Abstract
The ability to access alleles from unadapted germplasm collections is a long-standing problem for geneticists and breeders. Here we developed, characterized, and demonstrated the utility of a wild barley advanced backcross-nested association mapping (AB-NAM) population. We developed this population by backcrossing 25 wild barley accessions to the six-rowed malting barley cultivar Rasmusson. The 25 wild barley parents were selected from the 318 accession Wild Barley Diversity Collection (WBDC) to maximize allelic diversity. The resulting 796 BC2F4:6 lines were genotyped with 384 SNP markers, and an additional 4022 SNPs and 263,531 sequence variants were imputed onto the population using 9K iSelect SNP genotypes and exome capture sequence of the parents, respectively. On average, 96% of each wild parent was introgressed into the Rasmusson background, and the population exhibited low population structure. While linkage disequilibrium (LD) decay (r2 = 0.2) was lowest in the WBDC (0.36 cM), the AB-NAM (9.2 cM) exhibited more rapid LD decay than comparable advanced backcross (28.6 cM) and recombinant inbred line (32.3 cM) populations. Three qualitative traits: glossy spike, glossy sheath, and black hull color were mapped with high resolution to loci corresponding to known barley mutants for these traits. Additionally, a total of 10 QTL were identified for grain protein content. The combination of low LD, negligible population structure, and high diversity in an adapted background make the AB-NAM an important tool for high-resolution gene mapping and discovery of novel allelic variation using wild barley germplasm.
Keywords: wild barley, advanced backcross, nested association mapping population, association mapping, plant genetic resources, Multiparent Advanced Generation Inter-Cross (MAGIC), multiparental populations, MPP
DIVERSE germplasm collections are valuable resources for crop improvement. However, breeders often neglect these resources due to the time and effort required to identify and deploy beneficial exotic alleles. Breeding for complex traits requires balancing the introduction of genetic diversity with maintaining the selective progress obtained over many cycles of breeding (Bernardo 2002). Due to the malting quality requirements imposed by North American malting and brewing industries, barley (Hordeum vulgare subsp. vulgare) breeding has been restricted to a narrow germplasm base and focused on elite-by-elite crosses (Rasmusson and Phillips 1997). Over many cycles of breeding, extensive genome-wide linkage disequilibrium (LD) can develop in closed breeding populations (Fang et al. 2013), and the genetic diversity of these populations becomes reduced (Condón et al. 2008; Fu and Somers 2009; Muñoz-Amatriaín et al. 2010; Poets et al. 2015). The need to expand the genetic diversity of the breeding pool has become evident as breeders face disease and environmental pressures which are threatening crop production. Today, genomics technologies are advancing our ability to understand the genetic basis of variation across barley germplasm, and providing the opportunity to look beyond conventional sources of genetic diversity for sustained crop improvement.
Wild barley (Hordeum vulgare subsp. spontaneum), the progenitor of cultivated barley, is a rich source of genetic diversity. Resequencing-based estimates indicate that barley landraces retain ∼80% and modern cultivars retain ∼71% of the diversity found in the wild (Saisho and Purugganan 2007; Morrell et al. 2014). Despite low levels of outcrossing in both wild and cultivated barleys (∼0–2%) (Abdel-Ghani et al. 2004), wild barley populations exhibit much lower LD than typical breeding populations (Morrell et al. 2005; Caldwell et al. 2006; Hamblin et al. 2010). The combination of low LD and high diversity in wild barley germplasm collections presents an opportunity for high-resolution association mapping (Steffenson et al. 2007; Roy et al. 2010). But, assaying wild barley lines in field trials for agronomic traits is difficult because wild accessions exhibit a brittle rachis that causes seed shattering (Pourkheirandish and Komatsuda 2007; Pourkheirandish et al. 2015), and wild barley is generally not adapted to agronomic growing conditions.
The advanced backcross (AB) technique was developed to address the difficulties of using unadapted germplasm for trait mapping and cultivar improvement (Tanksley and Nelson 1996). Instead of developing typical F2-derived recombinant inbred line (RIL) mapping populations, AB populations are comprised of multiple-backcross derived RILs, with an exotic donor parent crossed to an adapted recurrent parent. With a much smaller portion of the exotic genome present in each line, the effects of agronomically-unadapted alleles are reduced, allowing estimates of the value of exotic alleles in the context of cultivated germplasm. AB populations have been developed and used successfully to identify beneficial alleles in several crops, including tomato, rice, wheat, maize, cotton (reviewed in Wang and Chee 2010), and barley (Matus et al. 2003; Pillen et al. 2003; von Korff et al. 2004; Li et al. 2006; Yun et al. 2006). AB analysis is limited by the genetic content of the parents selected and, the controlled crossing scheme leads to extensive LD, a minimal number of recombination events, and unbalanced allele frequencies (Tanksley and Nelson 1996).
Association mapping is a popular technique for high-resolution mapping of quantitative traits in crop germplasm (Rafalski 2002), but cryptic relatedness or population structure within association mapping populations can confound marker-trait associations leading to false positives if not properly controlled (Yu et al. 2006; Vilhjálmsson and Nordborg 2013). Furthermore, small-effect loci and traits associated with low-frequency alleles are difficult to detect (Korte and Farlow 2013). To address these limitations, populations developed from more complex crossing schemes that involve multiple parental lines and family-based association mapping approaches have been employed (reviewed in Guo et al. 2013). The nested association mapping (NAM) approach (Yu et al. 2008) combines the power of linkage mapping with the resolution of association mapping by crossing a diverse set of lines to a single reference genotype. This population design nests ancestral LD within novel recombination events, allowing for imputation of high-density genotypic data from parental lines, high-power and high-resolution mapping, and the use of diverse germplasm (Yu et al. 2008).
The maize NAM population is an invaluable resource for the maize community, and has been used for studies exploring genome dynamics, identifying trait associations, and improving the genomic and breeding resources of maize. Genetic characterization of the maize NAM population revealed patterns of recombination and segregation distortion within the maize genome (McMullen et al. 2009), and trait mapping has identified numerous QTL for many traits (Buckler et al. 2009; Poland et al. 2011; Tian et al. 2011; Peiffer et al. 2013, 2014). The NAM design is one of several multiparent mapping strategies that have been developed to dissect complex trait architecture. Population design varies based on considerations such as mating system and resource availability. Multiparent intercross populations that use more complex crossing designs have been developed in mouse (Churchill et al. 2004), Drosophila (Macdonald and Long 2007), Arabidopsis (Kover et al. 2009; Huang et al. 2011), rice (Bandillo et al. 2013), wheat (Rebetzke et al. 2014), and barley (Sannemann et al. 2015); and backcross-NAM designs have been developed in sorghum (Jordan et al. 2011) and barley (Schnaithmann et al. 2014; Maurer et al. 2015). Notably, with its wide variation in flowering time and disease response, the BC1-derived Halle exotic barley (HEB) population has been used to characterize the genetic architecture of flowering time (Maurer et al. 2015) and to map seedling leaf-rust resistance (Schnaithmann et al. 2014).
Here, we report the development and genetic characterization of a resource for barley breeders and geneticists that combines the development schematics of AB and NAM populations. By combining these designs, we were able to develop a population that minimizes the difficulties of assessing exotic germplasm, while providing a robust resource for high-power, high-resolution trait mapping. Our objectives were to: (1) develop a barley population that incorporates a large amount of exotic germplasm, but with sufficient agronomic adaptation to be analyzed in standard field trials; (2) identify regions of the barley genome that are subject to segregation distortion during wild barley introgression; (3) compare the LD of four different populations that use exotic germplasm for mapping [a wild barley advanced backcross-nested association mapping (AB-NAM) population, a diverse wild barley association mapping panel, awild × cultivated barley AB population, and a wild × cultivated barley RIL population]; and (4) use the AB-NAM population to map three qualitative traits with varying segregation patterns and a quantitative trait. This genetic characterization will inform future efforts to perform trait mapping within the AB-NAM population and improve our understanding of the challenges related to trait introgression from wild barley.
Materials and Methods
Plant materials
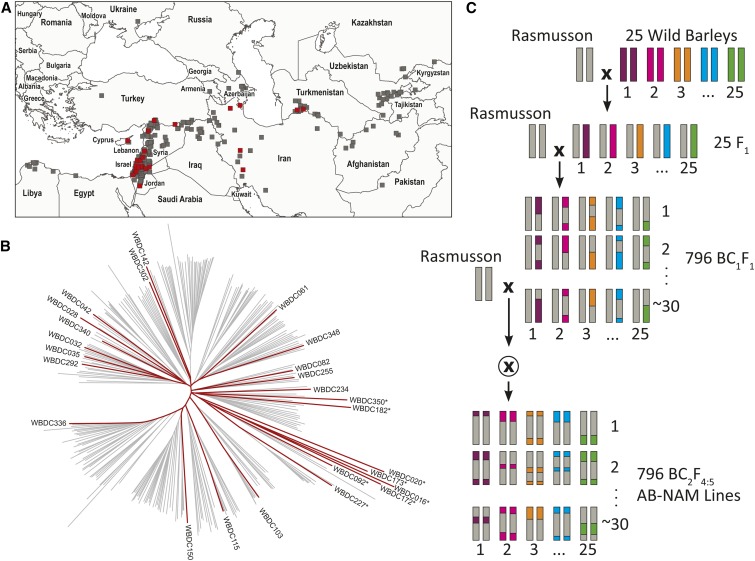
Twenty-five wild barley parents were chosen from the Wild Barley Diversity Collection (WBDC), a germplasm collection of 318 accessions that represent wild barley’s native range in the Middle East, Central Asia, North Africa, and Caucasus region (Table 1, Figure 1A) (Steffenson et al. 2007). Using the PowerMarker software’s core set function (Liu and Muse 2005), parental accessions were selected by maximizing the allele content of 25 chosen parents at 1402 SNP Barley Oligo Probe Assay 1 (BOPA1) (Close et al. 2009), 556 Diversity Arrays Technology (DArT) (Alsop et al. 2011), and 46 SSR (Fang et al. 2014) markers that were polymorphic in the WBDC. The recurrent parent Rasmusson is a six-rowed spring malting barley cultivar that was selected for its high yield and favorable malting quality characteristics, and descendant relation to the genome sequence reference cultivar Morex (Smith et al. 2010).
