h = 0.45 is associated with Hrel < 1 (acceleration of loss of neutral variability). This illustrates the conflict between AOD and BGS, noted previously in simulation results (Pamilo and Palsson 1998; Wang and Hill 1999); for sufficiently low h, AOD is the dominant force as far as the level of neutral variability is concerned, whereas BGS dominates when h is sufficiently close to 1/2. We provide some insights into the reasons for this behavior in the next but one section.
Approximation for the rate of loss of variability with weak selection:
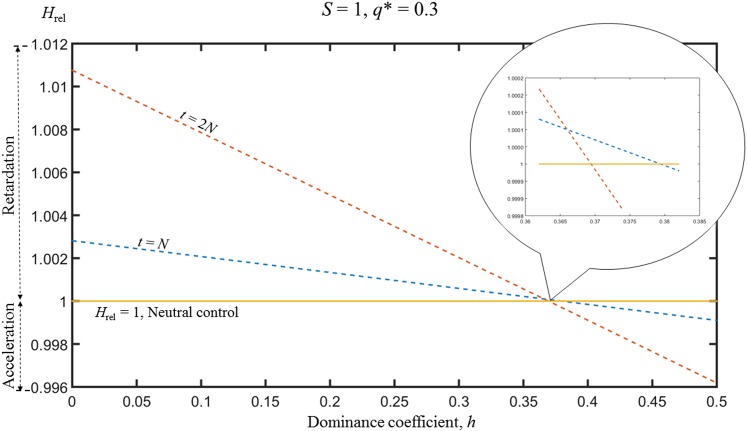
This conflict implies the existence of a critical value of h, hc, where the two effects exactly cancel each other; the properties of hc are examined in more detail below. Unfortunately, there is no exact fixed value of hc, since the extent to which Hrel is greater or less than one depends on the generation in question, as shown in Figure 2, which uses the matrix results. As h increases from 0 to 0.5, Hrel declines, at both generations N and 2N. The inset in Figure 2 shows that hc is larger for the earlier generations. However, hc is close to 0.37 for all generations for the selection and mutation parameters used in Figure 2.
Figure 2.
The heterozygosity at the A locus relative to neutral expectation, Hrel, as a function of the dominance coefficient, h, at generations N and 2N, for the case of mutation and selection. S = 1, C = 0.1, N = 50, and q* = 0.3. The results were obtained using the matrix approximation.
When q* is small and S is of order 1, agreement with the simulations is good only in earlier generations of the process, reflecting a high level of dispersion of q around q*, so that our neglect of higher-order terms in q – q* is inaccurate. In general, however, the matrix gives reasonably good approximations up to 2N generations, although the extent of retardation of the rate of loss of variability tends to be underestimated (or the rate of acceleration overestimated) by the matrix approximation with small q* (see Figure B in File S1, section S10). We will often use results for this generation as a standard, since it is similar to the duration of the Drosophila experiments on AOD described in the Discussion.
Apparent selection coefficients and their relation to the rate of loss of variability:
These conclusions are confirmed by the results displayed in Table 1 for an initial frequency of A1 of 0.1, over a range of recombination rates. Table 1 also shows the simulation results for an additional measure of variability, the proportion of segregating neutral loci, Ps, at 0.5N and 2N generations. In addition, the exact and approximate apparent selection coefficients are displayed, together with the matrix and simulation values for Hrel. More detailed results are shown in Table A of File S1, section S9.
Table 1. Simulation and theoretical results for a neutral locus linked to a locus subject to mutations to deleterious alleles (2Ns = 1, 2Nu = 0.1, 2Nv = 0.01, q* = 0.296, N = 50, x0 = 0.1).
| Generation 0.5N | Generation 2N | ||||||
|---|---|---|---|---|---|---|---|
| 2N | 2N | Hrel | Ps | 2N | 2N | Hrel | Ps |
| h = 0.1 | |||||||
| C = 0 | |||||||
| 0.150 (0.00007) | 0.030 (0.00002) | 1.0007 (0.0004) | 0.5534 (0.0002) | 0.157 (0.00015) | 0.113 (0.00013) | 1.0090 (0.0007) | 0.1967 (0.0001) |
| 0.404a | 0.075a | 1.0004a | 0.5536 (0.0002)b | 0.374a | 0.192a | 1.0083a | 0.1956 (0.0001)b |
| C = 0.1 | |||||||
| 0.147 (0.00008) | 0.030 (0.00002) | 1.0008 (0.0004) | 0.5531 (0.0002) | 0.145 (0.00015) | 0.105 (0.00012) | 1.0072 (0.0007) | 0.1964 (0.0001) |
| 0.394a | 0.071a | 1.0004a | 0.348a | 0.178a | 1.0078a | ||
| C = 1 | |||||||
| 0.127 (0.00007) | 0.025 (0.00002) | 0.9999 (0.0004) | 0.5531 (0.0002) | 0.085 (0.00011) | 0.058 (0.00008) | 1.0052 (0.0007) | 0.1958 (0.0001) |
| 0.322a | 0.058a | 1.0003a | 0.199a | 0.100a | 1.0048a | ||
| C = 10 | |||||||
| 0.053 (0.00004) | 0.008 (0.00001) | 1.0000 (0.0004) | 0.5532 (0.0002) | 0.018 (0.00004) | 0.011 (0.00002) | 1.0001 (0.0007) | 0.1956 (0.0001) |
| 0.087a | 0.015a | 1.0001a | 0.032a | 0.016a | 1.0005b | ||
| h = 0.45 | |||||||
| C = 0 | |||||||
| 0.019 (0.00001) | 0.004 (0.00002) | 1.0005 (0.0004) | 0.5531 (0.0002) | 0.019 (0.00002) | 0.013 (0.00002) | 0.9999 (0.0007) | 0.1953 (0.0001) |
| 0.05a | 0.009a | 0.9999a | 0.5536 (0.0002)b | 0.046a | 0.023a | 0.9974a | 0.1956 (0.0001)b |
| C = 0.1 | |||||||
| 0.015 (0.00001) | 0.003 (0.00002) | 0.9995 (0.0004) | 0.5534 (0.0002) | 0.015 (0.00002) | 0.010 (0.00002) | 0.9975 (0.0007) | 0.1951 (0.0001) |
| 0.049a | 0.009a | 0.9999a | 0.043a | 0.022a | 0.9976a | ||
| C = 1 | |||||||
| 0.016 (0.00001) | 0.003 (0.00002) | 0.9998 (0.0004) | 0.5528 (0.0002) | 0.010 (0.00001) | 0.007 (0.00001) | 0.9988 (0.0007) | 0.1954 (0.0001) |
| 0.041a | 0.007a | 0.9999a | 0.025a | 0.010a | 0.9985a | ||
| C = 10 | |||||||
| 0.007 (0.00001) | 0.001 (0.00001) | 0.9997 (0.0004) | 0.5531 (0.0002) | 0.002 (0.00001) | 0.001 (0.00001) | 0.9990 (0.0007) | 0.1952 (0.0001) |
| 0.012a | 0.002a | 1.0000a | 0.004a | 0.002a | 0.9999a | ||
Standard errors for 107 simulations are in parentheses, after the corresponding means.
Approximate values of r2 were obtained from the matrix recursions; neutral values were used for the expectations of x–1 and (1 – x) –1 (Equation A11 and following text); the linear approximation was used for heterozygosity at locus B (Equation A3).
Values from neutral simulations.
It can be seen that the approximate apparent selection coefficients are similar in magnitude to the exact ones, but tend to exceed them, especially with very close linkage and for the neutral allele with the lower initial frequency. This is mainly because of the inaccuracy of the assumption of independence between the probability distributions of allele frequencies at the neutral and selected loci used in deriving the equations for the apparent selection coefficients. The correlation coefficient r is a property of the genealogies at the two loci and so should be relatively independent of the allele frequencies. The simulations show, however, that the levels of variability at the two loci are positively correlated, so that the expectation of pqx–1 in Equation 3a is smaller than E{pq}E{x–1} in Equation 4a. If the exact simulation results for the apparent selection coefficients are compared with those using the simulated values for the expectations of r2, pq, and x–1 or (1 – x)–1 in Equations 4, as opposed to the approximations for these quantities used to obtain the values in Table 1, it is found that there is still a substantial discrepancy when linkage is tight, which can arise only from the inaccuracy of the independence assumption used to obtain Equations 4.
