Abstract
MicroRNAs (miRNAs) are implicated in the pathogenesis of oral squamous-cell carcinoma (OSCC). miR-101 is involved in the development and progression of OSCC, but the biological functions and underlying molecular mechanisms of this miRNA remain largely unknown. In this study, we showed that miR-101 was underexpressed in OSCC tissues and cell lines. miR-101 downregulation was inversely correlated with zinc finger E-box binding homeobox 1 (ZEB1) expression, lymph-node metastasis, and poor prognosis in OSCC patients. Enhanced expression of miR-101 significantly inhibited OSCC cell proliferation, apoptosis resistance, migration and invasion in vitro, and suppressed tumor growth and lung metastasis in vivo. Bioinformatics analyses showed that miR-101 directly targeted ZEB1, as confirmed by a dual-luciferase reporter assay. The inhibitory effects of miR-101 on OSCC growth and metastasis were attenuated and phenocopied by ZEB1 overexpression and knockdown, respectively. Overall, our findings indicated that miRNA-101 reduced OSCC growth and metastasis by targeting ZEB1 and provided new evidence of miR-101 as a potential therapeutic target for OSCC patients.
Keywords: microRNA-101, oral squamous-cell carcinoma, proliferation, metastasis, zinc finger E-box binding homeobox 1
Introduction
Oral squamous-cell carcinoma (OSCC) is the sixth most common malignancy worldwide [1]. Despite significant advances in diagnosis and therapy, the five-year survival rate of OSCC patients remains poor because of frequent loco-regional recurrences and neck lymph node metastases [2,3]. Cell proliferation, apoptosis resistance, migration, and invasion have been shown to play pivotal roles in the pathogenesis of OSCC [4,5]. Therefore, strategies aimed at reducing these malignant events should be urgently identified for OSCC therapy.
MicroRNAs (miRNAs) are a class of highly conserved small non-coding RNA molecules that regulate gene expression by binding to the 3’-untranslated regions (UTRs) of the complementary mRNAs, leading to translational repression and gene silencing [6]. miRNAs are involved in tumor cell processes, such as proliferation, apoptosis, migration, and invasion [7,8]. The dysregulation of miRNAs contributes to the tumorigenesis of OSCC [9,10]. Accumulating evidence has suggested that miR-101 is frequently underexpressed in multiple malignancies [11-15], including OSCC. miRNA-101 inhibits breast cancer growth and metastasis by targeting CX chemokine receptor 7 [12]. The loss of miR-101 promotes epithelial-mesenchymal transition (EMT) by targeting zinc finger E-box binding homeobox 1 (ZEB1) in hepatocytes [13]. miR-101 also inhibits EMT and tumor metastasis by targeting enhancer of zeste homolog 2 in oral tongue squamous carcinoma cells [14]. However, the precise mechanism by which miR-101 reduces the growth and metastasis of OSCC cells remains unknown.
ZEB1, originally identified as a DNA-binding protein containing a homeodomain and two zinc finger clusters, is proved to function as a transcriptional repressor by specifically binding to the E-box motif located in the E-cadherin promoter region [16,17]. As a well-known factor in the activation of EMT, ZEB1 causes the loss of E-cadherin and gain of mesenchymal markers and consequently facilitate cancer progression by increasing tumor cell migration and invasion [18]. Recent studies showed that ZEB1 may serve as a prognostic indicator for tumor patients and correlates with liver and lymph-node metastases [19-23]. A depletion of ZEB1 causes decreased proliferative, migratory, and invasive capabilities of bladder cancer cells [24]. Lei et al. [25] demonstrated that targeting ZEB1 inhibits OSCC growth. Nevertheless, the biological roles of ZEB1 in the growth and metastasis of OSCC remain unclear.
In this study, we investigated the expression and biological functions of miR-101 in OSCC and found that miR-101 expression was dramatically decreased in OSCC cell lines and tissues and was thus inversely correlated with ZEB1 level, lymph-node metastasis, and poor prognosis in OSCC patients. Dual-luciferase reporter assays revealed that miR-101 directly targeted ZEB1. The restoration of miR-101 significantly inhibited OSCC growth and metastasis in vitro and in vivo; such inhibition was counteracted and mimicked by the overexpression and depletion of ZEB1, respectively. Overall, the present study provided novel insights into the molecular mechanisms by which miR-101 inhibits OSCC growth and metastasis, indicating miR-101 as a promising target for OSCC therapy.
Materials and methods
Clinical samples
Clinical samples were obtained from OSCC patients who were diagnosed, treated, and followed up at the Department of Oral and Maxillofacial Surgery, School of Stomatology, Fourth Military Medical University between 2009 and 2013. An informed consent was signed by every participant. Ethics approval for the study was obtained from the Fourth Military Medical University (China). None of the patients had received chemotherapy or radiotherapy before surgery. All specimens were confirmed by pathological examinations. Fresh frozen tissues were stored in liquid nitrogen until use. Clinical parameters, including pathological features and TNM stage, were retrospectively collected by reviewing patients’ charts.
Cell culture
Human normal oral keratinocytes (hNOKs) were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in an oral keratinocyte medium (ScienCell Research Laboratories). Human OSCC cell lines, including Tca8113, OSCC-15, SCC-9, and SCC-25, as well as human embryonic kidney 293T (HEK293T) cells were obtained from American Type Culture Collection (Manassas, VA, USA). SCC-9-luciferase (luc) cells stably expressing highly efficient luciferase were purchased from PerkinElmer (Santa Clara, CA, USA). All the cells were used no later than 6 months after receipt. HEK293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Carlsbad, CA, USA). All OSCC cells were cultured in DMEM/Nutrient Mixture F12 (Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), 2 mM L-glutamine, and 100 μg/mL penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA) in a humidified incubator containing 5% CO2 at 37°C. All cells from passage 4 were used for all the experiments.
