Abstract
Glycan macro- and microheterogeneity have profound impacts on protein folding and function. This heterogeneity can be regulated by physiological or environmental factors. However, unregulated heterogeneity can lead to disease, and mutations in the glycosylation process cause a growing number of Congenital Disorders of Glycosylation. We systematically studied how mutations in the N-glycosylation pathway lead to defects in mature proteins using all viable Saccharomyces cerevisiae strains with deletions in genes encoding Endoplasmic Reticulum lumenal mannosyltransferases (Alg3, Alg9, and Alg12), glucosyltransferases (Alg6, Alg8, and Die2/Alg10), or oligosaccharyltransferase subunits (Ost3, Ost5, and Ost6). To measure the changes in glycan macro- and microheterogeneity in mature proteins caused by these mutations we developed a SWATH-mass spectrometry glycoproteomics workflow. We measured glycan structures and occupancy on mature cell wall glycoproteins, and relative protein abundance, in the different mutants. All mutants showed decreased glycan occupancy and altered cell wall proteomes compared with wild-type cells. Mutations in earlier mannosyltransferase or glucosyltransferase steps of glycan biosynthesis had stronger hypoglycosylation phenotypes, but glucosyltransferase defects were more severe. ER mannosyltransferase mutants displayed substantial global changes in glycan microheterogeneity consistent with truncations in the glycan transferred to protein in these strains. Although ER glucosyltransferase and oligosaccharyltransferase subunit mutants broadly showed no change in glycan structures, ost3Δ cells had shorter glycan structures at some sites, consistent with increased protein quality control mannosidase processing in this severely hypoglycosylating mutant. This method allows facile relative quantitative glycoproteomics, and our results provide insights into global regulation of site-specific glycosylation.
Protein glycosylation is a highly conserved co- and post-translocational modification of proteins that influences protein folding, stability, solubility, and function (1, 2). N-glycosylation of Asparagine (Asn)1 residues occurs in eukaryota, archaea, and some bacteria, although the biosynthetic pathways and glycan structures are diverse in these organisms (3). In eukaryotes, nascent polypeptides in the Endoplasmic Reticulum (ER) are the protein acceptor substrates for N-glycosylation, as folded proteins cannot be efficiently N-glycosylated and N-glycosylation is critical for efficient protein folding. After protein folding, glycan structures can be further truncated or extended by Golgi-resident glycosyltransferases. Glycan biosynthesis is inherently inefficient, resulting in structural diversity of mature glycoproteins. Diversity in the presence or absence of glycans on glycoproteins is termed macroheterogeneity, whereas diversity in the structures of glycans at a specific site is termed microheterogeneity. This structural diversity is key for the regulation of many biological functions of glycoproteins.
Eukaryotic N-glycosylation is catalyzed in the ER lumen, where the enzyme oligosaccharyltransferase (OTase) transfers donor glycans en bloc from a dolichol pyrophosphate (DolP) carrier (Lipid-linked oligosaccharide; LLO) to selected Asn in nascent polypeptides. Approximately 80% of secretory proteins are N-glycosylated, making this modification of high fundamental, medical, and biotechnological relevance (4, 5). In Bakers' yeast Saccharomyces cerevisiae, the LLO is synthesized in a defined, sequential, stepwise process by a series of enzymes integral or attached to the ER membrane: Alg1–14 (Fig. 1A) (6). LLO biosynthesis begins on the cytosolic face of the ER membrane, where Alg7 and the Alg13-Alg14 complex transfer the first and second N-acetylglucosamine (GlcNAc) residues to DolP. This is followed by the sequential addition of Mannose (Man) residues by Alg1, Alg2, and Alg11 to synthesize the Man5GlcNAc2 structure. This LLO is then flipped into the ER lumen by a process requiring Rft1 (7). Man residues are then successively added by Alg3, Alg9, Alg12, and again Alg9 to construct the Man9GlcNAc2 structure (Fig. 1A). The LLO structure is completed by addition of three Glucose (Glc) residues to branch A through the sequential action of Alg6, Alg8, and Die2/Alg10 (Fig. 1A). This final Glc3Man9GlcNAc2 structure is the preferred LLO donor transferred to proteins by OTase, compared with biosynthetic intermediates. Because of the sequential nature of LLO biosynthesis, defects in LLO biosynthesis because of deficiency of an Alg enzyme result in accumulation of the precursor LLO structure (8–10). For instance, in alg6Δ yeast the unglucosylated Man9GlcNAc2 LLO structure accumulates and is transferred to protein (11). Importantly, OTase transfers these truncated LLOs to proteins with reduced efficiency, resulting in hypoglycosylation of diverse proteins (11). In particular, the terminal α1,2 Glc on the A branch of the LLO is critical for efficient in vivo glycosylation (12). The Man content of the B and C branches also influences OTase function (13). Thus, although the glycan normally transferred by OTase has a canonical structure, OTase can transfer a variety of truncated glycans to polypeptides. This capability of OTase can increase the glycan heterogeneity of mature proteins and ensures that the essential process of protein glycosylation can continue even with perturbation of the glycan biosynthetic pathway.
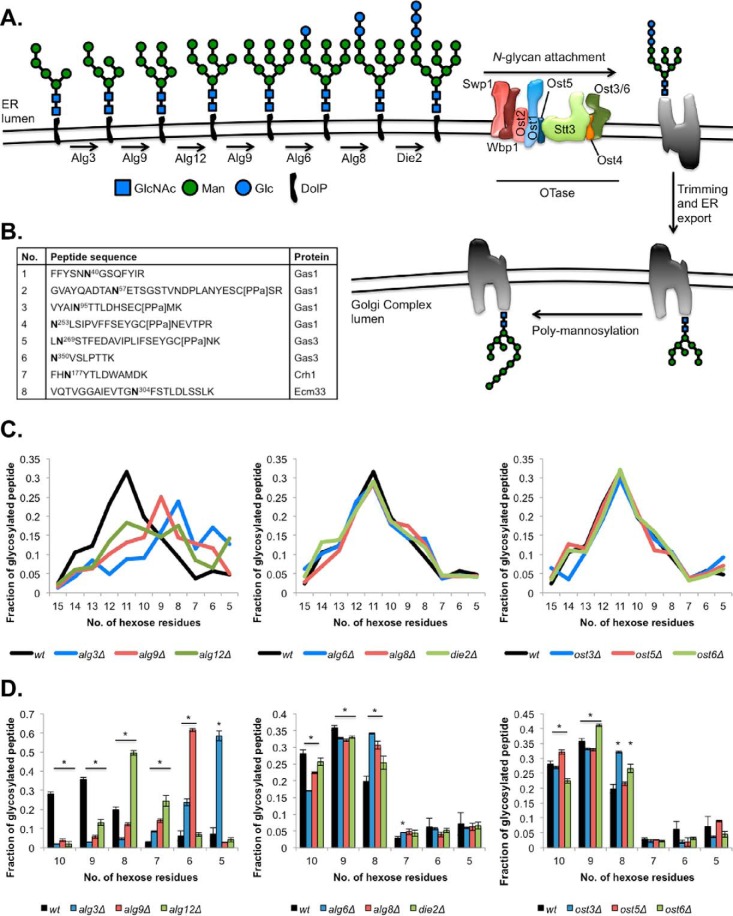
Fig. 1.
