Abstract
Previous studies have shown that infection of Prochlorococcus MED4 by the cyanophage P-SSP7 leads to increased transcript levels of host endoribonuclease (RNase) E. However, it has remained enigmatic whether this is part of a host defense mechanism to degrade phage messenger RNA (mRNA) or whether this single-strand RNA-specific RNase is utilized by the phage. Here we describe a hitherto unknown means through which this cyanophage increases expression of RNase E during phage infection and concomitantly protects its own RNA from degradation. We identified two functionally different RNase E mRNA variants, one of which is significantly induced during phage infection. This transcript lacks the 5′ UTR, is considerably more stable than the other transcript, and is likely responsible for increased RNase E protein levels during infection. Furthermore, selective enrichment and in vivo analysis of double-stranded RNA (dsRNA) during infection revealed that phage antisense RNAs (asRNAs) sequester complementary mRNAs to form dsRNAs, such that the phage protein-coding transcriptome is nearly completely covered by asRNAs. In contrast, the host protein-coding transcriptome is only partially covered by asRNAs. These data suggest that P-SSP7 orchestrates degradation of host RNA by increasing RNase E expression while masking its own transcriptome from RNase E degradation in dsRNA complexes. We propose that this combination of strategies contributes significantly to phage progeny production.
Keywords: antisense RNA, T7-like cyanophage P-SSP7, dsRNA fishing, RNase E, Prochlorococcus
A common theme of the lifestyle of viruses is the manipulation of host metabolism for the purpose of viral reproduction. However, different virus types have established different regulatory pathways for the takeover and manipulation of their particular hosts. For example, all genes of T4 and the early genes of T7 Escherichia coli bacteriophages are transcribed by the host RNA polymerase (RNAP) (Ueno and Yonesaki 2004; Molineux 2006). However, expression of middle and late T4 genes is mediated by host RNAP that has been modified by T4-encoded factors (Ueno and Yonesaki 2004). In contrast to T4, the subsequent T7 expression program is coordinated by the activity of a phage RNAP (Maniloff and Ackermann 1998). Nevertheless, direct manipulation of host proteins plays a crucial role for the gene expression program of T7.
The gene product of the early T7 0.7 gene codes for a protein kinase (Rahmsdorf et al. 1974) and is responsible for phosphorylation and, thus, functional manipulation of >90 host proteins (Robertson et al. 1994). For example, phosphorylation of host RNAP β′-subunit (Zillig et al. 1975) results in reduced processivity, which is believed to assist in the coordinated transition of expression from T7 cluster 1 to cluster 2 genes (Severinova and Severinov 2006). In addition, phosphorylation of translation factors such as IF1, IF2, and IF3 is required for expression of T7 cluster 3 genes (Robertson and Nicholson 1992).
Another protein of particular interest that is regulated by phages is the host endoribonuclease (RNase) E protein. Ueno and Yonesaki (2004) describe RNase E-dependent changes in the stability of host and phage messenger RNAs (mRNAs) upon infection of E. coli by phage T4. They found that the half-lives of host mRNAs were markedly reduced, whereas phage mRNA was stabilized (Ueno and Yonesaki 2004). However, the mechanism for the differential regulation of the stability of host and T4 phage mRNA is not yet known. Stability of phage mRNA is also of crucial importance in T7 as the T7 RNAP outpaces elongating ribosomes, such that RNase E recognition sites are exposed on nascent phage mRNAs, rendering phage mRNA vulnerable to RNase E-mediated cleavage (Makarova et al. 1995). However, the destabilizing effect on T7 mRNAs is neutralized by the 0.7 protein; phosphorylation of the C-terminal domain of E. coli RNase E attenuates RNase E activity on T7 mRNA (Marchand et al. 2001). The RNase E/G family from cyanobacteria lack the C-terminal domain (Kaberdin et al. 1998; Baginsky et al. 2001; Rott et al. 2003; Bollenbach et al. 2004; Stazic et al. 2011) excluding this type of regulatory modification in cyanobacterial host–phage systems.
Previous work implicated a potential role for mRNA:antisense RNA (asRNA) duplexes in protection of a number of host mRNAs from host RNase E degradation (Stazic et al. 2011) during infection of the cyanobacterium Prochlorococcus MED4 by a T7-like cyanophage, P-SSP7. Here, we investigated the functional relationship between host RNase E and phage asRNAs during MED4 infection by P-SSP7. We found extensive coverage of the phage protein-coding transcriptome (hereafter referred to as the transcriptome) with complementary asRNA, whereas only partial asRNA coverage was found for host mRNAs. We further describe a novel regulatory means responsible for increased cellular levels of the RNase E protein. The functional consequence of phage double-stranded RNAs (dsRNAs) is the stabilization of phage mRNAs at a global scale while shifting the elevated RNase E activity toward degradation of the host transcriptome.
Materials and Methods
Growth conditions of Prochlorococcus MED4 and infection with P-SSP7
Cells were grown in Pro99 medium as described (Stazic et al. 2011) and at 21° under a cycle of 14 hr of light (12–15 µmol quanta m−2/s−1) and 10 hr of dark. Prochlorococcus MED4 cultures (1600 ml) at 108 cells/ml−1 (in midlog stage of exponential growth) were infected with the podovirus P-SSP7 (108 phage/ml−1).
RNA extraction, Northern blot analysis, and probe radiolabeling
Cells were harvested by filtration onto Supor-450 membranes (Pall) or by centrifugation at 10,000 × g for 10 min at room temperature. Total RNA extractions were carried out as described (Voigt et al. 2014). Briefly, the aqueous phase was extracted twice, once with the PGTX buffer and chloroform and once with chloroform. RNA was recovered either by precipitation with 1/10 volume 3 M sodium acetate, three volumes 100% ethanol, and 0.2 µl glycogen at −20° overnight followed by centrifugation at 13,000 × g and resuspension in RNase-free H2O or by column purification using the RNA Clean and Concentrator Kit (Zymo Research). Synthesis of probes and Northern hybridization experiments were performed as described (Stazic et al. 2011), with 1–2 µg and 3 µg of total RNA used for probing of P-SSP7 asRNAs and MED4 rne, respectively. Oligonucleotides used for synthesis of templates are listed in Supplemental Material, Table S1.
