Abstract
Reconstructing genome history is complex but necessary to reveal quantitative principles governing genome evolution. Such reconstruction requires recapitulating into a single evolutionary framework the evolution of genome architecture and gene repertoire. Here, we reconstructed the genome history of the genus Lachancea that appeared to cover a continuous evolutionary range from closely related to more diverged yeast species. Our approach integrated the generation of a high-quality genome data set; the development of AnChro, a new algorithm for reconstructing ancestral genome architecture; and a comprehensive analysis of gene repertoire evolution. We found that the ancestral genome of the genus Lachancea contained eight chromosomes and about 5173 protein-coding genes. Moreover, we characterized 24 horizontal gene transfers and 159 putative gene creation events that punctuated species diversification. We retraced all chromosomal rearrangements, including gene losses, gene duplications, chromosomal inversions and translocations at single gene resolution. Gene duplications outnumbered losses and balanced rearrangements with 1503, 929, and 423 events, respectively. Gene content variations between extant species are mainly driven by differential gene losses, while gene duplications remained globally constant in all lineages. Remarkably, we discovered that balanced chromosomal rearrangements could be responsible for up to 14% of all gene losses by disrupting genes at their breakpoints. Finally, we found that nonsynonymous substitutions reached fixation at a coordinated pace with chromosomal inversions, translocations, and duplications, but not deletions. Overall, we provide a granular view of genome evolution within an entire eukaryotic genus, linking gene content, chromosome rearrangements, and protein divergence into a single evolutionary framework.
Eukaryotic genomes evolve through the accumulation of point mutations and chromosomal rearrangements that ultimately contribute to the evolution of the gene repertoire. Point mutations can promote gene inactivation by pseudogenization of coding sequences (Mighell et al. 2000; Lafontaine and Dujon 2010) but also participate in gene gain by de novo gene creation from noncoding sequences (Khalturin et al. 2009; McLysaght and Guerzoni 2015). Balanced rearrangements—including translocations, inversions, and chromosome fusion/fission—modify gene order and orientation. Although these rearrangements are often thought to occur mostly in intergenic regions (Peng et al. 2006; Poyatos and Hurst 2007; Berthelot et al. 2015), they have the potential to modify gene expression, create new gene combinations, and disrupt genes at their breakpoints (Rowley 1998; Perez-Ortin et al. 2002; Avelar et al. 2013; Quintero-Rivera et al. 2015). Unbalanced chromosomal rearrangements include deletions and duplications of the chromosome segments, which promote reduction and expansion of the gene repertoire, respectively (Llorente et al. 2000; Dujon et al. 2004; Wapinski et al. 2007; Butler et al. 2009; Souciet et al. 2009; Scannell et al. 2011; Gabaldon et al. 2013). Whole-genome duplication (WGD) and hybridization events also affect gene repertoire, as well-documented in yeasts (Semon and Wolfe 2007; Louis et al. 2012; Morales and Dujon 2012; Marcet-Houben and Gabaldon 2015). The impact of horizontal gene transfers (HGTs), although seemingly important in Pezizomycotina, is limited in Saccharomycotina, with only a few dozen reported events so far (Rolland et al. 2009; Galeote et al. 2010; Marcet-Houben and Gabaldon 2010; Wisecaver et al. 2014; Marsit et al. 2015).
Comparative genomics has been instrumental in identifying these mechanisms and deciphering their contribution to genome evolution. Notably, the study of synteny conservation across multiple species allowed critical conceptual advances in the understanding of genome dynamics. Comparative studies on synteny conservation revealed highly variable rates of chromosome rearrangements between individual lineages both in vertebrates and in yeasts (Bourque et al. 2005; Fischer et al. 2006). Interestingly, a comparative study between 12 Drosophila genomes reported that the disruption of synteny regions via chromosomal inversions approximated a linear process over time (Bhutkar et al. 2008). At a broader evolutionary scale, reconstructions of ancestral gene content in Proteobacteria and Archaea showed that gene losses and/or duplications correlated with amino acid substitution rates (Snel et al. 2002; Csuros and Miklos 2009). Similarly, linear correlations between the rates of genomic rearrangements such as gene duplications and losses, HGTs and gene creations, and the rate of nonsynonymous substitutions were recently reported in bacteria (Puigbo et al. 2014). By analogy to the traditional molecular clock (Zuckerkandl and Pauling 1962), the investigators coined the term “genomic clock” to describe the coordinated pace of fixation between point mutations and large-scale rearrangements. The first attempt to define a genomic clock in yeast was based on the correlation between synteny conservation and amino acid identity between orthologous genes (Rolland and Dujon 2011).
Reconstructing genome history is a rather difficult task requiring efficient reconstruction of ancestral genome organization and precise characterization of the chromosomal rearrangements that occurred along different lineages. Reconstruction of ancestral genome architecture has benefited from the development of several computational models (Ma et al. 2006; Faraut 2008; Muffato and Roest Crollius 2008; Alekseyev and Pevzner 2009; Ouangraoua et al. 2011; Gagnon et al. 2012). However, integrating the reconstruction of ancestral genome architecture into an evolutionary framework has only been achieved in a limited number of cases. In yeast, Wolfe and colleagues manually reconstructed the ancestral genome of Saccharomyces cerevisiae as it was before the WGD event and identified at least 144 structural rearrangements, as well as 124 genes that are present in the actual S. cerevisiae genome but absent from its ancestor (Gordon et al. 2009). These investigators also traced the complete rearrangement history of the Lachancea kluyveri genome since its common ancestor with S. cerevisiae (Gordon et al. 2011). In vertebrates, a recent study used ancestral genome reconstruction to explain the distribution pattern of rearrangement breakpoints in Boreoeutherian genomes (Berthelot et al. 2015). The investigators found a strong positive correlation between gene density and evolutionary rearrangement breakpoints and showed that this property could be extended to yeast genomes. Finally, Weng et al. (2014) recently reconstructed the ancestral genome organization of highly rearranged Geraniaceae plastid genomes and characterized the rearrangements unique to each genus. They found that the degree of plastid genome rearrangements was correlated with nonsynonymous substitution rates but not with synonymous substitution rates, compatible with the existence of a genomic clock in plastid genomes.
Based on genome comparison between three previously sequenced Lachancea species, we predicted that the number of rearrangements that reached fixation in this genus was sufficiently high, but not too high, to provide key information on the dynamics of chromosome evolution (Fischer et al. 2006; Payen et al. 2009; Souciet et al. 2009; Drillon and Fischer 2011; Gordon et al. 2011). Therefore, we undertook the reconstruction of genome history in this genus to seek for quantitative rules that govern the evolution of genomes. First, we sequenced, assembled, and annotated the genomes of seven additional Lachancea species. With 10 fully sequenced, assembled, and annotated genomes, the Lachancea clade is the most densely sampled yeast genus at the genomic level within the Saccharomycetaceae family. Second, we developed a new computational method called AnChro, to reconstruct ancestral genome organization and identify all balanced rearrangements that accumulated during evolution. We combined these reconstructions with an exhaustive survey of the gene repertoire and revealed general principles that govern genome evolution in this yeast genus.
Results
High-quality reference genomes of the Lachancea genus
We sequenced and assembled into one scaffold per chromosome the nuclear genomes of seven Lachancea species (see Methods). Haploid genome sizes range from 10.2 to 11.3 Mb (Fig. 1A). All Lachancea species have eight chromosomes, each containing one centromere with the three typical elements CDEI, CDEII, and CDEIII (Supplemental Fig. S1; Supplemental Table S1), except Lachancea fantastica that has only seven chromosomes because of a telomere-to-telomere fusion (Supplemental Fig. S2; Supplemental Table S2). The genomic GC content ranges from 41.2%–47.3% (Supplemental Table S2). In L. kluyveri, a region of 1 Mb containing the MAT locus and covering almost the whole left arm of chromosome C, has an average GC content of 52.9%, which is significantly higher than the 40.4% global GC content of the rest of the genome (Payen et al. 2009; Souciet et al. 2009). The orthologous counterpart of this chromosomal region is found in all other Lachancea species, but none of them presented the GC content heterogeneity characterized in L. kluyveri, reinforcing the hypothesis of an introgression event at the origin of this chromosomal arm (Friedrich et al. 2015).
Figure 1.
(A) Cumulative sequence length of the annotated genetic elements in the 10 Lachancea genomes. The percentages of protein-coding sequences are in parentheses. (B) Genome size in Lachancea positively correlates with intergene length (top) but not with cumulative gene length (bottom). (C) Phylogeny of 34 Saccharomycetaceae species inferred from a maximum likelihood analysis of a concatenated alignment of 756 families of syntenic homologs. The tree topology within the Lachancea clade remains identical for several reconstruction methods: concatenation tree, majority tree, and extended majority rule consensus (eMRC) tree (see Methods). Internal branches within the Lachancea clade are named b1 to b9. The corresponding internode certainty (IC) values, indicating the robustness of the eMRC tree topology, are given. (WGD) Whole-genome duplication. (D) Relationship between orthologous protein similarity and the number of conserved synteny blocks within different yeast genera. The Lachancea genus is the only clade showing a continuum of genome reorganization and pairwise protein similarities over a large evolutionary range.
