Abstract
Crop canopies create environments of highly fluctuating light intensities. In such environments, photoprotective mechanisms and their relaxation kinetics have been hypothesized to limit photosynthetic efficiency and therefore crop yield potential. Here, we show that overexpression of the Arabidopsis thylakoid K+/H+ antiporter KEA3 accelerates the relaxation of photoprotective energy-dependent quenching after transitions from high to low light in Arabidopsis and tobacco. This, in turn, enhances PSII quantum efficiency in both organisms, supporting that in wild-type plants, residual light energy quenching following a high to low light transition represents a limitation to photosynthetic efficiency in fluctuating light. This finding underscores the potential of accelerating quenching relaxation as a building block for improving photosynthetic efficiency in the field. Additionally, by overexpressing natural KEA3 variants with modification to the C-terminus, we show that KEA3 activity is regulated by a mechanism involving its lumen-localized C-terminus, which lowers KEA3 activity in high light. This regulatory mechanism fine-tunes the balance between photoprotective energy dissipation in high light and maximum quantum yield in low light, likely to be critical for efficient photosynthesis in fluctuating light conditions.
Keywords: Arabidopsis, KEA3, Non-photochemical quenching, PSII quantum efficiency, Thylakoid membrane
The nucleotide sequence of KEA3.3 presented in this paper has been submitted to GenBank under accession number KT581346.
Introduction
Sunlight drives plant growth and reproduction through photosynthesis. A large fraction of crop photosynthesis occurs in dense canopy structures, where light availability can undergo massive fluctuations on a short time scale. In such environments, photosynthetic efficiency is tightly linked to the speed with which plant cells can transition between photoprotection in excess light, and high quantum yield in limiting light periods (Murchie and Niyogi 2011, Demmig-Adams et al. 2012). In high light, non-photochemical quenching (NPQ) mechanisms, which dissipate excess absorbed light energy harmlessly as heat and thereby prevent photo-oxidative stress, are rapidly switched on (Müller et al. 2001). However, upon shifts to low light, these mechanisms are not instantly switched off, and the lag in their response time has been hypothesized to limit photosynthetic efficiency in crop canopies (Zhu et al. 2004, Zhu et al. 2010, Long et al. 2015).
Energy-dependent quenching (qE) is the major NPQ mechanism in plants (Demmig-Adams et al. 1996) and contributes significantly to plant fitness in fluctuating light and in the field (Külheim et al. 2002). The qE pathway is activated by a high proton concentration in the thylakoid lumen in periods of excess light, when proton translocation into the lumen by the photosynthetic electron transport chain exceeds proton consumption by ATP synthase (Horton et al. 1996). By dissipating excess absorbed light energy as heat, qE adjusts light energy input into photosynthesis to match the activity of downstream processes. Because qE is proportional to the proton concentration in the lumen, its relaxation kinetics upon transition to low light follow the kinetics of lumenal pH decay (Zaks et al. 2012). We recently showed that proton export from the lumen via the thylakoid K+/H+ antiporter KEA3 (K+ efflux antiporter 3) accelerates NPQ relaxation after a high light to low light transition in Arabidopsis thaliana (Armbruster et al. 2014). Thereby, KEA3 increases the light efficiency of photosynthesis in fluctuating light.
Here, we show that overexpression of the Arabidopsis major leaf KEA3 isoform (KEA3.2) further accelerates NPQ relaxation in Arabidopsis and tobacco. This acceleration is accompanied by an increase in PSII quantum efficiency immediately following a high to low light transition in both organisms. Additionally, analyses of two minor KEA3 isoforms with modifications to the C-terminus suggest that thylakoid K+/H+ antiport is regulated in response to light intensity. A regulatory mechanism involving the KEA3.2 C-terminus, which is located in the thylakoid lumen, may inhibit thylakoid K+/H+ antiport and thus down-regulation of energy-dependent quenching in high light. Our results show that NPQ relaxation kinetics respond to KEA3 protein levels in a highly specific manner, thus supporting the role of KEA3 as a promising target for efforts to improve photosynthesis in the field.
Results
Alternative splicing of the KEA3 transcript yields three protein isoforms
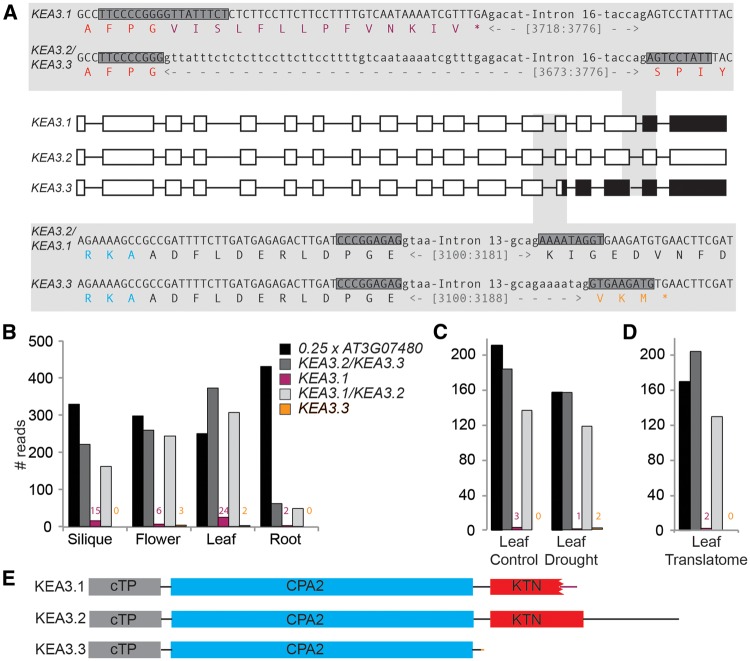
Two splice forms of KEA3 have been annotated. We have previously shown that KEA3.2 is the major protein isoform in Arabidopsis leaves (Armbruster et al. 2014). However, while attempting to clone the KEA3.2 cDNA from Arabidopsis leaves, we obtained mostly clones containing either the KEA3.1 splice form, which differs from KEA3.2 by alternative splicing of intron 16, or a third, not previously annotated splice form. This new splice form, which we have named KEA3.3, is spliced like KEA3.2 at intron 16, but has a different splicing acceptor site downstream of intron 13 (Fig. 1A). Of 20 KEA3 clones analyzed, eight were KEA3.1, 11 were KEA3.3 and only one was KEA3.2. In order to understand the abundance and distribution of the three different KEA3 splice forms in planta, we analyzed publicly available RNA sequencing (RNA-seq) data (Aubry et al. 2014, Clauw et al. 2015). This analysis demonstrated that KEA3.2 is the most abundant splice form in all plant organs (silique, flower, leaf and root), but it also revealed that, besides KEA3.1, KEA3.3 is another true minor variant that can be found in flowers and in leaves (Fig. 1B). No major differences in KEA3 splice form accumulation was found between watered control and mild drought-exposed leaves (here, KEA3.3 was only detected in the latter, Fig. 1C). Both KEA3.2 and KEA3.1 could be found in the leaf translatome in ratios similar to those in the leaf transcriptome obtained from the drought stress experiment (Fig. 1D), suggesting that the minor KEA3 variants are also translated into their respective protein isoforms. The three KEA3 protein isoforms all contain the chloroplast transit peptide (cTP) and the cation/proton antiport 2 (CPA2) domain, which mediates the K+/H+ antiport. However, the three isoforms differ with respect to their C-termini. The C-terminus of the major KEA3 isoform KEA3.2 contains a putative regulatory K+ transport, nucleotide-binding (KTN) domain. The KEA3.1-specific splicing event results in a truncated KTN domain with a KEA3.1-specific modification to the C-terminus, whereas the KEA3.3-specific splicing event leads to the production of a truncated protein that lacks the entire C-terminus including the KTN domain (Fig. 1A, E).