ER LLO glycosyltransferase defects alter glycan microheterogeneity on mature cell wall proteins. A, Schematic representation of the ER and Golgi lumenal glycan biosynthetic pathways. Monosaccharides are indicated in colors: blue squares, N-acetylglucosamine (GlcNAc); green circles, Mannose (Man); and blue circles, Glucose (Glc). The dolichol-pyrophosphate (DolP) lipid carrier to which the glycan is attached is shown in black. The glycan is transferred by oligosaccharyltransferase to an Asn in a sequon in a protein acceptor (gray). Glycans are processed in the ER by glucosidases and mannosidases (21), and the glycoprotein is then exported to the Golgi Complex, where α1,6 mannosyltransferases such as Och1 poly-mannosylate the glycan. B, Eight selected peptides used for the measurements presented in this figure. C, Average fraction of glycosylated peptides 1–8 containing glycans with 5–15 Man residues and 2 GlcNAc residues in each strain. D, Fraction of glycosylated Gas1p N40GS peptide containing glycans with 5–10 Man residues and 2 GlcNAc residues in each strain. Data is the mean ± S.E. of biological triplicates. * p < 0.05. Data from supplemental Table S2.
OTase catalyzes the transfer of glycans from LLO to selected Asn residues in nascent polypeptides. Asn are glycosylated with high efficiency if they are located in glycosylation sequons (NxS/T; x≠P), as this is the peptide acceptor-binding motif of Stt3, the catalytic subunit of OTase (14). The yeast OTase is a hetero-oligomeric complex composed of essential (Ost1, Ost2, Wbp1, Stt3, and Swp1) and nonessential (Ost3, Ost4, Ost5, and Ost6) subunits. There are two OTase isoforms in yeast containing one of either of the paralogous Ost3 or Ost6 subunits, which have different protein substrate specificity at the level of individual glycosylation sites (15–18). The function of the other OTase subunits is less well defined. However, lack of any of the nonessential subunits or mutations in essential subunits leads to inefficient N-glycosylation of diverse glycoproteins (15, 19, 20).
The structures of N-glycans are substantially processed after transfer to protein. Immediately after the glycan is transferred to nascent polypeptides by OTase, the terminal α1,2 Glc is trimmed by Glucosidase I, whereas the remaining Glc residues and some Man residues are removed during protein folding (21). N-glycans are further modified in a protein-, cell-, and species-specific manner as glycoproteins progress through the Golgi Complex. In yeast this results in high mannose N-glycans with extended polymannose structures (22, 23).
Despite the critical function of the presence and structure of N-glycans on key aspects of protein maturation, N-glycosylation by the OTase can proceed even with immature LLO structures. Remarkably, yeast is viable in the absence of genes encoding several Alg enzymes and OTase subunits (9). However, many reports have shown that these mutations result in inefficient glycosylation of diverse proteins (8, 11, 12, 20, 24–26). Here, we describe a simple, semi-quantitative, automated, and sensitive SWATH-mass spectrometry method to simultaneously measure macro- and microheterogeneity of diverse glycoproteins. We use this method to analyze the cell wall glycoproteome of the complete set of viable yeast deletion mutants in N-glycan biosynthesis to understand how OTase activity is affected by sub-optimal glycan donor structure at a systems level.
METHODOLOGY
Yeast Strains and Growth Conditions
All yeast strains used in this study derive from the BY4741 wild-type strain and were obtained from the genome deletion collection (Open Biosystems) (Table I). Yeast strains were grown in YPAD medium (1% yeast extract, 2% dextrose, 2% Bactopeptone) at 30 °C.
Table I. Strains used in this study.
| Name | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| alg3Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 alg3Δ∷KANMX | Open Biosystems |
| alg6Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 alg6Δ∷KANMX | Open Biosystems |
| alg8Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 alg8Δ∷KANMX | Open Biosystems |
| alg9Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 alg9Δ∷KANMX | Open Biosystems |
| die2Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 alg10Δ∷KANMX | Open Biosystems |
| alg12Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 alg12Δ∷KANMX | Open Biosystems |
| ost3Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ost3Δ∷KANMX | Open Biosystems |
| ost5Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ost5Δ∷KANMX | Open Biosystems |
| ost6Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ost6Δ∷KANMX | Open Biosystems |
Cell Wall Sample Preparation
Yeast were grown in 50 ml YPAD in an orbital shaker at 30 °C and 200 rpm, harvested at mid-log phase (OD600 nm 1.0) by centrifugation, and frozen at −20 °C. Proteins covalently linked to the polysaccharide cell wall were prepared following previously published protocols (15), but excluding endoglycosidase treatment. Briefly, cells were completely lysed, cysteines were reduced and alkylated with acrylamide, the insoluble cell wall polysaccharide with covalently attached proteins was thoroughly washed in strongly denaturing conditions, and proteins were digested with trypsin. Peptides and glycopeptides were desalted using C18 ZipTips (Millipore) prior to analysis by LC-ESI-MS/MS.
Mass Spectrometry and Data Analysis
Desalted peptides were analyzed by LC-ESI-MS/MS using a Prominence nanoLC system (Shimadzu) and TripleTof 5600 mass spectrometer with a Nanospray III interface (SCIEX) as previously described (27). Peptides were separated with buffer A (1% acetonitrile and 0.1% formic acid) and buffer B (80% acetonitrile with 0.1% formic acid) with a gradient of 10–60% buffer B over 45 min. Gas and voltage setting were adjusted as required. For information-dependent acquisition (IDA), an MS TOF scan from m/z of 350–1800 was performed for 0.5 s followed by IDA of MS/MS in high sensitivity mode with automated CE selection of the top 20 peptides from m/z of 40–1800 for 0.05 s per spectrum and dynamic exclusion of peptides for 5 s after 2 selections. Identical LC conditions were used for SWATH-MS, with an MS-TOF scan from an m/z of 350–1800 for 0.05 s followed by high-sensitivity information- independent acquisition with 26 m/z isolation windows with 1 m/z window overlap each for 0.1 s across an m/z range of 400–1250. Collision energy was automatically assigned by the Analyst software (SCIEX) based on m/z window ranges.
Peptide identification was performed essentially as previously described (27) using ProteinPilot 4.1 (SCIEX), searching the UniProt database (downloaded from http://uniprot.org on 15 January 2015; 16,818,973 sequences) with standard settings: sample type, identification; cysteine alkylation, acrylamide; instrument, TripleTof 5600; species yeast; ID focus, biological modifications; enzyme, trypsin; Search effort, thorough ID. False discovery rate analysis using ProteinPilot was performed on all searches with limits of 99% identification confidence and 1% local false discovery rate. The ProteinPilot search results were used as ion libraries for SWATH analyses. For glycopeptide analysis, ion libraries were manually created for each possible glycopeptide using fragment ions from the nonglycosylated version of each peptide and parent masses corresponding to various glycan structures ranging from GlcNAc2 to Man15GlcNAc2. Ion libraries used are included as Supporting Information (supplemental Table S1). The abundance of peptides was measured using PeakView Software with standard settings, summing the integrated areas of up to six fragment ions per peptide. Protein abundance was measured using the sum of the abundances of up to six peptides per protein. The accuracy of peak selection by PeakView was manually confirmed in each sample for all sequon-containing peptides. When detected, different charge states of sequon-containing peptides were considered independently. Glycopeptides were typically detected at +1 charge state relative to their corresponding unglycosylated version. Macroheterogeneity was measured by the fraction of the summed abundance of all glycosylated forms of a peptide to the summed abundance of all glycosylated and unglycosylated forms of that peptide. Microheterogeneity was measured by the fraction of a given glycosylated form of a peptide to the summed abundance of all glycosylated forms of that peptide. This allowed comparison of microheterogeneity between yeast strains independently of macroheterogeneity. All analyses were performed in biological triplicate. Statistical analyses for SWATH proteomics were performed using MSStats in R (27). Mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD003091.
Experimental Design and Statistical Rationale
All experiments using the wild-type strain as control and the 9 mutants (Table I) were performed in biological triplicates, as in previously published studies (15, 24, 27). Statistical analyses for SWATH-MS cell wall proteomics were performed using MSStats in R (27, 28). Statistical analysis of differences in micro- and macroheterogeneity were performed using t test in Microsoft excel. Comparisons with p < 0.05 were considered significant.