Table 1. Wild barley parental accessions used in this study.
| Parent accessiona | ICARDA genebank designator | Country of origin | Total BC2 individuals | Percentage of parent genome introgressed |
|---|---|---|---|---|
| WBDC016b | 38661 | Iran | 34 | 94.9 |
| WBDC020b | 38672 | Turkey | 36 | 92.0 |
| WBDC028 | 38840 | Israel | 26 | 96.4 |
| WBDC032 | 38869 | Israel | 33 | 96.0 |
| WBDC035 | 38981 | Israel | 34 | 98.0 |
| WBDC042 | 39673 | Israel | 27 | 89.3 |
| WBDC061 | 39910 | Syria | 29 | 98.3 |
| WBDC082 | 40009 | Jordan | 32 | 98.3 |
| WBDC092b | 40034 | Jordan | 34 | 95.1 |
| WBDC103 | 40071 | Jordan | 35 | 95.9 |
| WBDC115 | 40104 | Turkmenistan | 39 | 100.0 |
| WBDC142 | 40188 | Lebanon | 26 | 92.4 |
| WBDC150 | 40200 | Iran | 30 | 95.6 |
| WBDC172b | 112673 | Iran | 32 | 90.8 |
| WBDC173b | 112674 | Iran | 30 | 97.4 |
| WBDC182b | 115781 | Jordan | 31 | 98.9 |
| WBDC227b | 132552 | Azerbaijan | 36 | 99.1 |
| WBDC234 | 39884 | Cyprus | 30 | 96.6 |
| WBDC255 | 115792 | Jordan | 30 | 94.1 |
| WBDC292 | 38926 | Israel | 30 | 97.9 |
| WBDC302 | 38635 | Syria | 30 | 97.7 |
| WBDC336 | 126406 | Turkmenistan | 33 | 94.0 |
| WBDC340 | 116116 | Turkey | 34 | 99.0 |
| WBDC348 | Damon 11-11(B)c | Israel | 35 | 97.6 |
| WBDC350b | 41-1d | Israel | 30 | 97.0 |
ICARDA genebank or alternate designation, collection location, number of BC2-derived AB-NAM lines, and percentage of parental genome covered by introgression in BC2-derived individuals. ICARDA, International Center for Agricultural Research in the Dry Areas.
Accessions with probable cultivated or landrace barley introgression (Fang et al. 2014).
Obtained from University of Haifa collection (Fetch et al. 2003).
Unknown ICARDA germplasm number (Baum et al. 2003).
Figure 1.
Characteristics of wild barley parents and AB-NAM design. (A) Geographic distribution of the WBDC with selected parents in red. (B) Neighbor-joining tree diagram of 318 WBDC accessions based on BOPA 1 and 2 SNP genotypes. Genotypes with probable cultivated or landrace barley introgression are marked with an *. (C) Population development scheme.
Population development
The wild barley AB-NAM population was developed by crossing each of the 25 wild barley donor parents to Rasmusson. The wild parents underwent three generations of single seed descent from the seed source prior to crossing. F1 plants from each Rasmusson × wild parent cross were backcrossed to Rasmusson to create BC1F1 seed. A total of ∼40 BC1F1 seeds were planted for each wild parent, and a second backcross was attempted with all BC1F1 plants. A single BC2F1 seed was advanced for each successful BC1F1 backcross. We aimed for 30 independently-derived BC2F4 lines per family. This goal was based on our observation that randomly sampling varying numbers of individuals from the Harrington × OUH-602 AB population (Yun et al. 2006) led to an average of 90% of the wild parent genome introgressed into the recurrent parent, with diminishing returns beyond 30 individuals. In the BC2F2 generation, plants were selected for six-rowed spike morphology and selfed by single seed descent to the BC2F4 generation. Six-rowed spike morphology was selected to reduce the confounding effect of spike morphology on trait analysis. Rasmusson served as the female parent in all crosses. Individuals being crossed were vernalized as seedlings in a cold chamber for 4–6 weeks to ensure consistent flowering. Population development occurred in controlled greenhouse and growth chamber environments to minimize inadvertent selection.
Parental genotyping and exome capture sequencing
The 25 wild barley parents and Rasmusson were genotyped with the barley 9K SNP Illumina (San Diego, CA) iSelect platform (Comadran et al. 2012) and exome capture sequenced (Mascher et al. 2013a). Genomic libraries were constructed using the barley Roche (Madison, WI) NimbleGen SeqCap EZ Developer probe pool and Illumina HiSeq sequenced at the University of Kansas Medical Center Genome Sequencing Facility, Kansas City, KS. The exome capture assay is designed to capture 61.6 Mb of barley gene space (Mascher et al. 2013a). Sequence variants, including SNPs and indels, were called using the HaplotypeCaller function in the GATK 3.3 software package (https://www.broadinstitute.org/gatk/). Exome capture sequences for parent WBDC103 were discarded due to low quality sequence alignment. Therefore, sequence-derived analyses were performed on the 761 individuals derived from the remaining 24 wild barley parents and Rasmusson. Sequence variant calls were filtered by GATK genotyping quality score >10 and read depth >10 reads. Additional details of read mapping and SNP calling are available in Kono et al. (2015). Sequence variants with heterozygous Rasmusson genotype calls, >2 alleles, missing calls, or calls located on contigs without genetic map information were excluded from the marker set. Sequence variants that were located on Morex contigs with identical population sequencing (POPSEQ) (Mascher et al. 2013b) cM location and that contained identical parental genotype calls were binned, and only one variant was used for further analysis.
Population genotyping and marker imputation
Tissue from six to eight seedlings per BC2F4:5 line was bulk harvested, and DNA was extracted using the QIAGEN (Valencia, CA) DNAeasy Plant Mini Kits according to manufacturer’s instructions. Each of the 796 lines was genotyped using a custom 384-SNP Illumina VeraCode assay that contained markers selected from 2994 mapped SNPs on the Barley Oligo Probe Assays 1 and 2 (BOPA1 and 2) (Close et al. 2009; Muñoz-Amatriaín et al. 2011). SNP selection was based on even distribution throughout the genome according to map locations from the consensus map (Muñoz-Amatriaín et al. 2011) and the ability to distinguish Rasmusson from the 25 wild parents. A total of 5 of the 384 SNPs did not meet quality standards and were excluded from further analysis.
Scaffold markers as well as higher-density parental markers were coded relative to the Rasmusson genotype (0 = homozygous Rasmusson, 1 = homozygous non-Rasmusson). For each family, the subset of segregating genotyped markers (Supplemental Material, Table S1) was used as a scaffold to impute the higher-density iSelect and exome capture sequence variants of each respective wild parent. The probabilities of SNP calls were calculated based on the genetic map locations of typed scaffold marker calls (Guo and Beavis 2011). For each line, when two-typed flanking markers were derived from the same parent, the imputed marker took the value of the flanking markers. When the markers indicated a recombination event, the imputed marker was given a value between 0 and 1, as a function of the genetic distance between the imputed marker and each of the two flanking markers. The iSelect consensus map (Muñoz-Amatriaín et al. 2014) and the POPSEQ Morex × Barke sequence contig map (Mascher et al. 2013b) were used to impute scores for iSelect and exome capture markers, respectively.
Additionally, a set of artificial, interpolated marker values were constructed for each 1 cM in the Muñoz-Amatriaín et al. (2014) consensus map. Marker calls were generated by interpolating the presence or absence of wild barley introgression using the appropriate flanking, segregating genotyped markers. The interpolation was performed similarly to the imputation process. Instead of true parental genotype calls (0 or 1) at unevenly spaced intervals being imputed, an artificial segregating marker (1) was imputed at each cM in the consensus map. This provided a means to compare introgression frequencies and distributions across families with varying segregating, genotyped markers, and it allowed us to estimate the effect of wild barley introgression regardless of which wild barley allele was present.
Segregation distortion and recombination
Introgressed regions were determined based on the set of segregating markers for each family, with recombination events predicted halfway between two flanking markers. For each line, the proportion of wild barley introgression was calculated as the number of 1-cM intervals subject to wild introgression divided by the total map size. To test for segregation distortion from the expected 12.5% BC2 wild allele frequency, chi-square tests of significance were calculated at each marker, across the entire population (d.f. = 1) and within families (d.f. = 24). The number of recombination events was identified for 10-cM intervals throughout the genome.
Population structure and LD
Neighbor-joining trees were calculated using the nj function in the Analyses of Phylogenetics and Evolution (ape) R package using 2878 BOPA1 and BOPA2 SNP markers for the WBDC and 6976 iSelect markers for the AB-NAM parents. Tree diagrams were constructed in FigTree 1.2.0 (http://tree.bio.ed.ac.uk/software/figtree/). Principal component analysis was performed using the R eigenfunction with and without the parental genotypes included. To reduce the overrepresentation of high LD markers in low recombination regions of the genome, 20 SNPs from the imputed iSelect markers were randomly selected from each 5-cM interval throughout the genome, for a total of 1932 markers used in the analyses.