Plasmid and lentivirus constructs
The luciferase-3’-UTR-wild-type (WT) reporter or luciferase-3’-UTR-mutant (MUT) plasmids were prepared by inserting the ZEB1-3’-UTR-WT carrying a putative miR-101 binding site or its mutant sequence into the pGL3-control plasmid (Promega, Madison, WI, USA). The primers used to amplify the WT and MUT 3’-UTRs of ZEB1 are as follows: for WT, 5’-AAACTCGAGTACTTCAATTCCTCG GTATTG-3’ (forward), and 5’-AAATCTAGACACACTGTTCTACAGTCCAAGGC-3’ (reverse); for MUT, 5’-CTGTGCAACATTTTTTGTACAAATGTCTTCAAACCTGG-3’ (forward), and 5’-CCAGGTTTGAAGACATTTGTACAAAAAATGTTGCA CAG-3’ (reverse). The pUNOI-hZEB1 (open reading frame) plasmid was purchased from InvivoGen (Hong Kong, China). All constructs were confirmed by DNA sequencing. For the lentiviral expression of miR-101, cDNA strands corresponding to the pre-miR-101 sequence were synthesized and cloned into the AgeI/EcoRI sites of pGCsi-H1-CMV-GFP (GeneChem, Shanghai, China). The constructs were transfected into the HEK293T cells, along with pMD2.G and psPAX2 packaging plasmids. After 48 h transfection, the supernatant was collected, centrifuged, filtered, and used for infection of SCC-9-luc cells. A short hairpin RNA (shRNA) was designed based on the ZEB1 sequence, and a scrambled shRNA was used as a control. Paired deoxyribonucleotide oligos encoding the shRNAs were synthesized, annealed, and cloned into the EcoRI/NcoI sites of the pLKO.1 vector (Addgene, Cambridge, MA, USA). The constructs were co-transfected with pCMV-VSVG and pCMV-bA.9 packaging plasmid into HEK293T packaging cells. The viral supernatants were harvested, filtered, and transferred to SCC-9-luc cells. Cells were selected with 5 μg/mL puromycin (Sigma) to generate stable shRNA-expressing clones.
Cell transfection
The miR-101 and miR-NC mimics were synthesized by RiboBio (Guangzhou, China). Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) was used for transfection, according to the manufacturer’s instructions. When cell confluence reached 80%, 80 nM miR-101 or miR-NC mimics was transfected into the SCC-9 cells (1 × 106 cells/6 cm dish).
Dual-luciferase reporter assay
Plasmid pGL3-ZEB1-3’-UTR-WT or pGL3-ZEB1-3’-UTR-MUT was co-transfected with miR-101 and miR-NC mimics into SCC-9 cells. A luciferase assay was conducted 48 h after transfection using a dual-luciferase reporter assay kit (Promega). Renilla luciferase was co-transfected as a control for normalization.
Quantitative real-time polymerase chain reaction (qPCR)
For RNA extraction, the fresh tissues and cells were lysed using TRIzol reagent (Invitrogen) and purified using RNeasy Mini Kit (Qiagen). cDNA was synthesized using reverse transcriptase (Epicentre, Madison, WI, USA) or the miScript Reverse Transcription Kit (Qiagen) and then amplified using SYBR Premix Ex Taq™ (TaKaRa, Otsu, Shiga, Japan). The expression levels of mRNAs and miRNAs were determined using the 2-ΔΔCt method, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 as internal controls, respectively. The primers used for PCR amplification are listed as follows: for ZEB1, 5’-AGAGCAGTGAAAGAGAAGGGAATGC-3’ (forward) and 5’-GGTCCTCTTCAGGTGCCTCAG-3’ (reverse); for GAPDH, 5’-TGCACCACCAACTGCTTAGC-3’ (forward) and 5’-GGCATGGAC TGTGGTCATGAG-3’ (reverse); for miR-101, 5’-CGGCGGTACAGTACTGTGATA A-3’ (forward) and 5’-CTGGTGTCGTGGAGTCGGCAATTC-3’ (reverse); and for U6, 5’-CTCGCTTCGGCAGCACA-3’ (forward) and 5’-AACGCTTCACGAATTTGC GT-3’ (reverse).
Western blot analysis
Proteins were extracted from the fresh tissues and cultured cells with a protein extraction kit (KeyGEN, Nanjing, China), separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis, and transferred electrophoretically to polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). The membranes were blocked with 5% non-fat milk in TBS containing 0.1% Tween 20 at 37°C for 2 h, incubated with mouse anti-human ZEB1 and β-actin antibodies (all from Santa Cruz, Dallas, TX, USA) at 4°C overnight, washed, and probed with rabbit anti-mouse secondary antibody coupled with horseradish peroxidase (Sigma). The proteins were detected by enhanced chemiluminescence kit (Santa Cruz).
Cell proliferation assay
Cell proliferation was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay kit (Promega). In brief, the cells seeded in a 96-well plate (1 × 103 cells/well) were transfected with miR-101 mimics, miR-NC, or miR-101 mimics + ZEB1-expressing plasmid. The plates were incubated for 1, 2, 3, or 4 days after transfection. Each well was added with 25 μL of MTT (10 mg/mL) and incubated for 4 h. Then, the supernatant was removed, and 150 μL of dimethyl sulfoxide was added to each well (Sigma). The absorbance was detected at 490 nm with a microplate reader (Bio-Rad, Hercules, CA, USA).
Colony formation assay
A total of 1 × 103 SCC-9 cells transfected with miR-101 mimics, miR-NC, or miR-101 mimics + ZEB1-expressing plasmid were seeded in six-well plates and allowed to grow for 14 days to determine their colony-forming ability. After 70% ethanol fixation, the plate was stained with 0.5% crystal violet (Sigma), and cell colonies were counted under the microscope (Olympus, Tokyo, Japan). Five random fields were selected for each well to determine total number of colonies.
