Abstract
Introduction
Dead enzymes are gene products (proteins) that lack key residues required for catalytic activity. In the pre-genome era, dead enzymes were thought to occur only rarely. However, they now have been shown to represent upwards of 10% of the total enzyme population in many families. The aldehyde dehydrogenase (ALDH) gene family encodes proteins that, depending on the isozyme, may be either catalytically-active or -inactive. Importantly, several ALDHs exhibit biological activities independent of their catalytic activity. For many of these, the physiological and pathophysiological functions remain to be established.
Areas covered
This article reviews the non-enzymatic functions of the ALDH superfamily. In addition, a search for additional non-catalytic ALDH records is undertaken. Our computational analyses reveal that there are currently 182 protein records (divided into 19 groups) that meet the criteria for dead enzymes.
Expert Opinion
Dead enzymes have the potential to exert biological actions through protein-protein interaction and allosteric modulation of the activity of an active enzyme. In addition, a dead enzyme may also influence availability of substrate for other active enzymes by sequestering substrate, and/or anchoring the substrate to a particular subcellular space. A large number of putatively non-catalytic ALDH proteins exist that warrant further study.
Keywords: aldehyde dehydrogenase, computational analysis, dead enzymes
1. Introduction
The existence of catalytically-inactive homologues of enzymes has been known for more than half a century [1], and have been called inactive enzyme-homologues, nonenzymes, pseudoenzymes or dead enzymes [2, 3]. These can be defined as homologues of enzymes which are predicted to retain protein expression, subcellular localization and typical protein folding, but which have lost key residues required for catalytic activity. In addition, the dead enzymes of individual superfamilies sometimes receive their own monikers. For example, the dead enzymes of the kinase and phosphatase families may be referred to as pseudokinases or pseudophosphatases, and the dead enzymes of the rhomboid protease family have been dubbed the iRhoms, for inactive rhomboid proteases. Dead enzymes, originally thought to be evolutionary relics, have now been shown to exert regulatory functions [2] and there is a lot of interest in the “forgotten genes” that code for them.
Since the original identification of dead enzymes, examples of enzymes losing catalytic activity and yet exerting biological functions have been documented. However, this has been assumed to be a relatively rare occurrence. Whole genome sequencing has altered this contention dramatically. When the entire human genome was combed for protein kinases in 2002, nearly 10% (50 of 518) of all human kinases were found to lack at least one of the three conserved catalytic residues required for activity [4]. Rather than being unique defects, twenty-eight of these inactive kinases possessed inactivations that were conserved in human, fly, worm and yeast [4]. These enzymes were shown to have taken on new, evolutionarily-conserved, non-catalytic roles. It is now understood that such dead enzymes are present in a wide variety of enzyme families and play diverse roles in physiology and pathophysiology.
Dead enzymes have now been identified in most enzyme families and a number of commonalities have been found. Many of them are conserved across taxonomic groups (e.g., among vertebrates or insect species), consistent with functional significance and conservative selective pressure – otherwise, many of these proteins would have been lost over evolutionary time [5]. Dead enzymes disproportionately have regulatory functions [5], and, frequently, the processes modulated are those in which their active counterparts participate [6]. The mechanism by which such modulation occurs typically involves protein-protein interaction. The dead enzyme may affect the activity of an active homologous enzyme, or interact with a separate protein substrate by acting as an allosteric modulator. In some cases, dead enzymes interact with their natural substrates directly, sequestering them and preventing their processing by other enzymes, or anchoring them in a particular subcellular space [7]. Examples of some of these mechanisms are provided below.
The best studied dead enzymes belong to the kinase and phosphatase families (sometimes referred to as pseudokinases and pseudophosphatases). The Trib family of pseudokinases has three members, all of which have been implicated as modulators of tumorigenesis. TRIB1 allosterically interacts with MEK1, leading to greater ERK phosphorylation. This has been found to be a key factor in myeloid leukemias [8]. TRIB2, a downstream target of Wnt in liver cancer, stabilizes YAP by interacting with two E3 ligases. Binding of TRIB2 to βTrCP blocks its targeting of YAP for proteasomal degradation, and TRIB2 promotes degradation of C/EBPα, an inhibitor of YAP/TEAD transcriptional activation, likely by interaction with the E3 ligases COP1 or TRIM21 [9]. TRIB3 (also called TRB3) is the best studied member of this family. It has been found to interact with a range of partners from transcription factors, ubiquitin ligase, BMP type II receptor, and members of the MAPK and PI3K signaling pathways. In so doing, TRIB3 affects a wide range of physiological processes including energy homeostasis, apoptosis, differentiation, and stress response [10]. Knockdown of TRIB3 inhibited migration and invasion of tumor cells, as well as modulating proteins regulating epithelial-to-mesenchymal transition (EMT) [10]. ErbB3 is a pseudokinase that interacts as part of a heterodimer with its active enzyme counterpart to modulate its activity. ErbB receptors typically hetero- or homodimerize and undergo trans-phosphorylation [7]. However, despite ErbB3’s inability to bind ATP and lack of tyrosine kinase activity, ErbB2-ErbB3 dimers are considered the most potent pairing of ErbB dimers with regards to mitogenic effects. Expression of one or both of these partners has been seen in a number of cancers [11]. It is also important to note that mutations in key catalytic residues may, in fact, be atypical mechanisms of action rather than inactivating mutations. Both WNK1 and CASK are kinases that were originally thought to be inactive due to missing key residues or motifs, but have since been shown to be catalytically-active using non-canonical modes of action [12]. A list of known pseudokinases and phosphatases and the effects of knockdown / knockout in mice are presented by Reiterer et al. [7].