To compare genome-wide measures of LD in wild barley-derived mapping populations, genome-wide pairwise marker correlations were calculated using 967 typed or imputed BOPA1 SNP markers that were segregating in each of four populations: (1) the 796 line wild barley AB-NAM described here, (2) the 318 accession WBDC (Steffenson et al. 2007), (3) 98 BC2F8 AB lines (HOUH-AB), and (4) 92 RIL lines (HOUH-RIL) derived from the wild barley × two-row malting barley cross OUH-602 × Harrington (Yun et al. 2005, 2006). The HOUH-AB and HOUH-RIL populations were genotyped with the same 384-SNP platform used to genotype the AB-NAM.
To eliminate the effect of population size confounding LD comparisons, LD was also calculated after sampling 90 individuals from each population, 100 times. LD decay was measured as the genetic distance (cM) at which the squared correlation coefficient r2 decayed to 0.2 on a logarithmic regression. Background levels of LD were calculated as the average r2 between all pairwise combinations of markers on different chromosomes. The average pairwise r2 was calculated for 40-cM sliding windows shifting by 2 cM, which contained an average of 102 markers per window.
Trait phenotyping
The traits glossy spike and glossy sheath were scored visually at heading. Traits were scored as present = 1/absent = 0 in augmented field experiments in Crookston, MN in the summers of 2012 and 2013 (CR12, CR13). Field plots were two 1-m long rows. Rasmusson was planted as a repeated check in the center of each three by five plot block, and two additional check varieties were randomly placed within six blocks throughout the field. Black hull color was scored as present = 1/absent = 0 in the remnant seed from the CR12 and St. Paul, MN 2012 (SP12) field trials. The mean trait value was calculated across environments for each line. Grain protein levels were obtained using a Perten (Hägersten, Sweden) diode array near infrared spectroscopy (NIRS) instrument on 22-ml samples of cleaned grain from augmented field experiments in CR12; SP12; and Bozeman, MT and Fargo, ND in 2013 (MT13, ND13). Grain protein was analyzed in each environment and best linear unbiased predictors (BLUPs) were calculated for each line based on phenotypic data from all four environments using ASReml-R (VSN International).
Trait mapping
Genome-wide association analysis (GWAS) was performed for each trait using the ridge regression best linear unbiased prediction (rrBLUP) R package GWAS function (Endelman 2011), which implements an Efficient Mixed-Model Association eXpedited variance component model (Kang et al. 2010). The analysis used a kinship (K) model, controlling for K among individuals, with no additional covariates. Black hull was mapped using all individuals with genotypic information, and results were filtered with a minor allele frequency (MAF) threshold = 0.001 due to the low frequency of the trait in the population. For grain protein, a bootstrap approach was taken where 25 individuals from each family (600 total individuals) were sampled 100 times. Results for glossy sheath, glossy spike, and grain protein were filtered on a MAF = 0.013 threshold, corresponding to 10 individuals containing a wild barley allele.
Marker-trait associations were deemed significant when they were above a 0.05 false discovery rate (FDR) threshold. To obtain estimates of allele substitution effect, marker effects were calculated by passing a single marker to the mixed.solve function in the rrBLUP R package (Endelman 2011; Mohammadi et al. 2015). For protein, the frequency of detection of marker-trait associations was calculated across sampling subsets, and those associations observed <5% of samples are not reported. The reported marker effects and significance [−log(P-value)] values are averages across significant bootstrapped samples. Marker-trait associations were deemed independent loci if there was a ≥5-cM gap between significant markers. Markers with maximum significance in each direction are reported.
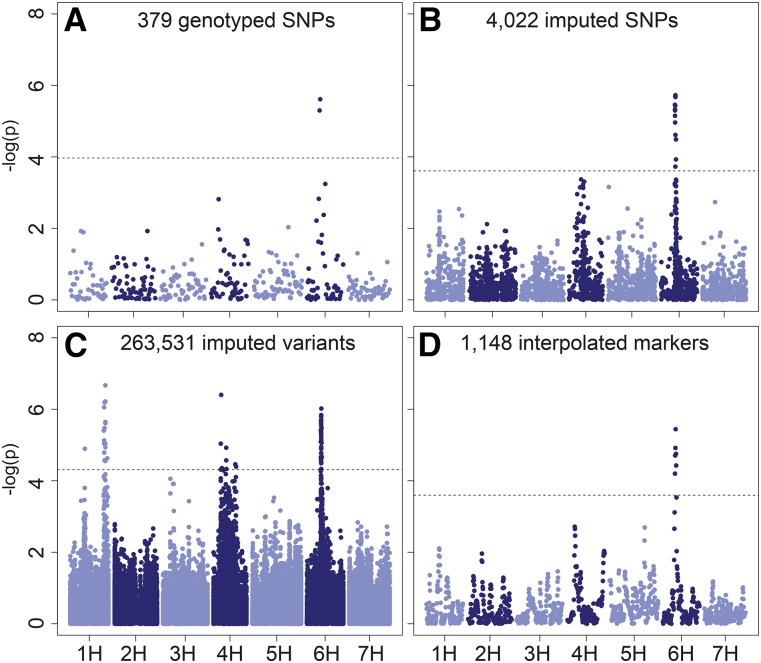
Mapping was performed using four marker sets: (1) 379 SNPs genotyped across the entire population, (2) 4022 SNPs (3520 unique bins) imputed from the parental iSelect SNP marker calls, (3) 263,531 variants (126,303 unique bins) imputed from exome capture sequencing of the parents, and (4) 1148 interpolated biparental calls at each 1 cM of the barley genome consensus map.
Data availability
The data sets supporting the results of this article are available in the Tritceae Toolbox repository, https://triticeaetoolbox.org/barley/. To access data, enter WBIP into the left-hand ‘Quick search…’ box. Clicking on ‘Trial’ will bring up links to phenotypic and genotypic data experiments. To locate exome capture variant calls for parents and imputed variant scores for AB-NAM lines, follow instructions in ‘Comments’ section of ‘WBIP384_ABNAM_2012’ genotyping experiment.
File S1 is an Excel file containing all significant exome capture sequence variant marker-trait associations for the traits: grain protein content, glossy spike, glossy sheath, and black hull.
Exome capture sequences from 24 wild barley parents and Rasmusson have been submitted to the NCBI Sequence Read Archive (PRJNA305889 and PRJNA305578).
Results
Development of a wild barley AB-NAM population
The wild barley AB-NAM population was developed by selecting a highly-diverse set of 25 wild barley accessions from the 318 accessions in the WBDC. These accessions were backcrossed twice to a common recurrent spring barley cultivar Rasmusson (Figure 1C). The resulting population encompasses 25 biparental families, each with 26–39 BC2F4-derived lines for a total of 796 AB-NAM lines. The selected donor parent accessions were pure lines (<1% heterozygosity as assayed by 6976 iSelect SNP markers) and contained 92% of the genetic variants assayed across all marker sets. In particular, these wild barley parents captured 96% of the BOPA1 and 2 SNP alleles, 99% of DArT alleles, and 57% of SSR alleles present in the 318 accessions of the WBDC. The lower percent of SSR alleles captured was due to an average of 19 alleles per SSR locus.
Genetic characterization
To determine the genomic regions of wild parent introgression in the Rasmusson background, the AB-NAM population was genotyped with 379 SNP markers distributed throughout the genome. The number of genotyped, segregating markers ranged from 233 to 326 per family and were distributed every ∼3.57 cM throughout the genome (Table S1). Areas with lower density of segregating markers were found in each family. The maximum gap between segregating markers ranged from 13.43 to 23.42 cM (Table S1), not including line WBDC020, which shows a significant region on chromosome 3HS that appears to be monomorphic with Rasmusson. The parents were genotyped using the barley 9K iSelect SNP platform and exome capture sequenced. We imputed 4022 SNP (3520 unique bins) and 263,531 sequence variants (126,303 unique bins) onto the population. Principal component analysis revealed minimal population structure with the first two components accounting for >5% of the genetic variation, indicating that population structure was successfully controlled by the crossing scheme (Figure S1B).