RESULTS
N-glycosylation is an enzymatic process catalyzed by the multiprotein subunit OTase enzyme. OTase displays preferences both for the acceptor protein to be glycosylated and the LLO glycan donor to be transferred to the protein (15, 29). Our goal was to quantify how differences in LLO structure affect the ability of OTase to glycosylate substrates in vivo and identify the repercussions of these changes on glycan macro- and microheterogeneity in mature cell wall proteins. To generate different LLO structures in vivo we utilized the full set of viable mutants in Alg enzymes responsible for the ER lumenal portion of the LLO biosynthetic pathway (Fig. 1A). The strains selected carry single mutations in genes encoding the ER lumenal mannosyltransferases (Alg3, Alg9, and Alg12) and glucosyltransferases (Alg6, Alg8, and Die2/Alg10) (Fig. 1A). Lack of these enzymes causes accumulation of the LLO precursor substrate of the enzyme, which is inefficiently transferred to proteins by OTase (8, 11, 12, 25, 26, 30). We focused our analysis on proteins covalently linked to the yeast cell wall, a well-defined subcellular fraction enriched in glycoproteins that has been previously extensively characterized by MS (glyco)proteomics (15, 24, 27, 31). To measure glycan macro- and microheterogeneity we designed and used a novel SWATH-MS glycoproteomic workflow to measure cell wall protein site-specific glycan structure and occupancy, and cell wall proteome abundance.
Defects in ER Mannosyltransferases lead to alterations in the structure of N-glycans on mature proteins
The structure of glycans on mature proteins can have profound impacts on glycoproteins' activity and stability. To be able to directly measure site-specific glycan occupancy and structure on peptides in a complex mixture we employed a variation of our previously published SWATH-MS glycoproteomic protocol (27), omitting the deglycosylation step to allow direct detection and relative quantification of glycopeptides. Peptide libraries of nonglycosylated but sequon-containing peptides were generated from Information Dependent Acquisition analysis of hypoglycosylating strains. Glycopeptides were not confidently identified by standard CID MS/MS and database searching, preventing automated identification and SWATH-MS quantification. To overcome this, we generated additional glycopeptide libraries containing experimentally determined fragment ions from nonglycosylated versions of sequon-containing peptides and parent ion masses corresponding to all theoretical glycopeptide masses. To allow use of the same fragment ions for glycosylated and nonglycosylated versions of the same peptide, we selected b- and y- ions that did not contain the glycosylated Asn residue. Although CID fragmentation of glycopeptides typically results in abundant glycan fragment ions, with few detectable peptide fragment ions (32, 33), we were able to use this approach to measure glycopeptides corresponding to eight glycosylation sites (Fig. 1, 2, and 3). Parent glycopeptide masses were constrained based on yeast glycan biosynthesis and instrument m/z detection limits. During N-glycosylation, OTase transfers a glycan with a maximum number of 14 monosaccharides (GlcNAc2Man9Glc3, Fig. 1A). All the Glc and some Man residues are trimmed during ER protein folding quality control, leaving glycans with a maximum of 11 monosaccharides (GlcNAc2Man9) as proteins exit the ER (21). If a protein is delayed in folding it will be retained in the ER and subjected to extensive Man trimming on the B and C branches, resulting in truncated glycan structures on the mature glycoprotein (21) (Fig. 1A). This means that proteins exiting the ER may contain a range of glycans with short B and C branches. These same branches are biosynthetically truncated in alg mutants (Fig. 1A). Secretory glycoproteins can be further mannosylated as they traffic through the Golgi complex (22). In yeast, Golgi polymannosylation of N-glycans occurs at branch A (34) (Fig. 1A). Because large glycopeptides may exceed the maximum detectable m/z for a given mass spectrometer instrument, the glycan size that can be measured is limited by the length of a particular glycopeptide. We therefore designed a matrix of glycopeptide masses considering that the glycans attached will have a Man0-XGlcNAc2 structure, where X was limited by the size of the particular peptide, up to a maximum total glycopeptide m/z of 1250, ranging from 9 to 15.
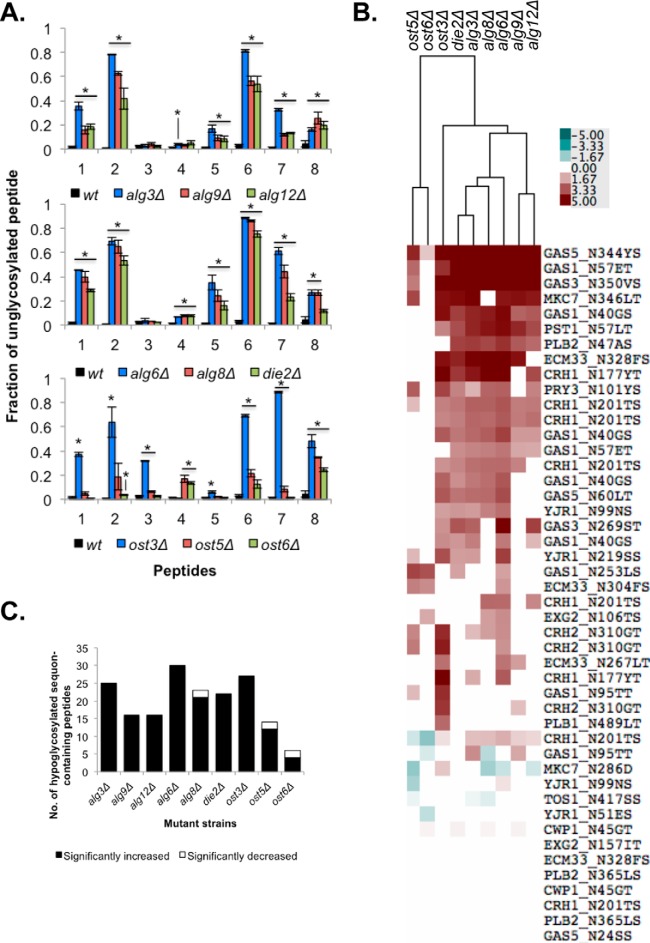
Fig. 2.
Early defects in LLO ER mannosylation or glucosylation lead to more severe hypoglycosylation defects. A, Fraction of unglycosylated peptide to the sum of un- and glycosylated forms of that peptide for each of the eight selected peptides from Fig. 1B in each strain. Data is the mean ± S.E. of biological triplicates. * p < 0.05. Data from supplemental Table S2. B, Heatmap showing the fraction of unglycosylated peptide to whole protein in each mutant strain relative to the wild-type strain. Each square represents the mean of biological triplicates. Only significant differences are displayed (p < 0.05). The data was clustered with Cluster 3.0 and presented using TreeView. Data from supplemental Table S3C). C, Number of sequon-containing peptides measured in each mutant strain with significantly (p < 0.05) increased (in white) or decreased (in black) glycosylation occupancy compared with wild type.
Fig. 3.

Simultaneous measurement of global site-specific glycan macro- and microheterogeneity. Heatmaps depicting the fraction of unglycosylated peptide (U) or peptide glycosylated with glycans containing two GlcNAc and an increasing number of Man (5–15), for each of the eight selected peptides in Fig. 1B. Fractions were calculated as in Figs. 1 and 2. The data was presented using TreeView. Each square represents the mean from biological triplicates. Data from supplemental Table S2.