Another well-studied group of dead enzymes is the pseudoenzyme rhomboid proteases, dubbed the iRhoms. Active rhomboid proteases are transmembrane proteins whose active site lies within the transmembrane domain. They bind and cut Type I transmembrane proteins and release them into the luminal or extracellular space. This action is especially critical in extracellular signaling. iRhoms are catalytically-inactive, but retain the transmembrane localization and binding to transmembrane proteins [12]. iRhom2 is located in the endoplasmic reticulum (ER) and interacts with TACE (TNFα-converting enzyme), allowing its exit from the ER to cleave and release tumor necrosis factor (TNF) from the cell surface. Together, these represent some of the well-known modes of action of dead enzymes.
The aldehyde dehydrogenase (ALDH) superfamily represents a group of enzymes that catalyze the NAD(P)+-dependent oxidation of a wide variety of aldehydes to their corresponding carboxylic acids [13]. This group is widely distributed and found in all kingdoms from archaea to mammals, including humans. Their known substrates include aldehydes involved in growth and development, differentiation, oxidative stress, osmoregulation, neurotransmission, and detoxification of dietary and environmental aldehydes [13, 14]. ALDH proteins usually comprise ≈ 500 amino acids, although some multifunctional or multi-domain members may be larger [15]. They are typically dimers or tetramers and consist of three domains: a substrate-binding (catalytic) domain, a cofactor (NAD(P)+) binding domain, and a dimerization/tetramerization domain [16].
Several amino acid residues are highly conserved and required for activity. These include CYS302, ASN169, and GLU285 in the catalytic domain, GLY262 and GLY267 in the Rossmann fold, and LYS209, GLU416, PHE418 in the cofactor binding domain [16]. In addition to their catalytic roles, ALDHs have been shown to possess non-catalytic roles (Table 1). Only two ALDH dead enzyme groups have been studied in any great detail. In the course of the present study, several more were identified. Here, we focus on the non-enzymatic roles of ALDH proteins, in both catalytically-active and -inactive proteins. This provides a starting point for understanding the possible roles of novel catalytically-inactive ALDH proteins.
Table 1.
Non-enzymatic functions of ALDHs
| ALDH isozyme | Function |
|---|---|
| ALDH3A1 | Scavenging of hydroxy radicals by CYS sulfhydryl groups |
| ALDH1A1 | Small molecule binding (e.g. T3, daunorubicin, androgen) |
| ALDH1, 2, 3 families | Lens and corneal crystallins |
| ALDH16A1 | Protein-protein interaction |
| ALDH2 | Regulation of active ALDH subunit by inactive subunit |
2. Small molecule binding and adduction by catalytically-active ALDHs
ALDH3A1 protects cellular proteins by the enzymatic detoxification of lipid peroxidation-derived aldehydes [17], and also by directly scavenging hydroxyl radicals via CYS sulfhydryl groups in a manner reminiscent of GSH-mediated quenching [18]. Thus, overexpression of these proteins can be protective regardless of catalytic function. Other ALDHs, such as ALDH1A1 and ALDH2, are targets of adduction by acetaminophen, often with partial enzyme inactivation being a result [19, 20]. In non-small lung cell carcinoma cell lines, the synthetic flavone flavopiridol, a cytotoxic drug, binds tightly to ALDH1A1 without inhibiting its ALDH catalytic activity or undergoing metabolism [21]. Xenopus ALDH1A1, first described as Cytosolic Thyroid Hormone-binding Protein (xCTBP), shows a high affinity for binding triiodothyronine (T3), which is disrupted by NAD(H)+ but not NADP(H)+ [22, 23]. This is consistent with reports that the T3 binding site is estimated to be in residues 93–114, i.e., in the cofactor-binding domain, as ALDH1A1 requires NAD+ (and cannot use NADP+) as a cofactor for catalytic oxidation of aldehydes [23]. Similarly, ALDH1A1 has been shown to bind daunorubicin, a chemotherapeutic agent, in a manner that is competitive with NAD+ binding [24]. In genital skin fibroblasts, ALDH1A1 has also been shown to bind androgen, specifically dihydrotestosterone bromoacetate [25].
3. Ocular ALDHs
The cornea and lens of the eye have the unique physiological requirements of maintaining high clarity (low light diffraction) while being resistant to incident UV radiation (UVR). The presence of specialized proteins, crystallins, helps these tissues accomplish these important tasks. Corneal and lens crystallins may be a single protein or set of proteins that compose up to 90% of the total water-soluble proteins within these tissues [26, 27]. Examples of crystallins include ALDH1A1, α-enolase, glutathione-S-transferase, lactic dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase (G3PDH), and arginino-succinate lyase [28]. They typically share several properties including i) being recruited from diverse, pre-existing cytoplasmic stress-response enzymes, ii) having a highly taxon-specific nature (different genes may serve the same function in different groups), and iii) accumulating to high levels in transparent tissues [26].
Members of the ALDH1, ALDH2, and ALDH3 families serve as lens and corneal crystallins [26]. ALDH3A1 is the predominant corneal crystallin in most mammals [29]; ALDH1A1 serves this purpose in human, rabbit, pig, chicken, and fish [30]. ALDH2 is also expressed in the cornea of rabbit and fish [31]. In the lens, ALDH1 family members serve as crystallins as follows: ALDH1A1 in most mammals, ALDH1A8 (η-crystallin) in the elephant shrew, ALDH1A9 (Ω-crystallin) in scallops and ALDH1C1/2 (Ω-crystallins) in cephalopods [32]. The catalytic activity of the ALDHs allows for the enzymatic detoxification of UVR-induced, ROS-mediated lipid peroxidation products. However, some important functions of the crystallins manifest independently of their catalytic properties. For example, high concentrations of these proteins provide short-range order within the cytoplasm of the lens fibers to reduce light scattering [28]. Another structural role involves direct absorption of UVB irradiation. The effectiveness of this process is illustrated in the bovine cornea in which ALDH3A1 accounts for 17% of the total cellular protein, but accounts for ≈50% of the total UVB absorptive capacity of the cornea [33].
