Abstract
Lung cancer and its metastasis is the leading cause of cancer-related mortality world-wide. Non-small cell lung cancer (NSCLC) accounts for about 90% of total lung cancer cases. Despite advancements in therapeutic approaches, only limited improvement has been achieved. Therefore, alternative strategies are required for the management of lung cancer. Here we report the chemotherapeutic effect of silymarin, a phytochemical from milk thistle plant (Silybum marianum L. Gaertn.), on NSCLC cell migration using metastatic human NSCLC cell lines (A549, H1299 and H460) together with the molecular targets underlying these effects. Using an in vitro cell migration assay, we found that treatment of human NSCLC cells (A549, H1299 and H460) with silymarin (0, 5, 10 and 20 µg/mL) for 24 h resulted in concentration-dependent inhibition of cell migration, which was associated with the inhibition of histone deacetylase (HDAC) activity and reduced levels of class 1 HDAC proteins (HDAC1, HDAC2, HDAC3 and HDAC8) and concomitant increases in the levels of histone acetyltransferase activity (HAT). Known HDAC inhibitors (sodium butyrate and trichostatin A) exhibited similar patterns of therapeutic effects on the lung cancer cells. Treatment of A549 and H460 cells with silymarin reduced the expression of the transcription factor ZEB1 and restored expression of E-cadherin. The siRNA knockdown of ZEB1 also reduced the expression of HDAC proteins and enhanced re-expression of the levels of E-cadherin in NSCLC cells. MicroRNA-203 (miR-203) acts as a tumor suppressor, regulates tumor cell invasion and is repressed by ZEB1 in cancer cells. Silymarin treatment restored the levels of miR-203 in NSCLC cells. These findings indicate that silymarin can effectively inhibit lung cancer cell migration and provide a coherent model of its mechanism of action suggesting that silymarin may be an important therapeutic option for the prevention or treatment of lung cancer metastasis when administered either alone or with standard cancer therapeutic drugs.
Keywords: Silymarin, non-small cell lung cancer cells, cancer cell migration, HDACs, miR-203, E-cadherin, ZEB1 transcription factor
Introduction
Lung cancer remains the leading cause of cancer-related deaths worldwide in both men and women, and is the second most common cancer accounting for about 13% of all newly diagnosed cancers. It was estimated that about 221,220 new cases of lung cancer were diagnosed in the year 2015 leading to 158,040 deaths from this disease in the United States [1,2]. Non-small cell lung cancer (NSCLC) and small cell lung cancer account for approximately 90% of all lung cancers. NSCLC represents about 80% of all lung cancers includes adenocarcinoma, squamous cell carcinoma and large cell carcinoma [3,4]. In clinical practice, a combination of chemotherapy and radiation therapy improves survival of the patients, but most of the patients die of disease progression due to acquired or intrinsic resistance to chemotherapeutic drug regimens [5] with metastasis being the major cause of mortality. Therefore, an approach that can prevent or block the metastatic capacity of lung cancer cells may facilitate the development of an effective strategy for the treatment and/or prevention of lung cancer and thus reduce the mortality due to lung cancer.
Epigenetic modification of DNA and histones plays a role in the control of gene expression and has been linked to the initiation, progression and metastasis of cancer cells [6,7]. Histone deacetylases (HDACs) regulate many biologic processes, including cell cycle progression and cell differentiation, etc. Among the four classes of HDACs, class I HDACs (HDACs 1-3 and 8) are most frequently overexpressed in human cancers and this overexpression correlates with poor prognosis and drug resistance [7,8]. Thus, class I HDACs are considered important candidate therapeutic targets for cancer [8,9], and several HDAC inhibitors (HDACi) have been identified. As HDACi modulate the expression of several genes that regulate multiple pathways associated with cancer cell growth and development [10,11], it is thought that inhibition of histone deacetylation may inhibit the epigenetic silencing of tumor suppressor genes that is frequently observed in cancer. This has driven the development of HDAC inhibitors for cancer therapy. Downregulation of E-cadherin expression also occur at the transcriptional level and plays a critical role in tumor progression and tumor cell metastasis. It has been demonstrated that epigenetic modifications are correlated with tumor suppressors, such as E-cadherin [6].
MicroRNAs (miRNAs) are a class of small non-coding RNAs that are approximately 19-24 nucleotides in length, and are capable of regulating about 20-30% of the genes in the human genome [12]. Experimental evidence indicates that miRNAs may function as tumor promoters or suppressors, regulating a wide range of biologic processes such as invasion, proliferation and apoptosis [13]. Several miRNAs families have been reported to be involved in the development of numerous cancers through regulation of cell proliferation, invasion and the epithelial-mesenchymal transition (EMT), etc. [14-16]. Studies have demonstrated that miRNAs are critical in the development of lung cancer [17]. miRNA-203 (miR-203) has been classified as a skin-specific miRNA but also is expressed in the squamous epithelium of cervix and esophagus [18-20]. It not only controls the skin’s protective barrier formation and epidermal differentiation and plays a role in skin disease but also acts as a tumor suppressor gene by regulating cell proliferation, differentiation, invasion, cell metastasis and apoptosis in certain type of cancers [19-23]. The miR-203 is downregulated in lung cancer cells and negatively regulates proliferation and the invasive potential of these cells [21]. In colorectal and pancreatic cells, miR-203 transcription is repressed specifically by the EMT activator ZEB1, thereby contributing to the invasive and metastatic behavior of these cells [24]. ZEB1 was found to be the most relevant repressor of E-cadherin expression by recruitment of HDAC1 and HDAC2 in cancer cells [25]. ZEB1 knock down was associated with prevention of HDAC binding to the CDH1 promoter, resulting in histone acetylation and re-expression of E-cadherin [25]. HDAC inhibitors, due to their ability to reactivate epigenetically tumor-silenced genes that are capable of inhibiting cancer cell migration, invasion and reversal of EMT are gaining interest as potential anticancer drugs [26,27]. Preclinical studies involving HDACi have shown a range of anticancer effects, such as tumor cell apoptosis, cell cycle arrest, modulation of immune response, and altered angiogenesis and have minimal toxicity against normal cells [28,29], however, there is less than convincing evidence that these agents are effective against solid tumors when used as a single agent [27]. Considering the limitations, the full therapeutic potential of HDACi will probably be best realized through their use in combination with other anticancer drugs, and certain phytochemicals that are non-toxic but possess anti-cancer activities.
Phytochemicals offer promising alternative approaches for the prevention of cancer cell metastasis [27,29]. As metastasis is a leading cause of cancer deaths in humans, in the present study we tested the chemotherapeutic effect of silymarin on the migration of human NSCLC cells. Silymarin, a flavonoid obtained from milk thistle (Silybum marianum L. Gaertn.) plant [30], has been shown to have significant anti-cancer activity in various tumor models but has non-significant toxic effects on normal non-malignant cells. In the present study, we explored the therapeutic effect of silymarin, on the migration of NSCLC cells. The A549, H1299 and H460 human NSCLC cell lines were used as in vitro models. We also tested whether the inhibitory effects of silymarin on the migration of NSCLC cells is associated with its effects on the multiple molecular targets that have been implicated in cancer cell metastasis, including class 1 HDACs, ZEB1 expression, and restoration of miR-203 and E-cadherin in NSCLC cells.
