Abstract
Circular RNAs (circRNAs) are a naturally occurring type of universal and diverse endogenous noncoding RNAs which unlike linear RNAs, have covalently linked ends. They are usually stable, abundant, conserved RNA molecules and often exhibit tissue/developmental-stage specific expression. Functional circRNAs have been identified to act as microRNA sponges and RNA-binding protein (RBP) sequestering agents as well as transcriptional regulators. These multiple functional roles elicit a great potential for circRNAs in biological applications. Emerging evidence shows that circRNAs play important roles in several diseases, particularly in cancer where they act through regulating protein expression of the pivotal genes that are critical for carcinogenesis. The presence of abundant circRNAs in saliva, exosomes and clinical standard blood samples will make them potential diagnostic or predictive biomarkers for diseases, particularly for cancer development, progression and prognosis. Here, we review the current literature and provide evidence for the impact of circRNAs in cancers and their potential significance in cancer prognosis and clinical treatment.
Keywords: RNA, circular RNA, expression, regulation, cancer
Introduction
The human transcriptome is very complex and diverse comprising of between 10,000-32,000 long non-coding RNAs (lncRNAs), around 9,000 small RNAs, ~11,000 pseudogenes, and close to 21,000 protein-coding mRNAs [1]. This suggests that a large portion of the mammalian genome is transcribed into non-coding RNAs. At present, circular RNAs, a naturally occurring class of non-coding RNAs have become an important area of research focus [2]. The high-throughput RNA sequencing (RNA-Seq) technology combined with bioinformatics method has been able to predict circRNAs. So far, several circRNA related databases have been developed that are available for public use, including find_circ [3], CIRCexplorer [4], circRNAFinder [5], Circ2Traits [6], CIRI [7], circBase [8], starBase [9] and CircNet [10]. Evidence is emerging that a large number of circRNAs are endogenous, stable, abundant and conserved RNA molecules with cell type- or developmental stage-specific expression patterns in eukaryotic cells [11-14]. CircRNAs are typically devoid of the terminal structures (i.e., 5’ cap or a poly A tail) and mostly non-translated, can occur in any genomic region including gene-bearing regions and intergenic regions, and range in length from few hundred to thousands of nucleotides [3,13,15]. Because of the lack of free ends, circular RNAs are resistant to exonucleases and more stable than linear RNA isoforms [3].
Regarding the function of circRNAs, recent publications have revealed that circRNAs serve as microRNA (miRNA) sponges which are believed to negatively regulate miRNAs by absorbing and sequestering miRNA molecules [3,16]. Subsequent reports proposed that circRNAs can also regulate their parental genes transcription [3,11,12], modulate processes of development and cell proliferation and interact with RNA-binding proteins [14,17-20]. Most recently, circRNAs have also been used as potential biomarkers of aging in Drosophila [5] and as potential biomarkers to detect diseases from human saliva [21,22]. The roles of circRNAs in the process of cancer initiation and progression has also gathered prominence [23-26]. Cancer remains as one of the leading causes of mortality worldwide [27], therefore it remains imperative to identify new diagnostic biomarkers and novel therapies to improve the survival rate of cancer patients. Deciphering circRNAs interplay with other RNA species in cancer would likely confer circRNAs great potential to become new diagnostic markers in early stages of cancer, and raise possibility for novel therapeutic interventions.
In this review, we provide a concise and up to date overview of circRNAs, and in particular discuss the context in which circRNAs play a role in cancer diagnosis and targeted therapy.
What is the nature of circRNA?
According to different biogenesis patterns from genomic regions, circRNAs can be divided into four categories: exonic circRNA (ecircRNA) [3,11,12], circular intronic RNA (ciRNAs) [17], exon-intron circRNAs [18] and intergenic circRNAs [3,28]. It has been reported that circRNAs are predominantly generated from the back-splice exons, where downstream 3’ splice donors are covalently linked to upstream 5’ splice acceptors in reverse order [29]. Jeck and colleagues proposed two models of ecircRNA: ‘lariat-driven circularization’ (Figure 1A) and ‘intron-pairing driven circularization’ (Figure 1B) [11]. Long flanking introns are believed to be essential for the exon circularization: they contain ALU repeats [11], and possibly help determine the production rate of circRNAs [20]. Finally, introns between the encircled exons are usually spliced out [13], but are retained in a few cases which Li termed exon-intron circRNAs or EIciRNAs [18]. Additionally, another class of intron-derived circRNAs in human cells, ciRNAs, were identified by Zhang and colleagues [17]. The biogenesis of such ciRNAs depends on a consensus motif containing a 7 nucleotide (nt) GU-rich element near the 5’ splice site and an 11 nt C-rich element near the branchpoint site [17] (Figure 1C). In general very little is known about these circularized transcripts, and the precise mechanism guiding the biogenesis of circRNAs remains unclear.