The Ω-crystallins (ALDH1A9 and ALDH1C1/2) are the only dead enzyme ALDH crystallins known to date (i.e., that do not have catalytic activity) [34], and arise from a single gene homologue. Ω-crystallin represents 14% of the total soluble lens protein in octopus eye, but is present in minor, non-crystallin amounts in squid eye lens [35]. However, this protein represents ≈ 70% of the total soluble protein of the photophore (light organ) lens in the squid [35].
4. ALDH16A1
ALDH16A1 is structurally unusual in that it contains two ALDH family domains (Pfam: aldedh) [36], as opposed to the single domain typical of other ALDH family members. One of these domains is full length and the other is truncated, i.e., missing most of the catalytic domain [3]. This protein has widely distributed homologues in bacteria, protists, fish, amphibians and mammals, but not in archaea, fungi, and plants [3]. However, in all vertebrates (with the unusual exception of the frog), ALDH16A1 is predicted to be a dead enzyme as it is missing a number of key catalytic and cofactor binding residues [3]. This makes ALDH16A1 a unique case among known ALDH family members in that it has a form that is predicted to be catalytically-inactive and yet conserved among a large phylogenetic group. Of interest, ALDH16A1 protein expression has been detected in human immortalized cell lines, demonstrating that a protein product of this gene is indeed produced [3].
ALDH16A1 interacts with a number of proteins, and it is likely that its physiological role lies in these interactions [3]. A nucleotide insertion resulting in the premature truncation of the protein maspardin underlies mast syndrome, a form of spastic paraplegia [37]. This protein has been shown to colocalize and interact with ALDH16A1 in neuronal cells, although it is not yet known what function this interaction plays [38]. Other proteins that have been found to interact with ALDH16A1 include S-phase kinase-associated protein 1 (SKIP-1) [39], proteasomal ATPase-associated factor 1 (PAAF1) [40], ubiquitin specific peptidase 1 (USP1) [41], and protein kinase, AMP-activated, gamma 2 non-catalytic subunit (PRKAG2) [42]. In addition, databases of interaction data either curated or automatically extracted from Pubmed predict that ALDH16A1 interacts with albumin (ALB), solute carrier family 2-facilitated glucose transporter (SLC2A4), heat shock protein 90 kDa alpha class B member 1 (cytosolic; HSP90AB1), hypoxanthine phosphoribosyltransferase 1 (HPRT1), betaine–homocysteine S-methyltransferase (BHMT), and glioblastoma amplified sequence (GBAS) [43, 44].
An epidemiological study in an Icelandic population identified a single nucleotide polymorphism (SNP) of ALDH16A1 (ALDH16A1*2) as a risk factor for hyperuricemia and gout [45]. Computational analyses suggest that this is likely due to altered interactions with hypoxanthine phosphoribosyltransferase 1 (HPRT1) [3]. Homology modeling and docking experiments with frog ALDH16A1 (resembling the ALDH16A1 of lower animals) predict the frog enzyme to be catalytically-active and bind aldehyde substrates normally, although substrate specificity has not been extensively investigated. This suggests that at some point, ALDH16A1 was recruited from an catalytically-active role in lower animals to a non-catalytic role which has been preserved in vertebrates.
5. Negative regulation of ALDH catalytic activity by ALDH mutants
The E487K mutant of ALDH2 (ALDH2*2) is inactive, showing a 150-fold increase in the Km for cofactor (NAD+) and a 2–10-fold increase in Vmax [46]. Structural studies have shown that this mutation, located in the oligomerization domain, results in a large disordered region at the dimer interface, which includes much of the coenzyme-binding cleft, as well as part of the catalytic cleft [47]. This shift leads to poor NAD binding and poor catalytic activity. Further, it has been demonstrated that ALDH2*2 can negatively regulate ALDH2*1 via dominant negative hetero-tetramerization [46]. Estimates of the reduction in activity in heterozygotes are ~85% [48, 49]. Although it has not been shown in vitro, heterodimerization between closely-related ALDH proteins has been predicted as well, based on colocalization and conserved interactions in dimer and tetramer interfaces [50]. It stands to reason that conservation of dimerization and tetramerization domains may allow crosstalk between active enzymes and closely-related dead enzymes.
6. Discovering new ALDH dead enzymes
There are a number of residues in both the catalytic and cofactor binding sites that are absolutely required for ALDH activity. One of these residues is the primary catalytic cysteine (CYS319 in ALDH2). Other residues that are either invariant or highly conserved include GLU285, ASN186 (for catalytic activity), GLY262 and GLY267 (in the Rossmann fold), LYS209, GLU416, PHE418 (for cofactor binding). Although there are countless ways in which an ALDH enzyme might become catalytically-inactive, for screening purposes, we have defined catalytically-inactive in this review as lacking a homologous catalytic cysteine in an alignment with a model ALDH, specifically human ALDH2. To find records with an ALDH domain, every record in the UniProt database [51] was scanned against an ALDH profile (Pfam: aldedh) from the Pfam 27.0 build [36] using the profile hidden Markov model tool HMMER [52]. Positive ALDH sequences were aligned individually using T-COFFEE [53] against a model ALDH (human ALDH2, UniProt ID: P05091) and classified as catalytically-active or -inactive at that site. Multiple records of the same gene and multiple isoforms were combined.
