Abstract
It has been hypothesized that Alzheimer disease (AD) is primarily a disorder of the synapse. However, assessment of the synaptic proteome in AD subjects has been limited to a small number of proteins and often included subjects with end-stage pathology. Protein from prefrontal cortex gray matter of 59 AD subjects with mild to moderate dementia and 12 normal elderly subjects was assayed using targeted mass spectrometry to quantify 191 synaptically expressed proteins. The profile of synaptic protein expression clustered AD subjects into two groups. One of these was characterized by reduced expression of glutamate receptor proteins, significantly increased synaptic protein network coexpression, and associated withApolipoprotein E*4 (APOE*4) carrier status. The second group, by contrast, showed few differences from control subjects. A subset of AD subjects had altered prefrontal cortex synaptic proteostasis for glutamate receptors and their signaling partners. Efforts to therapeutically target glutamate receptors in AD may have outcomes dependent on APOE*4 genotype.
An important hypothesis in Alzheimer disease (AD)1 posits that it is a disorder primarily of the synapse (1, 2). This hypothesis derives, in part, from evidence that the strongest correlate of cognitive impairment in AD is loss of synapses (3, 4), with excitatory synapses onto dendritic spines particularly affected (5–7). Conversely, in cognitively normal individuals who nevertheless have substantial AD pathology, neuron number and synaptic markers are largely preserved (8).
Direct studies of synaptic pathology in AD subjects have relied on evaluations of synaptic ultrastructure to yield estimates of synapse volume density in affected regions (9) or used antibody labeling of proteins with synaptic expression (4, 10, 11). Use of synapse ultrastructure provides an excellent estimation of synaptic density but does not provide information regarding underlying molecular pathology and is labor intensive, precluding studies of large numbers of subjects. In contrast, antibody-based approaches provide information of the abundance of the labeled proteins and when assessed via confocal microscopy can additionally provide estimates of synapse density and number (12). However, these approaches are typically limited to the assessment of only a limited number of proteins. Moreover, as a group, studies of synaptic pathology in AD have tended to include preclinical or severe AD stages, with only limited numbers of subjects in mild to moderate stages of AD studied to date.
Recently, proteomics approaches using liquid chromatography–tandem mass spectrometry (LC-MS/MS) have been utilized to interrogate the synaptic proteome in a number of model systems, highlighting the extent and complexity of the synaptic protein network (13, 14). We have recently validated an approach to LC-MS/MS, using selective reaction monitoring (LC-SRM/MS) in combination with a stable isotope labeled mammalian brain standard, to quantify more than 100 synaptically expressed proteins in postmortem human brain tissue (15) and successfully applied this approach to elucidate synaptic pathology in another neuropsychiatric illness characterized by cognitive impairment and synapse loss, schizophrenia (16). We therefore undertook in the current study to use this approach to interrogate the expression of synaptic proteins in a cohort of AD subjects in predominantly mild to moderate clinical and pathologic disease stages. Results were contrasted with elderly cognitively and neuropathologically normal subjects and with a comparison cohort with frontotemporal lobar degeneration (FTLD).
MATERIALS AND METHODS
(Please also see Supplementary Methods for additional details of subject characterization, sample preparation, assay, and statistical analysis.)
Subjects
AD Subjects and FTLD Subjects
Fifty-nine AD subjects and 10 FTLD subjects (Table I) were identified through the brain bank of the Alzheimer Disease Research Center (ADRC) at the University of Pittsburgh, using protocols approved by the University of Pittsburgh Institutional Review Board and Committee for Oversight of Research Involving the Dead. Individuals underwent neurologic, neuropsychological, psychiatric, and neuropathologic evaluations as part of the participation in the ADRC as previously described (17–19).
Table I. Subject characteristics.
| Variable | AD (n = 59) Mean (S.D.) or N (%) | Control (n = 12) Mean (S.D.) or N (%) | p |
|---|---|---|---|
| Age, years | 84.0 (7.3) | 70.7 (9.4) | <0.001 |
| Male | 30 (50.8%) | 8 (66.7%) | 0.31 |
| PMI, hours | 6.2 (3.4) | 12.2 (5.9) | 0.005 |
| Age of onseta, years | 75.6 (7.4) | ||
| Duration of illnessa, years | 8.3 (3.3) | ||
| Lewy body stage | |||
| Negative | 30 (50.8%) | 12 (100%) | |
| Brainstem/transitional | 15 (25.4%) | 0 (0%) | |
| Neocortical | 14 (23.7%) | 0 (0%) | |
| Braak stage | |||
| 0 - II | 0 (0%) | 12 (100%) | |
| III | 6 (10.2%) | 0 (0%) | |
| IV | 21 (35.6%) | 0 (0%) | |
| V | 32 (54.2%) | 0 (0%) |
a n = 58 for this variable. PMI—postmortem interval.
Normal Control Subjects
Twelve normal control subject brain specimens were obtained through the ADRC as described above and through the Allegheny County Medical Examiner's Office, with consent obtained from the subjects' next of kin. The protocol used to obtain consent was approved by the University of Pittsburgh Institutional Review Board and Committee for Oversight of Research Involving the Dead. An independent committee of experienced clinicians made consensus Diagnostic and Statistical Manual of Mental Disorders 4th edition (DSM-IV) diagnoses for each subject, using information obtained from clinical records and structured interviews with surviving relatives. Samples from subjects without any DSM-IV diagnosis (i.e. including no diagnosis of a cognitive disorder) were used in this study (Table I).
The right hemisphere was blocked coronally at 1–2 cm intervals and the resultant slabs snap frozen in 2-methyl butane on dry ice and stored at −80 °C. Tissue slabs containing either the dorsolateral prefrontal cortex (DLPFC) or the entorhinal cortex (ERC) were identified. From these slabs, DLPFC and ERC were removed as single blocks. Gray matter was collected by cutting 40 μm sections and frozen at −80 °C. Samples from the frontal pole, hippocampus, ERC, and cerebellum were collected, and an experienced neuropathologist reviewed sections stained using hematoxylin and eosin, Bielschowsky silver stain, amyloid β immunohistochemistry, and were determined to be without evidence of any neurodegenerative disease.
APOE3 and APOE4 Mice
APOE3 and APOE4 targeted replacement mice were originally purchased from Taconic on a C57BL/6 background and were maintained on the same background. Brains from five female and three male mice of each genotype, all 7–10 months old, were removed and snap frozen as described previously (7).
Sample Preparation
Tissue homogenates were prepared from fresh frozen human DLPFC and ERC gray matter, and total protein was extracted and mixed with the (13) C6 standard as previously described (15). To evenly distribute AD, FTLD, and normal control subjects throughout preparation and analysis, samples were organized in a balanced block distribution. For the DLPFC experiment, each block was composed of seven subjects and one pooled technical replicate, for a total of 12 blocks. For the ERC experiment, each block was also composed of seven subjects and one pooled technical replicate, for a total of four blocks. APOE mice were similarly prepared and analyzed, with one exception; trypsin digestion was performed by filter-aided sample preparation (20).