dsRNA purification
Duplicate P-SSP7-infected Prochlorococcus MED4 cultures (250 ml) were harvested 4 hr after inoculation with phage by centrifugation at 9000 × g for 8 min. Pellets were resuspended in AMP1 medium (Moore et al. 2007) supplemented with 200 mM sucrose and snap frozen in liquid nitrogen. After thawing on ice, cells were lysed with the Precellys homogenizer (PEQLAB Biotechnologie) at 6500 × g and 4° by applying three cycles each of 15-sec homogenization and 1-min incubation on ice. Cell debris was removed by centrifugation at 13,000 × g at 4° for 30 min. The supernatant was supplemented with 1.7 volumes 1× PBS, pH 7.0, and 1× complete protease inhibitor (Roche) and the final pH was adjusted to pH 7.0. The RNA-containing cell lysate was supplemented with 40 µg of the monoclonal antibody J2 that specifically interacts with dsRNA and the reaction mixture was incubated for 30 min at room temperature with gentle rotation. The J2 antibody complexed with dsRNA was selectively immobilized on prepacked protein A or G sepharose columns (GE Healthcare) by gentle rotation for 30 min at room temperature. In order to reduce nonspecific interaction with the column matrix, the column was blocked twice with 600 µl ribonuclease-free 1% BSA (Life Technologies) at room temperature for 4 min and washed with 600 µl protein A/G sepharose column 1× equilibration buffer (GE Healthcare). J2–RNA complexes bound by Protein A or G were eluted with 4 × 100 µl elution buffer (0.1 M glycine, pH 2.7) in a microcentrifuge tube containing 30 µl 1 M Tris-HCl, pH 9.0. Subsequently, dsRNA isolated with the J2 antibody was extracted using the PGTX method described above and was concentrated using the RNA Clean and Concentrator-5 Kit (Zymo Research). Further information about the selective purification of dsRNA is provided in File S1.
Complementary DNA library construction for RNA sequencing and data analysis
Residual DNA was removed from RNA samples using TURBO DNase (Life Technologies). In total, 4–6 units of DNase were added to the RNA samples and digestion of DNA was carried out in two or three consecutive incubation steps, each at 37° for 10 min. Reaction mixtures were purified using the RNA Clean and Concentrator-5 Kit according to standard manufacturer’s instructions (Zymo Research). RNA samples for total RNA libraries were depleted of ribosomal RNA using the MICROBExpress Kit (Life Technologies). Next, RNA samples were subjected to chemical fragmentation in 25 mM KOAc, 7.5 mM MgOAc, 50 mM Tris-HCl, pH 8.1 at 94° for 2.5 min or 8 min. The reactions were quenched by addition of one volume of 1× TE-buffer (10 mM Tris, 1 mM EDTA, pH 8.0) and briefly chilled on ice for 5 min. Purification of fragmented RNA was carried out according to the manufacturer’s alternative protocol, which selectively eliminates RNA species smaller than 100 nt (Zymo Research). All follow-up steps for total RNA complementary DNA (cDNA) library synthesis were performed according to a previously developed protocol (Sharma et al. 2010). Alternatively, construction of cDNA libraries was carried out by Vertis Biotechnologie. cDNA libraries were analyzed on an Illumina GAII sequencer and the results were deposited in National Center for Biotechnology Information’s Sequence Read Archive (SRA) under accession nos. SRR2079408, SRR2079426, SRR2079427, and SRR2079428. cDNA library LIB 7 was reamplified using Ion Torrent-specific adaptor primers and sequenced on an Ion Torrent Personal Genome Machine System. Sequenced reads were deposited in SRA under accession no. SRR3138052. After removing the 5′ hexamer indices, reads were mapped using segemehl (version 0.1.2, default parameters) and all libraries were normalized per number of base pairs (bp) sequenced. cDNA libraries used in this study are listed in Table S2.
RNase protection assay and 5′ RACE
Prior to employing RNase protection assays (RPAs) RNA samples were treated with terminator exonuclease (1 unit enzyme per 1 µg RNA, Epicentre), which selectively digests processed or degraded RNA fragments possessing a 5′ monophosphate or 5′ hydroxyl end, followed by column purification (RNA Clean and Concentrator-5 Kit; Zymo Research). Transcript probes were in vitro synthesized as described (Stazic et al. 2011). To eliminate truncated RNA originating from premature transcription abortion, probes were purified according to the small RNA elimination protocol of the RNA Clean and Concentrator-5 Kit. Using oligonucleotides RPA PMM1501 5′-UTR/CDS probe forward (fwd) and RPA PMM1501 5′-UTR/CDS probe reverse (rev) (Table S1), a 350-nt-long probe was synthesized that begins at nucleotide position 1,440,337 and extends to nucleotide position 1,440,700 in the MED4 genome so that it would hybridize to 363 nt of the full-length mRNA and 141 nt of the short rne mRNA variant. RPAs were carried out with the RPA III Kit (Invitrogen, Carlsbad, CA). Briefly, 2 fmol probes were hybridized to 4 µg TEX-treated RNA at 45° for 15 hr and cleavage was carried out with 1:50 RNase A/T1 mix for 30 min at 37°. The length of the protected fragments was determined by comparison with reference transcripts of known length. The transcriptional start site of MED4 rne short was determined by 5′ RACE as previously described (Steglich et al. 2008) using oligonucleotide PMM1501RT rev for reverse transcription and oligonucleotides PMM1501nestRT and HESWAdapt57 for amplification by PCR (Table S1).
Overexpression and purification of standard and N-terminally truncated MED4 recombinant RNase E
Expression and purification of a C-terminal hexahistidine fusion of recombinant standard RNase E was carried out as described (Stazic et al. 2011) using oligonucleotides PMM1501_C′K12 fwd and PMM1501_C′K12 rev (Table S1). A short variant of a C-terminal hexahistidine fusion of recombinant MED4 RNase E lacking the first 48 amino acids at the N terminus was amplified by PCR using oligonucleotides PMM1501 short −147 C′K12 fwd and PMM1501_C′K12 rev (Table S1). The resulting fragment was cloned into pQE70 (Qiagen, Valencia, CA) and the expression and purification was conducted as described (Stazic et al. 2011). All oligonucleotides were optimized for E. coli codon usage (Kazusa Codon Usage Database).
Western blot analysis
Total cellular protein was extracted from 50-ml cultures by centrifugation at 13,000 × g for 10 min at room temperature. Cell pellets were resuspended in protein lysis buffer (50 mM Tris, 100 mM NaCl, 1 mM DTT, pH 8.0) and snap frozen in liquid nitrogen. The homogenate was thawed on ice and diluted in 1/2 volume 12 M urea. Cell lysis was facilitated by two consecutive passages through a French press at 36,000 psi (Constant Systems). After removal of cell debris at 13,000 × g for 30 min at room temperature, 15 µg of total protein was electrophoretically separated on NuPAGE Bis-Tris 4–12% gradient polyacrylamide gels (Invitrogen) and subsequently transferred to Hybond ECL nitrocellulose membranes (GE Healthcare). Polyclonal antibodies raised in rabbits against MED4 RNase E and against Synechocystis PCC6803 AtpB (courtesy of Annegret Wilde) were used in 1:500 and 1:10,000 dilutions, respectively. Secondary antibodies conjugated with horseradish peroxidase were used in 1:10,000 dilutions. After addition of ECL solution, immunoreactive proteins were visualized and quantified using the ImageQuant LAS 4000 mini imager (GE Healthcare).