Figure 1.
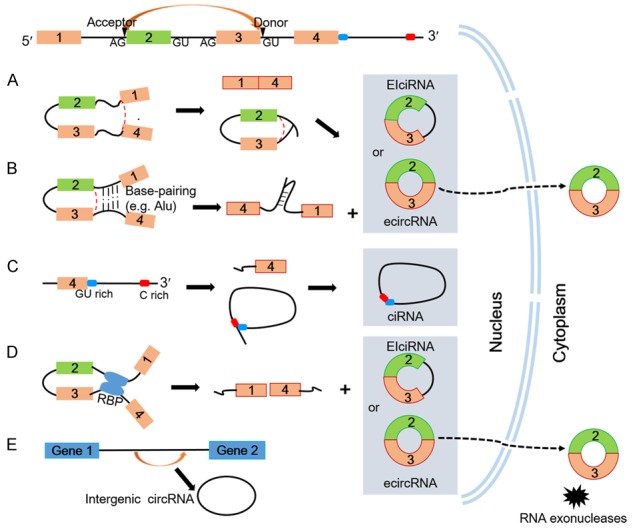
Possible models of circRNAs biogenesis. A. Lariat-driven circularization. Exon skipping event results in circular RNA formation where the 3’ splice donor site of exon 1 covalently links to the 5’ splice acceptor of exon 4, forming a lariat structure containing the skipped exons 2 and 3 as well as an mRNA consisting of exons 1 and 4. This lariat is spliced internally, removing or retaining the intronic sequence to generate ecircRNA or EIciRNA. B. Intron-pairing-driven circularization. Direct base-pairing of the introns flanking inverted repeats or ALU elements forms a circular structure. The introns are removed or retained to form ecircRNA or EIciRNA. C. ciRNAs. The ciRNAs are generated from lariat introns that can escape debranching. GU-rich sequences near the 5’ splice site (blue box) and C-rich sequences near the branch point (red box) are sufficient for an intron to escape debranching and become a stable circRNA. D. RBP-driven circularization. Interactions between RBPs form a bridge between the flanking introns to facilitate the exon circulation. A circRNA or an EIciRNA is generated due to the retention of internal introns. E. Intergenic circRNA. ecircRNAs are always located in the cytoplasm, where they have extraordinary stability due to their resistance to RNA exonucleases. In contrast, ciRNAs and EIciRNAs are predominantly located in the nucleus.
Emerging studies suggest an additional mode of circRNA biogenesis depend on RNA binding proteins (RBPs) (Figure 1D). The splicing factors Quaking (QKI) [30] and Muscleblind (MBL) [20] are able to promote the formation of circRNAs by binding to sequence motifs of flanking introns, forming a bridge to link two flanking introns close together. This process is similar to the model of intron-pairing driven circularization, except that RBPs regulate adjacent splice sites in place of direct base pairing between complementary motifs. In contrast, researchers have identified that RNA-editing enzyme ADAR1 is able to bind to double-stranded RNA to abolish circRNA formation by melting the stem structure [18]. Therefore, RBPs may either serve as activators of circularization by bridging complementary sequences or as inhibitors by inhibiting canonical splicing. Kramer and colleagues demonstrated that the Drosophila laccase2 circular RNA level is controlled by multiple heterogeneous nuclear ribonucleoprotein (hnRNP) and serine-arginine (SR) proteins in a combinatorial manner which is unlike the previously characterized MBL circular RNA [31]. Both hnRNP and SR proteins also regulate the expression of other Drosophila circular RNAs, such as Plexin A, suggesting that combinatorial control may be a common regulatory strategy for modulating circRNAs levels [31]. Indeed, in addition to the above elements, some other characteristics may be essential for the biogenesis of circRNAs. For example, genes that give rise to circRNAs present more active promoter regions, including significantly higher H3K27Ac and lower DNA methylation, compared to genes hosting only linear RNAs [32].