LC-SRM/MS
The selection process for the proteins included in this SRM assay has been extensively described (15) resulting in nonredundant peptides from synaptically expressed proteins, including glutamate receptors, kinases, phosphatases, and those with roles in vesicular fusion, energy and amino acid metabolism, protein trafficking, cytoskeleton, and scaffolding. We previously established the stability of peptides from 100 of these proteins up to 24 h postmortem in an animal model (15).
LC-MS/MS analyses of peptides derived from synaptically expressed proteins were conducted as previously described using a TSQ Quantiva triple-stage quadrupole mass spectrometer (Thermo Scientific, Pittsburgh PA USA) with an UltiMate 3000 Nano LC Systems (Thermo Scientific) (15). In total, 311 peptides from 192 proteins were assessed in DLPFC. In ERC, analysis was performed on a subset of 219 peptides from 138 proteins. Precision of the assay was high, with a mean coefficient of variation (CV) for peptide quantification of 8.6% in DLPFC and 11.2% in ERC. In DLPFC, 95.8% of all peptide CVs were < 20%, in ERC 92.2% of peptide CVs were < 20% (Supplemental Table 8). In the APOE mice, 314 peptides from 189 proteins were quantified with a mean peptide CV of 15.6% and 76.1% of peptides with a CV < 20%.
Statistical Analysis
Protein-level measures were derived by calculating the weighted average of all standardized (i.e. centered and scaled) peptide measures mapped to a protein, with weights inverse to the CV of the peptide measures. Between groups comparisons used analysis of variance (ANOVA), with the p value and the Benjamini–Hochberg adjusted q value reported for each protein. Unsupervised hierarchical clustering was used to construct self-organized heat maps of peptide and subject by peptide values with Cluster 3.0 and Treeview (21, 22). Values were log2 transformed, median centered by subject and peptide level, and then normalized by subject and peptide level. Uncentered Pearson correlation was used as the similarity metric and the clustering method was centroid linkage. Pathway analysis used the Database for Annotation, Visualization and Integrated Discovery (DAVID) Functional Annotation Tool (23) and searched for gene ontology term enrichment. For these analyses, the list of proteins was tested for enrichment relative to the specific proteins assayed (e.g. 192 proteins in DLPFC), rather than against the entire genome, to increase stringency. Protein coexpression networks were constructed by adapting the Weighted Gene Co-expression Network Analysis approach (24) to investigate synaptic protein network alternations as previously described in schizophrenia (16). The networks then were visualized using Cytoscape (25). The tuning parameters of networks were set to the same values for comparison of network topology measures between groups.
RESULTS
Protein Level Changes
Only minimal changes in protein levels were evident in the DLPFC of the AD subjects as compared with the controls. Fifteen proteins showed nominally significant (defined as p < 0.05) changes in AD (Table II). This resulted in only one protein, SEPT9, showing altered levels at a false discovery rate (FDR) < 0.1 (for all proteins see Supplemental Table 1). Several additional analyses were conducted to ascertain whether the small number of alterations were due to technical factors in our approach. To evaluate whether there was any impact of the inexact matching of the AD and control groups, we reran the contrasts adjusting for covariates age, sex, postmortem interval, and gel block. However, in these analyses, no proteins were altered at an FDR < 0.1. Second, to ensure that the small number of changes detected were not a result of limited sensitivity of our assay, we analyzed gray matter protein extracted from the DLPFC of a group of FTLD subjects (Supplemental Table 2). In these subjects, alterations in protein levels were widespread, with 114 proteins showing nominally significant changes and 95 proteins changed at an FDR < 0.05, compared with control subjects (Supplemental Table 3).
Table II. Proteins with nominally significant alterations in prefrontal cortex of subjects with Alzheimer disease. Protein level ratios, p, and q values are from comparisons of AD and control subjects unadjusted for covariates, all proteins with p < 0.05 are shown.
| Protein | AD: Control | p | q | p1 |
|---|---|---|---|---|
| SEPT9 | 2.12 | 0.0005 | 0.087 | 0.055 |
| ADD2 | 1.99 | 0.0012 | 0.111 | 0.004 |
| MAP2 | 0.56 | 0.0020 | 0.130 | 0.011 |
| FLOT2 | 1.75 | 0.0079 | 0.379 | 0.041 |
| STX1B | 1.62 | 0.0133 | 0.409 | 0.055 |
| RAPGEF4 | 0.59 | 0.0140 | 0.409 | 0.088 |
| PRDX1 | 1.62 | 0.0150 | 0.409 | 0.124 |
| CALB2 | 1.64 | 0.0238 | 0.557 | 0.027 |
| FLOT1 | 1.6 | 0.0266 | 0.557 | 0.116 |
| SEC22B | 1.57 | 0.0349 | 0.557 | 0.136 |
| GNAQ | 1.57 | 0.0349 | 0.557 | 0.419 |
| HSPA2 | 1.58 | 0.0360 | 0.557 | 0.522 |
| RDX | 1.59 | 0.0377 | 0.557 | 0.442 |
| DLGAP2 | 0.64 | 0.0442 | 0.580 | 0.144 |
| SUCLA2 | 1.52 | 0.0453 | 0.580 | 0.138 |
p1 are the corresponding p values from a model in which covariates age, sex, PMI, and gel block were adjusted.
The widespread reductions in the synaptic proteome present in FTLD, suggested that these changes in protein levels might be most reflective of neuron loss, which is prominent in the frontal lobes in FTLD and, thus, would similarly be present in the ERC in our AD subjects, as this region is affected by neuron loss more severely and earlier in AD than DLPFC (8, 26, 27). We evaluated this in a subset of our AD and control subjects (Supplemental Table 4). 103 proteins were changed at an FDR < 0.05 in the ERC of AD subjects. For 95 of these proteins, there was a significant interaction of region by diagnosis (Supplemental Table 5). All of the proteins, except PRDX1, had lower levels in ERC of AD than control subjects, consistent with alterations due to neuron loss. This interpretation is further supported by specifically examining the levels of MAP2, an established marker of neuronal apoptosis after brain insult, including Aβ toxicity (28). Although MAP2 levels were modestly reduced in the DLPFC of AD subjects compared with normal controls, the magnitude of reduction was significantly less than the profound reduction in MAP2 in the ERC of AD subjects, and significantly less (q = 0.014) than the profound reductions seen in DLPFC of FTLD subjects.