Cell cycle analysis
Forty-eight hours after transfection, the cells were fixed with 70% ethanol and treated with 1 mg/mL RNase at 37°C for 30 min. Then, the cells were stained with propidium iodide (PI; 50 μg/mL; Sigma) at 4°C for 30 min. Subsequently, the cells were analyzed with a FACS flow cytometer (BD Biosciences, San Jose, CA, USA). The percentages of cell populations in different stages of the cell cycle were determined using ModFit software (Verity Software House Inc., Topsham, ME, USA).
Apoptosis assay
Apoptosis was assessed using an annexin V-fluorescein isothiocyanate (FITC)/PI apoptosis detection kit (BD Biosciences) according to the manufacturer’s instructions. Briefly, the cells (5 × 105) were collected 48 h after transfection, washed twice with PBS, and then resuspended in 1× binding buffer. Annexin V-FITC (5 μL) and PI (5 μL) were added to 100 μL of the solution. After a 15 min incubation in the dark at room temperature, 400 μL of 1× binding buffer was added, and the cells positive for annexin V-FITC and/or PI were analyzed on a FACS flow cytometer (BD Biosciences) with ModFit software (Verity Software House Inc.).
Nucleosomal fragmentation assay
Cell apoptosis was quantified using a nucleosomal fragmentation kit (Cell Death Detection ELISA PLUS; Roche Applied Science, Indianapolis, IN, USA) 48 h after transfection according to the manufacturer’s instructions. The absorbance values were normalized to those of the control cells to derive a nucleosomal enrichment factor.
Caspase-3 activity assay
A caspase-3 activity assay was performed using a caspase-3 colorimetric protease assay kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Briefly, 48 h after transfection, 1 × 105 cells were lysed, and the supernatant was collected after centrifugation at 12,000 × g. Protein (40 μL per sample) was added to 50 μL of reaction buffer with 5 μL of N-acetyl-Asp-Glu-Val-Asp-pNA substrate and incubated at 37°C for 2 h. Caspase-3 activity was measured with a microplate reader (Bio-Rad) at an absorbance of 405 nm.
Wound healing assay
The transfected cells were plated in six-well plates at 2 × 105 cells/well. When the cells reached 80% confluence, an artificial wound was created with a 10 μL pipette tip. The cells were then rinsed with a medium to remove any free-floating cells and debris. The medium was then added, and the culture plates were incubated at 37°C. The wound width was monitored with a phase-contrast microscope (Olympus) at 0 and 24 h after scratch.
Invasion assay
An invasion assay was performed using a Transwell system (8 μm pore size, Matrigel-coated polycarbonate membrane; BD Biosciences). Briefly, 2 × 104 transfected cells per insert were seeded into the upper chambers. A complete medium containing 10% FBS was added to the lower chambers to act as a chemoattractant. After incubation for 24 h at 37°C, a cotton swab was used to scrape and remove the cells from the upper surface of the membrane. The cells that invaded and adhered to the bottom of the Matrigel membrane were stained with crystal violet and visualized with an inverted microscope (Olympus). The invaded cells were counted in five separate fields per membrane.
Terminal deoxynucleotidyl transferase mediated dUTP nick-end labeling (TUNEL) assay
The tissue specimens were fixed in 10% paraformaldehyde overnight and embedded in paraffin by employing a standard histological procedure. TUNEL staining was performed in accordance with manufacturer’s protocols (Roche, Mannheim, BW, Germany). Briefly, paraffin-embedded sections were deparaffinized, rehydrated, and then subjected to antigen retrieval with 0.1 M citrate buffer (Sigma). The sections were rinsed with PBS and treated with proteinase K followed by the incubation with terminal deoxynucleotidyl transferase (TdT) reaction mixture for 1 h at 37°C. After washing with PBS thrice, the sections were incubated in anti-digoxigenin-peroxidase at room temperature for 30 min, rinsed in PBS, and then treated with diaminobenzidine (DAB; Dako, Carpinteria, CA, USA) for 1-2 min at room temperature. Subsequently, the sections were counterstained with hematoxylin (Sigma) and stored in the mounting medium (Dako). Negative controls were incubated in a medium lacking TdT enzyme. TUNEL-positive (apoptotic) cells were ultimately expressed as a percentage of the total cells determined by hematoxylin staining.
Animals
Six-week-old CB17-severe combined immune deficiency (SCID) male mice were purchased from the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences (Beijing, China). The animals were bred under aseptic conditions and kept in a facility with constant humidity and temperature (25°C-28°C) and a 12 h light/dark cycle. The experiments were approved by the Institutional Animal Care and Use Committee of the Fourth Military Medical University.
Experimental studies in vivo
OSCC xenografts were established by subcutaneously injecting 1 × 106 SCC-9-luc cells that were infected with a recombinant lentivirus expressing a miR-101 precursor or shZEB1, or their control lentivirus, on the hind flanks of SCID mice (six mice per group). Tumor volumes were measured and calculated as follows: tumor volume = width2 × length/2. Six weeks after inoculation, the mice were sacrificed by euthanasia, and the tumors were weighed. For in vivo metastasis assays, the SCID mice were injected with 1 × 106 SCC-9-luc cells infected with or without various lentiviruses via their tail vein. The mice were sacrificed by euthanasia eight weeks after injection. The lung tissues were removed, fixed, paraffin-embedded, serially sectioned, and subjected to hematoxylin and eosin staining (Sigma). The metastases per section were counted. For the apoptosis assay, the sections were stained using TUNEL kits.
Bioluminescence imaging
The mice were anesthetized with 2% isoflurane and received a single intraperitoneal injection of D-luciferin (150 mg/kg; Promega) for bioluminescence imaging. The in vivo luciferase activity in the mice was recorded on IVIS Imaging System (Caliper Life Science, MA, USA) one week after subcutaneous injection. The signals in defined regions of interest were expressed as photon flux (photons/s/cm2) with the use of Living Image software (Xenogen Corporation, Berkeley, CA, USA).