With an initial frequency of A1 of 0.5, the apparent selection coefficients for A1A1 and A2A2 are always equal, as expected from Equations 3 and 4. In contrast, for an initial frequency of 0.1 there is initially a stronger apparent selection coefficient against A1A1, again as expected, but the selection coefficients converge on equality as the mean neutral allele frequency at segregating sites approaches 0.5; these are also similar to the selection coefficients for the case of an initial frequency of 0.5. The apparent selection coefficients at generation 2N are quite close to the asymptotic values given by Equations 4 when the B locus has reached mutation–selection–drift equilibrium, when E{pq} is calculated using the linear approximation described above. For example, with h = 0.1, C = 0.1, and the selection and mutation parameters in Table 1, the asymptotic apparent selection coefficient for populations segregating at the A locus is 0.0014 when the equilibrium neutral value of σd2 is used as the estimate of E{r2} in Equations 4, whereas the observed value at 2N generations for the case with x0 = 0.5 is 0.0013. In contrast, the asymptotic value predicted by Equation 7 of Ohta (1971) is 0.0024, a somewhat worse fit. This probably reflects the fact that Ohta assumed a fixed allele frequency at the neutral locus, as well as assuming q* << 1.
As expected, the magnitudes of the apparent selection coefficients decline sharply as the recombination rate increases, but always indicate heterozygote advantage, even with C = 10 and h = 0.45. But with h = 0.45, there is an acceleration of the loss of variability at the neutral locus when linkage is tight, despite the apparent selective advantage to heterozygotes. This confirms the above conclusion that the apparent selection coefficients are not relevant to the extent to which mutation and selection at one locus affect the rate of loss of variability at a linked neutral locus; they show apparent heterozygote advantage even when BGS causes an accelerated loss of variability. With C = 10, little effect of selection on the rate of loss of variability can be detected (Table 1).
Conflict between AOD and BGS with respect to the rate of loss of variability:
The existence of a conflict between AOD and BGS, and its evident dependence on the dominance coefficient, raises the question of how to make generalizations about the regions of parameter space in which one or the other force is dominant. One way of approaching this problem is to study the properties of the leading eigenvalue of the R matrix, λ0, as a function of the relevant variables. If we equate λ0 to 1 – 1/(2Ne), where Ne is the effective population size in the presence of selection at the B locus, 1/(1 – λ0) provides a measure of 2Ne. Consider first the limiting case when S = 2Ns >> 1 and u << hs, so that the expected frequency of A2 is close to u/(hs) (Equation 5b). The approximation for λ0 – 1 for this case (Equations S17 and S18 of section S2 of File S1) shows that Ne is given by the standard equation for BGS at a single locus (Charlesworth and Charlesworth 2010, p. 402), which assumes these conditions.
As argued by Whitlock and Barton (1997), the value of 2Ne obtained from 1/(1 – λ0) gives the asymptotic value of the mean coalescent time, T2, for a pair of alleles at the neutral locus. Under the infinite-sites model, the nucleotide diversity at mutation–drift equilibrium is equal to the product of the neutral mutation rate and 2Ne (Charlesworth and Charlesworth 2010, p. 211), so that we can use these results to predict the effect of selection at the B locus on the equilibrium level of variability at the neutral locus. (See section S2 of File S1, Equation S16, for a more rigorous derivation of this result.)
However, since the matrix approximation is inaccurate for later generations with S < 1 and small q*, this use of the leading eigenvalue to estimate Ne is limited in scope and is not necessarily accurate for general q* and small S. The approximate approach described in the next section deals with the case when S is of order ≤1.
Approximation for the rate of loss of variability with weak selection:
The starting point for this analysis is Equation 11, which shows that the change in expected heterozygosity due to selection, ΔHsel, is the sum of two terms, Δ1 = – 2shY4 and Δ2 = – 2s(1 – 2h)Y5, where Y4 = E{D(x – y)} and Y5 = E{D(x – y)q}. We know from previous work on BGS that an acceleration of loss of neutral variability under drift always occurs in haploid models with linked deleterious mutations. These are equivalent to diploid models with h = 0.5, for which Δ2 = 0, so that the process is entirely driven by Δ1; a necessary and sufficient condition for acceleration in this case is thus Δ1 < 0. This implies that, with h < 0.5, there is a conflict between the two terms when Δ1 < 0 and Δ2 > 0. Larger h gives more weight to Δ1 relative to Δ2, so that selection is more likely to reduce variability when h is large, in agreement with the numerical results just described. It is, however, unclear from this argument why small S should favor AOD over BGS, which we now proceed to investigate.
The complexity of the recursion relations means that it is impossible to obtain simple, exact expressions for Y4 and Y5, but useful approximations for weak selection (S < 1) can be obtained as follows. Using expressions (S4d) and (S4e) from the matrix approximation derived in section S1 of File S1, the changes per generation in these moments in the absence of selection are
| (14a) |
| (14b) |
In the absence of selection, Y4 thus tends to zero, and Y5 tends to – 2Y7/[ (C + U + V) + 5], which is <0 and <Y7 in magnitude. This suggests that the magnitude of Δ1 will be much smaller than that of Δ2 when selection is weak, which is confirmed by the following argument.
Approximate recursions for Y4 and Y5 with selection are given by Equations S4 in File S1, section S1. We make the simplification of dropping terms involving u, v, and products of s or D2 with q and q2, on the assumption that these are small relative to similar terms involving s or D2 alone. [These assumptions will be violated when selection is strong in relation to drift (S > 1) or when q* is >> 0.] In addition, for the neutral case, it is possible to show that Y6 and Y5 approach equality and Y8 approaches Y7/2 if q* is set to 0.5 (we neglect Y9, since it is considerably smaller than Y8): see Equations S27 of File S1, section S6. Our numerical results show that these relations are approximately true under much wider conditions, so that we can replace Y6 with Y5 and Y8 with Y7/2 in Equations S4 in File S1, section S1, allowing us to obtain a pair of coupled difference equations for Y4 and Y5, given the value of Y7.
It is also convenient to rescale all terms by multiplying by 2N. We can then write
| (15a) |
| (15b) |
where
| (16a) |
| (16b) |
| (16c) |
If the changes in Y4, etc., are sufficiently slow, we can set ΔY4 and ΔY5 to zero and solve the resulting linear equations to obtain approximations for Y4 and Y5 as functions of Y7:
| (17a) |
| (17b) |
| (17c) |
This shows that Y4 is O(S), whereas Y5 is of order 1, so that ΔHsel is dominated by Δ2 when S is small, provided that h < 1/2. The determinant (det) is always positive for S ≤ 60; this condition is not very restrictive given our assumption of relatively small S, and we assume positivity from now on. Given that X7 > 0, it can easily be shown that Y4 > 0 when h < 1/2; S < 2 is sufficient for Y5 < 0 when h = 0.5. For small values of h these conditions are less stringent; with h = 0, Y4 > 0 independently of S, and Y5 < 0 when S < 6. Furthermore, neither Y4 nor Y5 depends on q*, except through their common factor of Y7. Our numerical results show that these conclusions are correct, provided that S is of order 1.
From Equation 11, the sign of the change in heterozygosity at the neutral locus due to selection, ΔHsel, is the same as the sign of –[h Y4 + (1 – 2h) Y5]. Variability will be increased by selection if ΔHsel > 0 and reduced if ΔHsel < 0, so that this implies that the critical upper value of h, hc, required for an increase in neutral variability due to AOD, is independent of q* under the assumed conditions. The value of hc can be found by setting ΔHsel = 0. Using Equations 11 together with the numerators of Equations 17a and 17b, we find that hc is the value of h that satisfies the following cubic equation:
| (18) |
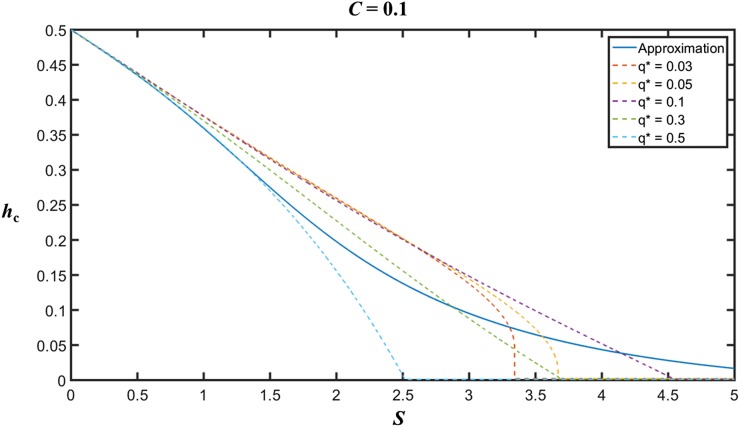
With S close to 0, Equation 18 reduces to 12h – 6 = 0, i.e., hc = 1/2, suggesting that variability will always be increased for realistic h values when selection is very weak. For other cases, some insights into the properties of hc can be obtained by assuming C = 0, which approximates cases with low recombination rates. With S = 1, the solution to the cubic is ∼hc = 0.36, slightly smaller than the value of 0.37 obtained from the numerical results described earlier. Figure 3 shows a comparison of the values obtained by solving Equation 18 using Matlab symbolic calculations with the values from the matrix approximation for generation 2N with C = 0.1 (very similar patterns are obtained for C values of up to 1). It will be seen that they agree quite well when S < 1, regardless of the value of q*, but Equation 18 mostly underestimates hc for S > 1, except for values of q* much smaller than or >>0.1. For S << 1, dominance coefficients close to 1/2 are compatible with an enhanced level of neutral variability, but BGS always prevails for values of S >> 3, even when h is very close to 0.
