Abstract
MSI (MSI) was introduced more than five decades ago with secondary ion mass spectrometry (SIMS) and a decade later with laser desorption/ionization (LDI) mass spectrometry (MS). Large biomolecule imaging by matrix-assisted laser desorption/ionization (MALDI) was developed in the 1990s and ambient laser MS a decade ago. Although SIMS has been capable of imaging with a moderate mass range at sub-micrometer lateral resolution from its inception, laser MS requires additional effort to achieve a lateral resolution of 10 μm or below which is required to image at the size scale of single mammalian cells. This review covers untargeted large biomolecule MSI using lasers for desorption/ionization or laser desorption and post-ionization. These methods include laser microprobe (LDI) MSI, MALDI MSI, laser ambient and atmospheric pressure MSI, and near-field laser ablation MS. Novel approaches to improving lateral resolution are discussed, including oversampling, beam shaping, transmission geometry, reflective and through-hole objectives, microscope mode, and near-field optics.
1. Introduction
Mass spectrometry imaging (MSI) was first developed over 50 years ago and there have been many developments in the succeeding years in both ionization and mass selection technology that have brought the field to new levels of spatial resolution, sensitivity, and mass resolution [1-4]. A mass spectrometry image typically comprises an array of mass spectra obtained sequentially at regular spatial intervals on an object and, for any m/z value in a mass spectrum, a heat map image can be created that represents the abundance of a particular ion. To obtain a mass spectrum image, samples are bombarded in vacuum by an energetic beam of ions [5, 6], or irradiated with a pulsed laser under vacuum [7, 8] or at atmospheric pressure (AP) [9]. Under ambient conditions, charged particles [9] and plasma [10] can also be used to generate ions or compounds can also be extracted by a solvent in contact with the surface.
The first approach to MSI was secondary ion mass spectrometry which was first reported by Castaing and Slodzian in 1962 [11]. In subsequent decades, the sensitivity and mass range of SIMS have been extended with time-of-flight (TOF) mass analyzers [12] and cluster ion sources [13, 14]. SIMS is capable of MSI of atoms and small molecules down to 35 nm lateral resolution [6] and molecules up to approximately 1500 Da at a spatial resolution of a few hundred nanometers [14]. The use of lasers for MSI was accomplished in the 1970s through the work of Hillenkamp and coworkers [15]. Laser desorption/ionization (LDI) MSI has a mass range similar to TOF SIMS up to around 1000 Da and a lateral resolution around 1 μm [16]. The mass range of laser-based MSI was significantly extended with the application of matrix-assisted laser desorption/ionization (MALDI) by Caprioli and coworkers in the late 1990s [17]. With current instrumentation, MALDI MSI has a working mass range up to 30 kDa [8]; the lateral resolution has improved from approximately 50 μm to the point where the latest commercial instruments are approaching 10 μm resolution [18].
The development of ambient and atmospheric pressure ionization methods over the past decade has led to a wide variety of new MSI methods [9]. Many of these imaging approaches use pulsed lasers to ablate material for ionization. MALDI, for example, can be performed at atmospheric pressure [19] and the use of infrared [20] or short-pulse lasers [21] can obviate the need for matrix addition. Novel matrix materials and conditions can be used to generate highly charged ions [22-24] and laser ablation can be combined with post-ionization by electrospray [25] or chemical ionization [26]. These AP/ambient laser ionization methods show promise as alternatives for fast and direct tissue imaging.
A critical dimension for all types of MSI is the size of a single mammalian cell, which is approximately 20 μm in diameter [27]. The ability to image at the single cell level is essential for probing the biochemical heterogeneity of individual cells and understanding their fundamental cellular processes [28-30]. SIMS imaging has achieved single cell spatial resolution although it faces fundamental challenges in achieving the mass range required for single cell protein analysis [29, 30]. Laser-based MSI such as MALDI and related ambient and atmospheric pressure ionization techniques are better positioned as general methods for MSI of large biomolecules at the single cell level. The challenges that the laser imaging techniques face are improving the lateral resolution while maintaining sufficient detection efficiency for the small sample quantities found at these scales.
