Abstract
Bone morphogenetic protein receptors (BMPRs) are multifunctional proteins; they have indispensible roles in the process of BMP signaling. However, their function in dedifferentiated chondrosarcoma is uncertain. It has been reported that BMPR2 is associated with chondrosarcoma. Moreover, the detection of BMPR2 is more frequent in dedifferentiated chondrosarcomas (DDCS) than in conventional chondrosarcomas (CCS). BMPR2, phospho-SMAD1/5 (pSMAD1/5), and runt-related transcription factor 2 (RUNX2) expressions were found to be associated with the pathological grades of chondrosarcoma and could be a promising target of treatment outcome. Moreover, BMPR2 was found to induce the RUNX2 expression via pSmad1/5. Knockdown of BMPR2 and pSmad1/5 results in the downregulation of RUNX2 expression in DDCS cells, while the upregulation of BMPR2 and Smad1/5 in CCS cells leads to increased RUNX2 expression. The luciferase reporter gene assay suggested that BMPR2 can induce the RUNX2 expression at the transcriptional level. By chromatin immunoprecipitation (ChIP) and electrophoresis mobility shift assay (EMSA), it was found that pSmad1/5 combined directly to RUNX2. The in vivo tumorigenicity assay in mice showed that the inhibition of BMPR2 or Smad1/5 in DDCS cell line reduced tumor growth, while the upregulation of BMPR2 or Smad1/5 in CCS cell line increased tumor growth. Furthermore, a BMPR signaling inhibitor, LDN-193189, was introduced to investigate its role as a potential drug to treat DDCS. Taken together, the present-study results suggest that BMPR2-pSmad1/5 signaling pathway has an important role in regulating not only the RUNX2 expression but also the tumorigenesis of DDCS.
Keywords: Dedifferentiated chondrosarcoma, conventional chondrosarcoma, BMPR2, RUNX2, p-Smad1/5, progression
Introduction
Dedifferentiated chondrosarcoma (DDCS) is a special type of sarcoma, accounting for approximately 10%-15% of conventional chondrosarcoma (CCS) [1], which is a neoplasm consisting of two different components, a high-grade malignant tumor adjacent to a low-grade CCS, with a clear border between the two parts [2]. DDCS has a poor prognosis, with a 5-year survival rate of 10%-24% because of the early metastasis and lack of response to chemotherapy [3-5]. Hence, there should be an urgent need to explore a novel understanding how CCS progresses into DDCS and find a new therapy to deal with it.
Bone morphogenetic proteins (BMPs) are members of transforming growth factor-beta (TGF-β) superfamily. They play crucial roles in regulating far-ranging developmental functions, such as proliferation, differentiation, migration, cell death, and so on [6]. Till date, approximately 20 BMP family members have been identified and defined [7]. BMP signaling is activated by ligands binding to type I and II receptors at the cell surface [8]. Ligand-induced receptor activation activates two signal transduction pathways: The Smad-dependent pathway, in which receptor-specific Smad1, 5, and 8 are activated, then combine with Smad4 to form complexes, and eventually the Smad4 complex translocates into the nucleus to regulate the transcription of target genes [9-12]. The other pathway is the Smad-independent pathway that is the mitogen-activated protein kinase pathway, which includes p38, Jun kinase, and extracellular signal-regulated kinase pathways [11,13-16]. Although the function of BMP-dependent signaling pathway in bone is well known, its function in cancer is somewhat nonconformity [17]. In some cancers, BMP seems to play a suppression role, such as colon cancers [18], prostate cancerand bone metastasis [19], while in some other cancers, it plays an important role in promoting tumor, especially in breast cancer [20-22]. Recently, a report demonstrated that the inhibition of bone morphogenetic protein receptor 2 (BMPR2) suppresses growth and viability of breast cancer cells [23]. Our previous study showed that the detection of BMPR2 was more frequent in DDCS than in CCS [24], and the inhibition of BMPR2 leads to apoptosis and autophagy in CCS [25]. The runt-related transcription factor 2 (RUNX2) is essential in osteoblastic differentiation and skeletal morphogenesis. It is also required for chondrocyte maturation in both mice and humans [26]. Our previous study demonstrated that RUNX2 was overexpressed in DDCS cell line NDCS-1 compared to CCS cell line SW1353 [27]. However, the mechanism of BMPR2 in regulating the RUNX2 expression, tumor growth, and progression in DDCS remains unclear.
First, with patients’ tissue samples, it was confirmed that the RUNX2 expression was significantly upregulated in DDCS than in CCS. Besides, the expression levels of BMPR2 and pSmad1/5 were detected both with clinical samples and with DDCS cell line (NDCS-1) and CCS cell lines (HCS2/8 and SW1353). Then, the luciferase report assay showed that the RUNX2 transcriptional activity was regulated by BMPR2, while chromatin immunoprecipitation (ChIP) and electrophoresis mobility shift assay (EMSA) showed the direct binding of pSmad1/5 with the RUNX2 gene. Finally, the effects of BMPR2 siRNA and Smad1/Smad5 siRNA were investigated on the human DDCS cell line NDCS-1; in addition, the overexpression of BMPR2 and the co-overexpression of Smad1 and Smad5 were used to clarify the worsening effects on human CCS cell lines HCS2/8 and SW1353. This study showed that the RUNX2 expression, cell growth, and cell invasion were suppressed after silencing the expression of BMPR2 or Smad1/5, while increased after the upregulation of BMPR2 or Smad1/5 in vitro and in vivo. Moreover, LDN-193189, an inhibitor of BMPR, also suppressed the expression of RUNX2 and progression in DDCS in vitro and in vivo.
Materials and methods
Patients, tissue samples, and follow-up
Fifteen fresh chondrosarcoma tissue samples were collected under the protocols approved by the ethics committee of Peking University People’s Hospital. Informed consents were acquired from all patients (written in the light of the ethical guidelines). Fifty-seven paraffin-embedded tissue specimens of different histopathologically diagnosed chondrosarcoma were acquired from the Department of Pathology and the Musculoskeletal Tumor Center, Peking University People’s Hospital (Beijing, China). Tissue samples were gathered after surgery, sectioned (4-μm thickness), and conserved properly at room temperature until experiments were performed. Clinical and histopathological information were recorded through retrospective chart review records.
Cell culture, siRNA, plasmid, and transfection
NDCS-1 and HCS2/8 were kindly provided by Dr Akira Ogose [28] and Dr. Takigawa, respectively, while SW1353 was purchased from ATCC. NDCS-1, SW1353, and HCS2/8 were grown in RPMI 1640, L-15, and DMEM/F12, respectively, supplemented with 10% fetal bovine serum and 1% antibiotics. The cultures were maintained at 37°C in 5% CO2 atmosphere.
siRNAs were constructed in pGpU6/GFP/Neo vector. The nucleotide target sequences were as follows: BMPR2, GCA GTA CTA GTT CTA GCT TGC; Smad1, AAC TGC AAC TAC CAT CAT GGA TT; Smad5, AAG CCG TTG GAT ATT TGT GAA TT, named as siBMPR2 and co-siSmad1/siSmad5, respectively. The open reading frame sequences of overexpression BMPR2, Smad1, and Smad5 were cloned into the pReceiver-M02 vector, and named pM/BMPR2 and co-pM/Smad1/pM/Smad5. The transfections were conducted with Lipofectamine 2000 Reagent (Invitrogen), following the manufacturer’s instruction.