Statistical analysis
Data are expressed as mean ± standard derivation (SD). The statistical significance of differences was determined by Student’s two-tailed t-test in two groups and one-way analysis of variance in multiple groups, and Fisher’s exact test was used for the comparisons of the qualitative variables. Chi-squared tests were used to evaluate frequencies. Kaplan-Meier method was used to draw the survival curve, and log-rank test was used for survival analysis. SPSS version 16.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analyses. Results are considered statistically significant when P < 0.05.
Results
Reduced miR-101 and increased ZEB1 levels in OSCC cells
miR-101 expression was significantly lower in the OSCC specimens than that in the corresponding adjacent non-cancerous specimens by qPCR assay (Figure 1A). The OSCC patients with low miR-101 expression had poor survival rate (Figure 1B) and high lymph-node metastasis (Table 1) compared with those with high miR-101 levels. Previous studies showed that miR-101 directly and negatively modulates ZEB1 and suppresses EMT in hepatocytes and ovarian cancer [13,26]. Here, we found that ZEB1 expression was elevated in OSCC specimens compared with the corresponding adjacent non-cancerous specimens by qPCR and Western blot assays (Figure 1C and 1D) and inversely correlated with miR-101 level in OSCC tissues (Figure 1E). In addition, miR-101 expression level was decreased in the four OSCC cell lines (Tca8113, OSCC-15, SCC-9, and SCC-25) compared with that in hNOK, whereas ZEB1 protein expression exhibited the opposite tendency (Figure 1F and 1G). The SCC-9 cell line was selected for further study because it had the lowest miR-101 level among all the tested OSCC cell lines. The results suggested that miR-101 could serve as a prognostic marker and play a key role in OSCC development.
Figure 1.
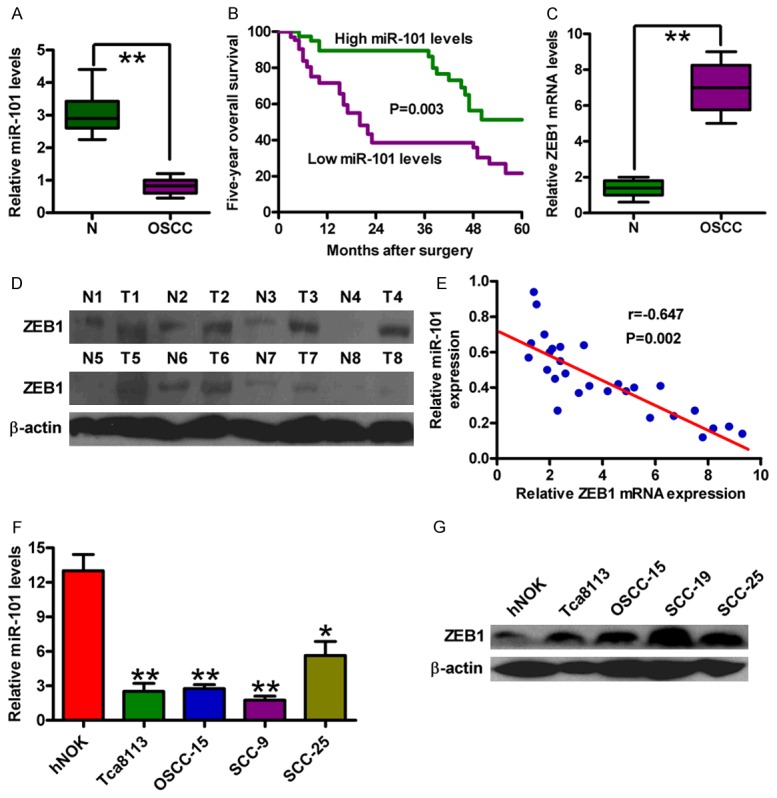
Expression levels of miR-101 and ZEB1 in OSCC tissues and cell lines. (A) qPCR analysis of miR-101 levels in OSCC specimens and the adjacent non-cancerous specimens (n = 30). U6 was used as the endogenous control. (B) Kaplan-Meier survival curves of 40 patients with OSCCs divided by miR-101 expression. Log-rank test was used for survival analysis. qPCR (C) and Western blot (D) analyses of the mRNA and protein levels of ZEB1 in OSCC specimens and the adjacent non-cancerous specimens. GAPDH and β-actin were used as the internal controls, respectively. (E) Spearman’s correlation analysis between miR-101 and ZEB1 in OSCC specimens analyzed by qPCR assay. qPCR (F) and Western blot (G) assays were performed to assess miR-101 expression in four OSCC cell lines (Tca8113, OSCC-15, SCC-9 and SCC-25) and the hNOKs. U6 and β-actin were used as the endogenous controls. All data are shown as mean ± SD of three separate experiments. *P < 0.05; **P < 0.01.
Table 1.
The association between miR-101 expression and lymph node metastatic status of 183 patients with OSCC
| Lymph node metastasis (n=183) | P value | ||
|---|---|---|---|
|
| |||
| + | - | ||
| High miR-101 | 3 | 95 | 0.001 |
| Low miR-101 | 15 | 70 | |
miR-101 inhibits the proliferation, apoptosis resistance, migration, and invasion of OSCC cells
To explore the biological functions of miR-101 in OSCC, we transfected miR-101 and miR-NC mimics into SCC-9 cells. Cell viability was significantly reduced in miR-101-treated cells in comparison with the miR-NC-treated cells (Figure 2A). miR-101 significantly decreased the colony formation of SCC-9 cells (Figure 1B and 1C). The miR-101-treated cells showed increased accumulation in G1 phase and decreased accumulation in S and G2/M phases (Figure 2D). As shown in Figure 2E, miR-101 introduction markedly increased the percentage of apoptotic cells. Increases in nucleosomal fragmentation (Figure 2F) and caspase-3 activity (Figure 2G) were also observed in SCC-9 cells with miR-101 mimic transfection. These results indicated that miR-101 inhibits the proliferation of OSCC cells and induces their apoptosis in vitro.
Figure 2.
