Abstract
Recent studies of protein evolution contend that the longer an amino acid substitution is present at a site, the less likely it is to revert to the amino acid previously occupying that site. Here we study this phenomenon of decreasing reversion rates rigorously and in a much more general context. We show that, under weak mutation and for arbitrary fitness landscapes, reversion rates decrease with time for any site that is involved in at least one epistatic interaction. Specifically, we prove that, at stationarity, the hazard function of the distribution of waiting times until reversion is strictly decreasing for any such site. Thus, in the presence of epistasis, the longer a particular character has been absent from a site, the less likely the site will revert to its prior state. We also explore several examples of this general result, which share a common pattern whereby the probability of having reverted increases rapidly at short times to some substantial value before becoming almost flat after a few substitutions at other sites. This pattern indicates a characteristic tendency for reversion to occur either almost immediately after the initial substitution or only after a very long time.
Keywords: weak mutation, fitness landscape, entrenchment, reversible Markov chain
IN the context of evolutionary theory, reversion describes a population that returns to an ancestral character state (Porter and Crandall 2003). While many early (Dollo 1893; Muller 1939; Simpson 1953; Gould 1970) and more recent (Teotónio and Rose 2001; Collin and Miglietta 2008; Bridgham et al. 2009; Tan et al. 2011) discussions of reversion consider an environmental change that confers a selective advantage to an ancestral phenotype, reversion may also occur at the level of nucleic acids or protein sequences, with evolution proceeding under long-term purifying selection (Kimura 1983). Such reversions occur both because of the strictly limited number of character states (four possible nucleotides or 20 possible amino acids, Jukes and Cantor 1969) and because selection on molecular function may constrain a given position to only a subset of these possible character states (Rokas and Carroll 2008; Breen et al. 2012).
It has long been hypothesized that epistatic interactions should lower the rate of reversion, rendering evolution effectively irreversible (Muller 1918, 1939). This issue has been especially important recently, due to ongoing debate in the field of protein evolution about how position-specific preferences for amino acids may change over time (Naumenko et al. 2012; Pollock et al. 2012; Ashenberg et al. 2013; Pollock and Goldstein 2014; Bazykin 2015; Doud et al. 2015; Goldstein et al. 2015; Risso et al. 2015; Shah et al. 2015; Usmanova et al. 2015). Specifically, several groups have suggested that once an amino acid substitution occurs at a particular position, epistatic interactions with subsequent substitutions at other positions should tend to increase the selective preference for the derived amino acid relative to the ancestral state (Naumenko et al. 2012; Pollock et al. 2012; Shah et al. 2015; cf. Fisher 1930, p. 95), a phenomenon known as entrenchment (Shah et al. 2015). This means that a mutation that was nearly neutral when it originally went to fixation may become increasingly deleterious to revert, which would cause a decreasing propensity to revert as time elapses.
However, the above verbal argument is not entirely convincing. While it is easy to imagine some forms of epistasis that would cause reversion rates to decrease over time, evolutionary dynamics on high-dimensional fitness landscapes can have many counterintuitive properties (Conrad 1990; Gavrilets 1997; Carneiro and Hartl 2010; McCandlish et al. 2013, 2015b; Kondrashov and Kondrashov 2015). Here we undertake a rigorous mathematical investigation into the relationship between the rate of reversion and the presence of epistasis, for arbitrary fitness landscapes. We study this problem under the assumption that mutation is weak relative to drift, so that the evolution of the population can be modeled as a Markov chain on the set of genotypes (for a review see McCandlish and Stoltzfus 2014). Our main result concerns the dynamics at stationarity, where we consider the probability distribution of waiting times until a focal substitution reverts, averaging appropriately over the genetic backgrounds in which this substitution could occur. Under these conditions, we show that for any site involved in at least one epistatic interaction, the rate of reversion is a strictly decreasing function of the time since the initial substitution.
Our first task is to provide a rigorous definition for the rate of reversion. For a population evolving under weak mutation, we can treat the population as a single particle that jumps from one genotype on the fitness landscape to another at each substitution event. We consider some focal set of genotypes that includes the starting state of the population. If we observe the population for long enough, the population will eventually leave this focal set and trace a path through the space of genotypes. At each point along this path, it has some propensity to fix a genotype in the focal set, that is, to revert. Eventually, this propensity is realized and the population returns to the focal set. If we continue to watch the population for long enough, this process will repeat itself many times, and we can ask the following question: Given that the population left the focal set t time units ago and has not yet returned to it, what is the expected instantaneous propensity for that population to return to the focal set; i.e., what is the rate of reversion as a function of time?
To study reversion it is helpful to note the following relationship between the rate of reversion and the distribution of waiting times until a reversion event occurs. As we observe the population evolving on the fitness landscape, every time the population leaves the focal subset, we can record the waiting time until it first returns. And again, if we observe the population for long enough, these waiting times will converge to a particular distribution. Such a distribution naturally averages over all of the possible mutational paths by which the population could leave the focal set, weighting each by its probability of occurring under long-term purifying selection, i.e., at stationarity. Importantly, the reversion rate described above is equal to the hazard function of this probability distribution of reversion times, that is, the probability density of this distribution at time t, conditioned on drawing a value t or greater. Thus, we can study how the rate of reversion changes the longer the population has been absent from the focal subset by studying the hazard function of this distribution of reversion times.
The case of a biallelic nonepistatic (i.e., additive) fitness landscape provides an instructive, introductory example. In this case, it is easy to show that the distribution of return times for a particular allele at a particular site is always exponentially distributed, corresponding to a constant hazard function. That is, for a biallelic site on a nonepistatic fitness landscape, the reversion rate does not change as a function of time. We want to understand how this simple situation changes in the presence of epistasis.
Here we show that if the focal site interacts epistatically with at least one other site, then the hazard function of the distribution of reversion times, and therefore the rate of reversion, is strictly decreasing in time. This implies that the longer a population has been away from the focal set of genotypes, the longer the expected waiting time until it returns to that set. Moreover, this decreasing reversion rate is due to two factors, each of which would individually result in a decreasing reversion rate.
The first factor is coevolution between sites as suggested by, e.g., Pollock et al. (2012). As long as the derived allele is resident at the focal site, it forms part of the genetic background for other substitutions, and this causes the population to tend to spend more time at genotypes where the derived allele is selectively favored.
To isolate the effect of this first factor, we consider a modified process where we do not allow the focal site to return to its original state after the initial substitution. Thus, the dynamics after the initial substitution capture the acclimatization of the rest of the genome to the derived state at the focal site. While reversion events cannot occur under this modified model, we nonetheless keep track of the reversion rate that would occur if we were to suddenly allow reversions. We show that the reversion rate for this modified process is decreasing for sites involved in at least one epistatic interaction, which demonstrates that coevolution between sites leads to reversion rates that decrease in time.
The second factor that produces decreasing rates of reversion is statistical in nature. This second factor arises because to focus on the first time a population returns to the ancestral state, we must condition on that return not having yet occurred when we calculate the rate of reversion. If the population is at a genotype with a high reversion rate, it tends to actually revert, so that the high reversion rate no longer contributes to the expectation. This alone results in reversion rates that decrease in time. To put this in a more biological light, populations that have been gone for a long time from the focal subset tend to have a low propensity to return to the focal subset, because if they had a high propensity, they would have returned already.
To isolate the effects of this second factor, we can consider a different, modified model in which the population never moves to another genotype outside the focal set once it leaves the focal set. Thus, each time the population leaves the focal set, its propensity to return to the focal set is constant. Nonetheless, the reversion rate under this model will be strictly decreasing in time if there is any variation among these genotypes in the propensity to return to the focal set.
In actuality, these two factors operate simultaneously, and their effects on the time evolution of the rate of reversion are coupled. Nonetheless, when both factors operate, we show that the reversion rate is still decreasing with the time since the substitution of the derived allele at the focal site.
We also consider what occurs when more than two alleles are available at a site and the more general case of reversion to subsets of genotypes, e.g., reversion to the set of codons corresponding to a particular amino acid. The key observation in this context is that the two factors above operate when there is any genotype-to-genotype variation in the propensity to return to the focal set. While for models with biallelic sites epistasis is the only way of producing variation in these propensities, for more general models with more than two alleles per site the rate of reversion may be decreasing even in the absence of epistasis.