Immunohistochemistry
Tissue sections were deparaffinized, while antigen was retrieved by an antigen retrieval solution. Then, paraffinized tissue sections were incubated with primary antibodies overnight at 4°C, and the next day followed by a biotinylated secondary antibody staining. Positive controls were included in each experiment, while negative controls were stained with nonimmune mouse serum (1:50) instead of primary antibody. When >10% of tumor cells were stained, the tumor was considered positive. Immunostaining was assessed by two independent pathologists who were unknown about the clinical data. The primary antibodies included in this study are the BMPR2 antibody (Abcam; 1:50) and the RUNX2 antibody (Santa Cruz; 1:50).
Real-time RT-PCR
Total RNA was extracted and purified from cell lines using TRIzol (Invitrogen), and then using RNeasyMinElute columns (Qiagen). Complementary DNA (cDNA) was synthesized using the First-Strand cDNA Synthesis Kit (Invitrogen), according to the manufacturer’s instructions. Finally, quantitative reverse transcription polymerase chain reaction (RT-PCR) was performed with GoTaq (Promega) using the following conditions: 95°C for 2 minutes; 40 cycles of 95°C for 15 seconds; and 60°C for 1 minute; followed by 72°C for 10 minutes, as described previously. Primer sequences for real-time PCR were as follows: BMPR2 forward, 5’-TCA AGA ACG GCT ATG TGC GT-3’/reverse, 5’-AAC TGG ACG CTC ATC CAA GG-3’; Smad1 forward, 5’-TGT ATT CGT GAG TTC GCG GT-3’/reverse, 5’-GCT GTT GGG TTG CTG GAA AG-3’; Smad5 forward, 5’-CGA AAA GGA AGC TGT TGA AGT T-3’/reverse, 5’-AAG GAG TGT TGT TGG GCT GG-3’; RUNX2 forward 5’-CAC GCU AUU AAA UCC AAA UTT-3’/reverse. 5’-AUU UGG AUU UAA UAG CGU GTT-3’, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5’-ACA ACT TTG GTA TCG TGG AAG G-3’/reverse, 5’-GCC ATC ACG CCA CAG TTT C-3’.
Western blotting
Whole cell lysates were made using cell lysis buffer (CST) supplemented with proteinase inhibitors. Protein concentrations were determined using the BCA Protein Assay. Equal amounts of proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidinedifluoride membrane (Amersham Bioscience). After being blocked with 5% bovine serum albumin, the primary antibody, human BMPR2 (Abcam), total-Smad1 (Abcam), p-SAMD1/5 (CST), human RUNX2 (Santa Cruz), and GAPDH (Santa Cruz) were incubated overnight at 4°C, at a dilution of 1:500, 1:1000, 1:1000, 1:200, and 1:2000, respectively. Afterward, the appropriate secondary antibody was incubated for 1 hour, and the blot was visualized by using the SuperSignal West Pico Trial Kit (Thermo). The band signals were quantified using the ImageJ software (Wayne Rasband).
Luciferase reporter assays
The RUNX2 promoter was constructed into the firefly luciferase reporter vector pGL4.14 (Promega). NDCS-1, HCS2/8, and SW1353 cells were co-transfected with pGL/RUNX2, pM/BMPR2, and pRL-TK (Promega), the renilla luciferase reporter plasmid. After 48 hours, the luciferase activity was assayed using the Dual-Glo Luciferase Assay System (Promega).
Chromatin immunoprecipitation assay
NDCS-1, SW1353, and HCS2/8 cells were fixed with 1% p-formaldehyde for 10 minutes at room temperature to make protein-DNA complexes. Following sonication, the supernatants were incubated with 10 mg/mL of anti-pSmad1/5 antibody. Then the A/G agarose beads were used to precipitate the fragments. After elution, the RUNX2 DNA was amplified by the gel electrophoresis assay. Anti-immunoglobulin G (anti-IgG) antibody and GAPDH primers were used in this assay as negative control, while inputs were used as positive control.
Electrophoretic mobility shift assay
Nuclear extracts were obtained from NDCS-1, HCS2/8, and SW1353 cells. Antibody supershift assay was performed using an EMSA kit, following the manufacturer’s instructions (Beyond, China). The nuclear extracts were incubated for 45 minutes with pSmad1/5 antibodies, and the RUNX2 probe was labeled with [g-32P] ATP before this reaction mixture was loaded on acrylamide gels.
Cell growth inhibition assay
The three cell lines were plated in 24-well plate (50,000 cells/well). After overnight, the siRNA or overexpression plasmids were transfected for 24, 48, 72, and 96 hours, following the 100-mL MTT addition. Then the absorbance was read at 570 nm.
Transwell invasion assay
The three cell lines were transfected as described in the preceding section, followed by 6 × 104 cells plating on top of transwell filters (in 24-well format, Corning). Cells were allowed to grow for 48 hours, and the cells in the lower side were counted by Cell-Titer Glo.
Xenograft tumorigenicity assays
Mice experiment were followed by a high standard of animal welfare. Six-week-old BALB/c female nude mice and SCID-CB17 female mice were subcutaneously injected in the right flank with 2 × 106/mL cells (HCS2/8 or NDCS-1) combined with Matrigel. Once chondrosarcoma cells developed palpable tumors, the mice were randomly divided into several groups, followed by 4 weeks’ treatment with an intratumoral injection. Mice bearing NDCS-1 or HCS2/8 cells were treated daily with 30 μg of overexpression plasmids or 2 μg of siRNA, respectively. PBS was used as the negative control, and 3 mg/kg of LDN-193189 was used as the positive control by daily intraperitoneal injection. For each injection, 15 L of the nucleic acid (siRNAs or plasmids) was dissolved in 5% glucose, then was mixed with 15 L of Entranster-in vivo (Engreen, China), according to the manufacturer’s instructions. The mixture was locally injected into the tumor. The tumor volume was measured once a week using the following expression: Volume = (Length × Width2)/2. The tumor samples were detected using the Western blot assay.
Statistical analysis
All statistical analyses were performed by the SPSS19.0 software package. The relationship between patient survival and indicated protein levels was assessed by the Kaplan-Meier analysis. The correlation between protein levels and clinicopathological tumor grading was analyzed using the standard χ 2 test. The Student t test was used to specify the differences with P < 0.05.
Results
Expressions of BMPR2 and RUNX2 associate with chondrosarcoma clinicopathological grades and predict the prognosis
Western blot analyses were used to investigate expressions of BMPR2 and RUNX2 in different grades of chondrosarcoma patients; there were four samples of each grade, except grade 3, which had three samples. Expressions of BMPR2 and RUNX2 were also detected in three chondrosarcoma cell lines. The Western blot analysis showed that BMPR2 and RUNX2 expressions increased with the deterioration in the clinicopathological level (Figure 1A-C). In addition, BMPR2 and RUNX2 expressions were evaluated in 57 patients with various grades of chondrosarcoma using an immunohistochemical (IHC) staining method (Figure 1D). The relationships between these protein expressions and clinicopathological factors were statistically analyzed (Table 1). Positive BMPR2 staining was detected more often in DDCS (12/17 patients) compared to grade I (6/24 patients) and grade II + III (11/16 patients). The RUNX2 level presents similar trends with 12/17 patients in DDCS, while 3/24 in grade I and 3/16 in grade II + III. Furthermore, using Kaplan-Meier survival analysis (P = 0.030), it was found that expression levels of BMPR2 or RUNX2 were related with disease-free survival of DDCS patients (Figure 1E, 1F). The aforementioned results revealed that BMPR2 and RUNX2 expressions were correlated with aggressive tumor behaviors and would be a potential marker for prognosis.