Fig. 1.
Alternative splicing events yield three KEA3 isoforms with differences in the putative C-terminal regulatory domain. (A) Overview and sequence details of the three KEA3 splice forms. Translated exons are depicted as white boxes, untranslated exons as black boxes and introns as black lines. Alternative splicing of intron #16 (upper shaded panel) yields KEA3.1 and KEA3.2/KEA3.3. Alternative splicing of intron #13 (lower shaded panel) yields KEA3.1/KEA3.2 and KEA3.3, respectively. Exons and introns are written in upper case and lower case, respectively. The encoded protein sequence is given as the single letter amino acid code. Amino acids of the KTN and CPA2 domains are colored red and blue, respectively. Amino acids specific for KEA3.1 and KEA3.3 are colored purple and orange, respectively. The 18-mers used for probing RNA-seq data for the specific splice variants are shaded dark gray. (B−D) KEA3.2 is the major splice form in all plant organs (B), under mild drought stress (C) and associated with ribosomes (D). Publicly available RNA-seq data were probed for the presence of splice form-specific 18-mers (marked by dark gray boxes in A) and a control (At3g07480). (E) Protein models of all three KEA3 isoforms.
Overexpression of KEA3.2 increases PSII quantum efficiency after transition from high to low light
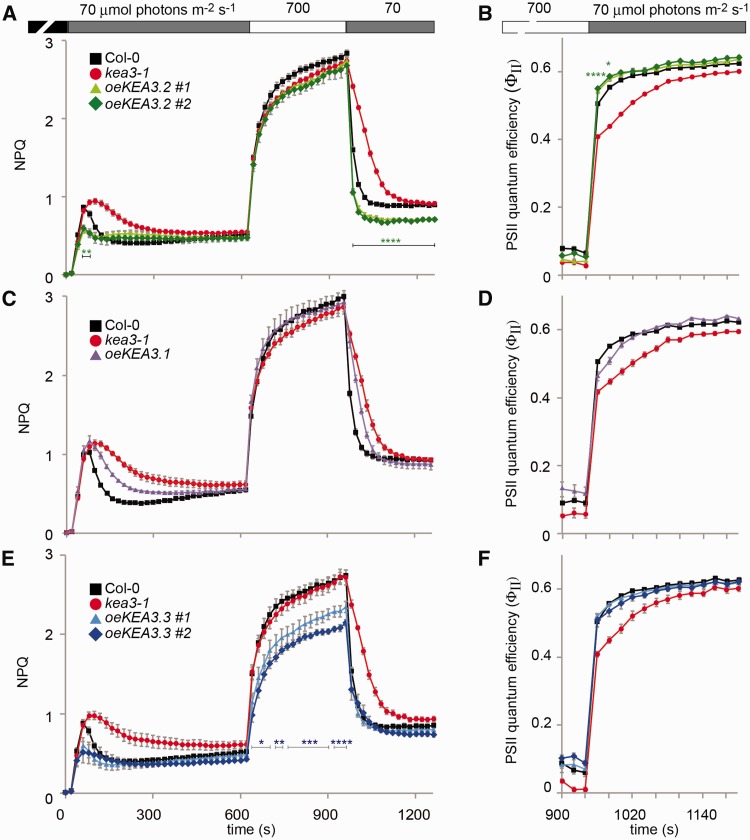
We have previously shown that loss of thylakoid K+/H+ antiport in kea3 knockout mutants resulted in an increased and extended transient NPQ after transition from the dark-acclimated state to low light conditions and slower NPQ relaxation kinetics after transition from high to low light (Armbruster et al. 2014). This strongly suggested that both the extent of the transient NPQ and NPQ relaxation kinetics are dependent on thylakoid K+/H+ antiport activity and therefore the abundance of KEA3. In order to test whether increasing KEA3 levels can lower the transient NPQ and accelerate NPQ relaxation kinetics, we selected plants that overexpressed a C-terminal green fluorescent protein (GFP)-tagged version of the main KEA3.2 isoform, which had been shown to complement the kea3-1 NPQ phenotype when expressed at wild-type levels (Armbruster et al. 2014). Out of 100 plants expressing the selection marker, five overexpressed KEA3 protein as compared with Col-0. The two T2 lines with the highest KEA3.2 expression (six and eight times more as compared with native KEA3 in Col-0, oeKEA3.2, Supplementary Fig. S1A) were analyzed for Chl fluorescence after dark acclimation during a time course of low light (10 min at 70 μmol photons m−2 s−1), high light (5 min at 700 μmol photons m−2 s−1) and low light (5 min at 70 μmol photons m−2 s−1), and NPQ was calculated. As we had hypothesized, the oeKEA3.2 lines displayed a phenotype opposite to that of kea3-1, with both a decrease in the transient NPQ after transition from dark to low light (Fig. 2A) and accelerated NPQ relaxation kinetics after transition from high to low light, as compared with the Col-0 wild type (Fig. 2A). In addition, NPQ levels of oeKEA3.2 stayed below those in Col-0 at the end of the second low light treatment (Fig. 2A). In order to understand whether the lower NPQ in oeKEA3.2 has a beneficial effect on quantum yield of photosynthesis, we calculated PSII quantum efficiency (ΦII) from the Chl a fluorescence data. This analysis demonstrated that PSII quantum efficiency was increased in oeKEA3.2 as compared with Col-0 between 40 and 80 s after the transition from dark to low light (Supplementary Fig. S2). Also after the high to low light shift, PSII quantum efficiency was significantly higher in oeKEA3.2 as compared with Col-0 for approximately 40 s after the light intensity shift [P < 0.001 at 20 s: +7.6% for oeKEA3.2 #1 and +9.0% for oeKEA3.2 #2; P = 0.02 at 40 s: +4.8% for oeKEA3.2 #1 and +6.1% for oeKEA3.2 #2 at 40 s; one-way analysis of variance (ANOVA) with Tukey’s post-hoc test; Fig. 2B; Supplementary Fig. S2]. After 60 s in low light, no significant differences in ΦII could be observed for both oeKEA3.2 lines.
Fig. 2.