To combine records into groups of homologous genes, this final list of non-catalytic ALDH records was aligned using T-COFFEE, and then neighbor-joining phylogenetic analysis with 8000 bootstrap replicates by PHYLIP [54]. Gene identification was approximated by comparing the record against the full UniProt database using HMMER and retrieving the closest named ALDH relative. In addition, to assist with classification and identification of genes, members of known ALDH families were included in the analysis as positive controls. Phylogenetic classification for each record was obtained through UniProt. Groups were combined by several factors including 1) consensus-tree bootstrap values, 2) contiguously or similarly named groups, and 3) phylogenetically contiguous groups.]
A summary of ALDH dead enzyme groups can be found in Table 2, and a full list of identified records is presented as Supplemental Table 1. One hundred and eighty-two unique records were found which met the conditions specified above. These were divided into 19 groups. Both known families of ALDH dead enzymes are represented, i.e., ALDH16A1 (group 1) and Ω-crystallins (group 19). ALDH dead enzyme records were found in Archaea, Bacteria, and Eukaryota (Table 3). Newly-discovered groups were especially prevalent in bacteria and fungi, and many of these records represent groups whose function is unstudied or poorly understood. Records were also discovered in vertebrates, but a majority of these are ALDH16A1 (group 1), which has been described previously. Several dead enzyme records were found from other groups including archaea (5), alveolata (2), oomycetes (2), and viridiplantae (7).
Table 2.
Summary of groups of ALDH dead enzyme records. Group number (group), ALDH family identification (identification), number of records (records), median protein length (in number of amino acids), and phylogenetic lineage are presented. The number of records in each lineage is presented in parentheses.
| Group | Identification | Records | Median Protein Length | Lineage |
|---|---|---|---|---|
| 1 | ALDH16A1 | 38 | 802 | Eukaryota - Metazoa - Vertebrata (38) |
| 2 | Unknown Function | 23 | 305 | Bacteria - Actinobacter - Actinomycetales (23) |
| 3 | ALDH4 | 16 | 556 | Bacteria - Bacteroidetes - Flavobacteriale (3) Bacteria - Actinobacteria - Streptomycetae (1) Eukaryota - Metazoa - Arthropoda (9) Eukaryota - Fungi - Ascomycota (3) |
| 4 | ALDH3 | 13 | 492 | Bacteria - Tenericutes - Mycoplasmatacea (2) Bacteria - Proteobacteria - Desulfovibriona (3) Eukaryota - Viridiplantae - Tracheophyta (1) Eukaryota - Oomycetes (2) Eukaryota - Metazoa - Vertebrata (5) |
| 5 | ALDH6 | 7 | 463 | Eukaryota - Viridiplantae (4) Eukaryota - Fungi - Ascomycota (1) Eukaryota - Metazoa - Arthropoda (2) |
| 6 | ALDH22 | 9 | 535 | Bacteria - Actinobacteria - Actinomycetales (7) Eukarya - Fungi (2) |
| 7 | ALDH24 | 9 | 467 | Archaea (2) Bacteria (3) Eukaryota - Alveolata (2) Eukaryota - Viridiplantae (1) Eukaryota - Fungi (1) |
| 8 | ALDH5 | 8 | 441 | Bacteria (2) Eukaryota - Fungi - Ascomycota (3) Eukaryota - Viridaplantae (1) Eukaryota - Metazoa - Chordata (2) |
| 9 | Unknown function | 8 | 381 | Bacteria - Actinobacteria - Actinomycetales (5) Bacteria - Proteobacteria (3) |
| 10 | Phenylacetaldehyde dehydrogenase | 9 | 436 | Bacteria - Actinobacteria - Actinomycetales (4) Bacteria - Proteobacteria - Rhodobacterales (1) Eukaryota - Fungi (4) |
| 11 | 2-hydroxymuconic semialdehyde dehydrogenase | 7 | 485 | Bacteria - Actinobacteria - Actinomycetales (7) |
| 12 | PutA | 6 | 1068 | Bacteria - Proteobacteria (6) |
| 13 | Unknown Function | 6 | 453 | Bacteria - Proteobacteria (1) Eukarya - Fungi - Ascomycota (5) |
| 14 | Urate Oxidase (bacterial) | 3 | 473 | Bacteria - Cyanobacteria - Nostocales (3) |
| 15 | 2-ketoglutaric semialdehyde dehydrogenase | 5 | 482 | Bacteria - Protobacteria (5) |
| 16 | Delta-1-pyrroline-5-carboxylate dehydrogenase 2 | 3 | 458 | Archaea (3) |
| 17 | Unknown Function | 4 | 350 | Eukarya - Fungi - Ascomycota (4) |
| 18 | Unknown Function | 6 | 381 | Eukarya - Fungi - Ascomycota (4) Eukarya - Fungi - Basidiomycota (2) |
| 19 | Ω-crystallin | 2 | 496 | Eukarya - Metazoa - Cephalopoda (2) |
Table 3.