Figure 1.
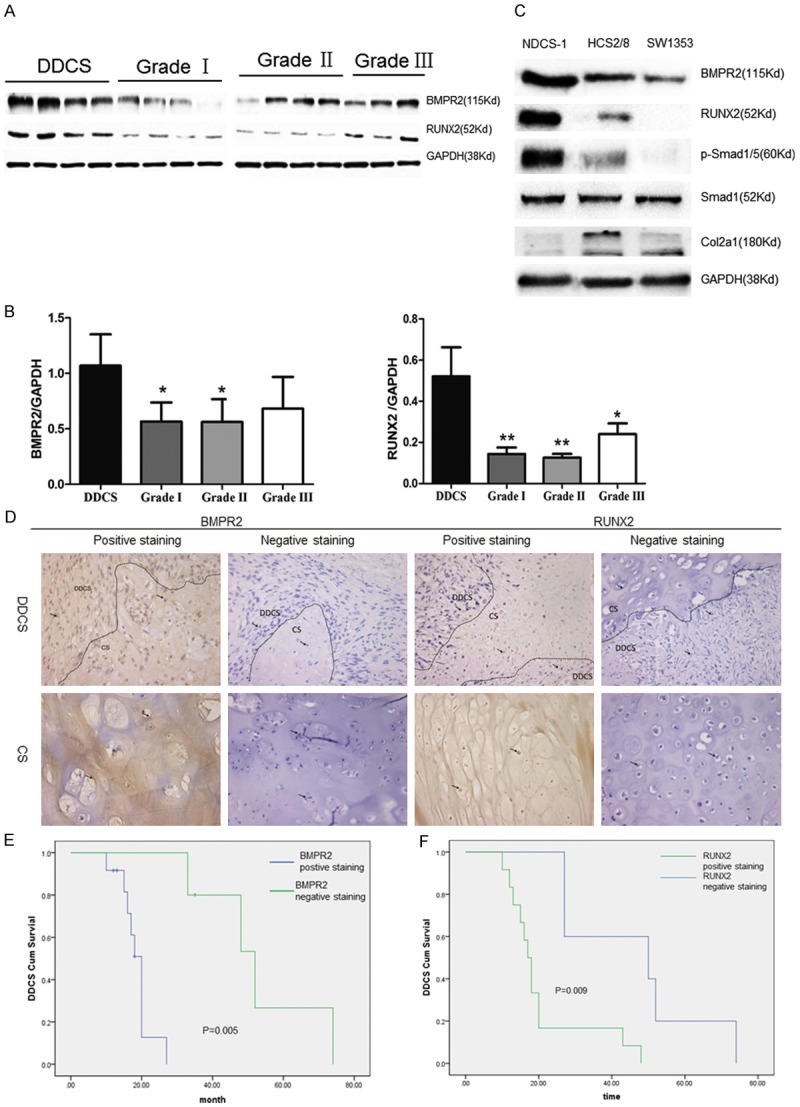
BMPR2 and RUNX2 expressions are correlated with clinicopathological features of chondrosarcomas and predict the prognosis. A, B. Chondrosarcoma specimens from patients ranked Grade I to DDCS were analyzed the BMPR2 and RUNX2 levels using Western blot. C. Western blot of BMPR2, RUNX2, pSmad1/5, Col2a1 and Smad1 in NDCS-1, SW1353, and HCS2/8 cell lines. D. IHC of clinicopathological sections with BMPR2 and RUNX2 expressions, followed by H&E staining. The tissue included DDCS and CCS. E. Kaplan-Meier analysis for relapse-free survival in DDCS patients with or without positive BMPR2 staining. Positive BMPR2 stainingin DDCS specimens ipredicted a poorer relapse-free survival (p = 0.005).
Table 1.
Association between clinicopathologic characteristics and BMPR2 or RUNX2 expression
| Clinicopathological variables | n | BMPR2 expression | p-value | Runx2 | p-value | |||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Positive | Negative | Positive | Negative | |||||
| Sex | Male | 30 | 16 | 14 | 0.696 | 13 | 17 | 0.843 |
| Female | 27 | 22 | 17 | 20 | 19 | |||
| Age (year) | ≥40 | 34 | 19 | 16 | 0.421 | 17 | 17 | 0.872 |
| <40 | 23 | 10 | 13 | 11 | 12 | |||
| Pathogenic site | Limb | 22 | 14 | 8 | 0.627 | 10 | 12 | 0.589 |
| Pelvis | 35 | 20 | 33 | 18 | 17 | |||
| Histopathological grading | Low Grade (I) | 24 | 6 | 18 | 0.004* | 3 | 21 | 0.000* |
| High Grade (II + III) | 16 | 11 | 5 | 3 | 13 | |||
| DDCS | 17 | 12 | 5 | 12 | 5 | |||
DDCS: dedifferentiated chondrosarcoma;
p<0.05.
Regulation of the RUNX2 expression by BMPR2 and pSmad1/5
As BMPR2 and RUNX2 expressions were related to the deterioration of chondrosarcoma, this study hypothesized that RUNX2 may be regulated by BMPR2 and pSmad1/5. To verify this assumption, first it was found that BMPR2 and pSmad1/5 expressions were higher in NDCS-1 than in HCS2/8 and SW1353 cell lines (Figure 1C). Then, the BMPR2 mRNA was knocked down into the DDCS cell line NDCS-1, and it was found that RUNX2 protein level and mRNA expression also decreased (Figure 2A-C). However, the upregulation of BMPR2 in the CCS cell lines, HCS2/8 and SW1353, revealed that the RUNX2 level increased too (Figure 2A-C).
Figure 2.