Overexpression of the different KEA3 splice forms reveals isoform-specific properties. Chl fluorescence of detached, dark-acclimated leaves from Col-0, kea3-1 and kea3-1 overexpressing KEA3.2-GFP (oeKEA3.2) (A, B), KEA3.1-GFP (oeKEA3.1) (C, D) and KEA3.3-GFP (oeKEA3.3) (E, F) was monitored during alternating low light (70 μmol photons m−2 s−1, gray bar), high light (700 μmol photons m−2 s−1, white bar) and low light. NPQ (A, C, E) and PSII quantum efficiency (B, D, F) were calculated. Asterisks indicate where NPQ is significantly lower and PSII quantum efficiency is significantly higher in all the measured lines of one construct as compared with Col-0 (*0.01 < P < 0.05; **0.005 < P < 0.01, ***0.001 < P < 0.005; ****P < 0.001, one-way ANOVA with Tukey post-hoc test). Error bars represent the SEM (n = 4−6).
The transiently higher ΦII in oeKEA3.2 as compared with the Col-0 could result in higher levels of CO2 fixation if electrons from H2O were transported to NADP+. However, they could alternatively be transported to O2 via the Mehler reaction, which would lead to an increase in the production of H2O2 (reviewed by Asada 1999). Therefore, we analyzed H2O2 levels in Col-0 and oeKEA3.2 20 s after shift from high to low light. This analysis showed that at this time point, H2O2 levels were slightly, but not significantly higher in oeKEA3.2 (+ 1.5% as compared with Col-0, Supplementary Fig. S3).
Thylakoid K+/H+ antiport activity is regulated by the KEA3 C-terminus
The observation that NPQ in high light does not markedly respond to KEA3 levels, with kea3-1 and oeKEA3.2 showing NPQ values similar to Col-0 (Fig. 2A), suggested that either KEA3 activity does not affect NPQ in high light or that KEA3 is inactive. Because the major KEA3.2 isoform contains a putative regulatory domain at its C-terminus, we decided to analyze the effect of changes to the C-terminus on KEA3 activity. Here, we chose to employ kea3-1 mutants overexpressing the two minor KEA3 isoforms, KEA3.1 and KEA3.3, which contain a modification to the C-terminus or completely lack the C-terminus, respectively (Fig. 1A, E). For oeKEA3.1, we screened 50 plants that expressed the selection marker, and could identify only one line that expressed KEA3.1 protein at higher levels in the T2 generation than the native KEA3.2 levels in Col-0 (oeKEA3.1, Supplementary Fig. S1B). This line expressed KEA3 at about 1.7 times the levels of KEA3 in Col-0, which is four times lower than the levels of KEA3 in the oeKEA3.2 lines. For KEA3.3, of which all KEA3-expressing lines displayed very high KEA3 levels, we selected T2 lines with expression levels similar to oeKEA3.2 (nine and 11 times more as compared with native KEA3 in Col-0, oeKEA3.3, Supplementary Fig. S1C). We monitored Chl fluorescence in oeKEA3.1 and oeKEA3.3 lines under the same low light, high light regime as for oeKEA3.2 and calculated NPQ. Interestingly, overexpression of KEA3.1 only partially rescued the kea3-1 NPQ and PSII quantum efficiency phenotype [while KEA3.2 expressed at wild-type levels fully restored the kea3-1 NPQ and PSII quantum efficiency phenotype (Armbruster et al. 2014)], suggesting that KEA3.1−GFP is a KEA3 version with intrinsically lower K+/H+ antiport activity (Fig. 2C, D). oeKEA3.3, in contrast to oeKEA3.2, displayed significantly lower NPQ during the entire high light period, suggesting that KEA3.3, but not KEA3.2, is active in high light (Fig. 2E). No significant difference in PSII quantum yield was detectable between oeKEA3.3 and Col-0 after transition from high to low light (Fig. 2F; Supplementary Fig. S2C). To analyze whether the specific differences in NPQ and PSII quantum yield between Col-0 and overexpressors of KEA3.2 and KEA3.3 could be due to alterations in the photosynthetic apparatus, we examined Chl content and complex composition and abundance by blue-native (BN)−PAGE. This analysis showed no differences between Col-0 and the KEA3 overexpressors (Supplementary Fig. S4).
Loss of KEA3 results in reduced growth upon a shift to fluctuating light
In order to analyze the effects of KEA3 activity and regulation on plant photosynthesis and growth in fluctuating light, we shifted constant light (150 μmol photons m−2 s−1) grown Col-0, kea3-1, oeKEA3.2 and oeKEA3.3, which had very similar rosette sizes (Fig. 3A), to alternating high and low light conditions (1 min at 900 μmol photons m−2 s−1, 4 min at 90 μmol photons m−2 s−1). We then calculated the growth rate during the fluctuating light treatment from increases in leaf area. Due to a much higher variance in oeKEA3.2 than in the other three genotypes (Fig. 3B), the data failed the ANOVA equal variance test. When we excluded oeKEA3.2 from the statistical analysis, the growth rate of kea3-1 was calculated to be significantly smaller as compared with Col-0 (P = 0.027, one-way ANOVA with Tukey’s posthoc test; Fig. 3A, B; Supplementary Fig. S5).
Fig. 3.
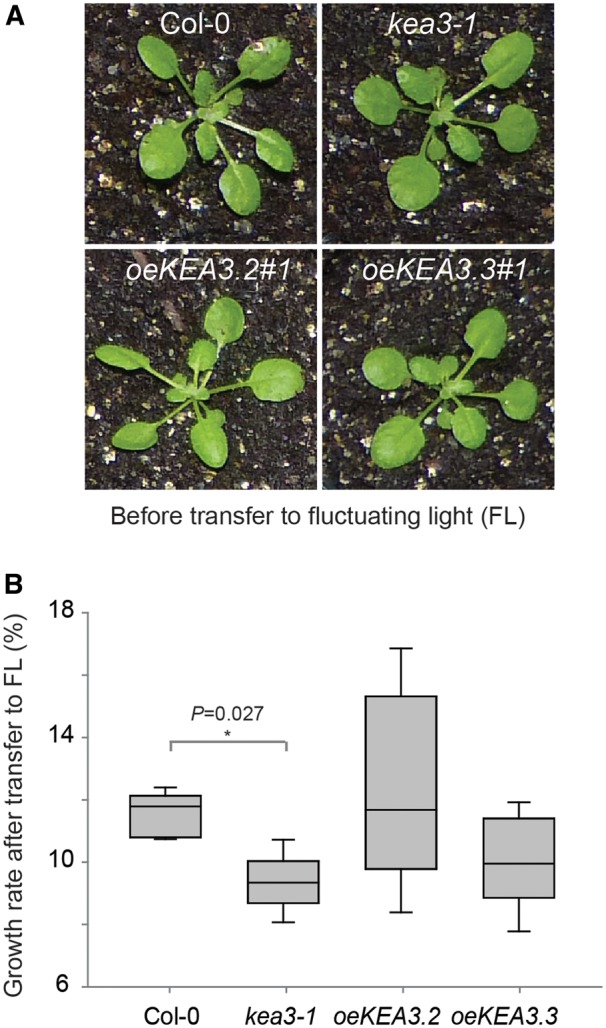
Loss of KEA3 results in reduced growth upon a shift to fluctuating light. (A) Four-week-old Col-0, kea3-1, oeKEA3.2#1 and oeKEA3.3#1 before shift to fluctuating high and low light conditions (1 min at 900 μmol photons m−2 s−1, 5 min at 90 μmol photons m−2 s−1). (B) Box plot depicting growth rates of Col-0, kea3-1, oeKEA3.2#1 and oeKEA3.3#1 in fluctuating light as determined by exponential fitting of increases in leaf area over time (n = 6; an asterisk indicates a significant difference with P = 0.027, excluding oeKEA3.2; one-way ANOVA with Tukey post-hoc test). Error bars represent the SEM between measurements.
