Abstract
UHRF1 is best known for its positive role in the maintenance of DNMT1-mediated DNA methylation and is implicated in a variety of tumor processes. In this paper, we provided evidence to demonstrate a role of UHRF2 in cell motility and invasion through the regulation of the epithelial-mesenchymal transition (EMT) process by acting as a transcriptional co-regulator of the EMT-transcription factors (TFs). We ectopically expressed UHRF2 in gastric cancer cell lines and performed multidimensional proteomics analyses. Proteome profiling analysis suggested a role of UHRF2 in repression of cell-cell adhesion; analysis of proteome-wide TF DNA binding activities revealed the up-regulation of many EMT-TFs in UHRF2-overexpressing cells. These data suggest that UHRF2 is a regulator of cell motility and the EMT program. Indeed, cell invasion experiments demonstrated that silencing of UHRF2 in aggressive cells impaired their abilities of migration and invasion in vitro. Further ChIP-seq identified UHRF2 genomic binding motifs that coincide with several TF binding motifs including EMT-TFs, and the binding of UHRF2 to CDH1 promoter was validated by ChIP-qPCR. Moreover, the interactome analysis with IP-MS uncovered the interaction of UHRF2 with TFs including TCF7L2 and several protein complexes that regulate chromatin remodeling and histone modifications, suggesting that UHRF2 is a transcription co-regulator for TFs such as TCF7L2 to regulate the EMT process. Taken together, our study identified a role of UHRF2 in EMT and tumor metastasis and demonstrated an effective approach to obtain clues of UHRF2 function without prior knowledge through combining evidence from multidimensional proteomics analyses.
UHRF (ubiquitin-like, containing PHD and RING finger domains, inverted CCAAT Box-Binding Protein of 90 kDa) family contains four members characterized with multiple domains in structure (1). The founding member of this family, UHRF1 is the best characterized. UHRF1 has been reported to play critical roles in various processes, including the maintenance of DNMT1-mediated DNA methylation. UHRF1 binds to hemi-methylated DNA through its SRA domain and therefore plays an important role in targeting DNMT1 to sites of replication (2–8). In addition, UHRF1 recognizes dimethylated/trimethylated lysine 9 of histone H3 (H3K9me2/3) via its PHD and TTD domains (8–15), and exerts ubiquitin E3 ligase activity on DNMT1 and histone substrates via its RING domain (9, 10, 16, 17). Importantly, several studies have demonstrated the oncogenic role of UHRF1 in tumors by promoting proliferation and metastasis of cancer cells (18–26).
UHRF2 has similar sequence and domain architectures as UHRF1. This similarity suggests a potential functional conservation between these two proteins. Like UHRF1, UHRF2 also recognizes hemimethylated DNA substrates and H3K9me2/3, and interacts with DNMTs and histone methyltransferase G9a in vitro (27). However, there are substantial differences between UHRF1 and UHRF2. UHRF2 cannot rescue the DNA methylation defect in Uhrf1−/− ES cells because of its inability to recruit DNMT1 to replication foci during S phase (28). In addition, UHRF2 was reported as a specific binder of 5hmC with its SRA domain, whereas UHRF1-SRA does not have this binding preference (29, 30). Unlike UHRF1, which is often found in ESCs, UHRF2 is more commonly expressed in differentiated cells. A function of UHRF2 in regulation of cell cycle was also speculated. UHRF2 was found to interact with cyclins, CDKs, p53, pRB, PCNA, and was able to induce G1 arrest by ubiquitinating cyclins D1 and E1 (31, 32). Other substrates of UHRF2 E3 Ub ligase include PCNP, nuclear aggregates containing polyglutamine repeats, hepatitis B virus core protein and zinc finger protein 131 (ZNF131) (33–36). Like UHRF1, UHRF2 was also implicated in tumors; but reports about the role of UHRF2 in tumors are contradictory and uncertain. Some studies demonstrated that UHRF2 behaves like a tumor suppressor to inhibit the inappropriate cell cycle progression (31, 32), whereas other studies suggested potential oncogenic characteristics of UHRF2 with up-regulated expression in cancers (37–40).
Metastasis is an important characteristics of cancer and responsible for more than 90% of cancer associated mortality. Metastasis of cancer cells is a complex process that is partly regulated by activation of epithelial to mesenchymal transition (EMT)1 to acquire the ability to invade and metastasize (41). During EMT, epithelial cells lose cell-cell contacts and cell polarity, and acquire mesenchymal-like characteristics with increased ability of migration and invasion. EMT is orchestrated by transcription factor cascades that regulate the expression of proteins involved in cell-cell contacts, cell polarity, cytoskeleton structure and extracellular matrix degradation. For instance, EMT-TFs repress one of the key epithelial genes E-cadherin through binding the promoter region of CDH1 directly or indirectly. The reported key EMT-TFs include SNAIL1/2, TWIST1/2, ZEB1/2, TCF3 and FOXC2 (42–45). Because of the limited number of studies on the involvement of UHRF2 in tumorigenesis, the precise biological functions of UHRF2 in cancer and whether it also functions like UHRF1 remain to be investigated.
MS-based proteomics is a powerful approach for large scale protein analysis in biological research (46, 47). Our lab has developed a fast, label-free quantification workflow (Fast-quan) for protein identification, in which 7,000 proteins can be identified and quantified with 12 h of MS running time (48). This has enabled analysis of multiple samples. We also developed a concatenated tandem array of transcription factor response elements (catTFRE) pull-down assay that allows for enrichment and identification of endogenous transcription factors (TFs) (49). The combination of measuring changes in DNA binding activity of TFs and proteome-wide profiling of protein abundance allows us to correlate TF activity with target genes in response to exogenous stimulation. Thus, the proteome-wide identification of activated TFs when cells are perturbed can provide important biological clues about the mechanisms and signal transduction pathways.
Current UHRF researches focused on how UHRF proteins impact genome DNA methylation. This direction is important in studying cancer initiation when changes in UHRF proteins can reprogram the epigenome. It is entirely not clear whether and what roles UHRF2 may play when cells become cancerous. We thus ectopically expressed UHRF2 in gastric cancer cell lines and performed multidimensional proteomics analyses to obtain clues for UHRF2 functions in a consistent manner. The MS profiling revealed down-regulation of a number of epithelial markers including CDH1, JUP, TJP1, DSG2, INADL, CXADR, SPINT1, and TJP2. The catTFRE-MS analysis also demonstrated up-regulation of multiple key transcription factors involved in EMT, including TWIST2, FOXC2, and TCF family of transcription factors. Furthermore, we demonstrated that silencing UHRF2 in gastric cancer cells could inhibit the ability of cell migration and invasion in vitro. Together, these results suggested that UHRF2 play a role in tumor metastasis.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
SGC7901, MKN74, N87, and MKN45 human gastric cancer cell lines were cultured in DMEM supplemented with 10% FBS (fetal bovine serum) (Gibco, Carlsbad, CA), 1% Penicillin-Streptomycin (Sigma-Aldrich, St. Louis, MO) and incubated at 37 °C and 5% CO2.
Generation of UHRF2 Antibody
Antibody specific to UHRF2 was generated against a recombinant GST-tagged UHRF2 (N-terminal 351aa) fragment. Specific antibody was purified from serum of immunized rabbit with His-tagged-UHRF2 protein. The specific recognition of endogenous UHRF2 by the antibody was confirmed by IP-MS.
Generation of UHRF2 Overexpression Stable Cell Lines with Lentivirus Infection
The human UHRF2 coding sequence was amplified by PCR from cDNA libraries. The amplified fragment was cloned into the pENTR-vector. The lentiviral vector containing UHRF2 cDNA was constructed by recombination of pHAGE-EF-ZsG-DEST with pENTR. The sequence was verified by DNA sequencing. Lentivirus supernatants were collected 48 h after transfecting pHAGE-EF-UHRF2-ZsG-DEST with the packaging vectors pMD2.G and psPAX2 into 293T cells using lipofectamine 2000 reagent (Invitrogen). The lentivirus packaged with the empty pHAGE-EF-ZsG-DEST vector was used as control. The concentrated and purified lentivirus from supernatants were used to infect cells with 8 μg/ml Polybrene to generated control or UHRF2-expressing stable cells of SGC7901, MKN74, N87, and MKN45. The expression of UHRF2 in different cell lines was analyzed by Western blot.
CatTFRE DNA Pull-down
CatTFRE was done as previously described (49). Briefly, the catTFRE DNA was prepared by PCR amplification with biotinylated primers. Nuclear extracts (NEs) were prepared with NE-PER nuclear extraction reagents (Thermo Fisher Scientific, Boston, MA). Two milligram of control or OE NEs was used to incubate with pre-immobilized biotinylated catTFRE on Dynabeads (Dynabeads M-280 Streptavidin, Invitrogen, Carlsbad, CA). The mixture was supplemented with EDTA to a final concentration of 1 mm and adjusted with NaCl to 200–250 mm total salt concentration and incubated for 2h at 4 °C. The supernatant was discarded and Dynabeads were washed with NETN [100 mm NaCl, 20 mm Tris-Cl, 0.5 mm EDTA, and 0.5% (v/v) Nonidet P-40] twice followed by PBS twice. Beads were added to 20 μl 2× loading buffer and incubated in 95 °C for 5 min. The supernatant was put on SDS-PAGE for separation. SDS-PAGE gels were stained with Coomassie Blue R-250 to visualize the protein bands and each lane was cut into 12 gel slices. In-gel trypsin digestion was performed. After incubation with rotation for overnight at 37 °C, the digested products were extracted with acetonitrile (ACN) and dried in vacuum.