In addition to our main results, which concern populations that have already been evolving on the same fitness landscape for a long time, we briefly explore how changes to the fitness landscape affect the dynamics of reversion. Finally, we explore several simple examples to gain intuition for the magnitude and evolutionary importance of reversion rates that decrease in time.
Materials and Methods
Population-genetic model
We consider a population evolving in continuous time under weak mutation on an arbitrary finite-state fitness landscape (e.g., Iwasa 1988; Sella and Hirsh 2005; McCandlish et al. 2015b). In this regime, we can model the population as a single particle that moves from genotype to genotype at each fixation event (see McCandlish and Stoltzfus 2014, for a review). More formally, we model evolution as a continuous-time Markov chain with a rate matrix
| (1) |
where is the scaled Malthusian fitness of genotype i and is the mutation rate from i to j. We further assume that the fitness landscapes is connected, so that there exists a mutational path between any two genotypes i and j and that the Markov chain defined by is reversible, so that there exists a stationary probability distribution of the chain defined by such that for all This latter condition will be satisfied whenever the neutral mutational dynamics produce a reversible Markov chain (see, e.g., Sella and Hirsh 2005; McCandlish et al. 2015b); a simple sufficient condition is that the mutation rates are pairwise symmetric, for all
We are interested in the situation where a population has just left some subset of states A and want to study the waiting time for the population to return to that subset A. Without loss of generality, we can order the states so that all states in A come after the states in the complement of A, so that we can write in block matrix form as
| (2) |
where is the complement of A and gives the transition rates from to A and gives the transition rates from A to
Our main object of study is the absorbing Markov chain with rate matrix where absorption corresponds to a return to the subset A. Because we assume that a population starts at time 0 having just left the subset of states A, this means that an absorption event is also a reversion event, so that we can study the dynamics of reversion by studying the waiting time until absorption for the Markov chain with rate matrix For brevity, we simply call this matrix
The row sums of (or equivalently, the row sums of ) give the propensity for a population currently fixed at genotype i to return to A, and we write the rate at which such an event occurs for a population fixed at genotype i as Let be the probability that the population is fixed for genotype i and has not yet reverted at time t. Then the time evolution of is given by
| (3) |
where gives the probability that the population initially left the subset A by becoming fixed for genotype i.
The hazard function of reversion times
Consider a population that first leaves subset A by fixing genotype i. The probability that the population reverts, that is, first becomes fixed for a genotype in the subset A, during the time interval is given by where
| (4) |
and is 1 for and 0 otherwise. Thus, is the probability density function of the distribution of reversion times for a population that initially leaves subset A by fixing genotype i; we note that is indeed a proper probability density since defines an ergodic Markov chain and so populations return to the subset A with probability 1.
Now, a population that has already been evolving on a fitness landscape for a long time is much more likely to leave the subset A by fixing some genotypes rather than others. To capture this effect, we can choose the initial distribution by considering a population whose genotype is described by the stationary distribution and then condition on leaving the subset A in the interval We thus specify the distribution as
| (5) |
| (6) |
| (7) |
With this choice of initial distribution, the probability density function for the distribution of reversion times is given by
| (8) |
Note that this is the same distribution that we would get if we watched a single population evolve for an infinite amount of time and recorded, each time the population left A, the waiting time to return to A.
We now turn to formally defining the rate of reversion. What we want to understand is how the rate at which a population first returns to some set of states changes the longer a population has been outside that set. This suggests that we should define the reversion rate as the probability density of a population returning to set A for the first time in the time interval given that the population has not already returned to set A before time t. In the more general context of nonnegative probability distributions, this quantity is known as the hazard function (or failure rate or force of mortality). Thus, we define the reversion rate to be the hazard function of the probability distribution of reversion times.
For instance, consider the distribution of reversion times at stationarity, with density given by cumulative distribution function and complementary cumulative distribution function Then the reversion rate at time t is given by the hazard function where
| (9) |
If is increasing in t, it means that populations that have been away from the set A for a long time on average have instantaneous rates of return to A larger than the average rate of return to A of populations that have been away only for a short time. Likewise, if is decreasing in t, it means that populations that have been away from the set A for a long time on average have instantaneous rates of return to A smaller than the average rate of return to A of populations that have been away only for a short time. Another quantity of interest is the expected remaining time until reversion, given that the population has not yet reverted at time t. This expected waiting time can be expressed in terms of the hazard function as
| (10) |
Furthermore, from the above equation it is easy to see that if the reversion rate is increasing in time, then the expected remaining waiting time is decreasing in time, whereas if the reversion rate is decreasing in time, then the expected remaining waiting time is increasing.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
General theory of reversions
Our first main result is that for a population that has already been evolving on the same fitness landscape for a long time, the rate of reversion to A, is a nonincreasing function of t, where t is the time since the population left A. Moreover, the reversion rate is strictly decreasing unless all genotypes in have the same reversion rate (i.e., is constant), in which case is also constant. This result shows that populations that have spent a longer time away from A cannot possibly have higher rates of return to the ancestral character state or shorter expected remaining waiting times until return.
This result is a simple consequence of the fact, well known in the mathematical literature (Kielson 1979; Aldous and Fill 2002), that the distribution of return times to a subset for a stationary, finite-state, reversible, continuous-time Markov chain takes the form of a mixture of exponential distributions (in the mathematical literature, such a distribution is known as a “completely monotone” distribution). It is easy to show that the hazard function for such a distribution is strictly decreasing except in the case of a pure exponential distribution. In Appendix A, we provide elementary proofs of these facts as well as the fact that that the distribution of return times is a pure exponential only in the case when is constant.
Connection to epistasis:
An immediate implication of this result is that the reversion rate for any site in a fitness landscape that is involved in an epistatic interaction must be strictly decreasing in time. This is because epistatic interactions result in variation in the genotype-specific rates of return to A, since different back mutations have different fitness consequences.
More formally, consider a biallelic fitness landscape with L sites and with forward and backward mutation rates of and respectively, at site l. Thus, if j is produced from i by a forward mutation at site l, if j is produced from i by a back mutation at site l, and otherwise. Furthermore, let us pick a focal site and order the states such that all genotypes that have the allele produced by the forward mutation are in set and indexed first. We are interested in the distribution of reversion times at site
We say a site is not involved in any epistatic interactions if its fitness effect is constant, in the sense that if genotype i is produced from genotype j by a forward mutation at site then the scaled selection coefficient is equal to some constant In this case is constant and equal to where is the function relating the scaled selection coefficient to the rate of evolution. Thus if the selection coefficient of the forward mutation is constant, then is also constant. On the other hand, suppose is involved in an epistatic interaction. Then there exist and such that (1) i is produced from j by a forward mutation at site (2) is produced from by a forward mutation at site and (3) But then because since is strictly increasing and hence invertible. Thus, in this case is strictly decreasing.
The above argument shows that is strictly decreasing for a biallelic fitness landscape if and only if the focal site is involved in at least one epistatic interaction. For fitness landscapes that include more than two alleles at a site, by contrast, the notation and argument become somewhat more involved (Appendix B), but the end result is weakened to the statement that epistasis is sufficient for to be strictly decreasing. Indeed, even for nonepistatic multiallelic fitness landscapes we generically expect to be strictly decreasing. This is because even if fitness is additive between sites, the different alleles within a site will have different fitness differences from the focal allele and therefore different probabilities of fixation that result in a nonconstant
Do reversions also become more deleterious?