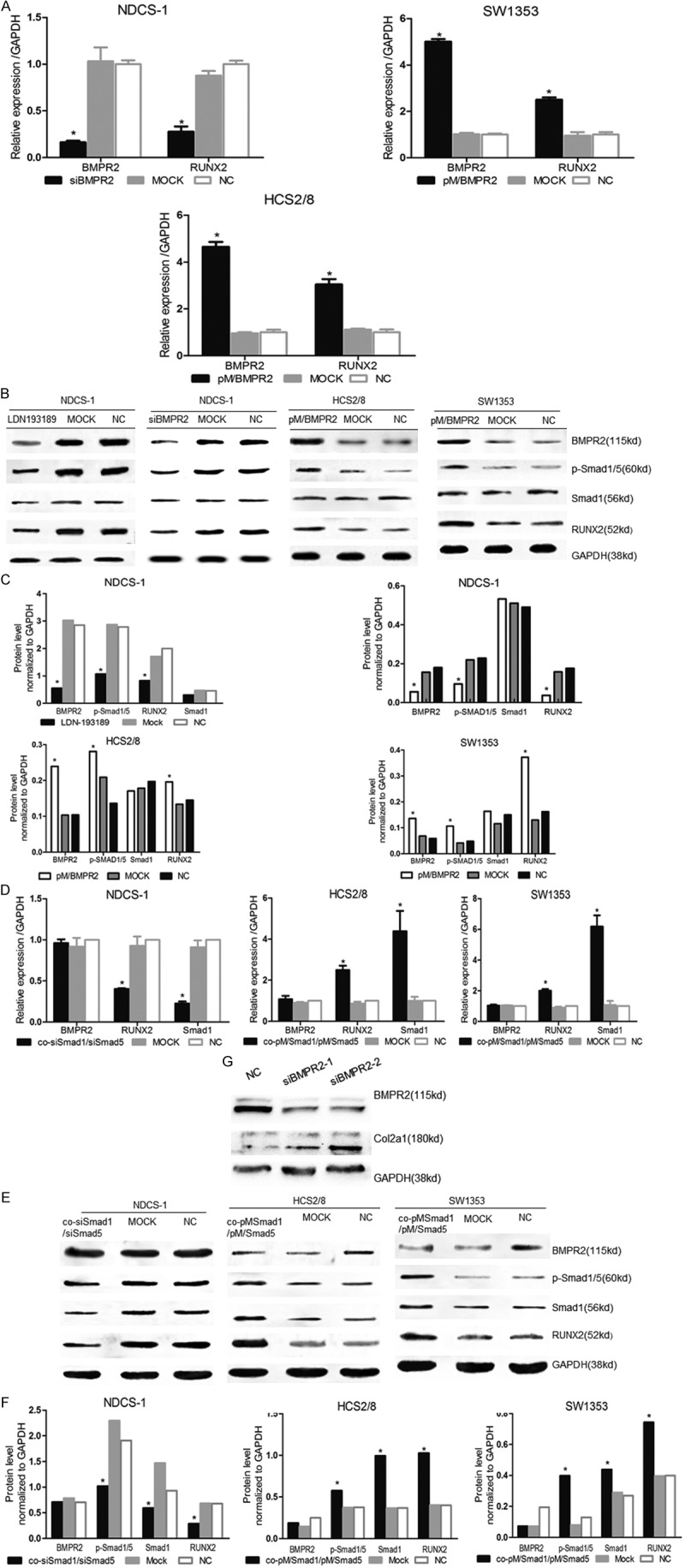
Inhibition of BMPR2 or p-Smad1/5 decreased RUNX2 expression and reversed cell phenotype, while upregulation enhanced RUNX2 expression. BMPR2 or Smad1/Smad5 was interfered in NDCS-1, whereas overexpressed in SW1353 and HCS2/8. The BMPR2, RUNX2, and Smad1 expressions were detected by qRT-PCR (A, D). Western blot showed BMPR2, pSmad1/5, Smad1 and RUNX2 levels (B, E). The densitometric analysis of Western blot (C, F). (G) 100 pmol (siBMPR2-1) and 150 pmol (siBMPR2-2) BMPR2 siRNA had effect on Col2a1 expression in NDCS-1 cell. Mock: scrambled siRNA or empty vector; NC: normal cells; LDN193189: 5 nM was treat NDCS-1 for 24 h. n = 3, mean ± S.D. *p<0.05.
Furthermore, BMPR2 had an effect on its downstream molecular by phosphorylation, activation, and nucleus translocation of Smad1/5. Hence, it was hypothesized that pSmad1/5 can also regulate the expression of RUNX2. siRNA constructs were introduced targeting Smad1 and Smad5 co-transfected into NDCS-1 cell line, and the pSmad1/5 protein level was confirmed to be significantly decreased in the protein level while Smad1 decreased in the mRNA level (Figure 2D-F), with a result that the RUNX2 level declined in response to pSmad1/5. In contrast, Smad1 and Smad5 were overexpressed by plasmid transfection in SW1353 and HCS2/8 cells. As a result, the RUNX2 level increased in response to the increase in pSmad1/5. These results demonstrated that BMPR2 and pSmad1/5 were upstream of RUNX2 and positively regulated RUNX2.
BMPR2 activated the RUNX2 gene expression by pSmad1/5 directly binding to the RUNX2 promoter
To assess whether BMPR2 can regulate the RUNX2 expression at the transcriptional level, luciferasereporter assays were performed. The BMPR2 overexpression in the three chondrosarcoma cell lines caused a significant upregulation of the RUNX2 promoter activity (Figure 3A).
Figure 3.
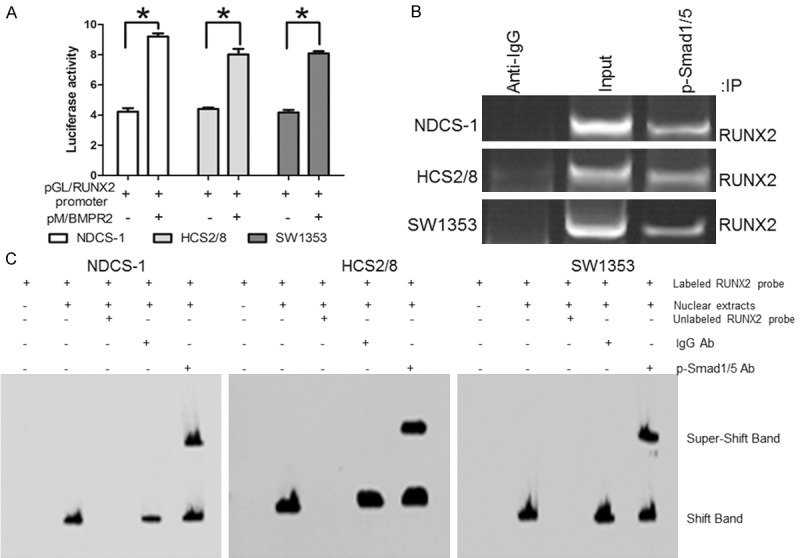
BMPR2 activated the RUNX2 gene expression by binding pSmad1/5 to the RUNX2 promoter. A. The plasmid pM/BMPR2 was co-transfected after 48 h into chondrosarcoma cell lines with plasmids encoding RUNX2 promoter and renilla luciferase. B. The ChIP assay. Protein-DNA complexes from NDCS-1 cell were incubated with pSmad1/5 antibody. The binding of RUNX2 DNA was detected by PCR amplification using RUNX2-specific primers. IgG antibody and GAPDH primers were used in this assay as negative controls. C. The super gel shift assay showed the binding activity of pSmad1/5 with RUNX2 DNA in nuclear extracts of chondrosarcoma cells. For experimental controls, the labeled RUNX2 probes were incubated alone, with nuclear extracts, in combination with nuclear extracts and unlabeled probe, or in combination with nuclear extracts and IgG antibody. Data are presented as mean ± S.D. (n = 3). *P < 0.05.
A ChIP assay was performed to verify whether BMPR2 bound directly to the RUNX2 gene using an anti-BMPR2 antibody. The PCR analysis of the BMPR2 immunoprecipitates showed no product in the three cell lines (data not shown); thus, it was hypothesized that pSmad1/5 may bind directly to RUNX2. Then an anti-pSmad1/5 antibody was used to verify this assumption. PCR analysis of the pSmad1/5 immunoprecipitates showed significant products in the three cell lines compared to IgG (Figure 3B).