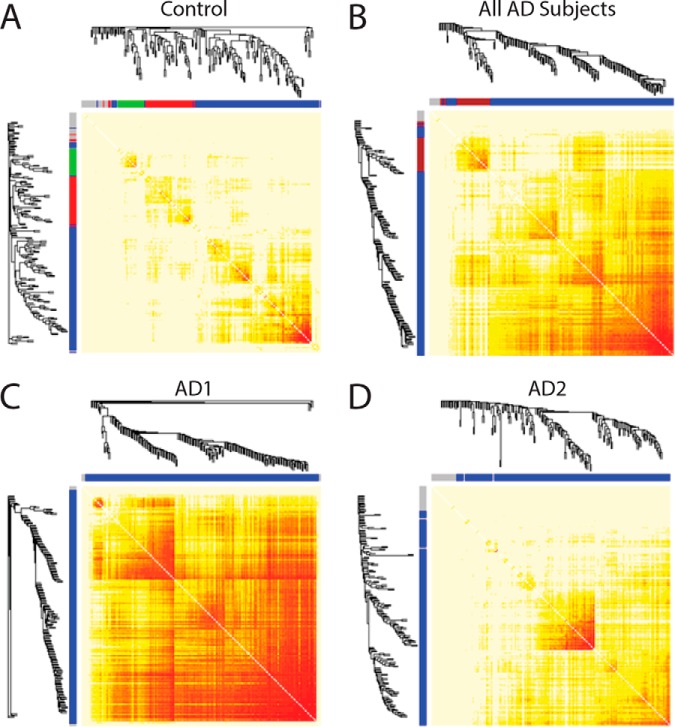
Hierarchical Clustering Based on the Expression of Synaptic Proteins in DLPFC Distinguished Two Groups of AD Subjects
We next used unsupervised hierarchical clustering to evaluate whether the levels of all peptides assayed in the DLPFC would identify biologic substructure within the AD subjects. Peptide levels clustered AD into two groups, referred to as AD1 and AD2 (Fig. 1). When we repeated the clustering with the control subjects included, 11/12 (91.7%) of the controls clustered with the AD2 group (Supplemental Fig. 3) and the separation of AD1 subjects and AD2 subjects was preserved in 51/55 (92.7%) cases.
Fig. 1.

Unsupervised hierarchical clustering of assayed peptides in AD subjects identifies two subgroups. A higher resolution version of this image is available as Supplemental Fig. 2.
Proteins with significant differential expression in AD1 relative to control subjects are shown in Table IIIA. Down-regulated proteins included glutamate receptor subunits (GRIN2B, GRIA2, GRIA3, GRIA4, GRM2, GRM3) and their interactors. Relative to the background of all assayed proteins, down-regulated proteins were significantly enriched for gene ontology terms related to the postsynaptic density (Table IV). In contrast, up-regulated proteins included a mix of cytoskeletal, metabolic, and signaling proteins, and were not significantly enriched for any gene ontology terms relative to the assay background. Fewer proteins showed nominally significant differential expression in the AD2 group relative to controls (Table 3B). These comprised a variety of cytoskeletal, vesicular, and calcium signaling related proteins and no gene ontology terms were significantly enriched relative to the assay background. These differences between the AD1 and AD2 groups did not appear to result from differential neuron loss, as MAP2 levels did not differ between AD1 and AD2 (p = 0.71).
Table III. A. Prefrontal cortex proteins with nominally significant alterations in subgroup AD1 versus control subjects.
| AD1: Control | p | q | |
|---|---|---|---|
| Down-regulated Proteins | |||
| RAPGEF4 | 0.47 | 3.63E-04 | 0.012 |
| GRM2 | 0.55 | 0.0017 | 0.030 |
| MAP2 | 0.54 | 0.0027 | 0.038 |
| SHANK3 | 0.58 | 0.004 | 0.044 |
| DLGAP2 | 0.53 | 0.0066 | 0.066 |
| GRM3 | 0.61 | 0.0079 | 0.076 |
| GRIN2B | 0.64 | 0.021 | 0.17 |
| GRIA3 | 0.72 | 0.022 | 0.17 |
| HOMER1 | 0.62 | 0.023 | 0.17 |
| NDUFS4 | 0.68 | 0.030 | 0.22 |
| GRIA2 | 0.66 | 0.039 | 0.26 |
| GRIA4 | 0.68 | 0.050 | 0.28 |
| Up-regulated Proteins | |||
| PRDX1 | 2.31 | 1.06E-07 | 2.04E-05 |
| GNAQ | 2.37 | 8.94E-05 | 0.0086 |
| VIM | 2.02 | 1.48E-04 | 0.0095 |
| SEPT2 | 2.18 | 2.07E-04 | 0.0099 |
| SEPT9 | 2.38 | 2.97E-04 | 0.011 |
| RDX | 1.98 | 5.70E-04 | 0.016 |
| RHOA | 2.08 | 0.001 | 0.024 |
| PPIA | 2.16 | 0.0012 | 0.027 |
| HSPA2 | 2.04 | 0.0015 | 0.029 |
| GAPDH | 2.08 | 0.0026 | 0.038 |
| SUCLA2 | 1.95 | 0.0028 | 0.038 |
| RHOB | 1.83 | 0.0035 | 0.044 |
| CNP | 1.87 | 0.0037 | 0.044 |
| ADD2 | 1.59 | 0.0041 | 0.044 |
| DCTN | 1.81 | 0.0083 | 0.076 |
| DPYSL2 | 1.73 | 0.009 | 0.077 |
| AK1 | 1.53 | 0.016 | 0.13 |
| AP2M1 | 1.52 | 0.039 | 0.26 |
| STX1B | 1.52 | 0.041 | 0.26 |
| YWHAQ | 1.64 | 0.045 | 0.27 |
| PTGES3 | 1.63 | 0.046 | 0.27 |
| CDC42 | 1.47 | 0.047 | 0.27 |
| RALA | 1.45 | 0.050 | 0.28 |
Table IV. Gene ontology (GO) enrichment terms for prefrontal cortex proteins with nominally significant downregulation in subgroup AD1 versus control subjects. No GO term was significantly enriched relative to the background of all assayed proteins for upregulated proteins in the comparison of AD1 versus control subjects, nor for up- and down-regulated proteins in the comparison of AD2 versus control subjects.
| Term | Genes | Fold enrichment | P value | Benjamini |
|---|---|---|---|---|
| GO:0014069∼postsynaptic density | GRIN2B, GRIA2, GRIA4, SHANK3, GRIA3, HOMER1, GRM3, DLGAP2 | 6.90 | 5.63E-06 | 2.62E-04 |
| GO:0045211∼postsynaptic membrane | GRIN2B, GRIA2, GRIA4, SHANK3, GRIA3, HOMER1, GRM3, DLGAP2 | 7.33 | 3.39E-06 | 3.15E-04 |
| GO:0044456∼synapse part | GRIN2B, GRIA2, GRIA4, SHANK3, GRIA3, HOMER1, GRM2, GRM3, DLGAP2 | 4.55 | 2.56E-05 | 7.92E-04 |
| GO:0044430∼cytoskeletal part | GRIN2B, GRIA2, GRIA4, SHANK3, GRIA3, HOMER1, GRM3, MAP2, DLGAP2 | 3.57 | 1.99E-04 | 0.0046 |
| GO:0045202∼synapse | GRIN2B, GRIA2, GRIA4, SHANK3, GRIA3, HOMER1, GRM2, GRM3, DLGAP2 | 3.14 | 5.52E-04 | 0.010 |
| GO:0030425∼dendrite | GRIN2B, GRIA4, GRIA3, GRM3, MAP2, DLGAP2 | 5.50 | 0.0011 | 0.016 |
| GO:0005856∼cytoskeleton | GRIN2B, GRIA2, GRIA4, SHANK3, GRIA3, HOMER1, GRM3, MAP2, DLGAP2 | 2.75 | 0.0016 | 0.020 |
| GO:0044463∼cell projection part | GRIN2B, GRIA4, GRIA3, GRM3, MAP2 | 6.67 | 0.0022 | 0.025 |
| GO:0043005∼neuron projection | GRIN2B, GRIA4, GRIA3, GRM2, GRM3, MAP2, DLGAP2 | 3.54 | 0.0031 | 0.031 |
| GO:0043228∼nonmembrane-bounded organelle | GRIN2B, GRIA2, GRIA4, SHANK3, GRIA3, HOMER1, GRM3, MAP2, DLGAP2 | 2.44 | 0.0038 | 0.035 |
| GO:0043232∼intracellular nonmembrane-bounded organelle | GRIN2B, GRIA2, GRIA4, SHANK3, GRIA3, HOMER1, GRM3, MAP2, DLGAP2 | 2.44 | 0.0038 | 0.035 |
| GO:0030054∼cell junction | GRIN2B, GRIA2, GRIA4, SHANK3, GRIA3, HOMER1, DLGAP2 | 3.31 | 0.0046 | 0.038 |
Table IIIB. Prefrontal cortex proteins with nominally significant alterations in subgroup AD2 versus control subjects.