The phylogenetic distribution of ALDH dead enzyme records found showing the name of major groups and the number of records (shown in parentheses) in each group.
| Kingdom (#) | Species (#) | Phylum (#) |
|---|---|---|
| Archaea (5) | ||
| Bacteria (79) | Acidobacteria (1) | |
| Actinobacteria (47) | ||
| Bacteroidetes (3) | ||
| Cyanobacteria (3) | ||
| Proteobacteria (23) | ||
| Tenericutes (2) | ||
| Eukaryota (98) | Alveolata (2) | |
| Fungi (29) | Ascomycota (23) | |
| Basidiomycota (6) | ||
| Metazoa (58) | Chordata (45) | |
| Arthropoda (11) | ||
| Mollusca (2) | ||
| Oomycetes (2) | ||
| Viridiplantae (7) | Chlorophyta (3) | |
| Streptophyta (4) |
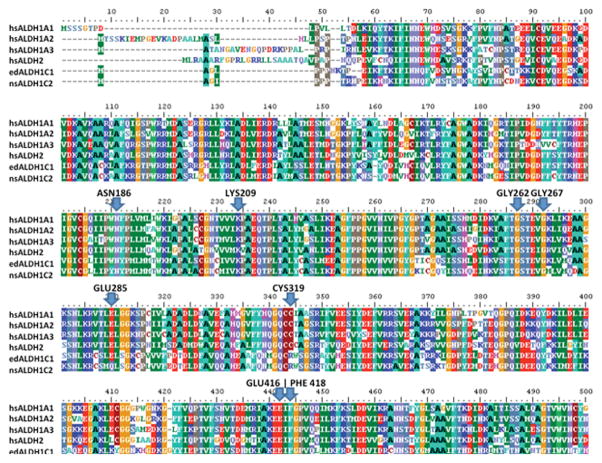
Figure 1 presents a representative amino acid alignment (group 19) and shows the location of key residues required for ALDH catalytic activity. The distribution of the most common inactivating mutations in these residues for each group is given in Table 4. In six of the groups, CYS319 was primarily deleted, and in another eight of the groups, various mutations were found. In only five of the groups was a single amino acid replacement of CYS319 dominant. Deletions were far less common in the rest of the residues surveyed. The residues of the Rossmann fold were highly conserved with 19/19 groups retaining GLY262 and 14/19 groups retaining GLY267. The residues of the NAD-binding pocket were moderately conserved with 15/19 groups retaining LYS209, 13/19 groups retaining GLU416 and 11/19 groups retaining PHE418. The remaining catalytic residues were the least well conserved with 9/19 groups retaining ASN186 and 10/19 groups retaining GLU285.
Figure 1.
Representative amino acid alignment showing the key residues required for enzymatic activity. The alignment compares closely-related human (hs) catalytically-active ALDHs (i.e., hsALDH1A1, hsALDH1A2, hsALDH1A3, and hsALDH2) with two ALDH dead enzymes (i.e., group 19 Ω-crystallins, ALDH1C1 from Enteroctopus dofleini (ed) and ALDH1C2 from Nototodarus sloanii (ns), UniProt IDs: P30841 and P30842, respectively). Amino acids are colored by residue and highlighted if 50% or more of the sequences are either identical or highly similar at that location. Proteins sequences for this alignment were downloaded from UniProt.
Table 4.
Summary of mutations in key residues of catalytic site, Rossmann fold and NAD(P)+ binding site in ALDH dead enzyme groups. The residues for the catalytically-active ALDH isozyme, ALDH2, is provided for comparison.
| ALDH | Catalytic Site * | Rossmann Fold * | NAD(P)+ Binding Site * | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA186 | % | AA319 | % | AA285 | % | AA262 | % | AA267 | % | AA209 | % | AA416 | % | AA418 | % | |
| ALDH2 | N | 100 | C | 100 | E | 100 | G | 100 | G | 100 | K | 100 | E | 100 | F | 100 |
| Group 1 | T | 66 | del | 95 | A | 76 | G | 92 | G | 84 | K | 71 | E | 84 | F | 92 |
| Group 2 | var | del | 100 | P | 100 | G | 100 | L | 74 | V | 65 | del | 139 | del | 139 | |
| Group 3 | N | 56 | var | E | 81 | G | 56 | Y | 56 | N | 81 | P | 50 | var | ||
| Group 4 | var | var | var | G | 92 | G | 62 | var | E | 62 | var | |||||
| Group 5 | N | 86 | del | 71 | N | 57 | G | 71 | G | 57 | K | 71 | E | 57 | F | 71 |
| Group 6 | var | var | var | G | 78 | var | K | 56 | E | 89 | var | |||||
| Group 7 | var | var | var | G | 78 | G | 78 | K | 78 | E | 67 | F | 78 | |||
| Group 8 | N | 88 | del | 50 | E | 50 | G | 75 | G | 75 | K | 63 | E | 75 | F | 63 |
| Group 9 | N | 75 | del | 63 | var | G | 88 | G | 50 | K | 100 | var | var | |||
| Group 10 | N | 67 | var | E | 67 | G | 100 | G | 89 | K | 67 | E | 78 | F | 67 | |
| Group 11 | N | 100 | S | 57 | E | 100 | G | 100 | G | 100 | K | 100 | E | 100 | L | 57 |
| Group 12 | N | 50 | Q | 50 | var | G | 83 | A | 83 | K | 100 | G | 50 | var | ||
| Group 13 | N | 67 | var | E | 67 | G | 83 | G | 83 | K | 50 | E | 67 | F | 50 | |
| Group 14 | var | var | E | 100 | G | 100 | G | 100 | K | 100 | Q | 67 | F | 67 | ||
| Group 15 | S | 100 | S | 60 | E | 60 | G | 100 | G | 100 | K | 100 | E | 100 | F | 100 |
| Group 16 | S | 100 | L | 100 | E | 100 | G | 100 | M | 100 | S | 67 | D | 100 | G | 100 |
| Group 17 | del | 75 | var | E | 100 | G | 100 | G | 100 | K | 75 | E | 100 | F | 75 | |
| Group 18 | var | del | 100 | E | 67 | G | 83 | G | 67 | K | 67 | E | 67 | F | 67 | |
| Group 19 | N | 100 | R | 100 | var | G | 100 | G | 100 | K | 100 | E | 100 | F | 100 | |
The amino acid (AA) indicated is the residue that is present at a majority (≥ 50%) of the sites, with the percentage of residues presented to the right. (del) indicates a deletion at that position. (var) indicates various mutations, i.e., no single amino acid residue was present at a majority of all sites. Shaded AAs are identical to the residue in ALDH2 at the same position.
