Abstract
Unconventional prefoldin RPB5 interactor (URI), a RNA polymerase II Subunit 5-Interacting protein, is known to participate in the regulation of nutrient-sensitive mTOR-dependent transcription programs. Multiple studies have recently demonstrated that URI functions as an oncoprotein, possibly through the mTOR pathway, and regulates tumor cell motility, invasion, and metastasis. However, whether and how URI plays a role in gastric oncogenesis has not been elucidated. Due to drug resistance, recurrence and metastasis, the prognosis of gastric cancer remains poor. This study aims to explore the effects of URI on gastric cancer cells by focusing on their migratory ability and resistance to adriamycin. URI was over-expressed or knocked-down in MGC-803 and HGC-27 gastric cancer cells using URI plasmid or siRNA transfection approach. The cell viability, apoptosis, and migration ability were then examined by the CCK-8 assay, flow cytometer Annexin V/PI staining, and the Transwell cell migration assay respectively. The protein levels of apoptosis and EMT related genes were detected by western blot. The results showed that overexpression of URI promoted while knock-down of URI inhibited gastric cancer cell proliferation. URI overexpression resulted in increased Bcl-2 expression but decreased levels of Bax, cleaved PARP-1 and cleaved caspase-3. Conversely, cells treated with URI siRNA showed increased adriamycin induced apoptosis, along with reduced Bcl-2, but increased Bax, cleaved PARP-1 and cleaved caspase-3 expression. We have also shown that overexpression of URI enhanced cancer cell proliferation and migration with higher levels of Snail and Vimentin, whereas knockdown of URI in MGC-803 and HGC-27 cells inhibited proliferation and migration with decreased Snail and Vimentin expression. Together, our results support that URI promotes cell survival and mobility and acts as a chemotherapeutics resistant protein in MGC-803 and HGC-27 cells. URI might be a potential biomarker for gastric cancer diagnostics and prognostics.
Keywords: URI, gastric cancer cell, drug resistance, adriamycin, migration
Introduction
URI, unconventional prefoldin RPB5 interactor, has previously been shown as a target of nutrient signaling and participates in its regulation via mTOR-dependent transcription programs [1,2]. Recently, several studies have demonstrated the involvement of URI in the pathogenesis of multiple malignancies. URI was found amplified and overexpressed in human ovarian cancer cell lines and carcinomas and may promote cancer cell survival through targeting the S6K1-BAD signaling pathway [3]. URI played an anti-apoptotic role in the growth of hepatocellular carcinoma cells (HCC) and is required for the proliferation of HCC both in vitro and in vivo [4]. URI was also shown to promote development of multiple myeloma and may contribute to its chemotherapeutic resistance through activating the IL-6/STAT3 pathway [5]. We have recently shown that URI promoted growth of cervical cancer cells that are either human papillomavirus (HPV) positive or negative. URI enhanced cisplatin resistance of cervical cancer cells and promoted their migration and invasive capacity, possibly by up-regulation of Vimentin [6]. Gastric cancer is the second leading cause of cancer-related deaths worldwide [7]. Besides surgical resection of the tumors at relatively early stages, chemotherapy remains the main method for treating advanced and metastatic gastric cancer patients so as to improve the quality of life and to extend survival [8]. Due to the obvious adverse effect and drug resistance of current regimen, extensive efforts have been made toward finding novel and more effective targets in gastric cancer therapy [9]. PI3K/Akt/mTOR is one of the representative signaling pathways and clinical dimensions of gastric cancer [10]. This pathway is activated and overexpressed in gastric cancer, with potential prognostic significance [11,12]. PI3K/Akt/mTOR, as well as human EGFR and angiogenic pathways, have shown promising for targeted therapy in advanced gastric cancer, although the effect of mTOR inhibitors on gastric cancer is still inconclusive in clinical trials [13,14]. Notably, URI has been associated with mTOR/S6K1 signaling pathway that is known to participate in multiple carcinogenesis as indicated above [3,11-14]. However, whether URI plays a role in gastric oncogenesis remain elusive. Here we investigated the effect of URI on biological characteristics of gastric cancer cell lines MGC-803 and HGC-27, with a focus on its influence on the resistance to adriamycin and the migratory ability of gastric cancer cells.
Materials and methods
Chemicals and antibodies
Adriamycin was purchased from Selleck and was dissolved at a concentration of 1 mM as a stock solution. β-actin (SC-47778) was obtained from Santa Cruz Biotechnology. Primary antibodies to RMP/URI (5844S) and Bcl-2 (2870P) were purchased from Cell Signaling Technology (Danvers, MA). Primary antibodies to cleaved caspase-3 (D120074) and cleaved poly (ADP-ribose) polymerase (PARP) (RLM3145) were purchased from Ruiyingbio. Antibodies for Bax (D220073), Snail (D221239), and Vimentin (D120268) were obtained from Sangon Biotech. The secondary antibodies containing anti-rabbit IgG (AB10058) and anti-mouse IgG (D111050) were also obtained from Sangon Biotech.
Cell culture
Human gastric carcinoma SGC-7901, BGC-823, HGC-27 and MGC-803 cells were gifts from Professor Wei Zhu at Jiangsu University. MGC-803 cells were grown in Dulbecco’s Modified Eagle Medium (DMEM, Corning, USA) medium and HGC-27 cells were grown in RPMI-1640 (Corning, USA) medium. The culture media used were all supplemented with 10% fetal bovine serum (Gibco, New Zealand) and 1% penicillin-streptomycin mixture (Invitrogen, CA, USA). Exponentially growing cell cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C.