Preparation of WCE and Manual Reversed Phase (sRP) Separation
Whole cellular lysates were extracted using 8 m urea. MKN74 control and UHRF2-OE cells (MKN74 is a gastric cancer cell line with low invasiveness, which expresses low level of UHRF2 and high level of CDH1 on its surface) were lysed with 8 m urea containing protease inhibitor PMSF for 30min at 4 °C. The lysate was centrifuged at 24,000 × g and the supernatant was collected as whole cell extracts (WCE). Protein concentration was determined by BCA assay. Twenty micrograms of control and OE proteins were digested with trypsin. Tryptic peptides were separated on a C18 column with acetonitrile of different percentage as 6%, 9%, 12%, 15%, 18%, 21%, 25%, 30%, and 35%. Nine separations were combined to six fractions and dried in vacuum. Peptides were stored at −80 °C until re-dissolved for MS analysis.
Immunoprecipitation
NEs were prepared with NE-PER Kit (Thermo) from control and UHRF2-OE cells of N87, MKN45, SGC7901, and MKN74. Equal amounts (1 mg∼10 mg) of NEs from control and OE cells were incubated with 5 μg UHRF2 antibody. The incubation solution was adjusted with NaCl to 200 mm total salt concentration and incubated at 4 °C for overnight. After the addition of 30 μl protein A/G-Sepharose for each IP reaction and incubation for another 2h, the immunoprecipitate was washed with NETN twice followed by PBS buffer twice. The Sepharose beads were re-suspended in 20 μl 2× loading buffer and incubated at 95 °C for 5 min. The supernatant was resolved by 10% SDS-PAGE and in-gel trypsin digestion was performed after the gel was sliced.
LC-MS/MS Analysis
Dried peptide samples were re-dissolved in solvent A (0.1% formic acid in water). Liquid chromatography - tandem mass spectrometry (LC-MS/MS) analysis was performed with Q-Exactive Plus or FUSION mass spectrometer (Thermo) equipped with an online Easy-nLC 1000 nano-HPLC system (Thermo). The injected peptides were separated on a reversed phase nano-HPLC C18 column (Pre-column: 5 μm, 300 Å, 2 cm × 100 μm ID; analytical column: 3 μm, 120 Å, 15 cm × 75 μm ID) at a flow rate of 350 nl/min with a 75-min gradient of 3 to 30% solvent B (0.1% formic acid in acetonitrile). For the detection with FUSION mass spectrometry, a precursor scan was measured in the Orbitrap by scanning from m/z 300–1400 with a resolution of 120,000. Ions selected under top-speed mode were isolated in Quadrupole and fragmented by higher energy collision-induced dissociation (HCD) with normalized collision energy of 35%, then measured in the linear ion trap. Typical mass spectrometric conditions were: AGC targets were 5e5 ions for Orbitrap scans and 5e3 for MS/MS scans; dynamic exclusion was employed for 18 s. For the Q-Exactive Plus, the instrument was operated in the data-dependent acquisition mode with a resolution of 70,000 at full scan mode and 17,500 at MS/MS mode. The full scan was processed in the Orbitrap from m/z 300–1400, the top 20 most intense ions in each scan were automatically selected for HCD fragmentation with normalized collision energy of 27% and measured in Orbitrap. Typical mass spectrometric conditions were: AGC targets were 3e6 ions for full scans and 5e4 for MS/MS scans; dynamic exclusion was employed for 18 s. The acquired MS/MS spectra were searched by Mascot 2.3 (Matrix Science Inc, MA) implemented on Proteome Discoverer 1.4 (Thermo) against the human National Center for Biotechnology Information (NCBI) RefSeq protein databases (updated on April 7, 2013, 32,015 protein entries). The parameter settings were: the mass tolerances were 20 ppm for precursor and 50mmu for product ions from Q-Exactive Plus and 20 ppm for precursor and 0.5 Da for product ions from FUSION respectively; two missed cleavages were allowed; the fixed modification was set as carbamidomethyl (C), dynamic modifications were protein acetyl (protein N-term), oxidation(M) for profiling data; dynamic modification for catTFRE were phosphor (Y), phosphor (ST), deStreak (C), acetyl (protein N-term), oxidation (M); A false discovery rate (FDR) of 1% was applied at the peptide level. Protein identification data (accession numbers, peptides observed, sequence coverage) and peptide identifications (sequence, charge, m/z, identification score) of Profiling, TFRE and IP data are in supplemental Tables S5, S6 and S7, respectively. All raw data and search results have been deposited to the iProX system (http://www.iprox.org/index) with the identifier IPX00067800. Please access the raw files with user ID 'reviewer678' and password 'rnhtgqt1'.
Experimental Design and Statistical Rationale
Proteins were assembled with unique and strict peptides (<1% FDR, usPepts). Relative protein quantitation of profiling and catTFRE data was performed using label free quantification (LFQ). For WCE profiling data, two biological repeats and two technical repeats of UHRF2 overexpression and corresponding control samples were processed; the protein area was normalized by total area in each sample to adjust for differences in overall protein levels between samples. This fraction of total (FOT) was then used to estimate protein abundance. Proteins with 1% FDR on protein level were used for statistical analysis. Proteins were considered to be significantly changed in abundance if there was a more than 1.5-fold difference between paired samples with a p value <0.05 using paired two-tailed t test (The data set showed a normal distribution). For catTFRE data, quantification was achieved using intensity based absolute quantification (iBAQ) for the label-free quantification (LFQ). As catTFRE was an enrichment experiment, the analysis method for enrichment data was performed (50), TFs which have 1.5-fold or greater difference in at least five out of six replicates were considered to have a significant DNA binding activity change. For IP data, UHRF2 interacting proteins were identified by PSM ratios of OE/control in duplicate experiments with more than fivefold change. GO term enrichment analysis was performed in WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/). The significance level was set to p < 0.05 at Benjamini p value using all identified proteins as background reference. Interactive network analysis was performed using the Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string.embl.de/).
Knockout UHRF2 with CRISPR/cas9
For CRISPR/cas9 experiment, two guide RNAs (gRNA) were designed according to the website http://crispr.mit.edu/and cloned into pLKO.1 lentiviral plasmid.
gRNA1:GTCCTCAATGGTGCACGTCT
gRNA2:ATACAGGTTCGCACCATTGA
The empty and gRNA-cloned pLKO.1 plasmids were transfected in 293T cells with packaging mix plasmids using Lipofectamine 2000 (Invitrogen). Lentivirus supernatants were collected 48 h after transfection and used to infect SGC7901 and BGC823 cells (two aggressive cancer cell lines that express high level of UHRF2). Lentivirus packaged with empty pLKO.1 plasmid was used as control. Cells were selected against 10 μg/ml puromycin for 10 days and then infected with adenovirus which express cas9 enzyme. After expression validation of cas9, cell clones were screened for UHRF2 knock-out by WB analysis.
Cancer Cell Migration and Invasion Assays
The migration assay was done with Transwell inserts that have 6.5-mm polycarbonate membranes with 8.0 mm pores (Sigma). The invasion assay was performed using inserts with membranes coated by 60 μl 1:8 diluted Matrigel matrix (BD Discovery Labware, Bedford, MA). DMEM medium with 10% FBS was added in 24-well plate at lower chamber and 2 × 105 cells were suspended in serum-free DMEM and seeded into the upper chamber. Cells were cultured in 37 °C incubator with 5% CO2 for 18 h. Cells on the membranes of inserts were fixed with methanol for 20 min and stained with crystal violet for 20 min. The number of cells that had migrated to the basal side of the insert membrane was quantified by counting 10 independent symmetrical visual fields under the microscope. Each experiment was performed in three replicates.
Western Blot
Whole-cell lysates were prepared with T-PER Tissue Protein Extraction Reagent (Thermo) and 50 μg lysate was resolved on 8% SDS-PAGE. Proteins were transferred onto nitrocellulose membranes. After blocking with 5% milk (BD Science) solution in TBST for 1 h, the membranes were incubated with 5% milk containing appropriate primary antibodies overnight at 4 °C followed by 2 h incubation with horseradish peroxidase-conjugated secondary antibodies. Signals of target protein bands were detected using Chemiluminescent detection reagent. UHRF2 antibody (custom made), E-cadherin antibody (BD Biosciences) and β-actin antibody were used at 1:1000 dilution.
Immunofluorescence
About 1 × 104 cells were seeded on an 8-well Millicell EZ Slide (EMD Millipore, Billerica, MA). After 72 h, cells were washed twice with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. Cells were then incubated with Rhodamine-conjugated phalloidin (50 μg/ml) for 45 min. Images were acquired with a confocal microscope.
Chromatin Immunoprecipitation and Quantitative PCR (q-PCR)
Gastric cancer cells (SGC7901 and N87) were crosslinked by incubation for 10 min in 1% formaldehyde and lysed in SDS lysis buffer (50 mm Tris HCl, pH 8.0, 10 mm EDTA, 0.1% SDS including protease inhibitors). Cells were fragmented with sonication at 3s on/3s off at 25% power. Cell lysates were then pre-cleared using rabbit IgG with protein-A agarose and small amount of cell lysates were saved as input. Immunoprecipitation was performed by addition of antibody against UHRF2 or rabbit IgG in the remaining lysates. The IP solution was incubated in 4 °C for overnight. Immunoprecipitate complex were collected using protein-A dynabeads (Thermo) and washed sequentially with low salt, high salt, LiCl and TE wash buffer. DNA was eluted with extraction buffer (1%SDS, 0.1 m NaHCO3) and extracted using phenol/chloroform/isoamyl alcohol. Immunoprecipitated and input DNA were subjected to PCR amplification. The target promoter sequences were amplified by 40 cycles at 94 °C for 10 s, 60 °C for 10 s, and 72 °C for 20 s.