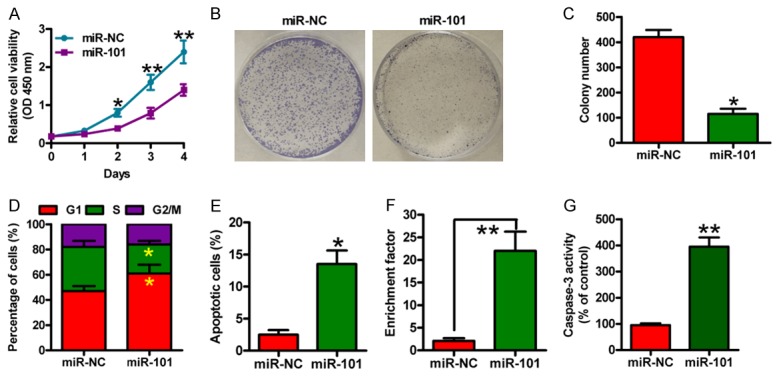
miR-101 reduced proliferation and apoptosis resistance of OSCC cells. SCC-9 cells were transfected with miR-NC or miR-101 mimics. (A) MTT assay was performed to determine cell viability. (B) Representative photographs of the colony formation of SCC-9 cells. (C) Quantitative analysis of colony formation of (B). (D) Flow cytometry analysis of the fraction of SCC-9 cells in G1, S and G2/M phases 48 h after transfection. (E) The percentages of apoptotic cells analyzed by flow cytometry. Nucleosomal fragmentation (F) and caspase-3 activity (G) assays of SCC-9 cells. All data are shown as mean ± SD of three separate experiments. *P < 0.05; **P < 0.01.
Subsequently, we evaluated whether miR-101 was able to suppress the migration and invasion of OSCC cells by wound healing and Transwell invasion assays. As expected, miR-101 significantly inhibited the migration of SCC-9 cells (Figure 3A and 3B). Similarly, the invasion of SCC-9 cells was significantly reduced by miR-101 (Figure 3C and 3D). These results suggested that miR-101 potently inhibits the migration and invasion of OSCC cells in vitro.
Figure 3.
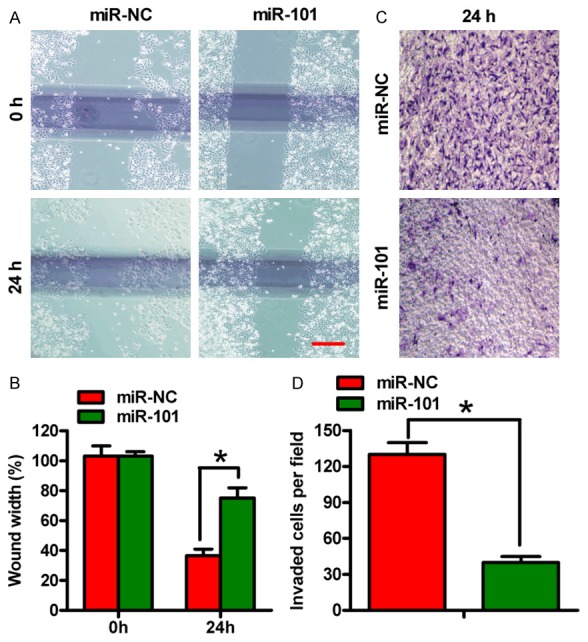
miR-101 inhibited migration and invasion of OSCC cells. SCC-9 cells were transfected with miR-NC or miR-101 mimics. (A) A wound healing assay was performed in SCC-9 cells. The images were captured at 0 and 24 h. (B) Percentage of wound widths in (A). (C) Cell invasion was determined using Transwell chambers. (D) The number of invaded cells in (C). All data are shown as mean ± SD of three separate experiments. *P < 0.05.
miR-101 directly targets ZEB1 in OSCC cells
miR-101 targets were predicted using the following three algorithms: TargetScan, PicTar, and miRBase (Figure 4A). A complementary sequence of miR-101 to the 3’-UTR of ZEB1 mRNA is shown in Figure 4B. The dual-luciferase reporter assay showed that miR-101 reduced the activity of the luciferase reporter fused to the 3’-UTR-WT of ZEB1 but did not suppress that of the reporter fused to the MUT version (Figure 4C). The introduction of miR-101 reduced ZEB1 expression both at the mRNA and protein levels in the SCC-9 cells (Figure 4D and 4E). The data suggested that miR-101 directly targeted ZEB1 in OSCC cells.
Figure 4.
Identification of ZEB1 as a direct target of miR-101. (A) The putative target genes of miR-101 were predicted using TargetScan, PicTar, and miRBase. (B) The predicted binding sites of miR-101 in the WT and MUT 3’-UTR of ZEB1. (C) Dual-luciferase reporter assays were performed 24 h after co-transfection of SCC-9 cells with miR-NC or miR-101 mimics and a pGL3 construct containing WT or MUT 3’-UTR of ZEB1. Data were normalized to those from cells co-transfected with miR-NC and pGL3 plasmid. The mRNA (D) and protein (E) levels of ZEB1 in SCC-9 cells transfected with miR-NC or miR-101 mimics were measured by qPCR and Western blot assays. GAPDH and β-actin were used as internal controls, respectively. All data are shown as mean ± SD of three separate experiments. **P < 0.01.
Inhibition of malignant phenotypes of OSCC cells by miR-101 is mediated by ZEB1
To confirm that ZEB1 is a functional target of miR-101, we performed ZEB1 rescue experiments. ZEB1 overexpression markedly counteracted the inhibition of cell viability of SCC-9 cells by miR-101 (Figure 5A). Similarly, cell proliferation reduction and cell cycle arrest caused by miR-101 restoration were inhibited by co-transfection with ZEB1-expressing plasmid (Figure 5B and 5C). ZEB1 rescue also reduced miR-101-induced increase in nucleosomal fragmentation, caspase-3 activity, and cell apoptosis (Figure 5D-F). ZEB1 overexpression could partially counteract the decrease of cell invasion and migration of SCC-9 cells by miR-101 (Figure 5G and 5H). These results indicated that ZEB1 is a key downstream mediator of miR-101 effects on OSCC cells.