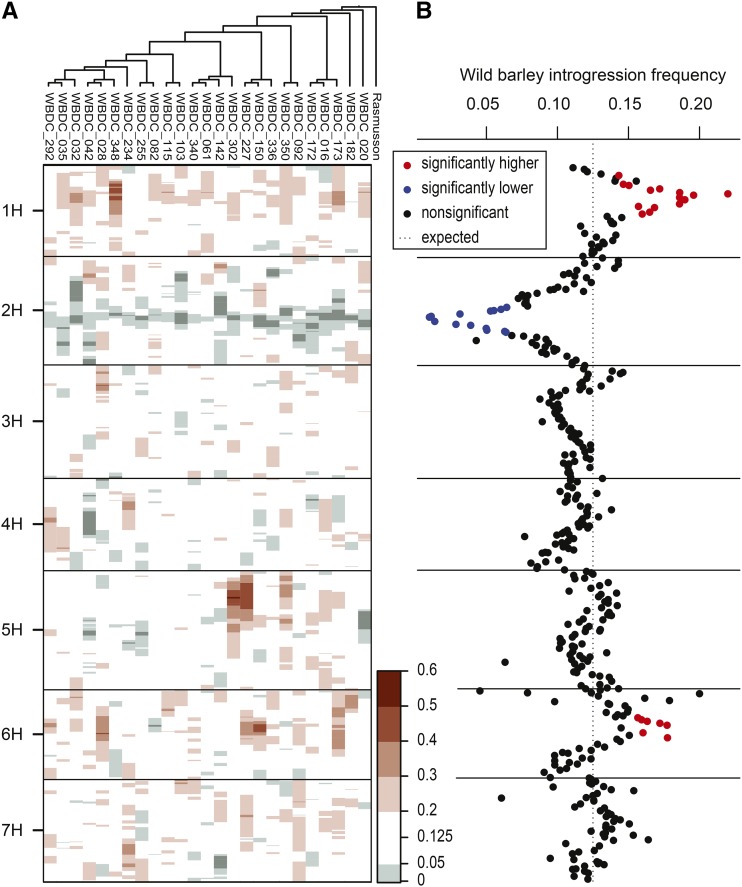
On average, 96% of each wild parent genome was introgressed into the Rasmusson background (Table 1). Individual lines contain 0.79–37.4% wild barley introgression with a mean of 13.5% wild barley introgression per line (Table S2). The mean introgression size is 27.98 cM, and the mean number of introgressions per line is 5.9 on an average of 4.3 chromosomes (Table S2). The expected donor allele frequency for a BC2 derived individual is 0.125. The expected segregation for a BC2F4 population is: 85.93% homozygous recurrent, 3.13% heterozygous, and 10.94% homozygous donor. Selection for six-row spike morphology in the BC2F2 led to a significant decrease in wild barley allele frequency surrounding the six-rowed spike morphology locus (vrs1) on chromosome 2H. Additional genomic regions of segregation distortion were found on chromosomes 1H and 6H, where wild barley introgression was more frequent than expected, both within and across families (Figure 2). Heterozygosity among the 379 genotyped SNP markers ranged from 0 to 6.54% with a mean of 3.18%, close to the expectation of 3.13%. Deviations from the expected heterozygosity for specific markers may be due to errors in genotyping calls.
Figure 2.
Wild barley introgression frequency across the genome. (A) Within families and (B) whole population. Introgression frequency was determined using the appropriate set of segregating markers within each family.
To determine whether it is appropriate to apply the previously developed genetic maps to this population, we compared the recombination frequency across the genome in the AB-NAM with the iSelect consensus map (Muñoz-Amatriaín et al. 2014). When the AB-NAM recombination events were binned in 10-cM intervals, we observed no areas of the genome where recombination frequency was substantially different from the genome-wide average of 67.1 recombination events per 10 cM (Figure S2). Recombination frequency deviated slightly across chromosomes with the lowest average recombination rate of 61.1 recombinations/10 cM on chromosome 4H and the highest rate of 76.8 on chromosome 1H (Table S3).
Patterns of LD in wild barley-derived populations
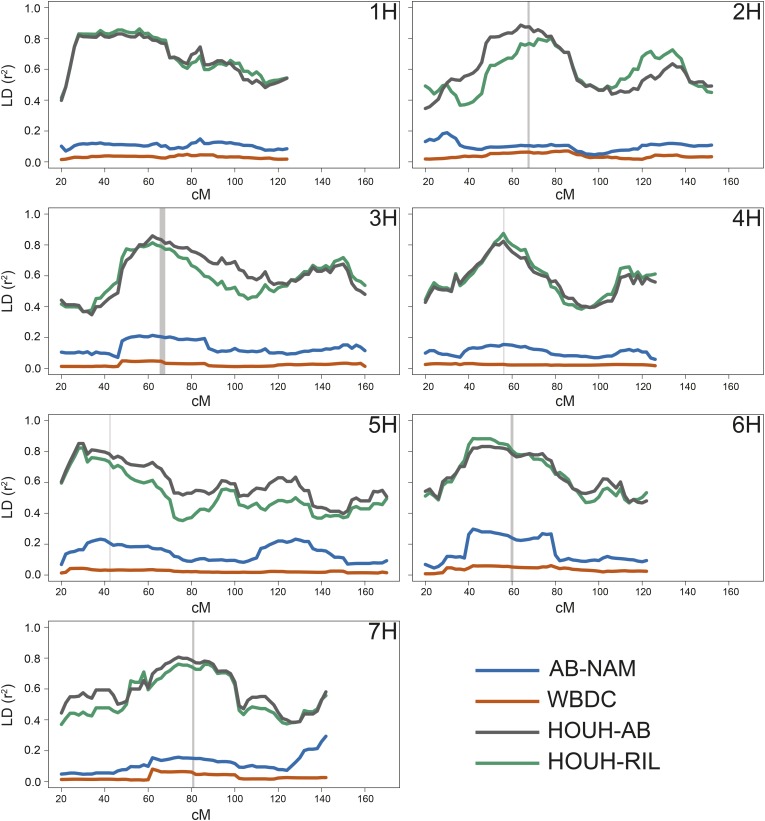
To assess the mapping utility of the AB-NAM population compared to other populations, we compared the LD in the AB-NAM, the WBDC, an RIL population, and an AB population. The parents of the RIL and AB populations were the wild barley accession OUH-602 and the two-rowed spring cultivar Harrington (Yun et al. 2005, 2006). In the WBDC population, LD decayed to an r2 = 0.2 within 1 cM (Figure S3B). In contrast, LD decay in the biparental HOUH-AB and HOUH-RIL populations extended to 28.6 and 32.3 cM, respectively (Figure S3, C and D). The AB-NAM had an intermediate level of LD decay of 9.2 cM when 90 individuals were sampled (Figure S3A), but the LD decay was more rapid (4.9 cM) when sampled across the entire 796 individual AB-NAM population (Figure S4A). In the biparental populations, LD appears to increase slightly in intervals containing the pericentromeric region. This trend is less distinct in the AB-NAM and WBDC populations (Figure 3). Interchromosomal measures of LD were highest in the WBDC population (0.0216), but similar in the AB-NAM (0.0121), AB (0.0137), and RIL (0.0117) populations (Table S4, Figure S5, and Figure S6), suggesting that the crossing design of the AB-NAM minimizes LD caused by factors other than linkage, such as population structure.
Figure 3.
Genome-wide pair-wise LD in wild barley mapping populations. Average pairwise LD (r2) calculated for all segregating BOPA1 markers in 40-cM interval moving windows, with windows calculated every 2 cM across each chromosome 1H–7H in the wild barley AB-NAM population, the WBDC association mapping panel, the OUH-602 × Harrington Advanced Backcross (HOUH-AB) mapping population, and the OUH-602 × Harrington Recombinant Inbred Line (HOUH-RIL) mapping population. Pericentromeric regions as identified in Muñoz-Amatriaín et al. (2011) are denoted by vertical gray bars.
Glossy, hull color, and grain protein content traits in the AB-NAM
Glossy spike, glossy sheath, and black hull color are all highly-heritable qualitative traits which were scored visually in the AB-NAM population, and grain protein content is a quantitative trait scored using NIRS. The glossy spike and glossy sheath phenotypes are characterized by reduced or absent wax that leads to a shiny, bright-green appearance of the spike or spike and sheath, respectively. The glossy spike phenotype is common in wild barley populations, and segregated in all families except WBDC173, for a total of 109 glossy spike AB-NAM lines. Glossy sheath segregated in three families derived from parents WBDC032, WBDC035, and WBDC348 and was observed in 12 individual AB-NAM lines. Black coloration of the mature hull segregated in a single family, WBDC042, and was observed in four individual AB-NAM lines.
Grain protein content exhibited transgressive segregation in the AB-NAM population (Figure S7). The BLUP values for grain protein (%) across the four measured environments (CR12, SP12, MT13, and ND13) ranged from 10.19 to 13.85% with a mean of 12.24% protein. The population mean was higher than the Rasmusson BLUP value of 11.83% protein. The estimated broad-sense heritability calculated on a line-mean basis for grain protein in the AB-NAM was 0.42.