We used our extended glycopeptide library in conjunction with SWATH-MS detection to determine the impact of defects in the LLO biosynthetic pathway on site-specific glycan structures of mature cell wall proteins. For these analyses, we selected eight peptides from four cell wall glycoproteins for which we could robustly detect glycopeptide microheterogeneity (Fig. 1B, supplemental Table S2). We then measured the effects of the alg mutations compared with the wild-type strain on the glycan microheterogeneity of the mature proteins. The glycopeptides detected in the wild-type strain had an average distribution of glycan structures ranging from 8 to 15 Man (Fig. 1C). Absence of any of the enzymes responsible for ER lumenal LLO mannosylation (Alg3, Alg9, or Alg12) had a profound effect on the Man content of glycans on mature glycoproteins (Fig. 1C). Deletion of any of the mannosyltransferases led to a decrease in the number of hexoses per glycan, consistent with the expected truncations in the LLO structures accumulating in these strains (Fig. 1A). This phenotype was more severe in the alg3Δ mutant than in the alg9Δ mutant, and in the alg9Δ mutant compared with the alg12Δ mutant (Figs. 1C, 1D, and 3). For example, for the Gas1 N40GS glycopeptide (Figs. 1B and 3) the Man9 glycan was the most abundant form in wild-type cells, Man8 in alg12Δ cells, Man6 in alg9Δ cells, and Man5 in alg3Δ cells (Figs. 1B, 1D, and 3). These results indicate that even after glycan extension in the Golgi complex, mannosyltransferase defects early in LLO biosynthesis lead to shorter glycan structures on mature glycoproteins (Figs. 1 and 3).
The nonessential OTase subunits Ost3 and Ost6 define two isoforms of OTase with different peptide acceptor substrate specificities, and deletion of the more abundant Ost3 paralog produces a substantially more severe hypoglycosylation defect than loss of Ost6 (15). Little is known about the precise function of Ost5, but this small protein links Stt3 to the other subcomplexes within the OTase multiprotein complex (20, 35) (Fig. 1A). Because mutations in the OTase are not likely to impact LLO biosynthesis, we predicted that glycans in the ost3Δ, ost5Δ, and ost6Δ strains would have similar structures to the wild-type strain. Indeed, the ost3Δ, ost5Δ, and ost6Δ strains showed the same average glycan structure distribution as wild-type cells (Fig. 1C). The same result was observed for the mutants in any of the three ER LLO glucosyltransferases (Fig. 1C). However, close inspection of site-specific glycan microheterogeneity revealed strain-specific differences at some glycosylation sites. For example, Man10 and Man9 glycans in Gas1 N40GS were significantly less abundant in alg6Δ, alg8Δ, and die2Δ cells than in wild type, whereas the Man8 glycan was significantly more abundant in these mutant cells at this site (Figs. 1D and 3). Together, this shows that the glycan structure at this site is significantly shorter in these mutant cells. A similar effect is apparent in ost3Δ cells at Gas1 N40GS (Fig. 1D), Gas1 N95TT, Gas1 N253LS, and Ecm33 N304FS (Fig. 3, supplemental Table S2).
Together, these data indicate that deletion of nonessential OTase subunits or the lack of efficient LLO glucosylation have site-specific effects on glycan structures on mature proteins, but these effects are minor compared with the defects caused by inefficient LLO mannosylation. Further, our data confirm that LLO with truncated B and C Man branches can be used as donors by OTase, and that although these glycans are still able to be extended with polymannose in the Golgi, they retain signatures of LLO truncation, resulting in shorter glycan structures attached to mature glycoproteins.
Mutations in the Glycan Biosynthetic Machinery Lead to Reduced Glycan Occupancy
Although OTase has a preference for Glc3Man9GlcNAc2 LLO, it is able to attach a variety of LLO structures to sequons, albeit inefficiently (Fig. 1) (6). To determine how different LLO structures affect the ability of the OTase to glycosylate its substrates in vivo, we determined the level of glycan occupancy at many sequons in cell wall proteins from the suite of alg and ost mutants (Table I). We performed this analysis of glycan macroheterogeneity in two different ways. In the first approach we used the data from the eight glycopeptides analyzed above for microheterogeneity (Fig. 1B). The fraction of each glycosylation site that was unglycosylated was calculated using the intensity of the unmodified peptide divided by the summed intensities of all detectable glycopeptides plus the unmodified peptide (Fig. 2A). This ratio gave a relative quantitative measurement of the level of hypoglycosylation in each strain. As expected, we observed that in the wild-type strain most sequons were efficiently glycosylated and the unglycosylated peptide:all peptide ratio was low (Fig. 2A). In contrast, all alg mutant strains showed a significant increase in the level of underglycosylation of at least six of the eight selected peptides compared with the wild-type strain. We observed that within mannosyltransferase mutants or within glucosyltransferase mutants, defects early in the biosynthetic pathway resulted in quantitatively more severe hypoglycosylation. For example, absence of the first (Alg3), second (Alg9), or third (Alg12) ER LLO mannosyltransferase resulted in underglycosylation of 78%, 63%, or 42% respectively at Gas1 N57ET (Fig. 2A, supplemental Table S2). Similarly, absence of the first (Alg6), second (Alg8), or third (Die2) ER LLO glucosyltransferase resulted in underglycosylation of 61%, 44%, or 23% respectively at Crh1 N177YT (Fig. 2A, supplemental Table S2). Thus, defects in both LLO mannosylation and glucosylation negatively impacted the efficiency of protein glycosylation by OTase.
To gain a more global perspective on the effect of alg mutations on macroheterogeneity, we compared glycan occupancy at all identifiable peptides containing sequons that were N-glycosylated in wild-type cells. Instead of comparing the unglycosylated peptide to the pool of glycosylated peptides for that glycosylation site (Fig. 2A), we calculated the ratio of the intensity of each unglycosylated peptide to the intensity of its respective whole protein as measured by SWATH-MS (27) (Figs. 2B and 2C and supplemental Table 3). Using this approach, glycosylation defects that lead to decreased glycan occupancy increase the unglycosylated peptide:protein ratio.
To determine how the alg mutations impact sequon occupancy, we measured 46 robustly detectable sequon-containing peptides belonging to 15 cell wall proteins (supplemental Table S3). In some cases, more than one sequon-containing peptide was identified per protein, and some of the peptides were detected with multiple charge states (supplemental Table S3). Although not strictly quantitative, the degree of hypoglycosylation measured by both our two approaches correlated well (supplemental Fig. S1). Again, most sequons were efficiently glycosylated in the wild-type strain, with low unglycosylated peptide:protein ratios (Figs. 2C and 3). Deletion of ER LLO mannosyltransferases significantly affected glycosylation of at least 1/3rd (19.0 ± 5.2 peptides) of the measured peptides compared with the wild-type strain (Figs. 2B and 2C). As observed above, deletion of ALG3 had the stronger phenotype, leading to the hypoglycosylation of 25 peptides, compared with 16 peptides for the mutants in ALG9 and ALG12 (Fig. 2C). Thus, accumulation of the Man5GlcNAc2 LLO led to a more severe hypoglycosylation defect than accumulation of the Man6–9GlcNAc2 LLOs. This result suggests that the ER LLO glucosyltransferases or OTase more efficiently recognize LLO structures with more Man in the B and C branches. All three mutants in the ER glucosyltransferases (alg6Δ, alg8Δ, and die2Δ) significantly affected the glycosylation levels of ∼ 1/2 (24.3 ± 4.9 peptides) of the measured peptides compared with the wild-type strain (Fig. 2C). Deletion of ALG6 showed the strongest phenotype, leading to the hypoglycosylation of 30 peptides, compared with 21 and 22 peptides for the mutants in ALG8 and DIE2, respectively (Fig. 2C). These results are in agreement with our previous data showing that defects in LLO biosynthesis lead to inefficient glycosylation by OTase (24). All together, these data indicate that the level of hypoglycosylation correlates with the severity of the LLO truncation—the fewer Man residues in branches B and C or Glc residues in branch A, the more severe the hypoglycosylation defect.