Cell transfection
To overexpress URI, URI expression plasmid pCMV6-URI and its vector control pCMV6-entry (OriGene) were transiently transfected into gastric cancer cells using the Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) and opti-MEM (Invitrogen, Carlsbad, CA). Transfection was performed according to the manufacturer’s protocol. Briefly, cells were transfected at around 80% confluence. After 6 hours’ transfection, cells were added with fresh medium. Meanwhile, to knock-down URI, three small interfering RNAs (siRNAs) targeting human URI with scrambled sequence as a control were synthesized by Origene Technologies. The sequences of the siRNAs and the scrambled control are as the following: rUrUrArArUTG; siRNA-C:rGrArArCrUrArGrArGrArGrArCrArGrGrArArGrArArUrUGC. Scrambled-rCrGrUrUrArArUrCrGrCrGrUrArUrArArUrArCrGrCrGrUAT.
siRNAs and the scrambled control were transfected into cells that were around 60-80% confluence using Hiperfect Transfection Reagent (Qiagen, USA) and Opti-MEM (Invitrogen, Carlsbad, CA). Untransfected cells were served as blank control. All experiments were repeated three times with duplicate templates.
Western blotting analysis
Cells were washed with ice-cold phosphate-buffered saline (PBS) twice after experimental treatment. Cells were then lysed with a RIPA buffer (Beyotime Biotechnology, CA, China) containing 10% phosphatase inhibitor and 10% protease inhibitor cocktail (KangChen, Shanghai, China). After ice cold for 20 minutes, the cell lysates were centrifuged at 14,000×g for 15 min at 4°C to discard the cellular debris. Total protein concentrations were determined using BCA-assay (Eppendorf, Hamburg, Germany). An equal amount of lysate proteins (50 μg) were separated with 10% or 12% SDS-polyacrylamide gel electrophoresis and transferred to the Immobilon-P membranes (Millipore, Billerica, USA). After blocking with 5% nonfat dry milk in TBS-T (TBS and 0.01% Tween-20) buffer, the membranes were then probed with specific primary antibodies overnight at 4°C, Immune complexes were washed three timesin 1XTBST, and incubated with HRP-conjugated IgG secondary antibodies at 37°C for 1 h. The immunoreactive protein bands were captured by an enhanced chemiluminescencesystem (Minichemi, China). To ensure equal protein loading, blots were stripped and re-probed with a specific antibody recognizing β-actin. Three independent experiments were performed.
Cell proliferation assay
Cell viability was measured using a cell counting kit-8 (CCK-8, Vazyme Biotech, Nanjing, China) according to the manufacturer’s instructions. In brief, cells were seeded in 96-well plates at a density of 4500 cells per well and incubated for 24 hours after 48 h’s transfection. The optical density (OD) of cells were detected at various time points (0, 1, 2, 3 and 4 days respectively). 10 μl of CCK-8 kit reagent was added to each well, and the cells were incubated at 37°C for an additional 2 h. The absorbance was then measured spectrophotometrically at a wavelength of 450 nm by a microplate reader (Bio-Rad Model 680, Richmond, CA, USA). In drug resistance experiment, cells after transfection were incubated with designated concentrations of adriamycin or DMSO for another 24 h. The mean value of each concentration was based on the data of five replicates and each experiment was repeated three times. The concentration for 50% inhibition of cell growth were calculated by GraphPad Prism software version 5.0. Data represent mean ± SEM from three independent experiments combined.
Apoptosis analysis by flow cytometry
The apoptotic ratios of cells were performed by the FITC Annexin V and propidium iodide (PI) double-staining apoptosis detection kit (BD PharmingenTM, CA, USA) through analyzing the membrane redistribution of phosphatidylserine using flow cytometry. Cells were treated with adriamycin (250 nM) for 12 h after 48 h’s transfection. Cells were then collected and washed twice in ice-cold PBS buffer, and subsequently stained with FITC-Annexin V/PI dual staining assay kit, according to the manufacturer’s instructions. Cells were resuspended in 500 µl of binding buffer at a concentration of 1×106 cells/ml. 100 µl of the cell suspension (1×105 cells) were incubated with 5 µl Annexin V-FITC and 3 µl propidium iodide (PI) for 15 min in dark followed by addition of 400 µl of 1X binging buffer. The percentage of early apoptotic cells were analyzed by the BD Accuri C6 flow cytometry system within 1 h. Data represent mean ± SEM from three independent experiments combined.
Transwell assay
Transwell cell migration assay was performed to analyze the effect of URI on migratory ability of the two cell lines. In short, cells were arranged to pass through the polycarbonate membrane (8 mm pore size) using a 24-well transwell chamber (Corning Costar, New York, USA), and then the numbers of the pierced cells were calculated. The relative number of pierced cells reflected the ability of migration. After 48 h’s transfection, cells were resuspended with 300 µl serum-free medium, and cultured in the upper transwell chamber. The lower chamber was filled with 600 µl complete medium for 18 h at 37°C. Then, the migrated cells on the bottom surface of membrane were fixed with 4% paraformaldehyde for 30 minutes and stained with 0.5% crystal violet for 25 min at room temperature. Cells were calculated under microscope at 6 random fields (200×).