Mapping in the wild barley AB-NAM
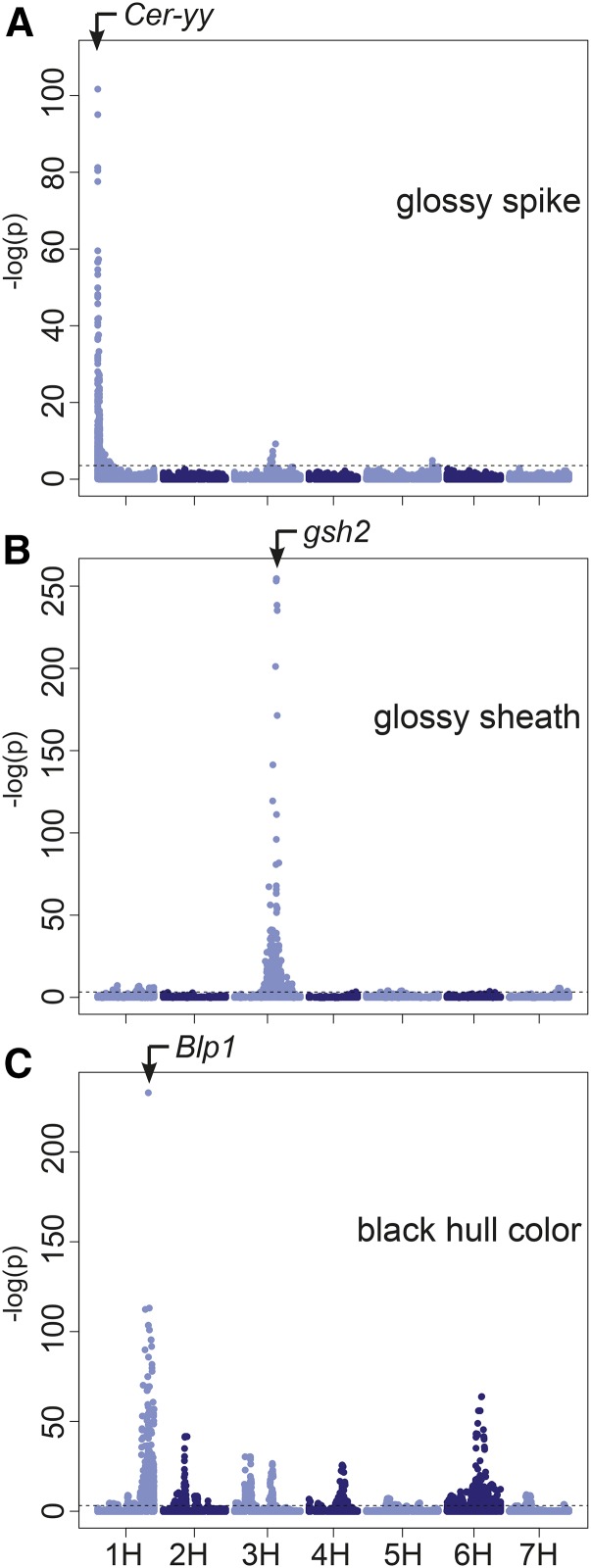
To determine the effectiveness of the wild barley AB-NAM population for mapping, four marker sets were used to map each trait: low-density genotyped SNP markers, medium-density imputed SNP markers, high-density imputed sequence variants, and biparental interpolated markers. For all three qualitative traits, marker-trait associations were detected in all marker sets, with the most significant results found using the high-density sequence-imputed markers. Associations for these traits with high-density sequence variants across 761 individuals in 24 AB-NAM families are shown in Figure 4. For each of these traits, a single highly-significant locus was identified that corresponded to mapped barley mutants for these traits, and an additional one or more loci were identified with lower significance, which we presume to be false positives. In the cases of glossy sheath and black hull, sequence variants were detected that are perfectly segregating with the trait.
Figure 4.
Manhattan plots of marker-trait associations. (A) Glossy spike, (B) glossy sheath, and (C) black hull color in the AB-NAM population using 263,531 exome capture sequence variant markers. Horizontal line indicates FDR = 0.05 significance threshold. Traits glossy sheath and black hull color had additional variants which perfectly segregated with the trait. These associations were not plotted due to an infinite −log(P) result.
For the quantitative trait grain protein, we detected a single variant on chromosome 6H in the three lower-density marker sets (Figure 5, A, B, and D). When mapped with high-density sequence-imputed variants, several other chromosomal regions appear to be significant (Figure 5C). To provide an additional measure of confidence for the detected associations, we used a bootstrap mapping approach to determine the frequency over 100 replicates that marker-trait associations were detected using subsets of AB-NAM individuals in the analysis. This value along with the MAF of the associated variant, the average non-Rasmusson relative effect size, and the average significance level are reported in Table 2 and File S1.
Figure 5.
Manhattan plots of marker-trait associations for grain protein content using various marker sets. (A) 379 genotyped SNP markers, (B) 4022 imputed SNP markers, (C) 263,531 imputed sequence variants, and (D) 1148 interpolated markers. Dashed horizontal line indicates FDR = 0.05 significance threshold. Plots shown are mapping results from all 761 individuals (not bootstrapped) with imputed sequence variants.
Table 2. Summary of marker-trait associations of maximum significance.
| Trait | Varianta | Chromb | cMb | MAFc | −log(P)d | Effecte | Freqf |
|---|---|---|---|---|---|---|---|
| Glossy spike | morex_contig_40051:2172_A/Gg | 1H | 0.1 | 0.12 | 101.7 | 0.46 | N/A |
| Glossy sheath | morex_contig_41718:9777_CG/Cg | 3H | 96.6 | 0.02 | Inf | 0.50 | N/A |
| Black hull | morex_contig_1573652:70_A/G | 1H | 116.5 | 0.01 | Inf | 0.51 | N/A |
| morex_contig_38907:1672_G/Ag | 1H | 116.6 | 0.01 | Inf | 0.50 | N/A | |
| morex_contig_53289:2443_G/Ag | 1H | 116.8 | 0.01 | Inf | 0.50 | N/A | |
| morex_contig_42987:7765_T/Cg | 1H | 117.7 | 0.01 | Inf | 0.50 | N/A | |
| morex_contig_5603:2020_T/Cg | 1H | 117.8 | 0.01 | Inf | 0.50 | N/A | |
| morex_contig_243581:952_C/Gg | 1H | 118.1 | 0.01 | Inf | 0.50 | N/A | |
| morex_contig_1576759:813_C/Tg | 1H | 118.3 | 0.01 | Inf | 0.50 | N/A | |
| morex_contig_5976:324_G/Ag | 1H | 119.0 | 0.01 | Inf | 0.50 | N/A | |
| morex_contig_39431:11917_G/Ag | 1H | 119.4 | 0.01 | Inf | 0.50 | N/A | |
| GPC | morex_contig_43675:8617_T/C | 1H | 50.6 | 0.17 | 4.9 | −0.14 | 0.13 |
| morex_contig_47454:1292_C/T | 1H | 119.7 | 0.01 | 6.8 | −0.37 | 0.95 | |
| morex_contig_44650:4992_C/T | 3H | 25.3 | 0.02 | 4.8 | −0.18 | 0.17 | |
| morex_contig_41236:9829_G/A | 3H | 37.4 | 0.02 | 4.9 | −0.10 | 0.06 | |
| morex_contig_99201:2611_C/T | 4H | 27.5 | 0.10 | 5.6 | 0.17 | 0.90 | |
| morex_contig_46131:3154_TA/T | 4H | 27.8 | 0.01 | 4.8 | −0.05 | 0.08 | |
| morex_contig_47914:307_C/T | 4H | 43.5 | 0.02 | 4.9 | 0.36 | 0.09 | |
| morex_contig_246786:1453_AACGTACGC/A | 4H | 44.9 | 0.03 | 5.0 | −0.20 | 0.59 | |
| morex_contig_156722:1830_G/A | 4H | 51.6 | 0.03 | 4.8 | 0.33 | 0.11 | |
| morex_contig_45564:4922_A/G | 4H | 52.2 | 0.07 | 4.5 | −0.07 | 0.06 | |
| morex_contig_52783:3343_C/T | 4H | 63.4 | 0.02 | 4.5 | −0.27 | 0.06 | |
| morex_contig_1575414:1899_A/G | 4H | 67.1 | 0.03 | 4.5 | 0.29 | 0.13 | |
| morex_contig_146580:705_T/A | 4H | 76.3 | 0.01 | 5.0 | −0.30 | 0.56 | |
| morex_contig_42454:4904_G/A | 4H | 78.7 | 0.04 | 4.8 | 0.23 | 0.11 | |
| morex_contig_7108:5074_G/Tg | 6H | 50.0 | 0.17 | 5.8 | 0.37 | 0.96 |
All significant marker-trait associations can be found in File S1. Markers for a single QTL of maximum significance are reported for qualitative traits. For grain protein content, QTL on the same chromosome were considered unique loci if there was a >5-cM gap between significant markers, and markers of maximum significance with both positive and negative effects are reported for each QTL. Chrom, chromosome; Freq, frequency; Inf, markers which cosegregate with the trait values; GPC, grain protein content.
Variants significantly associated with trait. Association determined by an FDR >0.05 significance threshold, and a frequency of detection >0.05 in 100 bootstrap samplings of 25 individuals from each family for the trait protein.
Chromosome and cM positions of Morex contig containing the variant as determined by the Mascher et al. (2013b) POPSEQ map.
MAF of variant in the AB-NAM population.
−log(P) averaged across significant bootstrap tests.
Relative effect of non-Rasmusson allele, averaged across significant bootstrap tests.
Frequency of significant association with variant detected in 100 bootstrap samples.
Additional variants in the same bin with identical segregation pattern can be found in File S1.
Three significant loci were identified for the glossy sheath phenotype. Two small regions of chromosome 1H at 46.5 cM [−log(P) = 7.32] and 97.5cM [−log(P) = 6.90] were identified, in addition to five variants on chromosome 3HL (96.6 cM), which cosegregate with the trait (File S1). These variants are located in a single gene of unknown function on Morex contig 41718 (Table 2). This map location corresponds to the glossy sheath 2 (gsh2) mutant that exhibits similar lack of wax on the leaf sheath (von Wettstein-Knowles 1990; Druka et al. 2011).
The glossy spike trait was mapped to the distal region of chromosome 1HS. Significant variants were found between position 0.11 and 9.92 cM on the POPSEQ map (Mascher et al. 2013b), with the maximum significance of −log(P) = 101.69 occurring at 0.11 cM (Table 2). An additional significant locus was detected on chromosome 3H with the peak significance of −log(P) = 9.21 occurring at 96.6 cM (File S1). This association falls in the region of the most significant glossy sheath locus, and is likely due to the interaction between glossy spike and glossy sheath. The highly-significant region on chromosome 1HS is coincident with the location of the Eceriferum-yy (Cer-yy) barley mutants, which exhibit a reduction of wax production on the barley spike (Lundqvist and Wettstein-Knowles 1982; Druka et al. 2011).