Lateral resolution in raster mode laser MSI is determined by a number of factors; foremost among these is the diameter of the focused laser. The minimum focus spot size diameter d is given by
where is the laser wavelength, n is the index of refraction of the medium and is the half-angle of the beam exiting the lens. The quantity n sin is known as the numerical aperture (NA) of the lens and can reach values around 1.5 in a high-quality optical system. High numerical aperture lenses tend to be wide and must be positioned close to the sample with high-index-of-refraction immersion oil, properties that are not suited to laser desorption and ablation mass spectrometry. Thus, the laser focus diameter used for MSI is often far from the diffraction limit. Other factors affecting imaging resolution are the raster step size and resulting image pixel size, which is typically in the same range as the laser diameter. For MALDI MSI, the resolution is also affected by the addition of matrix: diffusion of the analyte molecules in the sample as a result of matrix addition influences the lateral resolution of the image.
Detection sensitivity is of the utmost importance for MSI at high spatial resolution. The average concentration of proteins in a cell can be estimated to be roughly 3 million per cubic micrometer [31]. Thus, a 10 μm diameter laser spot that is irradiating a 10 μm-thick section of tissue has the potential to probe 2 × 109 proteins. The most abundant of these exist at approximately 0.1% of the total protein concentration [32] suggesting an upper bound of approximately one million copies of high-abundant proteins that may be accessible to mass spectrometry at this spatial resolution. This quantity is near the limits of detection for MALDI [33] and electrospray [34], clearly indicating the need for both highly efficient ionization as well as efficient transfer of ions into the mass spectrometer.
This review covers the use of lasers to create ions or ablate material for the creation of ions for MSI of biomolecules at 10 μm lateral resolution, a size range that has the potential to enable untargeted biomolecule imaging at the single cell level. Early developments in high-spatial resolution laser microprobe MS that are now being adapted to laser ablation biomolecule MSI are discussed as well as below-the-diffraction-limit methods using near-field laser ablation [35] and laser microscope mode [36, 37] MSI. Laser ablation coupled with inductively coupled plasma mass spectrometry (ICP-MS) for bioimaging have been discussed in recent reviews [38-40] and will not be covered here. For a broad discussion of MSI, the reader is directed to several recent reviews [2-4, 7].
2. Early LDI Microprobe and Imaging Analysis
Although the use of lasers to create ions from desorption and ablation of solids dates back to the late 1960s [41, 42], many of the concepts developed for these instruments are relevant to current laser-based MSI. Early laser desorption mass spectrometers used a pulsed laser focused onto the back (transmission geometry) or front (reflection geometry) of a thin film sample under vacuum. The original design for a laser desorption time-of-flight (TOF) mass spectrometer was described by Fenner in 1966 [43]. The instrument used a Q-switched ruby laser at 694 nm to ablate material in transmission geometry from a thin metal foil or glass slide. The laser had a 50 μm focal spot but a larger spot diameter was observed at the pulse energies used for MS. Nanogram quantities of material were detected using a 1 m TOF mass spectrometer, which used an electric sector energy filter and achieved a mass resolution of 30.
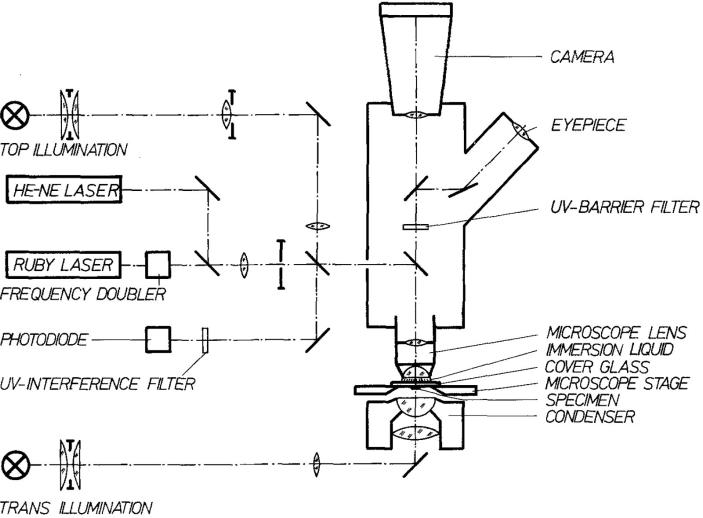
In the mid 1970s Hillenkamp and coworkers improved on the Fenner design, using a frequency-doubled ruby laser at 347 nm wavelength focused in transmission geometry onto a glass slide that also served as the vacuum window for the mass spectrometer [44, 45]. A schematic of the instrument is shown in Figure 1. A TOF MS was used to detect the ions with an m/z range for detected ions of less than 200. The optical configuration allowed a high numerical aperture lens to be used that resulted in a laser spot size of 0.5 μm diameter. The instrument could ablate a total mass of 400 fg and achieved a detection limit of 0.14 ag (14,000 atoms) for lithium. The biological sample analysis capabilities of the instrument were recognized early: single bacterial cells were analyzed, detecting compounds of up to m/z 150 [46].