Correlation between circRNAs and cancer
Recent research has indicated that circRNAs may play a crucial role in the initiation and development of some diseases such as atherosclerosis, Parkinson’s disease, diabetes, prion related diseases and many others [33,34]. It is possible that some circRNAs may have cancerogenic functions in cancer. Gene ontology (GO) enrichment analysis of the database Circ2Traits was performed on the set of protein-coding genes in miRNA- circRNA interactome of individual diseases to detect the enrichment of genes associated with particular biological processes. This study revealed enrichment of biological processes for mRNAs related to 90 diseases, including several kinds of tumors. Among these mRNAs, 22 genes are involved in responding to light stimulus and 43 genes are cell cycle-related genes associated with breast cancer. There are 194 and 68 genes involved in different pathological processes of cervical cancer and gastric cancer, respectively. Interestingly 12 genes that respond to DNA damage stimulus involved in oral carcinoma were also identified [6]. In addition, researchers [25] have reported a global reduction in the ratio of circular RNA to their corresponding linear transcripts in tumor tissue, especially in colorectal cancer cell lines compared to normal colon samples. Furthermore, there is a negative correlation between this ratio and the proliferation index which could infer that circRNAs accumulate in non-proliferating cells [25]. A similar phenomenon was found in yeast (Schizosaccharomyces pombe) following starvation where decreased cell proliferation was accompanied by a rise in circRNA abundance [35]. Another publication reported that the expression patterns of circRNAs are more diverse in cancer cells than in non-cancer cell types, whereas the reverse tendency was observed for their counterpart linear mRNA transcripts [7]. Qu and colleagues also identified that circRNA expression signatures are dysregulated in Pancreatic Ductal Adenocarcinoma (PDAC) through microarray platform analysis [36]. These findings implied that dysregulated circRNAs likely play profound roles in the progression of cancer. So far, our understanding of the biological function of circRNAs in tumorigenesis is limited. Therefore, further studies on the mechanism of circular RNA in cancer initiation and progression should be conducted.
CircRNAs as miRNA sponge in cancer
miRNAs are non-coding RNAs that have an important role in fine-tuning post-transcriptional gene expression and are associated with several diseases, particularly cancer, where they act through the repression of downstream tumor-suppressive mRNAs [37]. Because of the complementarity of circRNAs to their targets, they can bind to miRNAs and competitively suppress the activity of miRNAs. The human/mouse ciRS-7/CDR1as and mouse Sry are the two most representative circRNA, which function as miRNA sponges [3,16]. ciRS-7 originating from the transcript anti-sense to the CDR1 gene is ~1,500 nt in length and is found predominantly in human and mouse brain [38]. circRNA Sry has 16 miR-138 binding sites and CDR1as/ciRS-7 comprises as many as 74 miR-7 binding sites [39]. ciRS-7 is sensitive to miR-671 and can be endonucleolytically cleaved via binding of miR-671 in an AGO2-dependent manner. Hence, ciRS-7 may facilitate transport of miR-7 to a subcellular location where miR-671 activity would release miR-7 by cleavage of ciRS-7 [3,40]. Accumulated evidence shows that overexpression of circRNA CDR1as/ciRS-7 or Sry results in increased expression of miRNA target genes, while knockdown of these circRNAs has the opposite effect [3,16].
This establishes a novel approach whereby circRNAs can regulate the progression of cancer through sequestering specific miRNA species associated with proliferation, differentiation, migration and carcinogenesis process. For example, miR-7 can serve as a tumor suppressor that is down-regulated in a variety of cancers, such as breast cancer [41], cervical cancer [42], Schwannoma tumor [43], tongue cancer [44], lung neoplasm [45], gastric carcinoma (GC) [46], hepatocellular carcinoma (HCC) [47] and colorectal cancer [48]. It has conclusively been demonstrated that miR-7 is involved in many cancer-related signaling pathways via directly down-regulating expression of crucial oncogenic factors including EGF receptor, IRS-1 and IRS-2 [49], Pak1 [50], Raf1 [51], Ack1 [43], PA28γ [52], YY1 [48], PIK3CD and mTOR [47]. In addition, miR-7 indirectly down-regulates STAT3 by down-regulating SETDB1, resulting in partial reversal of epithelial to mesenchymal transition (EMT) and suppressing the invasion and metastasis in breast cancer stem cells [41]. miR-7 inhibits metastasis in HCC by directly targeting and attenuating RELA, subsequently leading to NF-κB activation [53].