End-point in vitro RNase E cleavage assay
In vitro RNase cleavage assays were carried out according to described protocols (Stazic et al. 2011) with the following modifications. The end-point assay consists of ∼0.5 or ∼21 pmol of recombinant full-length MED4 RNase E or ∼0.5 pmol of short MED4 RNase E and 1 pmol in vitro transcribed RNA in a total volume of 15 µl. Cleavage fragments were separated on 7 M urea-6% polyacrylamide gels and visualized by ethidium bromide staining. RNA was in vitro transcribed (Stazic et al. 2011) and full-length RNA was purified by gel extraction (Georg et al. 2014). For generation of templates for in vitro synthesis of isiA 5′ UTR, intcA and isrR, Synechocystis PCC6803 genomic DNA was used. For PMM0684 RNA, MED4 rne long and MED4 rne short RNA, MED4 genomic DNA was used. The corresponding oligonucleotides are listed in Table S1.
Continuous in vitro RNase E cleavage assay
The in vitro RNase cleavage assay was carried out according to the described protocol (Stazic et al. 2011). Briefly, continuous assays were carried out in 50-µl reaction volumes using ∼1 or ∼35 pmol of recombinant standard RNase E and ∼1 pmol of short RNase E, together with a synthetic, fluorogenic RNA oligonucleotide. Relative protein concentrations were estimated by Western blot analysis (data not shown). Cleavage reactions were monitored in a Victor X3 multilabel plate reader (Perkin-Elmer, Norwalk, CT).
In vitro transcript half-life measurement
In a 5-µl reaction mixture, 1 pmol of in vitro-transcribed MED4 rne full-length or MED4 rne short RNA was supplemented with 7 pmol of recombinant standard MED4 RNase E (Stazic et al. 2011). After incubation at 30° for 1, 2, 5, 10, 15, 20, 30, and 60 min, cleavage reactions were quenched with 1 µl 0.5 M EDTA and 1 volume RNA loading buffer. A reaction mixture omitting RNase E was incubated for 60 min at 30° and served as a no RNase E control. Samples were heated to 95° for 3–5 min and separated on 7 M urea-6% polyacrylamide gels. Following gel staining with ethidium bromide, the relative amounts of MED4 rne full-length and MED4 rne short RNA were quantified densitometrically using Quantity One software (Bio-Rad, Hercules, CA). Half-lives were estimated by fitting an exponential decay function to all time points.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Expression of phage asRNAs and formation of phage dsRNAs
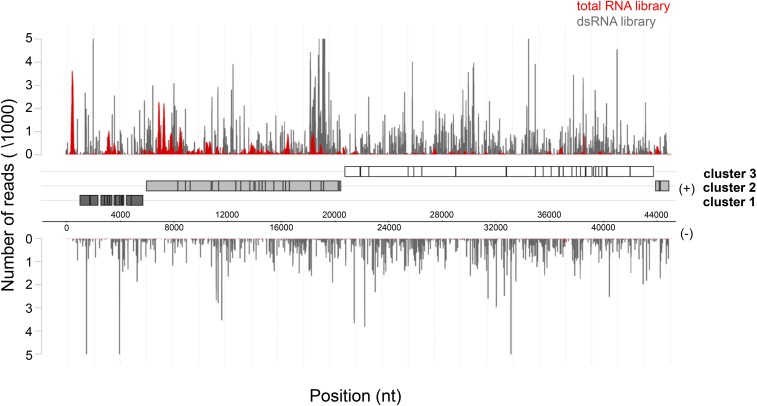
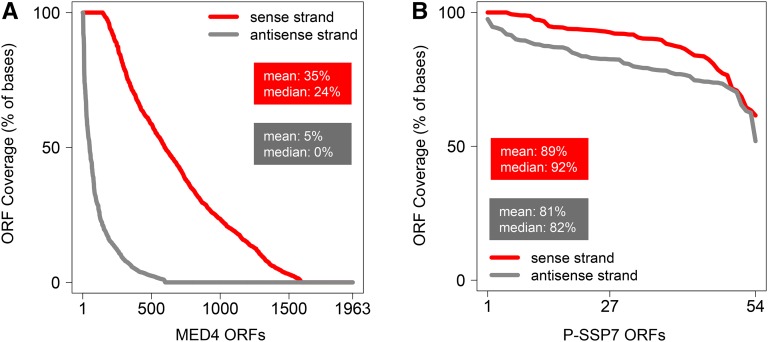
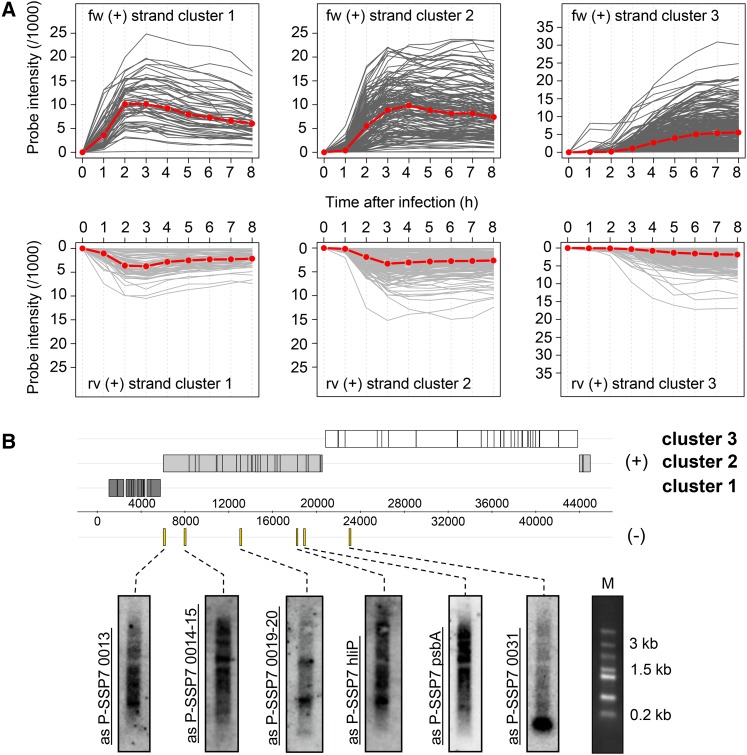
To assess the presence of asRNAs interacting with target mRNAs in the cell, we isolated dsRNA from 4-hr phage-infected MED4 cultures according to a modified protocol established by Lybecker et al. (2014) (Figure S1; see also File S1 and File S2). This time point is midway in the latent period of the P-SSP7 infection cycle (Lindell et al. 2007). Endogenous dsRNA was immunoprecipitated from cell lysates using the monoclonal anti-dsRNA J2 antibody followed by depletion of genomic DNA and ribosomal RNA. Resultant dsRNA was converted into cDNA preserving the information on transcript direction and was subjected to high-throughput sequencing (Table S2, LIB 1). Approximately 1.5-fold more sequence reads were obtained for the sense (553,019) than the antisense (353,387) strand of the P-SSP7 phage genome. Mapping of the reads displayed uniform genome-wide distribution for both strands (Figure 1). Furthermore, the number of bases covered on the sense and antisense strand within each ORF was extremely high, with a similar degree of coverage for both the sense (89%) and antisense (81%) strand (Figure 2). We observed a different expression profile for total RNA libraries, exhibiting enrichment of mRNA sequence reads (Figure 1; Figure S2; File S2). Similar results were obtained by Lybecker et al. (2014), who showed that asRNAs are likely to be depleted from total RNA-sequence (RNA-seq) libraries if the complementary mRNA is present in relative excess. We assume that this is attributable to the inherent structural bias in RNA-seq technology, i.e., the RNA-seq read frequency for a given RNA is constrained by its inter- and intramolecular secondary structure (van Dijk et al. 2014; Raabe et al. 2014). Therefore, we estimated the representation of phage transcripts by microarray analysis (Lindell et al. 2007), which indicated an average threefold excess of mRNA over asRNA (Figure 3A).