The primer sequences of CDH1 promoter were: forward 5-ACTCCAGGCTAGAGGGTCACC-3, reverse 5-CCGCAAGCTCACAGGTGCTTTGCAGTTCC-3.
RESULTS
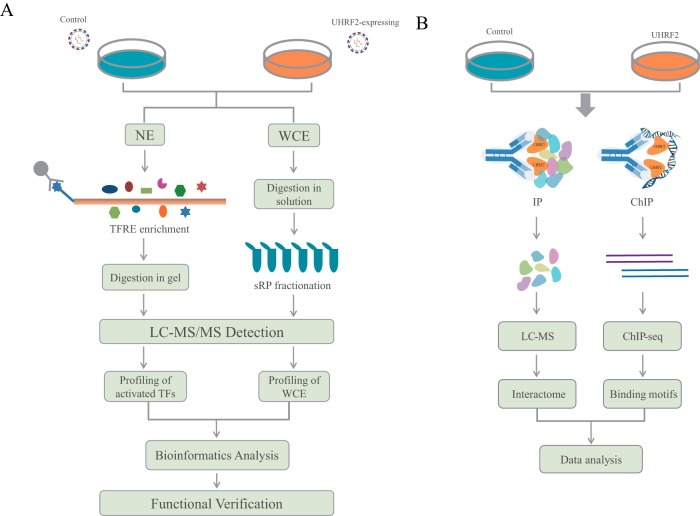
The Strategy for Dissection of UHRF2 Function with Multidimensional Proteomics
To explore the functions of UHRF2, we used multiple proteomics tools including whole proteome profiling, catTFRE pull-down-MS, IP-MS and ChIP-seq to investigate what UHRF2 does to the proteome when over-expressed in cancer cell lines (Fig. 1). We hypothesized that the overlapping or convergent functions inferred from multiple-dimensional analyses are likely to be the true cellular functions of UHRF2. Gastric cancer cell lines were infected with mock or UHRF2-overexpression (OE) lentivirus. Stably transduced UHRF2-OE and control cells were selected and the overexpression levels were determined by Western blotting (WB) and MS detection (supplemental Fig. S1). Cells were collected and processed for proteomics and functional analyses. Data from different proteomics measurements were subjected to bioinformatics analysis such as GO annotation, network and pathway analysis. UHRF2-induced differential proteins were identified and subsets of them were validated by the targeted and more accurate MS measurement parallel reaction monitoring (PRM) or WB. Functional validations were performed based on the information and clues obtained from proteomics results.
Fig. 1.
Experimental workflow for dissecting UHRF2 functions with multidimensional proteomics. A, Experimental strategy of catTFRE and profiling analysis. Gastric cancer cells were infected with mock or UHRF2 overexpression lentivirus. Control and UHRF2-OE stable cells were collected to extract nuclear extracts (NEs) and whole cell extracts (WCE) which were used for catTFRE enrichment and whole proteome profiling, respectively. MS data from different proteomics measurements were subjected to further bioinformatics analysis. Functional validation experiments were performed based on the information and clues provided by proteomics results. B, Experimental setup of UHRF2 interactome analysis and DNA binding motif analysis. Equal amount of NEs from control and UHRF2-OE cells were subjected to IP experiments using UHRF2 antibody followed by LC/MS analysis. UHRF2-OE cells were collected to do ChIP experiments with IgG and UHRF2 antibody followed by DNA sequencing.
Proteome Profiling of UHRF2 Overexpression Cells
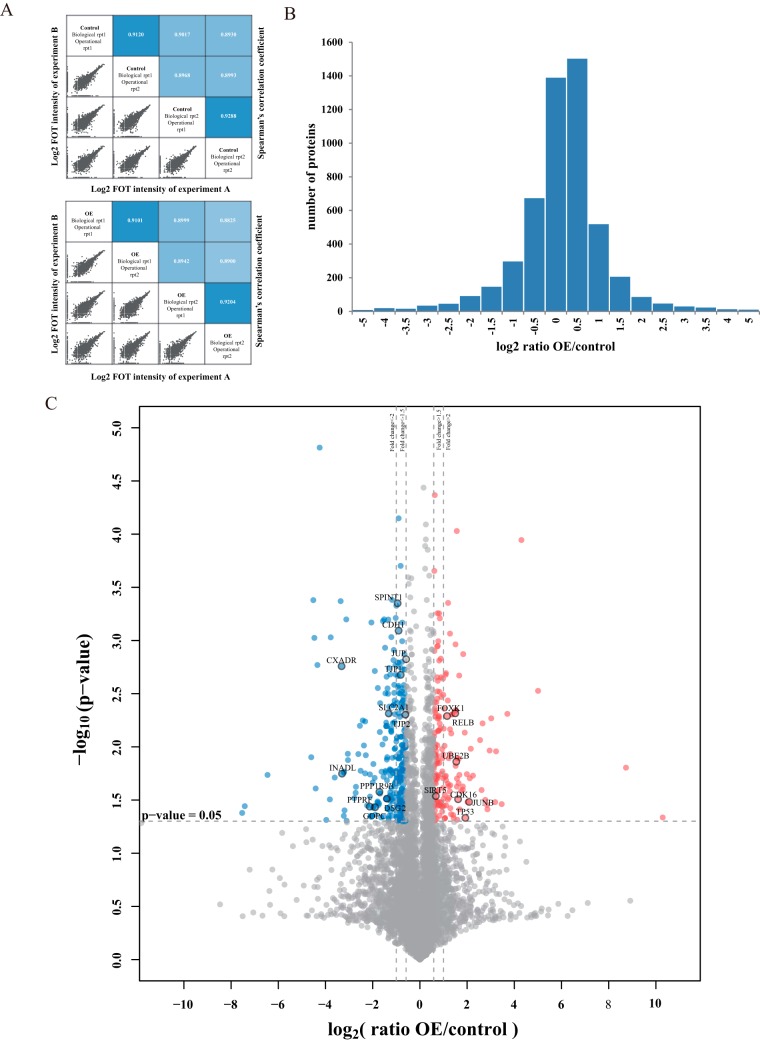
We profiled the proteome upon UHRF2 overexpression in a gastric cancer cell line MKN74 from two biological and two operational replicates. We used fraction of total (FOT) to normalize protein loading for MS profiling. The reproducibility between biological/operational replicates was good as we obtained high degree of correlation in LFQ intensity between each two replicates (Fig. 2A). A total of 7662 unique proteins were identified with the detection of at least 1 unique peptide at 1%FDR. Among them, 5856 proteins were identified at 1% protein FDR, and 5159 were shared in at least four out of eight experiments (supplemental Fig. S2A, supplemental Table S1). These 5159 proteins were used for subsequent bioinformatics analysis. The distribution of number of proteins in different ratio range is shown in Fig. 2B. A volcano plot (Fig. 2C) illustrates differential protein abundance (expressed as the mean ratio of OE/control of four replicates) against the corresponding p value obtained from t test. One hundred seventy eight (178) proteins (3.4% of the proteome) appeared to be significantly increased in their abundances upon UHRF2 overexpression (> 1.5-fold change, p value < 0.05, and marked as red dots). Similarly, 281 proteins (5.4% of the proteome) were considered as decreased (marked as blue dots). The rest of the 4700 proteins (91.2% of the proteome) were considered as not significantly changed (Fig. 2C, supplemental Fig. 2A). The 6 significantly changed proteins that were identified by least number of PSMs (2–7psms) were manually validated (supplemental Fig. S2B).
Fig. 2.
Whole proteome profiling of UHRF2 overexpression cells by label-free quantitative MS analysis. A, The up panel shows the correlation of control experiments and the down panel shows the correlation of OE groups. In each panel, the lower-left half depicts pairwise scatter plots of the experiments, with x and y axes representing log2 FOT intensity of experiments in corresponding columns and rows, respectively. The upper-right half depicts pairwise Spearman's correlation coefficients for the same comparisons. B, The distribution of protein abundance ratios is displayed with histogram. The log2 fold change of OE/control is shown on the x axis and the corresponding protein numbers are shown on the y axis. (C) Identification of the differentially expressed proteins. The log2 mean ratio of four replicates was plotted against the corresponding -log10 p value. The vertical dotted lines mark 1.5- and twofold change and the horizontal dotted line represent the cutoff of p value = 0.05. Proteins that exhibited fold change greater than 1.5-fold and p value<0.05 were considered up-regulated and marked with red. Proteins that decreased more than 1.5-fold and had p values<0.05 were considered downregulated and marked with blue. The gray proteins were considered with nonsignificant change.