Although increasing evidence supports miR-7’s role as a tumor suppressor, contrary evidence has also been presented where viral oncogene E6/E7 expression in the HPV-positive HeLa cell line is associated with miR-7 overexpression [54]. miR-7 expression is increased in advanced colorectal cancers and in selected cell lines (SW480, DLD-1, and COLO201) compared to normal mucosa specimens [55]. Furthermore, miR-7 overexpression by stable transfection induces proliferation and migration in naturally immortalized skin cells HaCaT and A549 cells, suggesting miR-7 can act as an oncomiR in an epithelial context [56]. In summary, miR-7 is closely coupled to ciRS-7 through multiple miRNA response elements (MREs) in the circular RNA. It has been implied that fine-tuning of the miR-7/miR-671/ciRS-7 axis will likely serve as profound factors involved in cancer-associated biological processes, which may either promote cancer progression or suppress carcinogenesis depending on the expression level of miRNA target genes. An overview of miR-7/ciRS-7/target gene regulatory network constructed by CircNet database [10] is shown in Figure 2.
Figure 2.
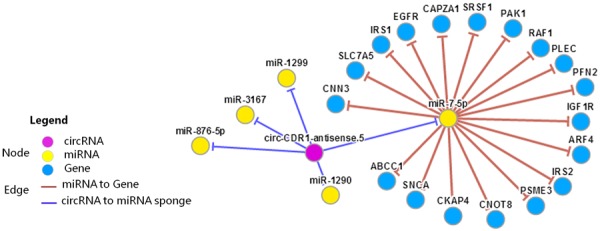
A miR-7/ciRS-7/target gene regulatory network analyzed through CircNet database (http://circnet.mbc.nctu.edu.tw/). The ciRS-7/CDR1as is also named circ-CDR1-antisense.5 in the CircNet database.
Considering the large a number of circRNAs that have been identified, it is possible that many circRNAs may function as microRNA sponges in regulating the proliferation, metastasis and invasion of cancer. A recent study reported that cir-ITCH expression is significantly decreased in esophageal squamous cell carcinoma (ESCC) compared to the peritumoral tissue. Cir-ITCH may have a tumor suppressive role in ESCC and serves as the sponge of miR-7, miR-17 and miR-214, resulting in an increase in the level of ITCH, which promotes ubiquitin-mediated Dvl2 degradation and reduces the expression of the oncogene c-myc, thereby inhibiting the Wnt/β-catenin signaling [24]. Analogously, Foxo3 circular RNA (circ-Foxo3) can also act as a sponge of potential miRNAs increasing Foxo3 translation thereby suppressing tumor growth, cancer cell proliferation and survival [57]. Expression patterns in CircNet indicated that four identified circRNAs--circ-ZEB1.5, circ-ZEB1.19, circZEB-1.17 and circ-ZEB1.33 were down-regulated in lung cancer specimens compared to the normal lung tissue samples, and all four circRNAs have the potential to sponge the miR-200 [10]. Zhang and colleagues also identified a highly expressed circRNA, CircRNA_1093, with a short sequence of 108 bp that has 4 miR-342-3p binding sites [15]. Notably, miR-342 has recently been reported to regulate BRCA1 expression in breast cancer [58]. Although the proposal of competing endogenous RNA (ceRNAs) as regulators is novel [59,60] and controversial [61], the stability and other properties of circRNAs may confer ceRNA activity with additional advantages. The circRNAs that act as miRNA sponges are providing us a novel perspective to treat cancer, so more circRNA-miRNA-gene regulatory networks in cancer initiation and progression are needed for further investigations.