The KEA3.2 C-terminus is localized in the thylakoid lumen
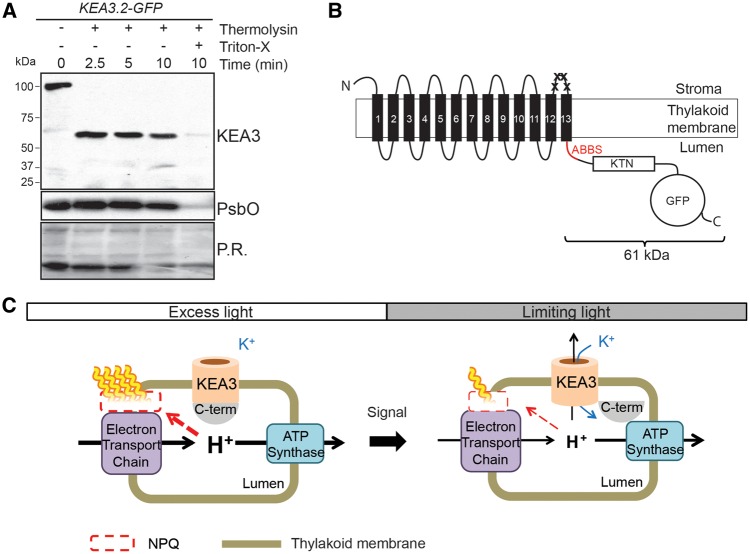
In order to identify the localization of the KEA3.2 C-terminus, we treated intact thylakoids of KEA3.2−GFP with thermolysin to digest protein regions exposed to the stroma. Addition of the protease to thylakoids caused detectable KEA3 to shift quantitatively from approximately 100 kDa to approximately 60 kDa, suggesting that the C-terminus, which bears the antibody-binding site, was protected from the protease. In order to investigate whether the protection was caused by the lumenal localization of the C-terminus, thylakoids were treated with low concentrations of the detergent Triton X-100 (Fig. 4A), which make the lumen accessible for the protease but do not impact protein−protein interactions (Brooks et al. 2013). After treatment with Triton X-100 and access of thermolysin to the lumen, very little KEA3 could be detected (Fig. 4A). The lumenal PSII subunit PsbO was immunodetected as a control for the integrity of the thylakoid membranes. PsbO levels remained largely unchanged by thermolysin treatment, except for when the thylakoid membrane was disrupted by detergent treatment (Fig. 4A). These experimental data strongly suggest that the KEA3 C-terminus is localized in the thylakoid lumen. This finding was corroborated by in silico thermolysin digestion of a KEA3.2−GFP topology with the lumenal C-terminus, which predicted a protected digestion product of 61 kDa detectable with the specific KEA3 antibody (Fig. 4B). The localization of the entire C-terminus to the thylakoid lumen was confirmed by protease treatment of oeKEA3.3-GFP thylakoids (Supplementary Fig. S6). In Fig. 4C, a model of the light-dependent regulation of KEA3 activity involving the lumenal C-terminus is shown.
Fig. 4.
The KEA3 C-terminus is localized in the thylakoid lumen. (A) Intact thylakoids of oeKEA3.2 were treated with thermolysin in the absence and presence of Triton X-100. Aliquots were removed at the specified time points, separated by SDS−PAGE, and immunodetection of KEA3 and PsbO was performed. Prior to immunodetection, membranes were stained with Ponceau Red (P.R.). (B) Schematic overview of the KEA3 topology resulting from the thermolysin treatment. The antibody-binding site (ABBS, red, containing five thermolysin sites) present in the C-terminus is protected from thermolysin treatment. Crosses mark stroma-accessible thermolysin sites between transmembrane helices 12 and 13 close to the antibody-binding site. In order to generate this topology model, transmembrane helices of KEA3 were predicted by Phyre2 (see the Materials and Methods). (C) Possible model of light-responsive regulation of KEA3 via the C-terminus.