7. Conclusions
Nineteen groups containing one hundred and eighty-two unique records were found that are predicted to be expressed as proteins but be catalytically-inactive and thus meet the criteria for dead enzymes. Review of the locations of potentially inactivating mutations (Table 4) suggests that in general, substrate-binding residues in the catalytic domain may be missing or degraded, but the structural elements of the Rossmann Fold are preserved strongly and the binding residues required for NAD(P)+ binding are moderately conserved. This suggests that the remaining function of these dead enzymes may be either structural (e.g. protein-protein binding) or in substrate (i.e. NAD(P)+) binding. Further analysis is suggested, especially in regard to the overall structure of the proteins via computational structural modeling or protein expression.
8. Expert opinion
Three examples of the non-catalytic roles have been shown to exist in the ALDH superfamily: 1) small-molecule binding, 2) structural, optical, and oxidative stress sinks, and 3) protein-protein interactions. Each one of these may play a role in the biological actions of dead enzymes. Dead enzymes often retain their cellular localization and substrate / cofactor binding potential, even if they lose catalytic ability. As such, they are well positioned for regulatory roles [2]. For example, they could be membrane proteins that bind to transmembrane proteins and thereby regulate their subsequent trafficking in a manner similar to iRhoms [12]. Even lacking catalytic activity, many enzymes may retain their substrate / cofactor binding regions. This may allow them to sequester substrates or bind alternate molecules, as in the case of Xenopus ALDH1A1 binding T3. Dual functional properties of ALDH1 have also been reported for other species, such as for elephant shrew η [32] and giant octopus crystallins as major component of lens proteins and human 56-kDa androgen-binding protein in genital skin fibroblast [25]. Further, they may act as structural or optical elements as has been shown for ALDH crystallins. They may also simply act as sinks for oxidative stress (also seen in ALDH crystallins). Finally, they make take on completely new roles in protein-protein binding. ALDH16A1’s conserved, yet catalytically-inactive structure is likely to act via interactions with other proteins. In addition, it is likely that a catalytically-active ALDH subunit may be repressed by the formation of dimers or tetramers with ALDH dead enzyme subunits. This work represents the novel discovery of groups of genes that were previously unknown to contain non-catalytic members; the functions of these members have not been investigated. The presence of multiple, closely-related records and contiguous phylogenetic groups is highly suggestive of conservative pressure to keep these dead enzymes and, thus, suggestive of a physiological role for these proteins. Given the increased information and interest in the role of dead enzymes, there is no doubt that their role in this enzyme superfamily will grow as well.
Supplementary Material
Article Highlights.
Dead enzymes are homologues of enzymes, which retain protein expression but not catalytic activity.
Dead enzymes have been found in many enzyme families including kinases, phosphatases, and rhomboid proteases.
The ALDH superfamily has members with non-enzymatic functions including radical scavenging, small molecule binding, crystallin activity, and protein-protein interactions.
In certain lineages. ALDH1A1 and ALDH16A1 lack catalytic functions.
Computational analysis suggests that many additional ALDH dead enzymes exist, especially in bacteria and fungi.
Footnotes
9. Declaration of Interest
The authors were supported by in part by NIH grants AA022057, AA021724 and EY017963 (V Vasiliou). Fellowship assistance for BC Jackson (F31 AA020728) is acknowledged. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
*=of importance,
**= of considerable importance
- 1.Brew K, Vanaman TC, Hill RL. Comparison of the amino acid sequence of bovine alpha-lactalbumin and hens egg white lysozyme. J Biol Chem. 1967 Aug 25;242(16):3747–9. [PubMed] [Google Scholar]
- 2.Leslie M. Molecular biology. 'Dead' enzymes show signs of life. Science. 2013 Apr 5;340(6128):25–7. doi: 10.1126/science.340.6128.25. [DOI] [PubMed] [Google Scholar]
- 3**.Vasiliou V, Sandoval M, Backos DS, Jackson BC, Chen Y, Reigan P, et al. ALDH16A1 is a novel non-catalytic enzyme that may be involved in the etiology of gout via protein-protein interactions with HPRT1. Chem Biol Interact. 2013 Feb 25;202(1–3):22–31. doi: 10.1016/j.cbi.2012.12.018. This report is the only detailed examination of ALDH16A1, the only dead enzyme in the human ALDH family. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4**.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002 Dec 6;298(5600):1912–34. doi: 10.1126/science.1075762. This is the first report which detailed the extent of dead enzymes in a human enzyme superfamily. [DOI] [PubMed] [Google Scholar]
- 5.Pils B, Schultz J. Inactive enzyme-homologues find new function in regulatory processes. J Mol Biol. 2004 Jul 9;340(3):399–404. doi: 10.1016/j.jmb.2004.04.063. [DOI] [PubMed] [Google Scholar]
- 6.Adrain C, Freeman M. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol. 2012 Aug;13(8):489–98. doi: 10.1038/nrm3392. [DOI] [PubMed] [Google Scholar]