The black hull phenotype was only identified in four individuals. Thus, there appear to be many spurious associations, including peaks on chromosomes 2H, 3H, 4H, and 6H with significance ranging from −log(P) = 7.03 to −log(P) = 30.41 (File S1). Because of this, caution must be taken when exploring such low-frequency traits. On chromosome 1HL, nine bins that include 43 variants on 15 contigs cosegregated with the black hull phenotype. These contigs are located in a 2.9-cM region between 116.5 and 119.4 cM. This region corresponds to the Black lemma and pericarp (Blp1) mutant (Druka et al. 2011).
Ten QTL were detected for grain protein content (Table 2). These QTL were located on chromosomes 1H, 3H, 4H, and 6H. The wild barley alleles for the QTL on chromosomes 1H and 3H conferred lower grain protein content, a beneficial quality for malting and brewing. Four significant loci were identified on chromosome 4H, each of which had both positive and negative wild barley effects, indicating that there may be multiple haplotypes segregating at these loci. The most-frequently detected QTL was located on chromosome 6H at 50.0 cM. The wild barley alleles at this locus conferred higher (unfavorable for malting) grain protein content (Figure S8).
Discussion
We developed a population that incorporates a large amount of diversity from wild barley into a six-rowed, cultivated spring barley background, which will serve as a resource for barley breeders and geneticists to efficiently screen a large amount of genetic diversity for many traits of interest. We examined the genetic composition of the population for factors which influence mapping: segregation distortion, distribution of recombination, population stratification, LD, population size, and marker set. Additionally, three qualitative traits: glossy spike, glossy sheath, and black hull color, and the quantitative trait grain protein content were mapped with high resolution in the population. The mapping resolution, coupled with the diverse genomic content of the AB-NAM has demonstrated that screening the AB-NAM for traits of interest is an effective initial step for cloning barley genes.
The wild barley AB-NAM increases the genetic diversity available to barley breeders
Because we aimed to maximize the amount of genetic diversity in the population, the parents of the AB-NAM are a set of 25 wild barley accessions selected solely on genetic data, without considering ecogeographic or phenotypic characteristics. This set of diverse lines approaches an ideal mini core of the larger wild barley population. But, the selection of lines was subject to marker ascertainment bias (Nielsen 2000). The identification of SNPs for the BOPA marker platforms used was based on ESTs and sequences primarily from cultivated barley (Close et al. 2009), and ascertainment bias using this panel has previously been demonstrated; wherein estimates of diversity were observed to be higher in cultivated barley germplasm than in landrace germplasm when assayed with BOPA1 markers (Moragues et al. 2010). Since the original selection of parental lines, the genetic signatures that differentiate wild and landrace barleys have been examined more extensively (Russell et al. 2011; Poets et al. 2015) and it is likely that ascertainment bias led to the inclusion of eight accessions which appear to contain varying levels of landrace and/or cultivated barley admixture (Fang et al. 2014). These eight accessions are those with larger genetic distances from the majority of the WBDC (as indicated in Figure 1B) and appear in between the AB-NAM population and the majority of the wild parents in the PCA plot in Figure S1A. Still, the phenotypic and geographic distribution of the selected parents is only slightly narrower than the WBDC range (Figure 1A). In addition to the BOPA SNP markers; the use of SSR markers, which have been shown in maize to be less influenced by ascertainment bias (Hamblin et al. 2007); and DArT markers, which were developed from wild barley germplasm (Alsop et al. 2011); may have helped mitigate the bias. Nevertheless, the presence of landraces in the population should not influence the effectiveness of mapping and gene discovery or the discovery of novel allelic variation in this population.
During population development, we attempted to minimize unintentional selection and therefore maximize the amount of wild barley genome introgressed. At the same time, we imposed selection for the Rasmusson six-rowed spike morphology. As expected, wild barley introgression is significantly reduced in the region of 2H surrounding the vrs1 locus responsible for the six-rowed phenotype (Figure 2). This selection will limit our ability to map loci on chromosome 2H, but it will also minimize the confounding effects of phenotyping a population that is segregating for a major morphological trait. This is especially true for assessment of Fusarium head-blight resistance and malting quality, which can be markedly affected by row type (Marquez-Cedillo et al. 2000; Choo et al. 2004). No other regions of the genome show reduced introgression frequency, but regions on chromosomes 1H and 6H show an overrepresentation of wild barley introgression across the population. Notably, vrs3, a known determinant of spike morphology, is located in the region of chromosome 1H that has an overrepresentation of wild barley introgression (Muñoz-Amatriaín et al. 2014). The vrs3 phenotype appears six-rowed at the top of the spike (Lundqvist and Franckowiak 1997), a characteristic that may have influenced the selection for six-rowed morphology in the population, and consequently, affected the allele frequency in this region. Segregation distortion in the HEB population was also observed, but generally it was restricted to specific crosses and none of the regions identified in the HEB population correspond with those identified here (Schnaithmann et al. 2014). The HEB population may have more subpopulation structure due to the fact that each HEB family was derived from only 20 BC1 individuals, then expanded to families of 22–75 individuals (Maurer et al. 2015).
Wide crosses can be subject to factors such as segregation distortion and deviations in recombination rate across the genome (Xu et al. 1997; Bauer et al. 2013). We found no regions of the genome that deviate substantially from the mean recombination rate, suggesting that fluctuations in recombination throughout the AB-NAM population are consistent with the genetic distances represented by the consensus map. We also observed that the percentage of introgressed regions ranged widely, with individuals containing 0.79–37.45% wild barley introgression, but the average introgression frequency of 13.55% was relatively consistent with the expectation of 12.5%. Collectively, these results corroborate previous studies (Yun et al. 2006; Schnaithmann et al. 2014) that showed only minor fluctuations in allele frequency occur when introgressing wild barley genomes.
The wild barley AB-NAM exhibits low population structure and low LD
Population stratification present in association mapping panels can lead to spurious associations if not appropriately controlled, and effective control of population structure can lower the power to detect marker-trait associations (Larsson et al. 2013). The NAM design attempts to eliminate this problem by nesting segments of diverse blocks in a controlled crossing structure (Yu et al. 2008). Principal component analysis shows only minimal population stratification in the wild barley AB-NAM (Figure S1B), with no consistent trends among families. Because of this, we did not include measures of population stratification (Q) in our association mapping, but we did use a K model to control for cryptic relatedness among individuals.
Because wild barley populations exhibit lower LD than cultivated barley populations, using wild barley populations presents an opportunity for high-resolution mapping. But phenotyping wild barley for adult plant traits can be difficult and population structure can confound mapping results. One of the aims of the AB-NAM was to develop a population that mitigates the difficulties of working directly with wild barley, while maintaining the high genetic diversity and high mapping resolution possible with wild barley populations. To understand how the AB-NAM compares to other wild barley-derived populations, we compared genome-wide LD in the AB-NAM, the WBDC association mapping panel, and the wild barley-derived biparental mapping populations HOUH-AB and HOUH-RIL. As expected, the rate of LD decay is much higher in the WBDC population than in the biparental populations (Figure S3). Because differences in the parental genotypes can be imputed onto the segregating families of the AB-NAM, LD in the AB-NAM approaches the low level of the WBDC (Figure 3).
The level of LD present in the mapping population is an indicator of potential mapping resolution, but it is also an indication of the level of marker coverage necessary for high-power trait mapping. When working with low LD populations, a higher density of markers is required to tag QTL with markers. Simulations have shown that power increases as markers are added to multifamily mapping experiments, and being able to impute high-density markers onto a lower-density genotyped panel appears to be an effective means to increasing marker density (Liu et al. 2013). Ultimately, the AB-NAM optimizes the trade-off between the practical difficulties of phenotyping unadapted material and high-resolution mapping in a diverse germplasm set.
The wild barley AB-NAM provides a new resource for trait mapping in barley
The NAM design provides a straightforward means to projecting high-density marker data onto a segregating population. We showed that the addition of medium- and high-density imputed sequence variants can improve the power of detecting marker-trait associations (Figure 5), in some cases allowing for the identification of variants that cosegregate with traits of interest (Table 2). These highly-significant associations were identified even when only a small number of individuals exhibited the trait. In the case of glossy sheath, the variants with the highest significance were localized to a single contig on the genome sequence. This indicates that there is a benefit to the NAM design for both power and resolution as compared to a biparental population. Furthermore, the size of the population allows for a bootstrapping approach that can provide additional information to support or filter marker-trait associations.
A major advantage of the NAM design is the increase in MAF of uncommon alleles to detectable levels as compared to association mapping panels. In the NAM design, MAF is a function of the number of parents containing the allele and the expected MAF in the individual crosses. In RIL-derived populations, the expected MAF is 0.5. In BC2 derived populations, the expected MAF is 0.125. This means that when private alleles are present in the AB-NAM population with 25 parents and 796 individuals, the expected MAF of that allele is 0.125/25 = 0.005, ∼4 individuals in the 796 individual wild barley AB-NAM. In contrast, a similarly sized RIL-derived NAM with 25 parents would have a private allele MAF of 0.5/25 = 0.02 or ∼16 individuals with the allele. This means that, except for very large effect traits like black hull, causative alleles segregating in a single family in the wild barley AB-NAM may not be detected. Furthermore, multiparent mapping models that assign unique alleles for each parent are unlikely to work with the AB-NAM design. Future population development should take this limitation into account.