To gauge the relative severity of the alg glycosylation phenotypes, we measured sequon occupancy in mutants lacking the Ost3, Ost5, or Ost6 subunits of OTase. As expected, absence of any of the three OTase subunits led to a significant hypoglycosylation defect compared with the wild-type strain (Figs. 2 and 3). Deletion of OST3 showed the strongest hypoglycosylation phenotype in both approaches (Figs. 2 and 3). In the first approach, lack of Ost3, Ost5, or Ost6 led to a respective average 20.8×, 6.8×, or 3.75× increased abundance of the unglycosylated forms of the eight selected peptides compared with the wild-type strain (Fig. 2A). In agreement with previous results, absence of the Ost3 subunit led to the significant hypoglycosylation of 7/8 peptides, absence of Ost5 to 5/8 peptides, and of Ost6 to 3/8 peptides (15). As previously observed, the more abundant Ost3-OTase isoform was critical for glycosylation of all peptides except Gas1 N253LS, which is a preferential substrate of Ost6-OTase (15). Interestingly, this same site also required Ost5 for efficient glycosylation (Figs. 2A, 2B, and 3). We also observed that only deletion of OST3 or OST5, but not of OST6 or any of the alg genes, led to the significant hypoglycosylation of Gas1 N95TT (Figs. 2A and 2B). Similarly, our second approach showed that lack of Ost3 showed hypoglycosylation of 27 peptides, compared with 12 and 4 for the mutants in OST5 and OST6, respectively, when comparing unmodified peptides:protein (Figs. 2B and 2C). These data agree with previous work indicating that the Ost3 subunit has a more prominent role in N-glycosylation than Ost6 (15) and that deletion of OST5 leads to a milder glycosylation defect (20). The hypoglycosylation levels observed in the ost3Δ strain were similar to the levels observed in the alg3Δ and in the alg6Δ strains (Fig. 2). Collectively, these results indicate that defects in LLO mannosylation and LLO glucosylation led to defects in peptide glycosylation as severe as defects in select OTase subunits. Thus, overall OTase glycosylation efficiency is similarly affected by absence of the Ost3 subunit or by the sole availability of Man5GlcNAc2 and Man9GlcNAc2 LLO structures as donors.
Mutations at different stages in the glycan biosynthetic pathway or in the glycosylation machinery had a wide impact on the glycan macroheterogeneity of mature proteins. Although the effect of each mutation on each specific glycosylation site varied, mutations that prevented the first ER lumenal mannosyltransferase step (Alg3), the first glucosylation step (Alg6), or that eliminated Ost3 led to the largest hypoglycosylation phenotypes across many sites (Fig. 2 and 3). Mutants in OTase subunits showed clear site-specific hypoglycosylation (Figs. 2 and 3), consistent with previous studies showing that Ost3-OTase and Ost6-OTase have distinct protein substrate preferences at the level of individual glycosylation sites (15, 18, 36). In contrast, alg mutations led to a consistent pattern of underglycosylation across different sites (Figs. 2 and 3). That is, in general the same sites were qualitatively affected in the alg strains, albeit with different quantitative degrees of hypoglycosylation. It appears that this effect is driven by general protein acceptor substrate binding affinity to OTase, as hypoglycosylated sites in alg3Δ mutants were enriched in the less efficiently modified NxS sequons, compared with the more efficiently modified NxT sequons (p < 0.01, Fisher's exact test).
Defects in LLO Biosynthesis and Glycosylation Lead to Changes in the Cell Wall Proteome
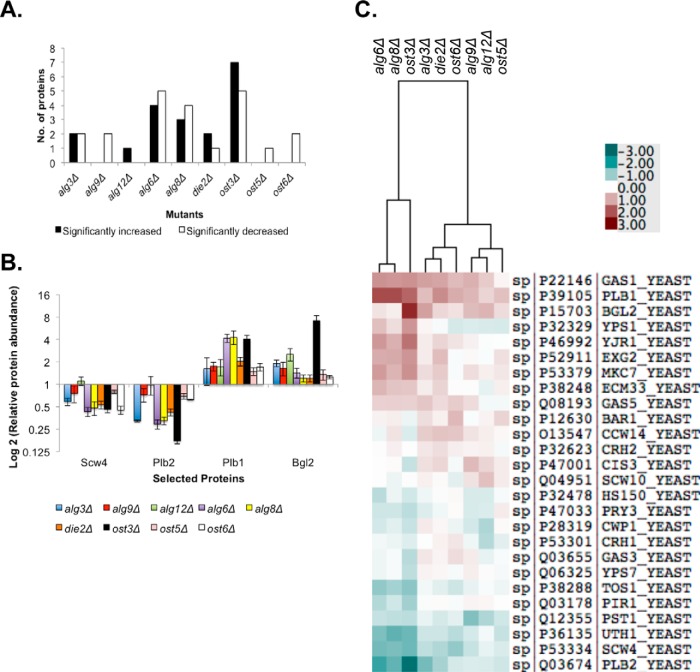
Global hypoglycosylation and alterations in glycan microheterogeneity would be expected to result in defects in glycoprotein folding, secretion, and function. To measure the severity of these effects we used the data from our SWATH-MS analyses to perform relative quantification of the abundance of cell wall proteins in our suite of alg and ost mutant strains. Changes in the cell wall proteome in these strains may be because of direct effects on the folding efficiency or stability of hypoglycosylated glycoproteins, or indirect effects compensating for cell wall damage because of defects in hypoglycosylated glycoprotein function. In either case, changes in the cell wall proteome can be used as a quantitative measure of the severity of defects in the glycosylation biosynthesis pathway. We selected peptides from 26 cell wall proteins that were confidently identified (Table II) and determined their relative abundance in each of the yeast strains (supplemental Table S4). The levels of 14 out of the 26 measured proteins were not significantly altered in any mutant strain compared with the wild type strain (at least twofold compared with wild-type, p < 0.05) (Table II and Fig. 4). The remaining 12 proteins showed either a significant increase in abundance (Bgl2, Gas1, Yps1, Plb1, Yjr1, Exg2, and Mkc7) (Table II, in bold, and Fig. 4B and 4C) or a significant decrease in abundance (Uth1, Tos1, Scw4, Plb2, and Pst1) in selected mutants (Table II and Fig. 4B and 4C). These results indicate that the cell wall proteome does not change uniformly in response to alterations in the glycosylation machinery.
Table II. Cell wall proteins and functions. In bold are proteins with significantly increased levels in mutant strains compared to the wild-type strain. * At least twofold difference compared to the wild-type strain, p < 0.05.
| Protein | Mutants affected* | Description |
|---|---|---|
| Gas1 | 5 | Glycosylphosphatidylinositol (GPI)-anchored beta-1,3-glucanosyltransferase; required for cell wall assembly |
| Plb1 | 4 | Phospholipase B (lysophospholipase) involved in lipid metabolism |
| Plb2 | 4 | Phospholipase B (lysophospholipase) involved in lipid metabolism |
| Scw4 | 4 | Cell wall protein with similarity to glucanases; paralog is SCW10 |
| Uth1 | 3 | Probable secreted beta-glucosidase; implicated in cell wall biogenesis, paralog is NCA3 |
| Bgl2 | 2 | Endo-beta-1,3-glucanase; major protein of the cell wall, involved in cell wall maintenance; involved in incorporation of newly synthesized mannoprotein molecules into the cell wall |
| Exg2 | 2 | Exo-1,3-beta-glucanase; involved in cell wall beta-glucan assembly; may be a (GPI) anchored protein |
| Mkc7 | 2 | GPI-anchored aspartyl protease; member of the yapsin family of proteases involved in cell wall growth and maintenance; paralog is YPS1 |
| Tos1 | 2 | Covalently-bound cell wall protein of unknown function; has sequence similarity to TOH1 |
| Toh1/Yjr1 | 2 | Also known as: JL171C, P46992. GPI-anchored cell wall protein; unknown function; sequence similarity to TOS1. |
| Yps1 | 1 | GPI-anchored aspartic protease; hyperglycosylated member of the yapsin family of proteases |
| Pst1 | 1 | Cell wall protein that contains a putative GPI-attachment site; up-regulated by activation of the cell integrity pathway, paralog is ECM33 |
| Bar1 | 0 | Aspartyl protease; secreted into the periplasmic space of mating type a cell |
| Ccw14 | 0 | Covalently linked cell wall glycoprotein; present in the inner layer of the cell wall |
| Cis3 | 0 | Mannose-containing glycoprotein constituent of the cell wall; member of the PIR (proteins with internal repeats) family |
| Crh1 | 0 | Chitin transglycosylase; functions in the transfer of chitin to beta(1–6) and beta(1–3) glucans in the cell wall; similar and functionally redundant to Utr2; expression induced by cell wall stress |
| Crh2 | 0 | Chitin transglycosylase; functions in the transfer of chitin to beta(1–6) and beta(1–3) glucans in the cell wall; similar to and functionally redundant with Crh1; GPI-anchored protein localized to bud neck |
| Cwp1 | 0 | Cell wall mannoprotein that localizes to birth scars of daughter cells; linked to a beta-1,3- and beta-1,6-glucan heteropolymer through a phosphodiester bond |
| Ecm33 | 0 | Paralog is PTS1 |
| Gas3 | 0 | Putative 1,3-beta-glucanosyltransferase; has similarity go other GAS family members; low abundance, possibly inactive member of the GAS family of GPI-containing proteins |
| Gas5 | 0 | 1,3-beta-glucanosyltransferase; has similarity to Gas1p; localizes to the cell wall |
| Hsp150 | 0 | O-mannosylated heat shock protein; secreted and covalently attached to the cell wall via beta-1,3-glucan and disulfide bridges; required for cell wall stability; paralog is PIR3 |
| Pir1 | 0 | O-glycosylated protein required for cell wall stability; attached to the cell wall via beta-1,3-glucan; expression regulated by the cell integrity pathway; paralog is YJL160C |
| Pry3 | 0 | Cell wall-associated protein involved in export of acetylated sterols |
| Scw10 | 0 | Paralog is SCW4 |
| Yps7 | 0 | Putative GPI-anchored aspartic protease; member of the yapsin family of proteases involved in cell wall growth and maintenance |
Fig. 4.