Figure 3.
The critical dominance coefficient, hc, as a function of the scaled selection coefficient, S, for the case of mutation and selection with C = 0.1 and N = 50. The initial frequency of A1 was 0.5. The blue line is the value of hc given by Equation 18. The dashed lines represent the values obtained from the matrix approximation at generation 2N, for several different values of the equilibrium frequency, q*, of the deleterious allele at the B locus.
The quantity 2N ΔHsel/H is equivalent to ε = Ne/N – 1, where Ne is the effective population size experienced by the neutral locus for the generation in question. For small S, we can approximate ε by a Taylor series around S = 0. We have
| (19) |
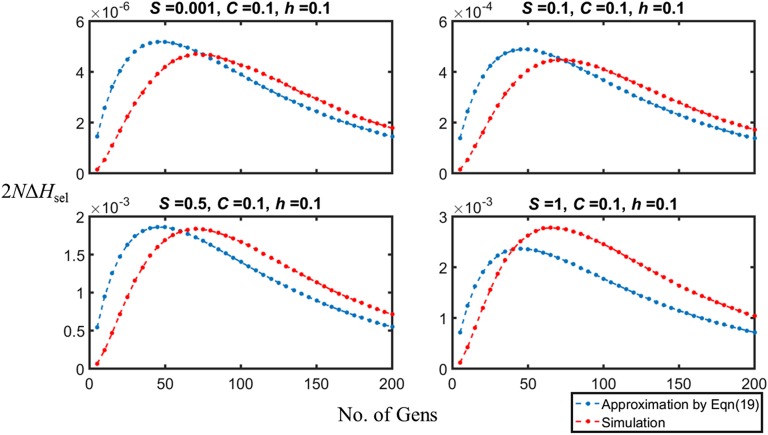
The first term in this equation is derived entirely from the contribution of Y5 to ΔHsel. It implies that selection will cause increased variability if h is slightly <1/2 when O(S2) terms are negligible, consistent with the conclusions reached above. With very weak selection, BGS is not effective unless mutations are close to being semidominant. The second-order term is always <0 and increases in magnitude as S increases; under the most favorable situation (h = 0), S < 4(5 + C)/10 is necessary for ε > 0, which is slightly more restrictive than is indicated by the results in Figure 3. Figure 4 shows some examples of the extent to which this approximation matches the simulation results; in general, it tends to underestimate the extent to which loss of variability is retarded by selection when S is of order 1.
Figure 4.
A comparison of the values of 2N ΔHsel obtained from the simulations and from Equation 19 (using values of Y7 = E{D2} obtained from the simulations), with C = 0.1, q* = 0.3, and N = 50. The initial frequency of A1 was 0.5. The panels display the approximate results from Equation 19 (blue solid circles) and the simulation results (red solid circles), for different values of S. The confidence intervals are too small to be easily seen.
These approximations also imply that the magnitude of ΔHsel is proportional to Y7 = E{D2}. If we approximate E{D2} by E{xy}E{pq}σ2d = HE{pq}σ2d/2, as was done when obtaining Equations 4 for the apparent selection coefficients, Equations 17 and 18 give
| (20) |
Because σ2dE{pq} ≤ 0.25, the quadratic dependence of the denominator on C, and the inverse relation between σ2d and C, implies that there will be only a very small effect of selection on neutral variability when C >> 1, unless S >> C. In addition, the size of the effect is strongly determined by S and by E{pq}, since σ2d takes a value between 0 and 1.
Overall, these results suggest that retardation of loss of neutral variability caused by a single selected locus is unlikely to be important except for S values of the order of ≤1. In such cases, if linkage is sufficiently tight, there can be a significant retardation of loss of neutral variability and an enhancement of the equilibrium level of variability even when h approaches 1/2.
Two selected loci with mutation and selection
From simulation studies of the effect of multiple, linked selected loci on the behavior of neutral loci located among them, we would expect to see larger values of both the apparent selection coefficients and the level of neutral heterozygosity than with a single selected locus (Ohta 1971; Latter 1998; Pamilo and Palsson 1998; Palsson and Pamilo 1999; Wang and Hill 1999; Bierne et al. 2000). However, it is unclear whether the effects of multiple selected loci are approximately additive or whether there is some degree of synergism.
With small population size, very close linkage, strong selection, and a sufficiently high level of recessivity of deleterious mutations, simulations have shown that “crystallization” of the population into two predominant and complementary haplotypes (+ – + – … and – + – +…, where + denotes wild type and – mutant) with respect to the selected loci can occur, such that the behavior of the system is very similar to that of a single locus with strong heterozygote advantage (Charlesworth and Charlesworth 1997; Palsson and Pamilo 1999; Palsson 2001). This can result in long-term maintenance of variability at the selected loci. This is strongly suggestive of a synergistic effect of multiple selected loci.
No analytical treatment of more than one selected locus has yet been done, so we have investigated this problem, using simulations of a small number of selected loci. The results of simulations with two selected loci surrounding a neutral locus are shown in Table 2, using the same set of recombination frequencies between the neutral and nearest selected locus as in Table 1. As far as the apparent selection coefficients are concerned, with h = 0.1 and the lower two recombination rates, there is clear evidence that the apparent selection coefficients in generation 2N (but not necessarily in generation 0.5N) are somewhat larger than twice the apparent selection coefficients in the single-locus case, indicating a degree of synergism. A similar pattern is found with four selected loci (results not shown).
Table 2. Simulation results for a neutral locus surrounded by two loci subject to mutation to deleterious alleles (2Ns = 1, 2Nu = 0.1, 2Nv = 0.01, q* = 0.296, N = 50, x0 = 0.5).
| Generation 0.5N | Generation 2N | ||||||
|---|---|---|---|---|---|---|---|
| 2N | 2N | Hrel | Ps | 2N | 2N | Hrel | Ps |
| h = 0.1 | |||||||
| C = 0 | |||||||
| 0.142 (0.00001) | 0.142 (0.00001) | 1.0008 (0.0001) | 0.9842 (0.00006) | 0.294 (0.00003) | 0.297 (0.00003) | 1.0180 (0.0004) | 0.5444 (0.0002) |
| 0.136a | 0.136a | 0.9841 (0.00004)b | 0.268a | 0.268a | 0.5375 (0.0002)b | ||
| C = 0.1 | |||||||
| 0.139 (0.00001) | 0.139 (0.00001) | 1.0001 (0.0001) | 0.9843 (0.00006) | 0.272 (0.00003) | 0.272 (0.00003) | 1.0159 (0.0005) | 0.5433 (0.0002) |
| 0.134a | 0.134a | 0.248a | 0.248a | ||||
| C = 1 | |||||||
| 0.115 (0.00001) | 0.115 (0.00001) | 1.0007 (0.0001) | 0.9843 (0.0006) | 0.151 (0.00002) | 0.151 (0.00002) | 1.0010 (0.0005) | 0.5412 (0.0002) |
| 0.110a | 0.110a | 0.142a | 0.142a | ||||
| C = 10 | |||||||
| 0.037 (0.00001) | 0.037 (0.00001) | 1.0002 (0.0001) | 0.9841 (0.0006) | 0.028 (0.00001) | 0.028 (0.00001) | 1.0000 (0.0005) | 0.5372 (0.0002) |
| 0.036a | 0.036a | 0.028a | 0.028a | ||||
| h = 0.45 | |||||||
| C = 0 | |||||||
| 0.018 (0.0001) | 0.018 (0.0001) | 0.9999 (0.0001) | 0.9842 (0.00006) | 0.036 (0.00003) | 0.036 (0.00003) | 0.9952 (0.0005) | 0.5352 (0.0002) |
| 0.016a | 0.016a | 0.9841 (0.00004)b | 0.032a | 0.032a | 0.5375 (0.0002)b | ||
| C = 0.1 | |||||||
| 0.018 (0.0001) | 0.018 (0.0001) | 0.9999 (0.0001) | 0.9841 (0.00006) | 0.034 (0.00003) | 0.034 (0.00003) | 0.9961 (0.0005) | 0.5356 (0.0002) |
| 0.016a | 0.016a | 0.030a | 0.030a | ||||
| C = 1 | |||||||
| 0.015 (0.0001) | 0.015 (0.0001) | 0.9996 (0.0001) | 0.9841 (0.0006) | 0.019 (0.00002) | 0.019 (0.00002) | 0.9975 (0.0005) | 0.5360 (0.0002) |
| 0.014a | 0.014a | 0.018a | 0.018a | ||||
| C = 10 | |||||||
| 0.005 (0.0001) | 0.005 (0.0001) | 0.9999 (0.0001) | 0.9842 (0.0006) | 0.004 (0.0001) | 0.004 (0.0001) | 0.9998 (0.0005) | 0.5337 (0.0002) |
| 0.004a | 0.004a | 0.004a | 0.004a | ||||
C is the scaled recombination rate between the neutral locus and each selected locus. The selection and mutation parameters are the same for each selected locus. Standard errors for 5 × 106 simulations with two selected loci are in parentheses, following the corresponding means.