Figure 1.
Schematic diagram of a transmission geometry laser microprobe mass spectrometer [45].
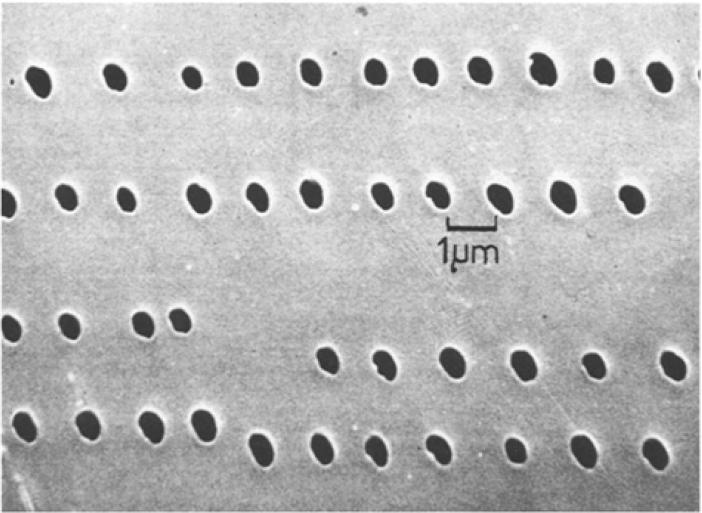
An example of transmission geometry ablated holes is shown in Figure 2, which shows an electron micrograph image of a 1 μm thick epoxy resin film that was irradiated with the frequency doubled ruby laser [45]. The numerical aperture of the lens was 1.25, which was reduced to an effective numerical aperture of 1.0 due to the 18 μm gap between the objective and the sample. The transmission geometry configuration was also demonstrated for ambient ionization: compounds deposited on the atmospheric pressure side of a thin epoxy film were perforated with a laser which resulted in the ionization of components of organic compounds on the atmospheric pressure side of the film [47].
Figure 2.
Laser perforations in a 1 μm-thick epoxy resin section from the transmission geometry laser microprobe mass spectrometer [45].
The Hillenkamp design was implemented commercially as the “Laser Microprobe Mass Analyzer” LAMMA 500 (Leybold Heraeus) [48]. The need to analyze thicker samples led to the development of the LAMMA 1000 [49] that used a frequency-tripled (355 nm) or quadrupled (266 nm) Nd:YAG laser focused in reflection geometry using a 50× lens with a numerical aperture of 0.6. The LAMMA 1000 configuration was the forerunner of the current MALDITOF mass spectrometer and was capable of imaging at a lateral resolution of 2.5 μm. In addition to TOF mass spectrometers, laser desorption ion probe instruments based on Fourier transform ion cyclotron resonance (FTICR) mass spectrometers were also developed [50]. Using conventional reflection geometry optics, laser spot sizes between 5 and 10 μm were realized for ion formation at the ICR cell [51, 52] as well as external to the cell [53, 54].
An alternative approach to reflection geometry surface irradiation is a Schwarzschild objective, which uses a pair of mirrors to focus the light. For this objective, a smaller diameter convex primary mirror is paired with a larger diameter concave secondary mirror with a center aperture [55]. The resulting objective is achromatic and has a long working distance that is useful for reflection geometry desorption and ionization. This configuration was used in the commercial laser induced mass analysis instrument (LIMA [56], Cambridge Consultants) that achieved a lateral resolution less than 10 μm [57, 58]. A Schwarzschild objective was used for laser desorption with 1 μm lateral resolution with an FTICR mass spectrometer [59]. The system used a 337 nm nitrogen laser for desorption and, for some applications, the fifth harmonic of a Nd:YAG laser at 213 nm was used for post-ionization. The nitrogen laser was focused with an objective that was 11 cm in diameter and 18 cm in length and had a numerical aperture of 0.2 and a 13 cm working distance. The desorption laser was focused onto the sample target that was placed within the FTICR magnet bore and adjacent to the ICR cell. The post-ionization laser was directed above the target using a set of steering prisms. The lateral resolution in line scan mode for direct laser desorption ionization of Cu+ ions from a copper grid and laser desorption and post-ionization of a C60 thin film on indium was less than 1 μm. A similar Schwarzschild microscope system was demonstrated with a TOF mass spectrometer [60]. For this system, a 524 nm Nd:YLF or 532 nm frequency-doubled Nd:YAG laser was used for desorption and post-ionization was accomplished with a femtosecond Ti:saphire laser at 800 nm wavelength. In this device, the objective was outside the vacuum and a right angle turning mirror with a central hole was used to direct the focused beam onto the target. MS images of rhodamine dye deposited on a copper grid were obtained at a lateral resolution of less than 2 μm.