CircRNAs as RBP sponges in cancer
A variety of functions have been elucidated regarding circRNAs, including “sponging” or sequestering of factors, such as RNA-binding proteins (RBPs) or ribonucleoprotein complexes (RNPs) [2]. Recent studies indicate that circRNAs can stably associate with AGO proteins [3,16], RNA polymerase II [17], MBL [20], Quaking [30], EIF4A3 [62], and perhaps other potential RBPs as well. Analysis of RBP binding sites on human circRNAs by a new web tool, CircInteractome, suggested that some circRNAs are ‘super-sponges’ with exceptionally high density of binding sites for a given RBP; for example, hsa_circ_0024707 could function as a super sponge for AGO2 with 85 predicted positions, and the mature circRNA hsa_circ_0000020 contains multiple binding sites for several RBPs like HuR (6 sites) and FMRP (10 sites) [62]. These circRNAs can be used as carriers to store, sort, or deliver RBPs to particular subcellular locations, and perhaps regulate the function of RBPs by acting as competing elements [3,63].
RBPs may be involved in a diversity of post-transcriptional regulatory processes such as RNA alternative splicing, stability, transport and translation. So by regulating gene expression, RBPs play important roles in cell proliferation, differentiation, motility, apoptosis, senescence, as well as the cellular responses to oxidative stress [64,65]. RBP dysregulation of transcription or expression plays a critical role in cancer development [66,67]. For example, in recent years, the RNA-binding protein quaking 5 (QKI-5) has been recognized as a novel tumor suppressor in many cancers. Some of QKI isoforms are frequently down-regulated in lung cancer, and QKI-5 inhibits the proliferation and transformation in lung cancer cell lines [68]. Similarly, in prostate cancer, QKI-5 can inhibit prostate cancer cell proliferation, cell cycle progression and invasion capability and experimentally induces cancer cell apoptosis [69]. While the formation of circRNAs is regulated by QKI proteins, which themselves are regulated during EMT. Furthermore, upregulation of most circRNAs during EMT suggests they carry out important functions in EMT [30]. Since EMT is widely known to have an important function in the progression of carcinomas to metastasis, further work in this area will bring insight to the therapeutic role of QKI-mediated circRNAs in cancer. In addition, as a multifunction player, AGO proteins have also revealed participation in tumorigenesis via miRNAs-dependent or independent pathways. AGO2 was found ectopically over-expressed in carcinomas and closely related to the progression of cancers by way of interacting with well-known tumor factors [70]. These examples imply that RBPs and circular RNAs might intricately intertwine to contribute to regulate the cancer progression.
CircRNAs as transcriptional regulators in cancer
Misregulation of cancer-related alternative splicing is frequently associated with an imbalance in the expression of splicing factors. As circRNAs can be regarded as a type of alternative splicing isoforms, they may function in regulating gene expression at the level of alternative splicing. Some ciRNAs such as ci-ankrd52, ci-mcm5 and ci-sirt7 can enhance the expression of their parental mRNAs [17,39]. Some of these parental genes can be associated with tumorigenesis, playing a tumor suppressor or promoter role. For example low expression of SIRT7 is associated with an aggressive tumor phenotype and poorer outcome in PDAC [71]. Similarly, over-expression of MCM5 is associated with the aggressive progression and poor prognosis of oral squamous cell carcinoma [72] and MCM5 can be considered as one candidate marker in colorectal cancer [73].
CircRNAs as potential diagnostic and prognostic biomarkers in cancer
According to recent publications, several remarkable characteristics of circRNAs can be highlighted. 1) High abundance: Salzman and colleagues firstly reported that circRNAs are the most universal molecules after linear RNAs distributed in human cells [74], and they appear more abundant than expected before. In some cases, the abundance of circRNAs exceeds those of the corresponding linear mRNAs by a factor of 10 [11]. 2) Stability: the covalently closed loop structures of circRNAs likely confer high resistance to RNA exonuclease or RNase R activity conferring much higher stability than linear RNAs [75]. The average half-life of circRNAs in most species is more than 48 h, while the half life on average of mRNAs is about 10 h [76]. 3) Conservatism: circRNAs show an ancient, evolutionarily conserved feature in different species [11,35]; For example, many circRNAs can be detected in both humans and mice including Drosophila. 4) Location: majority of circRNAs are ecircRNAs which are primarily located in the cytoplasm and possibly possess MREs [2,3]. Whereas intronic circRNAs both ciRNAs and EIciRNAs are primarily located in the nucleus in eukaryotes and may participate in regulation of gene expression [17,18]. 5) circRNAs often exhibit tissue/developmental-stage specific expression. For example, circRNAs are highly enriched in the mammalian brain, especially so in the synapses, and are dynamically up-regulated during neuronal differentiation [14]. Hence many of these properties confer a unique advantage to circRNAs as potential biomarkers of cancer diagnosis and prognosis in multiple organs including the brain.