Single-locus simulation values multiplied by two.
Values from neutral simulations.
It is more difficult to quantify the effect on Hrel, since this is strongly dependent on the generation in question. With close linkage and h = 0.1, the values of Hrel – 1 in generation 2N in Table 2 are slightly larger than twice the corresponding values in Table 2. Conversely, with h = 0.45 and close linkage, for which Hrel < 1, the absolute values of Hrel – 1 in generation 2N are less than twice the corresponding values in Table 1, although the apparent selection coefficients are slightly larger than twice the values with a single selected locus. A more rigorous test for additivity is described in section S5 of File S1; it suggests that the effects of two loci on Ne/N – 1, as estimated from ΔHsel, are somewhat greater than additive when there is a retardation of loss of variability and less than additive when there is an acceleration.
A single selected locus with heterozygote advantage
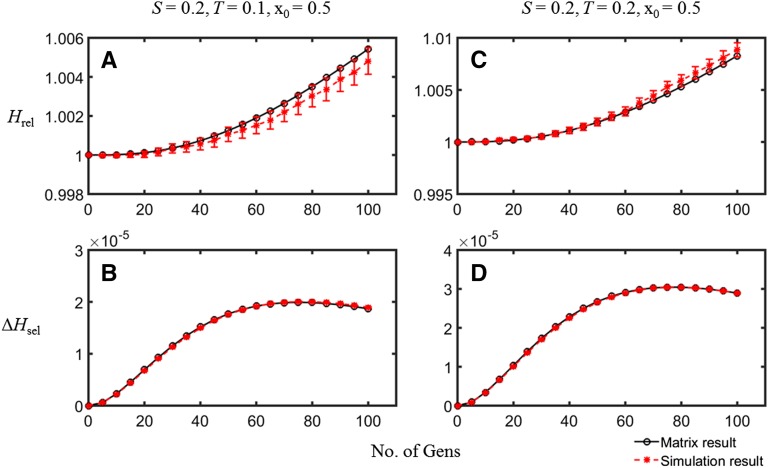
The methods used for the case of mutation and selection can be used to model the case of linkage to a single selected locus with heterozygote advantage, by making appropriate modifications to the terms in the R matrix and to the expressions for the changes in haplotype frequencies in the computer simulations (see Equations 9 and section S3 of File S1). Figure 5 and Table 3 compare some examples of matrix and simulation results; these again suggest that the matrix predictions are accurate for this parameter range (further details are shown in Table B, section S9 of File S1). Once again, the ratio of the rate of recombination to the strength of selection has to be sufficiently small for a substantial effect.
Figure 5.
A comparison of the simulation results (red symbols, with 95% confidence interval error bars) with the matrix approximation (black open circles) for the case of heterozygote advantage. A and B plot Hrel and ΔHsel against time for scaled selection coefficients S = 0.2 and T = 0.1; C and D give the values of these variables when S = 0.2 and T = 0.2. N = 50, C = 0.1, and the initial frequency of A1 was 0.5.
Table 3. Simulation and theoretical results for a single locus with heterozygote advantage.
| Generation 0.5N | Generation 2N | ||||||
|---|---|---|---|---|---|---|---|
| 2N | 2N | Hrel | Ps | 2N | 2N | Hrel | Ps |
| 2Ns = 0.2, 2Nt = 0.2, q* = 0.5, N = 50, x0 = 0.1 | |||||||
| C = 0 | |||||||
| 0.094 (0.00004) | 0.019 (0.00001) | 1.0002 (0.0004) | 0.5533 (0.0002) | 0.105 (0.00009) | 0.077 (0.00008) | 1.0082 (0.0007) | 0.1966 (0.0001) |
| 0.258a | 0.046a | 1.0003a | 0.5536 (0.0002)b | 0.212a | 0.108a | 1.0083a | 0.1956 (0.0001)b |
| C = 0.1 | |||||||
| 0.092 (0.00004) | 0.020 (0.00001) | 1.0005 (0.0004) | 0.5535 (0.0002) | 0.096 (0.00008) | 0.070 (0.00007) | 1.0311 (0.0007) | 0.1964 (0.0001) |
| 0.251a | 0.043a | 1.0003a | 0.179a | 0.092a | 1.0078a | ||
| C = 1 | |||||||
| 0.079 (0.00004) | 0.016 (0.00001) | 1.0008 (0.0004) | 0.5533 (0.0002) | 0.071 (0.00006) | 0.035 (0.00005) | 1.0053 (0.0007) | 0.1962 (0.0001) |
| 0.210a | 0.037a | 1.0003a | 0.103a | 0.061a | 1.0046a | ||
| C = 10 | |||||||
| 0.032 (0.00002) | 0.005 (0.00001) | 1.0004 (0.0004) | 0.5532 (0.0002) | 0.008 (0.00002) | 0.005 (0.00001) | 1.0004 (0.0007) | 0.1956 (0.0001) |
| 0.056a | 0.011a | 1.0001a | 0.016a | 0.008a | 1.0004a | ||
| 2Ns = 0.2, 2Nt = 0.1, q* = 0.667, N = 50, x0 = 0.1 | |||||||
| C = 0 | |||||||
| 0.062 (0.00003) | 0.013 (0.00001) | 1.0023 (0.0004) | 0.5534 (0.0002) | 0.069 (0.00006) | 0.051 (0.00006) | 1.0056 (0.0007) | 0.1962 (0.0001) |
| 0.171a | 0.031a | 1.0002a | 0.5536 (0.0002)b | 0.137a | 0.070a | 1.0054a | 0.1956 (0.0001)b |
| C = 0.1 | |||||||
| 0.061 (0.00003) | 0.013 (0.00001) | 1.0024 (0.0004) | 0.5531 (0.0002) | 0.063 (0.00006) | 0.046 (0.00005) | 1.0198 (0.0007) | 0.1961 (0.0001) |
| 0.171a | 0.032a | 1.0002a | 0.178a | 0.091a | 1.0051a | ||
| C = 1 | |||||||
| 0.053 (0.00003) | 0.010 (0.00001) | 1.0010 (0.0004) | 0.5532 (0.0002) | 0.033 (0.00004) | 0.023 (0.00003) | 1.0021 (0.0007) | 0.1959 (0.0001) |
| 0.137a | 0.025a | 1.0002c | 0.070a | 0.035a | 1.0030a | ||
| C = 10 | |||||||
| 0.021 (0.00001) | 0.003 (0.00001) | 1.0000 (0.0004) | 0.5531 (0.0002) | 0.006 (0.00001) | 0.003 (0.00001) | 0.9997 (0.0007) | 0.1953 (0.0001) |
| 0.035a | 0.006a | 1.0001c | 0.010a | 0.005a | 1.0002a | ||
Standard errors for 107 simulations are in parentheses, following the corresponding means.