Hypoglycosylation causes major alterations in the cell wall proteome. A, Number of cell wall proteins that show a significant increase (black) or decrease (white) in abundance in each mutant strain compared with wild-type, as measured by SWATH-MS. B, Change in abundance of selected cell wall proteins in each strain relative to wild-type. Data is mean ± S.E. of biological triplicates. C, Clustered heatmap showing changes in abundance of cell wall proteins in each strain relative to wild-type. Each square represents the mean of biological triplicates. The data was clustered with Cluster 3.0 and presented using TreeView. Data from supplemental Table S4.
Each alg and ost mutant showed quantitatively different changes in cell wall proteome (Fig. 4). The largest differences were observed in the alg6Δ and alg8Δ glucosyltransferase mutants, and in the ost3Δ mutant, with a total of 9, 7, and 12 proteins with significant differences in abundance compared with wild type, respectively (Fig. 4). These mutants resulted in similar overall effects on cell wall protein abundance across the 27 proteins measured, and the proteomes of these mutants clustered in heatmap analysis (Fig. 4C). Consistent with severe hypoglycosylation (Fig. 2 and 3), these changes in cell wall proteome suggest that early defects in LLO ER glucosylation have particularly severe consequences for OTase function (Figs. 1 and 2). However, also as observed above, the ost3Δ mutant showed the most severe defects both in terms of the number of proteins affected and the intensity of the defect (Fig. 4). The alg3Δ, alg9Δ, alg12Δ, die2Δ, ost5Δ, and ost6Δ mutants showed smaller changes in cell wall proteome, with the alg3Δ mutant being the most severely affected of these (Fig. 4).
Changes in abundance of proteins with similar function, similar sequence, or encoded by paralogous genes were often inversely correlated in the different mutants. For example, the abundance of phospholipase B Plb1 was significantly increased in the alg6Δ, alg8Δ, and ost3Δ mutants compared with wild type, whereas the abundance of the phospholipase B Plb2 was significantly decreased in these mutants (Fig. 4B). Similarly, the abundance of Tos1 was significantly reduced in the alg6Δ and ost3Δ mutants, whereas the level of Toh1/Yjr1 (Tos One Homolog) was increased in these same strains (Fig. 4C). In other examples, Gas1 was significantly more abundant in the alg3Δ, alg6Δ, alg8Δ, die2Δ, and ost3Δ mutants, whereas Gas3 and Gas5 remained unchanged in these strains. Similarly, the abundance of Scw4 and Pst1 were significantly affected in at least one of the mutants tested, but the expression levels of their respective paralogs Scw10 and Ecm33 showed no significant change (Fig. 4B and 4C). These differences cannot be simply explained by the number of glycosylation sequons present in each protein. For example: Scw4 and Scw10 have both 1 sequon; Pst1 and Ecm33 are both heavily N-glycosylated; Tos1 has two sequons but Toh1/Yjr1 has 11; and whereas Gas1 has 10 sequons, Gas3 and Gas5 have 6. These results also indicate that the number of sequons in a protein does not predict how defects in the glycosylation machinery will affect the abundance of the mature protein.
DISCUSSION
Biosynthesis of the LLO to be transferred to protein by OTase occurs through a series of sequential enzymatic reactions with high substrate specificity (4, 6) (Fig. 1A). Lack of any given LLO glycosyltransferase leads to the accumulation of the product of the preceding step, and transfer of this incomplete glycan structure to protein by OTase (Fig. 1A) (30). Our analysis of glycan microheterogeneity in the suite of N-glycan biosynthetic mutants confirmed this model, and further demonstrated that defects in LLO mannosylation are retained in N-glycan structures on mature glycoproteins (Figs. 1 and 3). Although strains lacking functional ER LLO glucosyltransferases or OTase subunits showed no overall change in glycan microheterogeneity compared with wild type, some site-specific differences were apparent (Figs. 1D and 3, and supplemental Table S2). In particular, several glycosylation sites had shorter glycans with fewer Man residues in ost3Δ cells compared with wild type. As ost3Δ cells showed the most severe hypoglycosylation phenotype of any strain (Figs. 2, 3, and 4), glycoprotein folding was likely particularly compromised in these cells. This would increase glycoprotein residence in the ER during folding, and result in increased mannosidase trimming, producing the shorter glycan structures at some sites in ost3Δ cells. Alternatively, hypoglycosylation in ost3Δ cells may affect Golgi glycosyltransferase function and result in less efficient polymannosylation. Analysis of global site-specific glycan macro- and microheterogeneity may be a valuable tool to investigate details of glycoprotein quality control.
Inhibition of the early stages of the LLO biosynthetic pathway either genetically (by deletion of ALG1, ALG2, ALG7, ALG13, or ALG14) or chemically (with tunicamycin) severely impacts cell growth (37–39), whereas mutations later in the pathway have milder effects. We therefore expected that more severe hypoglycosylation phenotypes would be observed in the biosynthetically early viable alg mutants. Indeed, we found that lack of Alg3 led to a more severe hypoglycosylation phenotype and shorter glycans on mature proteins than lack of Alg9 and Alg12 (Figs. 1–3). Similarly, lack of Alg6 led to a more severe hypoglycosylation phenotype than lack of Alg8 or Die2 (Figs. 1–3). Surprisingly, however, deletion of the glucosyltransferases led to a more severe defect than deletion of the mannosyltransferases. These results suggest that LLO glucosylation can proceed even when the B and C branches of the LLO are incompletely mannosylated (Man4–9GlcNAc2) (Figs. 1B and 2A). This is consistent with previous reports that hypoglycosylation in alg3Δ cells can be rescued by overexpression of the Alg6 glucosyltransferase (6). These data are also in agreement with previous reports showing that the OTase can transfer a wide range of LLO glycans with diverse numbers of Man (Fig. 3) (9). However, the observation that less LLO mannosylation led to more severe hypoglycosylation suggests that the Man structures on the B and C branches do affect LLO recognition by the ER LLO glucosyltransferases and/or by OTase. Thus, recognition of the donor LLO glycan by the Alg enzymes and OTase appears to involve many aspects of LLO glycan structure besides the integrity of the A branch or the presence of the terminal Glc residue. Specifically, our results show that recognition of donor LLO substrate by OTase is based on a combination of all three A-branch glucoses, and B- and C-branch mannoses.