To further determine whether the binding site of RUNX2 is a promoter, EMSA was performed using an oligonucleotide probe corresponding to the RUNX2 sequence from -314 to -189 bp. As shown in Figure 3C, nuclear extracts were supershifted by the anti-pSmad1/5 antibodies (Figure 3C), indicating the binding of pSmad1/5 with the RUNX2 region. These findings indicated that BMPR2 activated the RUNX2 gene expression by binding pSmad1/5 to the RUNX2 promoter.
Effect on growth behavior of siBMPR2 or siSmad1/siSmad5 in DDCS cells
As BMPR2 and pSmad1/5 expressions were higher in DDCS than in CCS, it was necessary to study whether the inhibition of BMPR2 or pSmad1/5 can kill human DDCS cell line (NDCD-1). First, to investigate the effect of BMPR2 on the cell viability in DDCS cell lines NDCS-1, BMPR2 siRNA was introduced. After 48 hours of transfection, the cell viabilities (detected by MTT) decreased significantly in the siBMPR2 transfection group compared with negative control (NC) groups (Figure 4A). However, to investigate the effects of Smad1 and Smad5 on cell viability, siRNA targeting Smad1 and Smad5 were co-transfected into the DDCS cell line. The results revealed that after 48 hours of co-transfection, the cell viabilities at 48 hours decreased significantly in the transfection group compared with NC groups (Figure 4A). Furthermore, the 12-, 24-, 48-, and 72-hour optical density values were tested, and the growth curve was drawn. The curve revealed that when an inhibition of BMPR2 or Smad1/Smad5 occurred, the growth curve was under the negative control curve (Figure 4B). Therefore, suppressing BMPR2 or Smad1/Smad5 can inhibit DDCS cell growth.
Figure 4.
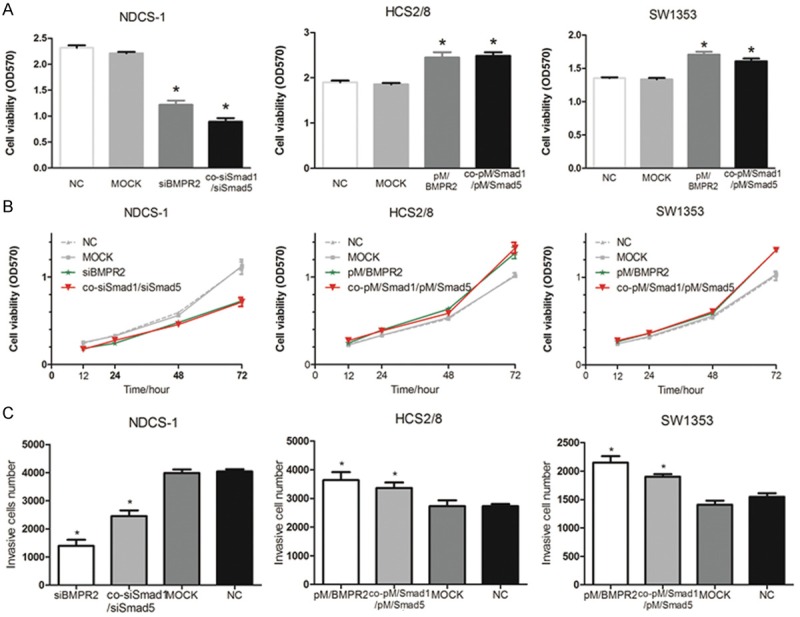
BMPR2 or p-Smad1/5 knockdown suppressed cell invasion and proliferation, while overexpression promoted cell invasion and proliferation. BMPR2 or Smad1/Smad5 siRNA were transfected into NCDS-1, whereas BMPR2 or Smad1/Smad5 plasmids were transfected into HCS2/8 and SW1353 cells. Mock: scrambled siRNA or empty vector; NC: normal cells. MTT assay showed the cell vialbility at 48 h (A) and the cell growth curve (B) after transfection against the three cell lines. (C) The invasive cell number was counted after 48 h by Cell-Titer Glo. *: p < 0.05, vs NC groups.
Effect on growth behavior of overexpression of BMPR2 or pSmad1/5 in CCS cells
As knockdown of BMPR2 or pSmad1/5 by siRNA results in an inhibitory growth of DDCS cells, it was interesting to investigate whether the upregulation of BMPR2 or pSmad1/5 would enhance the growth of chondrosarcoma cell lines (HCS2/8 and SW1353). The upregulation of BMPR2 or Smad1/Smad5 by plasmid was used to investigate the effect of BMPR2 and pSmad1/5 on the maintenance of cell viability in chondrosarcoma cell lines. The BMPR2 overexpression plasmids were introduced and the results showed that cell viabilities increased significantly at 48 hours compared with NC groups (Figure 4A). In addition, both Smad1 and Smad5 plasmids were transfected into HCS2/8 and SW1353 cell lines, and the results suggested that pSmad1/5 enhanced cell viabilities at 48 hours in the co-transfection group compared with NC groups (Figure 4A). Similarly, the growth curve was drawn, which showed that the enhanced curve was above the negative control curve (Figure 4B). Therefore, the overexpression of BMPR2 or pSmad1/5 can enhancechondrosarcoma cell growth.
Inhibition of BMPR2 or Smad1/5 by siRNA suppressed cell invasion, while overexpression of BMPR2 or pSmad1/5 promoted cell invasion
The most obvious characteristics of tumors are metastasis, and DDCS distant metastasis occurred frequently; hence, inhibition to metastasis may be a method to treat DDCS. Transwell assay was performed to determine whether a change in the BMPR2 or pSmad1/5 level would alter the invasion of DDCS cells. As shown in Figure 4C, the results revealed that after 48-hour transfection of siBMPR2 or co-siSmad1/Smad5, the invasion was significantly decreased compared with NC groups, while transfection of overexpression plasmids into CCS cell lines showed the inverted results (Figure 4C). As shown in Figure 4C, the invasion was significantly increased compared with NC groups in this two cell lines. Hence, the study concluded that the inhibition of BMPR2 or Smad1/5 suppressed cell invasion, while enhancement of BMPR2 or pSmad1/5 promoted it.
BMPR2-pSmad1/5 regulate cell progression via RUNX2
As shown in Figure 4, cell progression were regulated by BMPR2 or Smad1/5, while BMPR2-pSmad1/5 can regulate RUNX2 expression, hence, dose BMPR2 or Smad1/5 regulate cell progression via RUNX2? We first constructed the RUNX2 small interference RNA (siRUNX2), primer sequences for real-time PCR were as follows: RUNX2 forward 5’-CAC GCU AUU AAA UCC AAA UTT-3’/reverse. 5’-AUU UGG AUU UAA UAG CGU GTT-3’. MTT and Transwell assay were performed to determine whether a change in the RUNX2 level would alter the proliferation or invasion of DDCS cells. As shown in Supplementary Figure 1A, 1B, the results reveled that after 48-hour transfection of siRUNX2, the invasion and proliferation were significantly decreased compared with NC groups (Supplementary Figure 1A, 1B). Then, further study was needed to investigate that did BMPR2-pSmad1/5 regulate RUNX2 expression or RUNX2 regulate BMPR2-pSmad1/5 expression leads to the change of DDCS cell progression? Hence, qRT-PCR and Western-blot experiments were performed to verify the hypothesis. As shown in Supplementary Figure 1C-E, inhibition of RUNX2 did not change the expression of BMPR2 and pSmad1/5 both in protein or mRNA level. Therefore, combined with Figure 4 and Supplementary Figure 1, the study concluded that BMPR2-pSmad1/5 regulate cell progression via RUNX2.