- 7.Reiterer V, Eyers PA, Farhan H. Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol. 2014 Sep;24(9):489–505. doi: 10.1016/j.tcb.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Yokoyama T, Kanno Y, Yamazaki Y, Takahara T, Miyata S, Nakamura T. Trib1 links the MEK1/ERK pathway in myeloid leukemogenesis. Blood. 2010 Oct 14;116(15):2768–75. doi: 10.1182/blood-2009-10-246264. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Park JS, Wei Y, Rajurkar M, Cotton JL, Fan Q, et al. TRIB2 acts downstream of Wnt/TCF in liver cancer cells to regulate YAP and C/EBPalpha function. Mol Cell. 2013 Jul 25;51(2):211–25. doi: 10.1016/j.molcel.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua F, Mu R, Liu J, Xue J, Wang Z, Lin H, et al. TRB3 interacts with SMAD3 promoting tumor cell migration and invasion. J Cell Sci. 2011 Oct 1;124(Pt 19):3235–46. doi: 10.1242/jcs.082875. [DOI] [PubMed] [Google Scholar]
- 11.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009 Jul;9(7):463–75. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 12.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012 Jan 13;335(6065):225–8. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008 Jun;4(6):697–720. doi: 10.1517/17425250802102627. This report provides an extensive review of the roles and mechanisms of the ALDH superfamily. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brocker C, Vasiliou M, Carpenter S, Carpenter C, Zhang Y, Wang X, et al. Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta. 2013 Jan;237(1):189–210. doi: 10.1007/s00425-012-1749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, et al. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics. 2011 May;5(4):283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinmetz CG, Xie P, Weiner H, Hurley TD. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997 May 15;5(5):701–11. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- 17.Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. Human aldehyde dehydrogenase 3A1 (ALDH3A1): biochemical characterization and immunohistochemical localization in the cornea. Biochem J. 2003 Dec 15;376(Pt 3):615–23. doi: 10.1042/BJ20030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uma L, Hariharan J, Sharma Y, Balasubramanian D. Corneal aldehyde dehydrogenase displays antioxidant properties. Exp Eye Res. 1996 Jul;63(1):117–20. doi: 10.1006/exer.1996.0098. [DOI] [PubMed] [Google Scholar]
- 19.Landin JS, Cohen SD, Khairallah EA. Identification of a 54-kDa mitochondrial acetaminophen-binding protein as aldehyde dehydrogenase. Toxicol Appl Pharmacol. 1996 Nov;141(1):299–307. doi: 10.1006/taap.1996.0287. [DOI] [PubMed] [Google Scholar]
- 20.Lee YP, Liao JT, Cheng YW, Wu TL, Lee SL, Liu JK, et al. Inhibition of human alcohol and aldehyde dehydrogenases by acetaminophen: Assessment of the effects on first-pass metabolism of ethanol. Alcohol. 2013 Nov;47(7):559–65. doi: 10.1016/j.alcohol.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Schnier JB, Kaur G, Kaiser A, Stinson SF, Sausville EA, Gardner J, et al. Identification of cytosolic aldehyde dehydrogenase 1 from non-small cell lung carcinomas as a flavopiridol-binding protein. FEBS Lett. 1999 Jul 2;454(1–2):100–4. doi: 10.1016/s0014-5793(99)00773-5. [DOI] [PubMed] [Google Scholar]
- 22.Yamauchi K, Tata JR. Purification and characterization of a cytosolic thyroid-hormone-binding protein (CTBP) in Xenopus liver. Eur J Biochem. 1994 Nov 1;225(3):1105–12. doi: 10.1111/j.1432-1033.1994.1105b.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamauchi K, Nakajima J, Hayashi H, Horiuchi R, Tata JR. Xenopus cytosolic thyroid hormone-binding protein (xCTBP) is aldehyde dehydrogenase catalyzing the formation of retinoic acid. J Biol Chem. 1999 Mar 26;274(13):8460–9. doi: 10.1074/jbc.274.13.8460. [DOI] [PubMed] [Google Scholar]
- 24.Banfi P, Lanzi C, Falvella FS, Gariboldi M, Gambetta RA, Dragani TA. The daunorubicin-binding protein of Mr 54,000 is an aldehyde dehydrogenase and is down-regulated in mouse liver tumors and in tumor cell lines. Mol Pharmacol. 1994 Nov;46(5):896–900. [PubMed] [Google Scholar]
- 25.Pereira F, Rosenmann E, Nylen E, Kaufman M, Pinsky L, Wrogemann K. The 56 kDa androgen binding protein is an aldehyde dehydrogenase. Biochem Biophys Res Commun. 1991 Mar 29;175(3):831–8. doi: 10.1016/0006-291x(91)91640-x. [DOI] [PubMed] [Google Scholar]
- 26*.Chen Y, Thompson DC, Koppaka V, Jester JV, Vasiliou V. Ocular aldehyde dehydrogenases: protection against ultraviolet damage and maintenance of transparency for vision. Prog Retin Eye Res. 2013 Mar;33:28–39. doi: 10.1016/j.preteyeres.2012.10.001. This report provides an in-depth review of the role of ALDHs as crystallins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eguchi G. Crystalline lens. Tanpakushitsu Kakusan Koso. 1966 Oct;11(11):1083–4. [PubMed] [Google Scholar]
- 28.Jester JV. Corneal crystallins and the development of cellular transparency. Semin Cell Dev Biol. 2008 Apr;19(2):82–93. doi: 10.1016/j.semcdb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verhagen C, Hoekzema R, Verjans GM, Kijlstra A. Identification of bovine corneal protein 54 (BCP 54) as an aldehyde dehydrogenase. Exp Eye Res. 1991 Aug;53(2):283–4. doi: 10.1016/0014-4835(91)90085-s. [DOI] [PubMed] [Google Scholar]