One recent research reported that circRNAs are more abundant in exosomes compared to the cytoplasm of producer cells, and the abundance of tumor-derived exo-circRNA in serum of xenografted mice is related with tumor mass. In addition, they found that the expression profile of circRNAs in serum of cancer-bearing mice is significantly different from that in normal mice. As many as 67 circRNAs were absent and 257 circRNAs were newly identified in cancer samples, compared to healthy animals. Notably, many of the host genes of new circRNAs are significantly up-regulated in colorectal cancer tissues [77]. These results provide a new and unique reference for employing circRNAs as a class of exosome-based cancer biomarkers for the early diagnosis and prognosis. Moreover, attempts to identify other circRNAs as diagnostic biomarkers of cancer are emerging rapidly. Researchers have identified hsa_circ_002059 and hsa_circ_0001649 to be significantly down-regulated in gastric cancer and HCC. A decrease in the expression levels are considered responsible for the tumorigenesis and metastasis of the cancer and can potentially serve as a valuable biomarker for the diagnosis of gastric cancer and HCC [78,79]. Many well-studied cancer-associated circRNAs are summarized in Table 1.
Table 1.
Summary of well-studied cancer-related circular RNAs
| CircRNAs | Function | Cancer type | Reference |
|---|---|---|---|
| CDR1as | miR-7 sponge | Breast cancer | [41] |
| Cervical cancer | [42] | ||
| Schwannoma tumor | [43] | ||
| Tongue cancer | [44] | ||
| Lung neoplasm | [45] | ||
| Gastric carcinoma | [46] | ||
| Hepatocellular carcinoma | [47] | ||
| Colorectal cancer | [48] | ||
| circ-ZEB1.5 | miR-200 sponge | Lung cancer | [10] |
| circ-ZEB1.19 | |||
| circ-ZEB1.17 | |||
| circ-ZEB1.33 | |||
| circRNA_1093 | miR-342 sponge | Breast cancer | [15] |
| cir-ITCH | Inhibiting Wnt/β-catenin signaling | Esophageal squamous cell carcinoma | [24] |
| ci-sirt7 | Enhance sirt7 expression | Pancreatic ductal adenocarcinoma | [17,71] |
| ci-mcm5 | Enhance mcm5 expression | Oral squamous cell carcinoma | [17,72] |
| Colorectal cancer | [17,73] | ||
| circ-Foxo3 | Enhance Foxo3 translation | Breast cancer | [57] |
| hsa_circ_002059 | Potential value biomarker | Gastric carcinoma | [78] |
| hsa_circ_0001649 | Potential value biomarker | Hepatocellular carcinoma | [79] |
Perspective
In summary, functional roles of circRNAs in the regulation of protein-coding gene expression through acting as miRNA sponges, RBP sponges and transcriptional regulators confer a great variety of functional potential to circRNAs. The fact that circRNAs are found abundant in saliva [22], exosomes [77] and clinical standard blood samples [80] makes circRNA a promising diagnostic biomarker for diseases, such as cancer screening and susceptibility evaluation. In future studies identification of circRNA biomarkers will not only be possible in saliva and blood but also in other clinical samples such as urine and cerebrospinal fluid. More importantly, circRNAs studies provide a new avenue for disease therapy, by which circularization of RNA may be the future target of disease treatment. For example, a future scenario where overexpression of an artificial circRNA acting as a ‘super-sponge’ or silencing the circRNAs in cells to remodel and change the expression profile of miRNAs and other RNAs or RBP levels to increase the activities of a suppressor gene in the context of cancer therapy can be envisioned. Although there are several studies on circRNAs, relatively little is known of biological and molecular mechanisms of circRNAs in cancer progression. It will be necessary to identify many more circRNAs involved in diseases and explore their functional motifs and target sites to improve the diagnosis and treatment of circRNA-related diseases.
Acknowledgements
Current work is supported by the National Natural Science Foundation of China (No. 31371386) and a program for Excellent Talents in Henan Province (No. 124200510010). AJN is supported by Natural Sciences and Engineering Council of Canada.
Disclosure of conflict of interest
None.