We annotated the coding and noncoding elements of the seven newly sequenced genomes and reannotated the three previously sequenced genomes. Protein-coding genes range from 4997 in Lachancea meyersii to 5378 in L. kluyveri, and pseudogenes range from 52 in Lachancea cidri to 104 in Lachancea nothofagi (excluding Lachancea waltii where gaps in the original sequence [Kellis et al. 2004] artificially increase the number of pseudogenes to 295) (Supplemental Table S2). On average, coding sequences represent between 67% and 77% of the genome (Fig. 1A), similar to most Saccharomycotina sequenced genomes (Dujon et al. 2004). Finally, we found a small number of Class I retrotransposons (from one to 17) in all species except in L. cidri and L. meyersii, while Class II elements are more widespread, with at least one copy in L. cidri and Lachancea fermentati and up to 41 copies in L. fantastica (Supplemental Table S2; Sarilar et al. 2014).
We found no correlation between genome size and either the number of genes, the cumulative size of coding sequences, or the transposable element content. However, we found a clear positive correlation between genome sizes and the cumulative sizes of intergenic regions and introns (Fig. 1B; Supplemental Fig. S3). The largest size variation in intergenic regions equals a total of 1.38 Mb between L. fermentati and L. meyersii, showing that the differences between genome sizes are mainly due to variations in noncoding sequence length and not to differential gene or transposable element content. Interestingly, other studies also reported genome size changes targeted toward intergenic regions. A decrease in gene density with increasing genome size was observed in Ascomycota genomes, (Kelkar and Ochman 2012), and a similar correlation was observed for 81 Saccharomycotina mitochondrial genomes (Freel et al. 2015b).
A robust species tree to reliably reconstruct genome history
Establishing a robust species phylogenetic tree is a crucial prerequisite to any evolutionary reconstruction. The phylogenetic position of the Lachancea clade was first inferred from a maximum likelihood analysis of a concatenated alignment of 756 families of syntenic homologs (see Methods) shared by 36 species (Supplemental Table S3). The resulting tree shows that all Lachancea species share a monophyletic origin, supporting the existence of the genus (Fig. 1C).
We further reconstructed all gene trees for the 3598 sets of orthologs present in the 10 Lachancea species and in S. cerevisiae (see Methods). A total of 796 different topologies were observed among the 3598 individual gene trees. The most prevalent topology was shared by 472 gene trees (majority topology). The same topology was systematically retrieved from the concatenations of the corresponding 472 alignments, the 3598 alignments, or the 756 families of syntenic homologs (Supplemental Fig. S4). We also showed that the extended majority rule consensus (eMRC) tree (Felsentein 1995) topology was identical to both the concatenation and the majority topologies (see Methods). We applied the internode certainty (IC) measure (Salichos and Rokas 2013; Salichos et al. 2014) to estimate the robustness of the eMRC topology. Six of eight internal branches have good supporting values (IC higher than 0.8) (Fig. 1C). Branches b4 and b8 have the lowest IC values (0.47 and 0.22, respectively); however, their corresponding bipartitions are 7.2 and 3.2 times more frequent than their second most prevalent bipartitions, indicative of a weaker but still exploitable phylogenetic signal (Fig. 1C). Altogether these analyses show that the Lachancea phylogeny is robust and reliable for genome history reconstruction.
HGTs and gene creation events contributed to the gene repertoire evolution
We characterized 24 events of putative HGT that correspond to a total of 85 coding sequences (CDS) in Lachancea (0.2% of the CDS and pseudogenes; see Methods). Twenty-three events are novel compared to previously reported HGT cases (Rolland et al. 2009; Morel et al. 2015). The 24 HGT families are similar to proteins from Pezizomycotina species (10 cases), from other eukaryotes (three cases) and from bacteria (11 cases) (Supplemental Table S4). The phyletic patterns show that eight HGTs are common to several Lachancea species; therefore, the transfers would have happened along internal branches of the tree (Fig. 2). Nine HGT families are similar to proteins of unknown function; three are similar to proteins involved in DNA metabolism, i.e., a transcription regulator (pseudogene), an endonuclease, and a serine recombinase previously described (Rolland et al. 2009); and the 12 others are similar to proteins with catalytic activities mostly belonging to oxidation-reduction processes (Supplemental Table S4). Interestingly, homologs of the polysaccharide lyase family 3 of the phytopathogen fungus Botrytis cinerea (noble rot fungus) are present in L. fantastica, L. meyersii, Lachancea thermotolerans, and L. waltii (Family ID 4751) (Supplemental Table S4). Polysaccharide lyases are mostly found in phytopathogens because they catalyze the eliminative cleavage of pectin, which is a major component of the primary cell wall of many plants. L. fantastica, L. waltii, and L. thermotolerans were isolated from plant-associated habitats, while L. meyersii was isolated from seawater. Consistently with ecological distribution, the homolog in L. meyersii is highly diverged and only partially similar to the members in the three other species. Overall, our study suggests that the contribution of HGTs on the evolution of the gene repertoire in yeast is probably underestimated.
Figure 2.
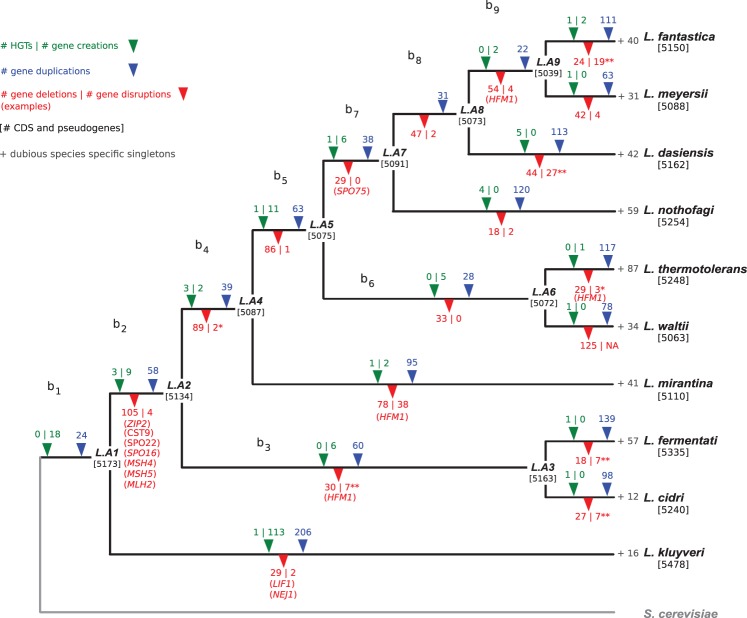
Evolution of the Lachancea gene repertoire. The number of gene duplications (in blue) and losses (in red) were estimated on each branch of the tree under a birth–death evolutionary model (Methods). The total number of gene losses indicated on the figure (1036) comprises the 107 cases of dubious loss (see text). The statistical significance of the enrichment of gene losses associated to rearrangements compared to the proportion of genes associated to rearrangements is indicated by an asterisk (P < 0.05) or double asterisk (P < 10−4). Notable examples of gene losses are in parentheses below their corresponding branches. The phyletic patterns of the 51,110 CDS and 1018 pseudogenes were used to map HGTs and gene creation events (in green) in the different branches of the tree (Methods). The number of species-specific singletons is in gray at the tip of each terminal branch. The total number of genes in each ancestral genome and the number of genes and pseudogenes in extant species are indicated between squared brackets.
We also characterized 596 taxonomically restricted gene (TRG) families that are specific to the Lachancea clade without any detectable homolog in the nonredundant sequence database or conserved domain in the PFAM database (see Methods). Sixty-six TRG families (encompassing 316 CDS and four pseudogenes) comprise members in at least two different Lachancea species and/or several paralogs within a given Lachancea species (Supplemental Table S5). Therefore, they could result from de novo gene formation events that occurred in the Lachancea clade. Their phyletic patterns were used together with a birth–death–innovation evolutionary model (see Methods) to map these events on the Lachancea tree (Fig. 2). The evolutionary rates for these 66 TRG families are generally above the median evolutionary rate of the set of orthologous genes (see Methods) but remain within the distribution, suggesting that no remote homolog would have been missed because of unusually high divergence. Four families show a nonsaturated rate of synonymous substitutions (dS < 1), and all of them have a mean pairwise ratio of nonsynonymous over synonymous substitution rates of dN/dS < 1, suggesting that they could be under purifying selection (Supplemental Table S5).
The remaining 530 singletons bear the usual characteristics of TRG (Khalturin et al. 2009); they globally have a lower GC content, a smaller size, and a lower codon adaptation index (CAI) value than orthologous gene sequences. With 131 predicted genes, the L. kluyveri branch encompasses the highest number of species-specific singleton genes. We used the available population genomic data (Friedrich et al. 2015) to check whether these CDS are conserved between the genomes of L. kluyveri isolates we recently sequenced. We found that 114 CDS have homologs conserved in several strains and, therefore, probably correspond to real genes. The remaining 17 CDS are absent or pseudogenized in all other sequenced genomes, suggesting that these genes should be considered dubious. For the other nine species for which no population data are available, all 403 species-specific singletons are also presently considered as dubious genes. Altogether, the nonvertically inherited genes in Lachancea would result from a minimum of 24 HGT and 159 gene creation events, which have enriched the genus’ gene repertoire.