Figure 5.
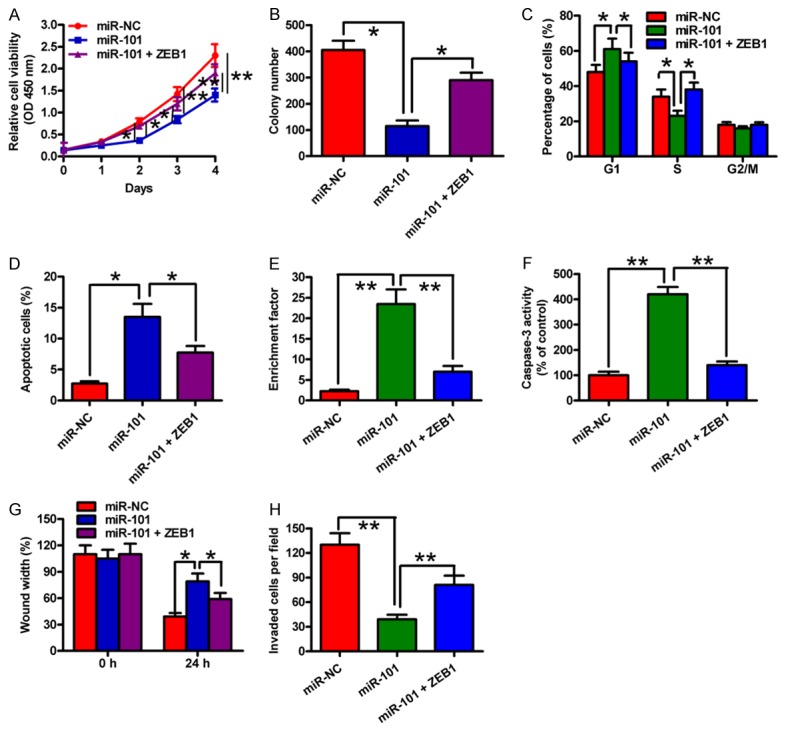
ZEB1 was a downstream functional mediator of miR-101 in OSCC cells. SCC-9 cells were transfected with miR-NC, miR-101 mimic or miR-101 mimic + ZEB1-expressing plasmid. (A) Cell viability was measured by MTT assay. (B) A colony formation assay was performed to assess cell proliferation, and the number of colonies was quantified. (C) Cell cycle distribution was analyzed by flow cytometry. (D) Cell apoptosis was detected by flow cytometry. Nucleosomal fragmentation (E) and caspase-3 activity (F) assays were used to evaluate cell apoptosis. (G) Cell migration was assessed by a wound healing assay, and the percentage of wound width was measured. (H) A transwell assay was performed to detect cell invasion, and the number of invaded cells was calculated. All data are shown as mean ± SD of three separate experiments. *P < 0.05; **P < 0.01.
miR-101 suppressed tumorigenesis and metastasis of OSCC in vivo
A xenograft or metastasis mouse model was established by subcutaneous or venous injection of SCC-9-luc cells stably expressing a miR-101 precursor or a shZEB1 or their vector controls to confirm the aforementioned findings in vitro. Compared with the vector control group, the miR-101 overexpression group or the ZEB1 knockdown group exhibited significantly reduced tumor growth (Figure 6A), volumes (Figure 6B) and weights (Figure 6C). miR-101 restoration or ZEB1 depletion also effectively reduced the lung metastases in vivo (Figure 6D). The apoptotic cells were considerably more in the tumors of ectopic miR-101 expression group or the ZEB1-silencing group than that in the tumors derived from the control groups (Figure 6E). These results suggested that the suppression of OSCC growth and lung metastasis by miR-101 is mediated by the reduction of ZEB1 in vivo.
Figure 6.
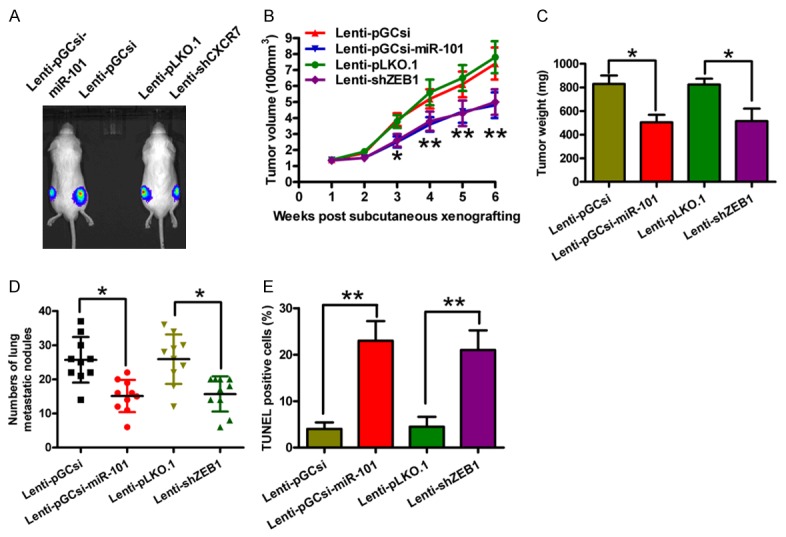
Overexpression of miR-101 or knockdown of ZEB1 suppressed tumor growth and metastasis and promoted apoptosis of OSCC cells in vivo. SCID mice were injected subcutaneously or via their tail vein with SCC-9-luc cells that were infected with a control lentivirus (Lenti-pGCsi or Lenti-pLKO.1) or a recombinant lentivirus expressing a miR-101 precursor (Lenti-pGCsi-miR-101) or shZEB1 (Lenti-shZEB1). A. In vivo luciferase image for the detection of the xenograft tumor growth in the mice implanted with various cells. B. Tumor volume was measured and calculated once a week. C. Tumor weight was measured after six weeks of implantation. D. The numbers of metastatic foci in the lungs of mice from various groups eight weeks after tail vein injection. E. A TUNEL assay was performed to detect the percentage of apoptotic cells. All data are shown as mean ± SD of three separate experiments. *P < 0.05; **P < 0.01.