Statistical analysis
All data were presented as mean ± SD (standard deviation) and were set up in triplicates. All statistical calculations were performed using GraphPad Prism software version 5.0. Significance of differences was determined by Student’s t test and a P value less than 0.05 was considered statistically significant. (*P < 0.05; **P < 0.01).
Results
URI expression of gastric cancer cells
We have selected four gastric cancer cell lines, SGC-7901, BGC-823, HGC-27 and MGC-803, to examine their levels of URI expression. Western blot results showed that URI expressed in all four gastric cancer cell lines (Figure 1A). We then chose the poorly differentiated MGC-803 and the undifferentiated HGC-27 cells for further experiments. The expression of URI was significantly increased in cells transfected with pCMV6-URI compared with the control cells. To knock-down URI, three candidate URI siRNA sequences (siRNA-A, -B, and -C) were used for transfection and compared with scrambled control sequence and untransfected cells. The results supported that siRNA-A sequence showed the strongest interfering effect for URI expression (Figure 1B).
Figure 1.
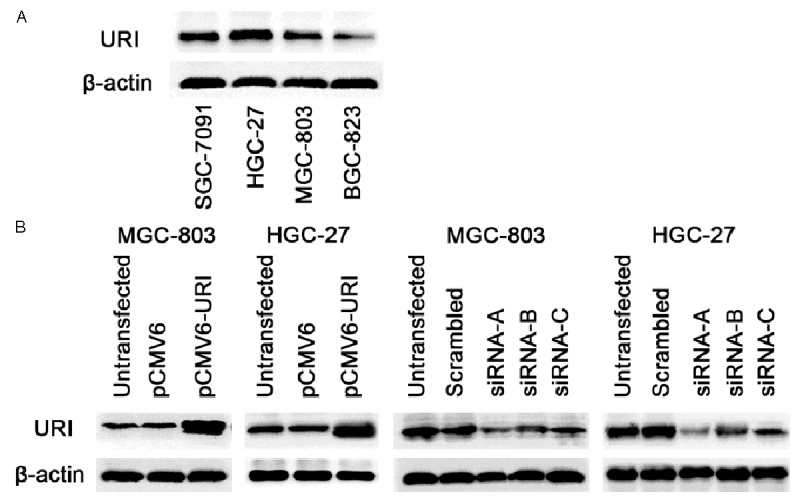
URI expression and transfection studies in gastric cancer cell lines. (A) Cells were harvested and lysed from gastric cancer cell lines SGC-7901, BGC-823, HGC-27, and MGC-803. Western blotting was performed. The basal expression of URI was shown in four gastric cancer cell lines. (B) Cells were harvested and lysed after transient transfection respectively with pCMV6-URI, URI siRNA-A, (B and C) for 48 hours in MGC-803 and HGC-27 cells, the expression of URI was detected by western blot, β-actin was used as an internal control. Left two panels: overexpression of URI; right two panels: knockdown of URI.
URI promotes gastric cancer cell proliferation
The cell viability was measured by the CCK-8 assay after transient transfection for 48 h in MGC-803 and HGC-27 cells. Compared with vector and untransfected controls, overexpression of URI by transient transfection of pCMV6-URI remarkably promoted cell proliferation in two cell lines (Figure 2A, 2B). Accordingly, knockdown of URI significantly decreased the cell proliferation compared with the control groups (Figure 2C, 2D).
Figure 2.
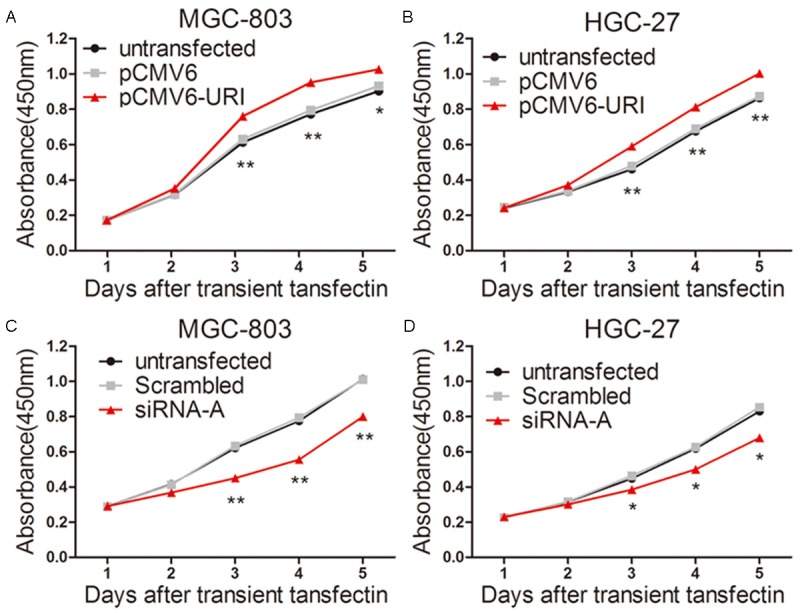
URI promotes cell proliferation of MGC-803 and HGC-27 cells. Cell viability was determined by a CCK-8 assay. A and B. After transiently transfected with pCMV6-URI and pCMV6-entry empty vector for 48 h, then cells were reseeded into 96 well plates, CCK-8 assay kit was used to detect the cell viabilities after five time points (1, 2, 3, 4, and 5 days respectively). Cell proliferation was significantly increased in MGC-803 and HGC-27 cells transfected with pCMV6-URI. C and D. Cells were transfected with URI siRNA-A and scrambled sequence for 48 hours. Cell viability markedly decreased in URI siRNA-A group compared with control groups. Untransfected cells were used as a control. Data was presented as mean ± SD. *p<0.05, **P<0.01, the representative result from three separate experiments was shown.