| AD2:Control | p | q | |
|---|---|---|---|
| Down-regulated Proteins | |||
| CNP | 0.51 | 2.96E-05 | 0.0057 |
| MAP1A | 0.52 | 0.00057 | 0.036 |
| MAP2 | 0.58 | 0.0052 | 0.13 |
| VIM | 0.67 | 0.019 | 0.28 |
| HSP90A | 0.61 | 0.024 | 0.33 |
| ATP1A3 | 0.66 | 0.035 | 0.45 |
| NDUFS4 | 0.71 | 0.044 | 0.52 |
| Up-regulated Proteins | |||
| ADD2 | 2.12 | 0.00041 | 0.036 |
| FLOT2 | 1.98 | 0.0019 | 0.089 |
| SEPT9 | 1.97 | 0.0029 | 0.089 |
| FLOT1 | 1.86 | 0.0032 | 0.089 |
| CALB2 | 1.93 | 0.0032 | 0.089 |
| STX1B | 1.71 | 0.0066 | 0.14 |
| VAMP2 | 1.76 | 0.0089 | 0.17 |
| HPCA | 1.66 | 0.012 | 0.21 |
| NCDN | 1.57 | 0.019 | 0.28 |
The separation of AD1 and AD2 groups did not result from a number of technical and demographic factors (Table V), suggesting that this molecular signature reflected a difference in underlying synaptic biology. Somewhat surprisingly given that interpretation, these subgroups also did not reflect several measures associated with greater synaptic pathology in AD, including illness duration and cognitive status (29), presence of psychosis (30), greater amyloid plaque burden (6, 31), greater neurofibrillary tangle pathology (32), and presence of comorbid Lewy body pathology (33). However, the AD1 group was significantly enriched for individuals with an APOE*4 genotype. A decision tree analysis to classify AD1 and AD2 membership revealed a primary effect of APOE*4 genotype, with additional effects of sex and illness duration (Supplemental Fig. 4).
Table V. Comparison of demographic and clinical characteristics of AD1 and AD2 subgroups.
| Variable | AD1 (n = 24) | AD2 (n = 31) | p |
|---|---|---|---|
| Age (S.D.), years | 85.0 (7.1) | 83.8 (7.1) | 0.56 |
| Male | 9 (37.5%) | 19 (61.3%) | 0.08 |
| PMI (S.D.), hours | 5.5 (2.6) | 6.9 (3.9) | 0.11 |
| Age of onseta (S.D.), years | 76.9 (6.9) | 75.4 (7.3) | 0.46 |
| Duration of illnessa (S.D.), years | 7.8 (3.1) | 8.4 (3.6) | 0.53 |
| Last MMSE (S.D.) | 15.3 (5.9) | 15.1 (6.3) | 0.87 |
| Psychosis | 8 (33.3%) | 12 (38.7%) | 0.68 |
| Lewy body stage | 0.86 | ||
| Negative | 13 (54.2%) | 16 (51.6%) | |
| Brainstem/transitional | 7 (29.2%) | 8 (25.8%) | |
| Neocortical | 4 (16.7%) | 7 (22.6%) | |
| CERAD plaque score of frequent | |||
| MFa | 22 (95.7%) | 28 (90.3%) | 0.46 |
| ERC | 23 (95.8%) | 28 (90.3%) | 0.16 |
| CERAD NFT score of frequent | |||
| MF | 11 (42.3%) | 15 (57.7%) | 0.39 |
| ERC | 23 (95.8%) | 30 (96.8%) | 0.85 |
| Braak stage | 0.30 | ||
| III | 1 (4.2%) | 5 (16.1%) | |
| IV | 10 (41.7%) | 9 (29.0%) | |
| V | 13 (54.2%) | 17 (54.8%) | |
| APOE genotype | 0.03b | ||
| 23 | 0 (0%) | 1 (3.2%) | |
| 24 | 0 (0%) | 2 (6.5%) | |
| 33 | 8 (33.3%) | 19 (61.3%) | |
| 34 | 14 (58.3%) | 8 (25.8%) | |
| 44 | 2 (8.3%) | 1 (3.2%) |
PMI—postmortem interval; MMSE—MiniMental State Exam; CERAD—Consortium to Establish a Registry for Alzheimer's Disease; NFT—neurofibrillary tangle.
a n = 54 for this variable.
b Fisher's exact test for the distribution of genotypes. For the comparison of APOE*4 carriers versus noncarriers, p = 0.02.
APOE3 and APOE4 Targeted Replacement Mice
Mice with targeted replacement of APOE4, in the absence of Aβ overproduction, have evidence for reductions in dendritic and synaptic markers (7, 34–36) and in cognition (37, 38), We therefore evaluated whether the reduced expression of glutamate receptor proteins in the AD1 group would be present in APOE4 versus APOE3 targeted replacement mice. APOE4 mice had down-regulation of numerous glutamate signaling and synaptic proteins, but there was little overlap with the specific subset of glutamate signaling proteins altered in the AD1 group (Supplemental Table 7 and Supplemental Fig. 5).
Synaptic Protein Coexpression Networks
Because synaptic function requires coordinated expression and activity of multiple proteins, we next examined the coexpression network topology of our protein panel. AD was characterized by an overall increase in coexpression relative to controls (Figs. 2 A and 2B), driven entirely by the AD1 cluster (Figs. 2C and 2D). Evaluation of the characteristics of the AD1, AD2, and control networks revealed significantly increased mean node weighted connectivity, node degree difference, and node clustering coefficient in the AD1 group in comparison to control subjects, whereas the AD2 group did not differ from controls on any of these parameters (Table VI).
Fig. 2.
Protein network co-expression heatmaps. Protein coexpression is shown for control subjects in (A) and all AD subjects in (B). Greater correlation between proteins is shown as intensity depth of red color. Clustering of proteins into multiple modules is evident in the controls subjects, as indicated by the dendrograms and corresponding X and Y axis color coding of the individual modules. The increase in coexpression in AD is evident. These effects are due largely to the AD1 but not the AD2, subgroup (C and D).