Overexpression of AtKEA3 can enhance photosynthetic efficiency in tobacco in fluctuating light
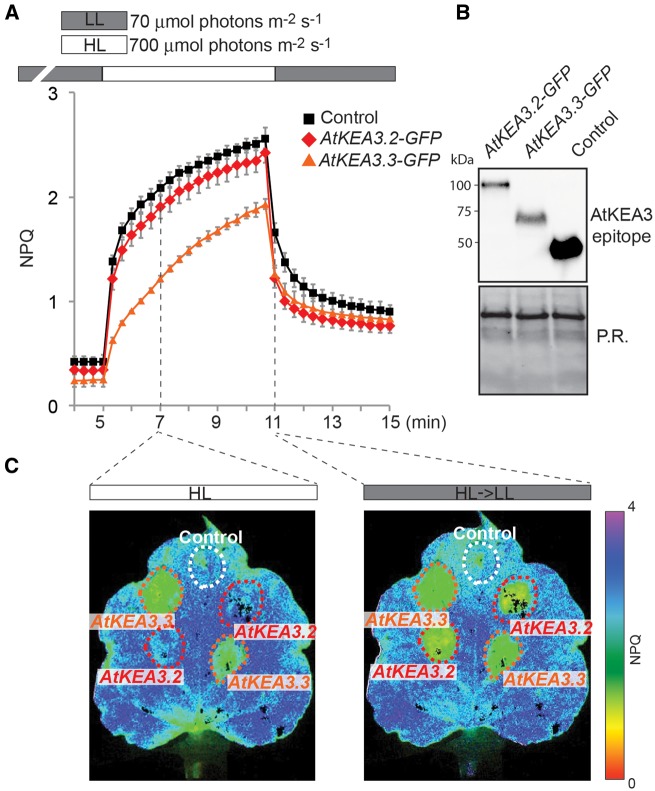
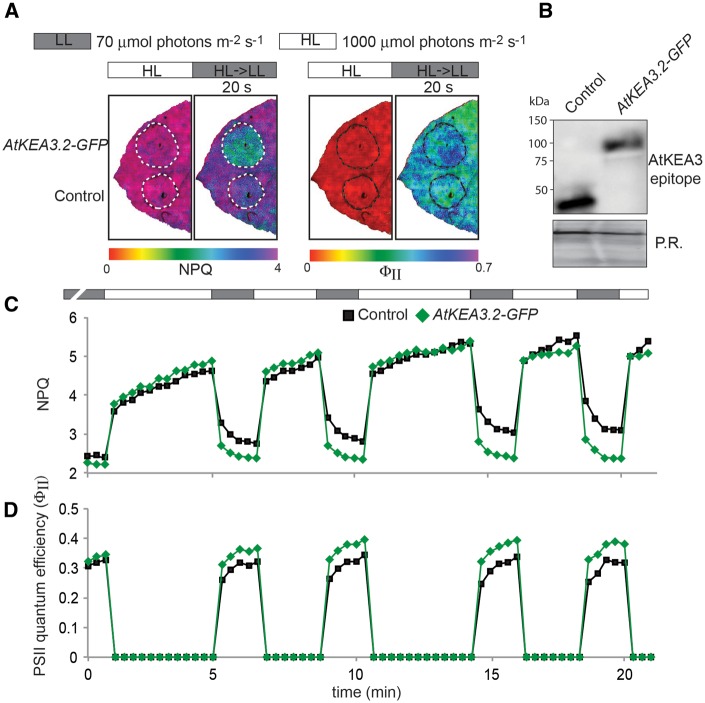
Crop plants experience high fluctuations in light energy availability, especially in their canopies. Because overexpression of KEA3.2 can transiently increase PSII quantum efficiency in Arabidopsis after a high to low light transition, we wanted to test whether the same was true for overexpression of Arabidopsis KEA3.2 (AtKEA3.2) in canopy-forming plants. Here we used Nicotiana benthamiana (tobacco), which can be transformed transiently with high efficiency (Sainsbury and Lomonossoff 2008) and is from the same genus as the tobacco crop plant Nicotiana tabacum. Expression from the 35S promoter of AtKEA3.2-GFP and AtKEA3.3-GFP in tobacco yielded very similar NPQ results to Arabidopsis (Fig. 5). In high light, AtKEA3.3-GFP-expressing leaf sections had low NPQ, whereas sections expressing AtKEA3.2-GFP were indistinguishable from the control (Fig. 5A, C). However, upon transition from high to low light, sections expressing either of the two AtKEA3 isoforms had significantly lower NPQ than the control (Fig. 5A, D). These data suggest that the signal(s) and potential transduction machinery that regulate KEA3 activity are conserved between Arabidopsis and tobacco. We then set out to test whether tobacco sections expressing AtKEA3.2-GFP would show repetitive higher PSII quantum efficiencies in fluctuating light conditions. Strikingly, sections expressing AtKEA3.2-GFP showed faster NPQ relaxation and higher PSII quantum efficiencies in each low light period of the fluctuating light (Fig. 6A−D; Supplementary Fig. S7).
Fig. 5.
Overexpression of KEA3.2 and KEA3.3 in tobacco. (A) Chl fluorescence of tobacco leaves transformed with KEA3.2−GFP, KEA3.3−GFP and control (GFP fused to the KEA3 antibody-binding site coding sequence at its N-terminus) was monitored during alternating low light (70 μmol photons m−2 s−1, gray bar), high light (700 μmol photons m−2 s−1, white bar) and low light. Error bars represent the SEM (n = 4). (B) Proteins extracted from the transformed tobacco sections were immunodetected with the specific KEA3 antibody. Ponceau red (P.R.) staining of the membrane prior to immunodetection is shown as a loading control. (C) Images of a tobacco leaf transformed with KEA3.2−GFP, KEA3.3−GFP and control 120 s after transition from low to high light (HL) and 20 s after transition from high to low light (HL→LL 20 s) are shown. False colors represent the NPQ as indicated on the color scheme.
Fig. 6.
Overexpression of KEA3.2 in tobacco enhances PSII quantum yield in fluctuating light. (A) Images of a tobacco leaf transformed with either KEA3.2-GFP or GFP fused to the KEA3 antibody-binding site coding sequence at its N-terminus (control) at the end of the high light period (1,000 μmol photons m−2 s−1, white bar; HL) and 20 s after transition from high to low light (70 μmol photons m−2 s−1, gray bar; HL > LL 20 s). False colors represent the NPQ in the left panel and PSII quantum efficiency in the right panel as indicated by the respective color bars. (B) Immunodetection of proteins extracted from the transformed tobacco sections with the specific KEA3 antibody. Ponceau red (P.R.) staining of the membrane prior to immunodetection is shown as a loading control. (C, D) Chl fluorescence of tobacco leaves transformed with KEA3.2-GFP and control was monitored under fluctuating high light (1,000 μmol photons m−2 s−1, white bar) and low light (70 μmol photons m−2 s−1, gray bar), and NPQ (C) and PSII quantum efficiency (D) were calculated. Because of the high background variation in PSII quantum efficiency between different tobacco leaves, a representative trace from one experiment is shown. More traces can be found in Supplementary Fig. S7.
Discussion
The three different KEA3 isoforms have distinct properties
Analysis of alternative splicing events at the KEA3 locus revealed that the KEA3 genomic DNA encodes at least three different protein isoforms. In the tested conditions, KEA3.2 is clearly the major splice form. This is in line with earlier work, which had shown that KEA3.2 is the only KEA3 protein isoform detectable in leaves (Armbruster et al. 2014). However, KEA3.1 and KEA3.3 are functional K+/H+ transporters as seen by their effect on NPQ (Fig. 2C, E). They differ from KEA3.2 by abolished regulation (KEA3.3) and potentially decreased activity (KEA3.1). Further analyses may reveal conditions where a specific accumulation of either of these two minor KEA3 isoforms has a physiological function.
Residual NPQ limits photosynthetic efficiency upon transition from high to low light
At least two factors have been proposed to limit photosynthetic efficiency immediately following high to low light transitions: (i) residual NPQ that continues to dissipate absorbed light energy as heat (Zhu et al. 2004); and (ii) an overshoot in sucrose synthesis that drains triose-phosphates from the chloroplast and thereby limits Calvin−Benson cycle activity (Prinsley et al. 1986). By overexpressing KEA3.2, we show that the acceleration of NPQ relaxation results in a faster recovery of higher PSII quantum efficiency (Figs. 2, 6; Supplementary Fig. S7). The increase in PSII quantum efficiency is accompanied by a slight, non-significant increase in the Mehler reaction product H2O2 (Supplementary Fig. S3). Thus, it appears possible that at least some of the higher PSII quantum efficiency is translated into a higher overall quantum efficiency of photosynthesis.