Approximate values of r2 were obtained from the matrix recursions; neutral values were used for the expectations of x–1 and (1 – x) –1 (Equation A11 and following text); the linear approximation was used for the heterozygosity at locus B (Equation A3).
Values from neutral simulations.
Approximate results can be obtained for the case when S = 2Ns and T = 2Nt are both <1, on the same lines as for Equations 15–20 for the mutation–selection balance case. The coefficients for the approximate recursion relations for Y4 and Y5 are now as follows:
| (21a) |
| (21b) |
| (21c) |
The equivalents of Equations 17 and 18 are given in section S3 of File S1 and can be used to calculate ΔHsel in the same way as for the mutation–selection case. The first-order term in S and T in ΔHsel gives
| (22) |
The inbred load in this case is (s + t)E{pq} (see Equations 9), so that this is identical in form to the first-order approximation for the mutation–selection balance case (see Equation 19). For sufficiently weak selection, therefore, heterozygote advantage should always lead to an enhancement of neutral variability at a linked locus.
It is known, however, that the asymptotic rate of loss of variability at a locus with heterozygote advantage has a complex dependence on the deterministic equilibrium allele frequency and the population size, with extreme allele frequencies (outside the range 0.2–0.8) leading to an acceleration of loss of variability when S + T is sufficiently large (Robertson 1962). This suggests that a similarly complex pattern should be found for the dependence of the measure of retardation/acceleration based on the leading eigenvalue of the recursion matrix. Figure C of File S1 (section S10) shows that this is indeed the case, although there is only a small region of parameter space where acceleration rather than retardation of loss of variability occurs.
Under most circumstances, some retardation of the loss of variability at a neutral locus is, therefore, likely to be observed as a result of linkage to a selected locus, although its magnitude is small unless linkage is tight and S + T >> 1. For C >> S + T it is unlikely that any experimentally detectable effects could be observed. The same applies to the apparent selection coefficients against homozygotes at the neutral locus.
Discussion
Some general considerations
Our results shed some new light on the old question of the properties of AOD at a neutral locus in a randomly mating population of finite size, resulting from randomly generated LD with respect to loci subject to selection. The pioneering work of Sved (1968, 1972), Ohta and Kimura (1970), and Ohta (1971) gave expressions for the apparent selection coefficients against homozygotes at a neutral locus, using the equivalents of our Equations 3 and 8. Our approximate estimates of the apparent selection coefficients for the case of a finite population founded from an infinite population at equilibrium with no LD between the selected and neutral loci (Equations 4 and 9) followed their approach in using only the terms involving D2 in these equations, which are the dominant terms.
However, as was noted by Sved (1968, p. 552) for the case of heterozygote advantage and by Latter (1998) for the case of selection against deleterious mutations, these selection coefficients do not result in any change in allele frequency at the neutral locus. As mentioned above (Equation 10), this is a completely general conclusion: application of the Price equation (Price 1970) shows that the change Δx per generation in the frequency of allele A1 at the neutral locus is equal to aD, where a is the average effect of A1 on fitness (see also Santiago and Caballero 1995, p. 1016), to the order of the approximations used here (neglect of second-order terms in s and 1/N). Neither the change in allele frequency nor the change in heterozygosity at the neutral locus is influenced by the D2 terms under these conditions.
There is thus no connection between apparent overdominance caused by linkage disequilibrium and any retardation of loss of variability by drift at a neutral locus. Such a connection seems to have been widely assumed (e.g., Latter 1998), although it was pointed out by Charlesworth (1991) that AOD caused by identity disequilibrium has no effect on allele frequencies at the neutral loci concerned. As was noted by Bierne et al. (2000), for this case it is necessary to treat apparent overdominance at the neutral locus as a phenomenon that is distinct from any retardation of loss of variability. This point is reinforced by the results in Table 1 and Table 2 for the case of selection against recurrent deleterious mutations, which show that apparent overdominance can accompany an acceleration of loss of neutral variability when the dominance coefficient h is sufficiently large (see the entries with h = 0.45). Of course, h < 1/2 is a necessary condition for both retardation of loss of variability and apparent overdominance. This is not restrictive, given the evidence that the dominance coefficients for slightly deleterious mutations are generally <1/2 (Crow 1993; Manna et al. 2011).
Selection against deleterious mutations
General conclusions:
Computer simulations of systems with many loci subject to mutation to deleterious alleles have shown that a noticeable retardation of loss of neutral variation at linked loci can occur (Latter 1998; Pamilo and Palsson 1998; Palsson and Pamilo 1999; Wang and Hill 1999; Wang et al. 1999), for certain ranges of values of selection and dominance coefficients. As pointed out by Pamilo and Palsson (1998), Palsson and Pamilo (1999), Wang and Hill (1999), and Wang et al. (1999), there is a conflict between the effect of AOD on neutral variability and the effect of BGS; the latter involves a reduction in the effective population size experienced by a neutral locus caused by its linkage to deleterious mutations (Charlesworth et al. 1993). Their multilocus simulations, as well as the matrix-based investigation by Wang and Hill (1999) of a model of a single selected locus linked to a neutral locus in a sib-mating and selfing population, showed that retardation vs. acceleration of the rate of loss of neutral variability is favored by relatively weak selection (2Ns values of order ≤4) and low dominance coefficients; see, for example, figure 1 of Wang and Hill (1999).
Our analytical and numerical methods, employing a 9 × 9 matrix of “moments” of functions of the allele frequencies at the two loci and D, as well as weak selection approximations for the change in heterozygosity, have allowed us to investigate in detail the regimes in which retardation vs. acceleration of loss of neutral variability occurs.
The main conclusions are as follows:
Retardation rather than acceleration of loss of variability is favored by sufficiently low values of the dominance coefficient, h, when S = 2Ns is in the range 0.5–4.5 (Figure 3). The commonly used estimate of h = 0.25 for slightly deleterious mutations (Manna et al. 2011; Charlesworth 2015) is consistent with retardation when S ≤ 2.5, except when q* is close to 0.5.
S < 0.5 allows retardation of loss even for h values approaching 1/2; this domain of S values can be thought of as the AOD limit.
In contrast, there is always an acceleration of loss of variability when S > 4.5, even with very low h values; this constitutes the BGS limit. Despite this acceleration, apparent heterozygote advantage at the neutral locus is always observed, provided that h < 1/2.
For q* << 1 and for S >> 1, the asymptotic behavior of the expected heterozygosity at the neutral locus is well predicted by the Ne value given by the standard equation for BGS.
The magnitude of the extent of retardation or acceleration of loss of variability is an increasing function of C/S, for a given h value. Equation 19 also shows that, when S << 1, the effect is proportional to the inbred load scaled by 2N, for a given value of C.
Implications for empirical results:
Population genomic analyses of levels of nonsynonymous site variability in a variety of organisms have consistently shown that there is a wide distribution of selection coefficients against new deleterious mutations and suggest a mean S for natural populations that is >>1 (reviewed by Charlesworth 2015). The large effective population sizes of most populations that have been studied in this way imply, however, that the mean selection coefficient against a new deleterious mutation is likely to be very small. For example, the current estimate of mean hs for new nonsynonymous mutations in Drosophila melanogaster is ∼0.001; with h = 0.25, the corresponding mean s is 0.004 (Charlesworth 2015). With N = 50, as used in the experimental and simulation studies discussed below, the mean S is 0.4. If there were no variation in s for new mutations, this would suggest that they fall well within the range where Equations 18 and 19 predict retardation of loss of variability. However, we also have to take into account the fact that there is a wide distribution of s, as we discuss below. A coefficient of variation of s for new mutations of ∼2 is suggested by the Drosophila polymorphism data (Kousathanas and Keightley 2013).
Effects of multiple loci subject to mutation and selection
We now consider how multiple loci subject to deleterious mutation affect a linked neutral locus; this question can be asked about both the apparent selection coefficients and the extent of retardation or acceleration of loss of variability.
Apparent selection coefficients:
With respect to the apparent selection coefficients, previous workers (Ohta 1971, 1973; Ohta and Cockerham 1974; Bierne et al. 2000) assumed that multiple loci combine approximately additively. Table 2, which describes results for a pair of selected loci, suggests that this assumption may be somewhat conservative; in most of the examples shown there, the selection coefficients are somewhat larger than twice the corresponding single-locus values, especially with a low initial frequency of the A1 allele.