3. MALDI MSI
MALDI MSI using a commercial MALDI TOF instrument with a lateral resolution of 25 μm and a reflection geometry configuration was first reported nearly two decades ago [17]. The resolution of MALDI MSI is influenced by the same factors as laser microprobe MSI with the additional constraint that a matrix must be added, which can result in the migration of biomolecules in tissue samples [8, 61]. The lateral resolution of commercial MALDI MSI instruments has remained in this range in the intervening years, although there is a recent move to bring imaging below the 10 μm range for routine analysis [18, 62]. Imaging at atmospheric pressure is also moving to the sub-10 μm regime [63]. Another notable if more incremental improvement in MALDI MSI has been the steady increase in data acquisition speed, which has increased due to improvements in signal digitization electronics and the replacement of relatively low-repetition rate 337 nm nitrogen lasers with frequency-tripled 355 nm Nd:YAG lasers with repetition rates up to 10 kHz [18] and 349 nm Nd:YLF lasers with a repetition rate of 5 kHz [64].
Improving the lateral resolution of MALDI MS has concentrated on several factors: (1) matrix addition, (2) oversampling, (3) beam focus in reflection geometry, and (4) transmission geometry. Matrix addition can be a limiting factor that can cause migration of sample components in excess of 10 μm even when the matrix is applied with an automated spray device [65]. Analyte migration can be reduced by dry matrix addition, for example by sublimation that produces matrix crystals as small as 1 μm in diameter [66, 67]. For application to MALDI MSI, the matrix can be sublimed for several minutes using a vacuum apparatus to achieve a coverage of approximately 0.2 mg/cm2 at a thickness of 2 μm [68]. The matrix and tissue are then incubated with solvent at low heat for several minutes to allow extraction of tissue components and matrix recrystallization. The resulting matrix crystals were smaller than 3 μm in diameter and allowed imaging at a lateral resolution of 10 μm. An alternative conventional dry matrix application is metal sputter coating. In a recent study, a 10 to 20 nm sputter coat of silver was deposited on tissue sections [69]. This metal thin-film matrix coating approach imaged small molecules in tissue sections with a lateral resolution of 5 μm.
Oversampling is a simple method for improving spatial resolution for MALDI MSI that does not require instrument modifications to achieve lateral resolution less than the laser beam diameter. To obtain higher spatial resolution through oversampling, the imaging step size is set to a value lower than the laser beam diameter and the spot is irradiated until the signal is depleted. A 25 μm step has been used to achieve 40 μm lateral resolution with a laser spot size of approximately 100 μm × 200 μm [70]. More recently, 5 μm lateral resolution was achieved with a 20 μm laser beam diameter and oversampling [71]. However, there are drawbacks, including longer data acquisition times and loss of signal [72].
An alternative to oversampling is beam shaping. The diameter of a focused laser beam can be expressed as
where is the laser wavelength, f is the focal length of the lens, M2 is the beam quality factor, and D(0) is the diameter of the unfocused laser beam [73]. The beam quality factor M2 is 1 for an ideal Gaussian beam and increases with decreasing beam quality. For a low-power continuous laser operating on the fundamental transverse oscillator mode, M2 can be as low as 1.1, whereas for a high-energy multi-mode laser M2 can be 10 or more.