BMPR inhibitor inhibited BMPR2-pSmad1/5 signaling pathway
LDN-193189, a BMPR inhibitor, was used to determine whether RUNX2 was regulated by BMPR2-pSmad1/5 signaling pathway and whether it would be a potential drug to treat DDCS. The study found that LDN-193189 (5 nM) reduced the expression of BMPR2, pSmad1/5, and RUNX2 (Figure 2B, 2C).
Inhibition of BMPR2 by siBMPR2 reversed the DDCS cell phenotype
Col2a1 was the collagen marker of cartilage, as shown in Figure 1C, its expression level was lower in DDCS compare to CCS (Figure 1C); hence, it was assumed that a loss of cartilage phenotype induced dedifferentiation. It was investigated whether the inhibition of BMPR2 would cause DDCS back to CCS. By siBMPR2 interfering, it was found that the col2a1 protein level increased compared with the NC group (Figure 2G). Hence, the study drew a conclusion that the inhibition of BMPR2 can reverse the cell phenotype.
Inhibition of BMPR2 or Smad1/5 suppressed chondrosarcoma tumor growth, while enhancement of BMPR2 or Smad1/5 promoted DDCS tumor growth in vivo
To confirm the aforementioned findings in vivo, xenograft models were established. SCID-CB17, which was injected in NDCS-1 cell lines, was on behalf of DDCS xenograft models. The group treated with siBMPR2 or co-siSmad1/Smad5 had a lower proliferation rate and a smaller tumor size compared with the NC group (Figure 5A). Furthermore, another group treated with LDN-193189 also showed a lower proliferation rate and a smaller tumor size compared with the NC group, as shown in Figure 5A. The tumor volume and weight at the time of death were (263.5405 ± 238.9176 mm3 and 0.2873 ± 0.2840 g) in the siBMPR2 group; (207.3295 ± 64.1769 mm3 and 0.2464 ± 0.0675 g) in the co-siSmad1/Smad5 group; and (189.6107 ± 126.9972 mm3 and 0.2332 ± 0.1514 g) in the LDN-193189 group, which were significantly less than those in the NC group (754.1285 ± 225.9596 mm3 and 1.2043 ± 0.3356 g) (Figure 5B-E). However, injection with HCS2/8 cells was on behalf of chondrosarcoma xenograft models. The mouse group injected with BMPR2 or co-Smad1/Smad5 overexpression plasmids had a higher proliferation rate and bigger tumor size compared with the NC group. The tumor volume and weight at the time of death were (2064.7124 ± 723.6145 mm3 and 1.1925 ± 0.3216 g in the upregulation BMPR2 group; (1468.1408 ± 728.1280 mm3 and 0.8225 ± 0.4373 g in the upregulation co-Smad1/Smad5 group, which were significantly higher than those in the NC group (399.6026 ± 82.5398 mm3 and 0.2650 ± 0.0332 g) (Figure 5B-E). These data suggest that siBMPR2 and siSmad1/5 reduce both tumor volume and growth rate of DDCS cells, while the upregulation of BMPR2 and Smad1/5 promotes tumor volume and growth rate of chondrosarcoma cells in vivo.
Figure 5.
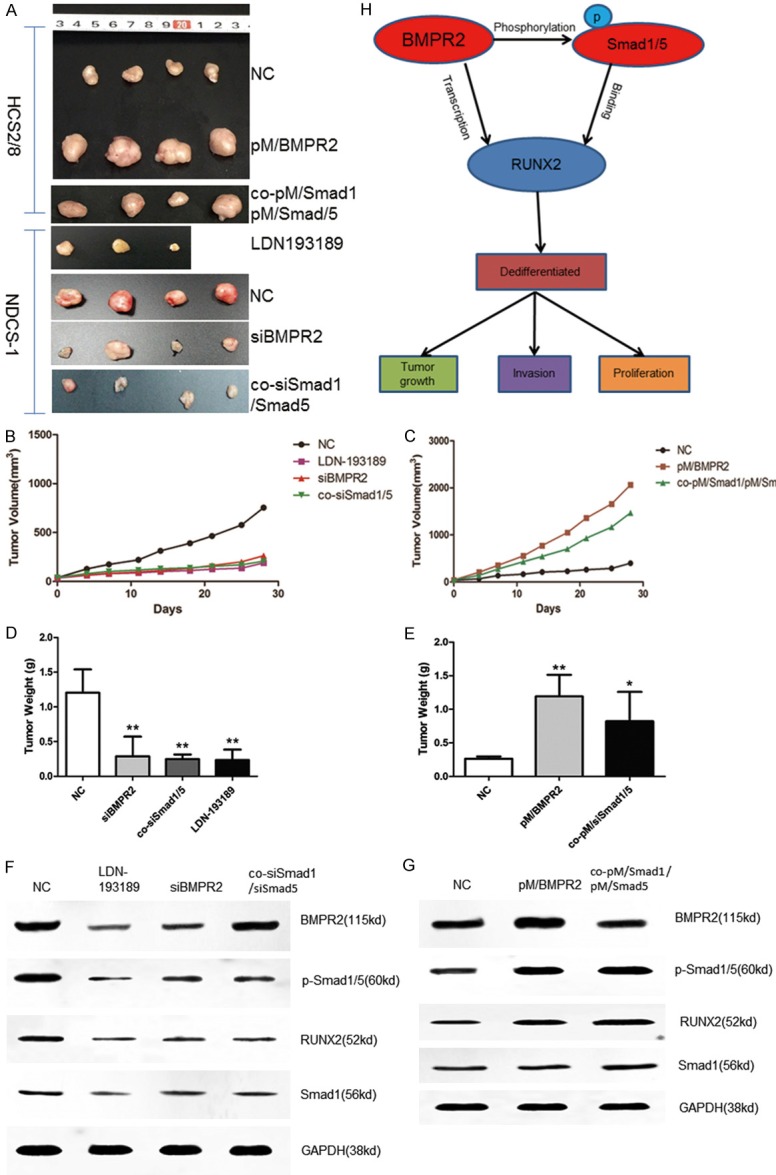
Inhibition of BMPR2 or Smad1/5 suppressed chondrosarcoma tumor growth, while enhancement of BMPR2 or Smad1/5 promoted DDCS tumor growth in vivo. SCID-CB17 mice and BALB/c nude mice were subcutaneously injected either NDCS-1 or HCS2/8 cells, followed by daily treatment at the onset of tumor detection. The groups included PBS (NC, subcutaneously), LDN-193189 (3 mg/kg, intraperitoneally), BMPR2 siRNA (2 μg/mouse, subcutaneously), Smad1/Smad5 siRNA (2 μg/mouse, subcutaneously), pM/BMPR2 (30 μg/mouse, subcutaneously), and pM/Smad1/pM/Smad5 (30 μg/mouse, subcutaneously). Entranster-in vivo was used as the transfection reagent. The tumor images, weight, and volume were acquired after 4 weeks’ treatment (A-E). Western blot (F, G) analysis of various proteins in the xenograft tumors. *: P < 0.05, vs NC groups. (H) Schematic mechanisms of BMPR2-pSmad1/5 signaling pathways induced effects in DDCS cells.