References
- 1.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 4.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Westholm Jakub O, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker Susan E, Graveley Brenton R, Lai Eric C. Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, Liu CC, Huang HD. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C, Wu H, Wang Y, Zhao Y, Fang X, Chen C, Chen H. Expression Patterns of Circular RNAs from Primary Kinase Transcripts in the Mammary Glands of Lactating Rats. J Breast Cancer. 2015;18:235–241. doi: 10.4048/jbc.2015.18.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular Intronic Long Noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 19.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Cui H, Wang W, Li L, Wang Z, Yang S, Zhang X. RETRACTED: Construction of circular miRNA sponges targeting miR-21 or miR-221 and demonstration of their excellent anticancer effects on malignant melanoma cells. Int J Biochem Cell Biol. 2013;45:2643–2650. doi: 10.1016/j.biocel.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer. Oncol Rep. 2015;33:2669–2674. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 27.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 28.Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Wilusz JE, Sharp PA. A Circuitous Route to Noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2015;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 35.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu S, Song W, Yang X, Wang J, Zhang R, Zhang Z, Zhang H, Li H. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data. 2015;5:385–387. doi: 10.1016/j.gdata.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero-Cordoba SL, Salido-Guadarrama I, Rodriguez-Dorantes M, Hidalgo-Miranda A. miRNA biogenesis: biological impact in the development of cancer. Cancer Biol Ther. 2014;15:1444–1455. doi: 10.4161/15384047.2014.955442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosik KS. Molecular biology: Circles reshape the RNA world. Nature. 2013;495:322–324. doi: 10.1038/nature11956. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, Jiang L, Zhang Y, Dou J. MiR-7, Inhibited Indirectly by LincRNA HOTAIR, Directly Inhibits SETDB1 and Reverses the EMT of Breast Cancer Stem Cells by Downregulating the STAT3 Pathway. Stem Cells. 2014;32:2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 42.Liu S, Zhang P, Chen Z, Liu M, Li X, Tang H. MicroRNA-7 downregulates XIAP expression to suppress cell growth and promote apoptosis in cervical cancer cells. FEBS Lett. 2013;587:2247–2253. doi: 10.1016/j.febslet.2013.05.054. [DOI] [PubMed] [Google Scholar]
- 43.Saydam O, Senol O, Würdinger T, Mizrak A, Ozdener GB, Stemmer-Rachamimov AO, Yi M, Stephens RM, Krichevsky AM, Saydam N. miRNA-7 attenuation in Schwannoma tumors stimulates growth by upregulating three oncogenic signaling pathways. Cancer Res. 2011;71:852–861. doi: 10.1158/0008-5472.CAN-10-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang L, Liu X, Chen Z, Jin Y, Heidbreder C, Kolokythas A, Wang A, Dai Y, Zhou X. MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. 2010;432:199–205. doi: 10.1042/BJ20100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Zheng Y, Sun G, Xiong S. Restoration of miR-7 expression suppresses the growth of Lewis lung cancer cells by modulating epidermal growth factor receptor signaling. Oncol Rep. 2014;32:2511–2516. doi: 10.3892/or.2014.3519. [DOI] [PubMed] [Google Scholar]
- 46.Kong D, Piao YS, Yamashita S, Oshima H, Oguma K, Fushida S, Fujimura T, Minamoto T, Seno H, Yamada Y. Inflammation-induced repression of tumor suppressor miR-7 in gastric tumor cells. Oncogene. 2012;31:3949–3960. doi: 10.1038/onc.2011.558. [DOI] [PubMed] [Google Scholar]
- 47.Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–1862. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 48.Zhang N, Li X, Wu C, Dong Y, Cai M, Mok M, Wang H, Chen J, Ng S, Chen M. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene. 2013;32:5078–5088. doi: 10.1038/onc.2012.526. [DOI] [PubMed] [Google Scholar]