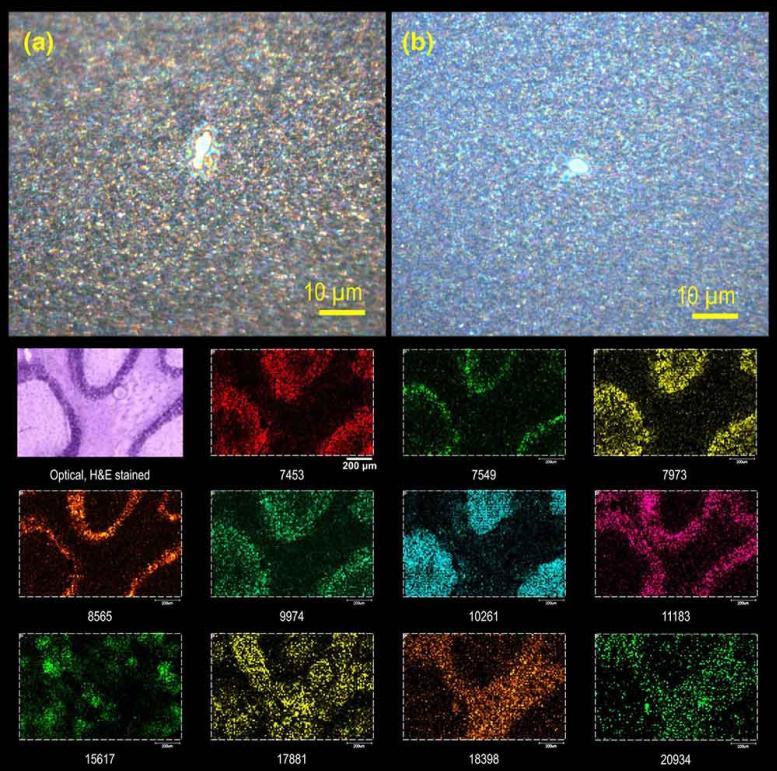
The laser beam quality in a MALDI system can be improved by sending the light through an optical fiber [74, 75], but this method is not ideally suited to high-energy pulsed laser applications. An alternative approach is to place a pinhole in the laser path. A 25 μm ceramic pinhole was used with a commercial MALDI MSI instrument to achieve 5 μm lateral resolution [71]. A Gaussian laser beam using both a pinhole aperture as well as an aspheric lens for MALDI MSI has also been described [76]. Results from this configuration are shown in Figure 3, which shows microscope images of laser spots in a 2,5-dihydroxybenzoic acid matrix film using 50 laser shots from a Gaussian beam without (Figure 3a) and with a 30 μm pinhole (Figure 3b). The lower half of Figure 3 shows protein images from a mouse brain tissue section using the shaped Gaussian laser beam. Laser beam shaping with a pinhole aperture has also been demonstrated with an intermediate pressure ion source Q-TOF mass spectrometer to obtain 10 μm lateral resolution [77].
Figure 3.
Optical microscope images of laser spots from 50 shots on a sublimed thin film of 2,5-dihydroxybenzoic acid matrix with a (a) Gaussian laser beam focused with an aspheric lens and (b) spatially filtered with a 30 μm pinhole [76]. Below are MALDI-TOF MS protein images from mouse cerebellum using a 5 μm step size and the Gaussian laser beam and aspheric lens. The middle left panel shows an optical microscope image of an H&E stained serial section.
As with the laser microprobe, transmission geometry allows a larger numerical aperture lens to be used, but this requires a fundamental reconfiguration of the MALDI MSI instrument. Furthermore, thin tissue sections must be used because the laser must penetrate the sample to irradiate the matrix and analyte at the surface. An additional complicating factor in transmission geometry irradiation is the propensity to produces larger particles compared to reflection geometry, [78] which can potentially lower the ionization efficiency. Nonetheless, there have been several transmission geometry MALDI MSI instruments demonstrated. In a recent report, the ion source of a commercial MALDI-TOF mass spectrometer with a 355 nm laser was modified to accept a 100× microscope objective and 0.1 μm resolution translation stage encoders [79]. The mass spectrometer was used to image tissue sections that were 5 μm thick as well as single mammalian cells. The transmission geometry configuration produced mass spectra comparable to those obtained in reflection geometry when both were used with a laser spot size of 5 μm. In transmission geometry, the minimum laser diameter was 1 μm and a step size down to 0.5 μm was used for imaging. The instrument was used to image tissue treated with photocleavable mass tags with a 2 μm laser spot and 2 μm step size [71]. A similar instrument was demonstrated with a 2 kHz Nd:YLF laser focused to a 1 μm spot and 2.5 μm raster step [62].
One of the difficulties in using a high numerical aperture lens with reflection geometry imaging is the small working distance. It is difficult to desorb and ionize with a conventional high numerical aperture objective because the lens itself blocks the avenue of escape for the ejected ions. A unique solution to this problem is achieved by the use of a lens with a central hole, through which the ions pass. An imaging mass spectrometer has been described that employs an objective lens with a numerical aperture of 0.6 and 16 mm working distance that has a central hole with a 6 mm inner diameter [80]. The objective is placed in the vacuum chamber above the sample and directs the focused laser onto the target. The desorbed ions pass through the central bore of the objective and are accelerated into the flight tube of the mass spectrometer [80]. The instrument can image down to a lateral resolution of 0.6 μm with a 0.25 μm step size. Controlled matrix deposition by sublimation and recrystallization allowed an imaging resolution of 2 μm to be achieved with biological samples [81], and imaging of proteins up to 27,000 Da in mass was demonstrated at better than 10 μm lateral resolution [82].