- 30.Holmes RS, Cheung B, VandeBerg JL. Isoelectric focusing studies of aldehyde dehydrogenases, alcohol dehydrogenases and oxidases from mammalian anterior eye tissues. Comp Biochem Physiol B. 1989;93(2):271–7. doi: 10.1016/0305-0491(89)90081-3. [DOI] [PubMed] [Google Scholar]
- 31.Pappa A, Sophos NA, Vasiliou V. Corneal and stomach expression of aldehyde dehydrogenases: from fish to mammals. Chem Biol Interact. 2001 Jan 30;130–132(1–3):181–91. doi: 10.1016/s0009-2797(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 32.Graham C, Hodin J, Wistow G. A retinaldehyde dehydrogenase as a structural protein in a mammalian eye lens. Gene recruitment of eta-crystallin. J Biol Chem. 1996 Jun 28;271(26):15623–8. doi: 10.1074/jbc.271.26.15623. [DOI] [PubMed] [Google Scholar]
- 33.Abedinia M, Pain T, Algar EM, Holmes RS. Bovine corneal aldehyde dehydrogenase: the major soluble corneal protein with a possible dual protective role for the eye. Exp Eye Res. 1990 Oct;51(4):419–26. doi: 10.1016/0014-4835(90)90154-m. [DOI] [PubMed] [Google Scholar]
- 34.Zinovieva RD, Tomarev SI, Piatigorsky J. Aldehyde dehydrogenase-derived omega-crystallins of squid and octopus. Specialization for lens expression. J Biol Chem. 1993 May 25;268(15):11449–55. [PubMed] [Google Scholar]
- 35.Tomarev SI, Chung S, Piatigorsky J. Glutathione S-transferase and S-crystallins of cephalopods: evolution from active enzyme to lens-refractive proteins. J Mol Evol. 1995 Dec;41(6):1048–56. doi: 10.1007/BF00173186. [DOI] [PubMed] [Google Scholar]
- 36.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014 Jan;42(Database issue):D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson MA, Cross H, Proukakis C, Pryde A, Hershberger R, Chatonnet A, et al. Maspardin is mutated in mast syndrome, a complicated form of hereditary spastic paraplegia associated with dementia. Am J Hum Genet. 2003 Nov;73(5):1147–56. doi: 10.1086/379522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanna MC, Blackstone C. Interaction of the SPG21 protein ACP33/maspardin with the aldehyde dehydrogenase ALDH16A1. Neurogenetics. 2009 Jul;10(3):217–28. doi: 10.1007/s10048-009-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster LJ, Rudich A, Talior I, Patel N, Huang X, Furtado LM, et al. Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC) J Proteome Res. 2006 Jan;5(1):64–75. doi: 10.1021/pr0502626. [DOI] [PubMed] [Google Scholar]
- 40.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009 Jul 23;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010 Jul 1;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niu Y, Otasek D, Jurisica I. Evaluation of linguistic features useful in extraction of interactions from PubMed; application to annotating known, high-throughput and predicted interactions in I2D. Bioinformatics. 2010 Jan 1;26(1):111–9. doi: 10.1093/bioinformatics/btp602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012 Jan;40(Database issue):D841–6. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sulem P, Gudbjartsson DF, Walters GB, Helgadottir HT, Helgason A, Gudjonsson SA, et al. Identification of low-frequency variants associated with gout and serum uric acid levels. Nat Genet. 2011 Nov;43(11):1127–30. doi: 10.1038/ng.972. [DOI] [PubMed] [Google Scholar]
- 46*.Xiao Q, Weiner H, Johnston T, Crabb DW. The aldehyde dehydrogenase ALDH2*2 allele exhibits dominance over ALDH2*1 in transduced HeLa cells. J Clin Invest. 1995 Nov;96(5):2180–6. doi: 10.1172/JCI118272. Although this represents a mutant form of ALDH2, the mechanism described in this paper represents a possible way that ALDH dead enzymes might regulate active counterparts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larson HN, Weiner H, Hurley TD. Disruption of the coenzyme binding site and dimer interface revealed in the crystal structure of mitochondrial aldehyde dehydrogenase “Asian” variant. J Biol Chem. 2005 Aug 26;280(34):30550–6. doi: 10.1074/jbc.M502345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yokoyama A, Muramatsu T, Omori T, Matsushita S, Yoshimizu H, Higuchi S, et al. Alcohol and aldehyde dehydrogenase gene polymorphisms influence susceptibility to esophageal cancer in Japanese alcoholics. Alcohol Clin Exp Res. 1999 Nov;23(11):1705–10. [PubMed] [Google Scholar]
- 49.Hammen PK, Allali-Hassani A, Hallenga K, Hurley TD, Weiner H. Multiple conformations of NAD and NADH when bound to human cytosolic and mitochondrial aldehyde dehydrogenase. Biochemistry. 2002 Jun 4;41(22):7156–68. doi: 10.1021/bi012197t. [DOI] [PubMed] [Google Scholar]
- 50.Jackson BC, Holmes RS, Backos DS, Reigan P, Thompson DC, Vasiliou V. Comparative genomics, molecular evolution and computational modeling of ALDH1B1 and ALDH2. Chem Biol Interact. 2013 Feb 25;202(1–3):11–21. doi: 10.1016/j.cbi.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.UniProt: a hub for protein information. Nucleic Acids Res. 2015 Jan;43(Database issue):D204–12. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011 Jul;39(Web Server issue):W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000 Sep 8;302(1):205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 54.Felsenstein J. Confidence-Limits on Phylogenies - an Approach Using the Bootstrap. Evolution. 1985;39(4):783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.