Figure 1.
In vivo phage dsRNA formation. Distribution of reads mapped to the sense (+) and antisense (−) strand of the P-SSP7 genome originating from total RNA (red plot) or enriched dsRNA (gray plot) after 4 hr of MED4 infection, respectively. Dark-gray, light-gray, and white boxes denote annotated genes located in expression clusters 1, 2, and 3, respectively. Genomic coordinates are given in bases on the x-axis. The number of reads is given on the y-axis.
Figure 2.
Coverage of bases in P-SSP7 and MED4 ORFs by mapped reads on the sense and antisense strand. Coverage of the sense and antisense strand for (A) phage P-SSP7 and (B) host MED4 plotted in descending order. The x-axis shows the number of ORFs. The y-axis denotes the number of bases covered by sense (red) and antisense (gray) reads, respectively, within each ORF at a single base resolution. The P-SSP7 genome has 54 ORFs and the MED4 genome has 1963 ORFs.
Figure 3.
Expression of phage genes and asRNAs during infection of MED4. (A) Temporal expression patterns for the coding and noncoding strand of each P-SSP7 ORF as determined from microarrays (Lindell et al. 2007). Microarrays were reanalyzed at single probe resolution and plotted for each expression cluster at indicated time points (x-axis). The red line specifies the median signal intensity. (B) Northern blot analysis showing expression of six P-SSP7 asRNAs 4 hr after infection. All but the 0031 gene had asRNAs >3 kb. Probed regions on the genome are marked with yellow bars. M denotes Fermentas high-range marker. The genome map is the same as in Figure 1.
Northern blot analysis for six phage ORFs confirmed the expression of asRNAs. Intriguingly, asP-SSP7 0014-15 and asP-SSP7 psbA each exhibited a distinct band of >3 kb in size and the upper band of asP-SSP7 0019-20 migrates >1.5 kb (Figure 3B). However, the smaller bands evident in the blot might result from processing or degradation of larger fragments.
In contrast to the phage genome, the dsRNA coverage of the MED4 host during infection was only partial, with 35% coverage on the sense strand and 5% coverage on the antisense strand (Figure 2). The unbalanced ratio of host sense and antisense reads likely resulted from MED4 asRNAs <100 nt (Voigt et al. 2014), which were depleted from the dsRNA libraries as part of the processing (Figure S3A; see also File S1 and File S2). Additionally, 5′ end- or 3′ end-overlapping mRNAs transcribed from neighboring genes are a source of dsRNA loci (Lybecker et al. 2014) during the immunoprecipitation procedure using the J2 antibody (Figure S3, B and C; see also File S1 and File S2).
Analysis of total RNA libraries from 2 and 4 hr after infection (Table S2, LIB 2 and LIB 7) yielded results congruent with those from the dsRNA library at 4 hr postinfection. Phage genes displayed an average coverage of 77 and 72%, 2 and 4 hr after infection, respectively (Figure S4; File S2). In comparison, coverage of host genes was lower, with an average of 58% 2 hr after infection (Figure S4; File S2) that dropped to 21% over the course of infection (Figure S4; File S2).
Together these data indicate that long phage asRNAs are expressed and form dsRNA with phage mRNAs that span the entire transcriptome, whereas dsRNAs only partially cover the host transcriptome. These findings suggest a pivotal role for asRNAs in mRNA stability during phage infection.
Expression of full-length and short RNase E mRNA variants
Transcript levels of MED4 RNase E increase upon infection by P-SSP7 (Lindell et al. 2007). This prompted us to investigate the mechanism regulating RNase E expression during phage infection. To this end, we performed differential RNA-seq (dRNA-seq) analysis according to Sharma et al. (2010) on RNA enriched in primary transcripts from infected MED4 cultures at 1, 2, and 4 hr after infection (Table S2, LIB 4) and on both primary and total transcripts from uninfected cells (Table S2, LIB 5).
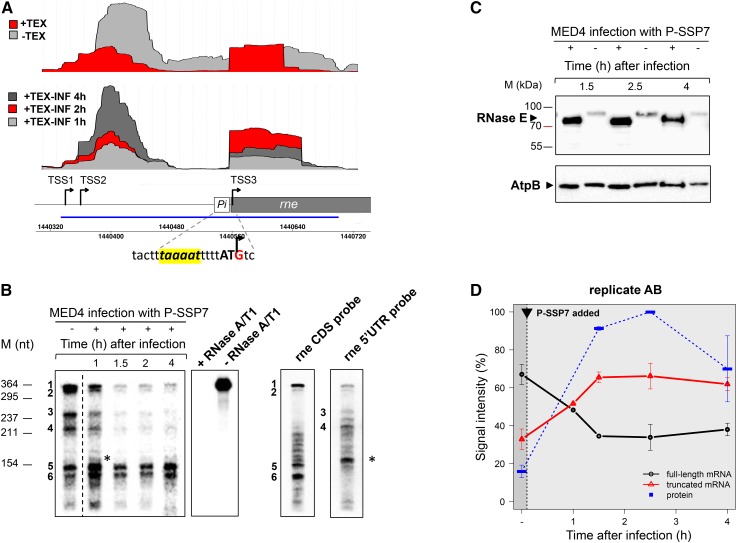
Three transcriptional start sites (TSSs) were identified for the RNase E-encoding gene, rne (ORF PMM1501 in the MED4 genome). TSS1 is located at position −220 relative to the ATG start codon (Figure 4A). Reads starting 20 nt downstream from TSS1 in primary transcript libraries indicate transcription initiation at another site, TSS2. This resembles the architecture of the RNase E gene in E. coli (Jain and Belasco 1995; Ow et al. 2002). In addition to the canonical TSS1/TSS2, we identified a third TSS internal to the rne gene, TSS3, not found previously for other rne genes. The internal site mapped to the G (+3) of the canonical ATG start codon and is preceded by a putative −10 element TAAAAT (Pi in Figure 4A). 5′ RACE experiments confirmed the presence of the internal TSS (Figure S5; File S2). Thus, expression of rne yields three variants of RNase E mRNAs. Two full-length mRNAs originate from TSS1/TSS2, and a short mRNA transcribed from the gene-internal TSS3, which lacks the 220-nt-long 5′ UTR present in the full-length mRNA variants (Figure 4A).