The genus Lachancea covers a unique continuous evolutionary range in Saccharomycotina
The number and size of conserved synteny blocks between Lachancea species reveal that they share a continuum of intermediate levels of genome reorganization, ranging from highly collinear genomes down to significantly reordered chromosome maps (Fig. 1D). This continuous range of relatedness is also recognizable through the pairwise protein similarities shared between Lachancea orthologs, ranging from 69%–83% (Fig. 1D). More importantly, divergence in the genus Lachancea, in both terms of protein sequences and chromosome reorganization, remains below the thresholds beyond which the accumulation of too many mutations and rearrangements leads to the progressive loss of detectable synteny blocks, which prevents any reliable reconstruction of genome history (Drillon and Fischer 2011). Such continuous evolutionary range is so far unique among sequenced Saccharomycotina species (Fig. 1D) and makes the genus Lachancea an ideal candidate for the evolutionary reconstruction of genome history.
AnChro, a new computational tool to reconstruct ancestral genome architecture
We developed a new computational method of ancestral genome reconstruction named AnChro. This tool is part of an integrated suite of software named CHROnicle (freely available at www.lcqb.upmc.fr/CHROnicle/). Briefly, in the first step of the reconstruction, SynChro identifies conserved synteny blocks between pairwise combinations of genomes (Drillon et al. 2014). In the second step, ReChro constructs cycles of linked breakpoints between adjacent synteny blocks, and in the last step, AnChro infers the ancestral gene order by comparison with external reference genomes (see Supplemental Information).
There are nine ancestral genomes in total in the Lachancea phylogenetic tree (named L.A1 to L.A9) (Figs. 2, 3). Genome reconstructions for these nine ancestors resulted in eight ancestral genomes composed of eight chromosomes and in one composed of nine scaffolds, probably because one ancestral adjacency was not reconstructed (Fig. 3, L.A4). The number of genes per ancestral genome varies between 4446 for L.A1 and 4799 for L.A9 (Fig. 3). Each ancestral genome is provided as a list of ordered ancestral genes with their corresponding orthologous genes in all 10 extant Lachancea species (Supplemental Table S6). The robustness of AnChro’s reconstructions was comprehensively tested by (1) calculating the probability of reconstructing ancestral genome organization with a single centromere by chromosome, given that AnChro does not use any information of centromere position to reconstruct ancestral genome organization (see Supplemental Information); (2) comparing the reconstructions to previously published ancestral genomes (Fig. 4; Supplemental Information; Gordon et al. 2009, 2011; Jones et al. 2012); and (3) benchmarking AnChro against four existing reconstruction software tools on both real and simulated genome data sets (Fig. 4; Supplemental Information). All these tests showed that AnChro achieved the most reliable and complete reconstruction of ancestral chromosome architecture.
Figure 3.
Chromosomal history of the Lachancea genomes. The chromosomal structures of the 10 extant species and the ancestral genomes L.A2 to L.A9 are represented as a function of the genome structure of L.A1, the last common ancestor of the clade. The number of genes of each ancestral genome is indicated with brackets. The total number of translocations and inversions accumulated between two genomes is indicated above each branch. Rearrangements involving MAT, HML, HMR, rDNA, or centromeres are indicated below each branch. The relocation of the rDNA array occurred in the branch between L.A4 and L.A5. This transposition event occurred intra-chromosomally from an ancestral site, represented as a purple region in L.A4, to a new genomic location close to the green centromere in L.A5. The relative orientation of the rDNA and the centromere was inverted between L.A4 and L.A5, suggesting that the rDNA relocation resulted from two intra-chromosomal inversions involving one breakpoint reuse (Supplemental Fig. S5; Supplemental Table S6). The interval between MAT and HML was never broken and was inherited intact from L.A1 in all extant species except L. kluyveri, which lost both HML and HMR. The HMR cassette underwent many rearrangements (three translocations, one inversion, and one duplication) but always remained subtelomeric. The HML, HMR, and the MAT loci were located on the same chromosome in the last common ancestor of the genus, L.A1, with one silent cassette at each chromosome end. The only Lachancea species that also harbors the three sexual loci on a single chromosome is L. fermentati, but this organization is not inherited from L.A1 as it results from additional translocations in the branch between L.A3 and L. fermentati. Therefore, none of the present-day Lachancea species has retained the original chromosomal organization of the sexual loci. The inset plot recapitulates the total number of translocations and inversions that accumulated since each extant species diverged from the last common ancestor, L.A1.
Figure 4.
(A) Comparison between the two versions of the L.A1 ancestral genome reconstructed by AnChro and by ANGES (Jones et al. 2012). Chromosome painting representations of ancestral genomes are colored relatively to the L. kluyveri chromosomes. The black triangles indicate the same 12 ancestral adjacencies that resulted from six translocations identically reconstructed by the two tools. (B) Comparison between the manually reconstructed (Gordon et al. 2009, 2011) and the AnChro version of the pre-WGD ancestral genome relative to the L. kluyveri genome. The only inter-chromosomal difference between the two reconstructions is indicated by the red triangles. (C) Comparison of the nine Lachancea ancestors (L.A1 to L.A9) reconstructed by AnChro, ANGES, GapAdj, MGRA, and PMAG+. Synteny blocks were computed with I-ADHoRe for the five reconstruction tools. For AnChro, a single default reconstruction is presented. Each column represents the ancestral chromosomes of a given ancestor as an alternation of gray and beige boxes, with size being proportional to the number of reconstructed ancestral genes. The small black circles indicate the centromere position. The small red circles indicate the centromere positions when an ancestral chromosome was reconstructed with two centromeres. (D) Ancestral genome reconstructions on simulated genomes. The figure presents 900 reconstructed ancestral genomes corresponding to nine different ancestors per simulation and 100 simulations, performed with AnChro (default reconstructions), ANGES, GapAdj, MGRA, and PMAG+. Each genome reconstruction is represented by a dot. The quality of the reconstructions is assessed by three criteria: the number of ancestral scaffolds (ideally eight), the number of ancestral genes (ideally 5000), and the proportion of scaffolds per reconstruction with a single centromere (ideally 100%). (E) Average proportions of correctly and incorrectly reconstructed adjacencies for the 900 reconstructions obtained by the five tools. Incorrect adjacencies are decomposed in single block inversions and intra- and inter-chromosomal contradictions. The average proportion of adjacencies that were not reconstructed by the different software is also indicated.
Unbalanced rearrangements outnumbered balanced rearrangements
We performed two independent analyses to identify both balanced chromosomal rearrangements, i.e., translocations, inversions, and chromosome fusion/fission, and unbalanced rearrangements, i.e., duplications and deletions, that occurred since divergence from the last common ancestor of the genus.
To identify balanced rearrangements, we used SynChro (Drillon et al. 2014) to compute the synteny blocks between consecutive ancestral genomes in internal branches of the tree and between an ancestor and an extant genome in the terminal branches. The SynChro stringency parameter was set to zero to allow building synteny blocks comprising a single inverted orthologous gene-pair (see Supplemental Information). These blocks subsequently served as inputs for ReChro to identify all the balanced rearrangements that occurred in each branch of the tree, including single gene inversions. We identified a total of 423 balanced rearrangements (Fig. 3). The number of rearrangements accumulated between L.A1 and the different Lachancea species was highly variable, from 22 in L. kluyveri up to 152 in Lachancea dasiensis (Fig. 3, inset plot). These rearrangements correspond to 136 inversions, including nine with endpoints at telomeres; 147 translocations, including 140 reciprocal translocations; seven telomeric nonreciprocal translocations; and 140 rearrangements for which it was not possible to discriminate between inversions and translocations because of breakpoint reuse. We identified 102 cases of inversion corresponding to individual chromosomal events with no overlap or breakpoint reuse with other rearrangements. The size distribution of these 102 inversions fits a power law, clearly showing that small inversions are favored over longer ones (Fig. 5A). Only two very large inversions of 318 and 351 genes were found.
Figure 5.
(A) Distribution of inversion sizes (in number of genes) accumulated since the divergence from the last common ancestor of the genus (L.A1). The red line symbolizes a power law fit to the data (, with C = 106.4 and α = 1.86), which represents the probability of an inversion having its two end-points at s genes apart. The inset plot shows a cumulative histogram of the same data plotted with logarithmic scale. (B) Correlations between the number of genes and pseudogenes in extant genomes and number of balanced rearrangements, i.e., inversions and translocations (left y-axis) and with the numbers of gene losses, i.e., gene disruptions and deletions (right y-axis). (**) P < 10−4. No correlation was found between the number of genes in extant genomes and the number of gene duplications (data not shown). (C) Correlation between the number of balanced rearrangements (inversions and translocations) and the corresponding individual branch lengths from the Lachancea species tree based on the concatenation of 3598 orthologous genes corresponding to 1,983,702 aligned positions (Supplemental Fig. S4). (**) P < 10−4. (D) Correlation between the number of gene disruptions resulting from balanced rearrangements and branch lengths estimated as in C. (**) P < 10−4. (E) Correlation between the number of gene duplications and branch lengths estimated as in C. (*) P < 0.05. (F) No significant correlation between the number of gene deletions and branch lengths estimated as in C.