To examine a quantitative trait with lower heritability, we used the wild barley AB-NAM population to map grain protein content. Grain protein is an important trait for both malting (requiring low protein) and animal feed (requiring high protein) purposes (Blake et al. 2010). A single QTL was identified in each of the marker sets, corresponding to the grain protein content QTL on chromosome 6HS (Distelfeld et al. 2008; Lacerenza et al. 2010). When mapped with higher-density, sequence-imputed markers, additional QTL were detected throughout the genome (Table 2). QTL identified on chromosome 1H exhibited negative marker effects, indicating that wild barley alleles may have beneficial effects for malting quality. Additionally, QTL identified on chromosome 4H exhibit both negative and positive effects of wild barley alleles, indicating that either multiple haplotypes or multiple genes are influencing protein at these loci. Additional analyses that take marker haplotypes into account may improve the interpretability of these results.
While the use of exotic germplasm resources continues to be a challenge for breeders, the AB-NAM population design serves as a resource that bridges the gap between the germplasm of gene banks and breeding programs. Creating AB populations within the context of a NAM design expands the utility of NAM populations to more distantly-related germplasm. The effort expended to produce the AB design is balanced by the added ease and accuracy of phenotyping lines containing exotic and unadapted alleles. Furthermore, the high mapping resolution achieved in the AB-NAM population provides a quick means to fine map and identify candidate genes, particularly for highly-heritable qualitative traits.
Acknowledgments
We thank Shane Heinen for greenhouse support and Ed Schiefelbein, Guillermo Velasquez, and Martin Hochhalter for field management. Funding provided by the United States Department of Agriculture (USDA) National Needs Fellowship 2007-38420-17749 and USDA Triticeae Coordinated Agricultural Project 2011-68002-30029.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.190736/-/DC1.
Communicating editor: A. Charcosset
Literature Cited
- Abdel-Ghani A. H., Parzies H. K., Omary A., Geiger H. H., 2004. Estimating the outcrossing rate of barley landraces and wild barley populations collected from ecologically different regions of Jordan. Theor. Appl. Genet. 109: 588–595. [DOI] [PubMed] [Google Scholar]
- Alsop B. P., Farre A., Wenzl P., Wang J. M., Zhou M. X., et al. , 2011. Development of wild barley-derived DArT markers and their integration into a barley consensus map. Mol. Breed. 27: 77–92. [Google Scholar]
- Bandillo N., Raghavan C., Muyco P. A., Sevilla M. A. L., Lobina I. T., et al. , 2013. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice (N Y) 6: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer E., Falque M., Walter H., Bauland C., Camisan C., et al. , 2013. Intraspecific variation of recombination rate in maize. Genome Biol. 14: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M., Grando S., Backes G., Jahoor A., Sabbagh A., et al. , 2003. QTLs for agronomic traits in the Mediterranean environment identified in recombinant inbred lines of the cross ‘Arta’ × H. spontaneum 41–1. Theor. Appl. Genet. 107: 1215–1225. [DOI] [PubMed] [Google Scholar]
- Bernardo R., 2002. Breeding for quantitative traits in plants. Stemma Press, Woodbury, MN. [Google Scholar]
- Blake, T., V. C. Blake, J. G. P. Bowman, and H. Abdel-Haleem, 2010 Barley Feed Uses and Quality Improvement, pp. 522–531 in Barley: Production, Improvement and Uses, edited by S.E. Ullrich, Wiley-Blackwell, Oxford. [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J., et al. , 2009. The Genetic Architecture of Maize Flowering Time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Caldwell K. S., Russell J., Langridge P., Powell W., 2006. Extreme Population-Dependent Linkage Disequilibrium Detected in an Inbreeding Plant Species, Hordeum vulgare. Genetics 172: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo T. M., Vigier B., Shen Q. Q., Martin R. A., Ho K. M., et al. , 2004. Barley Traits Associated with Resistance to Fusarium Head Blight and Deoxynivalenol Accumulation. Phytopathology 94: 1145–1150. [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Close T. J., Bhat P. R., Lonardi S., Wu Y., Rostoks N., et al. , 2009. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10: 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comadran J., Kilian B., Russell J., Ramsay L., Stein N., et al. , 2012. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 44: 1388–1392. [DOI] [PubMed] [Google Scholar]
- Condón F., Gustus C., Rasmusson D. C., Smith K. P., 2008. Effect of Advanced Cycle Breeding on Genetic Diversity in Barley Breeding Germplasm. Crop Sci. 48: 1027–1036. [Google Scholar]
- Distelfeld A., Korol A., Dubcovsky J., Uauy C., Blake T., et al. , 2008. Colinearity between the barley grain protein content (GPC) QTL on chromosome arm 6HS and the wheat Gpc-B1 region. Mol. Breed. 22: 25–38. [Google Scholar]
- Druka A., Franckowiak J., Lundqvist U., Bonar N., Alexander J., et al. , 2011. Genetic Dissection of Barley Morphology and Development. Plant Physiol. 155: 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J. B., 2011. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome J. 4: 250. [Google Scholar]
- Fang Z., A. Eule-Nashoba, C. Powers, T. Y. Kono, S. Takuno et al., 2013 Comparative Analyses Identify the Contributions of Exotic Donors to Disease Resistance in a Barley Experimental Population. G3 (Bethesda) 3: 1945–1953. [DOI] [PMC free article] [PubMed]
- Fang Z., A. M. Gonzales, M. T. Clegg, K. P. Smith, G. J. Muehlbauer et al., 2014 Two Genomic Regions Contribute Disproportionately to Geographic Differentiation in Wild Barley. G3 (Bethesda) 4: 1193–1203. [DOI] [PMC free article] [PubMed]
- Fetch T. G., Steffenson B. J., Nevo E., 2003. Diversity and Sources of Multiple Disease Resistance in Hordeum spontaneum. Plant Dis. 87: 1439–1448. [DOI] [PubMed] [Google Scholar]
- Fu Y.-B., Somers D. J., 2009. Genome-Wide Reduction of Genetic Diversity in Wheat Breeding. Crop Sci. 49: 161–168. [Google Scholar]
- Guo B., Beavis W. D., 2011. In silico genotyping of the maize nested association mapping population. Mol. Breed. 27: 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Wang D., Guo Z., Beavis W. D., 2013. Family-based association mapping in crop species. Theor. Appl. Genet. 126: 1419–1430. [DOI] [PubMed] [Google Scholar]
- Hamblin M. T., Warburton M. L., Buckler E. S., 2007. Empirical Comparison of Simple Sequence Repeats and Single Nucleotide Polymorphisms in Assessment of Maize Diversity and Relatedness. PLoS One 2: e1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M. T., Close T. J., Bhat P. R., Chao S., Kling J. G., et al. , 2010. Population Structure and Linkage Disequilibrium in U.S. Barley Germplasm: Implications for Association Mapping. Crop Sci. 50: 556–566. [Google Scholar]
- Huang X., Paulo M. J., Boer M., Effgen S., Keizer P., et al. , 2011. Analysis of natural allelic variation in Arabidopsis using a multiparent recombinant inbred line population. Proc. Natl. Acad. Sci. USA 108: 4488–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan D. R., Mace E. S., Cruickshank A. W., Hunt C. H., Henzell R. G., 2011. Exploring and Exploiting Genetic Variation from Unadapted Sorghum Germplasm in a Breeding Program. Crop Sci. 51: 1444–1457. [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T. J. Y., Fu F., Mohammadi M., Hoffman P. J., Liu C., et al. , 2015. The role of deleterious substitutions in crop genomes. bioRxiv DOI: http://dx.doi.org/10.1101/033175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Korff M., Wang H., Léon J., Pillen K., 2004. Development of candidate introgression lines using an exotic barley accession (Hordeum vulgare ssp. spontaneum) as donor. Theor. Appl. Genet. 109: 1736–1745. [DOI] [PubMed] [Google Scholar]
- Korte A., Farlow A., 2013. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A Multiparent Advanced Generation Inter-Cross to Fine-Map Quantitative Traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerenza J. A., Parrott D. L., Fischer A. M., 2010. A major grain protein content locus on barley (Hordeum vulgare L.) chromosome 6 influences flowering time and sequential leaf senescence. J. Exp. Bot. 61: 3137–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson S. J., Lipka A. E., Buckler E. S., 2013. Lessons from Dwarf8 on the Strengths and Weaknesses of Structured Association Mapping. PLoS Genet. 9: e1003246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Z., Huang X. Q., Heinrichs F., Ganal M. W., Röder M. S., 2006. Analysis of QTLs for yield components, agronomic traits, and disease resistance in an advanced backcross population of spring barley. Genome 49: 454–466. [DOI] [PubMed] [Google Scholar]
- Liu K., Muse S. V., 2005. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128–2129. [DOI] [PubMed] [Google Scholar]
- Liu W., Maurer H. P., Reif J. C., Melchinger A. E., Utz H. F., et al. , 2013. Optimum design of family structure and allocation of resources in association mapping with lines from multiple crosses. Heredity 110: 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist U., von Wettstein-Knowles P., 1982. Dominant mutations at Cer-yy change barley spike wax into leaf blade wax. Carlsberg Res. Commun. 47: 29–43. [Google Scholar]
- Lundqvist U., Franckowiak J. D., 1997. Six-rowed spike 3, vrs3. Barley Genet. Newsl. 315: 264–265. [Google Scholar]