Figure 4.
Regulation of Prochlorococcus MED4 RNase E during the course of P-SSP7 infection. (A) cDNA reads from total RNA (−TEX) or RNA enriched in primary transcripts of noninfected (+TEX) or infected cells (+TEX-INF) mapped to the rne locus encoding RNase E under standard growth conditions (top) and 1, 2, and 4 hr after infection by PSSP7 (middle). The schematic drawing of the rne gene indicates the location of TSSs (black arrows). Pi corresponds to the promoter associated with TSS3. The sequence of the putative −10 element (yellow box) and the transcription initiator G (within the first ATG codon of the full-length ORF) are given. The blue line denotes the position of the probe used for RNase protection assays depicted in B. (B) RNase protection assay of RNA enriched in primary transcripts before (−) and after (+) P-SSP7 infection. Protected fragments 1, 2, 3, and 4 correspond to the full-length rne mRNA originating from TSS1/TSS2. Protected fragments 5 and 6 correspond to the short rne mRNA originating from TSS3. The probe is stable during the assay (−RNase A/T1) and is completely digested in the presence of RNase A/T1 (+RNase A/T1) when no complementary RNA is present. Northern hybridizations of total RNA from uninfected MED4 cells using the rne CDS probe that would detect both the full-length and short rne mRNA (rne CDS probe), and using an rne 5′ UTR probe that would only detect the full-length rne mRNA (rne 5′ UTR probe). The asterisk marks the smallest band detected with the rne 5′ UTR probe. (C) Western blot analysis of temporal expression of MED4 RNase E and AtpB in infected (+) and noninfected (−) cultures. The same membrane was used for the detection of either RNase E or AtpB. (D) Transcript and protein levels of the MED4 full-length and short rne mRNA and the RNase E protein during P-SSP7 infection. The black arrow indicates the time point of P-SSP7 addition. Protein and mRNA quantification was performed by densitometry of the bands from Western and RPA blots, respectively. Data shown are mean ± range of two biological replicates. For protein data (blue line) the band with the highest signal intensity (2.5 hr after infection) was arbitrarily set to 100% and other bands were quantified proportionally. The mean signal intensity of the RNase E protein from all time points in the noninfected control was used for the time point before infection (−). For transcript data, signal intensities of the total RPA fragments for each time point were set as 100% transcription activity and relative signal intensities corresponding to full-length (black line) and short (red line) rne mRNA fragments were quantified proportionally.
In order to further evaluate the rne transcripts, we conducted RPAs on RNA enriched for primary transcripts, i.e., mRNA produced by active transcription. An RPA probe that spans the first 143 nt of the rne-coding region as well as the 220-nt 5′ UTR region of the full-length mRNA was used. Six protected fragments were identified (Figure 4B). The upper four protected fragments are longer than the 143-nt region of the CDS and therefore likely derived from the full-length rne mRNA variants initiated from TSS1 or TSS2, while the lower two protected fragments are shorter than 143 nt and thus likely resulted from TSS3. This was confirmed from Northern blot analysis of the protected fragments using probes that hybridize either to the coding region or the 5′ UTR region of rne mRNA (Figure 4B). The shorter two protected fragments hybridized only to the CDS probe, while the longest two fragments hybridized to both the CDS and 5′ UTR probes. The short fragment that hybridized to the 5′ UTR probe (Figure 4B, black asterisk) migrates well above the lower two protected fragments from the RPA. Note that the middle two fragments hybridized only to the 5′ UTR probe, indicating that they are digestion fragments derived from the full-length mRNA.
Quantification of the protected fragments resulting from the two rne mRNA variants (full-length vs. short mRNA) demonstrated preferential transcription initiation at TSS1/TSS2 in noninfected MED4 cells (Figure 4, B and D), which is consistent with dRNA-seq data (Figure 4A). However, transcription from TSS1/TSS2 decreased significantly with the onset of phage infection. In comparison, no selective downregulation in transcription from TSS3 was observed. Consequently, this results in a switch in promoter usage with preferential rne transcription initiation at TSS3 and a short rne mRNA dominating the rne transcript pool from 1.5 hr after phage infection onwards (Figure 4, A, B, and D).
Increased stability of the short RNase E mRNA
RNA stability is well known to be affected by structural and sequence-based elements, such as 5′ UTR stem-loop domains (Rauhut and Klug 1999; Rochat et al. 2013). The 5′ UTR of E. coli rne is involved in mRNA destabilization in an RNase E-dependent fashion (Jain and Belasco 1995). This raises the question as to whether the full-length and short MED4 RNase E messages are subject to accelerated and attenuated decay rates, respectively, in the presence of the RNase E enzyme. To test this hypothesis, we conducted in vitro cleavage assays using recombinant MED4 RNase E (Stazic et al. 2011) and rne transcripts containing or lacking the 5′ UTR region. Rapid decay was observed for the 5′ UTR-containing transcript with a half-life of 1.5 min, as opposed to a highly attenuated half-life of 20.7 min for the short variant lacking the 5′ UTR segment (Figure S6; File S2). These findings are consistent with Northern blot analysis, displaying the strongest signals of rne cleavage fragments in RNA from noninfected cells (Figure S7; File S2). Indeed an average reduction in rne cleavage by 66% was found for MED4 cells infected by P-SSP7 relative to the noninfected MED4 control cells (Figure S8; see also File S1 and File S2). These data, together with those showing preferential transcription from TSS3, indicate that a short, stable rne transcript accumulates during infection while the full-length variant declines over the course of phage infection.