- 49.Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 50.Reddy SDN, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68:8195–8200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 52.Xiong S, Zheng Y, Jiang P, Liu R, Liu X, Qian J, Gu J, Chang L, Ge D, Chu Y. PA28gamma emerges as a novel functional target of tumour suppressor microRNA-7 in non-small-cell lung cancer. Br J Cancer. 2014;110:353–362. doi: 10.1038/bjc.2013.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ning BF, Ding J, Liu J, Yin C, Xu WP, Cong WM, Zhang Q, Chen F, Han T, Deng X, Wang PQ, Jiang CF, Zhang JP, Zhang X, Wang HY, Xie WF. Hepatocyte nuclear factor 4α-nuclear factor-κB feedback circuit modulates liver cancer progression. Hepatology. 2014;60:1607–1619. doi: 10.1002/hep.27177. [DOI] [PubMed] [Google Scholar]
- 54.Honegger A, Schilling D, Bastian S, Sponagel J, Kuryshev V, Sültmann H, Scheffner M, Hoppe-Seyler K, Hoppe-Seyler F. Dependence of Intracellular and Exosomal microRNAs on Viral E6/E7 Oncogene Expression in HPV-positive Tumor Cells. PLoS Pathog. 2015;11:e1004712. doi: 10.1371/journal.ppat.1004712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakagawa Y, Akao Y, Taniguchi K, Kamatani A, Tahara T, Kamano T, Nakano N, Komura N, Ikuno H, Ohmori T. Relationship between Expression of Onco-Related miRNAs and the Endoscopic Appearance of Colorectal Tumors. Int J Mol Sci. 2015;16:1526–1543. doi: 10.3390/ijms16011526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meza-Sosa KF, Pérez-García EI, Camacho-Concha N, López-Gutiérrez O, Pedraza-Alva G, Pérez-Martínez L. MiR-7 Promotes Epithelial Cell Transformation by Targeting the Tumor Suppressor KLF4. PLoS One. 2014;9:e103987. doi: 10.1371/journal.pone.0103987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2015 doi: 10.1038/onc.2015.460. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 58.Crippa E, Lusa L, De Cecco L, Marchesi E, Calin GA, Radice P, Manoukian S, Peissel B, Daidone MG, Gariboldi M. miR-342 regulates BRCA1 expression through modulation of ID4 in breast cancer. PLoS One. 2014;9:e87039. doi: 10.1371/journal.pone.0087039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng DL, Xiang YY, Ji LJ, Lu XJ. Competing endogenous RNA interplay in cancer: mechanism, methodology, and perspectives. Tumour Biol. 2015;36:479–488. doi: 10.1007/s13277-015-3093-z. [DOI] [PubMed] [Google Scholar]
- 61.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim MY, Hur J, Jeong S. Emerging roles of RNA and RNA-binding protein network in cancer cells. BMB Rep. 2009;42:125–130. doi: 10.5483/bmbrep.2009.42.3.125. [DOI] [PubMed] [Google Scholar]
- 67.Calabretta S, Richard S. Emerging Roles of Disordered Sequences in RNA-Binding Proteins. Trends Biochem Sci. 2015;40:662–672. doi: 10.1016/j.tibs.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 68.Zong FY, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, Fang R, Lu D, Ji H. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014;10:e1004289. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Y, Zhang G, Wei M, Lu X, Fu H, Feng F, Wang S, Lu W, Wu N, Lu Z, Yuan J. The tumor suppressing effects of QKI-5 in prostate cancer: A novel diagnostic and prognostic protein. Cancer Biol Ther. 2014;15:108–118. doi: 10.4161/cbt.26722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye Z, Jin H, Qian Q. Argonaute 2: A Novel Rising Star in Cancer Research. J Cancer. 2015;6:877. doi: 10.7150/jca.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McGlynn LM, McCluney S, Jamieson NB, Thomson J, MacDonald AI, Oien K, Dickson EJ, Carter CR, McKay CJ, Shiels PG. SIRT3 & SIRT7: Potential Novel Biomarkers for Determining Outcome in Pancreatic Cancer Patients. PLoS One. 2015;10:e0131344. doi: 10.1371/journal.pone.0131344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu SY, Wang YP, Chang JYF, Shen WR, Chen HM, Chiang CP. Increased expression of MCM5 is significantly associated with aggressive progression and poor prognosis of oral squamous cell carcinoma. J Oral Pathol Med. 2014;43:344–349. doi: 10.1111/jop.12134. [DOI] [PubMed] [Google Scholar]
- 73.de Wit M, Kant H, Piersma SR, Pham TV, Mongera S, van Berkel MP, Boven E, Pontén F, Meijer GA, Jimenez CR, Fijneman RJ. Colorectal cancer candidate biomarkers identified by tissue secretome proteome profiling. J Proteomics. 2014;99:26–39. doi: 10.1016/j.jprot.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 74.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331–9342. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 79.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 80.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]