Besides the relationship between decreasing reversion rates and epistasis, it is interesting to ask about the relationship between decreasing reversion rates and the mean selection coefficient of reversions. This relationship is not necessarily simple. First, this is because different genotypes might have different mutation rates to the subset A, so that the decrease in might be realized by the nonreverting subset of the population becoming concentrated at genotypes with low mutation rates to the subset A rather than by having more negative selection coefficients for such mutations. However, even if all genotypes in produce mutations to subset A at the same rate, the decrease in might not correspond to reversion mutations becoming more deleterious. This is due to the nonlinearity of the function relating the scaled selection coefficient S to the rate of evolution. However, using the fact that is convex (McCandlish et al. 2015a), if each genotype in produces mutations at rate μ to a single corresponding genotype in set A, then Jensen’s inequality tells us that where is the average scaled selection coefficient of the reversion at time t and this average is taken with respect Thus, if for any t we have then the mean selection coefficient of a reversion among those populations that have not yet reverted is strictly less than the mean selection coefficient of a reversion immediately after the initial substitution away from A. This provides a sufficient condition for the mean selection coefficient of reversions to have decreased in time.
Expected reversion times:
If the reversion rate is nonincreasing over time, then the expected waiting time until reversion must be greater than what would be predicted based on the initial reversion rate alone; i.e., we must have In fact, there is a simple formula for the expected time until reversion that holds even if the Markov chain describing the weak mutation dynamics is not reversible. In particular, consider the expected rate of returns to A at stationarity when we condition on a population being in the subset i.e., Then is simply the reciprocal of this rate (see Appendix C).
However, because the reversion rate is nonincreasing in time, this mean value does not necessarily provide a very informative summary of the dynamics of reversion. For instance, consider the case where is initially high, but drops to a very low value. Under these circumstances, it can be the case that a large proportion of populations revert rapidly, but the subset of populations that do not revert rapidly may have extremely long expected reversion times, so that the mean reversion time is really an average over two very different subsets of populations. Intuitively, this situation will often arise when the set A is the basin of attraction of a fitness peak. Most populations that cross into the fitness valley around A will return to A rather than cross the valley, but a small subset will cross the fitness valley and end up at another fitness peak. The expected return time for this subset of populations might then be extremely long.
Why is the reversion rate decreasing?
To gain an intuitive understanding for why the reversion rate is decreasing in time, it is helpful to distinguish between two different phenomena that each contribute to this decrease.
The first phenomenon is the acclimatization of the rest of the genotype to being in the subset For instance, in the context of protein evolution, once a mutation has fixed at a focal site, it forms part of the genetic background that determines the fitness effects of mutations at other sites. For sites that interact epistatically with the focal site, this will tend to favor substitutions whose effects are more positive when the derived allele is present at the focal site.
Such acclimatization of the genome tends to result in a decreasing reversion rate at the focal site. To make this idea precise, let us consider a modified process where we do not allow populations to revert to A after they enter the subset Following this initial entrance, the dynamics are thus governed by a rate matrix where and is the diagonal matrix whose th entry is For ease of exposition, let us also assume that the subset is connected (i.e., one can always evolve from any state in to any other state in without returning to A; the more general case is handled in Appendix A).
Even though we do not allow reversions to occur back to A under this modified process, we can still keep track of the reversion rate that would occur were we to allow reversions. When the population initially leaves the subset A, it is distributed as However, as time elapses under the modified process, the probability that the population is fixed for genotype tends to a distribution that is Now, the initial reversion rate under the modified process is equal to the expected value of with respect to the first of these distributions, while the asymptotic reversion rate under the modified process is equal to the expected value of with respect to the second one. Clearly, the reversion rate is higher under the first distribution, since this distribution differs from the second one only in that it is more concentrated at values with high Indeed in Appendix A we show the stronger result that the reversion rate is strictly decreasing for the modified process unless is constant, in which case the reversion rate is also constant.
The second phenomenon is a statistical phenomenon having to do with the fact that we are interested in the first return to A. Even if there was no acclimatization to the initial substitution (and therefore no coevolution), the reversion rate would tend to be decreasing in time due to genotype-specific variation in the rates of return to A, i.e., the This effect occurs because populations that have not reverted even after a long time have likely spent most of this time at genotypes where returns to A are unlikely. To put this another way, if the population had spent a great deal of time at genotypes with high return rates, then it would have returned already. This phenomenon is well known in the reliability and demography literature, where unaccounted for heterogeneity can result in failure or mortality rates that decrease in time even when individual failure or mortality rates are constant (Vaupel and Yashin 1985).
To isolate this second phenomenon, we consider a second modified process in which a population that leaves A by fixing genotype i experiences no additional substitutions until it returns to A; i.e., the return rate for such a population is always The rate matrix for this process is thus The derivative of the reversion rate of this process is equal to times the variance in conditional on not having yet reverted. Since variances are nonnegative, the derivative of the reversion rate is nonpositive, so that the reversion rate is nonincreasing.
While we have constructed these two modified processes to separate the effects of acclimatization of the genome and the effects of conditioning on not having yet reverted, these two effects interact to determine the dynamics of the original process. The fact that both phenomena tend to lead to decreasing reversion rates when there is variation in (or more precisely for the second phenomenon, variation in its nonzero elements) helps clarify why the reversion rate is nonincreasing for the original process.
Reversion rates under nonstationary evolution:
So far, we have concentrated on the case of a population that has already been evolving on a fixed fitness landscape for a long time, so that each possible way of leaving the subset A is weighted by its stationary probability. However, we can also gain some strong intuitions for the behavior of the reversion rate in the more general case, where we allow the initial probability vector to be arbitrary.
In particular, in this more general setting we can show that the time evolution of the (sign-reversed) reversion rate is isomorphic to the time evolution of the mean fitness of an infinite population on a suitable fitness landscape. The basic idea is that the time evolution of the probability distribution describing the genotype of a population conditional on not having yet reverted can be viewed as the time evolution of the frequencies of genotypes in an infinite population whose mutational dynamics are specified by the off-diagonal entries of the matrix and where the Malthusian fitness of genotype i is given by That is, plays the role of a genotype-specific death rate.
More formally, let be the probability that a population is fixed for genotype i at time t given that it has not returned to the subset A by time t. Then Writing for the identity matrix, 1 for the vector of all 1’s, and for the diagonal matrix with down its main diagonal, we have
| (11) |
| (12) |
| (13) |
This is simply the “parallel” version of the replicator equation (i.e., where mutation occurs independently from reproduction), with as the rate matrix of the mutational process and as the vector of fitnesses. Furthermore, the reversion rate at time t is then given by which is simply the mean fitness with its sign reversed. The derivate of the reversion rate is then
| (14) |
| (15) |
where is the variance in the return rate with respect to the probability distribution Here, the term gives the effect on the reversion rate due to acclimatization of the genotype to being in the subset while captures the effects of conditioning on not having yet reverted. Unlike under stationarity, the effects of acclimatization for the nonstationary case can either increase or decrease the reversion rate. However, conditioning on not having yet reverted still always produces a bias toward a decreasing reversion rate, since is nonpositive.
Examples
The preceding results describe a general tendency for the reversion rate—quantified as the hazard function of the distribution of return times—to be decreasing in time. Here we present some simple examples to explore the magnitude and evolutionary consequences of this effect.
Crossing fitness valleys:
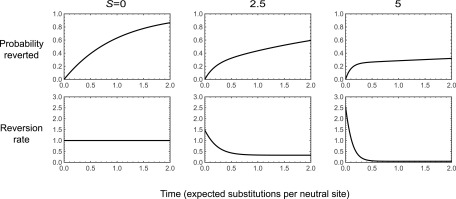
Consider the case of a biallelic fitness landscape with a symmetric fitness valley. In particular, consider the case where genotypes ab and AB have equal fitnesses and scaled selection coefficient S with respect to the valley genotypes Ab and aB and mutations occur independently at each locus at rate 1. Let us now examine the rate of reversion at the first site, so that the set and leaving A corresponds to an a A substitution. Given that the population has already been evolving on this fitness landscape for a long time and that an a A substitution has just occurred, we want to understand the distribution of times until an A a substitution occurs as a function of the depth of the fitness valley S.