Induction of a short protein isoform of RNase E during phage infection
The shorter mRNA variant of rne resulting from TSS3 is expected to yield a smaller RNase E isoform than that resulting from TSS1/TSS2, since the former transcript begins within the ATG start codon of the full-length protein (Figure 4A). Western blot data confirmed that indeed a 5–10 kDa smaller RNase E isoform was produced during infection, whereas the AtpB host protein did not show such a variation in migration (Figure 4C), indicating that the smaller RNase E isoform was not due to migration artifacts. A number of potential alternative start codons downstream may be used to produce this shorter isoform. These range from +147 to +291 relative to the +1 of the ATG of the full-length protein: +147 AUU; +162 AUA; +168 AUU; +195 AUU; +246 AUU; +282 GUG; and +291 ATG.
Since gene expression does not necessarily correlate well at mRNA and protein levels in Prochlorococcus (Waldbauer et al. 2012), we assessed host RNase E protein levels during infection. Western blot analysis revealed that the short RNase E isoform was induced six- to sevenfold from 1.5 hr after phage infection onwards relative to the full-length protein in uninfected cells (Figure 4C). This corresponds with the increase in relative amounts of the short rne mRNA from 1 hr in the P-SSP7 lytic cycle onwards and is due to the switch in transcription from TSS1/TSS2 to TSS3 (Figure 4D). However we cannot rule out the possibility that enhanced expression of RNase E is also due to an increased translation rate of the short mRNA.
To test whether the shorter isoform of the protein has altered catalytic properties, we produced an amino-terminal deletion version of the recombinant RNase E from MED4 lacking the first 48 amino acids (corresponding to the +147 AUU potential start codon). A comparison of enzyme cleavage properties revealed that the N-terminally shortened form maintains specificity of cleavage sites for some substrates (for example, the isiA 5′ UTR substrate), whereas differences in cleavage site selectivity were observed for other sites (for example, rne full-length, rne short, PMM0684, iNtcA sRNA, and IsrR sRNA); Figure S9A; File S2). Furthermore, cleavage activity of the short isoform was greater for a fluorescently labeled synthetic RNA, containing a canonical RNase E cleavage site (Stazic et al. 2011), which we used to monitor RNase E cleavage kinetics in real time (Figure S9B; File S2). These findings indicate that the short RNase E isoform retains intrinsic ribonuclease activity.
Discussion
In this study, we report for the first time transcriptome-wide asRNA expression and dsRNA formation for a phage during lytic infection. Based on this and our previous finding that long dsRNA duplexes protect selected host mRNAs from RNase E activity (Stazic et al. 2011), we propose that phage dsRNAs prevent RNase E degradation of the phage transcriptome. We further propose that this “phage dsRNA block,” together with the increase in RNase E protein levels expressed from an alternative promoter, serves to direct RNase E-mediated ribolysis toward the host transcriptome during phage infection.
Our data reveal the molecular basis for a novel regulation mechanism of RNase E during phage infection of Prochlorococcus MED4 for which we propose the model depicted in Figure 5. In E. coli and related bacteria, production of RNase E is regulated in cis by the rne 5′ UTR, which senses the cellular level of this enzyme and controls the decay rate of the rne message in response to changes in RNase E activity (Jain and Belasco 1995; Diwa et al. 2000; Schuck et al. 2009). Results from Northern blot analysis and in vitro half-life studies clearly support an E. coli-like autoregulation mechanism in the MED4 RNase E system in noninfected cells. In contrast, phage infection changes rne promoter usage, leading to enhanced transcriptional initiation of a short mRNA variant that lacks the cis-acting 5′ UTR regulator. We hypothesize that this differential promoter usage disables the negative feedback regulation on rne mRNA degradation during phage infection. This results in synthesis of a more stable RNase E message and, as a consequence, in higher protein yields. At present, it remains unclear whether this switch in promoter usage is actively regulated by a phage factor or if it is an indirect effect of the lack of a host factor due to the shutdown of host gene expression during infection. Based on the observation that the short mRNA variant is expressed at a basal level in noninfected cells, we suggest that synthesis of the short transcript is driven by the host RNAP.
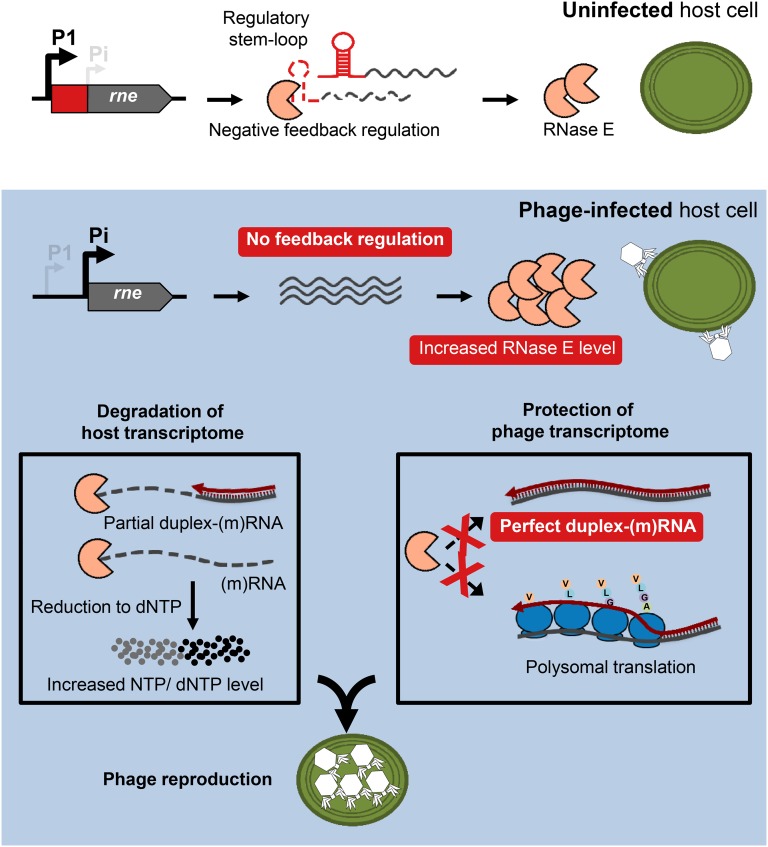
Figure 5.
Model for the P-SSP7 infection of MED4. In uninfected host cells, the level of RNase E is regulated via a negative feedback mechanism, in which a regulatory stem loop in the rne 5′ UTR provides an entry site for RNase E-mediated cleavage of the mRNA. During phage infection, rne transcription proceeds from an alternative transcription initiation site, resulting in a short mRNA variant that lacks the regulatory 5′ UTR. This leads to inactivation of the negative feedback regulation and concomitant increase in RNase E protein level. Genome-wide transcription of phage asRNAs drives the formation of extensive duplex RNA complexes that along with polysomal translation protect from RNase E activity, while host transcripts are only partially covered with asRNAs, thus rendering the transcriptome vulnerable to RNase E-mediated cleavage. Consequently, the increased pool of host-derived nucleic acids is likely to stimulate phage reproduction.