Materials and methods
NSCLC cell lines and cell culture
Human NSCLC cell lines, A549, H460 and H1299, and the normal human bronchial epithelial cell line (BEAS-2B) were obtained from the American Type Culture Collection (Manassas, VA). The cell lines were cultured as monolayers in RPMI 1640 culture medium supplemented with 10% heat-inactivated fetal bovine serum (Hyclone, Logan, UT), 100 µg/mL penicillin, and 100 µg/mL streptomycin and maintained in a humidified incubator. A stock solution of silymarin was prepared by dissolving the silymarin in a small amount of acetone (100 µL), which was then diluted as required in cell culture medium and added to sub-confluent cells (60-70%). Cells treated with the corresponding concentration of acetone (0.1%, v/v) served as the vehicle-treated control.
Chemicals, reagents and antibodies
Purified silymarin (>98%) was purchased from Sigma Chemical Co. Primary antibodies were purchased as follows: antibodies for class I HDAC (anti-HDAC1, anti-HDAC2, anti-HDAC3, anti-HDAC8); anti-histone H3; ZEB1; E-cadherin; β-actin; and secondary antibodies, horseradish peroxidase-linked anti-mouse immunoglobulin G and anti-rabbit immunoglobulin G, were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The HDAC Activity Assay Kit was purchased from Active Motif (Catalog # 56210, Carlsbad, CA). The enhanced chemiluminescence western blotting detection reagents were purchased from Amersham Pharmacia Biotech (Piscataway, NJ). ZEB1 siRNA transfection kit and transfection reagents were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz). Other chemicals and reagents were of analytical grade.
Measurement of migration of lung cancer cells
The migration of NSCLC cells was determined in vitro using Boyden Chamber (Neuroprobe, Inc., Gaithersburg, MD) fitted with Millipore membranes (8 µM pore size), as detailed previously [31,32]. Migratory cells were detected on the membrane after staining with crystal violet dye. The cell migration per treatment group was determined by counting the number of stained cells on at least three to four randomly selected areas/images using an Olympus BX41 microscope. Data are reported as the mean number of the migrating cells ± SD per image field. Representative photomicrographs showing the migratory cells were collected using a Qcolor5 digital camera system fitted to an Olympus BX41 microscope.
HDAC activity assay
HDAC activity was measured in nuclear lysates using the colorimetric HDAC Activity Assay Kit (Active Motif, Carlsbad, CA), which includes a positive control (a HeLa nuclear extract), a deacetylated HDAC assay standard, and a control inhibitor (trichostatin A; TSA) as well as the colorimetric HDAC substrate, following the manufacturer’s protocol. The absorbance was measured using a microplate reader at 405 nm, and the HDAC activity in treatment groups is presented as a percent of non-treated control group.
Histone acetyltransferase (HAT) activity assay
HAT activity in lung cancer cells was determined in nuclear lysates using the EpiQuikTM HAT Activity/Inhibition Assay Kit (Epigentek Group Inc.) following the manufacturer’s instructions. The color absorbance was read using a microplate reader at 450 nm and the HAT activity is reported as percent of control (non-treatment) group. Enzyme reaction quantifies the levels of acetylated histones colorimetrically through ELISA like reactions.
Western blot analysis
The NSCLC cells were harvested in the presence of a cocktail of protease inhibitors and whole cell lysates or nuclear lysates prepared, as detailed previously [31,32]. The protein concentration was determined and then equivalent amounts of total proteins resolved on 8-12% SDS-PAGE gels and transferred onto nitrocellulose membranes. The membrane was incubated with the specific primary antibody at 4°C overnight after blocking the non-specific binding sites with blocking buffer. The membrane was then incubated with the appropriate peroxidase-conjugated secondary antibody and the immune-reactive bands were visualized using the enhanced chemiluminescence reagents. The membrane was used 2-3 times after stripping and re-probed with any protein-specific antibody, or anti-β-actin or histone H3 antibody, to ensure equal protein loading on the gel. Western blot experiments were repeated once and the samples from two experiments were run in parallel.
miRNA extraction and RT-PCR
The extraction of miRNA from the cells and RT-PCR analysis were carried out as described previously [33]. Briefly, total RNA, which includes 95% of miRNA, were extracted from various lung cancer cell lines using the TRIZOL-chloroform method. The cDNA was prepared using the iScript cDNA synthesis kit (Bio-Rad) according to manufacturer’s protocol. RT-PCR was performed using Platinum Taq DNA Polymerase (Invitrogen, Carlsbad, CA) with a human-specific primer for miR-203: Forward primer: TCCAGTGGTTCTTAACAGTTCA, reverse primer: GGTCTAGTGGTCCT AAACATTTC and U6 forward primer CTCGCTTCGGCAGCACA. Reverse primer: AACGCTTCACGAATTTGCGT, as detailed [34]. The PCR reaction conditions were as follows: stage I: 95°C for 3 min, 53°C for 1 min, 72°C for 30 sec (2 cycles); Stage II: 95°C for 3 min, 53°C for 1 min, 72°C for 30 sec (55 cycles); Stage III: 72°C for 5 min. The PCR products were run on a 2.5-3% agarose gel prepared in 1x Tris-acetate EDTA buffer containing ethidium bromide and analyzed using Gel-Doc equipment.
MiR-203 mimic/inhibitor transfection in NSCLC cells
MiR-203 transfection in NSCLC cells was done using Mission® microRNA mimic from Sigma-Aldrich Co. (Cat No #HMI0357) following the manufacturer’s instructions. Briefly, 100 nM miR-203 mimic was used for cell transfection in six-well culture dishes. After transfection, cells were harvested and utilized in the cell migration assays. Cell lysates were prepared for western blot analyses and total RNA was extracted and used in RT-PCR for confirmation of upregulation of miR-203 in A549 and H460 NSCLC cells.
The A549 NSCLC cells were transfected with miR-203 inhibitor (Life Technologies, Cat. No. MH21541). Briefly, after 24 h of silymarin (20 µg/mL) treatment, transfection with anti-miR-203 (50 nM) was performed followed by manufacture’s protocol. After transfection, cells were harvested and migration was performed using Boyden chambers. Before transfection, silymarin was removed and data were compared with time matched migratory potential of lung cancer cells with and without silymarin treatment.