The procedure of summing contributions over all sites when there are many selected loci, as used by Ohta (1971, 1973), may thus underestimate the apparent selection coefficients, although the departure from additivity is likely to be small for the very weak selection that is probably most common. An alternative to Equation 13 of Ohta (1973), based on Equations 3, is derived in section S6 of File S1 (Equations S31). We assume that the population is being studied at a sufficiently long time after establishment that its LD is close to equilibrium. We also assume a single chromosome with a uniform rate of recombination along the chromosome, as well as a linear genetic mapping function (this somewhat overestimates the amount of recombination, compared with the true situation with partial crossover interference). For a randomly placed marker, we have the asymptotic result
| (23) |
where Btot is the inbred load associated with homozygosity for the whole chromosome, and C* = 2NM, with M denoting the map length of the chromosome (taking into account any sex differences in recombination rates, by averaging over the values for each sex).
It is useful to compare this formula with the multilocus simulation results of Latter (1998), who modeled a D. melanogaster autosome evolving in a population maintained at a size of 50 for 200 generations after its foundation from an equilibrium population. The chromosome was 100 cM in length; the absence of crossing over in males means that the effective value of M is 0.5 M, so that C* = 50 with N = 50. For a randomly placed marker, Equation 23 gives an approximate apparent selection coefficient of 0.312Btot.
Btot can be estimated as follows. The initial value of the net fitness (relative to a balancer chromosome) for homozygous D. melanogaster second chromosomes was 0.4, after purging them of major-effect mutations by extraction from a population maintained for many generations at an approximate size of 50 (Latter et al. 1995; Latter 1998). This can be equated to the fitness of a homozygous chromosome relative to that of an outbred genotype in the initial population; the value of Btot for the initial population is then –ln(0.4), ∼0.90, assuming multiplicative fitness effects. This assumption is consistent with the log-linear decline in fitness with inbreeding coefficient F in the small laboratory populations of D. melanogaster described by Latter et al. (1995).
The simulations of Latter (1998) adjusted the number of mutable loci to produce an initial Btot of 0.90. However, we need to take into account the reduction in Btot as variability is lost at the selected loci. A minimum estimate of Btot at generation t is provided by assuming that the ratio of E{pq} to its initial value follows the standard neutral result, 1 – Ft = exp(–t/2N). For N = 50 and t = 200, this procedure yields Btot = 0.90 × (1 – F200) = 0.122 and a value of 0.038 for the apparent selection coefficient on a random marker, somewhat lower than the value of 0.05 given in figure 6 of Latter (1998). The discrepancy probably reflects an overestimate of the loss of variability by use of the standard neutral value of Ft. Latter’s figure 4 shows F200 = 0.7 for the selected loci in his simulations, giving Btot = 0.270 and an apparent selection coefficient of 0.084. The single-locus approximations for the apparent selection coefficients often overestimate the true values (see Table 1), so that it is not surprising that Equation 23 overestimates the apparent selection coefficient.
Since the numerator of Equation 23 involves a product of C* and a logarithmic function of C*, the apparent selection coefficients are roughly inversely proportional to population size. Species with larger numbers of chromosomes than Drosophila, and with crossing over in both sexes, will have much smaller apparent selection coefficients for the same value of N. Detectable apparent selection coefficients arising from AOD in randomly mating populations are likely to be found only in very small populations, especially in organisms with large numbers of chromosomes such as most vertebrates. This suggests that AOD arising from identity disequilibrium (Szulkin et al. 2010) is a more likely explanation of heterozygosity/fitness relations in natural populations than LD due to drift in a randomly mating population (Hansson and Westerberg 2002), unless the population has been reduced to a very small number of breeding individuals.
Rate of loss of variability:
This approach can also be applied to the rate of loss of neutral variability, using the weak selection approximation of Equation 19, which includes only the first- and second-order terms in S in the expression for ε = Ne/N – 1 = 2N ΔHsel, again assuming that LD is close to neutral equilibrium. The derivation of the approximate expected value of ε for a neutral marker placed randomly on a chromosome, using the same assumptions as before, is given in section S7 of File S1 (Equations S32–S35). The final result is
| (24) |
where CVs is the coefficient of variation of the distribution of s for new mutations. With an exponential distribution, CVs = 1.
We can compare the predictions of this equation with the simulation results for a mean S of 0.5 in Latter’s (1998) table 6, which is similar to the value suggested by the population genomic analyses for a population size of 50 and meets the weak selection requirement. Latter used a measure (ΔF) derived from the net change in F for neutral loci over t generations, relative to the corresponding value without selection; this is approximately equivalent to the ratio of N to the harmonic mean of Ne, i.e., to 1/(1 + ε). For the case of an exponential distribution of s, he estimated ΔF as ∼0.6 for t = 200, for three different dominance coefficients, 0, 0.1, and 0.2. With h = 0.2, the estimate of ε for a random marker with a population size of 50 is 1.35Btot; with the above estimate of Btot = 0.270 for generation 200, this gives ε = 0.365 and ΔF = 0.73, which is somewhat higher than Latter’s (1998) value. But the comparisons of the single-locus simulation results with Equation 19 suggested that the approximation underestimates ε by ∼30% (Figure 4); increasing ε by this amount to 0.474 gives ΔF = 0.68. If the effects of different loci on Ne combine multiplicatively rather than additively, as is known to be the case for BGS (Charlesworth and Charlesworth 2010, p. 402), ε should be replaced by exp(ε) – 1 = 0.606, and ΔF = 0.62, which is very close to the simulation value. The value of h has only a small effect on the results; for example, with h = 0, the uncorrected ε = 0.454 and ΔF = 0.69.
We can also use Equation 24 to determine the expectations with the population genomics estimates of a mean s of 0.004 and CVs = 2, with h = 0.25. Substituting these values into Equation 24 with the same N and recombination parameters as before, Equation 24 gives a negative value of ε = –0.0663, and ΔF = 1.07. This implies that BGS would cause a reduction in Ne, to ∼93% of the neutral value. This conclusion should be treated with caution, since the very wide distribution of selection coefficients in this case means that there will be a substantial range of values that violate the assumption of S < 1 required for the validity of Equation 19.
Relation to the data on loss of variability in small populations
Most of the data that are relevant to the question of whether AOD generated by linkage to deleterious mutations can retard the loss of neutral variability in small randomly mating populations come from a few studies of the behavior of putatively neutral markers and quantitative traits in small laboratory populations of D. melanogaster. To avoid the complications of interpretation associated with high levels of inbreeding, we do not discuss results from studies of sib-mating or first-cousin mating, although these suggest a significant retardation of loss of variability (Rumball et al. 1994), consistent with the conclusion that retardation is favored when S is small. Latter (1998) analyzed the results of Latter et al. (1995) for two allozyme markers and found a ΔF value of 0.52 over 200 generations in populations maintained at a size of ∼50 (see Latter’s table 6), consistent with his multilocus simulation results based on an exponential distribution of s.
Experiments using seven allozyme markers and two bristle traits in pedigreed populations ranging in size from sib-mating to 500, and maintained for 50 generations, showed that the regression coefficients of heterozygosity and genetic variance on pedigree inbreeding coefficient were significantly lower than the value of 1 expected with neutrality, suggesting a retardation of loss of variability (Gilligan et al. 2005). The facts that the results from populations with different sizes were pooled, and that there is a nonlinear relation between heterozygosity and inbreeding coefficient when AOD is acting, mean that a quantitative analysis of these results in terms of our model is impossible. In addition, a later analysis of eight microsatellite loci over 48 generations for the same set of populations showed an acceleration of loss of variability by this criterion (Montgomery et al. 2010), which the authors interpreted as evidence for selective sweeps related to adaptation to the laboratory environment.
Different experiments have, therefore, given very different patterns, and it is certainly possible that any retardation of loss of variability due to AOD may be obscured by selective sweeps, even if N is of a size that would otherwise lead to a retardation of loss of variability. However, a basic problem with experiments of this type is that the heterozygosity for a single neutral locus has a very high stochastic variance, even if allele frequencies are estimated with complete accuracy (Avery and Hill 1977); see Equation S39 of File S1, section S8. This yields the asymptotic expression for large t:
| (25) |
This equation implies that the relative error increases as mean heterozygosity decreases. For example, at generation 100, if the initial heterozygosity was at the maximal value of 0.5 for a biallelic locus, the mean neutral H for N = 50 is 0.184, yielding an expected coefficient of variation for a single locus in a single population of 1.47. To obtain a coefficient of variation of 0.1, (1.47/0.1)2 = 188 independent heterozygosities would need to be measured, from either independent loci or populations or from a combination of the two. Given that figure 6 of Latter (1998) shows a difference of ∼0.2 between the expected neutral F value and the value from simulations with AOD due to deleterious mutations, which corresponds to the difference in scaled H values, it is clear that many more replications than this would be necessary to obtain statistically significant results, especially as the distribution of H is far from normal. The differences between the different experiments could thus be purely stochastic, especially as the autocorrelations between H values in different generations mean that the regression tests used by Gilligan et al. (2005) and Montgomery et al. (2010) are problematic.