Table VI. Comparison of network topology between AD1, AD2, and control subjects.
| Network topology measure | AD1 mean (S.D.) | AD2 mean (S.D.) | Control mean (S.D.) | AD1 vs. controlap |
|---|---|---|---|---|
| Node weighted connectivity | 37.07 (21.39) | 5.25 (5.33) | 3.53 (3.55) | < 0.001 |
| Node degree | 114.05 (50.49) | 3.25 (7.02) | 2.69 (5.96) | < 0.001 |
| Node clustering coefficient | 0.35 (0.06) | 0.12 (0.05) | 0.07 (0.07) | 0.001 |
a No comparison of AD2 vs control had p < 0.05.
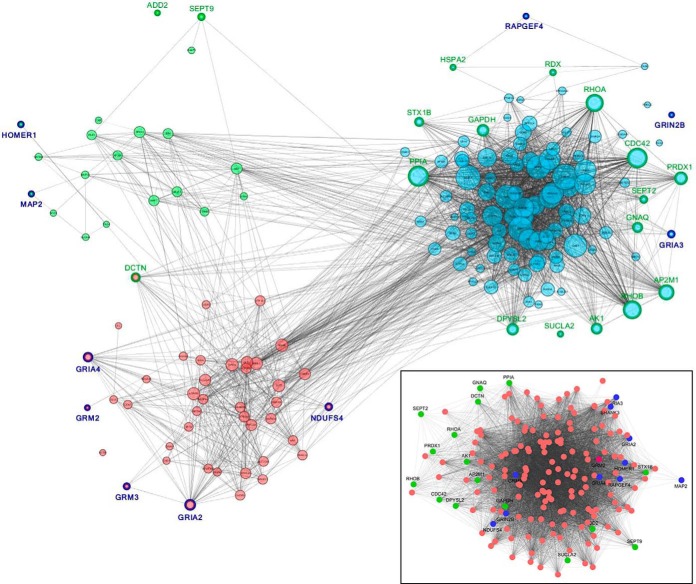
Visualization of the network structure (Fig. 3) further elucidated the nature of the changes in the AD1 group. In control subjects, most proteins clustered into one of three modules with functions as defined by their predominant constituent proteins as cytoskeletal (green module), postsynaptic (red module), and presynaptic/vesicular (blue module). In AD1, a single module of all highly connected proteins was present. Examination of the position of proteins that were up- and down-regulated in AD1 reveals them under normal conditions to be located in different modules, with up-regulated proteins more frequent in the blue module and down-regulated proteins more frequent in the red and green modules (p = 0.0002). Moreover, down-regulated proteins were more likely to be on the periphery of the control network modules, as evidenced by lower mean connectivity than proteins that were unchanged in AD1 (down-regulated mean (S.D.) connectivity 0.93 (1.45), other proteins 3.78 (3.61), p = 9.7E-06). However, in the AD1 network, these down-regulated glutamate signaling proteins had now developed increased connectivity at a level not differing from other proteins in the network (p > 0.05).
Fig. 3.
Protein networks in control subjects and AD1 subgroup. Proteins in the control network grouped into three modules (Green, red, and blue colors). Proteins that were up- (green borders) or down-regulated (dark blue borders) in the AD1 subjects are indicated. The inset shows the AD1 network, with increased connectivity of all proteins, forming a single module (all node sizes were set to be equal to enhance visibility of this image). The new position of the up- (green) and down-regulated (blue) proteins are shown.
DISCUSSION
Altered Synaptic Protein Expression in AD
We found profound reductions across the synaptic proteome in ERC of subjects in early to middle pathologic stages of AD. In contrast, in the DLPFC of these patients, there was no global loss of the synaptic proteome. However, in a subset of subjects (AD1), there were both reduced expression of glutamate signaling proteins and increased coexpression of synaptic proteins more globally. It should be noted that although we assayed a synaptically expressed proteome, it cannot be concluded that the observed protein level changes seen in the AD1 subgroup reflect altered levels of these proteins within synapses. The changes observed can reflect proteins at any point during synthesis, trafficking, localization in the synapse, and internalization and could thus result from synapse loss, neuronal loss, or disruption of homeostatic regulation of these proteins across compartments. Nevertheless, the most likely interpretation of our findings is that, in ERC, neuronal loss leads to global loss of their associated synapses and proteins. In contrast, in DLPFC, our findings are most consistent with altered synaptic protein homeostasis, driven by the subset of subjects defined by the AD1 group. This interpretation is supported by the finding of selective, rather than global alterations in protein levels. It is further consistent with the finding of significant changes in synaptic protein coexpression in this region, as coexpression results from the combined input of multiple factors involved in maintaining synaptic proteins (see below). While the observation of altered DLPFC protein homeostasis in AD is novel in scope, reflecting the concurrent assessment of nearly 200 proteins, it is consistent with earlier observations of altered DLPFC homeostasis of select synaptic proteins in subjects transitioning into the early stages of AD that have included elevations of synaptic proteins (e.g. as we observed for syntaxin 1B) that are then lost in late disease stages (10, 39–42).
APOE*4 and Synaptic Pathology
APOE*4 genotype might contribute to greater reductions in glutamate signaling protein expression in AD subjects by directly reducing glutamate signaling protein expression in a manner independent of, and therefore additive with, AD pathology. Prior studies had demonstrated that in the absence of overexpression of APP and Aβ, APOE4 targeted replacement mice have significant loss of synaptophysin-immunoreactive presynaptic terminals (34), postsynaptic dendritic spines (35), and dendritic structure (7), the latter evident by 2 months of age. How APOE4 targeted replacement causes a direct synaptic effect is not established, but reduction of synaptic NMDA and AMPA receptor expression via sequestration within intracellular compartments has been described (36). Consistent with these prior observations, we observed reductions in multiple synaptically expressed proteins, including GRIN1 and the obligatory NMDA receptor subunit. However, APOE4 targeted replacement alone did not reduce GRIN2B or recapitulate the broader pattern of reduction in AMPA, and metabotropic glutamate receptor subunits we observed in our AD1 cohort. Thus, our findings suggest it is unlikely that we observed an independent, additive effect of APOE*4 genotype on glutamate signaling protein expression in our AD subjects.
Alternatively, APOE*4 may interact with Aβ to alter synaptic proteostasis in AD. It has previously been shown that aggregation of Aβ into soluble oligomers causes synaptotoxicity, even prior to neuronal death (1, 43–45). APOE*4 interacts with this process via reducing the clearance of soluble Aβ from brain (46–47) and by promoting Aβ oligomer binding to synapses (11). Because we had previously determined soluble (monomeric and oligomeric) levels of Aβ1–40 and Aβ1–42 in superior frontal cortex for 20 of the current subjects (19), we were able to evaluate the accumulation of soluble Aβ in this subset. AD1 subjects had significantly lower levels of Aβ1–40 and Aβ1–42 than AD2 subjects (Supplemental Table 6), although there was no significant difference between these groups in ratio of Aβ1–42:Aβ1–40 and no difference in severity of neuritic plaques consisting of insoluble Aβ aggregates (Table V). Thus, it does not appear that APOE*4 genotype led to the synaptic disruption evident in the AD1 cohort via increased concentrations of soluble Aβ or increased insoluble Aβ plaque burden. However, whether the effects of APOE*4 on enhanced synaptic binding of toxic soluble Aβ species account for the alterations we observed in the synaptic proteome cannot be concluded from the current experiment.