Assessment of ZEB1 binding with HDAC1 and HDAC2
For binding assays, cells were treated with or without silymarin (10, 20 µg/mL), sodium butyrate (1, 10 mM) or TSA (0.1 and 1 µM) for 24 h [25], and nuclear lysates were prepared. Lysates containing 400 µg of protein were cleared with protein A/G-plus agarose beads (Santa-Cruz, CA). Pre-cleaned lysates from each treatment group were then mixed with anti-ZEB1 antibody/protein A/G-plus agarose beads mixture (100 µL) that had been pre-cleaned at 4°C for 4 h and washed to remove unbound antibody. The mixture was incubated overnight at 4°C with rotary agitation, then the beads were washed and the bound protein recovered in lysis buffer and subjected to 10% SDS-PAGE followed by immunoblotting using antibodies specific for the HDAC1 and HDAC2.
ZEB1-siRNA transient transfection of NSCLC cells
For functional analyses, ZEB1 was knocked down using the human-specific ZEB1 siRNA Transfection Reagent Kit (Santa Cruz Biotechnology, Santa Cruz, CA) following the manufacturer’s protocol. Briefly, cells were seeded onto 6-well culture plates (2x105 cells per well). The ZEB1 siRNA mix with transfection reagents or scrambled siRNA reagents were overlaid on the cells for 7 h at 37°C and then transferred into 2x growth medium. After 24 h of transfection, cells were kept in a culture medium containing 2% FBS for up to 48 h. Cells treated with scrambled probe were used as controls for the transfected cells.
Statistical analyses
The statistical significance of the difference between the values of control and treatment groups was calculated either by using the Student t test or simple one-way ANOVA using the GraphPad Prism program (San Diego, CA). In each case P<0.05 was considered statistically significant.
Results
Silymarin inhibits migration of human NSCLC cells
As the migration, invasion and/or metastasis of cancer cells is a major cause of human deaths due to cancer, we first checked the effect of silymarin on the migration of three major metastasis-specific NSCLC cell lines (A549, H1299, H460) using the in vitro Boyden chamber assay, as we have described previously [31,32]. The concentrations of silymarin (0, 5, 10 and 20 µg/mL) used in this analysis were selected on the basis that they did not significantly affect the viability of NSCLC cells within the time-frame of the experiments (24 h). As shown in Figure 1A, relative to cells that were not treated with silymarin (control group), treatment of cells with silymarin reduced the migration of A549, H1299 and H460 cells in a concentration-dependent manner. Representative photomicrographs of the filters are provided in Figure 1A and numbers of migrating cells/panel are summarized in Figure 1B. Silymarin had a significant inhibitory effect on all three cell lines: A549 cells, 40-74% (P<0.5-0.001); H1299 cells, 33-75% (P<0.5-0.001); H460 cells, 28-68% (P<0.5-0.001). As the effects of silymarin on the migration of the NSCLC cell lines were similar, we selected two prominent NSCLC cell lines (A549 and H460) for further exploration of molecular targets and the mechanisms involved in the silymarin suppression of cell migration.
Figure 1.
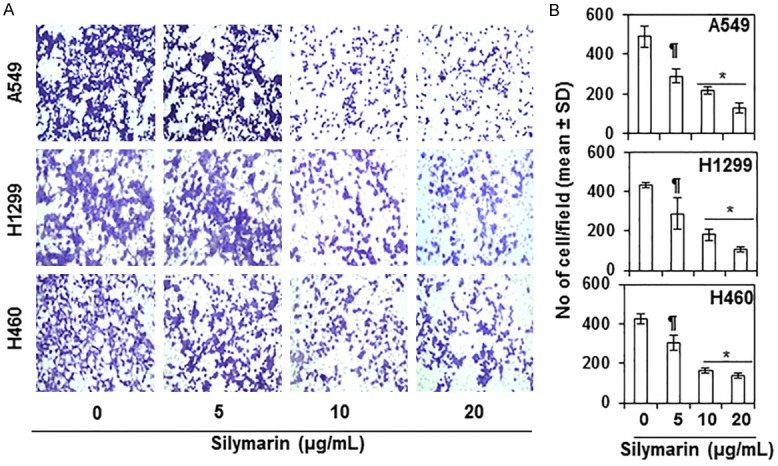
Effect of silymarin on migration of human non-small cell lung cancer (NSCLC) cells. Treatment of A549, H1299 and H460 NSCLC cells with silymarin (0, 5, 10 and 20 µg/mL) for 24 h inhibited the migration of the cells in a concentration-dependent manner. A. Representative photomicrographs of the Millipore filters used in the cell migration assays for each treatment group are shown. Migratory cells appear purple-blue. B. The number of migratory cells were counted on each photomicrograph for each treatment group and data are summarized as the mean number of migrating cells ± SD. Significant inhibition by silymarin versus vehicle-treated control cells, ¶P<0.05; *P<0.001.
Treatment with silymarin reduces the expression of class 1 HDACs proteins and HDAC activity while increasing HAT activity in NSCLC cells
Epigenetic regulators, in particular class 1 HDACs, have been shown to play a crucial role in tumor progression and tumor cell metastasis [6,7,9]. We therefore tested whether silymarin has any direct effect on class 1 HDAC protein (HDAC1, HDAC2, HDAC3 and HDAC8) expression and HDAC activity in NSCLC cells and whether any such silymarin-mediated changes in HDACs play a role in the silymarin inhibition of NSCLC cell migration. A549 and H460 cells were treated with silymarin (0, 5, 10 and 20 µg/mL) for 24 h and cell lysates were prepared for western blot analysis. Western blot analysis revealed that treatment with silymarin resulted in a dose-dependent reduction in the levels of the HDAC1, HDAC2, HDAC3 and HDAC8 proteins in both cell lines as compared with the vehicle-treated control cells (Figure 2A). Treatment with silymarin also significantly inhibited (P<0.05-0.01) HDAC activity in both cell lines as compared with cells treated with vehicle alone (Figure 2B). HAT activity also was determined in the same set of experiments using nuclear extracts and the EpiQuikTM HAT Activity Assay Kit. We found that the levels of HAT activity were significantly higher (P<0.05- 0.001) in silymarin-treated NSCLC cells as compared with the vehicle-treated control cells (Figure 3A & 3B). The increase in HAT activity induced by silymarin treatment ranged from 2-4 fold in A549 cells and ~1.5 to 2.5 fold in H460 cells.
Figure 2.