The Western blot assay was used to verify whether the BMPR2 or pSmad1/5 expression was suppressed or enhanced in vivo. The results showed that BMPR2 and pSmad1/5 changed in vivo (Figure 5F, 5G) and RUNX2 also changed later on. The results, taken together, indicate that a loss of BMPR2 or pSmad1/5 inhibited the growth in DDCS, while overexpression promoted the growth in CCS.
Discussion
According to this research study, the BMPR2 and pSmad1/5 signaling pathways were found to be dysfunctional in DDCS, and the proposed mechanisms responding to BMPR2-pSmad1/5 signaling pathways induced effects in DDCS cells was shown in Figure 5H. Although BMPR2 has been studied in many other tumors [23,29,30], that BMPR2 regulates the expression of RUNX2 in chondrosarcoma and DDCS was not reported. According to our previous report, BMPR2 was more likely to be detected in DDCS than in CCS [24]. Moreover, the inhibition of BMPR2 could induce apoptosis and autophagy via XIAP [25]. This study showed that both BMPR2 and RUNX2 were associated with histopathological grading and poor prognosis. It also clarified the effects of BMPR2-pSmad1/5 signaling in regulating the RUNX2 expression and influencing the DDCS progression both in vitro and in vivo.
DDCS is a special type of chondrosarcoma, accounting for approximately 10%-15% of CCS. While most previous researches were case reports and the analysis of molecular mechanisms in DDCS is little, espically in BMPR2, Therefore we emphasized on the BMPR2-RUNX2 signaling and explained the signaling pathways involved in the progression and prognosis of DDCS.
BMPR2, as one of the BMP receptors, is essential in BMP signaling pathway, so dysfunction of BMPR2 may lead to diseases such as pulmonary arterial hypertension [31]. Moreover, BMPR2 was dysfunctional in malignant tumor such as breast cancer and colorectal cancer [18,23,32]. Although most researches considered that BMPR2 was a tumor suppressor and inhibited tumor progression and metastasis [30], this study showed that BMPR2 can be a tumor promoter in chondrosarcoma in vitro and in vivo. BMPR2 could trigger tumor invasion and progression. Further, the BMP receptor inhibitor LDN-193189 was introduced and, interestingly, it was found that LDN-193189 could suppress tumor growth in vitro and in vivo.
Stephane Boeuf showed that phosphorylated Smad1/5/8 were significantly higher in high-grade samples compared to low-grade chondrosarcoma samples [33]. Therefore, we focus on the cell lines and want to know that if cell lines show the same results, the results showed that p-Smad1/5 increased with the pathological grades in cell lines, especially, in DDCS cell line, While we did not further test them in patients’ samples due to our samples were too long.
RUNX2 plays important roles in osteoblast differentiation and bone formation, while in chondrosarcoma, Tang showed that the RUNX2 expression was higher in DDCS than in CCS, and the same conclusion was drawn in the present study in clinic samples [27]. Although Tang did not study by which signaling pathways RUNX2 was regulated, this study found that BMPR2-pSmad1/5 signaling pathways could regulate RUNX2. The present study also found that BMPR2 can regulate RUNX2 in the transcriptional level by using the luciferase reporter assay while pSmad1/5 binds directly to RUNX2 by using ChIP and EMSA assays.
In the current study, the focus was on how the Smad-dependent pathway regulates the RUNX2 expression and progression in chondrosarcoma, but whether the Smad-independent pathway regulates RUNX2 was not studied. Besides, as the high-grade parts of the cell lines used in this study were osteosarcoma, whether other high-grade components such as fibrosarcoma and malignant fibrous sarcoma could show the same conclusion also need to be studied. Finally, more research is needed to investigate the exact mechanism why BMPR2 was overexpressed in DDCS, and how-by mutation or by microRNA.
Acknowledgments
The NDCS-1 and HCS2/8 cell line were generous gifts from Dr. Akira Ogose (Niigata University Graduate School of Medical and Dental Sciences Niigata Japan)and Dr. Takigawa (Osaka University Faculty of Dentistry, Japan), respectively. This work was supported by the Program for New Century Excellent Talents in University (NCET-12-0007) and the Natural Science Foundation of China (No. 81172544).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Sakamoto A. The molecular pathogenesis of dedifferentiated chondrosarcoma. Indian J Orthop. 2014;48:262–265. doi: 10.4103/0019-5413.132506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahlin DC, Beabout JW. Dedifferentiation of low-grade chondrosarcomas. Cancer. 1971;28:461–466. doi: 10.1002/1097-0142(197108)28:2<461::aid-cncr2820280227>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 3.Bharath G, Burrah R, Shivakumar K, Manjunath S, Bhanumathi R. Dedifferentiated chondrosarcoma: an aggressive variant of chondrosarcoma. Asian Cardiovasc Thorac Ann. 2015;23:221–223. doi: 10.1177/0218492314522253. [DOI] [PubMed] [Google Scholar]
- 4.Matsumoto Y, Takahashi Y, Harimaya K, Nakagawa T, Kawaguchi K, Okada S, Hayashida M, Doi T, Sakamoto A, Matsunobu T, Oda Y, Iwamoto Y. Dedifferentiated chondrosarcoma of the cervical spine: a case report. World J Surg Oncol. 2013;11:32. doi: 10.1186/1477-7819-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokota K, Sakamoto A, Matsumoto Y, Matsuda S, Harimaya K, Oda Y, Iwamoto Y. Clinical outcome for patients with dedifferentiated chondrosarcoma: a report of 9 cases at a single institute. J Orthop Surg Res. 2012;7:38. doi: 10.1186/1749-799X-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 7.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 8.von Bubnoff A, Cho KW. Intracellular BMP signaling regulation in vertebrates: pathway or network? Dev Biol. 2001;239:1–14. doi: 10.1006/dbio.2001.0388. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto N, Akiyama S, Katagiri T, Namiki M, Kurokawa T, Suda T. Smad1 and smad5 act downstream of intracellular signalings of BMP-2 that inhibits myogenic differentiation and induces osteoblast differentiation in C2C12 myoblasts. Biochem Biophys Res Commun. 1997;238:574–580. doi: 10.1006/bbrc.1997.7325. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura R, Kato Y, Chen D, Harris SE, Mundy GR, Yoneda T. Smad5 and DPC4 are key molecules in mediating BMP-2-induced osteoblastic differentiation of the pluripotent mesenchymal precursor cell line C2C12. J Biol Chem. 1998;273:1872–1879. doi: 10.1074/jbc.273.4.1872. [DOI] [PubMed] [Google Scholar]