This distribution of times is easy to understand intuitively. For S less than 2 the dynamics are approximately neutral and the waiting time for reversion is approximately exponential with mean 1. For larger S, the dynamics are substantially affected by the fitness valley. The best way to understand these dynamics is by considering where the population is immediately after an a → A substitution. At stationarity, we know that the frequency of substitutions into the fitness valley must be equal to the frequency of substitutions out of the valley. This means that at stationarity, half the time the population will have just fixed the valley genotype Ab and half the time it will have fixed the peak genotype Populations that fix the peak genotype are unlikely to revert in the short term, whereas populations that fix the valley genotype are likely to move to one or the other peak in the short term, with equal probability. Thus, roughly speaking, after a short time the population will either have reverted (with probability 1/4, since half the time it fixes the valley genotype and then immediately reverts half the time) or be fixed at the fitness peak (with probability 3/4). This means that reversions happen either very shortly after the initial substitution—and this occurs 1/4 of the time—or only after the long waiting time needed for deleterious fixations to occur.
Figure 1 shows these dynamics as a function of S. The top row shows the probability that an A → a reversion has occurred as a function of time and the bottom row shows the reversion rate, The leftmost column shows the neutral case. The reversion rate is constant and the probability that a reversion has occurred by time t is given by The center and right columns show the case where and respectively. In both cases, the reversion rate is high initially, starting at (probability 1/2 of starting at the valley genotype in which case returns to A occur at roughly rate S). The reversion rate drops rapidly as populations leave the valley genotype, approaching an asymptotic rate of the substitution rate of a deleterious fixation (populations that have not yet reverted are likely at the AB fitness peak; reversion occurs either via a direct substitution of a at rate or via fixation of the valley genotype Ab, which occurs at rate followed by reversion with probability 1/2). Thus, the reversion rate is initially high but rapidly approaches a much lower rate, which results in a characteristic “knee” in the probability that a population has reverted by time t, where a population has a substantial probability of having reverted at short times (corresponding to a steep initial increase in the fraction reverted as a function of time), but populations that do not revert during this initial period have much lower reversion rates (resulting in a slow increase in the fraction reverted after this initial period); see Figure 1, top row and center and right columns.
Figure 1.
Reversion rates for a biallelic fitness landscape with a symmetric fitness valley. Genotypes ab and AB have equal fitness and scaled selective advantage S relative to genotypes Ab and aB, and mutations occur at each site at rate 1 in each direction. The dynamics are shown for returns to the focal subset assuming that the population has just left the subset A at stationarity. Each column corresponds to a different value of S. The top row shows the probability that the population has reverted as a function of time, and the bottom row shows the reversion rate,
Mutational robustness and the rate of reversion:
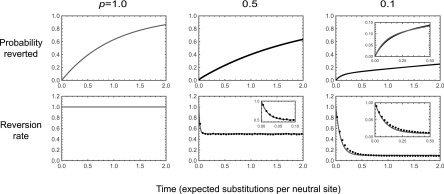
As a second example, we consider a biallelic fitness landscape with one focal site and L other sites, where each genotype is viable with probability p and all viable genotypes are neutral relative to each other. All sites experience forward and reverse mutations at rate 1. This example is motivated by the model used by Kondrashov and colleagues to study long-term purifying selection (Breen et al. 2012; Usmanova et al. 2015). We want to understand the dynamics of reversion at the focal site.
We proceed with a heuristic treatment based on the assumption that In this regime, we treat each genotype as having a constant number of neutral neighbors accessible by mutations at nonfocal sites. The key idea is that immediately after a substitution at the focal site, the back substitution is viable and so reversions initially occur at rate 1. However, once an additional substitution at a nonfocal site accrues, the probability that the reversion mutation at the focal site is viable is only p. Thus, the reversion rate drops very rapidly from 1 to ∼p.
In particular, after a substitution at the focal site, the expected substitution rate is and the probability that the first substitution is a reversion is Thus, we can approximate the probability distribution of waiting times until reversion as a mixture of two exponential distributions. The first distribution has rate 1 and is weighted by the probability that the population reverts before any other substitution occurs, The second exponential distribution has rate p and is weighted by the probability that a population experiences a substitution at another site before the reversion event occurs, This suggests that the probability of having reverted by time t can be approximated by
| (16) |
Based on a similar argument, we can approximate the reversion rate as
| (17) |
where the first term corresponds to the initial reversion rate of 1, which applies with probability (the probability of not having yet having experienced the first substitution), and the second term corresponds to the expected reversion rate given that one substitution has occurred, p, weighted by the probability that a substitution has already occurred,
Although an exact treatment of any particular realization of the model is possible using the associated rate matrix the construction of this matrix is not computationally feasible for large L. We thus proceeded by simulation. A population that has just experienced a substitution at the focal site must be at a viable genotype and have just come from a viable genotype that differs only at the focal site. We simulated the subsequent evolution of such a population under weak mutation, keeping track of the genotypes produced by mutation and whether they are viable (with probability p) or inviable (with probability ) until the first reversion event at the focal site. For each choice of parameters, we repeated this procedure 1,000,000 times.
The resulting reversion times are shown in Figure 2 for The top row shows the probability of having reverted by time t, and the bottom row shows the reversion rate as a function of time. The left column shows the exact theoretical results for the case where all genotypes are viable (shaded lines). The center and right columns show the results for and respectively, with the approximate theoretical results given by Equations 16 and 17 shown as shaded lines and the simulation results shown as solid lines. For the case the reversion rate drops rapidly from 1 to ∼p. Indeed, this drop is so rapid that relatively few populations actually revert at the higher initial rate, which is as we expect, since the probability that a population reverts before experiencing another substitution is only On the other hand, for the case we see a modest knee in the time evolution of the fraction of populations that have reverted. This is because a larger fraction of populations revert before experiencing another substitution, and the subsequent substitution rate is lower, ∼0.1 instead of 0.5.
Figure 2.
Dynamics of reversion for a biallelic fitness landscape with 100 sites other than the focal site, where each genotype is viable with probability p, all viable genotypes are neutral relative to each other, and each site experiences forward and backward mutations at rate 1. The top row shows the probability that a population has reverted as a function of time, and the bottom row shows the reversion rate. Theoretical results are shaded lines, and simulations are solid lines. Solid circles in the bottom row calculate the reversion rate in terms of temporal bins of width 0.05, where the reversion rate in a bin is calculated as the number of simulations that revert during that bin divided by the average of the number of simulations that have not yet reverted at the beginning of that bin and the number of simulations that have not yet reverted at the end of that bin. Insets show details of dynamics at short times. Simulation results are based on 1,000,000 runs for each value of p.
Intuitively, the knee shape illustrated in Figure 2 for small p and large arises because for small p there are fewer other loci at which a substitution can occur, and so populations stay at the genotype they initially arrived at—with its elevated reversion rate—longer, which leads to a substantial probability of having reverted at short times. However, eventually such populations leave the initial genotype, at which time they experience lower reversions rates, This produces a pattern where the probability of having reverted becomes substantial at short times but thereafter grows very slowly. Although the theory is complicated and so we do not discuss it in detail here, it is worth noting that when p is small enough that becomes small the waiting time until reversion again becomes short. Intuitively, this occurs because for very small the only viable mutation after a substitution at the focal site is the back mutation at the focal site, and thus essentially all reversions occur at the initial elevated rate.
More generally, this simple model suggests that the dynamics of reversion should depend on the degree of mutational robustness even when fitness is allowed to take more than two values. The key insight is that just after a substitution at a focal site, the population is not at a random genotype, but rather at one where the back substitution rate is likely to be unusually high. Now, if there is a high degree of mutational robustness, then the neutral substitution rate is high, and the population will leave this special genetic background quickly (cf. McCandlish 2013), so that when most reversions occur, the substitution rate for the back mutation is much lower than it was initially. When the degree of mutational robustness is very low, by contrast, the population will often stay at the initial genotype until the reversion occurs by a direct back substitution, which occurs at the initial rate. When the degree of mutational robustness is intermediate, we see a combination of these two dynamics: a substantial fraction of populations revert via the direct back substitution from the initial genotype, and another substantial fraction revert at the lower rates that are typical of other genetic backgrounds. This intermediate degree of robustness thus produces a characteristic knee in the time evolution of the probability that the population has reverted.