Mutations of any of the ER lumenal mannosyltransferases or glucosyltransferases caused a qualitatively similar pattern of hypoglycosylation (Fig. 2). That is, in general the same sequons were hypoglycosylated in the different alg mutants, but to quantitatively different extents (Fig. 2B). This hypoglycosylation phenotype was especially strong for NxS sequons, which have lower affinity for the OTase peptide-binding site (14). These results suggest that the OTase activity toward any glycosylation site is similarly affected by deviations from the canonical Glc3Man9GlcNAc2 LLO structure. This is in contrast to known defects in OTase, which show strong site-specificity.
Of the nonessential OTase subunits, deletion of OST3 led to the most severe hypoglycosylation (Fig. 2B). Deletion of OST6 on the other hand showed a minor hypoglycosylation phenotype, and resulted in hypoglycosylation of few sites, including the Gas1 N253LS sequon, a site previously observed to require Ost6, but not Ost3, for efficient glycosylation (15). Interestingly, this same sequon was also hypoglycosylated in the ost5Δ mutant. Lack of Ost5 also resulted in a limited effect on glycosylation, although its absence impacted glycosylation at more sequons than Ost6 (Fig. 2). As with Ost3 and Ost6, Ost5 is not a catalytic subunit of OTase, and it appears to be a small linker subunit tethering other OTase subunits. Ost5 appears to form a subcomplex with Ost1, and this subcomplex associates with the catalytic Stt3 subunit of OTase via Ost5 (20, 35). Ost5 is required for optimal OTase function in vitro, but OST5 deletion has no effect on the stability of Ost1 or Stt3 subunits, and has been previously reported to cause only a minor hypoglycosylation phenotype in yeast (20, 35). Our results here suggest a role for Ost5 in determining some aspect of site-specific glycosylation by OTase, as not all sites were equally affected by its deletion.
Inefficient protein glycosylation leads to ER stress and induction of the unfolded protein response. In yeast, glycosylation defects also lead to the activation of the cell wall integrity pathway, leading to alterations in the expression of certain cell wall proteins (40–46). Consistent with this, our SWATH-MS relative quantification of the cell wall proteome showed qualitatively consistent changes in protein abundance across the glycosylation mutants, but with larger changes in protein abundance in strains with severe hypoglycosylation defects (Figs. 2 and 4). Lack of glycosylation at specific sites can affect the folding and traffic of glycoproteins, and can lead to the premature degradation of hypoglycosylated folding intermediates (21, 47). However, we only measured mature proteins that had successfully trafficked to the cell wall. Importantly, our results nonetheless indicate that the majority of the mature proteins present at the cell wall in the alg and ost mutant strains were hypoglycosylated and may thus be partially functional or have an altered stability, in addition to changes in protein abundance. Glycosylation defects are likely to lead to similar global effects on glycoprotein macroheterogeneity, microheterogeneity, abundance, and function in other systems including human CDGs. Further, we observed that the strains with the most severe defects in microheterogeneity (the mannosyltransferase mutants) did not show the most severe hypoglycosylation defects across all strains nor the largest changes in cell wall proteome compared with wild type (Figs. 1–4). These results demonstrate that defects in microheterogeneity do not necessarily correlate with defects in macroheterogeneity or in the levels of mature proteins at their final destination. This emphasizes the importance of analytical approaches that consider macroheterogeneity, microheterogeneity, and protein abundance, as these have profound physiological effects and may fluctuate independently.
In this work we describe a sensitive SWATH-MS based glycoproteomic workflow that allows simultaneous characterization and relative quantification of global site-specific glycan macro- and microheterogeneity in complex protein mixtures. We have successfully used this approach to measure global changes in glycan macro- and microheterogeneity in the full set of viable yeast mutants in the N-glycosylation biosynthesis pathway. We also anticipate that the logic and rationale of the workflow we describe can be adapted to the study of other post-translational modifications.
Supplementary Material
Acknowledgments
We thank Julio J. Caramelo for helpfuldiscussions.
Footnotes
Author contributions: L.F.Z. and B.L.S. designed research; L.F.Z. and B.L.S. performed research; L.F.Z. and B.L.S. analyzed data; L.F.Z. and B.L.S. wrote the paper.
* This work was supported by Australian Research Council Discovery Project grant DP160102766 to BLS. LFZ holds a Post-doctoral Fellowship from CONICET, Argentina. BLS holds an Australian National Health and Medical Research Council RD Wright Biomedical (CDF Level 2) Fellowship APP1087975.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Asn
- Asparagine
- ER
- Endoplasmic Reticulum
- Glc
- Glucose
- GlcNAc
- N-acetyl-glucosamine
- LLO
- Lipid-linked oligosaccharide
- Man
- Mannose
- OTase
- Oligosaccharyltransferase
- Ser
- Serine
- SWATH
- Sequential window acquisition of all theoretical fragment ion spectra
- Thr
- Threonine.
REFERENCES
- 1. Schulz B. L. (2012) Beyond the sequon: sites of N-glycosylation. In: Petrescu S., ed. Glycosylation, pp. 21–40, Intech, Rijeka, Croatia [Google Scholar]
- 2. Schwarz F., and Aebi M. (2011) Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582 [DOI] [PubMed] [Google Scholar]
- 3. Spiro R. G. (2002) Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12, 43R–56R [DOI] [PubMed] [Google Scholar]
- 4. Aebi M. (2013) N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 [DOI] [PubMed] [Google Scholar]
- 5. Apweiler R., Hermjakob H., and Sharon N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 [DOI] [PubMed] [Google Scholar]
- 6. Burda P., and Aebi M. (1999) The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426, 239–257 [DOI] [PubMed] [Google Scholar]
- 7. Helenius J., Ng D. T., Marolda C. L., Walter P., Valvano M. A., and Aebi M. (2002) Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415, 447–450 [DOI] [PubMed] [Google Scholar]
- 8. Huffaker T. C., and Robbins P. W. (1983) Yeast mutants deficient in protein glycosylation. Proc. Natl. Acad. Sci. U.S.A. 80, 7466–7470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burda P., and Aebi M. (1999) The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426, 239–257 [DOI] [PubMed] [Google Scholar]
- 10. Cipollo J. F., and Trimble R. B. (2002) The Saccharomyces cerevisiae alg12delta mutant reveals a role for the middle-arm alpha1,2Man- and upper-arm alpha1,2Manalpha1,6Man- residues of Glc3Man9GlcNAc2-PP-Dol in regulating glycoprotein glycan processing in the endoplasmic reticulum and Golgi apparatus. Glycobiology 12, 749–762 [DOI] [PubMed] [Google Scholar]
- 11. Reiss G., te, Heesen S., Zimmerman J., Robbins P. W., and Aebi M. (1996) Isolation of the ALG6 locus of Saccharomyces cerevisiae required for glucosylation in the N-linked glycosylation pathway. Glycobiology 6, 493–498 [DOI] [PubMed] [Google Scholar]
- 12. Burda P., and Aebi M. (1998) The ALG10 locus of Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum: the terminal glucose of the lipid-linked oligosaccharide is required for efficient N-linked glycosylation. Glycobiology 8, 455–462 [DOI] [PubMed] [Google Scholar]
- 13. Izquierdo L., Mehlert A., and Ferguson M. A. (2012) The lipid-linked oligosaccharide donor specificities of Trypanosoma brucei oligosaccharyltransferases. Glycobiology 22, 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lizak C., Gerber S., Numao S., Aebi M., and Locher K. P. (2011) X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 [DOI] [PubMed] [Google Scholar]