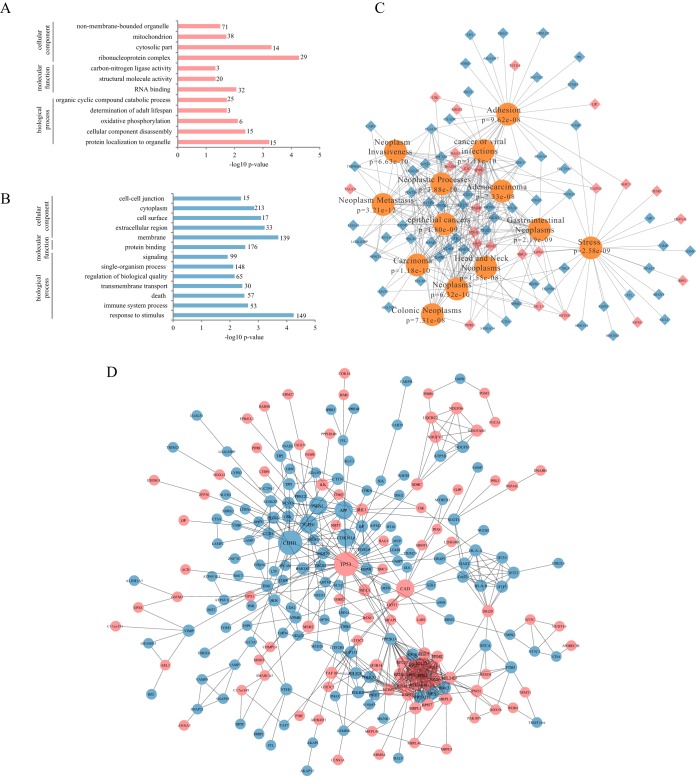
The up and down regulated proteins were subjected to GO term enrichment analysis in WebGestalt (51). The enrichment in cellular component, molecular function and biological process of up-regulated and down-regulated proteins are shown in Figs. 3A–3B, respectively. Notably, in GO terms, increased proteins are mainly annotated as residing in nonmembrane-bounded organelle and cytosolic part whereas decreased proteins are mainly annotated as residing in cell-cell junction, cell surface, extracellular region and membrane. It demonstrates that UHRF2 represses a number of epithelial markers including CDH1, JUP, TJP1, DSG2, INADL, CXADR, SPINT1, and TJP2 and stabilizes some important nuclear proteins including p53 (Fig. 2C). We next investigated disease association of up- and down- regulated proteins in WebGestalt. The results are displayed as a network using Cytoscape. The top 13 associated diseases with most significant p values are displayed with corresponding proteins marked as red or blue as up- or down- regulated respectively (Fig. 3C). Neoplasms (p = 6.32e-10) including adenocarcinoma, carcinoma, gastrointestinal neoplasms, colorectal neoplasms, and epithelial cancers as well as neoplasm metastasis (p = 3.21e-12), neoplasm invasiveness (p = 6.63e-10) and adhesion (p = 9.62e-08) are significantly associated with UHRF2 overexpression.
Fig. 3.
Bioinformatic analysis of differentially expressed proteins upon UHRF2 overexpression. A, Gene Ontology (GO) enrichment analysis for biological process, cell component and molecular function of up-regulated proteins. The -log10 p value of enrichment is shown on x axis; the numbers represent the number of associated proteins for each term. B, GO enrichment analysis of down-regulated proteins. C, Diseases associated with UHRF2-altered proteins were shown according to the enrichment analysis in WebGestalt. D, Network of proteins regulated by UHRF2. Two hubs of p53 and CDH1 were shown. Red nodes represent increased proteins and blue nodes represent decreased ones; nodes of more than ten interacting neighbors were drawn in large size; and nodes with 5–10 neighbors were in median size.
We then utilized the STRING database to uncover relationships between the up and down regulated proteins. Interactions between them with high confidence scores (above 0.7) are displayed as a network (Fig. 3D). The up-regulated nodes are filled with red color and the downregulated nodes are filled with blue. Nodes with more than ten interacting neighbors are displayed in large size and nodes with 5–10 neighbors are in medium size. We found that p53 and CDH1 represent two major hubs in the regulated network.
Targeted analysis of proteins with MS is becoming popular as an alternative to WB validation. We carried out PRM quantification to validate the above preliminary analysis (supplemental Fig. S3, supplemental Table S3). The overexpression of UHRF2 was quantified by PRM (supplemental Fig. S3A) and CDH1, DSG2, and OCLN were all validated as downregulated in UHRF2-OE cells (supplemental Fig. S3B–S3D). Down-regulation of CDH1 upon UHRF2 overexpression was also demonstrated by WB (supplemental Fig. S3E).
Taken together, the above results show that increased proteins upon UHRF2 expression mainly reside in cytosolic part and may be related to tumor suppressor protein p53 functions; decreased proteins upon UHRF2 overexpression mainly reside in cell membrane and are implicated in cell-cell junctions. A number of epithelial markers are repressed by UHRF2. Overall, altered expression of proteins by UHRF2 overexpression suggests an association of UHRF2 with cancers and metastasis.
Analysis of TF-DNA Binding Activity Change Upon UHRF2 Overexpression
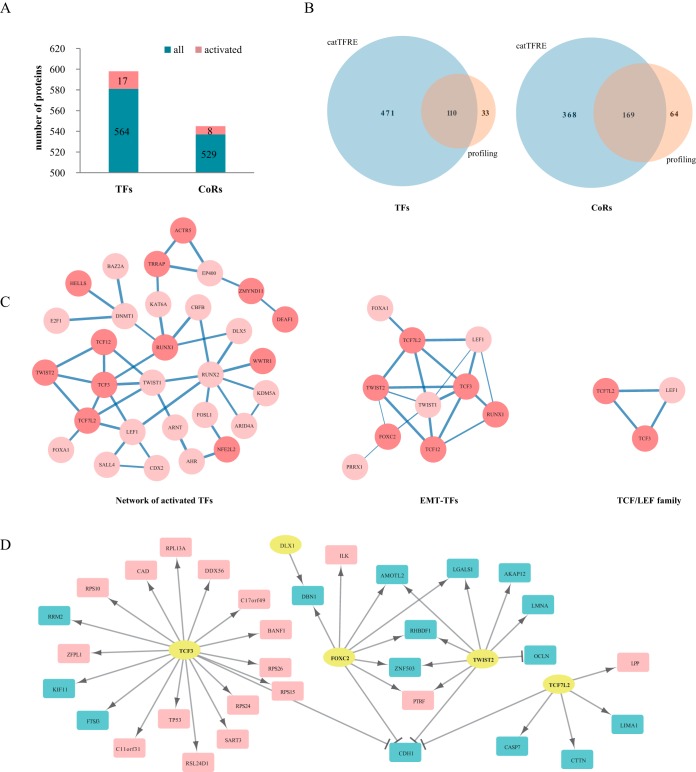
To gain further insights into the driving force for altered protein expression upon UHRF2 overexpression, we used catTFRE to measure TF-DNA binding activity. This allowed us to measure a group of low abundance transcription factors that were not detected in the previous profiling experiments. We performed three independent biological replicates and three operational replicates of catTFRE measurements of UHRF2 overexpression and control cells. We detected 581 TFs and 537 coregulators (CoRs) out of six replicates with unique peptides (usPepts, 1% FDR at peptide level) (supplemental Fig. S4A, supplemental Table S2). Among them, 471 TFs and 368 CoRs were only detected by catTFRE enrichment but were not detected in profiling experiments (Fig. 4B). Using iBAQ to represent protein abundance, we defined more than 1.5-fold intensity change in at least five out of six experiments as significantly changed and found 17 TFs and 8 CoRs were up-regulated upon UHRF2 overexpression as compared with controls (Fig. 4A).
Fig. 4.
Analysis of global changes of TF-DNA binding activity upon UHRF2 overexpression. A, Number of TFs and CoRs identified in six replicates. The numbers of proteins with increased TF-DNA binding activities are marked in red on the histogram. B, The comparison of TFs and CoRs identification between catTFRE and profiling. C, The network of altered TFs with interaction score>0.7; subnetworks of EMT-TFs and TCF/LEF family are shown in more details. Width of edges represents the interaction confidence score; activated TFs are colored dark red; nodes in light red represent TFs up-regulated in four out of six experiments. D, Activated TFs and their regulated targets from profiling data. Yellow circle represent TFs; red rectangles indicate up-regulated targets and blue rectangles indicate downregulated ones in protein level.
Table I summarizes the TF families, transcription functions, pathways and biological processes the activated TFs and CoRs are involved in and their fold changes. Five TFs (TFEB, TWIST2, TCF12, TCF3, and MXD4) belong to the bHLH family and three (SATB2, HNF1B, DLX1) contain the homeodomain. These up-regulated proteins are known to be involved in Wnt, TGFβ and MAPK signaling pathways and cancer. Notably, five TFs are annotated as EMT-TFs according to previous literature reports (42–44).
Table I. TFs and coregulators activated upon UHRF2 expression.