- Macdonald S. J., Long A. D., 2007. Joint Estimates of Quantitative Trait Locus Effect and Frequency Using Synthetic Recombinant Populations of Drosophila melanogaster. Genetics 176: 1261–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez-Cedillo L. A., Hayes P. M., Jones B. L., Kleinhofs A., Legge W. G., et al. , 2000. QTL analysis of malting quality in barley based on the doubled-haploid progeny of two elite North American varieties representing different germplasm groups. Theor. Appl. Genet. 101: 173–184. [Google Scholar]
- Mascher M., Richmond T. A., Gerhardt D. J., Himmelbach A., Clissold L., et al. , 2013a Barley whole exome capture: a tool for genomic research in the genus Hordeum and beyond. Plant J. 76: 494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., Muehlbauer G. J., Rokhsar D. S., Chapman J., Schmutz J., et al. , 2013b Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J. 76: 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus I., Corey A., Filichkin T., Hayes P. M., Vales M. I., et al. , 2003. Development and characterization of recombinant chromosome substitution lines (RCSLs) using Hordeum vulgare subsp. spontaneum as a source of donor alleles in a Hordeum vulgare subsp. vulgare background. Genome 46: 1010–1023. [DOI] [PubMed] [Google Scholar]
- Maurer A., Draba V., Jiang Y., Schnaithmann F., Sharma R., et al. , 2015. Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics 16: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M. D., Kresovich S., Villeda H. S., Bradbury P., Li H., et al. , 2009. Genetic properties of the maize nested association mapping population. Science 325: 737–740. [DOI] [PubMed] [Google Scholar]
- Mohammadi M., Blake T. K., Budde A. D., Chao S., Hayes P. M., et al. , 2015. A genome-wide association study of malting quality across eight U.S. barley breeding programs. Theor. Appl. Genet. 128: 705–721. [DOI] [PubMed] [Google Scholar]
- Moragues M., Comadran J., Waugh R., Milne I., Flavell A. J., et al. , 2010. Effects of ascertainment bias and marker number on estimations of barley diversity from high-throughput SNP genotype data. Theor. Appl. Genet. 120: 1525–1534. [DOI] [PubMed] [Google Scholar]
- Morrell P. L., Toleno D. M., Lundy K. E., Clegg M. T., 2005. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc. Natl. Acad. Sci. USA 102: 2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P. L., Gonzales A. M., Meyer K. K. T., Clegg M. T., 2014. Resequencing Data Indicate a Modest Effect of Domestication on Diversity in Barley: A Cultigen With Multiple Origins. J. Hered. 105: 253–264. [DOI] [PubMed] [Google Scholar]
- Muñoz-Amatriaín M., Xiong Y., Schmitt M. R., Bilgic H., Budde A. D., et al. , 2010. Transcriptome analysis of a barley breeding program examines gene expression diversity and reveals target genes for malting quality improvement. BMC Genomics 11: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Amatriaín M., Moscou M. J., Bhat P. R., Svensson J. T., Bartoš J., et al. , 2011. An Improved Consensus Linkage Map of Barley Based on Flow-Sorted Chromosomes and Single Nucleotide Polymorphism Markers. Plant Genome J. 4: 238–249. [Google Scholar]
- Muñoz-Amatriaín M., Cuesta-Marcos A., Endelman J. B., Comadran J., Bonman J. M., et al. , 2014. The USDA Barley Core Collection: Genetic Diversity, Population Structure, and Potential for Genome-Wide Association Studies. PLoS One 9: e94688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., 2000. Estimation of Population Parameters and Recombination Rates From Single Nucleotide Polymorphisms. Genetics 154: 931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer J. A., Flint-Garcia S., deLeon N., McMullen M. D., Kaeppler S. M., et al. , 2013. The Genetic Architecture of Maize Stalk Strength. PLoS One 8: e67066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer J. A., Romay M. C., Gore M. A., Flint-Garcia S. A., Zhang Z., et al. , 2014. The Genetic Architecture Of Maize Height. Genetics 196: 1337–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillen K., Zacharias A., Léon J., 2003. Advanced backcross QTL analysis in barley (Hordeum vulgare L.). Theor. Appl. Genet. 107: 340–352. [DOI] [PubMed] [Google Scholar]
- Poets A. M., M. Mohammadi, K. Seth, H. Wang, T. J. Y. Kono et al., 2015 The Effects of Both Recent and Long-Term Selection and Genetic Drift Are Readily Evident in North American Barley Breeding Populations. G3 (Bethesda) 6: 609–622. [DOI] [PMC free article] [PubMed]
- Poland J. A., Bradbury P. J., Buckler E. S., Nelson R. J., 2011. Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci. USA 108: 6893–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkheirandish M., Komatsuda T., 2007. The Importance of Barley Genetics and Domestication in a Global Perspective. Ann. Bot. (Lond.) 100: 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkheirandish M., Hensel G., Kilian B., Senthil N., Chen G., et al. , 2015. Evolution of the Grain Dispersal System in Barley. Cell 162: 527–539. [DOI] [PubMed] [Google Scholar]
- Rafalski A., 2002. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 5: 94–100. [DOI] [PubMed] [Google Scholar]
- Rasmusson D. C., Phillips R. L., 1997. Plant Breeding Progress and Genetic Diversity from De Novo Variation and Elevated Epistasis. Crop Sci. 37: 303–310. [Google Scholar]
- Rebetzke G. J., Verbyla A. P., Verbyla K. L., Morell M. K., Cavanagh C. R., 2014. Use of a large multiparent wheat mapping population in genomic dissection of coleoptile and seedling growth. Plant Biotechnol. J. 12: 219–230. [DOI] [PubMed] [Google Scholar]
- Roy J. K., Smith K. P., Muehlbauer G. J., Chao S., Close T. J., et al. , 2010. Association mapping of spot blotch resistance in wild barley. Mol. Breed. 26: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J., Dawson I. K., Flavell A. J., Steffenson B., Weltzien E., et al. , 2011. Analysis of >1000 single nucleotide polymorphisms in geographically matched samples of landrace and wild barley indicates secondary contact and chromosome-level differences in diversity around domestication genes. New Phytol. 191: 564–578. [DOI] [PubMed] [Google Scholar]
- Saisho D., Purugganan M. D., 2007. Molecular Phylogeography of Domesticated Barley Traces Expansion of Agriculture in the Old World. Genetics 177: 1765–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannemann W., Huang B. E., Mathew B., Léon J., 2015. Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol. Breed. 35: 1–16. [Google Scholar]
- Schnaithmann F., Kopahnke D., Pillen K., 2014. A first step toward the development of a barley NAM population and its utilization to detect QTLs conferring leaf rust seedling resistance. Theor. Appl. Genet. 127: 1513–1525. [DOI] [PubMed] [Google Scholar]
- Smith K. P., Rasmusson D. C., Schiefelbein E., Wiersma J. J., Wiersma J. V., et al. , 2010. Registration of “Rasmusson” Barley. J. Plant Regist. 4: 167–170. [Google Scholar]
- Steffenson B. J., Olivera P., Roy J. K., Jin Y., Smith K. P., et al. , 2007. A walk on the wild side: mining wild wheat and barley collections for rust resistance genes. Aust. J. Agric. Res. 58: 532–544. [Google Scholar]
- Tanksley S. D., Nelson J. C., 1996. Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 92: 191–203. [DOI] [PubMed] [Google Scholar]
- Tian F., Bradbury P. J., Brown P. J., Hung H., Sun Q., et al. , 2011. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43: 159–162. [DOI] [PubMed] [Google Scholar]
- Vilhjálmsson B. J., Nordborg M., 2013. The nature of confounding in genome-wide association studies. Nat. Rev. Genet. 14: 1–2. [DOI] [PubMed] [Google Scholar]
- Wang B., Chee P. W., 2010. Application of advanced backcross quantitative trait locus (QTL) analysis in crop improvement. J. Plant Breed. Crop Sci. 2: 221–232. [Google Scholar]
- von Wettstein-Knowles P., 1990. New alleles of Cer-yy and cer-b. Barley Genet. Newsl. 20: 66–68. [Google Scholar]
- Xu Y., Zhu L., Xiao J., Huang N., McCouch S. R., 1997. Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol. Gen. Genet. MGG 253: 535–545. [DOI] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Vroh Bi I., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Yu J., Holland J. B., McMullen M. D., Buckler E. S., 2008. Genetic Design and Statistical Power of Nested Association Mapping in Maize. Genetics 178: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S. J., Gyenis L., Hayes P. M., Matus I., Smith K. P., et al. , 2005. Quantitative Trait Loci for Multiple Disease Resistance in Wild Barley. Crop Sci. 45: 2563–2572. [Google Scholar]
- Yun S. J., Gyenis L., Bossolini E., Hayes P. M., Matus I., et al. , 2006. Validation of Quantitative Trait Loci for Multiple Disease Resistance in Barley Using Advanced Backcross Lines Developed with a Wild Barley. Crop Sci. 46: 1179–1186. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the results of this article are available in the Tritceae Toolbox repository, https://triticeaetoolbox.org/barley/. To access data, enter WBIP into the left-hand ‘Quick search…’ box. Clicking on ‘Trial’ will bring up links to phenotypic and genotypic data experiments. To locate exome capture variant calls for parents and imputed variant scores for AB-NAM lines, follow instructions in ‘Comments’ section of ‘WBIP384_ABNAM_2012’ genotyping experiment.
File S1 is an Excel file containing all significant exome capture sequence variant marker-trait associations for the traits: grain protein content, glossy spike, glossy sheath, and black hull.
Exome capture sequences from 24 wild barley parents and Rasmusson have been submitted to the NCBI Sequence Read Archive (PRJNA305889 and PRJNA305578).