Network Topology as an Indicator of Protein Homeostasis
When the expression levels of two proteins are correlated across a cohort, they are referred to as coexpressed. Groups of proteins with high coexpression form modules within the larger network. Modular identify often reflects proteins that share cellular compartments and/or functions, as seen in the composition of the three distinct modules we observed, defined by cytoskeletal, postsynaptic, and presynaptic/vesicular proteins. In addition, coexpression can result from any of several relevant biological functions, including chromosomal proximity, shared promoters, shared protein degradation, and shared expression within specific cell populations (48). Conversely, alterations in network features between control and disease cohorts can result from effects of disease on one or more of these functions. Network analysis can therefore complement traditional analyses of complex data sets and have provided valuable insight in a number of neuropsychiatric diseases (16, 49, 50).
Using this approach, we found greater connectivity of the synaptic protein coexpression network in AD than in control subjects, driven by the AD1 subgroup. This pattern of connectivity differed, for example, from our prior observations in schizophrenia (16), suggesting that increased coexpression is not merely a nonspecific consequence of constructing synaptic protein networks in the disease state. The observation of increased synaptic protein connectivity, i.e. that synaptic protein coexpression was greater in AD than in control subjects, could indicate that the normal diversification of synaptic protein homeostasis was overwhelmed by the effect of pathology in the AD subjects. As discussed above, this effect was driven by the AD1 subgroup, possibly reflecting increased synaptotoxicity conferred by APOE*4 status. Additionally, we observed a nonrandom location of proteins down-regulated in the AD1 subjects within the network. Down-regulated glutamate signaling proteins were located at the periphery of the postsynaptic module. This pattern has been previously described in psychiatric and other complex disorders (49) and is perhaps not surprising as changes to highly connected hub proteins can have catastrophic effects (51).
Potential Caveats
One potential limitation of this study was the significantly younger normal control group. This confound arose in part via our effort to obtain a neuropathologically normal control cohort, as alterations to the synaptic proteome may be present in cognitively normal individuals with AD pathologies (8), which would have further complicated interpretation of our findings. Several observations, however, make it unlikely that age mismatch accounts for the lack of global protein level alterations in DLPFC or the profound protein level changes in ERC in AD. First, because we observed a significant region (DLPFC versus ERC) by diagnosis interaction for most proteins tested, one would have to postulate age-specific effects that differed by region. Second, we observed global protein alterations in DLPFC of FTLD subjects who were substantially younger than our AD cohort and comparable to controls. Finally, the specific differences in DLPFC synaptic protein levels between the AD1 and AD2 groups was present despite lack of age differences between these groups. Another potential confound in human tissue studies is matching as closely as possible by post-mortem interval (PMI). Our study included AD subjects with significantly lower PMIs than normal controls, however, we had previously established the stability of the majority of proteins and peptides assayed for up to 24 h in an animal model (15), well beyond the PMIs of our study groups. A final concern is whether the quality of the sample preps of DLPFC versus ERC samples may have been responsible for the differences in protein reductions between these regions in AD. All SRM chromatograms from the DLPFC and the ERC were manually evaluated blind to diagnostic group. Peak shape and intensity were highly comparable for all peptides quantified in both the DLPFC and ERC, indicating that sample degradation does not account for the differences in protein expression between the two brain areas in the AD group.
CONCLUSIONS
Whereas global reductions in synaptic protein levels reflected neuron loss, in the frontal cortex (a region with limited neuron loss in mild to moderate stages of AD), alterations in synaptic protein expression and coexpression were largely restricted to a subgroup of individuals who were APOE*4 allele carriers. These observations are likely indicative of an effect of APOE genotype on synaptic protein homeostasis within the context of Aβ accumulation and AD pathology, particularly for postsynaptic glutamate receptor subunits and their signaling partners. As receptor proteins located at the periphery of the synaptic protein network, these glutamate signaling proteins may be possible to target therapeutically. However, our results suggest that efforts to enhance signaling at these receptors for therapeutic benefit in AD, e.g. via ampakines, metabotropic glutamate receptor agonists, or other glutamatergic agents, would have outcomes dependent on APOE*4 status of the recipients. A stratification of benefit by APOE*4 status is consistent with limited available data for the NMDA receptor antagonist, memantine (52), and a similar effect could contribute to lack of benefit seen in some initial studies of other glutamatergic agents (53).
Primary data can be accessed via the following links: DLPFC Link: https://chorusproject.org/anonymous/download/experiment/17ccb8ef19624aaaad00bf9d92cdbbfa
ERC Link: https://chorusproject.org/anonymous/download/experiment/fb292cffb5014aeb9f70e9993f4340fc
Supplementary Material
Footnotes
Author contributions: R.A.S., M.L.M., C.M.K., Y.D., and N.A.Y. designed the research; M.L.M., C.M.K., T.S., J.J., J.K., M.D.I., O.L.L., M.E.G., N.F.F., and R.K. performed the research; N.F.F. and R.K. contributed new reagents or analytic tools; M.L.M. and Y.D. analyzed data; R.A.S., M.L.M., J.K., and M.D.I. wrote the paper; and N.F.F. and R.K. reviewed manuscript.
* This work was supported by grants MH16804 (MLM), AG05133 (OLL), AG014449 (MDI), AG027224 (RAS), AG037481 (RK), AG044490 (NFF), and VAPHS grant BX000452 (RAS). The Biomedical Mass Spectrometry Center and UPCI Cancer Proteomics Facility are supported in part by award P30CA047904.The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health, the National Institutes of Health, the Department of Veterans Affairs, or the United States Government.
 This article contains supplemental material.
This article contains supplemental material.
R.A.S., M.L.M., C.M.K., Y.D., T.S., J.J.L., J.K., M.D.I., O.L.L., N.F.F., R.K., and N.A.Y. have no biomedical financial interests or potential conflicts of interest to disclose.