We used a birth–death evolutionary model on the gene family classification of the complete set of protein-coding genes from the 10 Lachancea genomes to identify unbalanced rearrangements (see Methods). We characterized 1503 gene duplications and 1036 gene losses. We checked all gene losses by looking for syntenic homologs that would have been missed during either genome annotation or gene family clustering because of a level of divergence that could have exceeded the threshold. We filtered out 107 cases of such dubious losses, leaving a total of 929 gene losses, clearly outnumbered by the 1503 gene duplications. We then determined their positions in the phylogenetic tree (Fig. 2). For 132 gene families where the phyletic patterns clearly indicated which members of the family corresponded to the duplicated copies, we found 94 inter- and 38 intra-chromosomally duplicated copies. The distribution of the distances between intra-chromosomally duplicated copies is bimodal, with 20 events separated by <10 kb, possibly resulting from tandem duplications (Supplemental Fig. S6).
At the level of the entire clade, unbalanced rearrangements are six times more abundant than inversions and translocations. Note that this ratio might be overestimated because the number of gene duplications and losses characterized in this work does not necessarily correspond to the number of events that occurred since some duplications and deletions could have involved several genes at the same time. Altogether, this detailed and exhaustive catalog of balanced and unbalanced chromosomal rearrangements positioned on the different branches of the phylogenetic tree provides the opportunity to identify quantitative principles governing genome evolution.
The number of genes in extant genomes is driven by the number of ancestral gene losses
The Lachancea gene repertoire underwent 1686 expansion events due to 1503 gene duplications, 159 putative gene creations, 24 HGTs, and 1947 reduction events, corresponding to 1018 pseudogenizations by point mutations or small indels and 929 gene losses by deletion or disruption of the coding sequence by a rearrangement breakpoint.
By integrating all these gene expansions and reductions, we estimated that the last common ancestor of the genus Lachancea, L.A1, contained about 5173 genes (Fig. 2). The number of genes in extant genomes ranges from 4768 in L. waltii to 5378 in L. kluyveri, not very different from the estimated ancestral number of genes. These comparable figures might give the impression that the total gene number in Lachancea genomes has reached equilibrium where gene expansion events roughly equal gene reduction events. However, we observed between 200 and 400 gene gains per lineage since the divergence from L.A1, while gene losses were highly variable, ranging from 31 in L. kluyveri to 466 in L. fantastica. As a result, we found a negative correlation between the number of genes in extant genomes and the number of gene losses that occurred since the divergence from the last common ancestor of the clade (R2 = 0.85, P = 1.4 × 10−4) (Fig. 5B), but not with the number of gene duplications or HGTs/de novo creations. Therefore, despite more abundant gene gains, the variation of the number of genes between extant genomes mainly results from the number of gene losses that occurred along the different branches of the tree. Remarkably, the especially low level of gene losses in L. kluyveri raises the exciting possibility that most gene losses occur by nonhomologous end joining (NHEJ), since essential components of this DSB repair pathway have been specifically lost in L. kluyveri (Gordon et al. 2011, and see below).
Finally, we found a negative correlation between the number of genes and pseudogenes present in extant genomes and the number of balanced rearrangements (inversions and translocations) that accumulated since the divergence from the last ancestor L.A1 (R2 = 0.70, P = 1.7 × 10−5) (Fig. 5B). This relationship was rather unexpected and suggested that fixed balanced rearrangements could be responsible for a significant proportion of gene losses.
Balanced rearrangements frequently disrupt genes at their breakpoints
We tested whether some gene losses could result from gene disruption caused by inversions or translocations with endpoints within coding sequences. From our initial estimate of 929 gene losses, we excluded the 125 losses specific to L. waltii because the sequencing gaps in the current genome assembly artificially increase the number of gene losses in this branch (Kellis et al. 2004; Di Rienzi et al. 2011), yielding 804 gene losses, which were positioned in the different branches of the tree. For each lost gene, we considered its two flanking genes in the species that did not undergo the loss. We then determined the positions of the orthologs of these flanking genes in the genome that underwent the loss. For each position, we looked at whether at least one of these orthologs was at the extremity of a synteny block involved in a balanced rearrangement predicted by ReChro on the corresponding branch. We found that 109 losses colocalized with a rearrangement breakpoint in a given branch of the tree (Fig. 2). This result suggests that up to 14% of all gene losses (109/804) could result from the disruption of a coding sequence by an inversion or a translocation event. We calculated the statistical significance of this result by testing the global enrichment of gene losses associated to rearrangements compared with the proportion of genes associated to rearrangements (χ2 test, P < 1.9 × 10−30). The same calculation performed on each branch of the tree showed significativity in only seven out of the 17 branches because of the small sample size in each branch (Fig. 2). Remarkably, we found three gene relics, i.e., highly degenerated remnants of genes (Lafontaine et al. 2004), within intergenic sequences corresponding to rearrangement breakpoints. Such relics could correspond to ancient genes disrupted by a rearrangement (Supplemental Fig. S7). These three cases correspond to gene disruptions that occurred in terminal branches of the tree and could therefore correspond to the most recent events that were not yet erased by the accumulation of subsequent point mutations and indels. Overall, these three gene relics support our finding that balanced rearrangements contributed to a significant number of gene losses.
Nonsynonymous substitution rates correlate with the number of inversions, translocations, and duplications, but not deletions
We tested whether the accumulation of large-scale chromosomal mutations and small-scale point mutations were coordinated during evolution, individually for each type of rearrangement. We correlated the number of rearrangements on each branch with branch lengths in the concatenation phylogenetic tree that represent the rates of fixed nonsynonymous substitutions. The number of balanced rearrangements per branch shows a significant positive correlation with the branch lengths (R2 = 0.69, P = 1.7 × 10−5) (Fig. 5C). This correlation holds when inversions and translocations are treated separately (R2 = 0.57 and R2 = 0.52, respectively; data not shown). Remarkably, the 109 gene disruptions resulting from balanced rearrangements show a similar positive correlation with branch lengths (R2 = 0.61, P = 3.5 × 10−4) (Fig. 5D). We also found that the number of gene duplications positively correlated with branch lengths (R2 = 0.31, P = 0.016) (Fig. 5E). In contrast, the subset of 695 (=804 − 109) gene deletions that are not associated with breakpoints show no significant correlation with the branch lengths (R2 = 0.04, P = 0.39) (Fig. 5F). We identified 45 gene relics among these 695 losses, resulting from the accumulation of point mutations and/or small deletions rather than from large deletions of entire ORFs. Removing these events from the analysis does not result in a significant correlation between DNA deletions and branch lengths (R2 = 0.05, P = 0.37; data not shown). These 45 gene relics are equivalent to the 723 annotated pseudogenes (excluding the L. waltii genome). We tested the correlation between these losses and branch lengths, focusing on terminal branches of the tree encompassing 103 out of 723 species-specific pseudogenes and 35 out of the 45 detected relics. No correlation was observed between those 138 events and terminal branch lengths (R2 = 0.008, P = 0.82), similarly to what was found for gene deletions.
Overall, these observations suggest the existence of a conserved genomic clock that applies to nonsynonymous substitutions, inversions, translocations, and duplications along each branch of the tree. However, deletions and pseudogenizations seem to accumulate independently from the other types of mutational events.
Functional consequences of gene repertoire evolution in Lachancea include the loss of the NHEJ and the crossover interfering pathways
Identifying all events that contributed to the evolution of the Lachancea gene repertoire allowed us to establish which main functional categories could be affected by gene losses and gains. We found that all essential genes in the S288c S. cerevisiae reference strain are conserved in all 10 Lachancea species, except 46 cases in L. waltii probably due to sequencing gaps (Kellis et al. 2004). No major change was observed in the gene repertoire involved in DNA replication, cell cycle checkpoints, or DNA repair, except the NHEJ pathway that is missing from L. kluyveri (Gordon et al. 2011). We confirmed that the orthologs of LIF1 and NEJ1 were missing from the L. kluyveri reference genome (Fig. 2) and from the 28 sequenced strains of L. kluyveri (Friedrich et al. 2015). A relic of NEJ1 was found next to a rearrangement breakpoint in the L. kluyveri genome, suggesting that the loss of NEJ1 resulted from a gene disruption event (Supplemental Fig. S7). DNL4 was found as a pseudogene, while a truncated copy of POL4 of 104 amino acids was annotated as a gene even if the average length in the other Lachancea species is 561 amino acids. All NHEJ factors LIF1, NEJ1, DNL4, and POL4 were found in the other nine Lachancea species (except LIF1 in L. waltii that is annotated as a pseudogene because of a sequencing gap) (Supplemental Table S7). Interestingly, the lower number of gene losses in L. kluyveri explains its larger gene complement of about 250 genes compared with the other Lachancea species (Fig. 2). This raises the exciting possibility that most gene losses occurred by NHEJ in the other species. Moreover, L. kluyveri underwent the smallest number of inversions and translocations among all Lachancea species since they diverged from their last common ancestor (Fig. 3, inset plot), suggesting that the NHEJ pathway could also participate in the formation of balanced rearrangements. On the contrary, the L. kluyveri lineage shows no clear deficit of duplication events compared with other Lachancea species such as Lachancea mirantina, L. cidri, or L. fermentati (Fig. 2), which is consistent with previous evidence that segmental duplications result from a replicative mechanism independent from the NHEJ pathway (Payen et al. 2008).