Amino acid reversion in codon models:
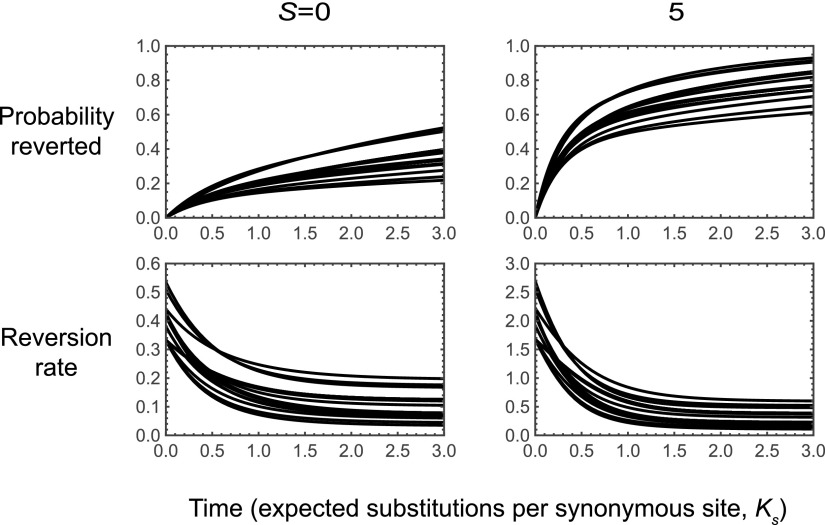
The two examples we have considered so far describe the dynamics of reversion at a single site in a biallelic fitness landscape. For our final example we consider the dynamics of a single amino acid under a codon model. Stop codons are treated as inviable, and mutations occur at each site with equal probability to each of the three alternative nucleotides (Jukes and Cantor 1969) with a total rate of 1 so that time is measured in the expected number of substitutions per synonymous site (). We implement selection by assuming that genotypes with the focal amino acid are neutral relative to each other and have a scaled selective advantage S over all other amino acids.
Figure 3 shows the dynamics of reversion under this model for the neutral case, and also for the case where the focal amino acid has a moderate selective advantage, (each amino acid corresponds to one line; the fraction reverted is shown in the first row and is shown in the second row). The key insight for understanding these dynamics is that, just after a population leaves the set of codons that code for the focal amino acid, it can still mutate back to the focal amino acid. However, after additional substitutions accrue, the population is likely to be at a genotype that is no longer mutationally adjacent to the focal amino acid, leading to a decreasing reversion rate over time. In particular, for both and the reversion rate drops from its initial value to nearly its asymptotic value by In the case of this produces a pronounced knee in the shape of the curve describing the time evolution of the probability that a population has reverted: more than one-third of populations are expected to revert almost immediately, whereas the remaining populations take a long time before they eventually revert.
Figure 3.
Dynamics of reversion for a codon model where the focal amino acid has scaled selection coefficient S over all other amino acids and mutations occur at rate 1 at each site with an equal probability of producing each of the alternative nucleotides. The top row shows the probability of having reverted by time t and the bottom row shows the reversion rate, where each of the 20 curves corresponds to a different choice of one of the 20 amino acids as the focal amino acid. Under moderate selection for the focal amino acid (), the time evolution of the fraction of populations that have already reverted shows a pronounced knee.
Discussion
We have analyzed reversions in a population that has just left some focal set of genotypes A. Our main interest has been the rate at which the population first returns to the set A—that is, the rate at which the population experiences a reversion—and how this rate changes over time. For a population that has already been evolving on a constant fitness landscape for a very long time, we have shown that the rate of reversion to A is nonincreasing in time. Furthermore, when the set A consists of all genotypes with a particular allele at a focal site in a biallelic fitness landscape, we have shown that the reversion rate is strictly decreasing if and only if epistasis is present at the focal site. We explored some simple examples of these reversion dynamics, to gain intuition about their magnitude and evolutionary impact.
We have also analyzed the case of a population that has been evolving on the fitness landscape only for a short time and whose initial state is therefore not drawn from the stationary distribution of the evolutionary dynamics. In this case we have shown that the time evolution of the reversion rate is, after a change in sign, mathematically identical to the time evolution of the mean Malthusian fitness for an infinite population evolving on an altered fitness landscape where the substitution rates of the original fitness landscape play the role of mutation rates and the rates of returns to A play the role of genotype-specific death rates.
One consequence of reversion rates that decrease in time is a distinctive pattern, whereby a population either reverts very quickly after the initial substitution with some moderate probability or else takes a very long time to revert. If we consider the probability of having reverted plotted as a function of time, such a pattern can be seen as a “knee”, where this curve rises rapidly at short times until the probability of having reverted is substantial and then shifts to rising much more slowly. This pattern occurs because the expected rate of substitutions back to A at stationarity is particularly high if we condition on just having left A. As time passes and other substitutions accrue, the features of the genetic background that resulted in the unusually high substitution rate to A are lost. One consequence of this knee-like phenomenon is that the mean waiting time until reversion may not be very informative about the actual dynamics of reversion.
Our results show that at stationarity, a strictly decreasing reversion rate occurs whenever there is genotype-to-genotype variation in the rate of substitutions to the set A. When the set A corresponds to the set of genotypes with a particular allele at a particular site, the presence of an epistatic interaction between the focal site and at least one other site is sufficient to produce such genotype-specific variation and hence sufficient for the reversion rate to be strictly decreasing. However, other factors besides epistasis can also be responsible for genotype-to-genotype variation in the rate of substitutions to A. For instance, in our codon example, which has more than two alleles per site, the structure of the genetic code itself results in variation in the substitution rate to any particular amino acid, because the focal amino acid will not be mutationally accessible from all other codons. More generally, the genotype–phenotype map will tend to produce genotype-to-genotype variation in the rate at which the focal phenotype becomes fixed in the population, so that the reversion rate for phenotypes should typically be decreasing. Finally, it is worth mentioning that while we define a site at the genotypic level by the feature that its mutational dynamics are independent of the states of the other sites, context-dependent mutation could also produce genotype-to-genotype variation in rates of substitution to A, which would again be sufficient for decreasing reversion rates (provided the resulting Markov chain is still reversible).
It is also interesting to ask the converse question: What conditions could possibly produce reversion rates that increase in time? To do this, it is helpful to consider a way of restating our main result: if evolution can be modeled as a reversible Markov chain on the set of genotypes, then at stationarity the reversion rate to any subset of genotypes is nonincreasing with the time since the population left that subset. A reversible Markov chain is a Markov chain such that the probability of moving through any cycle of states in one direction is equal to the probability of moving around that cycle in the reverse direction (this is known as Kolmogorov’s criterion; see, e.g., Kelly 1979, section 1.5). Thus, a reversible Markov chain is simply a Markov chain with no cyclic biases. This means we can restate our main result as saying that reversion rates can increase in time, starting from stationarity, only if some factor induces a cyclic bias in the Markov chain describing evolution among genotypes.
It is known that, under weak mutation, adding frequency- and time-independent natural selection does not add any cyclic bias if no such bias is present in the mutational dynamics (i.e., the weak mutation Markov chain under constant frequency-independent selection is reversible if the mutational dynamics are reversible; see, e.g., Sella and Hirsh 2005). However, we may expect reversion rates to sometimes increase in time under conditions where fitness is nontransitive (e.g., Kerr et al. 2002) or when environments change in a cyclic manner (e.g., Leslie et al. 2004; Hensley et al. 2009; Bergland et al. 2014). Under these types of conditions, natural selection tends to push populations through a cycle of genotypic states in a periodic manner, which means that reversion becomes more likely as time passes.
Another condition that might produce increasing reversion rates is when the evolutionary dynamics are not stationary, for instance at the beginning of an adaptive transient. We have provided intuition for such circumstances by noting that, under time- and frequency-independent selection, the time evolution of the reversion rate can be recast as the time evolution of the mean fitness of an infinite population on an alternative fitness landscape. This intuition suggests that the reversion rate should tend to go down just as the mean fitness tends to go up, unless the infinite population starts at an unusually high fitness or the reversion rate starts at an unusually low value. In the case of an adaptive transient, early adaptive substitutions will tend to be strongly favored by natural selection, meaning that the initial reversion rate is unusually low. In these circumstances we may expect the reversion rate to increase, particularly in the case of diminishing-returns epistasis, where mutations that have strong positive effects early in adaptation have much smaller effects toward the end of adaption (Draghi and Plotkin 2013; Kryazhimskiy et al. 2014; cf. Hartl et al. 1985; Hartl and Taubes 1996).