| TF families | Function | Pathway | Fold change (OE/control) | Biological roles involved ina | |
|---|---|---|---|---|---|
| TFAP2C | AP2 | Transcriptional activator and repressor | 9.74 | Activate genes in development; suppress MCAM/MUC18,C/EBP alpha and MYC; MTA1-mediated epigenetic regulation of ESR1 expression in breast cancer | |
| EYA1 | TC | Transcriptional Coactivator; | 7.03 | DNA repair; organogenesis; embryonic development | |
| histone phosphatase | |||||
| ZNF451 | ZNF | Coactivator for steroid receptors | 6.18 | ||
| DLX1 | Homeobox | Transcriptional regulator | TGFβ–SMAD2/3 | 5.00 | Development of the ventral forebrain |
| RUNX1 | p53 | 4.74 | Binds to the core element of many enhancers and promoters; development of normal hematopoiesis; leukemia | ||
| CHD7 | Probable transcription regulator | 4.22 | Maybe involved in the in 45S precursor rRNA production | ||
| TCF12 | bHLH | Transcriptional regulator; EMT-TF | ERK | 3.70 | Initiation of neuronal differentiation; activates transcription by binding to the E box (5-CANNTG-3) |
| WWTR1 | TC | Transcriptional coactivator | Hippo | 3.70 | downstream regulatory target in the Hippo; organ size control and tumor suppression |
| DEAF1 | SAND | Transcriptional regulator | 3.30 | Embryonic development; inhibitor of cell proliferation; expression of peripheral tissue antigens in pancreatic lymph nodes | |
| CDCA7L | TC | transcriptional repressor | 3.05 | Oncogenic role; downstream of P38-kinase and BCL-2; upstream of CASP3/caspase-3 as well as CCND1/cyclin D1 and E2F1 | |
| TWIST2 | bHLH | EMT-TF | MAPK | 2.89 | Cancers; metastsis |
| DBP | bZIP | Transcriptional activator | 2.65 | Circadian rhythm generation | |
| ACTR5 | TC | 2.60 | Proposed core component of INO80 complex; DNA replication and probably DNA repair; double-strand break repair and UV-damage excision repair | ||
| HNF1B | Homeobox | Transcriptional regulator | 2.43 | Nephron development; development of the embryonic pancreas diabetes; cancer | |
| HELLS | TC | 2.16 | Normal development and survival; regulation of the expansion or survival of lymphoid cells; de novo or maintenance DNA methylation; formation and organization of heterochromatin; regulation of transcription and mitosis | ||
| MXD4 | bHLH | Transcriptional repressor | 2.10 | A member of the MAD gene family; tumor suppressors; regulation of cell growth | |
| NFE2L2 | bZIP | Transcription activator | 2.08 | Regulates genes which contain ARE in promoters; response to oxidative stress | |
| TCF3 | bHLH | Transcriptional regulator; EMT-TF | Wnt | 1.93 | Lymphopoiesis; ymphoid malignancies; tumor metastasis |
| TFEB | bHLH | Transcriptional coactivator | Wnt | 1.85 | Humoral immunity; expression of lysosomal genes; autophagy; migratory phenotype; cancer |
| FOXC2 | Forkhead | Transcriptional activator; EMT-TF | TGFβ–SMAD3 | 1.83 | Formation of special mesenchymal tissues |
| ZMYND11 | TC | Transcriptional corepressor | 1.78 | Chromatin reader that specifically recognizes and binds H3.3K36me3 and regulates RNA polymerase II elongation; acts as a tumor-suppressor | |
| SATB2 | Homeobox | Transcriptional regulator | 1.72 | Chromatin remodeling | |
| TCF7L2 | HMG | Repressor in the absence of CTNNB1, activator in its presence; EMT-TF | Wnt | 1.72 | Blood glucose homeostasis; diabetes; tumor metastasis |
| ZMYND8 | TC | Transcriptional corepressor | 1.67 | Receptor for activated C-kinase (RACK) protein | |
| TRRAP | TC | Epigenetic transcription activation | Wnt | 1.56 | Component of many histone acetyltransferase (HAT) complexes; transcription and DNA repair; p53/TP53-, E2F1- and E2F4-mediated transcription activation |
a Reference from GeneCards human gene database.
The enrichment of the bHLH family of TFs upon UHRF2 overexpression led us to consider TF interactions, as it is known that bHLH TFs interact with other transcription factors to exert their functions (52). We analyzed the interactions of activated TFs by relaxing the constraint to include TFs that were up-regulated by 1.5-fold in at least four out of six replicates. The interactions were annotated with the STRING database and displayed as a network using Cytoscape. The network with interaction score>0.7 is shown in the left panel of Fig. 4C. The TCF3-centered subnetwork consists of ten TFs including TCF7L2, TCF3, TWIST2, TCF12, FOXC2, RUNX1 TWIST1, LEF1, FOXA1, and PRRX1 (Fig. 4C). They shared the common features as the key regulators of the EMT process. For instance, TWIST2 was reported to form homodimers with TWIST1 and then forms heterodimers with TCF3 to regulate E-box binding and transcription associated with EMT and cancer metastasis (42). This EMT subnetwork strongly suggested a role of UHRF2 in regulation of EMT. In particular, a smaller network composed of TCF7L2, TCF3, and LEF1 indicates the activation of TCF/LEF family which is the downstream of Wnt signaling pathway (Fig. 4C). The above analysis shows that overexpression of UHRF2 is correlated with an increase in DNA-binding activities of multiple TFs involved in the EMT process.
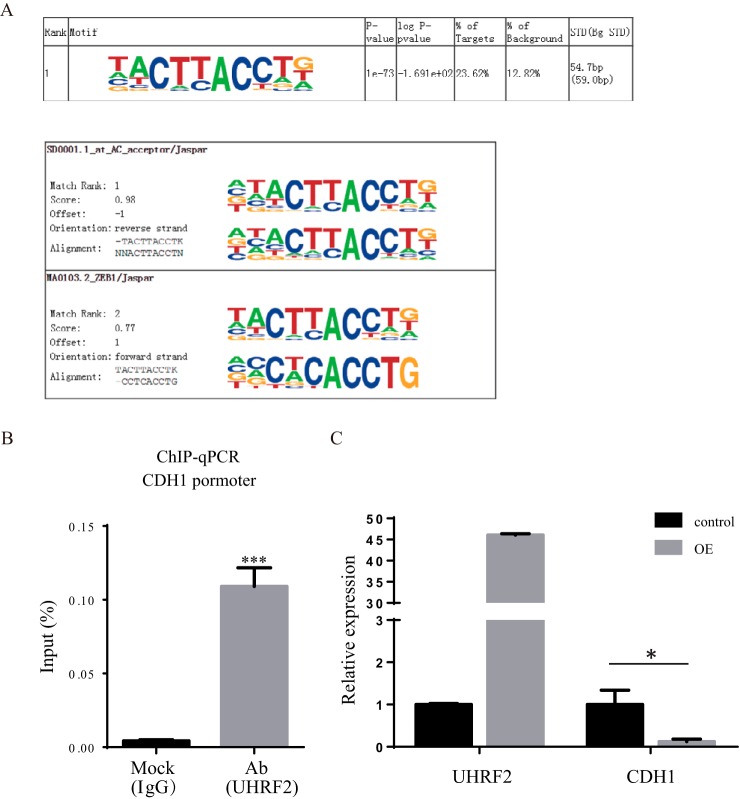
We next integrated the profiling results with catTFRE pull-down data to identify TF-targets. Using the CellNet database (53, 54), we linked targets of activated TFs with up- and down-regulated proteins in profiling (Fig. 4D, supplemental Fig. S4D). There appears to be two modules that are regulated by EMT-TFs, namely the TCF3 module that mainly positively regulate its target genes, and the TWIST2/TCF7L2/FOXC2 module that mainly negatively regulates their target genes. CDH1 appears to be repressed by four of the five TFs. To confirm this hypothesis, we measured CDH1 RNA levels by qPCR in control and UHRF2-OE cells and found that CDH1 was repressed by more than 80% when UHRF2 was overexpressed (Fig. 6C). In summary, the TF DNA binding activity data and protein profiling data correlate well and both support a model in which overexpression of UHRF2 leads to the activation of EMT-TFs and an altered expression of EMT proteins.
Fig. 6.
UHRF2 is recruited to the E-cadherin promoter. A, The most enriched motif of UHRF2 binding sequences and its best matches to known TF binding motifs. B, UHRF2 associates with CDH1 promoter at the chromatin level. Cells expressing UHRF2 were subjected to ChIP analyses using antibodies of UHRF2 and IgG. Human CDH1 promoter fragment (–179 to +39) was amplified and quantified with q-PCR. C, RNA expression level of CDH1 was significantly repressed by more than 80% (p = 0.01) upon UHRF2 overexpression.
Knockout UHRF2 Inhibits Migration and Invasion in Gastric Cancer Cell Lines and Induces Spheroid Formation in SGC7901 Cells
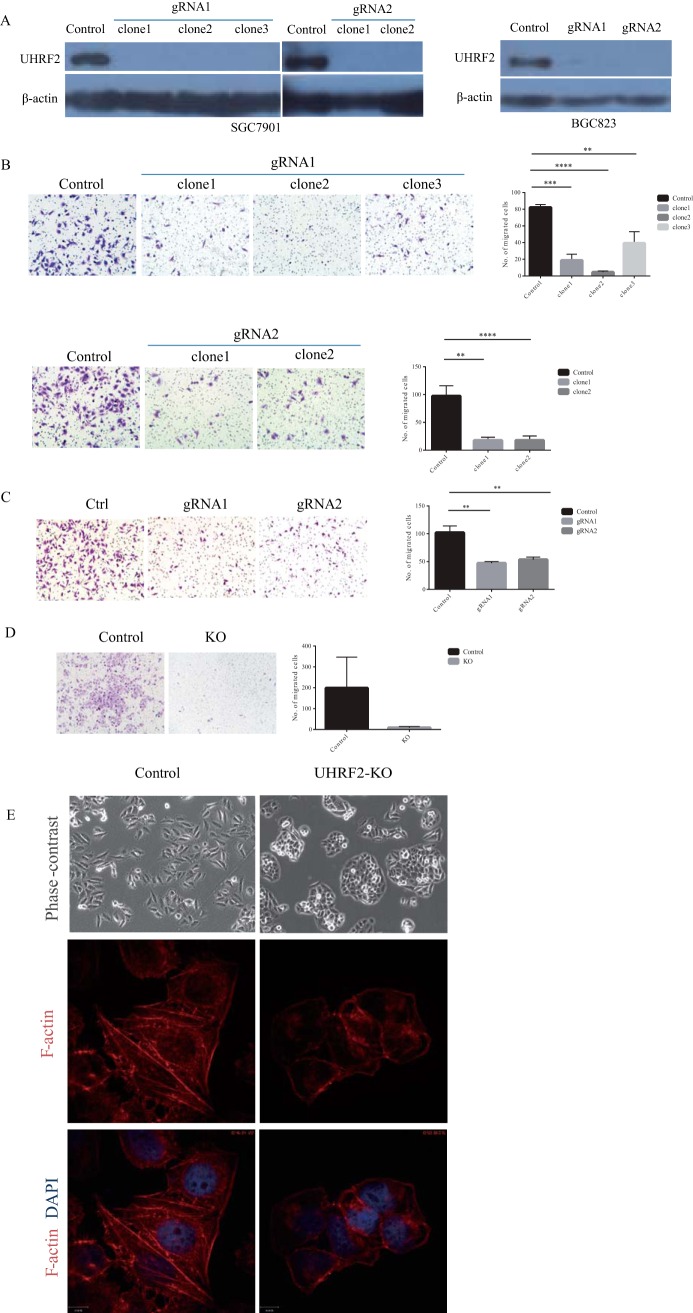
To provide further evidence for the involvement of UHRF2 in EMT process, we depleted UHRF2 in aggressive gastric cell lines SGC7901 and BGC823 using CRISPR-Cas9 system with two different guide RNAs (gRNAs) (supplemental Fig. S5A). Multiple clones of each line stably expressing each gRNA were selected and the decrease of UHRF2 expression in each was confirmed by WB (Fig. 5 A).