1 The abbreviations used are:
- AD
- Alzheimer disease
- LC-MS/MS
- liquid chromatography–tandem mass spectrometry
- LC-SRM/MS
- selective reaction monitoring
- FTLD
- frontotemporal lobar degeneration
- ADRC
- Alzheimer Disease Research Center
- DSM-IV
- Diagnostic and statistical manual of mental disorders 4th edition
- DLPFC
- dorsolateral prefrontal cortex
- ERC
- entorhinal cortex
- CV
- coefficient of variation
- ANOVA
- analysis of variance
- DAVID
- Database for annotation, visualization and integrated discovery
- FDR
- False discovery rate
- PMI
- Post-mortem interval
- GO
- Gene ontology
REFERENCES
- 1. Selkoe D. J. (2002) Alzheimer's disease is a synaptic failure. Science 298, 789–791 [DOI] [PubMed] [Google Scholar]
- 2. Koffie R. M., Hyman B. T., and Spires-Jones T. L. (2011) Alzheimer's disease: Synapses gone cold. Mol. Neurodegener. 6, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DeKosky S. T., and Scheff S. W. (1990) Synapse loss in frontal cortex biopsies in Alzheimer's disease: Correlation with cognitive severity. Ann. Neurol. 27, 457–464 [DOI] [PubMed] [Google Scholar]
- 4. Terry R. D., Masliah E., Salmon D. P., Butters N., DeTeresa R., Hill R., Hansen L. A. and Katzman R. (1991) Physical basis of cognitive alterations in Alzheimer's disease: Synapse loss is the major correlate of cognitive impairment. Ann. Neurol. 30, 572–580 [DOI] [PubMed] [Google Scholar]
- 5. Baloyannis S. J., Costa V., Mauroudis I., Psaroulis D., Manolides S. L., and Manolides L. S. (2007) Dendritic and spinal pathology in the acoustic cortex in Alzheimer's disease: Morphological and morphometric estimation by Golgi technique and electron microscopy. Acta Otolaryngol. 127, 351–354 [DOI] [PubMed] [Google Scholar]
- 6. Grutzendler J., Helmin K., Tsai J., and Gan W. B. (2007) Various dendritic abnormalities are associated with fibrillar amyloid deposits in Alzheimer's disease. Ann. N.Y. Acad. Sci. 1097, 30–39 [DOI] [PubMed] [Google Scholar]
- 7. Mounier A., Georgiev D., Nam K. N., Fitz N. F., Castranio E. L., Wolfe C. M., Cronican A. A., Schug J., Lefterov I., and Koldamova R. (2015) Bexarotene-activated retinoid X receptors regulate neuronal differentiation and dendritic complexity. J. Neurosci. 35, 11862–11876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perez-Nievas B. G., Stein T. D., Tai H. C., Dols-Icardo O., Scotton T. C., Barroeta-Espar I., Fernandez-Carballo L., de, Munain E. L., Perez J., Marquie M., Serrano-Pozo A., Frosch M. P., Lowe V., Parisi J. E., Petersen R. C., Ikonomovic M. D., López O. L., Klunk W., Hyman B. T., and Gómez-Isla T. (2013) Dissecting phenotypic traits linked to human resilience to Alzheimer's pathology. Brain 136, 2510–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scheff S. W., and Price D. A. (2003) Synaptic pathology in Alzheimer's disease: A review of ultrastructural studies. Neurobiol Aging 24, 1029–1046 [DOI] [PubMed] [Google Scholar]
- 10. Counts S. E., Nadeem M., Lad S. P., Wuu J., and Mufson E. J. (2006) Differential expression of synaptic proteins in the frontal and temporal cortex of elderly subjects with mild cognitive impairment. J. Neuropathol Exp. Neurol 65, 592–601 [DOI] [PubMed] [Google Scholar]
- 11. Koffie RM, Hashimoto T, Tai HC, Kay KR, Serrano-Pozo A, Joyner D., Hou S., Kopeikina K. J., Frosch M. P., Lee V. M., Holtzman D. M., Hyman B. T., and Spires-Jones T. L. (2012) Apolipoprotein E4 effects in Alzheimer's disease are mediated by synaptotoxic oligomeric amyloid-beta. Brain 135, 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sweet R. A., Fish K. N., and Lewis D. A. (2010) Mapping synaptic pathology within cerebral cortical circuits in subjects with schizophrenia. Front Hum. Neurosci. 4, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trinidad J. C., Thalhammer A., Specht C. G., Lynn A. J., Baker P. R., Schoepfer R., and Burlingame A. L. (2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, 684–696 [DOI] [PubMed] [Google Scholar]
- 14. Emes R. D., Pocklington A. J., Anderson C. N., Bayes A., Collins M. O., Vickers C. A., Croning M. D., Malik B. R., Choudhary J. S., Armstrong J. D., and Grant S. G. (2008) Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat. Neurosci. 11, 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Macdonald M. L., Ciccimaro E., Prakash A., Banerjee A., Seeholzer S. H., Blair I. A., and Hahn C. G. (2012) Biochemical fractionation and stable isotope dilution liquid chromatography-mass spectrometry for targeted and microdomain-specific protein quantification in human postmortem brain tissue. Mol. Cell. Proteomics 11, 1670–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. MacDonald ML, Ding Y, Newman J, Hemby S, Penzes P, Lewis DA, Yates N. A., and Sweet R. A. (2015) Altered Glutamate Protein Co-Expression Network Topology Linked to Spine Loss in the Auditory Cortex of Schizophrenia. Biol. Psychiatry 77, 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sweet R. A., Hamilton R. L., Lopez O. L., Klunk W. E., Wisniewski S. R., Kaufer D. I., Heily M. T., and DeKosky S. T. (2000) Psychotic symptoms in Alzheimer's disease are not associated with more severe neuropathologic features. Int. Psychogeriatr 12, 547–558 [DOI] [PubMed] [Google Scholar]
- 18. Lopez O. L., Becker J. T., Chang Y. F., Sweet R. A., Aizenstein H., Snitz B., Saxton J., McDade E., Kamboh M. I., DeKosky S. T., Reynolds C. F. 3rd, and Klunk W. E. (2013) The long-term effects of conventional and atypical antipsychotics in patients with probable Alzheimer's disease. Am. J. Psychiatry 170, 1051–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murray P. S., Kirkwood C. M., Gray M. C., Ikonomovic M. D., Paljug W. R., Abrahamson E. E., Henteleff R. A., Hamilton R. L., Kofler J. K., Klunk W. E., Lopez O. L., Penzes P., and Sweet R. A. (2012) beta-Amyloid 42/40 ratio and kalirin expression in Alzheimer disease with psychosis. Neurobiol Aging 33, 2807–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wiśniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 21. Page R. D. (1996) TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- 22. Eisen M. B., Spellman P. T., Brown P. O., and Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 24. Langfelder P., and Horvath S. (2008) WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C. Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., and Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gómez-Isla T., Price J. L., McKeel D. W. Jr, Morris J. C., Growdon J. H., and Hyman B. T. (1996) Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer's disease. J. Neurosci. 16, 4491–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kirkwood C. M., Macdonald M. L., Schempf T. A., Vatsavayi A. V., Ikonomovic M. D., Koppel J. L., Ding Y., Sun M., Kofler J. K., Lopez O. L., Yates N. A., Sweet R. A. (2016) Altered levels of visinin-like protein 1 correspond to regional neuronal loss in Alzheimer disease and frontotemporal lobar degeneration. J. Neuropathol Exp. Neurol. 75, 175–182 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fifre A., Sponne I., Koziel V., Kriem B., Yen, Potin F. T., Bihain B. E., Olivier J. L., Oster T., and Pillot T. (2006) Microtubule-associated protein MAP1A, MAP1B, and MAP2 proteolysis during soluble amyloid beta-peptide-induced neuronal apoptosis. Synergistic involvement of calpain and caspase-3. J. Biol. Chem. 281, 229–240 [DOI] [PubMed] [Google Scholar]