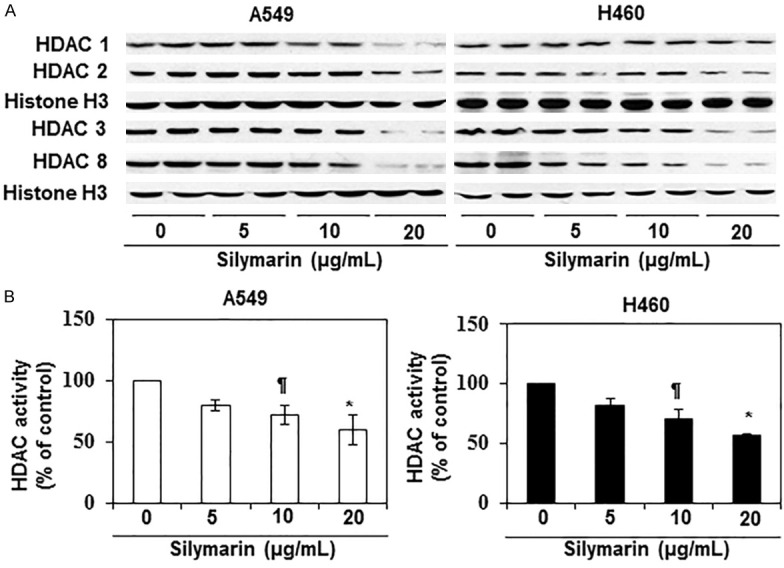
Effect of silymarin on class I HDAC proteins and HDAC activity in human NSCLC cells. A549 and H460 cell lines were treated with various concentrations of silymarin (0, 5, 10 and 20 µg/mL) for 24 h, nuclear lysates were then prepared and subjected to either western blot analysis or HDAC activity assay. A. Treatment of cells with silymarin reduced the levels of class 1 HDAC proteins (HDAC1, HDAC2, HDAC3 and HDAC8) in a concentration-dependent manner in both cell lines. Equal loading of proteins was verified using blotting with an antibody against histone H3. B. The effect of silymarin on HDAC activity was measured using the HDAC Activity Assay Kit (Active Motif), following manufacturer’s instructions. Significant inhibition was observed versus vehicle-treated control cells, ¶P<0.05, *P<0.01.
Figure 3.
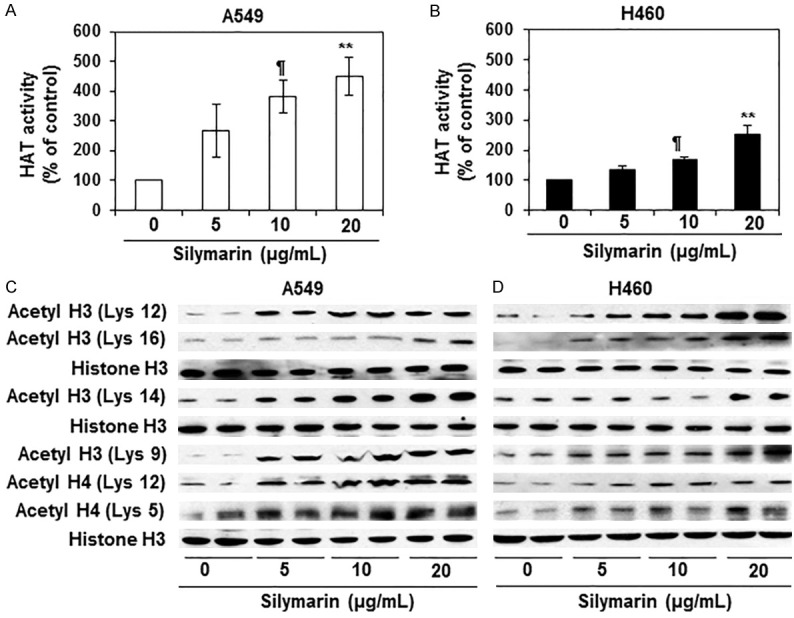
Effect of silymarin on HAT activity and acetylated histones H3 and H4 in A549 and H460 human NSCLC cells. (A and B) A549 and H460 cell lines were treated with various concentrations of silymarin for 24 h and HAT activity was measured using HAT Activity/Inhibition Assay Kit (Epigentek Group, Inc.). Treatment of NSCLC cells with silymarin enhanced the levels of HAT activity in both A549 (A) and H460 (B) cell lines in a dose-dependent manner. Data on HAT activity are presented in terms of percent of control as the mean ± SD, n=3. Significant difference versus vehicle-treated controls, ¶P<0.01, **P<0.001. (C and D) A549 and H460 cell lines were treated with various concentrations of silymarin for 24 h, and lysates were subjected to western blot analysis for acetylated forms of histones at various lysine residues. As shown, silymarin increased the expression levels of histone acetylation at various lysine residues in A549 and H460 cells. Equal loading of protein samples was verified using anti-histone H3 antibody.
Silymarin increases the levels of acetylated histones H3 and H4 at different lysine residues in NSCLC cells
We next determined whether silymarin affects the acetylation status of histones in the A549 and H460 cells. Cells were treated with silymarin (0, 5, 10 and 20 µg/mL) for 24 h, then harvested and nuclear lysates prepared for the analysis of acetylation of histones at specific lysine residues by western blot analysis. It was observed that treatment of A549 and H460 cells with silymarin resulted in a concentration-dependent increase in acetylation at various lysine residues of the acetylated histone H3 (Lys 12, Lys 16, Lys 14 and Lys 9) and histone H4 (Lys 12, Lys 5) as compared with the vehicle treated control cells (Figure 3C, 3D). The patterns of silymarin-induced lysine acetylation were similar in both cell lines.
Effects of silymarin on ZEB1 transcription factor and E-cadherin expression
ZEB1 is a transcription factor known to recruit HDACs. It forms a ZEB1/HDAC complex that binds to the CDH1 promoter, which results in histone deacetylation and suppression of E-cadherin levels [25]. E-cadherin is an important mediator of intercellular adhesion and tissue integrity that contributes to prevention of cancer cell migration [25]. Therefore, we determined the effect of silymarin on the expression profile of ZEB1 and E-cadherin in A549 and H460 cell lines following the treatment protocol described above. Western blot analysis revealed that treatment of A549 and H460 cell lines with silymarin for 24 h resulted in reduced expression level of ZEB1 and a concomitant increase in the expression levels of E-cadherin in a concentration-dependent manner (Figure 4A).
Figure 4.