Our analysis of plant growth rates in fluctuating light suggested that lack of KEA3 imposes a penalty on plant growth in such conditions (Fig. 3B). Thus, decreases in PSII quantum efficiency and CO2 assimilation in kea3 upon transition from high to low light (Armbruster et al. 2014) may be translated into reduced growth rate in fluctuating light. We could not detect an increase in growth rate of oeKEA3.2 as compared with Col-0 (Fig. 3B). However, oeKEA3.2 showed much higher variance compared with the other genotypes, and had the two highest growth rates observed in the experiments. The basis of the high variance awaits further elucidation.
KEA3 activity is regulated in a light intensity-responsive manner
Overexpression of the KEA3.3 isoform, which only consists of the CPA2 domain, causes slow NPQ induction and low levels of NPQ in high light, while overexpression of the major KEA3 isoform KEA3.2, which contains an additional C-terminal extension, results in wild-type-like NPQ induction and levels in high light. Plants overexpressing either of the two isoforms show fast NPQ relaxation kinetics upon transition from high to low light. These findings strongly argue for a regulatory function of the C-terminus by inhibiting K+/H+ antiport in high light. Intriguingly, such regulation of thylakoid K+/H+ antiport activity may constitute a novel regulatory loop in photosynthesis that increases photosynthetic efficiency in fluctuating light: in high light periods, K+/H+ antiport via KEA3.2 is inhibited, which allows the plant to build up a high lumenal proton concentration and dissipate excess energy as heat; upon transition to low light, the inhibition is released and the resulting activity of the KEA3 thylakoid K+/H+ antiporter facilitates the rapid relaxation of NPQ and recovery of high photosynthetic efficiency (Fig. 4C).
KEA3 activity may be regulated via the C-terminal KTN domain and changes in photosynthetic intermediates
A likely candidate involved in the light intensity-responsive gating of KEA3.2 K+/H+ antiport is its C-terminal KTN domain. KTNs (RCKs) are highly conserved protein domains found ubiquitously in prokaryotes and eukaryotes, which regulate K+ transporters and channels in response to changes in NADH/NAD+ or ATP/ADP (Roosild et al. 2002, Kröning et al. 2007, Roosild et al. 2009, Cao et al. 2013). Sequence alignments of KTN domains from Arabidopsis KEA3 and KEA2 (Aranda-Sicilia et al. 2012, Kunz et al. 2014), Escherichia coli KefC (Roosild et al. 2009) and Vibrio parahaemolyticus TrkA (Cao et al. 2013) show high sequence conservation of the amino acids surrounding and comprising the specific nucleotide-binding site (Supplementary Fig. S8A), suggesting that the KTN domains of KEA3 (and KEA2) also bind nucleotides. The ratios of both nucleotide pairs ATP/ADP and NADPH/NADP+ have been shown to decrease in response to a transition from excess light to limiting light conditions (Stitt et al. 1989) and thus might constitute or contribute to the signal that triggers KEA3 activation. Both photosynthetic intermediates are present in the chloroplast stroma, and ATP/ADP has been reported to cross the thylakoid membrane into the lumen (Spetea et al. 2004). The localization of the KEA3.2 C-terminus to the thylakoid lumen suggests that the Rossman fold of the KTN domain binds nucleotides that can enter the thylakoid lumen. According to our current knowledge of lumenal nucleotides, these may be either ATP/ADP or GTP/GDP (Spetea et al. 2004).
There is ample experimental evidence suggesting that KTN domains gate K+ fluxes by reversibly interacting with a gating structure of the ion transporter domain (Ness and Booth 1999, Roosild et al. 2002). This gating structure is moderately conserved in KEA3.2 (two out of three potentially critical acidic amino acid residues are conserved; Supplementary Fig. S8B). Because KEA3 is activated within secomds of the transition from high to low light, we propose that KEA3 regulation is triggered by certain photosynthesis intermediates, the levels of which change in response to excitation energy. Besides ATP/ADP ratios, which may regulate KEA3 activity via binding to the luminal KTN domain, other photosynthetic intermediates immediately affected by changes in excitation energy include the plastoquinone pool (Stitt et al. 1989, Grieco et al. 2012) and the electric potential across the thylakoid membrane (Kramer et al. 2003). Because activation of KEA3 seems to precede large changes in NPQ, and NPQ is proportional to the proton concentration in the lumen, we can probably exclude lumenal pH as a potential signal.
Conclusion
Results demonstrating that PSII quantum efficiency can be increased by accelerating NPQ relaxation via overexpression of KEA3.2 (Figs. 2A, B, 6) suggest rapid NPQ relaxation in general and enhancement of KEA3 activity in particular as promising building blocks for improving photosynthesis in the field. In future, understanding the specific signal(s) and regulatory mechanism that allow KEA3 activity to respond to changes in light intensity may unravel the molecular basis to an additional feedback loop of photosynthesis involved in optimizing photosynthesis in natural, fluctuating light environments.
Materials and Methods
Identification and expression analysis of alternative KEA3 splice forms
cDNA was synthesized from total leaf RNA using superscript III (Invitrogen) and oligo(dT) primers according to the manufacturer’s instruction. Primers specific for the KEA3.2 transcript (Armbruster et al. 2014) were used for amplification and Gateway- (Invitrogen) mediated recombination into pDONR201. Besides KEA3.2, two further splice forms, KEA3.1 and KEA3.3, were identified in the resulting pDONR201-KEA3 constructs. Large-scale analysis of splice form abundances was performed by filtering publicly available Col-0 RNA-seq data deposited in the sequence read archive (SRA) of NCBI (http://www.ncbi.nlm.nih.gov/sra) for the splice form-specific 18-mers TTCCCCGGGGTTATTTCT (KEA3.1), TTCCCCGGGAGTCCTATT (KEA3.2/KEA3.3), CCCGGAGAGGTGAAGATG (KEA3.3), GAGAGAAAATAGGTGAAG (KEA3.1/KEA3.2) and CAGAAACTCTCCTCTCAA (At3g04780, control).
Plant material, propagation and growth conditions
kea3-1 plants overexpressing the KEA3 variants with a C-terminal GFP tag were generated as described previously (Armbruster et al. 2014). KEA3.1 and KEA3.3 cDNA sequences were amplified using the same forward primer as for KEA3.2, and 5'-GGGGACCACTTTGTACAAGAAAGCTGGGTTAACGATTTTATTGACAAAA-3' (KEA3.1) and 5'-GGGGACCACTTTGTACAAGAAAGCTGGGtTCATCTTCACCTCTCCGGGAT-3' (KEA3.3), and introduced into pB7FWG2 (Karimi et al. 2002).
Col-0, kea3-1 and kea3-1 plants overexpressing KEA3.1-GFP, KEA3.2-GFP and KEA3.3-GFP were grown on Sunshine Mix 4 potting mix (Sun Gro Horticulture Distribution) in controlled conditions of 10 h dark, 22 °C/14 h light, 23 °C, with a light intensity of 120 μmol photons m − 2 s−1.