Remarkably, most genes from the ZMM pathway that generates interfering crossovers during meiosis are lost in Lachancea species except in L. kluyveri (Supplemental Table S8), suggesting a major change in the regulation of meiotic crossover within the genus Lachancea. While ZIP1 is ubiquitous, ZIP2, CST9, SPO22, SPO16, and the MutS homologs MSH4/5 are present in L. kluyveri only. In addition, MLH2, whose function seems to be related to meiotic recombination and to mismatch repair, is also absent from all the Lachancea species except L. kluyveri. These seven genes were probably lost after the divergence of L. kluyveri from the rest of the clade (along the b2 branch in Fig. 2). The ZMM pathway also comprises HFM1/MER3 that has homologs in L. kluyveri, L. waltii, L. dasiensis, and L. nothofagi (Supplemental Table S8). These genes are conserved in synteny in these species, suggesting that they were inherited vertically from their last common ancestor. Therefore, the phyletic pattern of HFM1 involves four independent losses (Fig. 2). A gene relic corresponding to HFM1 (also known as MER3) was only found in L. meyersii; none of the ZMM gene losses were found associated to a rearrangement breakpoint. Altogether, this suggests a major change in the regulation of meiotic crossover distribution between L. kluyveri and the other species of the clade. Interestingly, the ZMM pathway is found in many eukaryotes, including S. cerevisiae, plants, and mammals, but it has been lost independently several times during evolution, notably in yeasts, where it is absent from Schizosaccharomyces pombe, Yarrowia lipolytica, and Debaryomyces hansenii (Munz 1994; Richard et al. 2005).
Discussion
Our work combined a significant methodological contribution and a comprehensive comparative genomic analysis on a high-quality genome data set that we generated to achieve a detailed reconstruction of genome history in the model Lachancea yeast genus. We discovered relationships between genome size, gene content, chromosomal rearrangements, and rates of protein divergence that suggest the existence of several evolutionary principles so far uncharacterized.
Our methodological contribution consists in the development of AnChro for the reconstruction of ancestral genome architecture. AnChro proposes a new conceptual framework based on two original principles. First, our algorithm uses synteny blocks resulting from pairwise comparisons between extant genomes. This preserves the information of synteny conservation shared between closely related genomes even if more distantly related species are present in the clade. In contrast, for algorithms that use universal blocks, the presence of more distant species in the analysis restricts the synteny information to the highest common denominator to all species. Second, AnChro combines the advantage of reconstructing reliable adjacencies as in synteny-based methods and of identifying the balanced rearrangements on the branches of the tree as in distance-based methods. The combination of these two approaches presents the additional advantage of allowing the reconstruction of more ancestral adjacencies than by each method alone (Supplemental Information). The association of AnChro’s reconstructions with an independent inference of gene duplications and losses under a birth–death evolutionary model using a third-party tool and the identification of candidates for HGT and de novo gene creation events allowed us to achieve a detailed reconstruction of genome history in the model yeast genus Lachancea.
Gene duplication is a major driving force in genome evolution as previously anticipated by Ohno (1970). Yeast has exemplified the evolutionary importance of gene duplications and losses since the demonstration of an ancestral WGD in the S. cerevisiae lineage (Wolfe and Shields 1997; Fischer et al. 2001; Dietrich et al. 2004; Kellis et al. 2004; Scannell et al. 2007). Interestingly, a study performed at a larger evolutionary scale in fungi reported an excess of gene losses over gene duplications in lineages that diverged before the WGD (Wapinski et al. 2007). However, this analysis relied on published genomes with highly heterogeneous annotations, which may have had a deep impact on the inference of the number of evolutionary events, as acknowledged by the investigators themselves. In our case, the high-quality genome data set coupled with accurate annotations across more closely related species allowed a more precise quantification of the different types of genomic events. We found that gene duplications outnumbered gene losses, suggesting that gene duplication would also be the dominant evolutionary process in a protoploid genus that diverged from the S. cerevisiae lineage before the WGD. A similar trend was observed in the CTG yeast clade that did not undergo WGD and comprises most of the Candida species, including Candida albicans (Butler et al. 2009). These findings confirm the previously anticipated quantitative importance of segmental duplications in yeast genomes (Llorente et al. 2000; Dujon et al. 2004; Souciet et al. 2009).
We characterized 102 chromosomal inversions at single gene resolution. Previous estimates of the distribution of inversion length were constrained by the size of the synteny blocks into which inverted segments were identified (Fischer et al. 2006; Bhutkar et al. 2008). Furthermore, we found that the distribution of inversion lengths fits a power law of coefficient α = 1.86 (Fig. 5A). It is tempting to speculate that the power law relationship between the number and the length of fixed inversions indicates that inversions preferentially occur between regions coming into 3D contact in the nucleus since the 3D contact probability between two regions in the yeast nucleus decays with increasing genomic distance as a power law of coefficient 1.5 (Wong et al. 2012). Obviously, other parameters are likely to influence inversions such as chromatin accessibility as suggested by a recent study showing that the distribution of evolutionary breakpoints between five mammalian genomes depends on the 3D contact probability but also on the DNA accessibility in regions of open chromatin (Berthelot et al. 2015). Another parameter could be a higher selective cost associated with large heterozygous inversions compared with small inversions that could be better tolerated during the pairing of homologous chromosomes during meiotic prophase.
Our reconstruction of the Lachancea genome history sheds light on several genome evolution principles. We found that the gene number in extant genomes is negatively correlated to both the number of gene losses and the number of balanced rearrangements (inversions and translocations) that were fixed since divergence from the last common ancestor (Fig. 5B). On the contrary, while gene duplications were more abundant than gene losses, their number remained relatively homogeneous in all lineages, and therefore, they do not correlate with the gene complement in extant species. Remarkably, we found that gene losses are significantly enriched at balanced rearrangement breakpoints, representing 14% of the total gene losses. This strongly suggests that translocations and inversions contribute to the reduction of the gene repertoire by disrupting genes at their breakpoints. Further support comes from the identification of three truncated gene relics present at rearrangement breakpoints (Supplemental Fig. S7). In humans, numerous abnormal phenotypes, including intellectual disability and congenital anomalies, are caused by gene disruptions resulting from balanced rearrangements (Fruhmesser et al. 2013; Schluth-Bolard et al. 2013; Moyses-Oliveira et al. 2015; Schneider et al. 2015). This would occur in 6% of de novo reciprocal translocations and 9% of de novo inversions, but these events are detrimental and therefore remain rare in the population. By opposition, balanced rearrangements that reach fixation in populations are thought to be less detrimental because they are usually considered to occur in intergenic regions (Peng et al. 2006; Poyatos and Hurst 2007; Berthelot et al. 2015). Here, we show that numerous balanced rearrangements that occurred within coding sequences reached fixation in yeast populations. Our study provides the first genome-scale quantification of this phenomenon in a eukaryotic genus.
Finally, we showed that the number of balanced and unbalanced rearrangements varies greatly between lineages, leading to genomes in extant species that were differently rearranged compared with the ancestral genome of the genus (Figs. 2, 3). Such variable rates of genome rearrangements were already described in vertebrates and in yeasts (Bourque et al. 2005; Fischer et al. 2006). Furthermore, we found that nonsynonymous substitutions and inversions, translocations, and duplications reach fixation at a coordinated pace within each branch of the phylogenetic tree (Fig. 5). Previous works reported comparable correlations in Drosophila, bacteria, Archaea, and plastid genomes (Snel et al. 2002; Bhutkar et al. 2008; Csuros and Miklos 2009; Puigbo et al. 2014; Weng et al. 2014). Puigbo et al. (2014) coined the term genomic clock to describe the concept of coordinated pace of fixation between amino acid substitutions and large-scale rearrangements. This term might be misleading in the sense that a clock-like process is expected to follow a constant rate in time. This is clearly not the case here because the rates of substitution and number of rearrangements vary between branches. In bacteria, gene loss has been reported to be a more uniform, “clock-like” process than gene gain, suggesting that gene loss would be mostly neutral, whereas gene gain would be under positive selection or controlled by genetic drift enabled by population bottlenecks (Puigbo et al. 2014). In contrast, we found that gene deletion and pseudogenization are the only types of events that show no apparent correlation with protein sequence divergence. Overall, our findings open new questions on the respective selective value of various mutational events in eukaryotes. Further work is now needed to determine whether a genomic clock can be observed in a wider number of taxa. So far, the complete genome of approximately 100 yeast species have been published, and this number is still increasing. There are about 1200 known Saccharomycotina yeast species (Hittinger et al. 2015), and the project to sequence and analyze their genomes was recently initiated (http://www.y1000plus.org/). This will allow testing in this entire yeast subphylum of the existence of the evolutionary principles that we uncovered in the genus Lachancea. Further work will be needed to determine whether these principles also apply in other organisms such as vertebrates.