In other host–phage systems, such as E. coli and T7, RNase E substrate specificity is regulated by a phage-encoded protein kinase, which phosphorylates RNase E in the C-terminal domain (Marchand et al. 2001). Similarly, a T4 phage-encoded factor is responsible for the regulation of host RNase E substrate specificity during infection of E. coli (Ueno and Yonesaki 2004). We propose that in the MED4–P-SSP7 system differential promoter usage constitutes an alternative mode of gene expression regulation of RNase E and that the indirect regulation of substrate specificity through phage asRNAs compensates for the lack of the C-terminal domain in cyanobacterial RNase E enzymes.
Previously we reported that long, perfect duplex RNAs can protect host mRNAs from RNase E degradation in vitro (Stazic et al. 2011). We hypothesized that this dsRNA formation provides a means through which transcript levels of a selected number of host genes increased during infection relative to the dramatic decline in the rest of the host transcriptome (Lindell et al. 2007). Here we show that duplex mRNA formation is a general strategy employed by the P-SSP7 phage that is likely to provide transcriptome-wide protection from host RNase E activity during infection. However, it should be noted that a threefold excess of phage mRNA over phage asRNAs was found. This suggests that sequestration of a subset of the total mRNA body in dsRNA complexes provides sufficient protection from RNase E activity. It is possible that stabilization of the remaining mRNA pool is afforded by active translation due to an intimate synchronization of transcription and translation in bacteria (Miller et al. 1970; Grunberg-Manago 1999). Alternatively, this portion of the P-SSP7 transcriptome may be prone to RNase E degradation. If the latter is the case, this degradation does not prevent production of phage progeny. At this stage, we can only speculate about phage duplex RNA being vulnerable to turnover by the host dsRNA-specific RNase III. However, downregulation of the RNase III gene during P-SSP7 infection (Lindell et al. 2007) may well render the protein unavailable. Intriguingly, dRNA-seq data show a reciprocal expression pattern of RNase E and the DEAD-box type RNA helicase PMM1101 with the RNA helicase being upregulated 1 hr and 4 hr after infection (Figure S10 and File S2; compare with Figure 4A). Proteins of the DEAD-box family exert their function by rearranging RNA structures and unwinding long dsRNAs (Tanner and Linder 2001). As such they are associated with the translation process (López-Ramírez et al. 2011). We suggest that the RNA helicase could assist in translation of phage mRNAs embedded in dsRNA complexes.
The initial finding of increased rne transcript levels during infection on the backdrop of the decline of the vast majority of the host transcriptome, raised the question as to whether this constitutes a host defense strategy for degrading phage RNA or reflects exploitation by the phage of host RNase E to degrade host mRNA and thus supply nucleic acids for phage reproduction (Lindell et al. 2007). The findings reported here strongly suggest that the second hypothesis is the more likely scenario. We propose several means through which selective destabilization of the host transcriptome by RNase E aids phage reproduction: it would allow for a global switch in gene expression from the host to the phage genome through the removal of competing host mRNAs; and RNase E-triggered ribolysis, which ultimately leads to degradation of RNA to ribonucleotides (NTPs) (Stazic and Voss 2015), could stimulate efficient transcription of phage genes. This is consistent with the observation that transcription of T4 genes is impaired if infection takes place in an E. coli rne mutant (Otsuka et al. 2003). It should be noted that cleavage-mediated recycling of NTPs would require one or more exoribonucleases to work in concert with RNase E, as the latter is not capable of degrading RNA to mononucleotides (Callaghan et al. 2005; Mackie 2013). The dRNA-seq data for transcript levels of RNase J and RNase II suggest upregulation of both exoribonucleases over the course of phage infection (Figure S10; File S2), both of which have the intrinsic capacity to degrade RNA and release mononucleotides (Mackie 2013; Liu et al. 2014). However, the suggestion that either of the two exo-type RNases impact differential RNA stability during phage infection requires further investigation.
RNase E activity is also likely to stimulate the production of dNTPs and hence enables high efficiency of phage genome replication. The latter is consistent with the presence of a small number of auxiliary metabolic genes in P-SSP7 that include psbA (encoding D1, the photosystem II reaction center protein), nrd (the ribonucleotide reductase gene), and talC (encoding transaldolase) (Sullivan et al. 2005). These genes are expressed in concert and are thought to provide the capacity to reduce NTPs to dNTPs (Lindell et al. 2007; Thompson et al. 2011). Taken together, these three activities would augment the supply of nucleic acid resources available for phage reproduction, which may markedly expedite the lytic program of P-SSP7 or enable the production of more phage progeny.
Our findings for the cyanophage P-SSP7 (this study), combined with previous findings for the enteric phages T2, T4, and T7 (Marchand et al. 2001; Ueno and Yonesaki 2004), suggest that infection-induced RNase E-dependent turnover of host mRNA is a widespread phenomenon among bacteriophages. However, the P-SSP7 cyanophage has evolved a unique combination of means through which this is achieved: a transcriptional switch in promoter usage, providing a post-transcriptional increase in transcript stability and increased protein levels of RNase E, together with the protection of its mRNA pool through global dsRNA formation.
Acknowledgments
We thank René Schroeder for the idea of using the J2 antibody and Wolfgang Hess for critical reading of the manuscript. This work was supported by a Minerva short-term research grant to D.S., Deutsche Forschungsgemeinschaft grant STE 1119/4-1, the University of Freiburg’s Research Innovation Fund, a Ministry of Science, Research, and the Arts, Baden-Wuerttemberg Research Seed Capital grant to C.S., and the European Research Council starting grant 203406 to D.L.
Footnotes
Communicating editor: A. Hochschild
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.183475/-/DC1.