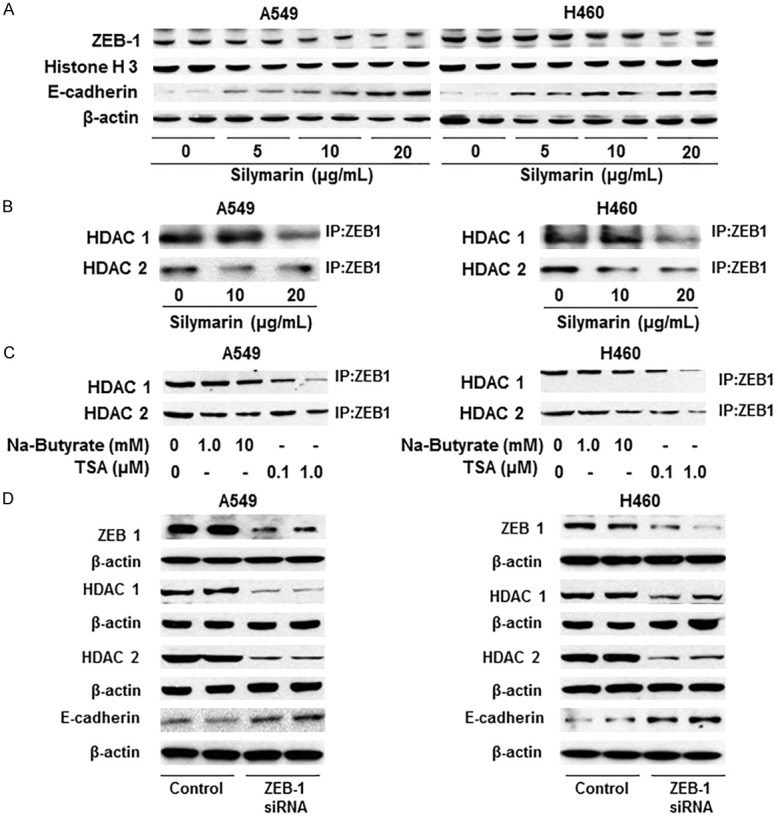
Effect of silymarin on the levels of ZEB1 and HDACs in NSCLC cells. A. A549 and H460 cells were treated with silymarin (0, 5, 10, 20 µg/mL) for 24 h, nuclear lysates were prepared and subjected to western blot analyses for ZEB1 and E-cadherin protein. Silymarin treatment decreased the levels of ZEB-1 while increasing or restoring the levels of E-cadherin in both the A549 and H460 cell lines. Histone H3 and β-actin antibodies were used to verify equal protein loading. B. Treatment of silymarin decreased binding or complex formation of HDAC1 or HDAC2 with the transcription factor ZEB1 in lung cancer cells. After treatment of cells with silymarin, ZEB1 was immunoprecipitated and immunoblotting was performed using antibodies against HDAC1 and HDAC2. C. Treatment of A549 and H460 cells with HDAC inhibitors (sodium butyrate and TSA) also decreased or inhibited the binding of HDAC1 and HDAC2 with ZEB1. Cells were treated with HDAC inhibitors for 24 h, then nuclear lysates were prepared and subjected to immunoprecipitation of ZEB1, as detailed in the Materials and methods section. Western blot analyses was performed using anti-HDAC-1 and anti-HDAC-2 antibodies. Sodium butyrate and TSA were used as the positive controls for HDAC inhibition. D. Effect of siRNA knockdown of ZEB1 on the levels of HDAC1, HDAC2 and E-cadherin in A549 and H460 cells. Knockdown of ZEB1 in cells decreased the levels of HDAC1, HDAC2 proteins and restored the levels of E-cadherin.
Silymarin and inhibitors of HDACs (sodium butyrate and TSA) block the binding of HDAC1 and HDAC2 to ZEB1 in NSCLC cells
To test the effect of silymarin on the binding of HDACs and ZEB1, A549 and H460 cells were treated with silymarin for 24 h. Cells were harvested and ZEB1 was immunoprecipitated from the nuclear extract, as detailed in the Materials and methods section, followed by immunoblotting using antibodies against HDAC1 and HDAC2. As shown in Figure 4B, silymarin treatment reduced the amount of HDAC1 or HDAC2 bound to ZEB1 in both cell lines. As a positive control for demonstration of HDAC inhibition, A549 and H460 cells were treated with known HDAC inhibitors (sodium butyrate and TSA) (Figure 4C). To further verify the role of ZEB1 on HDAC and E-cadherin expression in NSCLC cells, we knocked down ZEB1 using siRNA in A549 and H460 cells. Western blot analysis confirmed that the knockdown of ZEB1 in these cells reduced the levels of HDAC1 and HDAC2 proteins and induced re-expression of E-cadherin as compared with control cells that were treated with scrambled siRNA (Figure 4D).
Effects of HDAC inhibitors on E-cadherin and HDAC protein expression, HDAC and HAT activity in NSCLC cells
To further verify and explore whether the effects of silymarin on the NSCLC cells are similar to those induced by known inhibitors of HDAC, we treated the A549 and H460 cells with sodium butyrate and TSA separately for 24 h. Whole cell lysates and nuclear lysates were prepared and subjected to immunoblotting. Immunoblotting analysis revealed that treatment of NSCLC cells with these inhibitors reduced the expression of HDAC1 and HDAC2, while enhancing the expression of acetylated H3 (Lys 9) and E-cadherin in both cell lines. The patterns of the effects of these HDAC inhibitors on these biomarkers were identical in both cell lines (Figure 5A). Additionally, treatment with these HDAC inhibitors also significantly decreased (P<0.05-0.01) HDAC activity (Figure 5B) while significantly enhancing (P<0.05-0.001) HAT activity (Figure 5C) in both cell lines.
Figure 5.
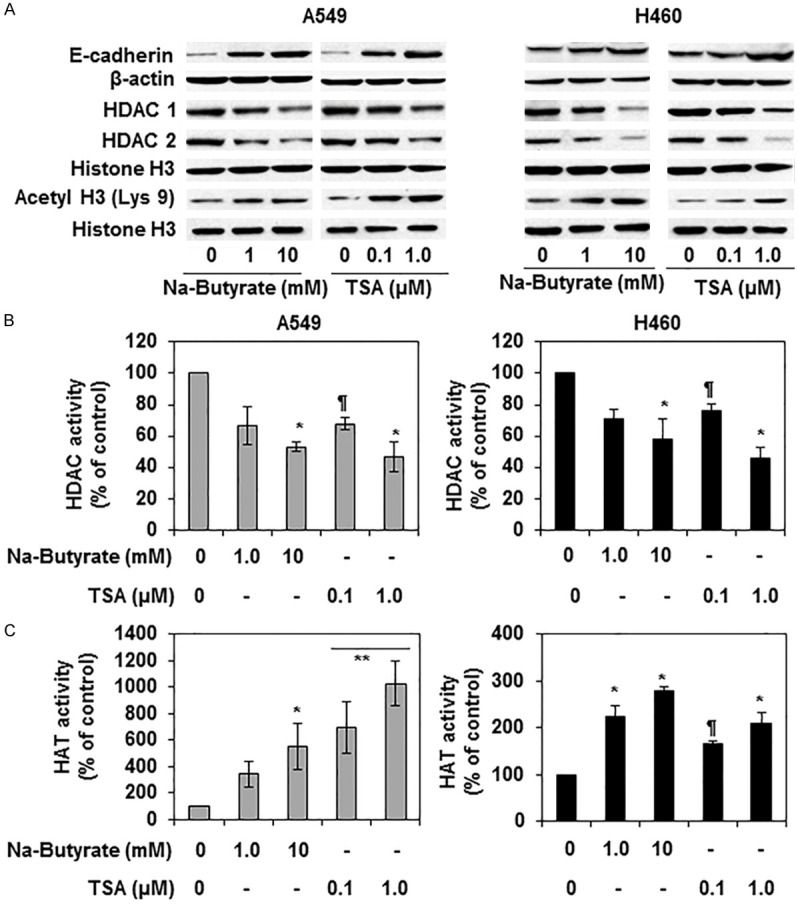
Effect of HDAC inhibitors (sodium butyrate and TSA) on E-cadherin and various biomarkers of epigenetic regulators in A549 and H460 cells. (A) Treatment of cells with HDAC inhibitors restored the levels of E-cadherin and acetylated histone H3 (Lys 9) while decreasing the levels of HDAC 1, HDAC2 in A549 and H460 cells. Cells were treated with sodium butyrate (0, 1, 10 mM) and TSA (0. 0.1 and 1 µM) in culture for 24 h. Whole cell lysates or nuclear lysates were prepared and used for western blot analyses. Histone H3 antibody was used as a loading control. (B) Treatment with HDAC inhibitors decreased HDAC activity in cells. Cells were treated as detailed above, and nuclear lysates were used to measure the activity using HDAC Activity Assay Kit following the manufacture’s protocol. Significant difference vs. control group, *P<0.01, ¶P<0.05. (C) HDAC inhibitor treatment enhanced the levels of HAT activity in both A549 and H460 cell lines. Treatment protocols were identical to those detailed in (A and B). Significant increase vs non-treated control cells, **P<0.001; *P<0.01; ¶P<0.05.