Methods
Strain selection, ploidy, karyotypes, and culture conditions
We selected seven Lachancea species: L. cidri, L. fermentati (both formerly called Zygosaccharomyces species) (Kurtzman 2003), L. meyersii (Fell et al. 2004), L. dasiensis (Lee et al. 2009), L. mirantina (Pereira et al. 2011), L. nothofagi (Mestre et al. 2010), and L. fantastica nomen nudum (Fig. 1). We renamed the strain CBS6924 as L. fantastica nomen nudum because it was erroneously classified as L. thermotolerans. These species were isolated worldwide often in association with plants, plant products, or insects. Several isolates from all different species were collected except for L. fantastica and L. mirantina, which were represented only by one strain. Electrophoretic karyotyping was performed for all strains as previously described (Neuveglise et al. 2000) (Supplemental Fig. S8). The ploidy of each strain was assessed using flow cytometry as previously described (Agier et al. 2013). Natural isolates were mainly haploids in all 10 species, while diploids were found in five species only (Supplemental Table S9). One haploid strain per species was selected for sequencing: L. meyersii CBS 8951T, L. fantastica nomen nudum CBS 6924, L. nothofagi CBS 11611T, L. dasiensis CBS 10888, L. fermentati CBS 6772, L. cidri CBS 2950, and L. mirantina CBS 11717 (Supplemental Table S10). Note that two other species, Lachancea lanzarotensis and Lachancea quebecensis, were described and sequenced during the course of this work and are not taken into consideration in this study (Gonzalez et al. 2013; Freel et al. 2015a, 2016; Sarilar et al. 2015).
DNA extraction, sequencing, and assembly of Lachancea genomes
Nuclear DNA was separated from mitochondrial and plasmid DNA by CsCl gradient (Supplemental Methods). Sequencing was carried out with a combination of Roche 454 in single-read and paired-end 8 kb on a GS-Flex+ apparatus, and Illumina in single read of 50 bp on a HiSeq2000 apparatus. Illumina reads allowed the correction of sequencing errors in homopolymer blocks that generated erroneous frameshifted genes. Genome assemblies were achieved with Celera Assembler version 6.1 (Myers et al. 2000) and Newbler v2.7 (454 Life Sciences) (Supplemental Methods).
Annotation of Lachancea genomes
The genomes of L. kluyveri and L. thermotolerans were used as references for gene structure annotation in the seven newly sequenced genomes and for the reannotation of L. waltii. We first completed the two reference genome annotations by detecting genes in intergenic regions through BLASTX against the UniProt fungi database. Gene models were annotated for the seven newly sequenced genomes with an annotation transfer pipeline that we developed with the AMADEA Biopack platform (Isoft, http://www.isoft.fr/bio/biopack_en.htm) (Supplemental Methods). Manual curation of gene models consisted of resolving gene models with missing start or stop codons, with not properly defined introns or with frameshifts. Additional CDSs were identified in intergenic regions of the 10 species by BLASTX search against the nr database and manual curation. Moreover, ORFs longer than 150 amino acids without any homologs in the nr database were predicted with Orffinder (NCBI). tRNA genes were predicted with tRNAscan-SE (v.1.3.1) (Lowe and Eddy 1997) with default searching parameters of tRNAscan and EufindtRNA; covariance model: tRNA2-euk.cm. The snRNA genes were searched by BLASTN using L. kluyveri and L. thermotolerans known snRNA sequences as query. We identified complete and partial transposable elements as well as solo-LTRs using BLAST against known transposable elements of Ty1/Copia, Ty3/Gypsy, and class-II superfamilies. Elements of the Rover and Roamer families are described elsewhere (Sarilar et al. 2014). The position of centromeres in the seven newly sequenced Lachancea genomes and in L. waltii was inferred from synteny conservation with already annotated centromeres in L. thermotolerans, L. kluyveri, and Zygosaccharomyces rouxii (Souciet et al. 2009). CDEI, CDEII, and CDEIII motifs were identified with the MEME program (Bailey and Elkan 1994), using the oops mode on both strands (Supplemental Methods).
The functional annotation of protein-coding genes has been established on the basis of homology with S. cerevisiae genes (SGD S288C ORF translations, release February 3, 2011, available at http://downloads.yeastgenome.org/sequence/S288C_reference/orf_protein/) or the NCBI Reference Sequence (RefSeq) database (release 58 of March 11, 2013, available at http://www.ncbi.nlm.nih.gov/refseq/) for putative genes without homologs in S. cerevisiae (Supplemental Methods).
Phylogenetic analyses
Orthologous genes were defined as syntenic homologs. Synteny block reconstructions were computed with the SynChro algorithm (Drillon et al. 2014) for all pairwise combinations between 36 yeast species (Supplemental Table S3). We inferred by transitivity 756 and 3598 groups of syntenic homologs composed of only one gene per species in the 36 yeast and 10 Lachancea species, respectively.
A multiple alignment of each group of orthologs was generated at the amino acid level with the MAFFT algorithm (linsi implementation, default parameters) (Katoh and Toh 2008). The best substitution model was determined by ProtTest (Abascal et al. 2005). PhyML reconstructions were performed from the concatenation of the multiple alignments for the Saccharomycotina set (756 orthologous groups: 486,399 aligned positions) and for the Lachancea set (either all 3598 orthologous genes—1,983,702 aligned positions—or the 472 orthologous groups whose individual trees have the eMRC topology—387,091 aligned positions), using the LG model and a gamma-law distribution with four categories of evolution rates (Guindon and Gascuel 2003). In all cases, 500 bootstrap replicates were performed.
We selected the 15,227 most strongly supported bipartitions (bootstrap value >0.95) out of the 32,391 bipartitions present in the 3598 individual gene trees to construct an unrooted eMRC tree that displays the most prevalent bipartitions in the data set. Each internal branch in the eMRC tree is associated with its gene support frequency (GSF), i.e., the number of gene trees supporting it (Gadagkar and Kumar 2005). To estimate the level of incongruence in this set of gene trees, we calculated the IC as recently proposed by Salichos et al. (2014). Tree certainty (TC) values are the sum of the IC values for all internodes. TC values range from zero (maximum conflict among individual gene trees) to eight (total number of internal nodes in our 11 taxon eMRC tree, no conflict among the individual gene trees). The eMRC tree, IC, and TC values were calculated with RAxML V8 (Stamatakis 2014). All phylogenetic reconstructions were achieved by considering all aligned positions as homologous characters, i.e., no removal of gap positions because identical tree topologies with negligible variations of TC values were obtained with or without considering gapped positions.
Gene families
An all-against-all BLASTP (version 2.2.28+) comparison was performed between amino acid translations of all CDS from the 10 Lachancea species and S. cerevisiae, with default options and Smith-Waterman alignment (Altschul et al. 1997). Hits with an E-value lower than 10−3 were clustered with TribeMCL with an inflation value I = 6.5 (Supplemental Methods; Enright et al. 2003). The detailed composition of all gene families and singletons is provided in Supplemental Table S12; their repartition among the 10 species, in Supplemental Table S2.
Systematic search for homologs to the Lachancea protein families was performed in the nr database with PSI-BLAST using a position-specific scoring matrix (PSSM) built from the family multiple alignments (only one iteration is performed). A search for homologs to Lachancea singletons was performed in the nr database with BLASTP (Altschul et al. 1997). Hits with an E-value lower than 10−3 with at least 25% sequence identity and coverage of the longest sequence of at least 50% were considered as significant. Similarity search against the PFAM database (version 27.0) was performed with hmmsearch from the HMMER3 package (Mistry et al. 2013), and hits with an E-value lower than 10−5 were considered significant. Search for conserved protein domains was also performed with rpsblast from the BLAST 2.2.29+ distribution, against the CDD database, version 01/10/2014.
Gene content evolution
Gene acquisitions and losses were inferred with the BadiRate program (Librado et al. 2012). For nonvertically inherited gene families (HGTs and TRGs), we used the birth, death, and innovation (BDI) model with free rates (FRs) estimated by the Wagner parsimony method (CWP). For vertically inherited families, we used the birth and death model with FR and CWP, assuming that all families derived from ancestral genes present in the common ancestor of all Lachancea species.
CAI values were calculated for all TRGs using CAIJava, (Carbone et al. 2003). Rates of synonymous substitutions (dS) and rates of nonsynonymous substitutions (dN) were estimated with the yn00 program from the PAML package (Yang 1997).