URI inhibited adriamycin-induced apoptosis and enhanced adriamycin resistance
The expression of apoptosis-related proteins Bax, cleaved caspase-3 and cleaved PARP-1 (one of the main cleavage targets of caspase-3 in vivo) significantly decreased while the level of the anti-apoptotic Bcl-2 increased in pCMV6-URI transfected cell lines that were treated with 250 nM adriamycin for additional 12 hours (Figure 3A, 3C). On the contrary, the levels of Bax, cleaved caspase-3 and cleaved PARP-1 were increased and Bcl-2 level decreased in URI siRNA-A transfected cells after adriamycin treatment for 12 h (Figure 3B, 3D). We evaluated the effect of URI knockdown on adriamycin-induced apoptosis in MGC-803 and HGC-27 cells. The percentage of early apoptotic cells (Annexin-V(+)/PI(-)) induced by adriamycin for 12 hours, was significantly higher in URI siRNA treated cells than in control groups (Figure 3E). We further assessed the effect of URI on adriamycin resistance in pCMV6-URI and URI siRNA-A transfected MGC-803 and HGC-27 cells by CCK-8 cell viability assay. After transient transfection for 48 hours, cells were treated for 24 hours with adriamycin at different concentrations ranging from 200 μM to 300 μM in MGC-803 cells and 200 μM to 400 μM in HGC-27 cells. The results showed that URI overexpression resulted in moderately increased resistance to adriamycin (Figure 4A) while knock-down of URI markedly decreased cell resistance to adriamycin in a dose-dependent manner (Figure 4B). The mean IC50 was significantly higher in pCMV6-URI transfected cells (Figure 4C) and markedly decreased in URI knock-down cells (Figure 4D) compared with cells of control groups. These results clearly indicated that URI enhanced the resistance of gastric cancer cells to adriamycin.
Figure 3.
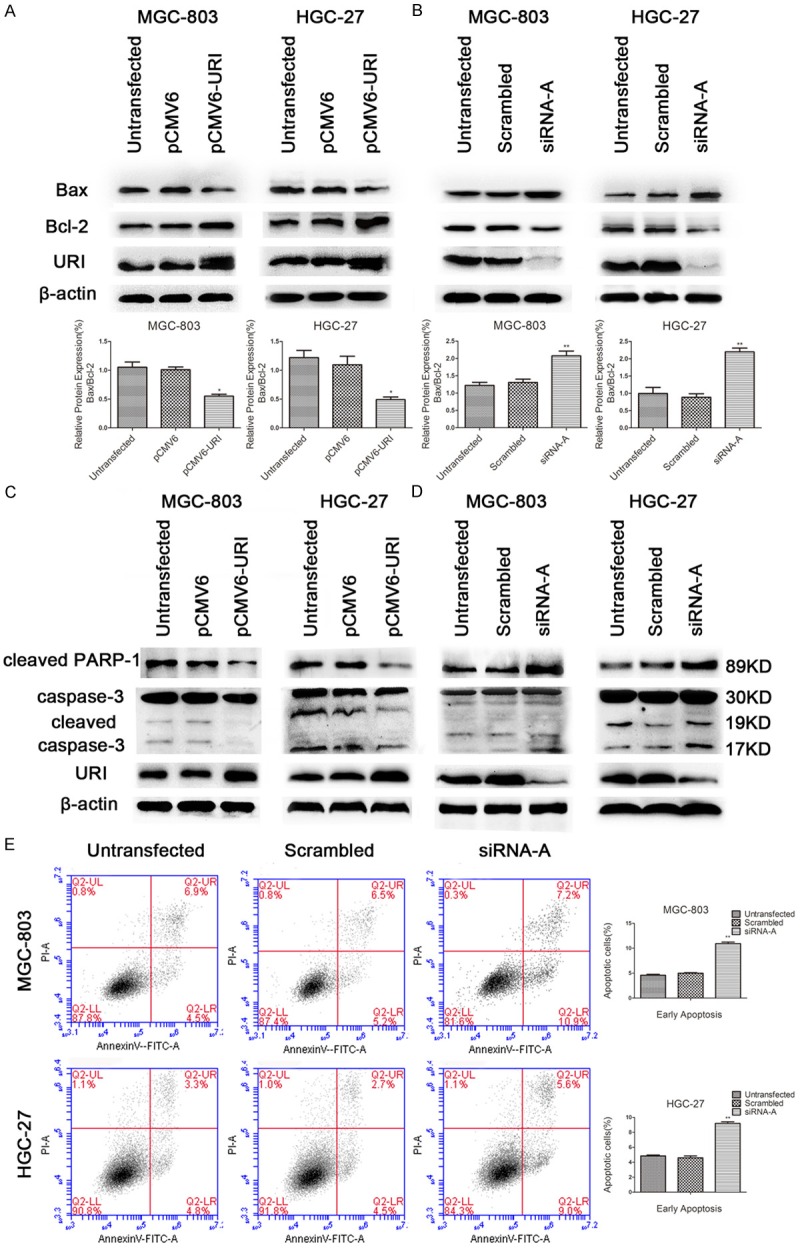