Discussion
In this study, we found that miR-101 functioned as a tumor suppressor of OSCC by targeting ZEB1. The key findings were as follows: first, miR-101 was significantly underexpressed in OSCC tissues and cell lines. Second, miR-101 expression was inversely correlated with ZEB1 expression, as well as lymph-node metastasis and poor survival rate in OSCC patients. Third, miR-101 directly targeted ZEB1. Fourth, miR-101 restoration inhibited OSCC cell proliferation, apoptosis resistance, migration, and invasion in vitro, and tumor growth and lung metastasis in vivo. Lastly, the inhibitory effects of miR-101 on malignant phenotypes of OSCC were attenuated and mimicked by the overexpression and knockdown of ZEB1, respectively. Overall, these results suggest the potential diagnostic and prognostic roles of miR-101 in OSCC and indicate that miR-101 is a tumor suppressor of OSCC by targeting ZEB1.
miRNAs possess several features that make them attractive candidates as new prognostic biomarkers and powerful tools for the early diagnosis of cancer [27]. miR-101 has been shown to be underexpressed and to act as a tumor suppressor in multiple cancers [11,12]. In the current study, we found that miR-101 was lowly expressed in OSCC tissues and cell lines. OSCC patients with high miR-101 levels generally had longer survival than patients with low miR-101 expression had. miR-101 expression was also able to distinguish patients with or without lymph-node metastasis, indicating the diagnostic and prognostic significance of miR-101 in OSCC. Several studies have revealed that miR-101 may target multiple effectors to inhibit the tumorigenesis and metastasis of malignancies [12-14]. Here, we demonstrated that the ectopic expression of miR-101 inhibited OSCC cell proliferation, apoptosis resistance, migration, and invasion in vitro, and tumor growth and lung metastasis in vivo.
The relevance of ZEB1 to tumor progression has been studied in several human cancers [28-31]. In the present study, we found that ZEB1 was significantly upregulated in OSCC tissues and cell lines and inversely correlated with miR-101 expression level. A recent study has shown that targeting ZEB1 can inhibit colorectal cancer cell proliferation [32]. Majid et al. [24] reported that the knockdown of ZEB1 suppresses proliferation, migration, and invasion of bladder cancer cells and induces their apoptosis. The inhibition of ZEB1 expression can also suppress OSCC cell growth [25]. As shown here, ZEB1 was directly targeted by tumor suppressor miR-101 in OSCC. Furthermore, the overexpression and silencing of ZEB1 attenuated and mimicked, respectively, the inhibitory effects of miR-101 on OSCC cell proliferation, apoptosis resistance, migration, and invasion in vitro, and tumor growth and lung metastasis in vivo, indicating that the inhibitory effects of miR-101 on malignant phenotypes of OSCC are partly mediated by ZEB1 depletion.
In summary, we demonstrated that miR-101 is lowly expressed in OSCC tissues and cell lines and inversely correlated with ZEB1 expression, lymph-node metastasis, and poor prognosis of OSCC patients. In vitro and in vivo assays showed that miR-101 inhibited OSCC growth and metastasis by targeting ZEB1. Therefore, miR-101 may be a potential therapeutic target for OSCC.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Jerjes W, Upile T, Petrie A, Riskalla A, Hamdoon Z, Vourvachis M, Karavidas K, Jay A, Sandison A, Thomas GJ, Kalavrezos N, Hopper C. Clinicopathological parameters, recurrence, locoregional and distant metastasis in 115 T1-T2 oral squamous cell carcinoma patients. Head Neck Oncol. 2010;2:9. doi: 10.1186/1758-3284-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao CT, Hsueh C, Lee LY, Lin CY, Fan KH, Wang HM, Huang SF, Chen IH, Kang CJ, Ng SH, Tsao CK, Huang YC, Yen TC. Neck dissection field and lymph node density predict prognosis in patients with oral cavity cancer and pathological node metastases treated with adjuvant therapy. Oral Oncol. 2012;48:329–336. doi: 10.1016/j.oraloncology.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Hu J, He Y, Yan M, Zhu C, Ye W, Zhu H, Chen W, Zhang C, Zhang Z. Dose dependent activation of retinoic acid-inducible gene-I promotes both proliferation and apoptosis signals in human head and neck squamous cell carcinoma. PLoS One. 2013;8:e58273. doi: 10.1371/journal.pone.0058273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo WL, Yu CC, Chiou GY, Chen YW, Huang PI, Chien CS, Tseng LM, Chu PY, Lu KH, Chang KW, Kao SY, Chiou SH. MicroRNA-200c attenuates tumour growth and metastasis of presumptive head and neck squamous cell carcinoma stem cells. J Pathol. 2011;223:482–495. doi: 10.1002/path.2826. [DOI] [PubMed] [Google Scholar]
- 10.Yu T, Liu K, Wu Y, Fan J, Chen J, Li C, Yang Q, Wang Z. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/beta-catenin signaling pathway. Oncogene. 2014;33:5017–5027. doi: 10.1038/onc.2013.448. [DOI] [PubMed] [Google Scholar]