The anti-lung cancer cell migration activity of silymarin is correlated with the re-expression of miRNA-203 in NSCLC cells
To examine the role of miR-203 in lung cancer cell migration, we first examined the basal levels of miR-203 in several lung cancer cell lines, including H1299, A549, A427, H460, HCC827 and SK-MES 1, and compared these with the level of expression in normal human bronchial epithelial (NHBE) cells. miRNA was extracted from all the cell lines and subjected to RT-PCR analysis as detailed in the Materials and methods section. As shown in Figure 6A, gel electrophoresis data indicated that the levels of miR-203 were markedly lower in the lung cancer cell lines as compared with the NHBE control cells. Next we examined the effect of silymarin on the levels of miR-203 in two selected NSCLC cell lines, A549 and H460. Cells were treated with silymarin (5, 10 and 20 µg/mL) for 24 h. miRNA was then isolated from the cells and subjected to RT-PCR analysis with the RT-PCR products being separated by agarose gel electrophoresis. The resultant data indicated that the treatment of cells with silymarin increased the levels of miR-203 in a dose-dependent manner in both the A549 and H460 cell lines (Figure 6B).
Figure 6.
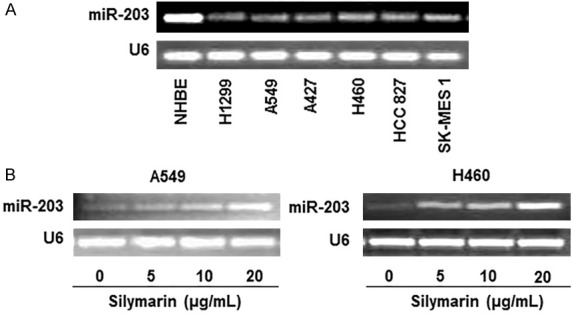
Analysis of miRNA-203 level in various human NSCLC cell lines using RT-PCR. A. Levels of miR-203 in various NSCLC cell lines (H1299, A549, A427, H460, HCC827 and SK-MES1) were lower as compared to normal human bronchial epithelial (NHBE) cells. B. Treatment of silymarin restored the levels of miR-203 in A549 and H460 cells. Cells were treated with silymarin (0, 5, 10, 20 µg/mL) in culture for 24 h and total RNA was isolated, cDNA was synthesized and subjected to miR-203 specific PCR analysis. U6 was used as a loading control, as detailed in the Materials and methods section.
Treatment of NSCLC cells with miR-203 mimic reduces the migration of lung cancer cells and decreases the levels of HDACs and ZEB1 while restoring the expression of E-cadherin
To explore whether miR-203 has a role in silymarin-mediated inhibition of cancer cell migration, A549 and H460 NSCLC cells were treated with a mimic of miR-203 and the effects on cell migration determined. It was observed that treatment with the miR-203 mimic inhibited the migration of lung cancer cells as is evident by the density of migrating cells in blue/purple on the membrane (Figure 7A). Quantitative analysis of the migrating cells indicated that treatment of cells with mimic miR-203 significantly inhibited (45-47%, P<0.01) the migration of cells, as shown in Figure 7B. RT-PCR data confirmed that treatment of A549 and H460 cells with the mimic miRNA-203 enhanced the expression of miR-203 in cancer cells (Figure 7C). Collectively, these results suggest that silymarin might inhibit lung cancer cell migration through induction of the expression of miR-203 in lung cancer cells. To examine whether treatment with the mimic miR-203 has any effect on HDACs and ZEB-1 expression or E-cadherin expression in lung cancer cells, NSCLC cells were treated with mimic miR-203 for 24 h, and cell lysates prepared for analysis of these biomarkers. Western blot analysis revealed that treatment of A549 and H460 cells with mimic miR-203 suppressed the expression of ZEB-1, HDAC1 and HDAC2 while increasing the expression of the epithelial biomarker, E-cadherin, as compared with the control cells which were not treated with mimic miR-203 (Figure 7D).
Figure 7.
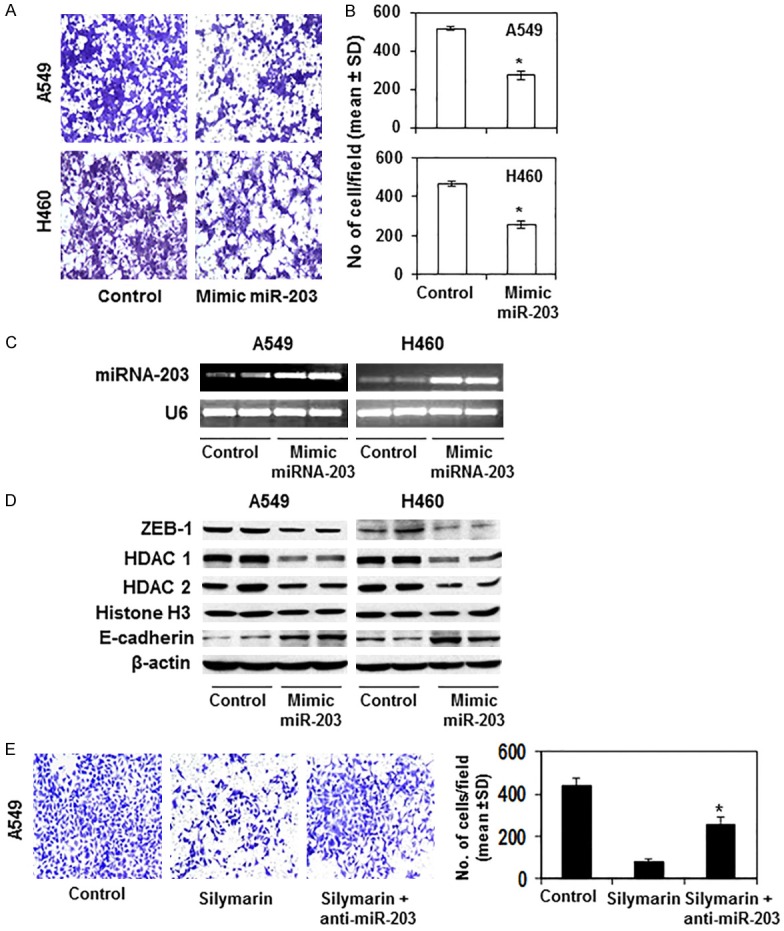
Effect of transient transfection of cells with miR-203 mimic on cell migration, ZEB1, HDACs and E-cadherin proteins in A549 and H460 cells. A and B. miR-203 transfection resulted in decreased cell migration in A549 and H460 cells. The migrating cells were counted and the number expressed as the mean number of migratory cells ± SD per photomicrograph. Significant inhibition by miR-203 mimic versus control cells, *P<0.001. C. Confirmation of expression of miR-203 after transfection of cells with miR-203 mimic. D. Protein levels of ZEB1, HDAC1, 2 and E-cadherin were assessed after transfection of cells with mimic miR-203. E. Depletion of miR-203 in A549 cells using anti-miR-203 decreases the anti-cell migration ability of silymarin (20 µg/mL). Significant difference versus silymarin treatment alone group of cells, *P<0.01.