Our results help bring clarity to a recent controversy concerning site-specific amino acid preferences during protein evolution (Naumenko et al. 2012; Pollock et al. 2012; Ashenberg et al. 2013; Pollock and Goldstein 2014; Bazykin 2015; Doud et al. 2015; Goldstein et al. 2015; Risso et al. 2015; Shah et al. 2015; Usmanova et al. 2015). First, our results show that in the presence of epistasis, reversion rates will be decreasing in time and that the longer a population has left a set of genotypic states, the longer the expected time until reversion. However, our results show that when considering reversion to an amino acid state, these results already hold even if there is no epistasis at the amino acid level, because of the structure of the genetic code itself. Indeed, it is worth noting that the epistasis already present in the genetic code is sufficient to produce many of the dynamical signatures of epistatic evolution even if fitnesses are additive at the amino acid level. For instance, it is easy to specify site-specific amino acid preferences that produce multipeaked fitness landscapes at individual codons; such codons can have extremely long equilibration times, contrary to the analyses of Kondrashov et al. (2010) and Breen et al. (2013) who consider only the case where every amino acid can mutate to every other amino acid. Overall, decreasing reversion rates are a generic feature of evolution under long-term purifying selection and not a definitive signature of epistasis at the amino acid level.
Second, it is helpful to distinguish between what is expected when we consider a population that is conditioned to remain outside a subset of states vs. one that is simply restricted from entering that subset (cf. Ashenberg et al. 2013; Pollock and Goldstein 2014). The difference is whether, after a substitution, one considers only populations that have not yet reverted or whether all populations are prevented from reverting (but where we nonetheless keep track of the average propensity to return to the focal subset if such substitutions were to be permitted). We have shown that these two processes have different, but related, mathematical characteristics. If there is any variation in the genotype-specific rates of return to the focal subset, then the reversion rate will be strictly decreasing for both processes. While acclimatization of the rest of the genome to being in a new region of the fitness landscape contributes to decreasing reversion rate for both processes, for the conditioned process there is also a statistical effect because populations that spend more time at genotypes with a rapid rate of return to the focal subset are likely to revert quickly. In addition, there is a simple mathematical relationship between these two processes: the asymptotic rate of transitions back to the focal subset when populations are restricted from entering it is the rate of an exponential distribution with mean equal to the expected reversion time, and this rate is bounded between the initial reversion rate and the asymptotic reversion rate when conditioning. Thus, while Ashenberg et al. (2013) have criticized simulations where reversions are prevented from occurring as being unrealistic and misleading, such simulations are in fact more relevant to understanding the evolutionary process than would appear at first glance.
Finally, our results help clarify the relationship between various quantities observed in the literature and the entrenchment phenomenon described here. Observed changes in site-specific amino acid preferences (Bazykin 2015) detected either by direct measurement (Doud et al. 2015) or through comparative sequence analysis (Naumenko et al. 2012) are sufficient to produce reversion rates that decrease in time under the assumption that sequence evolution occurs as a stationary reversible Markov chain. Furthermore, the nonlinear mapping between protein stability and fitness means that even if the stability effects of amino acid substitutions are more or less conserved (Risso et al. 2015), fitness effects will often be background dependent (Ashenberg et al. 2013), which is again sufficient to produce reversion rates that decrease in time. Fits of covarion-like models (Fitch and Markowitz 1970; Galtier 2001; Penny et al. 2001; Usmanova et al. 2015) also suggest that rates of reversion should be decreasing in time, although as emphasized above the same qualitative effect can occur simply due to the structure of the genetic code (Figure 3). It is also natural to ask about the predictions of the theory developed here for quantities observed in nature. Our predictions for the expected change in the selection coefficient of reversion are relatively weak. This is because the dynamics of the reversion rate and the dynamics of the expected selection coefficient of a reversion mutation may have qualitatively different shapes as a result of the nonlinearity in the probability of fixation and genotype-to-genotype variation in mutation rates. Nonetheless, it is the substitution rate that is most closely related to the evolutionary dynamics. This is why we can derive strong results on the reversion rate, but not on the time evolution of the average selection coefficient of reversions.
Our results are also relevant to a variety of studies that consider the temporal relationships between substitutions on phylogenies (Rogozin et al. 2008; Povolotskaya and Kondrashov 2010; Naumenko et al. 2012; Goldstein et al. 2015; Zou and Zhang 2015). Importantly, by viewing sequence evolution as a stationary reversible Markov chain on the branches of a phylogeny, we see that waiting times for reversion events in the sense described here are very closely related to waiting times for parallel changes in sister taxa because both events involve the Markov chain leaving and then reentering a focal subset as we look along the branches of the phylogeny. However, more work is needed to extend our results to a phylogenetic setting. This is because the distribution of waiting times observed on a phylogeny is different from the distribution considered here where we condition on a substitution having just occurred. In addition, there are a variety of practical problems with inferring substitution histories such as apparent homoplastic substitutions due to incomplete lineage sorting (Mendes and Hahn 2016) and the fact that substitution histories are typically inferred using site-independent models even when there is substantial evidence for epistasis (e.g., Goldstein et al. 2015). Nonetheless, multiple studies in this broader literature (Rogozin et al. 2008; Naumenko et al. 2012; Soylemez and Kondrashov 2012; Goldstein et al. 2015; Zou and Zhang 2015) support our qualitative prediction that reversions and parallel substitutions should occur either very rapidly or only after a long waiting time.
Acknowledgments
We thank A. S. Kondrashov and an anonymous reviewer for their sage and helpful comments on the manuscript. This work was funded by the Burroughs Wellcome Fund, the David and Lucile Packard Foundation, U.S. Department of the Interior grant D12AP00025, National Institutes of Health training grant 2T32AI055400-11, and U.S. Army Research Office grant W911NF-12-1-0552.
Appendix A: The Distribution of Reversion Times at Stationarity
Our analysis of the distribution of reversion times at stationarity relies on the fact that the matrix which gives the rates for the absorbing Markov chain describing the evolution dynamics until the time of reversion, admits an eigendecomposition. We develop this eigendecomposition and its properties and then use these results to show that the distribution of reversion times at stationarity can be expressed as a mixture of exponential distributions. We then use a very similar analysis to understand the modified process where reversion events are not permitted to occur.
We derive the eigendecomposition of Q by first noting some features of the larger matrix Because the Markov chain defined by is reversible, it satisfies the detailed balance relation for all As a consequence, the matrix is symmetric, where denotes the diagonal matrix with the vector y as its main diagonal.
Now, define the vector π of length such that for The matrix is thus symmetric, since it is simply a constant times a diagonal block of the symmetric matrix We can thus expand in terms of its eigenvalues and eigenvectors as
| (A1) |
where are the eigenvalues of and the eigenvectors are orthonormal (the eigenvalues are real because is symmetric and negative because they are same as those of where Q is the generator of an absorbing Markov chain so that all of its eigenvalues have negative real parts).
We can now use this decomposition of to likewise decompose In particular, multiplying Equation A1 by from the left and from the right gives us
| (A2) |
where and are the left and right eigenvectors of associated with
Using this decomposition, we can then write the probability density function of the distribution of reversion times at stationarity as
| (A3) |
| (A4) |
| (A5) |
| (A6) |
| (A7) |
where we have used the fact that at stationarity (Equation 7). Thus, is a mixture of exponential densities with rates where the exponential density with rate has weight
We just showed that the distribution of reversion times at stationarity is a finite mixture of exponential distributions. We now show that the hazard function for any finite mixture of exponential distributions is nonincreasing. In particular, if is a finite mixture of exponential densities
| (A8) |
with then the hazard function is
| (A9) |
Differentiating the hazard function, we see that that the sign of the derivative depends only on the sign of
| (A10) |
| (A11) |
| (A12) |
| (A13) |
| (A14) |
which is nonpositive since each term in the sum is a product of nonnegative quantities and hence nonnegative. Thus, the hazard function is nonincreasing.