- 11.Konno Y, Dong P, Xiong Y, Suzuki F, Lu J, Cai M, Watari H, Mitamura T, Hosaka M, Hanley SJ, Kudo M, Sakuragi N. MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation, invasion and stem cell-like phenotype of aggressive endometrial cancer cells. Oncotarget. 2014;5:6049–6062. doi: 10.18632/oncotarget.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JT, Jia LT, Liu NN, Zhu XS, Liu QQ, Wang XL, Yu F, Liu YL, Yang AG, Gao CF. MiRNA-101 inhibits breast cancer growth and metastasis by targeting CX chemokine receptor 7. Oncotarget. 2015;6:30818–30830. doi: 10.18632/oncotarget.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao S, Zhang Y, Zheng X, Tu X, Li H, Chen J, Zang Y, Zhang J. Loss of MicroRNA-101 Promotes Epithelial to Mesenchymal Transition in Hepatocytes. J Cell Physiol. 2015;230:2706–2717. doi: 10.1002/jcp.24995. [DOI] [PubMed] [Google Scholar]
- 14.Zheng M, Jiang YP, Chen W, Li KD, Liu X, Gao SY, Feng H, Wang SS, Jiang J, Ma XR, Cen X, Tang YJ, Chen Y, Lin YF, Tang YL, Liang XH. Snail and Slug collaborate on EMT and tumor metastasis through miR-101-mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget. 2015;6:6797–6810. doi: 10.18632/oncotarget.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miao L, Wang L, Yuan H, Hang D, Zhu L, Du J, Zhu X, Li B, Wang R, Ma H, Chen N. MicroRNA-101 polymorphisms and risk of head and neck squamous cell carcinoma in a Chinese population. Tumour Biol. 2016;37:4169–74. doi: 10.1007/s13277-015-4249-6. [DOI] [PubMed] [Google Scholar]
- 16.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 17.Fontemaggi G, Gurtner A, Damalas A, Costanzo A, Higashi Y, Sacchi A, Strano S, Piaggio G, Blandino G. deltaEF1 repressor controls selectively p53 family members during differentiation. Oncogene. 2005;24:7273–7280. doi: 10.1038/sj.onc.1208891. [DOI] [PubMed] [Google Scholar]
- 18.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 19.Zhou YM, Cao L, Li B, Zhang RX, Sui CJ, Yin ZF, Yang JM. Clinicopathological significance of ZEB1 protein in patients with hepatocellular carcinoma. Ann Surg Oncol. 2012;19:1700–1706. doi: 10.1245/s10434-011-1772-6. [DOI] [PubMed] [Google Scholar]
- 20.Okugawa Y, Toiyama Y, Tanaka K, Matsusita K, Fujikawa H, Saigusa S, Ohi M, Inoue Y, Mohri Y, Uchida K, Kusunoki M. Clinical significance of Zinc finger E-box Binding homeobox 1 (ZEB1) in human gastric cancer. J Surg Oncol. 2012;106:280–285. doi: 10.1002/jso.22142. [DOI] [PubMed] [Google Scholar]
- 21.Lemma S, Karihtala P, Haapasaari KM, Jantunen E, Soini Y, Bloigu R, Pasanen AK, Turpeenniemi-Hujanen T, Kuittinen O. Biological roles and prognostic values of the epithelial-mesenchymal transition-mediating transcription factors Twist, ZEB1 and Slug in diffuse large B-cell lymphoma. Histopathology. 2013;62:326–333. doi: 10.1111/his.12000. [DOI] [PubMed] [Google Scholar]
- 22.Zhang GJ, Zhou T, Tian HP, Liu ZL, Xia SS. High expression of ZEB1 correlates with liver metastasis and poor prognosis in colorectal cancer. Oncol Lett. 2013;5:564–568. doi: 10.3892/ol.2012.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, Li S, Huang K, Zhang Q, Wang J, Li X, Hu T, Wang S, Yang R, Jia Y, Sun H, Tang F, Zhou H, Shen J, Ma D, Wang S. The nuclear protein expression levels of SNAI1 and ZEB1 are involved in the progression and lymph node metastasis of cervical cancer via the epithelial-mesenchymal transition pathway. Hum Pathol. 2013;44:2097–2105. doi: 10.1016/j.humpath.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Majid S, Dar AA, Saini S, Deng G, Chang I, Greene K, Tanaka Y, Dahiya R, Yamamura S. MicroRNA-23b functions as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS One. 2013;8:e67686. doi: 10.1371/journal.pone.0067686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei W, Liu YE, Zheng Y, Qu L. MiR-429 inhibits oral squamous cell carcinoma growth by targeting ZEB1. Med Sci Monit. 2015;21:383–389. doi: 10.12659/MSM.893412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo F, Cogdell D, Hu L, Yang D, Sood AK, Xue F, Zhang W. MiR-101 suppresses the epithelial-to-mesenchymal transition by targeting ZEB1 and ZEB2 in ovarian carcinoma. Oncol Rep. 2014;31:2021–2028. doi: 10.3892/or.2014.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 28.Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, Jung A, Kirchner T, Brabletz T. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 29.Matsubara D, Kishaba Y, Yoshimoto T, Sakuma Y, Sakatani T, Tamura T, Endo S, Sugiyama Y, Murakami Y, Niki T. Immunohistochemical analysis of the expression of E-cadherin and ZEB1 in non-small cell lung cancer. Pathol Int. 2014;64:560–568. doi: 10.1111/pin.12214. [DOI] [PubMed] [Google Scholar]
- 30.Bronsert P, Kohler I, Timme S, Kiefer S, Werner M, Schilling O, Vashist Y, Makowiec F, Brabletz T, Hopt UT, Bausch D, Kulemann B, Keck T, Wellner UF. Prognostic significance of Zinc finger E-box binding homeobox 1 (ZEB1) expression in cancer cells and cancer-associated fibroblasts in pancreatic head cancer. Surgery. 2014;156:97–108. doi: 10.1016/j.surg.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Zhang N, Wang Y, Xu M, Liu N, Pang X, Cao J, Ma N, Pang H, Liu L, Zhang H. Zinc finger E-box binding homeobox 1 promotes invasion and bone metastasis of small cell lung cancer in vitro and in vivo. Cancer Sci. 2012;103:1420–1428. doi: 10.1111/j.1349-7006.2012.02347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quan Y, Xu M, Cui P, Ye M, Zhuang B, Min Z. Grainyhead-like 2 Promotes Tumor Growth and is Associated with Poor Prognosis in Colorectal Cancer. J Cancer. 2015;6:342–350. doi: 10.7150/jca.10969. [DOI] [PMC free article] [PubMed] [Google Scholar]