For fluctuating light experiments, plants were grown for 4 weeks in controlled short-day conditions of 8 h light, 20 °C/16 h dark, 16 °C, with a light intensity of 150 μmol photons m − 2 s−1 and then shifted to alternating high and low light (1 min at 900 μmol photons m − 2 s−1, 4 min at 90 μmol photons m − 2 s−1) in the short-day light period for 6 d. Leaf area of whole plants was determined by using ImageJ. Growth rates were determined by exponential fitting.
Transient expression of KEA3 isoforms in Nicotiana benthamiana
For use as a control, a KEA3 fragment containing the KEA3 antibody-binding site was fused to the N-terminus of GFP, employing the primers 5'-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGAACCAACTTGGAAGAAAAGccg-3' and 5'-GGGGACCACTTTGTGAAAGCTGGGTTTGCATCTGTGGCTCCTGCTTT-3', and introduction of the amplified region into pB7FWG2 (Karimi et al. 2002). Colonies of Agrobacterium tumefaciens strain GV3101 transformed with the KEA3−GFP constructs or the control were resuspended in induction medium (0.1 mM MES pH5.6, 0.1 mM MgCl2, 0.1 mM acetosyringone) to an OD600 of 0.5. After 2 h at 28 °C, suspensions were inoculated onto sections of N. benthamiana (tobacco) leaves. Transfected plants were grown for 2−3 d in room light before detached leaves were scored for Chl fluorescence.
Chl fluorescence and H2O2 measurements
For NPQ and ΦII measurements, detached leaves of 5-week-old Arabidopsis plants were placed onto wet filter paper. Detached leaves of tobacco were placed with their petioles in centrifuge tubes containing water and sealed with parafilm. Leaves were dark acclimated for 30 min prior to measurement. Room temperature Chl a fluorescence of these leaves was monitored using the Walz MAXI IMAGING-PAM with blue actinic LEDs set to approximately 70 and 700/1,000 μmol photons m − 2 s−1 (low and high light, respectively). To measure Fm and Fm', white light pulses (4,000 μmol photons m − 2 s−1, duration 0.8 s) were applied. NPQ was calculated as (Fm − Fm′)/Fm′ and ΦII as (Fm′ − Fs)/Fm′. For H2O2 measurements, leaves were rapidly frozen 20 s after transition from high to low light employing the Imaging PAM as light source with the same program used for the NPQ and ΦII measurements. Hydrogen peroxide was detected by resorufin (Strand et al. 2015). In detail, leaves were ground in liquid nitrogen and extracted in 50 mM potassium phosphate buffer (pH 7.5). Extracts were incubated in a reaction buffer containing 10 U ml−1 horseradish peroxidase (Sigma) and 5 μM Amplex Red (Invitrogen) for 30 min in the dark. Peroxide concentration of the sample was estimated by comparison with a standard curve and relative to Chl. Depicted values were calculated by normalizing to the Col-0 average. Chl content was determined according to Porra et al. (1989).
For Fv/Fm [(Fm − F0)/Fm] measurements, whole plants were dark acclimated for 30−45 min prior to measurement with the Walz MAXI IMAGING-PAM. Fv/Fm values are averages of the whole plant. Fv/Fm recovery for each plant was measured as the Fv/Fm ratio after 6 d in fluctuating light and Fv/Fm before shift.
Statistical analyses on data were performed by employing one-way ANOVA and Tukey’s multiple comparison tests.
Protease protection assay, immunoblot analyses and BN−PAGE
Thylakoids were isolated as described (Armbruster et al. 2014). For protease protection assays, thylakoids were resuspended at 0.5 mg Chl per ml−1 in 0.3 M sorbitol, 2.5 mM EDTA, 5 mM MgCl2, 0.5% (w/v) bovine serum albumin (BSA), 20 mM HEPES (pH 7.6). Reactions were started by the addition of thermolysin (EMD Millipore) at 10 μg ml−1 to a final volume of 300 μl. At the indicated times, the reaction was stopped by transferring 50 μl to a tube containing EDTA so that the final concentration of EDTA was 50 mM. The tubes were vortexed immediately and sample buffer was added. For BN−PAGE, thylakoid membranes were solubilized with 0.7% β-n-dodecyl-d-maltoside (w/v) and separated by BN gels (Invitrogen) according to Peng et al. (2008).
Total protein was extracted from liquid nitrogen frozen leaf tissue (20 mg) supplemented with approximately 50 μl of lysing matrix D (MP Biomedicals). The Fastprep 24 tissue homogenizer (MP Biomedicals) was set to 6.5 s−1 and tissue was disrupted for 1 min prior to addition of 200 μl of protein extraction buffer [200 mM Tris, pH 6.8, 8% SDS (w/v), 40% glycerol and 200 mM dithiothreitol (DTT)]. Samples were heated at 65 °C for 10 min and proteins were separated on SDS−PAGE, blotted onto nitrocellulose, visualized with Ponceau Red [0.1% Ponceau S (w/v) in 5% (v/v) acetic acid] and detected with antibodies specific for KEA3 (Armbruster et al. 2014) or PsbO (Agrisera). KEA3 signals were quantified by densitometric analysis of Western bands and Ponceau stain using NIH ImageJ software and associated plug-ins (http://imagej.nih.gov/ij/).
Computational analyses
Arabidopsis KEA3.1 and KEA3.2 (At4g04850.1 and At4g04850.2) DNA and protein sequences were retrieved from TAIR (The Arabidopsis Information Resource; www.arabidopsis.org). The newly identified KEA3.3 splice form was translated into protein sequence by using Expasy Translate (web.expasy.org/translate). Amino acid sequences were aligned using ClustalOmega (http://www.ebi.ac.uk/Tools/msa/clustalo). Transmembrane helices of KEA3 were predicted by homology modeling using the Phyre2 server (www.sbg.bio.ic.ac.uk/∼phyre2; Kelley and Sternberg 2009). Thermolysin digestion sites were predicted with Expasy Peptide Cutter (web.expasy.org/peptide_cutter). For sequence comparisons, sequences of the KTN domain of E. coli KefC (EcKefC, P03819), AtKEA3, Arabidopsis KEA2 (AtKEA2, Q65272) and the first RCK domain of V. parahaemolyticus TrkA (VcTrkA, A0A072LGS4) were aligned using ClustalOmega and shaded according to amino acid identity (black) and similarity (gray) using the Box-shade server (http://www.ch.embnet.org/software/BOX_form.html).
Accession numbers
Sequence data determined for the KEA3.3 cDNA have been submitted to GenBank under accession number KT581346. Accession numbers for RNA-seq data used in this analysis were SRR656215, SRR656216, SRR656217, SRR656218; ERR754059, ERR754064, ERR754066, ERR754069, ERR754077, ERR754084, ERR754087 (Clauw et al. 2015) and ERR377676, ERR377677, ERR377678 (Aubry et al. 2014).
Supplementary data
Supplementary data are available at PCP online
Funding
This work was supported by the Carnegie Institution for Science; the Max Planck Society; the Bill & Melinda Gates Foundation RIPE project at the University of Illinois [a subaward]; the Howard Hughes Medical Institute; the Gordon and Betty Moore Foundation [grant GBMF3070 to K.K.N.]; the Deutsche Forschungsgemeinschaft [AR 808/1-1, 1-2 to U.A.].