URI inhibited apoptosis induced by adriamycin via caspase-dependent manner in MGC-803 and HGC-27 cells. After transfection of pCMV6-URI and URI siRNA-A for 48 hours, cells were treated with 250 nM adriamycin for 12 hours, apoptosis related protein Bax, Bcl-2, cleaved PARP-1, and cleaved caspase-3 were detected by western blotting. β-actin was used as an internal control. A and C. The relative value of Bax/Bcl-2 expression and the expression of cleaved caspase-3 and cleaved PARP-1 were significantly decreased in pCMV6-URI transfected cells compared with control cells. B and D. On the contrary, the expression of Bax/Bcl-2 and cleaved PARP-1 and cleaved caspase-3 were increased in URI siRNA-A transfected cells compared with scrambled control sequence and untransfected cells. E. After transfection of URI siRNA-A for 48 hours, cells were treated with 250 nM adriamycin for 12 hours. Apoptotic cells were detected by Annexin V/PI double staining via flow cytometer. The ratio of early apoptotic cells (Annexin V+/PI-) were significantly increased compared with control cells. A representative result from three independent experiments was shown. Values are expressed as means ± SD of three individual experiments. *P<0.05 or **P<0.01 relative to the control group.
Figure 4.
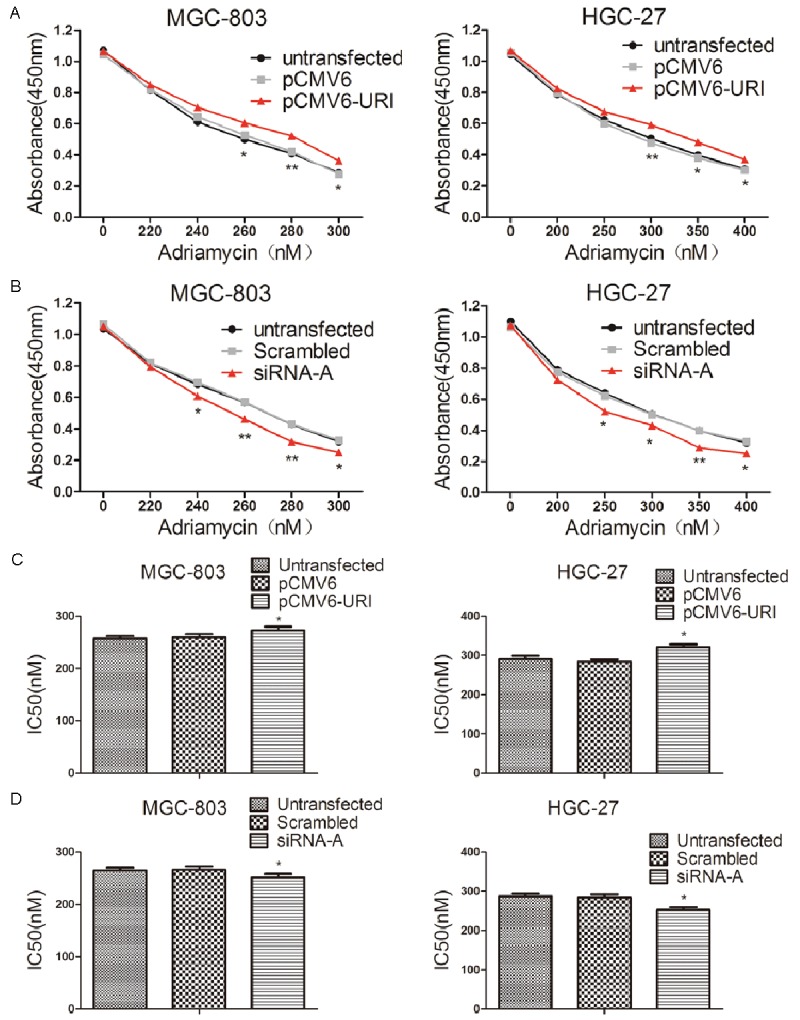
URI attenuated the effect of proliferative inhibition by adriamycin in MGC-803 and HGC-27 cells. After transfection of pCMV6-URI or URI siRNA-A for 48 hours, cells were treated with various concentrations of adriamycin for 24 hours. Cell viability was determined by CCK-8 assay at 450 nm. A. The pCMV6-URI transfected MGC-803 and HGC-27 cells both enhanced the drug resistance to adriamycin. B. The URI siRNA-A transfected MGC-803 and HGC-27 cells both showed reduced drug resistance to adriamycin. C and D. Graphic representation of the mean IC50 values of adriamycin. All data were representative of three independent experiments. Data represent mean ± SD.*p < 0.05; **p < 0.01.