Fig. 5.
Knockout UHRF2 inhibits migration and invasion in gastric cancer cell lines and induces spheroid formation in SGC7901 cells. A, The knockout of UHRF2 in gastric cancer cell lines SGC7901 and BGC823 with CRISPR/cas9 was determined by WB. B, Transwell migration assay using SGC7901 control cells or 2–3 clones of UHRF2-knockout cells from each guide RNA (gRNA). Representative images are shown on the left, and the quantification of three replicates (10 fields were randomly selected in each replicate) is shown on the right. C, Transwell migration assay using BGC823 control cells and UHRF2-KO cells from different gRNA sequence. D, Invasion assay of SGC7901 control or UHRF2 KO cells. E, UHRF2 knockout in SGC7901 induces spheroid formation and a mesenchymal to epithelial transition. Control and UHRF2 KO cells were stained of F-actin to show the changes in morphology and actin filament reorganization. DAPI staining was used to show nuclei.
We used transwell assays to evaluate the cell migration ability. Cells that had traversed the membrane were stained with crystal violet and counted. Compared with the control group, UHRF2 knockout with either gRNA1 or gRNA2 caused a significant decrease in cell migration in different clones of SGC7901 cells (Fig. 5B). The reduction in migration ability upon UHRF2 knockout was also observed in another GC cell line BGC823 (Fig. 5C). Furthermore, we determined the invasion ability of cells using transwell assay with Matrigel on the insert surface. We found that silencing of UHRF2 significantly suppressed the invasion of SGC7901 (Fig. 5D). These observations suggest that knockout UHRF2 can suppress GC cell migration and invasion in vitro.
We found that loss of UHRF2 caused a profound morphological change for SGC7901 cells from mesenchymal-like morphology to epithelial morphology and induced the formation of spheroid. The actin filament reorganization was observed in UHRF2 knockout cells by Phalloidin staining (Fig. 5E), suggesting that loss of UHRF2 makes cells undergo a MET in culture. Overall, these data suggest that UHRF2 plays a causal role in cell motility.
UHRF2 is Recruited to the CDH1 Promoter for Epigenetic Silencing of CDH1 Expression
Because UHRF2 binds histone and DNA through TTD-PHD and SRA domains, respectively, we set out to determine if UHRF2 functions through chromatin-mediated gene regulation. To this end, we performed chromatin immunoprecipitation (ChIP) experiments with UHRF2 antibody using IgG as control. The complete list of 16 UHRF2-enriched binding motifs is shown in supplemental Fig. S6A. The top ranked motif matches one of ZEB1-binding sequences with a significance score of 0.77 (Fig. 6A). This matching sequence contains a specific subclass of E-boxes (-CACCTG-). In addition to ZEB1, others TFs such as ZEB2, SNAI1/2 and TCF3 were also reported to bind E-box, which is located in the promoter region of CDH1 to repress the expression of E-cadherin (55–59). We next verified that UHRF2 was physically associated with CDH1 promoter by ChIP-qPCR assay. ChIP was performed using UHRF2 antibody and IgG in UHRF2 overexpression cells. The results showed a significant association of UHRF2 with CDH1 promoter which was not detected in the IgG control group (Fig. 6B). To verify the functional consequence of UHRF2 binding to CDH1 promoter, we quantified the RNA level of CDH1 in control and UHRF2 overexpressing cells by qPCR. As shown in Fig. 6C, CDH1 RNA was significantly repressed by more than 80%. We also measured several other EMT makers and found that epithelial gene TJP1 was also repressed whereas mesenchymal makers VIM and FN1 were not significantly changed (supplemental Fig. 6B). Taken together, we show that UHRF2 physically associates with the promoter region of CDH1 and may therefore function as a transcriptional co-regulator for its expression.
UHRF2 physically interacts with EMT-TFs
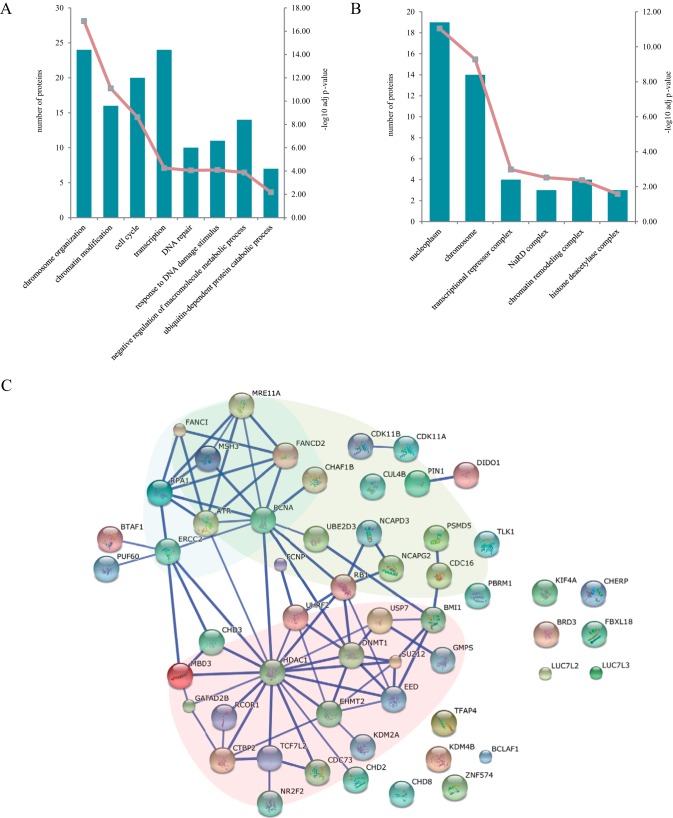
To gain more mechanistic insights into the action of UHRF2 in regulating the EMT process, we carried out IP-MS experiments to discover proteins that physically associate with UHRF2. We constructed UHRF2-OE stable cell lines in gastric cancer cell lines N87, MKN45, SGC7901 and MKN74 (supplemental Fig. S7A). Immunoprecipitations were done with nuclear extracts prepared from UHRF2-OE and their paired control cells. Fifty-four (54) proteins that were present in two out of four cell lines with unique peptide and at least fivefold higher PSM (peptide spectral match) values than the corresponding control experiment were considered as UHRF2-interacting proteins (supplemental Table S4). Among them, DNMT1, HDAC1, PCNA, EHMT2 (G9a), RB1, and PCNP are annotated as UHRF2 interactors in the STRING PPI database. USP7 is the most enriched interactor that was detected with 267 PSMs. This interaction was validated by the detection of UHRF2 in USP7 IP-MS data (supplemental Fig. S7B). GMPS, which was detected with 145 PSMs, was reported to form a complex with USP7 to catalyze deubiquitylation of histone H2B and stabilize the expression of p53 (60, 61). Other interactors include seven TFs, 25 CoRs, eight ubiquitin related proteins, five repair proteins, and two kinases. Notably, TCF7L2, which is also found to be up-regulated in catTFRE pull-down from UHRF2 overexpression cells, is among the 7 TFs. GO analysis demonstrates that main biological processes that the UHRF2 interactome involves in are chromosome organization/modification, cell cycle, transcription, DNA repair and ubiquitin-dependent protein catabolic process (Fig. 7A). Three subnetworks are derived from the known PPI which are mainly enriched in chromatin/histone modifications, cell cycle and DNA repair (Fig. 7C, Supplemental Fig. 7C). The complex analysis suggested the association of UHRF2 with some components of PRC1, PRC2, and NuRD complex (Fig. 7B, supplemental Fig. S7D). Overall, the IP-MS data revealed physical interaction of UHRF2 with several epigenetic regulation complexes, consistent with its known biological function; furthermore the identification of TCF7L2, a TF that regulates EMT, provides additional evidence for the involvement of UHRF2 in EMT processes.
Fig. 7.
Analysis of the UHRF2 interactome. A, GO enrichment analysis of UHRF2 interactors for biological process. The bar plot represents the proteins involved in each term and the numbers are displayed in left y axis. The -log10 p value of each function class are shown using line plot in right y axis. B, Cellular Component analysis of interactome. C, Protein interaction network of 54 UHRF2 interacting proteins were analyzed with STRING database. Three subnetworks which mainly enriched in chromatin/histone modifications, cell cycle and DNA repair are indicated by red, green and blue background respectively. Interactions shown have high confidence cutoff score of 0.7.