For the 127 L. kluyveri–specific TRGs, the CDS were considered physically absent from a given L. kluyveri strain if their sequencing coverage (estimated by mpileup in samtools) in the BAM files from the 28 sequenced L. kluyveri strains (Friedrich et al. 2015) was lower than the mean coverage minus two standard deviations for the core genome of that strain (L. kluyveri syntenic homologs).
Ancestral genome reconstruction
Ancestral genome reconstruction was performed with the CHROnicle suite of programs freely available at www.lcqb.upmc.fr/CHROnicle/ that comprises SynChro, ReChro, and AnChro. All the details about the ancestral gene order reconstruction steps, the identification of chromosomal rearrangements in the different branches of the tree, and the validation of the reconstructions are in the Supplemental information file. AnChro source code can also be found in the Supplemental Material.
Data access
Accession numbers and/or website sources for all yeast species used in this work are listed in Supplemental Table S3. Genome sequences and (re)annotations of the 10 Lachancea species are available on the GRYC website: http://gryc.inra.fr. The sequencing reads and the seven new genome assemblies from this study have been submitted to the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under the following accession numbers: PRJEB12910, PRJEB12928, PRJEB12929, PRJEB12930, PRJEB12931, PRJEB12932, and PRJEB12933.
Acknowledgments
This work was supported by the Agence Nationale de la Recherche (GB-3G, ANR-10-BLAN-1606). We thank Guillaume Achaz, Gilles Charvin, Frédéric Devaux, Cécile Fairhead, Romain Koszul, Gianni Liti, Marie-Claude Marsolier Kergoat, and Conrad Nieduszynski for fruitful discussions.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.204420.116.
References
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- Agier N, Romano OM, Touzain F, Lagomarsino MC, Fischer G. 2013. The spatio-temporal program of replication in the genome of Lachancea kluyveri. Genome Biol Evol 5: 370–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyev MA, Pevzner PA. 2009. Breakpoint graphs and ancestral genome reconstructions. Genome Res 19: 943–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avelar AT, Perfeito L, Gordo I, Ferreira MG. 2013. Genome architecture is a selectable trait that can be maintained by antagonistic pleiotropy. Nat Commun 4: 2235. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36. [PubMed] [Google Scholar]
- Berthelot C, Muffato M, Abecassis J, Roest Crollius H. 2015. The 3D organization of chromatin explains evolutionary fragile genomic regions. Cell Rep 10: 1913–1924. [DOI] [PubMed] [Google Scholar]
- Bhutkar A, Schaeffer SW, Russo SM, Xu M, Smith TF, Gelbart WM. 2008. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics 179: 1657–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Zdobnov EM, Bork P, Pevzner PA, Tesler G. 2005. Comparative architectures of mammalian and chicken genomes reveal highly variable rates of genomic rearrangements across different lineages. Genome Res 15: 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone A, Zinovyev A, Kepes F. 2003. Codon adaptation index as a measure of dominating codon bias. Bioinformatics 19: 2005–2015. [DOI] [PubMed] [Google Scholar]
- Csuros M, Miklos I. 2009. Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol 26: 2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Lindstrom KC, Lancaster R, Rolczynski L, Raghuraman MK, Brewer BJ. 2011. Genetic, genomic, and molecular tools for studying the protoploid yeast, L. waltii. Yeast 28: 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pohlmann R, Luedi P, Choi S, et al. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304: 304–307. [DOI] [PubMed] [Google Scholar]
- Drillon G, Fischer G. 2011. Comparative study on synteny between yeasts and vertebrates. C R Biol 334: 629–638. [DOI] [PubMed] [Google Scholar]
- Drillon G, Carbone A, Fischer G. 2014. SynChro: a fast and easy tool to reconstruct and visualize synteny blocks along eukaryotic chromosomes. PLoS One 9: e92621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, et al. 2004. Genome evolution in yeasts. Nature 430: 35–44. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Kunin V, Ouzounis CA. 2003. Protein families and TRIBES in genome sequence space. Nucleic Acids Res 31: 4632–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraut T. 2008. Addressing chromosome evolution in the whole-genome sequence era. Chromosome Res 16: 5–16. [DOI] [PubMed] [Google Scholar]
- Fell JW, Statzell-Tallman A, Kurtzman CP. 2004. Lachancea meyersii sp. nov., an ascosporogenous yeast from mangrove regions in the Bahama Islands. Stud Mycol 50: 359–363. [Google Scholar]
- Felsentein J. 1995. Phylogenetic inference package (PHYLIP), version 3.5. University of Washington, Seattle, WA. [Google Scholar]
- Fischer G, Neuveglise C, Durrens P, Gaillardin C, Dujon B. 2001. Evolution of gene order in the genomes of two related yeast species. Genome Res 11: 2009–2019. [DOI] [PubMed] [Google Scholar]
- Fischer G, Rocha EP, Brunet F, Vergassola M, Dujon B. 2006. Highly variable rates of genome rearrangements between hemiascomycetous yeast lineages. PLoS Genet 2: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel KC, Charron G, Leducq JB, Landry CR, Schacherer J. 2015a. Lachancea quebecensis sp. nov., a yeast species consistently isolated from tree bark in the Canadian province of Quebec. Int J Syst Evol Microbiol 65: 3392–3399. [DOI] [PubMed] [Google Scholar]
- Freel KC, Friedrich A, Schacherer J. 2015b. Mitochondrial genome evolution in yeasts: an all-encompassing view. FEMS Yeast Res 15: fov023. [DOI] [PubMed] [Google Scholar]
- Freel KC, Friedrich A, Sarilar V, Devillers H, Neuveglise C, Schacherer J. 2016. Whole-genome sequencing and intraspecific analysis of the yeast species Lachancea quebecensis. Genome Biol Evol 8: 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich A, Jung P, Reisser C, Fischer G, Schacherer J. 2015. Population genomics reveals chromosome-scale heterogeneous evolution in a protoploid yeast. Mol Biol Evol 32: 184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruhmesser A, Blake J, Haberlandt E, Baying B, Raeder B, Runz H, Spreiz A, Fauth C, Benes V, Utermann G, et al. 2013. Disruption of EXOC6B in a patient with developmental delay, epilepsy, and a de novo balanced t(2;8) translocation. Eur J Hum Genet 21: 1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T, Martin T, Marcet-Houben M, Durrens P, Bolotin-Fukuhara M, Lespinet O, Arnaise S, Boisnard S, Aguileta G, Atanasova R, et al. 2013. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics 14: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadagkar SR, Kumar S. 2005. Maximum likelihood outperforms maximum parsimony even when evolutionary rates are heterotachous. Mol Biol Evol 22: 2139–2141. [DOI] [PubMed] [Google Scholar]
- Gagnon Y, Blanchette M, El-Mabrouk N. 2012. A flexible ancestral genome reconstruction method based on gapped adjacencies. BMC Bioinformatics 13(Suppl 19): S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeote V, Novo M, Salema-Oom M, Brion C, Valerio E, Goncalves P, Dequin S. 2010. FSY1, a horizontally transferred gene in the Saccharomyces cerevisiae EC1118 wine yeast strain, encodes a high-affinity fructose/H+ symporter. Microbiology 156(Pt 12): 3754–3761. [DOI] [PubMed] [Google Scholar]
- Gonzalez SS, Alcoba-Florez J, Laich F. 2013. Lachancea lanzarotensis sp. nov., an ascomycetous yeast isolated from grapes and wine fermentation in Lanzarote, Canary Islands. Int J Syst Evol Microbiol 63(Pt 1): 358–363. [DOI] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH. 2009. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet 5: e1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH. 2011. Mechanisms of chromosome number evolution in yeast. PLoS Genet 7: e1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Rokas A, Bai FY, Boekhout T, Goncalves P, Jeffries TW, Kominek J, Lachance MA, Libkind D, Rosa CA, et al. 2015. Genomics and the making of yeast biodiversity. Curr Opin Genet Dev 35: 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BR, Rajaraman A, Tannier E, Chauve C. 2012. ANGES: reconstructing ANcestral GEnomeS maps. Bioinformatics 28: 2388–2390. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9: 286–298. [DOI] [PubMed] [Google Scholar]
- Kelkar YD, Ochman H. 2012. Causes and consequences of genome expansion in fungi. Genome Biol Evol 4: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624. [DOI] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. 2009. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet 25: 404–413. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res 4: 233–245. [DOI] [PubMed] [Google Scholar]
- Lafontaine I, Dujon B. 2010. Origin and fate of pseudogenes in Hemiascomycetes: a comparative analysis. BMC Genomics 11: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine I, Fischer G, Talla E, Dujon B. 2004. Gene relics in the genome of the yeast Saccharomyces cerevisiae. Gene 335: 1–17. [DOI] [PubMed] [Google Scholar]
- Lee CF, Yao CH, Liu YR, Hsieh CW, Young SS. 2009. Lachancea dasiensis sp. nov., an ascosporogenous yeast isolated from soil and leaves in Taiwan. Int J Syst Evol Microbiol 59(Pt 7): 1818–1822. [DOI] [PubMed] [Google Scholar]