URI promoted migration of gastric cancer cells
The effect of URI on gastric cancer cell migratory ability were analyzed by Transwell assay. The result showed that the number of cells passed through the polycarbonate membrane significantly increased in pCMV6-URI transfected cells compared with untransfected or the pCMV6-entry vector treated cells (Figure 5A). On the contrary, for cells transfected with siRNA, the number of cells passed through the well significantly decreased compared with the control groups of cells of the two gastric cancer cell lines (Figure 5B). The expression of migration-related EMT markers Snail and Vimentin significantly increased in URI transfected cells compared with control cells (Figure 5C), while URI knockdown by siRNA caused significantly decreased expression of Snail and Vimentin in two gastric cancer cell lines (Figure 5D).
Figure 5.
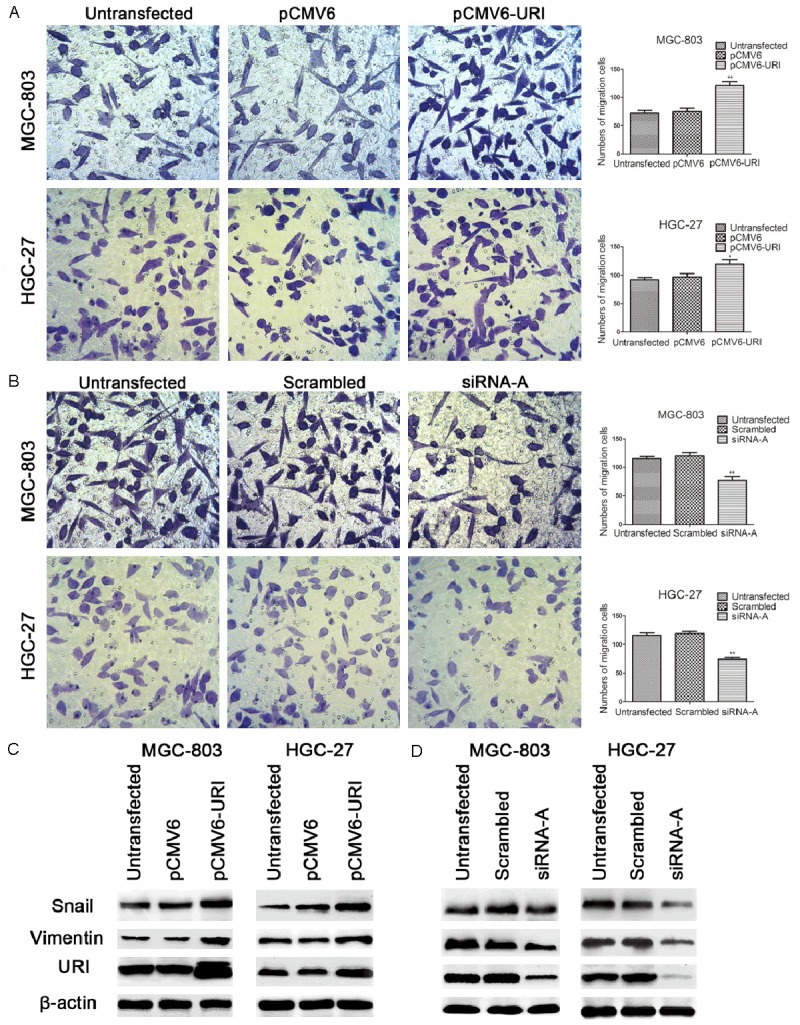
URI promoted migratory ability of MGC-803 and HGC-27 cells. Transwell assay was used to monitor the migration of gastric cancer cells. A. The number of migrated cells significantly increased in MGC-803 and HGC-27 cells with pCMV6-URI transfection. B. The number of migrated cells significantly decreased in MGC-803 and HGC-27 cells with URI siRNA-A transfection compared with control groups. All experiments were performed for three times and representative data were presented. The expression of migration related protein Snail and Vimentin were detected by western blotting in MGC-803 and HGC-27 cells transfected with pCMV6-URI and URI siRNA-A, β-actin was used as an internal control. C. Snail and Vimentin expression was significantly increased in MGC-803 and HGC-27 cells that showed upregulation of URI. D. Snail and Vimentin expression was significantly decreased in siRNA-A transfected MGC-803 and HGC-27 cells that also showed down-regulation of URI, compared with control groups. A representative result from three separate experiments was shown. Values are expressed as means ± SD (*P < 0.05; **P < 0.01 vs. untreated control group).
Discussion
URI functions as a prosurvival protein that supports cell proliferation and contributes to drug resistance of gastric cancer cells
Chemotherapy has been the mainstream treatment for gastric cancer given the fact that gastric cancer is often diagnosed at an advanced stage which is not suitable or ineffective for a surgical removal of the tumor. Unfortunately, chemotherapeutic failure is not unusual in gastric cancer treatment due to the frequently occurred drug resistance. Adriamycin, an anthracycline-based antibiotic, which induces DNA damage and apoptosis, has been widely utilized for the treatment of solid tumors, including gastric cancer [15]. The resistance to adriamycin occurs often in gastric cancer treatment and therefore, limits its effectiveness. The mechanisms of chemoresistance involving adriamycin remain to be fully elucidated. Previous studies have demonstrated the association of URI with cisplatin resistance in ovarian and cervical cancer cells [3,6]. It has also been shown that URI knock-down enhanced bortezomib induced cell death in multiple myeloma [5]. However, until now, the effect of URI on gastric cancer cells and its relation with adriamycinin treatment have never been elucidated.