We turn now to the analysis of the modified process with rate matrix where is the diagonal matrix with g on its main diagonal. Let us start by assuming that is mutationally connected; we will return to the case of disconnected momentarily. First, note that differs from Q only on its diagonal entries. Thus, following our analysis for is symmetric and can be expanded in terms of its eigenvalues and eigenvectors as
| (A15) |
where are the eigenvalues of and the eigenvectors are orthonormal (we have because is connected and so the continuous-time Markov chain generated by is ergodic). With this decomposition in hand, we can write and as the left and right eigenvectors of associated with and write the reversion rate under the modified process as
| (A16) |
| (A17) |
| (A18) |
| (A19) |
where we have used the fact that the row sums of are all 0 so that is the vector of all 1’s and thus Because for this expression is clearly strictly decreasing in t unless (in which case is obviously constant) or = 0 for In the latter case, we have = 0 for all so that must be orthogonal to all the for Since the are an orthonormal basis of the only direction remaining for is so that g must be in the direction of which is the vector of all 1’s. Thus, in that case g is also constant.
So far, we have shown that provided that is mutationally connected, at stationarity the reversion rate under is constant if is constant and otherwise it is strictly decreasing. Now we consider the case where has several disconnected components, Clearly the total reversion rate is a weighted average of the reversion rates for each disconnected component when analyzed separately as above. If is nonconstant within any one of these components, then that term in the average is strictly decreasing in time and hence the reversion rate as a whole is strictly decreasing. Thus, for general the reversion rate at stationarity under is constant if is piecewise constant in the sense of being constant for each of the disconnected components of otherwise it is strictly decreasing.
Appendix B: Role of Epistasis in Multiallelic Fitness Landscapes
In the main text, we showed that for biallelic fitness landscapes, the reversion rate at stationarity for a site is a decreasing function of time if and only if that site is involved in an epistatic interaction with another site. Here we show that for multiallelic fitness landscapes the presence of an epistatic interaction with another site is a sufficient condition under stationarity for reversion rates at the focal site to be decreasing.
First, we must establish some notation for multiallelic fitness landscapes. If we have a fitness landscape over L sites, we can label the alleles at the lth site, and write the mutation rate between and as Thus, if genotypes i and j differ at more than one site, we have otherwise they differ at exactly one site l in which case where genotype i has the allele at site l and genotype j has the allele at site l.
Without loss of generality we can define the focal set A to be the set of genotypes with allele in site 1. We want to show that if site 1 has an epistatic interaction with at least one other site, then the are not constant, since this is sufficient to show that the reversion rate is decreasing at stationarity.
Now, either for all alleles at site 1 or not. If not, then we can find an allele at site 1 such that But since the Markov chain defined by is reversible, we know there is also an allele at site 1 such that since otherwise populations could never return from to A. Thus g is nonconstant because it is zero for genotypes with allele at site 1, but positive for genotypes with allele at site 1.
We thus turn to the case where for all k. Our strategy is to show that if all alleles at a site can mutate to all the other alleles, then the presence of any epistatic interaction between one site and another is sufficient, by the transitivity of fitness differences, to guarantee the existence of a set of four genotypes to which we can apply the argument in the main text for biallelic sites. Now, if site 1 is epistatic with some other site, then without loss of generality we can let this other site be site 2. Because site 1 is epistatic with site 2, there exist, by the definition of epistasis, genotypes and alleles such that the following holds:
Genotypes j and are both single mutants with respect to genotype i, and is the corresponding double mutant. In particular, genotype j is formed from genotype i by a single mutation, genotype is formed from i as a single mutation, and the double-mutant genotype is formed from genotype i by the combination of an mutation and an mutation.
If either or then the selection coefficient of the reversion mutation to depends on the genetic background and so g is nonconstant by the same argument as given for the two-allele case in the main text. Therefore suppose and In that case we can find genotypes h and such that h is identical to i and j except that it has the allele at site 1 and likewise is identical to and except that it has the allele at site 1. Now, if it were the case that both and then we would have
But so either or Thus, when choosing a set of four genotypes to demonstrate the existence of an epistatic interaction between sites 1 and 2, we could have chosen h and instead of either i and or j and However, this new choice of four genotypes contains two different reversion mutations to with different selection coefficients and the same mutation rates, so that the nonconstancy of g follows by the same argument given for the two-allele case in the main text.
Appendix C: Expected Time Until Reversion
We want to derive the expected time until reversion to a set A at stationarity given that the population has just left A. To study this time, it is helpful to define a modified Markov chain on a state space where the new state a takes the place of the set A in the original chain.
In particular, consider the chain with rate matrix
| (C1) |
where k is the expected substitution rate from A to at stationarity for conditional on the population being in A, and is the probability of the population being at genotype under at stationarity, conditional on a substitution from A to having just occurred. By construction, this new chain with rate matrix has the same distribution of reversion times to a at stationarity as the original chain at stationarity did to A.
Furthermore, if contains n elements, it is easy to verify that the stationary distribution of is given by the vector where since the new chain conserves the stationary rate of substitutions both from A to i and from i to A for all states
Analyzing this new chain, a straightforward application of the reward–renewal theorem for the renewal process defined by returns to a and reward equal to the waiting time before leaving a after each return gives us
| (C2) |
(Grimmett and Stirzaker 2001, theorem 10.5.10), where is the expected waiting time for a population currently at a to leave a and then return to a for the first time.
Now, a population leaving a and then returning to a must first leave a and then revert to a. The expected time before the population leaves a is and then the expected waiting time to revert to a is the quantity that we are trying to develop an expression for. Thus, we have
| (C3) |
Plugging Equation C3 into Equation C2 and solving for gives us
| (C4) |
At stationarity, the total rate that populations arrive at a must be equal to the rate that they leave a, so we have and of course Substituting these expressions into Equation C4 yields
| (C5) |
i.e., the expected waiting-time reversion is equal to the reciprocal of the expected substitution rate to A at stationarity conditional on the population being in as required.
Footnotes
Communicating editor: N. A. Rosenberg
Literature Cited
- Aldous, D., and J. A. Fill, 2002 Reversible Markov chains and random walks on graphs. Available at: http://www.stat.berkeley.edu/∼aldous/RWG/book.html.