- 11.Nohe A, Hassel S, Ehrlich M, Neubauer F, Sebald W, Henis YI, Knaus P. The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J Biol Chem. 2002;277:5330–5338. doi: 10.1074/jbc.M102750200. [DOI] [PubMed] [Google Scholar]
- 12.Nohe A, Keating E, Knaus P, Petersen NO. Signal transduction of bone morphogenetic protein receptors. Cell Signal. 2004;16:291–299. doi: 10.1016/j.cellsig.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Gallea S, Lallemand F, Atfi A, Rawadi G, Ramez V, Spinella-Jaegle S, Kawai S, Faucheu C, Huet L, Baron R, Roman-Roman S. Activation of mitogen-activated protein kinase cascades is involved in regulation of bone morphogenetic protein-2-induced osteoblast differentiation in pluripotent C2C12 cells. Bone. 2001;28:491–498. doi: 10.1016/s8756-3282(01)00415-x. [DOI] [PubMed] [Google Scholar]
- 14.Kozawa O, Hatakeyama D, Uematsu T. Divergent regulation by p44/p42 MAP kinase and p38 MAP kinase of bone morphogenetic protein-4-stimulated osteocalcin synthesis in osteoblasts. J Cell Biochem. 2002;84:583–589. [PubMed] [Google Scholar]
- 15.Lai CF, Cheng SL. Signal transductions induced by bone morphogenetic protein-2 and transforming growth factor-beta in normal human osteoblastic cells. J Biol Chem. 2002;277:15514–15522. doi: 10.1074/jbc.M200794200. [DOI] [PubMed] [Google Scholar]
- 16.Guicheux J, Lemonnier J, Ghayor C, Suzuki A, Palmer G, Caverzasio J. Activation of p38 mitogen-activated protein kinase and c-Jun-NH2-terminal kinase by BMP-2 and their implication in the stimulation of osteoblastic cell differentiation. J Bone Miner Res. 2003;18:2060–2068. doi: 10.1359/jbmr.2003.18.11.2060. [DOI] [PubMed] [Google Scholar]
- 17.Groeneveld EH, Burger EH. Bone morphogenetic proteins in human bone regeneration. Eur J Endocrinol. 2000;142:9–21. doi: 10.1530/eje.0.1420009. [DOI] [PubMed] [Google Scholar]
- 18.Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet. 2001;28:184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi A, Okuda H, Xing F, Pandey PR, Watabe M, Hirota S, Pai SK, Liu W, Fukuda K, Chambers C, Wilber A, Watabe K. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J Exp Med. 2011;208:2641–2655. doi: 10.1084/jem.20110840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng L, Shang L, Bai S, Chen J, He X, Martin-Trevino R, Chen S, Li XY, Meng X, Yu B, Wang X, Liu Y, McDermott SP, Ariazi AE, Ginestier C, Ibarra I, Ke J, Luther T, Clouthier SG, Xu L, Shan G, Song E, Yao H, Hannon GJ, Weiss SJ, Wicha MS, Liu S. MicroRNA100 inhibits self-renewal of breast cancer stem-like cells and breast tumor development. Cancer Res. 2014;74:6648–6660. doi: 10.1158/0008-5472.CAN-13-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alarmo EL, Kuukasjarvi T, Karhu R, Kallioniemi A. A comprehensive expression survey of bone morphogenetic proteins in breast cancer highlights the importance of BMP4 and BMP7. Breast Cancer Res Treat. 2007;103:239–246. doi: 10.1007/s10549-006-9362-1. [DOI] [PubMed] [Google Scholar]
- 22.Davies SR, Watkins G, Douglas-Jones A, Mansel RE, Jiang WG. Bone morphogenetic proteins 1 to 7 in human breast cancer, expression pattern and clinical/prognostic relevance. J Exp Ther Oncol. 2008;7:327–338. [PubMed] [Google Scholar]
- 23.Pouliot F, Blais A, Labrie C. Overexpression of a dominant negative type II bone morphogenetic protein receptor inhibits the growth of human breast cancer cells. Cancer Res. 2003;63:277–281. [PubMed] [Google Scholar]
- 24.Guo W, Gorlick R, Ladanyi M, Meyers PA, Huvos AG, Bertino JR, Healey JH. Expression of bone morphogenetic proteins and receptors in sarcomas. Clin Orthop Relat Res. 1999:175–183. doi: 10.1097/00003086-199908000-00023. [DOI] [PubMed] [Google Scholar]
- 25.Jiao G, Guo W, Ren T, Lu Q, Sun Y, Liang W, Ren C, Yang K, Sun K. BMPR2 inhibition induced apoptosis and autophagy via destabilization of XIAP in human chondrosarcoma cells. Cell Death Dis. 2014;5:e1571. doi: 10.1038/cddis.2014.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JH, Park BH, Kim HK, Park TS, Baek HS. Hypoxia decreases Runx2/Cbfa1 expression in human osteoblast-like cells. Mol Cell Endocrinol. 2002;192:197–203. doi: 10.1016/s0303-7207(02)00036-9. [DOI] [PubMed] [Google Scholar]
- 27.Tang X, Lu X, Guo W, Ren T, Zhao H, Zhao F, Tang G. Different expression of Sox9 and Runx2 between chondrosarcoma and dedifferentiated chondrosarcoma cell line. Eur J Cancer Prev. 2010;19:466–471. doi: 10.1097/CEJ.0b013e32833d942f. [DOI] [PubMed] [Google Scholar]
- 28.Kudo N, Ogose A, Hotta T, Kawashima H, Gu W, Umezu H, Toyama T, Endo N. Establishment of novel human dedifferentiated chondrosarcoma cell line with osteoblastic differentiation. Virchows Archiv. 2007;451:691–699. doi: 10.1007/s00428-007-0426-3. [DOI] [PubMed] [Google Scholar]
- 29.Kim IY, Lee DH, Lee DK, Ahn HJ, Kim MM, Kim SJ, Morton RA. Loss of expression of bone morphogenetic protein receptor type II in human prostate cancer cells. Oncogene. 2004;23:7651–7659. doi: 10.1038/sj.onc.1207924. [DOI] [PubMed] [Google Scholar]
- 30.Owens P, Pickup MW, Novitskiy SV, Chytil A, Gorska AE, Aakre ME, West J, Moses HL. Disruption of bone morphogenetic protein receptor 2 (BMPR2) in mammary tumors promotes metastases through cell autonomous and paracrine mediators. Proc Natl Acad Sci U S A. 2012;109:2814–2819. doi: 10.1073/pnas.1101139108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gangopahyay A, Oran M, Bauer EM, Wertz JW, Comhair SA, Erzurum SC, Bauer PM. Bone morphogenetic protein receptor II is a novel mediator of endothelial nitric-oxide synthase activation. J Biol Chem. 2011;286:33134–33140. doi: 10.1074/jbc.M111.274100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, Willson JK, Markowitz S, Hamilton SR, Kern SE, Kinzler KW, Vogelstein B. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 33.Boeuf S, Bovée JV, Lehner B, van den Akker B, van Ruler M, Cleton-Jansen AM, Richter W. BMP and TGFbeta pathways in human central chondrosarcoma: enhanced endoglin and Smad 1 signaling in high grade tumors. BMC Cancer. 2012;12:488. doi: 10.1186/1471-2407-12-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