- Librado P, Vieira FG, Rozas J. 2012. BadiRate: estimating family turnover rates by likelihood-based methods. Bioinformatics 28: 279–281. [DOI] [PubMed] [Google Scholar]
- Llorente B, Durrens P, Malpertuy A, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, Casaregola S, et al. 2000. Genomic exploration of the hemiascomycetous yeasts: 20. Evolution of gene redundancy compared to Saccharomyces cerevisiae. FEBS Lett 487: 122–133. [DOI] [PubMed] [Google Scholar]
- Louis VL, Despons L, Friedrich A, Martin T, Durrens P, Casaregola S, Neuveglise C, Fairhead C, Marck C, Cruz JA, et al. 2012. Pichia sorbitophila, an interspecies yeast hybrid, reveals early steps of genome resolution after polyploidization. G3 (Bethesda) 2: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Zhang L, Suh BB, Raney BJ, Burhans RC, Kent WJ, Blanchette M, Haussler D, Miller W. 2006. Reconstructing contiguous regions of an ancestral genome. Genome Res 16: 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldon T. 2010. Acquisition of prokaryotic genes by fungal genomes. Trends Genet 26: 5–8. [DOI] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldon T. 2015. Beyond the whole-genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the Baker's yeast lineage. PLoS Biol 13: e1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit S, Mena A, Bigey F, Sauvage FX, Couloux A, Guy J, Legras JL, Barrio E, Dequin S, Galeote V. 2015. Evolutionary advantage conferred by an eukaryote-to-eukaryote gene transfer event in wine yeasts. Mol Biol Evol 32: 1695–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Guerzoni D. 2015. New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos Trans R Soc Lond B Biol Sci 370: 20140332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre MC, Ulloa JR, Rosa CA, Lachance MA, Fontenla S. 2010. Lachancea nothofagi sp. nov., a yeast associated with Nothofagus species in Patagonia, Argentina. Int J Syst Evol Microbiol 60: 2247–2250. [DOI] [PubMed] [Google Scholar]
- Mighell AJ, Smith NR, Robinson PA, Markham AF. 2000. Vertebrate pseudogenes. FEBS Lett 468: 109–114. [DOI] [PubMed] [Google Scholar]
- Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res 41: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales L, Dujon B. 2012. Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiol Mol Biol Rev 76: 721–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel G, Sterck L, Swennen D, Marcet-Houben M, Onesime D, Levasseur A, Jacques N, Mallet S, Couloux A, Labadie K, et al. 2015. Differential gene retention as an evolutionary mechanism to generate biodiversity and adaptation in yeasts. Sci Rep 5: 11571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyses-Oliveira M, Guilherme RS, Meloni VA, Di Battista A, de Mello CB, Bragagnolo S, Moretti-Ferreira D, Kosyakova N, Liehr T, Carvalheira GM, et al. 2015. X-linked intellectual disability related genes disrupted by balanced X-autosome translocations. Am J Med Genet B Neuropsychiatr Genet 168: 669–677. [DOI] [PubMed] [Google Scholar]
- Muffato M, Roest Crollius H. 2008. Paleogenomics in vertebrates, or the recovery of lost genomes from the mist of time. Bioessays 30: 122–134. [DOI] [PubMed] [Google Scholar]
- Munz P. 1994. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 137: 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, et al. 2000. A whole-genome assembly of Drosophila. Science 287: 2196–2204. [DOI] [PubMed] [Google Scholar]
- Neuveglise C, Bon E, Lepingle A, Wincker P, Artiguenave F, Gaillardin C, Casaregola S. 2000. Genomic exploration of the hemiascomycetous yeasts: 9. Saccharomyces kluyveri. FEBS Lett 487: 56–60. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Springer Verlag, New York. [Google Scholar]
- Ouangraoua A, Tannier E, Chauve C. 2011. Reconstructing the architecture of the ancestral amniote genome. Bioinformatics 27: 2664–2671. [DOI] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet 4: e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Fischer G, Marck C, Proux C, Sherman DJ, Coppee JY, Johnston M, Dujon B, Neuveglise C. 2009. Unusual composition of a yeast chromosome arm is associated with its delayed replication. Genome Res 19: 1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q, Pevzner PA, Tesler G. 2006. The fragile breakage versus random breakage models of chromosome evolution. PLoS Comput Biol 2: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LF, Costa CR Jr, Brasileiro BT, de Morais MA Jr. 2011. Lachancea mirantina sp. nov., an ascomycetous yeast isolated from the cachaca fermentation process. Int J Syst Evol Microbiol 61: 989–992. [DOI] [PubMed] [Google Scholar]
- Perez-Ortin JE, Querol A, Puig S, Barrio E. 2002. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res 12: 1533–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyatos JF, Hurst LD. 2007. The determinants of gene order conservation in yeasts. Genome Biol 8: R233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P, Lobkovsky AE, Kristensen DM, Wolf YI, Koonin EV. 2014. Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol 12: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero-Rivera F, Xi QJ, Keppler-Noreuil KM, Lee JH, Higgins AW, Anchan RM, Roberts AE, Seong IS, Fan X, Lage K, et al. 2015. MATR3 disruption in human and mouse associated with bicuspid aortic valve, aortic coarctation and patent ductus arteriosus. Hum Mol Genet 24: 2375–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Lafontaine I, Dujon B. 2005. Comparative genomics of hemiascomycete yeasts: genes involved in DNA replication, repair, and recombination. Mol Biol Evol 22: 1011–1023. [DOI] [PubMed] [Google Scholar]
- Rolland T, Dujon B. 2011. Yeasty clocks: dating genomic changes in yeasts. C R Biol 334: 620–628. [DOI] [PubMed] [Google Scholar]
- Rolland T, Neuveglise C, Sacerdot C, Dujon B. 2009. Insertion of horizontally transferred genes within conserved syntenic regions of yeast genomes. PLoS One 4: e6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. 1998. The critical role of chromosome translocations in human leukemias. Annu Rev Genet 32: 495–519. [DOI] [PubMed] [Google Scholar]
- Salichos L, Rokas A. 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497: 327–331. [DOI] [PubMed] [Google Scholar]
- Salichos L, Stamatakis A, Rokas A. 2014. Novel information theory-based measures for quantifying incongruence among phylogenetic trees. Mol Biol Evol 31: 1261–1271. [DOI] [PubMed] [Google Scholar]
- Sarilar V, Bleykasten-Grosshans C, Neuveglise C. 2014. Evolutionary dynamics of hAT DNA transposon families in Saccharomycetaceae. Genome Biol Evol 7: 172–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarilar V, Devillers H, Freel KC, Schacherer J, Neuveglise C. 2015. Draft genome sequence of Lachancea lanzarotensis CBS 12615T, an ascomycetous yeast isolated from grapes. Genome Announc 3: pii:e00292-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Frank AC, Conant GC, Byrne KP, Woolfit M, Wolfe KH. 2007. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci 104: 8397–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT. 2011. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 (Bethesda) 1: 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluth-Bolard C, Labalme A, Cordier MP, Till M, Nadeau G, Tevissen H, Lesca G, Boutry-Kryza N, Rossignol S, Rocas D, et al. 2013. Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations. J Med Genet 50: 144–150. [DOI] [PubMed] [Google Scholar]
- Schneider A, Puechberty J, Ng BL, Coubes C, Gatinois V, Tournaire M, Girard M, Dumont B, Bouret P, Magnetto J, et al. 2015. Identification of disrupted AUTS2 and EPHA6 genes by array painting in a patient carrying a de novo balanced translocation t(3;7) with intellectual disability and neurodevelopment disorder. Am J Med Genet A 167: 3031–3037. [DOI] [PubMed] [Google Scholar]
- Semon M, Wolfe KH. 2007. Reciprocal gene loss between Tetraodon and zebrafish after whole genome duplication in their ancestor. Trends Genet 23: 108–112. [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. 2002. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res 12: 17–25. [DOI] [PubMed] [Google Scholar]
- Souciet JL, Dujon B, Gaillardin C, Johnston M, Baret PV, Cliften P, Sherman DJ, Weissenbach J, Westhof E, Wincker P, et al. 2009. Comparative genomics of protoploid Saccharomycetaceae. Genome Res 19: 1696–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. 2007. Natural history and evolutionary principles of gene duplication in fungi. Nature 449: 54–61. [DOI] [PubMed] [Google Scholar]
- Weng ML, Blazier JC, Govindu M, Jansen RK. 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol 31: 645–659. [DOI] [PubMed] [Google Scholar]
- Wisecaver JH, Slot JC, Rokas A. 2014. The evolution of fungal metabolic pathways. PLoS Genet 10: e1004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713. [DOI] [PubMed] [Google Scholar]
- Wong H, Marie-Nelly H, Herbert S, Carrivain P, Blanc H, Koszul R, Fabre E, Zimmer C. 2012. A predictive computational model of the dynamic 3D interphase yeast nucleus. Curr Biol 22: 1881–1890. [DOI] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling LB. 1962. Molecular disease, evolution, and genetic heterogeneity. In Horizons in biochemistry (ed. Kasha M, Pullman B), pp. 189–225. Academic Press, New York. [Google Scholar]