DISCUSSION
The innovative aspect of this work is to demonstrate the power of multidimensional proteomics analyses in the elucidation of UHRF2 functions “de novo”—without prior knowledge. One single proteomics measurement (for example, profiling), although is informative, may not be sufficient to provide clear cut directions to test for its functions; independent evidence obtained from a multitude of proteomics approaches (including catTFRE, ChIP-seq and IP-MS) that point to same direction can be integrated to generate a solid model. Here, we performed multidimensional proteomics measurements to uncover the function of UHRF2 “de novo.” Using UHRF2 overexpression cell lines, we first measured the altered proteomes and obtained clues for a role of UHRF2 in cell-cell adhesion. Through the analysis of proteome-wide TF DNA binding activities, we found that many EMT-TFs are up-regulated and their target gene products are among the group of proteins that are up and down regulated by UHRF2 overexpression. These data suggest that UHRF2 is a regulator for cell motility and the EMT program. Indeed, in vitro cell invasion experiments demonstrated that silence of UHRF2 in aggressive cells impaired their abilities of migration and invasion in vitro, supporting our molecular measurements.
We used additional unbiased, discovery-driven assays such as ChIP-seq and IP-MS to gain insight for the molecular mechanism of UHRF2 action. These data suggest that genomic binding motifs of UHRF2 coincide well with binding motifs of several TFs including EMT-TFs. The binding of UHRF2 to CDH1 promoter was validated by ChIP-qPCR measurements. The IP-MS identified TCF7L2 as a physical interacting protein with UHRF2 along with several protein complexes that regulate chromatin remodeling and histone modifications, suggesting that UHRF2 acts as transcription co-regulator together with TCF7L2 to promote EMT.
UHRF2 was found to interact with USP7 in IP-MS; together with the interaction with DNMT1 reported previously, it suggests that UHRF2 may also function in DNA methylation like its cousin UHRF1. Although UHRF2 and DNMT3 physical interaction was not observed under the condition where UHRF2 and DNMT1 interaction could be readily observed, UHRF2 overexpression led to decreased DNMT3 protein abundance in the profiling measurement. This indicates that the mode of interaction between UHRF2 and DNMT3 with that of DNMT1 is different. Because UHRF2 is also an E3 ligase, it raises an intriguing possibility that DNMT3 might be a substrate of UHRF2, which would predict a negative regulation of DNA methylation by UHRF2, in contrast to UHRF1, which plays a positive role.
It is clear that UHRF2 resides in the nucleus and binds to chromatin, which places UHRF2 in a position for regulating transcription. Although attention has been focused on its role in DNA methylation, our data raises a possibility of a direct role of UHRF2 in transcription regulation, specifically as a transcription co-regulator. It is conceivable that during tumor initiation, reprograming of the genomic DNA methylation through the action of DNMT3 plays an important part, but in tumor maintenance and metastasis it is not clear whether reprograming of genomic DNA methylation needs to be actively maintained. Then what is the function of UHRF2 in this process? We speculate that UHRF2 plays a role as a transcription coregulator in this regard.
TFs altered by UHRF2 overexpression in our data are notably related to tumor metastasis and the EMT process. TWIST2, TCF3 (also known as E47, E2A), TCF7L2 (also known as E2–2, TCF4), FOXC2, and TCF12 are all EMT-TFs and have central roles in the EMT progression (42, 44, 62). Their activation controls the expression of genes involved in cell polarity, cell-cell contact, cytoskeleton structure and extracellular matrix degradation and contributes to the repression of the epithelial phenotype and induction of the mesenchymal phenotype. TCF7L2, which was shown to interact with UHRF2 and was also detected with an increased DNA-binding activity in catTFRE pull-down, forms a complex with CTNNB1 and induces the EMT-activator ZEB1 to regulate tumor invasiveness (63).
Our exploratory proteomics studies of UHRF2 provide a starting point for better understanding the biological or pathological roles of UHRF2. Our experimental setting was in a gastric cancer cell line. If overexpression of UHRF2 could be detected in tumors, particularly metastasis gastric tumors, then our studies would implicate an important role of UHRF2 in tumor metastasis. In fact, UHRF2 has been reported up-regulated at both mRNA and protein levels in colon cancer tissues and positively correlated with clinical stage, depth of invasion, positive lymph node and metastasis. Patients with UHRF2 positive in tumors were associated with shorter survival (38, 39).
UHRF2 contains several protein domains that are associated with E3 ligase activity, hydroxyl-methyl-C DNA binding activity and H3K9me2/3 binding activity; it is not clear what the impact of these activities on EMT. It will be interesting to dissect the role of each individual activity in the future.
Supplementary Material
Footnotes
Author contributions: M. Lai and J.Q. designed research; M. Lai, L.L., and S.J. performed research; S.G. contributed new reagents or analytic tools; M. Lai, J.C., and N.Q. analyzed data; M. Lai, Y.W., and J.Q. wrote the paper; T.S. provided expertise on data analysis; B.Z., M. Liu, and C.D. provided expertise on experiments.
* This work is supported in part by National Program on Key Basic Research Project (973 Program, 2014CBA02002, 2012CB910300).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- EMT
- Epithelial to mesenchymal transition
- WCE
- whole cell extracts
- NE
- nucleus extracts
- TFRE
- transcription factor response elements
- sRP
- manual RP
- LC-MS/MS
- Liquid chromatography - tandem mass spectrometry
- FDR
- false discovery rate
- iBAQ
- intensity based absolute quantification
- FOT
- fraction of total
- LFQ
- label-free quantification
- TFs
- transcription factors
- CoRs
- coregulators
- GO
- gene ontology
- gRNA
- guide RNA
- q-PCR
- quantitative real time PCR
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1. Bronner C., Achour M., Arima Y., Chataigneau T., Saya H., and Schini-Kerth V. B. (2007) The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol. Therapeutics 115, 419–434 [DOI] [PubMed] [Google Scholar]
- 2. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T. A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., Tajima S., Mitsuya K., Okano M., and Koseki H. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 [DOI] [PubMed] [Google Scholar]
- 3. Bostick M., Kim J. K., Estève P.-O., Clark A., Pradhan S., and Jacobsen S. E. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 [DOI] [PubMed] [Google Scholar]
- 4. Hashimoto H., Horton J. R., Zhang X., Bostick M., Jacobsen S. E., and Cheng X. (2008) The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455, 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avvakumov G. V., Walker J. R., Xue S., Li Y., Duan S., Bronner C., Arrowsmith C. H., and Dhe-Paganon S. (2008) Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455, 822–825 [DOI] [PubMed] [Google Scholar]
- 6. Arita K., Ariyoshi M., Tochio H., Nakamura Y., and Shirakawa M. (2008) Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818–821 [DOI] [PubMed] [Google Scholar]
- 7. Qian C., Li S., Jakoncic J., Zeng L., Walsh M. J., and Zhou M. M. (2008) Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J. Biol. Chem. 283, 34490–34494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rottach A., Frauer C., Pichler G., Bonapace I. M., Spada F., and Leonhardt H. (2010) The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 38, 1796–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Citterio E., Papait R., Nicassio F., Vecchi M., Gomiero P., Mantovani R., Di Fiore P. P., and Bonapace I. M. (2004) Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 24, 2526–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karagianni P., Amazit L., Qin J., and Wong J. (2008) ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol. 28, 705–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nady N., Lemak A., Walker J. R., Avvakumov G. V., Kareta M. S., Achour M., Xue S., Duan S., Allali-Hassani A., Zuo X., Wang Y. X., Bronner C., Chedin F., Arrowsmith C. H., and Dhe-Paganon S. (2011) Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J. Biol. Chem. 286, 24300–24311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arita K., Isogai S., Oda T., Unoki M., Sugita K., Sekiyama N., Kuwata K., Hamamoto R., Tochio H., and Sato M. (2012) Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc. Natl. Acad. Sci. U. S. A. 109, 12950–12955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rothbart S. B., Krajewski K., Nady N., Tempel W., Xue S., Badeaux A. I., Barsyte-Lovejoy D., Martinez J. Y., Bedford M. T., Fuchs S. M., Arrowsmith C. H., and Strahl B. D. (2012) Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Structural Mol. Biol. 19, 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie S., Jakoncic J., and Qian C. (2012) UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail. J. Mol. Biol. 415, 318–328 [DOI] [PubMed] [Google Scholar]
- 15. Cheng J., Yang Y., Fang J., Xiao J., Zhu T., Chen F., Wang P., Li Z., Yang H., and Xu Y. (2013) Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J. Biol. Chem. 288, 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qin W., Leonhardt H., and Spada F. (2011) Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J. Cell. Biochem. 112, 439–444 [DOI] [PubMed] [Google Scholar]
- 17. Du Z., Song J., Wang Y., Zhao Y., Guda K., Yang S., Kao H.-Y., Xu Y., Willis J., and Markowitz S. D. (2010) DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signaling 3, ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mudbhary R., Hoshida Y., Chernyavskaya Y., Jacob V., Villanueva A., Fiel M. I., Chen X., Kojima K., Thung S., Bronson R. T., Lachenmayer A., Revill K., Alsinet C., Sachidanandam R., Desai A., SenBanerjee S., Ukomadu C., Llovet J. M., and Sadler K. C. (2014) UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 25, 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou L., Zhao X., Han Y., Lu Y., Shang Y., Liu C., Li T., Jin Z., Fan D., and Wu K. (2013) Regulation of UHRF1 by miR-146a/b modulates gastric cancer invasion and metastasis. FASEB J. 27, 4929–4939 [DOI] [PubMed] [Google Scholar]