In this study, we have shown that overexpression or knockdown of URI promoted or inhibited proliferation of MGC-803 and HGC-27 gastric cancer cells respectively. We also analyzed the effects of URI on gastric cancer cell death and proliferation after adriamycin treatment. As detected by CCK-8 assay and Annexin V/PI staining, cell proliferation was inhibited and apoptotic cell number was increased in siRNA-A transfected cells after adriamycin induction. The levels of proapoptotic protein Bax, cleaved caspase-3, cleaved PARP-1 were upregulated, whereas the antiapoptotic protein Bcl-2 was downregulated in URI siRNA-A transfected cells treated with adriamycin. On the contrary, these apoptosis related genes showed the opposite changes in URI overexpressed cells. These results indicated that URI was a pro-survival protein and acts as a drug resistant protein in MGC-803 and HGC-27 cells. In mammalian cells, URI has been shown as a phosphorylated target of the mTOR/S6K1 pathway and contributes to rapamycin-sensitive transcription [1,2]. It is well known that activation of the mTOR/S6K1 axis stimulates protein synthesis, cell growth and proliferation, and activation of metabolism. Increased mTOR/S6K1 signaling has been seen in colorectal cancer [16]. Intriguingly, it was previously reported that, many genes, whose products involved primarily in the regulation of cell growth and maintenance of cell communication, were significantly altered in URI-silenced HeLa cells treated with rapamycin [1]. Our results showed that URI knockdown facilitated cell apoptosis induced by adriamycin in an caspase-dependent manner, and attenuated the resistance to adriamycin in gastric cancer cells. We speculate that the changes of the apoptotic protein Bcl-2 and Bax caused by URI knockdown may be via the mTOR/S6K1 downstream signaling pathway. However, the mechanism remains to be determined.
The molecular mechanism that URI promoted migration of gastric cancer cells may be related with up-regulated expression of Vimentin and Snail
In addition to the role of regulating growth and survival, the role of mTOR signaling in epithelial-mesenchymal-transition (EMT), motility, and metastasis of cancers has become a rising concern. mTORC1-mediated S6K1 and 4EBP1 pathways may play a role through phosphorylation of focal adhesion proteins and reorganization of F-actin. The mTOR may also promote metastasis by altering the tumour microenvironment, and inhibition of mTOR can prevent tumour cell dissemination to lymph nodes in head and neck cancer [17]. Elevated mTOR signaling has been demonstrated to regulate EMT, motility, and metastasis of colorectal cancer [16]. We have recently shown that URI promoted migration and increased Vimentin mRNA expression in cervical cancer cells [6]. In this study, we further confirmed that URI promoted tumor cell migration, and increased the expression of migration-related EMT markers Vimentin and Snail in MGC-803 and HGC-27 cells. Vimentin is an intermediate filament protein normally expressed in cells of mesenchymal origin. The gain of mesenchymal markers Vimentin and fibronectin has been demonstrated as hallmarks of EMT [18]. Vimentin expression in cancer epithelial cells has been associated with metastasis and poor survival [19,20]. In tumours of the gastrointestinal tract, Vimentin expression usually correlates with advanced stage of tumour, lymph node metastasis, and patient survival. Vimentin expression might contribute to the high invasive phenotype of gastric cancer, and may be a useful biomarker to determine the biological aggressiveness of gastric cancer [21,22]. Our results suggest that URI may be a novel regulator of Vimentin expression. Meanwhile, as an EMT regulator, Snail induces the EMT via repression of E-cadherin expression [23,24], and is considered as a center of transcription factors to control the process of EMT [25]. Snail significantly affects the migration and invasion ability of gastric cancers, and may be used as a predictive biomarker for prognosis or aggressiveness of gastric cancers [26]. It has previously been shown that cell size and invasion in TGF-beta-induced EMT is regulated by activation of the mTOR/S6K1 pathway [27]. Together, our results suggest that URI enhances migratory ability of gastric cancer cells by upregulating Snail and Vimentin expression. However, whether and how this process involves the mTOR signaling pathway needs further investigation. A more thorough understanding of the molecular mechanisms underlining URI’s role in gastric cancer will eventually help us improve the intervention and treatment outcomes for this disease.
Acknowledgements
This work was supported by the 2014 Rush University Medical Center Pilot Project (Q.Z., J.G. Y.L.), the Innovation Program of Jiangsu Province (No. 480, 2013, Q.Z.). We thank all members of the laboratories for helpful discussions.
Disclosure of conflict of interest
None.