It is possible that tight linkage of neutral markers to sets of very strongly and highly recessive deleterious variants that are in repulsion LD with each other could maintain variation as a result of the pseudo-overdominance that can arise in this circumstance: see the section on effects of multiple loci subject to mutation and selection, and Charlesworth and Charlesworth (1997), Palsson and Pamilo (1999), and Palsson (2001). This is most likely to occur in very small populations; major-effect mutations are relatively sparsely distributed, so that their effects on neutral variability are likely to be restricted to specific genomic regions, rather than evenly spread across the genome. Recent genomic investigations of the well-known Chillingham population of cattle, which have been maintained at a very small population size for 350 years, are consistent with this interpretation. Genome-wide diversity is very low, and residual variability is localized to a number of specific genomic locations (Williams et al. 2016).
A full assessment of the possibility that a realistic distribution of mutational effects on fitness (DFE), which are currently becoming available from genome-wide polymorphism data (Charlesworth 2015), can cause significant retardation of the loss of neutral variability will require simulations that use realistic DFEs, coupled with experiments on the effects of reduced population size that exploit the high levels of replication that can be achieved using modern genomic technology for generating large numbers of markers. It would also be desirable to use populations that have been maintained for a long period in the laboratory at a large size as the initial population, to avoid possible confounding effects of selective sweeps. Current theory and data cannot convincingly answer the question of whether AOD due to deleterious mutations is a credible explanation for the presence of more than expected levels of variability in small populations.
Selection in favor of heterozygotes
Our main conclusion is that retardation of loss of variability is nearly always observed when there is heterozygote advantage, except when the allele frequency at the selected locus is close to the boundaries 0 or 1, and there is relatively strong selection, when it is known that heterozygote advantage tends to accelerate the loss of variability (Robertson 1962). As shown in Figure 4 and Table 3, a substantial degree of retardation can occur when c < < 10(s + t), especially with intermediate equilibrium allele frequencies. As with mutation and selection, with S + T < 1 and a fixed value of C the magnitude of retardation is proportional to the inbred load scaled by 2N (see Equation 22).
Genome scans for signatures of balancing selection suggest that this is a relatively rare phenomenon compared with the number of genes in the genome (Charlesworth 2006; Gao et al. 2015), so that it is unlikely that a given neutral site will be closely linked to a locus maintained by heterozygote advantage. The exception is inversion polymorphisms in organisms such as Drosophila; these can cover relatively large proportions of the genome and also suppress crossing over to a considerable extent outside their breakpoints (Krimbas and Powell 1992). They could thus have a substantial impact on the behavior of neutral variants in small populations. Our conclusions concerning heterozygote advantage are, therefore, relevant to experimental results on small populations where inversions are segregating, as is often the case with D. melanogaster.
Acknowledgments
We thank Nick Barton and Bill Hill for useful discussions. Nick Barton, Reinhard Bürger, Bill Hill, Jinliang Wang, and an anonymous reviewer made helpful suggestions for improving this article. L.Z. was supported by a China Scholarship Council scholarship (no. 201506100068).
Appendix
Approximations for the Expected Frequency of B2 and Expected Heterozygosity at the Selected Locus
With weak selection, the deterministic change in the frequency q of allele B2 under selection and mutation (neglecting terms of order s2) is given by
| (A1) |
Because Δq is third degree in q, it is impossible to obtain exact, closed equations for the changes in the expectations of q and q2, E{q} and E{q2}, under drift, selection, and mutation. However, if it is assumed that second-order terms in the deviation of q from its deterministic equilibrium value q*, given by Equation 5a, can be ignored, approximate recursion relations can be obtained as follows. [The matrix calculations described in section S1 of File S1, which neglect third-order terms in (q – q*), yield better approximations.]
Taking the first derivative of Equation A1 with respect to q yields a linear deterministic recursion relation for the departure of q in generation t from the equilibrium frequency q* under mutation and selection, which is given by Equations 5,
| (A2a) |
where
| (A2b) |
Since this is linear in q, the mean of q under drift, selection, and mutation remains at q*.
The equilibrium expected heterozygosity at the selected locus is given by H2* = 2q*(1 – q*)A/(1 + A), where A = –4Neκ (e.g., Charlesworth and Charlesworth 2010, p. 355). The change in the expected heterozygosity at the selected locus at time t, H2t, is approximately equal to the expectation of 2(1 – 2qt)Δq(qt); combining this with Equations A2 yields the following recursion relation:
| (A3) |
Because H2 is twice the expectation of pq, Equation A3 was used in Equations 4 for determining the approximate apparent selection coefficients.
Approximate Neutral Recursion Relations for the Expectation of r2
For the purely neutral case, Sved (1971) proposed the following approximation for the recursion relation for the expected correlation coefficient in generation t, E{rt2},
| (A4a) |
where c is the frequency of recombination between the two loci, assumed to be <<0.5. Since mutation at the B locus reduces the coefficient of linkage disequilibrium D with the A locus by u + v each generation, this equation should be modified as follows:
| (A4b) |
This yields the equilibrium solution
| (A4c) |
where C′ = 2N(c + u + v) and E*{} indicates the expectation at statistical equilibrium.
If the initial population is in linkage equilibrium, Equations A4 yield the following homogeneous recursion relation:
| (A5) |
This suggests the following expression for σd2 = E{D2}/E{xypq}:
| (A6a) |
From Ohta and Kimura (1971), the equilibrium value of σd2 is given by
| (A6b) |
The simulation results show that Equation A6a tends to overestimate E{r2} for t ≤ 0.5N; it is quite accurate for later generations up to t = N and tends to underestimate E{r2} thereafter. Equation A5 gives a better approximation for the earlier and later generations, but a worse approximation for the intermediate generations.
Expectations of the Reciprocals of the Allele Frequencies at the Neutral Locus
The expectations of x–1 and y–1 = (1 – x) –1, conditioned on segregation at locus A, for use in Equations 3 and 9, can be derived using the diffusion equation solution for the case of pure drift (Kimura 1955), under the assumption that departures from strict neutrality can be neglected as a first-order approximation. For this purpose, it is convenient to rescale time in units of 2N generations, such that T = t/(2N). Using the first three terms in the series expansion for the unconditional probability density of the frequency x of A1 at time T, given an initial frequency x0, we have
| (A7) |
The expected frequency of heterozygotes at locus A at time T is
| (A8) |
To determine the conditional probability of x for a segregating population, we need the probability that the population is still segregating for A1 and A2 at time T, given by equation 8.4.9 of Crow and Kimura (1970):
| (A9) |
The conditional probability density of x at time T is then given by φ/Ps, which can be used to determine the mean of x–1 at time T for segregating populations,
| (A10) |
where γ is Euler’s constant (∼0.5772), which is equal to the difference between the sum and integral of 1/x.
An equivalent expression for the expectation of y–1 can be obtained by interchanging x and y and x0 and y0 in these equations.
For sufficiently large T, it is well known that Equation A7 implies that the conditional probability distribution becomes uniform (Fisher 1930), regardless of the initial allele frequency, so that its mean should be well approximated by
| (A11) |
As was pointed out by Fisher (1930), there is a slight departure of the asymptotic distribution from a uniform distribution, due to the inaccuracy of the diffusion approximation near the frequencies 0 and 1, with lower frequencies in the subterminal classes close to either boundary, although the distribution is still symmetrical about 0.5. Using the table of exact values computed by Fisher for the first 10 classes from 1/(2N) upward and from 1 – 1/(2N) downward, the asymptotic conditional means are slightly greater than the above value, by 2.299 – [2.6556 × 2N/(2N – 1)]. Addition of this term to Equation A11 provides a slightly more accurate approximation for the long-term expectation of the reciprocals of the allele frequencies, which was used to calculate the asymptotic expected selection coefficients.
Footnotes
Communicating editor: R. Nielsen
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.188912/-/DC1.