- 29. Scheff S. W., and Price D. A. (2006) Alzheimer's disease-related alterations in synaptic density: Neocortex and hippocampus. J. Alzheimers Dis. 9, 101–115 [DOI] [PubMed] [Google Scholar]
- 30. Murray P. S., Kumar S., Demichele-Sweet M. A., and Sweet R. A. (2014) Psychosis in Alzheimer's disease. Biol. Psychiatry 75, 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsai J., Grutzendler J., Duff K., and Gan W. B. (2004) Fibrillar amyloid deposition leads to local synaptic abnormalities and breakage of neuronal branches. Nat. Neurosci. 7, 1181–1183 [DOI] [PubMed] [Google Scholar]
- 32. Nelson P. T., Alafuzoff I., Bigio E. H., Bouras C., Braak H., Cairns N. J., Castellani R. J., Crain B. J., Davies P., Del Tredici K., Duyckaerts C., Frosch M. P., Haroutunian V., Hof P. R., Hulette C. M., Hyman B. T., Iwatsubo T., Jellinger K. A., Jicha G. A., Kövari E., Kukull W. A., Leverenz J. B., Love S., Mackenzie I. R., Mann D. M., Masliah E., McKee A. C., Montine T. J., Morris J. C., Schneider J. A., Sonnen J. A., Thal D. R., Trojanowski J. Q., Troncoso J. C., Wisniewski T., Woltjer R. L., and Beach T. G. (2012) Correlation of Alzheimer disease neuropathologic changes with cognitive status: A review of the literature. J. Neuropathol. Exp. Neurol. 71, 362–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boyle P. A., Wilson R. S., Yu L., Barr A. M., Honer W. G., Schneider J. A., and Bennett D. A. (2013) Much of late life cognitive decline is not due to common neurodegenerative pathologies. Ann. Neurol. 74, 478–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buttini M., Yu G. Q., Shockley K., Huang Y., Jones B., Masliah E., Mallory M., Yeo T., Longo F. M., and Mucke L. (2002) Modulation of Alzheimer-like synaptic and cholinergic deficits in transgenic mice by human apolipoprotein E depends on isoform, aging, and overexpression of amyloid beta peptides but not on plaque formation. J. Neurosci. 22, 10539–10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ji Y., Gong Y., Gan W., Beach T., Holtzman D. M., and Wisniewski T. (2003) Apolipoprotein E isoform-specific regulation of dendritic spine morphology in apolipoprotein E transgenic mice and Alzheimer's disease patients. Neuroscience 122, 305–315 [DOI] [PubMed] [Google Scholar]
- 36. Chen Y., Durakoglugil M. S., Xian X., and Herz J. (2010) ApoE4 reduces glutamate receptor function and synaptic plasticity by selectively impairing ApoE receptor recycling. Proc. Natl. Acad. Sci. U.S.A. 107, 12011–12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raber J., Wong D., Yu G. Q., Buttini M., Mahley R. W., Pitas R. E., Mucke L. (2000) Apolipoprotein E and cognitive performance. Nature 404, 352–354 [DOI] [PubMed] [Google Scholar]
- 38. Bour A., Grootendorst J., Vogel E., Kelche C., Dodart J. C., Bales K., Moreau P. H., Sullivan P. M., and Mathis C. (2008) Middle-aged human apoE4 targeted-replacement mice show retention deficits on a wide range of spatial memory tasks. Behav. Brain Res. 193, 174–182 [DOI] [PubMed] [Google Scholar]
- 39. Masliah E., Mallory M., Alford M., DeTeresa R., Hansen L. A., McKeel D. W. Jr, and Morris J. C. (2001) Altered expression of synaptic proteins occurs early during progression of Alzheimer's disease. Neurology 56, 127–129 [DOI] [PubMed] [Google Scholar]
- 40. DeKosky S. T., Ikonomovic M. D., Styren S. D., Beckett L., Wisniewski S., Bennett D. A., Cochran E. J., Kordower J. H., and Mufson E. J. (2002) Upregulation of choline acetyltransferase activity in hippocampus and frontal cortex of elderly subjects with mild cognitive impairment. Ann. Neurol. 51, 145–155 [DOI] [PubMed] [Google Scholar]
- 41. Mukaetova-Ladinska E. B., Garcia-Siera F., Hurt J., Gertz H. J., Xuereb J. H., Hills R., Brayne C., Huppert F. A., Paykel E. S., McGee M., Jakes R., Honer W. G., Harrington C. R., and Wischik C. M. (2000) Staging of cytoskeletal and beta-amyloid changes in human isocortex reveals biphasic synaptic protein response during progression of Alzheimer's disease. Am. J. Pathol. 157, 623–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Minger S. L., Honer W. G., Esiri M. M., McDonald B., Keene J., Nicoll J. A., Carter J., Hope T., and Francis P. T. (2001) Synaptic pathology in prefrontal cortex is present only with severe dementia in Alzheimer disease. J. Neuropathol. Exp. Neurol. 60, 929–936 [DOI] [PubMed] [Google Scholar]
- 43. Selkoe D, J. (2008) Soluble oligomers of the amyloid beta-protein impair synaptic plasticity and behavior. Behav. Brain Res. 192, 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Walsh D. M., and Selkoe D. J. (2007) A beta oligomers—A decade of discovery. J. Neurochem. 101, 1172–1184 [DOI] [PubMed] [Google Scholar]
- 45. Lue L. F., Kuo Y. M., Roher A. E., Brachova L., Shen Y., Sue L., Beach T., Kurth J. H., Rydel R. E., and Rogers J. (1999) Soluble amyloid beta peptide concentration as a predictor of synaptic change in Alzheimer's disease. Am. J. Pathol. 155, 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deane R., Sagare A., Hamm K., Parisi M., Lane S., Finn M. B., Holtzman D. M., and Zlokovic B. V. (2008) apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J. Clin. Invest. 118, 4002–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hudry E., Dashkoff J., Roe A. D., Takeda S., Koffie R. M., Hashimoto T., Scheel M., Spires-Jones T., Arbel-Ornath M., Betensky R., Davidson B. L., and Hyman B. T. (2013) Gene transfer of human Apoe isoforms results in differential modulation of amyloid deposition and neurotoxicity in mouse brain. Sci. Transl. Med. 5, 212ra161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gaiteri C., Ding Y., French B., Tseng G. C., and Sibille E. (2014) Beyond modules and hubs: The potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 13, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gaiteri C., and Sibille E. (2011) Differentially expressed genes in major depression reside on the periphery of resilient gene coexpression networks. Front. Neurosci. 5, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Parikshak N. N., Luo R., Zhang A., Won H., Lowe J. K., Chandran V., Horvath S., and Geschwind D. H. (2013) Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jeong H., Mason S. P., Barabási A. L., and Oltvai Z. N. (2001) Lethality and centrality in protein networks. Nature 411, 41–42 [DOI] [PubMed] [Google Scholar]
- 52. Han H. J., Kim B. C., Lee J. Y., Ryu S. H., Na H. R., Yoon S. J., Park H. Y., Shin J. H., Cho S. J., Yi H. A., Choi M. S., Heo J. H., Park K. W., Kim K. K., and Choi S. H. (2012) Response to rivastigmine transdermal patch or memantine plus rivastigmine patch is affected by apolipoprotein E genotype in Alzheimer patients. Dement. Geriatr. Cogn. Disord. 34, 167–173 [DOI] [PubMed] [Google Scholar]
- 53. Chappell A. S., Gonzales C., Williams J., Witte M. M., Mohs R. C., and Sperling R. (2007) AMPA potentiator treatment of cognitive deficits in Alzheimer disease. Neurology 68, 1008–1012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.