References
- 1.Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, Vigneron M, Peter M, Krek W. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302:1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- 2.Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R, Hess D, Krek W. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell. 2007;28:28–40. doi: 10.1016/j.molcel.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Theurillat JP, Metzler SC, Henzi N, Djouder N, Helbling M, Zimmermann AK, Jacob F, Soltermann A, Caduff R, Heinzelmann-Schwarz V, Moch H, Krek W. URI is an oncogene amplified in ovarian cancer cells and is required for their survival. Cancer Cell. 2011;19:317–332. doi: 10.1016/j.ccr.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Yang H, Gu J, Zheng Q, Li M, Lian X, Miao J, Jiang J, Wei W. RPB5-mediating protein is required for the proliferation of hepatocellular carcinoma cells. J Biol Chem. 2011;286:11865–11874. doi: 10.1074/jbc.M110.136929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan JL, Zhang J, Dong LW, Fu WJ, Du J, Shi HG, Jiang H, Ye F, Xi H, Zhang CY, Hou J, Wang HY. URI regulates tumorigenicity and chemotherapeutic resistance of multiple myeloma by modulating IL-6 transcription. Cell Death Dis. 2014;5:e1126. doi: 10.1038/cddis.2014.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu J, Liang Y, Qiao L, Lu Y, Hu X, Luo D, Li N, Zhang L, Chen Y, Du J, Zheng Q. URI expression in cervical cancer cells is associated with higher invasion capacity and resistance to cisplatin. Am J Cancer Res. 2015;5:1353–1367. [PMC free article] [PubMed] [Google Scholar]
- 7.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 8.Wagner AD, Grothe W, Haerting J, Kleber G, Grothey A, Fleig WE. Chemotherapy in advanced gastric cancer: a systematic review and meta-analysis based on aggregate data. J. Clin. Oncol. 2006;24:2903–2909. doi: 10.1200/JCO.2005.05.0245. [DOI] [PubMed] [Google Scholar]
- 9.De Vita F, Giuliani F, Silvestris N, Rossetti S, Pizzolorusso A, Santabarbara G, Galizia G, Colucci G, Ciardiello F, Orditura M. Current status of targeted therapies in advanced gastric cancer. Expert Opin Ther Targets. 2012;16(Suppl 2):S29–34. doi: 10.1517/14728222.2011.652616. [DOI] [PubMed] [Google Scholar]
- 10.Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643–655. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tapia O, Riquelme I, Leal P, Sandoval A, Aedo S, Weber H, Letelier P, Bellolio E, Villaseca M, Garcia P, Roa JC. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 2014;465:25–33. doi: 10.1007/s00428-014-1588-4. [DOI] [PubMed] [Google Scholar]
- 12.Byeon SJ, Han N, Choi J, Kim MA, Kim WH. Prognostic implication of TSC1 and mTOR expression in gastric carcinoma. J Surg Oncol. 2014;109:812–817. doi: 10.1002/jso.23585. [DOI] [PubMed] [Google Scholar]
- 13.Cheung DY, Kim JK. [Perspectives of the stomach cancer treatment: the introduction of molecular targeted therapy and the hope for cure] . Korean J Gastroenterol. 2013;61:117–127. doi: 10.4166/kjg.2013.61.3.117. [DOI] [PubMed] [Google Scholar]
- 14.Wong H, Yau T. Targeted therapy in the management of advanced gastric cancer: are we making progress in the era of personalized medicine? Oncologist. 2012;17:346–358. doi: 10.1634/theoncologist.2011-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 16.Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M, Lee EY, Weiss HL, O’Connor KL, Gao T, Evers BM. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011;71:3246–3256. doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel V, Marsh CA, Dorsam RT, Mikelis CM, Masedunskas A, Amornphimoltham P, Nathan CA, Singh B, Weigert R, Molinolo AA, Gutkind JS. Decreased lymphangiogenesis and lymph node metastasis by mTOR inhibition in head and neck cancer. Cancer Res. 2011;71:7103–7112. doi: 10.1158/0008-5472.CAN-10-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, Thompson EW. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 19.Al-Saad S, Al-Shibli K, Donnem T, Persson M, Bremnes RM, Busund LT. The prognostic impact of NF-kappaB p105, vimentin, E-cadherin and Par6 expression in epithelial and stromal compartment in non-small-cell lung cancer. Br J Cancer. 2008;99:1476–1483. doi: 10.1038/sj.bjc.6604713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu LK, Jiang XY, Zhou XX, Wang DM, Song XL, Jiang HB. Upregulation of vimentin and aberrant expression of E-cadherin/beta-catenin complex in oral squamous cell carcinomas: correlation with the clinicopathological features and patient outcome. Mod Pathol. 2010;23:213–224. doi: 10.1038/modpathol.2009.160. [DOI] [PubMed] [Google Scholar]
- 21.Brzozowa M, Wyrobiec G, Kolodziej I, Sitarski M, Matysiak N, Reichman-Warmusz E, Zaba M, Wojnicz R. The aberrant overexpression of vimentin is linked to a more aggressive status in tumours of the gastrointestinal tract. Prz Gastroenterol. 2015;10:7–11. doi: 10.5114/pg.2014.47502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuyuhiro Y, Yashiro M, Noda S, Kashiwagi S, Matsuoka J, Doi Y, Kato Y, Kubo N, Ohira M, Hirakawa K. Clinical significance of vimentin-positive gastric cancer cells. Anticancer Res. 2010;30:5239–5243. [PubMed] [Google Scholar]
- 23.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 24.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 25.Lee MY, Shen MR. Epithelial-mesenchymal transition in cervical carcinoma. Am J Transl Res. 2012;4:1–13. [PMC free article] [PubMed] [Google Scholar]
- 26.Shin NR, Jeong EH, Choi CI, Moon HJ, Kwon CH, Chu IS, Kim GH, Jeon TY, Kim DH, Lee JH, Park do Y. Overexpression of Snail is associated with lymph node metastasis and poor prognosis in patients with gastric cancer. BMC Cancer. 2012;12:521. doi: 10.1186/1471-2407-12-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]