Literature Cited
- Baginsky S., Shteiman-Kotler A., Liveanu V., Yehudai-Resheff S., Bellaoui M., et al. , 2001. Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA 7: 1464–1475. [PMC free article] [PubMed] [Google Scholar]
- Bollenbach T. J., Schuster G., Stern D. B., 2004. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 78: 305–337. [DOI] [PubMed] [Google Scholar]
- Callaghan A. J., Marcaida M. J., Stead J. A., Mcdowall K. J., Scott W. G., et al. , 2005. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437: 1187–1191. [DOI] [PubMed] [Google Scholar]
- Diwa A., Bricker A. L., Jain C., Belasco J. G., 2000. An evolutionarily conserved RNA stem-loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes Dev. 14: 1249–1260. [PMC free article] [PubMed] [Google Scholar]
- Georg J., Dienst D., Schürgers N., Wallner T., Kopp D., et al. , 2014. The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria. Plant Cell 26: 3661–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg-Manago M, 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33: 193–227. [DOI] [PubMed] [Google Scholar]
- Jain C., Belasco J. G., 1995. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes Dev. 9: 84–96. [DOI] [PubMed] [Google Scholar]
- Kaberdin V. R., Miczak A., Jakobsen J. S., Lin-Chao S., McDowall K. J., et al. , 1998. The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl. Acad. Sci. USA 95: 11637–11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D., Jaffe J. D., Coleman M. L., Futschik M. E., Axmann I. M., et al. , 2007. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449: 83–86. [DOI] [PubMed] [Google Scholar]
- Liu B., Deikus G., Bree A., Durand S., Kearns D. B., et al. , 2014. Global analysis of mRNA decay intermediates in Bacillus subtilis wild-type and polynucleotide phosphorylase-deletion strains. Mol. Microbiol. 94: 41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ramírez V., Alcaraz L. D., Moreno-Hagelsieb G., Olmedo-Álvarez G., 2011. Phylogenetic distribution and evolutionary history of bacterial DEAD-Box proteins. J. Mol. Evol. 72: 413–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker M., Zimmermann B., Bilusic I., Tukhtubaeva N., Schroeder R., 2014. The double-stranded transcriptome of Escherichia coli. Proc. Natl. Acad. Sci. USA 111: 3134–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A, 2013. RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 11: 45–57. [DOI] [PubMed] [Google Scholar]
- Makarova O. V., Makarov E. M., Sousa R., Dreyfus M., 1995. Transcribing of Escherichia coli genes with mutant T7 RNA polymerases: stability of lacZ mRNA inversely correlates with polymerase speed. Proc. Natl. Acad. Sci. USA 92: 12250–12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniloff J., Ackermann H. W., 1998. Taxonomy of bacterial viruses: establishment of tailed virus genera and the other Caudovirales. Arch Virol. 143: 2051–2063. [DOI] [PubMed] [Google Scholar]
- Marchand I., Nicholson A. W., Dreyfus M., 2001. Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol. Microbiol. 42: 767–776. [DOI] [PubMed] [Google Scholar]
- Miller O. L., Hamkalo B. A., Thomas C. A., 1970. Visualization of Bacterial Genes in Action. Science 169: 392–395. [DOI] [PubMed] [Google Scholar]
- Molineux I, 2006. The T7 Group, pp 277–301 in The bacteriophages, edited by Calendar R. Oxford University Press, New York. [Google Scholar]
- Moore L. R., Coe A., Zinser E. R., Saito M. A., Sullivan M. B., et al. , 2007. Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr Methods 5: 353–362. [Google Scholar]
- Otsuka Y., Ueno H., Yonesaki T., 2003. Escherichia coli Endoribonucleases Involved in Cleavage of Bacteriophage T4 mRNAs. J. Bacteriol. 185: 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M. C., Liu Q., Mohanty B. K., Andrew M. E., Maples V. F., et al. , 2002. RNase E levels in Escherichia coli are controlled by a complex regulatory system that involves transcription of the rne gene from three promoters. Mol. Microbiol. 43: 159–171. [DOI] [PubMed] [Google Scholar]
- Raabe C. A., Tang T. H., Brosius J., Rozhdestvensky T. S., 2014. Biases in small RNA deep sequencing data. Nucleic Acids Res. 42: 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmsdorf H. J., Pai S. H., Ponta H., Herrlich P., Roskoski R., et al. , 1974. Protein kinase induction in Escherichia coli by bacteriophage T7. Proc. Natl. Acad. Sci. USA 71: 586–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut R., Klug G., 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23: 353–370. [DOI] [PubMed] [Google Scholar]
- Robertson E. S., Nicholson A. W., 1992. Phosphorylation of Escherichia coli translation initiation factors by the bacteriophage T7 protein kinase. Biochem. 31: 4822–4827. [DOI] [PubMed] [Google Scholar]
- Robertson E. S., Aggison L. A., Nicholson A. W., 1994. Phosphorylation of elongation factor G and ribosomal protein S6 in bacteriophage T7-infected Escherichia coli. Mol. Microbiol. 11: 1045–1057. [DOI] [PubMed] [Google Scholar]
- Rochat T., Bouloc P., Repoila F., 2013. Gene expression control by selective RNA processing and stabilization in bacteria - Rochat - 2013 - FEMS Microbiology Letters - Wiley Online Library. FEMS Microbiol. Lett. [DOI] [PubMed] [Google Scholar]
- Rott R., Zipor G., Portnoy V., Liveanu V., Schuster G., 2003. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. J. Biol. Chem. 278: 15771–15777. [DOI] [PubMed] [Google Scholar]
- Schuck A., Diwa A., Belasco J. G., 2009. RNase E autoregulates its synthesis in Escherichia coli by binding directly to a stem-loop in the rne 5' untranslated region. Mol. Microbiol. 72: 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinova E., Severinov K., 2006. Localization of the Escherichia coli RNA Polymerase ' Subunit Residue Phosphorylated by Bacteriophage T7 Kinase Gp0.7. J. Bacteriol. 188: 3470–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C. M., Hoffmann S., Darfeuille F., Reignier J., Findeiß S., et al. , 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250–255. [DOI] [PubMed] [Google Scholar]
- Stazic D., Voss B., 2015. The complexity of bacterial transcriptomes. J. Biotechnol., DOI:10.1016/j.jbiotec.2015.09.041. [DOI] [PubMed] [Google Scholar]
- Stazic D., Lindell D., Steglich C., 2011. Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Research 39: 4890–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich C., Futschik M. E., Lindell D., Voss B., Chisholm S. W., Hess W. R., et al. , 2008. The Challenge of Regulation in a Minimal Photoautotroph: Non-Coding RNAs in Prochlorococcus. PLoS Genet. 4: e1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. B., Coleman M. L., Weigele P., Rohwer F., Chisholm S. W., 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N. K., Linder P., 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8: 251–262. [DOI] [PubMed] [Google Scholar]
- Thompson L. R., Zeng Q., Kelly L., Huang K. H., Singer A. U., et al. , 2011. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA 108: E757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H., Yonesaki T., 2004. Phage-induced change in the stability of mRNAs. Virology 329: 134–141. [DOI] [PubMed] [Google Scholar]
- van Dijk E. L., Jaszczyszyn Y., Thermes C., 2014. Library preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 322: 12–20. [DOI] [PubMed] [Google Scholar]
- Voigt K., Sharma C. M., Mitschke J., Lambrecht S. J., Voss B., et al. , 2014. Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J. 8(10): 8: 2056–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldbauer J. R., Rodrigue S., Coleman M. L., Chisholm S. W., 2012. Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS ONE 7: e43432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W., Fujiki H., Blum W., Janeković D., Schweiger M., et al. , 1975. In vivo and in vitro phosphorylation of DNA-dependent RNA polymerase of Escherichia coli by bacteriophage-T7-induced protein kinase. Pro Nat Aca Sc USA 72: 2506–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.