Supplementary Material
Acknowledgements
We thank Elisabeth Schmidtmann for excellent technical assistance, Mark Aurel Schöttler and Ralph Bock for help with the fluctuating light chamber, and anonymous reviewers for constructive comments and suggestions.
Glossary
Abbreviations
- CPA2 domain
cation proton antiport 2 domain
- cTP
chloroplast transit peptide
- GFP
green fluorescent protein
- KEA3
K+ efflux antiporter 3
- KTN domain
K+ nucleotide-binding domain
- NPQ
non-photochemical quenching
- qE
energy-dependent quenching
- RNA-seq
RNA sequencing
Disclosures
The Carnegie Institution for Science has submitted a patent form on behalf of U.A., M.C.J. and K.K.N. on aspects of the finding.
References
- Aranda-Sicilia M.N., Cagnac O., Chanroj S., Sze H., Rodriguez-Rosales M.P., Venema K. (2012) Arabidopsis KEA2, a homolog of bacterial KefC, encodes a K(+)/H(+) antiporter with a chloroplast transit peptide. Biochim. Biophys. Acta 1818: 2362–2371. [DOI] [PubMed] [Google Scholar]
- Armbruster U., Carrillo L.R., Venema K., Pavlovic L., Schmidtmann E., Kornfeld A, et al. (2014) Ion antiport accelerates photosynthetic acclimation in fluctuating light environments. Nat. Commun. 5: 5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. (1999) The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 601–639. [DOI] [PubMed] [Google Scholar]
- Aubry S., Smith-Unna R.D., Boursnell C.M., Kopriva S., Hibberd J.M. (2014) Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant J. 78: 659–673. [DOI] [PubMed] [Google Scholar]
- Brooks M.D., Sylak-Glassman E.J., Fleming G.R., Niyogi K.K. (2013) A thioredoxin-like/beta-propeller protein maintains the efficiency of light harvesting in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: E2733–E2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Pan Y., Huang H., Jin X., Levin E.J., Kloss B., et al. (2013) Gating of the TrkH ion channel by its associated RCK protein TrkA. Nature 496: 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauw P., Coppens F., De Beuf K., Dhondt S., Van Daele T., Maleux K., et al. (2015) Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol. 167: 800–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B., Adams W.W., III, Barker D.H., Logan B.A., Bowling D.R., Verhoeven A.S. (1996) Using chlorophyll fluorescence to assess the fraction of absorbed light allocated to thermal dissipation of excess excitation. Physiol. Plant. 98: 253–264. [Google Scholar]
- Demmig-Adams B., Cohu C.M., Muller O., Adams W.W., 3rd (2012) Modulation of photosynthetic energy conversion efficiency in nature: from s to seasons. Photosynth. Res. 113: 75–88. [DOI] [PubMed] [Google Scholar]
- Grieco M., Tikkanen M., Paakkarinen V., Kangasjarvi S., Aro E.M. (2012) Steady-state phosphorylation of light-harvesting complex II proteins preserves photosystem I under fluctuating white light. Plant Physiol. 160: 1896–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Ruban A.V., Walters R.G. (1996) Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 655–684. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Kramer D.M., Cruz J.A., Kanazawa A. (2003) Balancing the central roles of the thylakoid proton gradient. Trends Plant Sci. 8: 27–32. [DOI] [PubMed] [Google Scholar]
- Kröning N., Willenborg M., Tholema N., Hanelt I., Schmid R., Bakker E.P. (2007) ATP binding to the KTN/RCK subunit KtrA from the K+-uptake system KtrAB of Vibrio alginolyticus: its role in the formation of the KtrAB complex and its requirement in vivo. J. Biol. Chem. 282: 14018–14027. [DOI] [PubMed] [Google Scholar]
- Külheim C., Agren J., Jansson S. (2002) Rapid regulation of light harvesting and plant fitness in the field. Science 297: 91–93. [DOI] [PubMed] [Google Scholar]
- Kunz H.H., Gierth M., Herdean A., Satoh-Cruz M., Kramer D.M., Spetea C., et al. (2014) Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 7480–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.P., Marshall-Colon A., Zhu X.G. (2015) Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161: 56–66. [DOI] [PubMed] [Google Scholar]
- Müller P., Li X.P., Niyogi K.K. (2001) Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125: 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie E.H., Niyogi K.K. (2011) Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol. 155: 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness L.S., Booth I.R. (1999) Different foci for the regulation of the activity of the KefB and KefC glutathione-gated K+ efflux systems. J. Biol. Chem. 274: 9524–9530. [DOI] [PubMed] [Google Scholar]
- Peng L., Shimizu H., Shikanai T. (2008) The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J. Biol. Chem. 283: 34873–34879. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Prinsley R.T., Hunt S., Smith A.M., Leegood R.C. (1986) The influence of a decrease in irradiance on photosynthetic carbon assimilation in leaves of Spinacia oleracea L. Planta 167: 414–420. [DOI] [PubMed] [Google Scholar]
- Roosild T.P., Castronovo S., Miller S., Li C., Rasmussen T., Bartlett W., et al. (2009) KTN (RCK) domains regulate K+ channels and transporters by controlling the dimer-hinge conformation. Structure 17: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosild T.P., Miller S., Booth I.R., Choe S. (2002) A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch. Cell 109: 781–791. [DOI] [PubMed] [Google Scholar]
- Sainsbury F., Lomonossoff G.P. (2008) Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148: 1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spetea C., Hundal T., Lundin B., Heddad M., Adamska I., Andersson B. (2004) Multiple evidence for nucleotide metabolism in the chloroplast thylakoid lumen. Proc. Natl. Acad. Sci. USA 101: 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M., Scheibe R., Feil R. (1989) Response of photosynthetic electron transport and carbon metabolism to a sudden decrease of irradiance in the saturating or the limiting range. Biochim. Biophys. Acta 973: 241–249. [Google Scholar]
- Strand D.D., Livingston A.K., Satoh-Cruz M., Froehlich J.E., Maurino V.G., Kramer D.M. (2015) Activation of cyclic electron flow by hydrogen peroxide in vivo. Proc. Natl. Acad. Sci. USA 112: 5539–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaks J., Amarnath K., Kramer D.M., Niyogi K.K., Fleming G.R. (2012) A kinetic model of rapidly reversible nonphotochemical quenching. Proc. Natl. Acad. Sci. USA 109: 15757–15762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.G., Long S.P., Ort D.R. (2010) Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61: 235–261. [DOI] [PubMed] [Google Scholar]
- Zhu X.G., Ort D.R., Whitmarsh J., Long S.P. (2004) The slow reversibility of photosystem II thermal energy dissipation on transfer from high to low light may cause large losses in carbon gain by crop canopies: a theoretical analysis. J. Exp. Bot. 55: 1167–1175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.