- Ashenberg O., Gong L. I., Bloom J. D., 2013. Mutational effects on stability are largely conserved during protein evolution. Proc. Natl. Acad. Sci. USA 110: 21071–21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazykin G. A., 2015. Changing preferences: deformation of single position amino acid fitness landscapes and evolution of proteins. Biol. Lett. 11: 20150315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Behrman E. L., O’Brien K. R., Schmidt P. S., Petrov D. A., 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10: e1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen M. S., Kemena C., Vlasov P. K., Notredame C., Kondrashov F. A., 2012. Epistasis as the primary factor in molecular evolution. Nature 490: 535–538. [DOI] [PubMed] [Google Scholar]
- Breen M. S., Kemena C., Vlasov P. K., Notredame C., Kondrashov F. A., 2013. Breen et al. reply. Nature 497: E2–E3. [DOI] [PubMed] [Google Scholar]
- Bridgham J. T., Ortlund E. A., Thornton J. W., 2009. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461: 515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M., Hartl D. L., 2010. Adaptive landscapes and protein evolution. Proc. Natl. Acad. Sci. USA 107: 1747–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin R., Miglietta M. P., 2008. Reversing opinions on Dollo’s Law. Trends Ecol. Evol. 23: 602–609. [DOI] [PubMed] [Google Scholar]
- Conrad M., 1990. The geometry of evolution. Biosystems 24: 61–81. [DOI] [PubMed] [Google Scholar]
- Dollo L., 1893. The laws of evolution. Bull. Soc. Bel. Geol. Paleontol 7: 164–166. [Google Scholar]
- Doud M. B., Ashenberg O., Bloom J. D., 2015. Site-specific amino acid preferences are mostly conserved in two closely related protein homologs. Mol. Biol. Evol. 33: 2944–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghi J. A., Plotkin J. B., 2013. Selection biases the prevalence and type of epistasis along adaptive trajectories. Evolution 67: 3120–3131. [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, London. [Google Scholar]
- Fitch W. M., Markowitz E., 1970. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem. Genet. 4: 579–593. [DOI] [PubMed] [Google Scholar]
- Galtier N., 2001. Maximum-likelihood phylogenetic analysis under a covarion-like model. Mol. Biol. Evol. 18: 866–873. [DOI] [PubMed] [Google Scholar]
- Gavrilets S., 1997. Evolution and speciation on holey adaptive landscapes. Trends Ecol. Evol. 12: 307–312. [DOI] [PubMed] [Google Scholar]
- Goldstein R. A., Pollard S. T., Shah S. D., Pollock D. D., 2015. Nonadaptive amino acid convergence rates decrease over time. Mol. Biol. Evol. 32: 1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., 1970. Dollo on Dollo’s law: irreversibility and the status of evolutionary laws. J. Hist. Biol. 3: 189–212. [DOI] [PubMed] [Google Scholar]
- Grimmett, G., and D. Stirzaker, 2001 Probability and Random Processes. Oxford University Press, London/New York/Oxford. [Google Scholar]
- Hartl D. L., Taubes C. H., 1996. Compensatory nearly neutral mutations: selection without adaptation. J. Theor. Biol. 182: 303–309. [DOI] [PubMed] [Google Scholar]
- Hartl D. L., Dykhuizen D. E., Dean A. M., 1985. Limits of adaptation: the evolution of selective neutrality. Genetics 111: 655–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley S. E., Das S. R., Bailey A. L., Schmidt L. M., Hickman H. D., et al. , 2009. Hemagglutinin receptor binding avidity drives influenza a virus antigenic drift. Science 326: 734–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa Y., 1988. Free fitness that always increases in evolution. J. Theor. Biol. 135: 265–281. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Cantor C. R., 1969. Evolution of protein molecules, pp. 21–132 in Mammalian Protein Metabolism, Vol. 3, edited by Munro H. N. Academic Press, New York. [Google Scholar]
- Kelly, F. P., 1979 Reversibility and Stochastic Networks (Series in Probability and Mathematical Statistics). Wiley, Chichester, UK. [Google Scholar]
- Kerr B., Riley M. A., Feldman M. W., Bohannan B. J., 2002. Local dispersal promotes biodiversity in a real-life game of rock–paper–scissors. Nature 418: 171–174. [DOI] [PubMed] [Google Scholar]
- Kielson, J., 1979 Markov Chain Models—Rarity and Exponentiality (Applied Mathematical Sciences, Vol. 28). Spring-Verlag, New York. [Google Scholar]
- Kimura M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Kondrashov D. A., Kondrashov F. A., 2015. Topological features of rugged fitness landscapes in sequence space. Trends Genet. 31: 24–33. [DOI] [PubMed] [Google Scholar]
- Kondrashov A. S., Povolotskaya I. S., Ivankov D. N., Kondrashov F. A., 2010. Rate of sequence divergence under constant selection. Biol. Direct 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S., Rice D. P., Jerison E. R., Desai M. M., 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344: 1519–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A., Pfafferott K., Chetty P., Draenert R., Addo M., et al. , 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10: 282–289. [DOI] [PubMed] [Google Scholar]
- McCandlish D. M., 2013. On the findability of genotypes. Evolution 67: 2592–2603. [DOI] [PubMed] [Google Scholar]
- McCandlish D. M., Stoltzfus A., 2014. Modeling evolution using the probability of fixation: history and implications. Q. Rev. Biol. 89: 225–252. [DOI] [PubMed] [Google Scholar]
- McCandlish D. M., Rajon E., Shah P., Ding Y., Plotkin J. B., 2013. The role of epistasis in protein evolution. Nature 497: E1–E2. [DOI] [PubMed] [Google Scholar]
- McCandlish D. M., Epstein C. L., Plotkin J. B., 2015a Formal properties of the probability of fixation: identities, inequalities and approximations. Theor. Popul. Biol. 99: 98–113. [DOI] [PubMed] [Google Scholar]
- McCandlish D. M., Otwinowski J., Plotkin J. B., 2015b Detecting epistasis from an ensemble of adapting populations. Evolution 69: 2359–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes F. K., Hahn M. W., 2016. Gene tree discordance causes apparent substitution rate variation. Syst. Biol. DOI: 10.1093/sysbio/syw018. [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1918. Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics 3: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1939. Reversibility in evolution considered from the standpoint of genetics. Biol. Rev. Camb. Philos. Soc. 14: 261–280. [Google Scholar]
- Naumenko S. A., Kondrashov A. S., Bazykin G. A., 2012. Fitness conferred by replaced amino acids declines with time. Biol. Lett. 8: 825–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny D., McComish B. J., Charleston M. A., Hendy M. D., 2001. Mathematical elegance with biochemical realism: the covarion model of molecular evolution. J. Mol. Evol. 53: 711–723. [DOI] [PubMed] [Google Scholar]
- Pollock D. D., Goldstein R. A., 2014. Strong evidence for protein epistasis, weak evidence against it. Proc. Natl. Acad. Sci. USA 111: E1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock D. D., Thiltgen G., Goldstein R. A., 2012. Amino acid coevolution induces an evolutionary Stokes shift. Proc. Natl. Acad. Sci. USA 109: E1352–E1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M. L., Crandall K. A., 2003. Lost along the way: the significance of evolution in reverse. Trends Ecol. Evol. 18: 541–547. [Google Scholar]
- Povolotskaya I. S., Kondrashov F. A., 2010. Sequence space and the ongoing expansion of the protein universe. Nature 465: 922–926. [DOI] [PubMed] [Google Scholar]
- Risso V. A., Manssour-Triedo F., Delgado-Delgado A., Arco R., Barroso-delJesus A., et al. , 2015. Mutational studies on resurrected ancestral proteins reveal conservation of site-specific amino acid preferences throughout evolutionary history. Mol. Biol. Evol. 32: 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin I. B., Thomson K., Csürös M., Carmel L., Koonin E. V., 2008. Homoplasy in genome-wide analysis of rare amino acid replacements: the molecular-evolutionary basis for Vavilov’s law of homologous series. Biol. Direct 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A., Carroll S. B., 2008. Frequent and widespread parallel evolution of protein sequences. Mol. Biol. Evol. 25: 1943–1953. [DOI] [PubMed] [Google Scholar]
- Sella G., Hirsh A. E., 2005. The application of statistical physics to evolutionary biology. Proc. Natl. Acad. Sci. USA 102: 9541–9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P., McCandlish D. M., Plotkin J. B., 2015. Contingency and entrenchment in protein evolution under purifying selection. Proc. Natl. Acad. Sci. USA 112: E3226–E3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G. G., 1953. The Major Features Of Evolution. Columbia University Press, New York. [Google Scholar]
- Soylemez O., Kondrashov F. A., 2012. Estimating the rate of irreversibility in protein evolution. Genome Biol. Evol. 4: 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Serene S., Chao H. X., Gore J., 2011. Hidden randomness between fitness landscapes limits reverse evolution. Phys. Rev. Lett. 106: 198102. [DOI] [PubMed] [Google Scholar]
- Teotónio H., Rose M. R., 2001. Perspective: reverse evolution. Evolution 55: 653–660. [DOI] [PubMed] [Google Scholar]
- Usmanova D. R., Ferretti L., Povolotskaya I. S., Vlasov P. K., Kondrashov F. A., 2015. A model of substitution trajectories in sequence space and long-term protein evolution. Mol. Biol. Evol. 32: 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel J. W., Yashin A. I., 1985. Heterogeneity’s ruses: some surprising effects of selection on population dynamics. Am. Stat. 39: 176–185. [PubMed] [Google Scholar]
- Zou Z., Zhang J., 2015. Are convergent and parallel amino acid substitutions in protein evolution more prevalent than neutral expectations? Mol. Biol. Evol. 32: 2085–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.