- 15. Schulz B. L., and Aebi M. (2009) Analysis of Glycosylation Site Occupancy Reveals a Role for Ost3p and Ost6p in Site-specific N-Glycosylation Efficiency. Mol. Cell Proteomics 8, 357–364 [DOI] [PubMed] [Google Scholar]
- 16. Schwarz M., Knauer R., and Lehle L. (2005) Yeast oligosaccharyltransferase consists of two functionally distinct sub-complexes, specified by either the Ost3p or Ost6p subunit. FEBS Lett. 579, 6564–6568 [DOI] [PubMed] [Google Scholar]
- 17. Knauer R., and Lehle L. (1999) The oligosaccharyltransferase complex from Saccharomyces cerevisiae. Isolation of the OST6 gene, its synthetic interaction with OST3, and analysis of the native complex. J. Biol. Chem. 274, 17249–17256 [DOI] [PubMed] [Google Scholar]
- 18. Jamaluddin M. F., Bailey U. M., and Schulz B. L. (2014) Oligosaccharyltransferase subunits bind polypeptide substrate to locally enhance N-glycosylation. Mol. Cell Proteomics 13, 3286–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. te Heesen S., Knauer R., Lehle L., and Aebi M. (1993) Yeast Wbp1p and Swp1p form a protein complex essential for oligosaccharyl transferase activity. EMBO J. 12, 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reiss G., te Heesen S., Gilmore R., Zufferey R., and Aebi M. (1997) A specific screen for oligosaccharyltransferase mutations identifies the 9 kDa OST5 protein required for optimal activity in vivo and in vitro. EMBO J. 16, 1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zacchi L. F., Caramelo J. J., McCracken A. A., and Brodsky J. L. (2016) Endoplasmic Reticulum Associated Degradation and Protein Quality Control. In: Stahl R. B. a. P., ed. The Encyclopedia of Cell Biol., 1st Ed., pp. 596–611, Academic Press, Waltham, MA [Google Scholar]
- 22. Fabre E., Hurtaux T., and Fradin C. (2014) Mannosylation of fungal glycoconjugates in the Golgi apparatus. Curr. Opin. Microbiology 20, 103–110 [DOI] [PubMed] [Google Scholar]
- 23. Nakayama K., Nagasu T., Shimma Y., Kuromitsu J., and Jigami Y. (1992) OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 11, 2511–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bailey U. M., Jamaluddin M. F., and Schulz B. L. (2012) Analysis of congenital disorder of glycosylation-Id in a yeast model system shows diverse site-specific under-glycosylation of glycoproteins. J. Proteome Res. 11, 5376–5383 [DOI] [PubMed] [Google Scholar]
- 25. Frank C. G., and Aebi M. (2005) ALG9 mannosyltransferase is involved in two different steps of lipid-linked oligosaccharide biosynthesis. Glycobiology 15, 1156–1163 [DOI] [PubMed] [Google Scholar]
- 26. Runge K. W., and Robbins P. W. (1986) A new yeast mutation in the glucosylation steps of the asparagine-linked glycosylation pathway. Formation of a novel asparagine-linked oligosaccharide containing two glucose residues. J. Biol. Chem. 261, 15582–15590 [PubMed] [Google Scholar]
- 27. Xu Y., Bailey U. M., and Schulz B. L. (2015) Automated measurement of site-specific N-glycosylation occupancy with SWATH-MS. Proteomics 15, 2177–2186 [DOI] [PubMed] [Google Scholar]
- 28. Choi M., Chang C. Y., Clough T., Broudy D., Killeen T., MacLean B., and Vitek O. (2014) MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 [DOI] [PubMed] [Google Scholar]
- 29. Kelleher D. J., and Gilmore R. (2006) An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R [DOI] [PubMed] [Google Scholar]
- 30. Burda P., Jakob C. A., Beinhauer J., Hegemann J. H., and Aebi M. (1999) Ordered assembly of the asymmetrically branched lipid-linked oligosaccharide in the endoplasmic reticulum is ensured by the substrate specificity of the individual glycosyltransferases. Glycobiology 9, 617–625 [DOI] [PubMed] [Google Scholar]
- 31. Yin Q. Y., de Groot P. W., de Jong L., Klis F. M., and de Koster C. G. (2007) Mass spectrometric quantitation of covalently bound cell wall proteins in Saccharomyces cerevisiae. FEMS Yeast Res. 7, 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zacchi L. F., and Schulz B. L. (2015) N-glycoprotein macroheterogeneity: biological implications and proteomic characterization. Glycoconj J. 33, 359–376 [DOI] [PubMed] [Google Scholar]
- 33. Wang B., Tsybovsky Y., Palczewski K., and Chance M. R. (2014) Reliable determination of site-specific in vivo protein N-glycosylation based on collision-induced MS/MS and chromatographic retention time. J. Am. Soc. Mass Spectrom. 25, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakanishi-Shindo Y., Nakayama K., Tanaka A., Toda Y., and Jigami Y. (1993) Structure of the N-linked oligosaccharides that show the complete loss of alpha-1,6-polymannose outer chain from och1, och1 mnn1, and och1 mnn1 alg3 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 268, 26338–26345 [PubMed] [Google Scholar]
- 35. Mueller S., Wahlander A., Selevsek N., Otto C., Ngwa E. M., Poljak K., Frey A. D., Aebi M., and Gauss R. (2015) Protein degradation corrects for imbalanced subunit stoichiometry in OST complex assembly. Mol. Biol. Cell 26, 2596–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schulz B. L., Stirnimann C. U., Grimshaw J. P., Brozzo M. S., Fritsch F., Mohorko E., Capitani G., Glockshuber R., Grütter M. G., and Aebi M. (2009) Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc. Natl. Acad. Sci. U.S.A. 106, 11061–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Albright C. F., and Robbins R. W. (1990) The sequence and transcript heterogeneity of the yeast gene ALG1, an essential mannosyltransferase involved in N-glycosylation. J. Biol. Chem. 265, 7042–7049 [PubMed] [Google Scholar]
- 38. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Guldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kotter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., and Johnston M. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 39. Takatsuki A., Arima K., and Tamura G. (1971) Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J. Antibiot. 24, 215–223 [DOI] [PubMed] [Google Scholar]
- 40. Chen Y., Feldman D. E., Deng C., Brown J. A., De Giacomo A. F., Gaw A. F., Shi G., Le Q. T., Brown J. M., and Koong A. C. (2005) Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Cancer Res. 3, 669–677 [DOI] [PubMed] [Google Scholar]
- 41. Babour A., Bicknell A. A., Tourtellotte J., and Niwa M. (2010) A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell 142, 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arias P., Diez-Muniz S., Garcia R., Nombela C., Rodriguez-Pena J. M., and Arroyo J. (2011) Genome-wide survey of yeast mutations leading to activation of the yeast cell integrity MAPK pathway: novel insights into diverse MAPK outcomes. BMC Genomics 12, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scrimale T., Didone L., de Mesy Bentley K. L., and Krysan D. J. (2009) The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 20, 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li K., Ouyang H., Lu Y., Liang J., Wilson I. B., and Jin C. (2011) Repression of N-glycosylation triggers the unfolded protein response (UPR) and overexpression of cell wall protein and chitin in Aspergillus fumigatus. Microbiology 157, 1968–1979 [DOI] [PubMed] [Google Scholar]
- 45. Cullen P. J., Xu-Friedman R., Delrow J., and Sprague G. F. (2006) Genome-wide analysis of the response to protein glycosylation deficiency in yeast. FEMS Yeast. Res. 6, 1264–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arroyo J., Hutzler J., Bermejo C., Ragni E., Garcia-Cantalejo J., Botias P., Piberger H., Schott A., Sanz A. B., and Strahl S. (2011) Functional and genomic analyses of blocked protein O-mannosylation in baker's yeast. Mol. Micro. 79, 1529–1546 [DOI] [PubMed] [Google Scholar]
- 47. Aebi M. (2013) N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.