Depletion of miRNA-203 in A549 cells reduces the anti-cell migration capacity of silymarin
To verity that upregulation of miR-203 by silymarin in NSCLC cells contributes to the inhibition of cell migration, additional experiments were conducted. The increased expression of miR-203 by silymarin treatment in A549 cells was depleted using anti-miR-203 transfection, and thereafter cell migration potential was measured using Boyden chamber assay. Cell migration data (Figure 7E) revealed that depletion of miR-203 expression restored migratory potential of A549 cells by >40% (p<0.01) in presence of silymarin, while it was observed that silymarin treatment (20 µg/mL) inhibits lung cancer migration in A549 cells up to 82% (p<0.001). These data suggest that deletion of miR-203 decreases the anti-cell migration effect of silymarin.
Discussion
Available efficacious cancer therapeutic drugs commonly have serious toxicities and debilitating side effects and their long-term use is often compromised by the emergence of resistance. It has been shown that silymarin exhibits cytotoxic effects against cancerous cells, but does not exhibit toxic effects on the growth and viability of normal (non-neoplastic) cells [30,32]. Because of its anti-carcinogenic potential, we have assessed its effects on the migration of NSCLC cells and the underlying molecular mechanism involved. In the current study, we took into account that phytochemicals in general and silymarin specifically can affect multiple molecular pathways and, based on the findings sought to identify primary potential molecular targets that would explain the multiplicity of responses.
Cancers are manifestation of genetic and epigenetic alterations. Modifications in epigenetic regulators, such as histones, affect chromatin structure and play an important role in malignancies. Acetylation and deacetylation modifications of histones have been identified clinically as predictors of cancer progression [35-37]. We have found that silymarin treatment inhibits the migration of NSCLC cells (A549, H1299 and H460) and this inhibitory potential of silymarin is associated with the inhibition of HDAC activity and reduces the levels of class 1 HDAC proteins in a dose-dependent manner, while increasing HAT activity. It has been recognized that class 1 HDACs are responsible for deacetylation of the catalytic core for different co-repressor complexes resulting in transcriptional repression. Our study also reveals that treatment of silymarin enhances the expression levels of acetylated histones H3 and H4 at different lysine residues which is due to enhanced HAT activity in NSCLC cells.
E-cadherin is an important epithelial mediator of intercellular adhesion and tissue integrity. Loss of E-cadherin expression in tumor tissues promotes the epithelial-to-mesenchymal transition (EMT) thereby promoting cancer cell migration and invasion. In our study model, we found that silymarin treatment reactivates or restores the expression levels of E-cadherin in A549 and H460 cells. The effects of silymarin on several different regulatory mechanisms can contribute to the reactivation of E-cadherin expression in the silymarin-treated NSCLC cells. As we show in this study, these include functional inactivation of HDAC and/or activation of HAT activity in lung cancer cells by silymarin. Enhanced E-cadherin expression correlates with the reduced migration of NSCLC cells treated with silymarin. Therefore, it appears that silymarin-mediated histone modifications is a regulatory mechanism of re-expression or restoration of E-cadherin in NSCLC cells, which is associated with inhibition of lung cancer cell migration. These observations were supported and verified when NSCLC cells were treated with HDAC inhibitors, such as sodium butyrate and TSA. Treatment of A549 and H460 cells with these HDAC inhibitors also reduced the expression of HDAC1 and HDAC2 and HDAC activity while increases the expression of E-cadherin. Silymarin also targets ZEB1, a transcription factor, in NSCLC cells. Our study reveals that treatment of silymarin downregulates or reduces the expression of ZEB1 in A549 and H460 cells. HDAC inhibition and siRNA knockdown of ZEB1 in NSCLC cells resulted in re-expression of E-cadherin in these lung cancer cell lines. It has been shown that the transcriptional repressor ZEB1 binds to the E-cadherin promoter and has been found to be associated with downregulation of HDAC1 and HDAC2 [25]. Therefore, ZEB1 may be a critical regulatory mechanism for E-cadherin re-expression in silymarin-treated lung cancer cells.
Several miRNA families have been implicated in the EMT transition, and EMT regulators modulate the invasive potential of tumor cells [16]. ZEB1, which is an EMT activator, has been shown to repress the transcription of miR-203 that contributes to the metastatic behavior of cancer cells [21]. To explore whether miR-203 is involved in the silymarin-mediated inhibition of lung cancer cell migration, we tested the levels of miR-203 in NSCLC cells. We found that the expression levels of miR-203 were lower in the several NSCLC cell lines we studied as compared to NHBE control cells. Treatment of two of these NSCLC cell lines (A549 and H460) with silymarin was found to enhance the expression of miR-203. miR-203 acts as a tumor suppressor by regulating proliferation, invasion and metastasis via actions on its target genes [20,23]. Bueno et al. [38] have demonstrated that the genetic and epigenetic silencing of miR-203 enhanced the expression of oncogenes, and suggest that miR-203 can act as a tumor suppressor and provides an alternative targeting strategy for the treatment of malignancies. Our study suggests that miR-203 also plays a role in the anti-migratory effects of silymarin on lung cancer cells. The role of miR-203 was evident and further supported by the effects of transfection of NSCLC cells with miR-203 mimic as well as inhibitor of miR-203. This resulted in restoration of miR-203 and suppressed the migration of A549 and H460 cells. Concomitantly, it decreased the levels of ZEB1, and HDAC1 and HDAC2 while restoring the levels of E-cadherin in NSCLC cells. These novel findings suggest that miR-203 may be a useful indicator of the metastatic potential of NSCLC cells and that silymarin is a competent agent that has the ability to target this potential therapeutic target in lung malignancy.