- 20. Guan D., Factor D., Liu Y., Wang Z., and Kao H. Y. (2013) The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene 32, 3819–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bronner C., Krifa M., and Mousli M. (2013) Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis. Biochem. Pharmacol. 86, 1643–1649 [DOI] [PubMed] [Google Scholar]
- 22. Li X. L., Xu J. H., Nie J. H., and Fan S. J. (2012) Exogenous expression of UHRF1 promotes proliferation and metastasis of breast cancer cells. Oncol. Rep. 28, 375–383 [DOI] [PubMed] [Google Scholar]
- 23. Babbio F., Pistore C., Curti L., Castiglioni I., Kunderfranco P., Brino L., Oudet P., Seiler R., Thalman G. N., Roggero E., Sarti M., Pinton S., Mello-Grand M., Chiorino G., Catapano C. V., Carbone G. M., and Bonapace I. M. (2012) The SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene 31, 4878–4887 [DOI] [PubMed] [Google Scholar]
- 24. Unoki M., Daigo Y., Koinuma J., Tsuchiya E., Hamamoto R., and Nakamura Y. (2010) UHRF1 is a novel diagnostic marker of lung cancer. Br. J. Cancer 103, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Unoki M., Kelly J., Neal D., Ponder B., Nakamura Y., and Hamamoto R. (2009) UHRF1 is a novel molecular marker for diagnosis and the prognosis of bladder cancer. Br. J. Cancer 101, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Unoki M., Brunet J., and Mousli M. (2009) Drug discovery targeting epigenetic codes: the great potential of UHRF1, which links DNA methylation and histone modifications, as a drug target in cancers and toxoplasmosis. Biochem. Pharmacol. 78, 1279–1288 [DOI] [PubMed] [Google Scholar]
- 27. Mori T., Ikeda D. D., Yamaguchi Y., Unoki M., and Project N. (2012) NIRF/UHRF2 occupies a central position in the cell cycle network and allows coupling with the epigenetic landscape. FEBS Lett. 586, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 28. Zhang J., Gao Q., Li P., Liu X., Jia Y., Wu W., Li J., Dong S., Koseki H., and Wong J. (2011) S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 21, 1723–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spruijt C. G., Gnerlich F., Smits A. H., Pfaffeneder T., Jansen P. W., Bauer C., Munzel M., Wagner M., Muller M., Khan F., Eberl H. C., Mensinga A., Brinkman A. B., Lephikov K., Muller U., Walter J., Boelens R., van Ingen H., Leonhardt H., Carell T., and Vermeulen M. (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 [DOI] [PubMed] [Google Scholar]
- 30. Zhou T., Xiong J., Wang M., Yang N., Wong J., Zhu B., and Xu R. M. (2014) Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Mol. Cell 54, 879–886 [DOI] [PubMed] [Google Scholar]
- 31. Li Y., Mori T., Hata H., Homma Y., and Kochi H. (2004) NIRF induces G1 arrest and associates with Cdk2. Biochem. Biophys. Res. Commun. 319, 464–468 [DOI] [PubMed] [Google Scholar]
- 32. Mori T., Ikeda D. D., Fukushima T., Takenoshita S., and Kochi H. (2011) NIRF constitutes a nodal point in the cell cycle network and is a candidate tumor suppressor. Cell Cycle 10, 3284–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iwata A., Nagashima Y., Matsumoto L., Suzuki T., Yamanaka T., Date H., Deoka K., Nukina N., and Tsuji S. (2009) Intranuclear degradation of polyglutamine aggregates by the ubiquitin-proteasome system. J. Biol. Chem. 284, 9796–9803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mori T., Li Y., Hata H., and Kochi H. (2004) NIRF is a ubiquitin ligase that is capable of ubiquitinating PCNP, a PEST-containing nuclear protein. FEBS Lett. 557, 209–214 [DOI] [PubMed] [Google Scholar]
- 35. Qian G., Jin F., Chang L., Yang Y., Peng H., and Duan C. (2012) NIRF, a novel ubiquitin ligase, interacts with hepatitis B virus core protein and promotes its degradation. Biotechnol. Lett. 34, 29–36 [DOI] [PubMed] [Google Scholar]
- 36. Oh Y., and Chung K. C. (2013) UHRF2, a ubiquitin E3 ligase, acts as a small ubiquitin-like modifier E3 ligase for zinc finger protein 131. J. Biol. Chem. 288, 9102–9111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu J., Liu S., Liu G., Dombkowski A., Abrams J., Martin-Trevino R., Wicha M. S., Ethier S. P., and Yang Z. Q. (2012) Identification and functional analysis of 9p24 amplified genes in human breast cancer. Oncogene 31, 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu S., Yan D., Wu Z., Jiang T., Chen J., Yuan L., Lin J., Peng Z., and Tang H. (2014) Ubiquitin-like with PHD and ring finger domains 2 is a predictor of survival and a potential therapeutic target in colon cancer. Oncol. Reports 31, 1802–1810 [DOI] [PubMed] [Google Scholar]
- 39. Wang F., Zhang P., Ma Y., Yang J., Moyer M. P., Shi C., Peng J., and Qin H. (2012) NIRF is frequently upregulated in colorectal cancer and its oncogenicity can be suppressed by let-7a microRNA. Cancer Lett. 314, 223–231 [DOI] [PubMed] [Google Scholar]
- 40. Wu T.-F., Zhang W., Su Z.-P., Chen S.-S., Chen G.-L., Wei Y.-X., Sun T., Xie X.-S., Li B., Zhou Y.-X., and Du Z.-W. (2012) UHRF2 mRNA Expression is Low in Malignant Glioma but Silencing Inhibits the Growth of U251 Glioma Cells in vitro. Asian Pacific J. Cancer Prevention 13, 5137–5142 [DOI] [PubMed] [Google Scholar]
- 41. Spano D., Heck C., De Antonellis P., Christofori G., and Zollo M. (2012) Molecular networks that regulate cancer metastasis. Semin. Cancer Biol. 22, 234–249 [DOI] [PubMed] [Google Scholar]
- 42. Lamouille S., Xu J., and Derynck R. (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nature reviews. Mol. Cell Biol. 15, 178–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Puisieux A., Brabletz T., and Caramel J. (2014) Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 16, 488–494 [DOI] [PubMed] [Google Scholar]
- 44. De Craene B., and Berx G. (2013) Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 13, 97–110 [DOI] [PubMed] [Google Scholar]
- 45. Tam W. L., and Weinberg R. A. (2013) The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 19, 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aebersold R., and Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 47. Cravatt B. F., Simon G. M., and Yates J. R. Iii (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 48. Ding C., Jiang J., Wei J., Liu W., Zhang W., Liu M., Fu T., Lu T., Song L., Ying W., Chang C., Zhang Y., Ma J., Wei L., Malovannaya A., Jia L., Zhen B., Wang Y., He F., Qian X., and Qin J. (2013) A fast workflow for identification and quantification of proteomes. Mol. Cell. Proteomics 12, 2370–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ding C., Chan D. W., Liu W., Liu M., Li D., Song L., Li C., Jin J., Malovannaya A., Jung S. Y., Zhen B., Wang Y., and Qin J. (2013) Proteome-wide profiling of activated transcription factors with a concatenated tandem array of transcription factor response elements. Proc. Natl. Acad. Sci. U.S.A. 110, 6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van den Berg D. L. C., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., and Poot R. A. (2010) An Oct4-Centered Protein Interaction Network in Embryonic Stem Cells. Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., Davies S. R., Wang S., Wang P., Kinsinger C. R., Rivers R. C., Rodriguez H., Townsend R. R., Ellis M. J., Carr S. A., Tabb D. L., Coffey R. J., Slebos R. J., Liebler D. C., and Nci C. (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Atchley W. R., and Fitch W. M. (1997) A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 94, 5172–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Morris S. A., Cahan P., Li H., Zhao A. M., San Roman A. K., Shivdasani R. A., Collins J. J., and Daley G. Q. (2014) Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 158, 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cahan P., Li H., Morris S. A., da Rocha E. L., Daley G. Q., and Collins J. J. (2014) CellNet: network biology applied to stem cell engineering. Cell 158, 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Remacle J. E., Kraft H., Lerchner W., Wuytens G., Collart C., Verschueren K., Smith J. C., and Huylebroeck D. (1999) New mode of DNA binding of multi-zinc finger transcription factors: δEF1 family members bind with two hands to two target sites. EMBO J. 18, 5073–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., and de Herreros A. G. (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 57. Hajra K. M., Chen D. Y., and Fearon E. R. (2002) The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 62, 1613–1618 [PubMed] [Google Scholar]
- 58. Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., and van Roy F. (2001) The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 7, 1267–1278 [DOI] [PubMed] [Google Scholar]
- 59. Perez-Moreno M. A., Locascio A., Rodrigo I., Dhondt G., Portillo F., Nieto M. A., and Cano A. (2001) A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J. Biol. Chem. 276, 27424–27431 [DOI] [PubMed] [Google Scholar]
- 60. Reddy B. A., van der Knaap J. A., Bot A. G., Mohd-Sarip A., Dekkers D. H., Timmermans M. A., Martens J. W., Demmers J. A., and Verrijzer C. P. (2014) Nucleotide biosynthetic enzyme GMP synthase is a TRIM21-controlled relay of p53 stabilization. Mol. Cell 53, 458–470 [DOI] [PubMed] [Google Scholar]
- 61. van der Knaap J. A., Kumar B. R., Moshkin Y. M., Langenberg K., Krijgsveld J., Heck A. J., Karch F., and Verrijzer C. P. (2005) GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 17, 695–707 [DOI] [PubMed] [Google Scholar]
- 62. Lee C. C., Chen W. S., Chen C. C., Chen L. L., Lin Y. S., Fan C. S., and Huang T. S. (2012) TCF12 protein functions as transcriptional repressor of E-cadherin, and its overexpression is correlated with metastasis of colorectal cancer. J. Biol. Chem. 287, 2798–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sánchez-Tilló E., de Barrios O., Siles L., Cuatrecasas M., Castells A., and Postigo A. (2011) β-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc. Natl. Acad. Sci. U. S. A. 108, 19204–19209 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.