Literature Cited
- Avery P. J., Hill W. G., 1977. Variability in genetic parameters among small populations. Genet. Res. 29: 193–213. [DOI] [PubMed] [Google Scholar]
- Balick D. J., Do R., Cassa C. A., Reich D., Sunyaev S., 2015. Dominance of deleterious alleles controls the response to a population bottleneck. PLoS Genet. 11: e1005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N., Tsitrone A., David P., 2000. An inbreeding model of associative overdominance during a population bottleneck. Genetics 155: 1981–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2015. Causes of natural variation in fitness: evidence from studies of Drosophila populations. Proc. Natl. Acad. Sci. USA 12: 1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Campos J. L., 2014. The relations between recombination rate and patterns of molecular evolution and variation in Drosophila. Annu. Rev. Genet. 48: 383–403. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 1997. Rapid fixation of deleterious alleles by Muller’s ratchet. Genet. Res. 70: 63–73. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2010. Elements of Evolutionary Genetics. Roberts and Company, Greenwood Village, CO. [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., 1991. The apparent selection on neutral marker loci in partially inbreeding populations. Genet. Res. 57: 159–175. [Google Scholar]
- Charlesworth D., 2006. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham C. C., Weir B. S., 1968. Sib-mating with two loci. Genetics 60: 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., 1993. Mutation, mean fitness, and genetic load. Oxf. Surv. Evol. Biol. 9: 3–42. [Google Scholar]
- Crow J. F., Kimura M., 1970. An Introduction to Population Genetics Theory. Harper & Row, New York. [Google Scholar]
- Cutter A. D., Payseur B. A., 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat. Rev. Genet. 14: 262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David P., 1998. Heterozygosity-fitness correlations: new perspectives on old problems. Heredity 80: 531–537. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics. 1. Theoretical Introduction. Springer-Verlag, New York. [Google Scholar]
- Fisher R. A., 1930. The distribution of gene ratios for rare mutations. Proc. R. Soc. Edinb. 50: 205–220. [Google Scholar]
- Frydenberg O., 1963. Population studies of a lethal mutant in Drosophila melanogaster. I. Behaviour in populations with discrete generations. Hereditas 50: 89–116. [Google Scholar]
- Gao Z., Przeworski M., Sella G., 2015. Footprints of ancient-balanced polymorphisms in genetic variation: data from closely related species. Evolution 69: 431–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilligan D. M., Briscoe D. A., Frankham R., 2005. Comparative losses of quantitative and molecular genetic variation in finite populations of Drosophila melanogaster. Genet. Res. 85: 47–77. [DOI] [PubMed] [Google Scholar]
- Glémin S., Ronfort J., Bataillon T., 2003. Patterns of inbreeding depression and architecture of the load in subdivided populations. Genetics 165: 2193–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg R., Crow J. F., 1960. A comparison of the effect of lethal and detrimental chromosomes from Drosophila populations. Genetics 45: 1153–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P. R., Loewe L., Charlesworth B., 2010. Estimating the parameters of selection on nonsynonymous mutations in Drosophila pseudoobscura and D. miranda. Genetics 185: 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection. Part V. Selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844. [Google Scholar]
- Haldane J. B. S., 1949. The association of characters as a result of inbreeding. Ann. Eugen. 15: 15–23. [DOI] [PubMed] [Google Scholar]
- Hansson B., Westerberg L., 2002. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 11: 2467–2474. [DOI] [PubMed] [Google Scholar]
- Hill W. G., 1977. Correlation of gene frequencies between neutral linked genes in finite populations. Theor. Popul. Biol. 11: 239–248. [DOI] [PubMed] [Google Scholar]
- Hill W. G., 2010. Understanding and using quantitative genetic variation. Philos. Trans. R. Soc. B 365: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 226–231. [DOI] [PubMed] [Google Scholar]
- Hoffman J. I., Simpson F., David P., Rijks J., Kuiken T., et al. , 2014. High-throughput sequencing reveals inbreeding depression in a natural population. Proc. Natl. Acad. Sci. USA 111: 3775–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., Barton N. H., 2005. Theoretical models of selection and mutation on quantitative traits. Philos. Trans. R. Soc. B 360: 1411–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1955. Solution of a process of random genetic drift with a continuous model. Proc. Natl. Acad. Sci. USA 41: 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A., Keightley P. D., 2013. A comparison of models to infer the distribution of fitness effects of new mutations. Genetics 193: 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas C. B., Powell J. R. (Editors), 1992. Drosophila Inversion Polymorphism. CRC Press, Boca Raton, FL. [Google Scholar]
- Latter B. D. H., 1998. Mutant alleles of small effect are primarily responsible for the loss of fitness with slow inbreeding in Drosophila melanogaster. Genetics 148: 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latter B. D. H., Mulley J. C., Reid D., Pascoe L., 1995. Reduced genetic load revealed by slow inbreeding in Drosophila melanogaster. Genetics 139: 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna F., Martin G., Lenormand T., 2011. Fitness landscapes: an alternative theory for the dominance of mutation. Genetics 189: 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Montgomery M. E., Woodworth L. M., England P. R., Briscoe D. A., Frankham R., 2010. Widespread selective sweeps affecting microsatellites in Drosophila populations adapting to captivity: implications for captive breeding programs. Biol. Conserv. 143: 1842–1849. [Google Scholar]
- Neher R. A., 2013. Genetic draft, selective interference and population genetics of rapid adaptation. Annu. Rev. Ecol. Evol. Syst. 44: 195–215. [Google Scholar]
- Ohta T., 1971. Associative overdominance caused by linked detrimental mutations. Genet. Res. 18: 277–286. [PubMed] [Google Scholar]
- Ohta T., 1973. Effect of linkage on behavior of mutant genes in finite populations. Theor. Popul. Biol. 4: 145–172. [Google Scholar]
- Ohta T., Cockerham C. C., 1974. Detrimental genes with partial selfing and effects on a neutral locus. Genet. Res. 23: 191–200. [DOI] [PubMed] [Google Scholar]
- Ohta T., Kimura M., 1970. Development of associative overdominance through linkage disequilibrium in finite populations. Genet. Res. 18: 277–286. [DOI] [PubMed] [Google Scholar]
- Ohta T., Kimura M., 1971. Linkage disequilibrium between two segregating nucleotide sites under steady flux of mutations in a finite population. Genetics 68: 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson S., 2001. The effects of deleterious mutations in cyclically parthenogenetic organisms. J. Theor. Biol. 208: 201–214. [DOI] [PubMed] [Google Scholar]
- Palsson S., Pamilo P., 1999. The effects of deleterious mutations on linked neutral variation in small populations. Genetics 153: 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamilo P., Palsson S., 1998. Associative overdominance, heterozygosity and fitness. Heredity 81: 381–389. [DOI] [PubMed] [Google Scholar]
- Price G. R., 1970. Selection and covariance. Nature 227: 520–521. [DOI] [PubMed] [Google Scholar]
- Robertson A., 1962. Selection for heterozygotes in small populations. Genetics 47: 1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumball W., Franklin I. R., Frankham R., Sheldon B. L., 1994. Decline in heterozygosity under full-sib and double first-cousin inbreeding in Drosophila melanogaster. Genetics 136: 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago E., Caballero A., 1995. Effective size of populations under selection. Genetics 139: 1013–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J. A., 1968. The stability of linked systems of loci with a small population size. Genetics 59: 543–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J. A., 1971. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor. Popul. Biol. 2: 125–141. [DOI] [PubMed] [Google Scholar]
- Sved J. A., 1972. Heterosis at the level of the chromosome and at the level of the gene. Theor. Popul. Biol. 3: 491–506. [DOI] [PubMed] [Google Scholar]
- Szulkin M., Bierne N., David P., 2010. Heterozygosity fitness correlations: a time for reappraisal. Evolution 64: 1202–1217. [DOI] [PubMed] [Google Scholar]
- Wang J., Hill W. G., 1999. Effect of selection against deleterious mutations on the decline in heterozygosity at neutral loci in closely inbreeding populations. Genetics 153: 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Hill W. G., Charlesworth D., Charlesworth B., 1999. Dynamics of inbreeding depression due to deleterious mutations in small populations: mutation parameters and inbreeding rate. Genet. Res. 74: 165–178. [DOI] [PubMed] [Google Scholar]
- Whitlock M. J., Barton N. H., 1997. The effective size of a subdivided population. Genetics 146: 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. L., Hall S. J. G., Del Corvo M., Ballingall K. T., Colli L., et al. , 2016. Inbreeding and purging at the genomic level: the Chillingham cattle reveal extensive, non-random SNP heterozygosity. Anim. Genet. 47: 19–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.