In summary, we have identified that silymarin inhibits the migration of NSCLC cells and that this is mediated through: (i) inhibition of class 1 HDAC protein expression and HDAC activity and upregulation of HAT activity, (ii) a reduction in ZEB1 expression, and (iii) re-expression of miR-203 in NSCLC cells. These silymarin-induced alterations in NSCLC cells lead to re-expression of the epithelial marker E-cadherin in NSCLC cells. The reversal of epigenetic effects (e.g., class HDAC proteins), and re-expression of miR-203 and E-cadherin by silymarin may allow new therapeutic options for the prevention and/or treatment of non-small cell lung cancer in human patients.
Acknowledgements
This work was financially supported by the Veterans Administration Merit Review Award (1IO1 BX001410) to S.K.K. The funding agency had no roles in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of conflict of interest
None.
References
- 1.American Cancer Society. Cancer facts and figures. Available: http://www.cancer.org/. Accessed 2015, June 20.
- 2.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 3.Maziak DE, Markman BR, MacKay JA, Evans WK Cancer Care Ontario Practice Guidelines Initiative Lung Cancer Disease Site Group. Photodynamic therapy in nonsmall cell lung cancer: a systematic review. Ann Thorac Surg. 2004;77:1484–1491. doi: 10.1016/j.athoracsur.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 5.Ferrigno D, Buccheri G. Second-line chemotherapy for recurrent non-small cell lung cancer: do new agents make a difference? Lung Cancer. 2000;29:91–104. doi: 10.1016/s0169-5002(00)00112-4. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 7.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 8.Weichert W, Röske A, Niesporek S, Noske A, Buckendahl AC, Dietel M, Gekeler V, Boehm M, Beckers T, Denkert C. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008;14:1669–1677. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- 9.Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett. 2009;280:168–176. doi: 10.1016/j.canlet.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 10.Ellis L, Hammers H, Pili R. Targeting tumor angiogenesis with histone deacetylase inhibitors. Cancer Lett. 2009;280:145–153. doi: 10.1016/j.canlet.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava RK, Kurzrock R, Shankar S. MS-275 sensitizes TRAIL resistant breast cancer cells, inhibits angiogenesis and metastasis, and reverses epithelial-mesenchymal transition in vivo. Mol Cancer Ther. 2010;9:3254–3266. doi: 10.1158/1535-7163.MCT-10-0582. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Liu C, Liu X, Tang DG, Wang J. The microRNA miR-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44hi stem-like NSCLC cells. PLoS One. 2014;9:e90022. doi: 10.1371/journal.pone.0090022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Xu G, Yin C, Jin W, Zhang G. Down-regulation of miR-203 induced by Helicobacter pylori infection promotes the proliferation and invasion of gastric cancer by targeting CASK. Oncotarget. 2014;5:11631–11640. doi: 10.18632/oncotarget.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ju SY, Chiou SH, Su Y. Maintenance of the stemness in CD44(+) HCT-15 and HCT-116 human colon cancer cells requires miR-203 suppression. Stem Cell Res. 2014;12:86–100. doi: 10.1016/j.scr.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 17.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 18.Sonkoly E, Wei T, Janson PC, Sääf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Ståhle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Li A, Hong SM, Hruban RH, Goggins M. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res. 2012;18:981–992. doi: 10.1158/1078-0432.CCR-11-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luzna P, Gregar J, Uberall I, Radova L, Prochazka V, Ehrmann J Jr. Changes of microRNAs-192, 196a and 203 correlate with Bar-rett’s esophagus diagnosis and its progression compared to normal healthy individuals. Diagn Pathol. 2011;6:114. doi: 10.1186/1746-1596-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin J, Deng J, Wang F, Xia X, Qiu T, Lu W, Li X, Zhang H, Gu X, Liu Y, Cao W, Shao W. The expression and function of microRNA-203 in lung cancer. Tumour Biol. 2013;34:349–357. doi: 10.1007/s13277-012-0556-3. [DOI] [PubMed] [Google Scholar]
- 22.Bian K, Fan J, Zhang X, Yang XW, Zhu HY, Wang L, Sun JY, Meng YL, Cui PC, Cheng SY, Zhang J, Zhao J, Yang AG, Zhang R. MicroRNA-203 leads to G1 phase cell cycle arrest in laryngeal carcinoma cells by directly targeting survivin. FEBS Lett. 2012;586:804–809. doi: 10.1016/j.febslet.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 23.Yu J, Li A, Hong SM, Hruban RH, Gogins M. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res. 2012;18:981–992. doi: 10.1158/1078-0432.CCR-11-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schüler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 25.Aghdassi A, Sendler M, Guenther A, Mayerle J, Behn CO, Heidecke CD, Friess H, Büchler M, Evert M, Lerch MM, Weiss FU. Recruitment of histone deacetylase HDAC1 and HDAC2 by transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut. 2012;61:439–448. doi: 10.1136/gutjnl-2011-300060. [DOI] [PubMed] [Google Scholar]
- 26.Song SH, Han SW, Bang YJ. Epigenetic-based therapies in cancer: progress to date. Drugs. 2011;71:2391–2403. doi: 10.2165/11596690-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 27.Bots M, Johnstone RW. Anticancer activities of histone deacetyalse inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 28.Marsoni S, Damia G, Camboni G. A work in progress: the clinical development of histone deacetylase inhibitors. Epigenetics. 2008;3:164–171. doi: 10.4161/epi.3.3.6253. [DOI] [PubMed] [Google Scholar]
- 29.Singh T, Prasad R, Katiyar SK. Inhibition of class I histone deacetylases in non-small cell lung cancer by honokiol leads to suppression of cancer cell growth and induction of cell death in vitro and in vivo. Epigenetics. 2013;8:54–65. doi: 10.4161/epi.23078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones V, Katiyar SK. Emerging phytochemicals for prevention of melanoma invasion. Cancer Lett. 2013;335:251–258. doi: 10.1016/j.canlet.2013.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Punathil T, Katiyar SK. Inhibition of non-small cell lung cancer cell migration by grape seed proanthocyanidins is mediated through the inhibition of nitric oxide, guanylate cyclase, and ERK1/2. Mol Carcinog. 2009;48:232–242. doi: 10.1002/mc.20473. [DOI] [PubMed] [Google Scholar]
- 32.Vaid M, Prasad R, Sun Q, Katiyar SK. Silymarin targets β-catenin signaling in blocking migration/invasion of human melanoma cells. PLoS One. 2011;6:e23000. doi: 10.1371/journal.pone.0023000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Prasad R, Katiyar SK. Down regulation of miRNA-106b inhibits growth of melanoma cell by promoting G1-phase cell cycle arrest and reactivation of p21/WAF1/Cip1 protein. Oncotarget. 2014;5:10636–10649. doi: 10.18632/oncotarget.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis CD, Ross SA. Dietary components impact histone modifications and cancer risk. Nutr Rev. 2007;65:88–94. doi: 10.1111/j.1753-4887.2007.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 36.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Pérez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 37.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 38.Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernández-Piqueras J, Malumbres M. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]