All of the MSI methods described to this point are examples of microprobe mode, which is the sequential acquisition of mass spectra across an array of points on a sample surface [83]. An alternative approach is microscope mode MSI where ions are created simultaneously across an area of the sample with the detection maintaining their spatial orientation. Microscope mode MALDI MSI has been demonstrated using a TOF mass spectrometer originally developed for SIMS imaging [83]. A TRIFT II triple focusing TOF mass spectrometer was modified to accept a 337 nm pulsed nitrogen laser. The mass spectrometer was equipped with three hemispherical electrostatic analyzers, a dual microchannel plate phosphor screen detector assembly and CCD camera. A 5× microscope objective was installed under vacuum, and the target was irradiated at 200 J/m2 fluence on a 200 μm diameter spot. The detector was not sufficiently fast to detect all of the ions, therefore beam blanking of all but the ion of interest was used to obtain an image. When the sample was irradiated, the resulting ions were magnified and imaged on the detector with their spatial orientation intact. The system was demonstrated with a lateral resolution of 4 μm for peptide and protein test samples.
Microscope mode MSI was used with a mid-infrared laser to demonstrate MSI below the diffraction limit of the laser. For example, an Er:YAG laser at 2.94 μm wavelength was used to obtain a lateral resolution of 4 μm for a peptide standard [36], and more recently, an active pixel detector was used for multiple m/z detection using a mid-IR OPO laser system at lateral resolution of 10 μm for a tissue sample [37]. Figure 4 shows IR microscope mode MSI of a tissue section. Figure 4a shows mass spectra integrated over 2000 shots from the liver and stomach of fish tissue. Figure 4b shows a light microscope image of the sample overlaid with a mass spectrum heat map of the detected phospholipids.
Figure 4.
Microscope mode IR MALDI MSI of a bait fish cryo-section showing (a) IR MALDI integrated mass spectrum obtained without external matrix using all detector pixels and 2000 laser shots and (b) an overlay of an optical microscopy image of the sample and mass spectrometry images of several phosphatidylcholines [37]. The white dotted lines depict the boundary between the liver (left) and stomach (right).
4. Ambient and Atmospheric Pressure Laser Imaging Mass Spectrometry
Ambient MS is the direct analysis of materials in their native environment with ionization by methods that include a wide variety of approaches involving charged droplets, ions, or metastable molecules impinging on a sample [9, 84-87]. Imaging can be accomplished by translating the sample to create a two dimensional MS image [25, 88-91]. and laser ablation can be used in conjunction with post-ionization by inductively coupled plasma [92-94], chemical ionization [95, 96], photoionization [97], or electrospray ionization [21, 98-100]. AP MALDI with an IR laser and endogenous matrix [19, 20, 101] and femtosecond pulse width laser matrix-free ionization [102] can be considered ambient ionization methods, but AP MALDI [24, 103] is more accurately described as an AP technique rather than an ambient technique due to the requirement for adding a matrix.
One example of high lateral resolution AP MALDI MSI uses the high numerical aperture through-hole reflection mode design described above [104]. The electrospray ion source of an FTICR mass spectrometer was replaced with a high lateral resolution AP MALDI ion source. A 337 nm nitrogen laser was focused onto a target using a three-element quartz objective lens with a numerical aperture of 0.43 and a 40 mm working distance. The lens had a 7 mm hole through the bore that accommodated a 13 cm long ion and 0.6 mm inner diameter transfer capillary that was directly connected to the mass spectrometer inlet. The sample was mounted on a three-axis translation stage with 1 μm precision and 50 mm travel. Using this objective, the laser was focused to a spot size of 0.5 μm × 0.6 μm. Imaging of peptide standards was achieved with a laser diameter of 8 μm due to sensitivity limitations with smaller spot sizes.
Spatial resolution of 5 μm was reported with an AP MALDI MSI system coupled with an Orbitrap mass spectrometer [105]. A 337 nm nitrogen laser was focused to an 8 μm spot diameter using a high numerical aperture reflection geometry configuration. Images of neuropeptides in mouse pituitary were imaged at 5 μm lateral resolution. The instrument has also been used to image lipids in whole body sections of mosquitos [106].
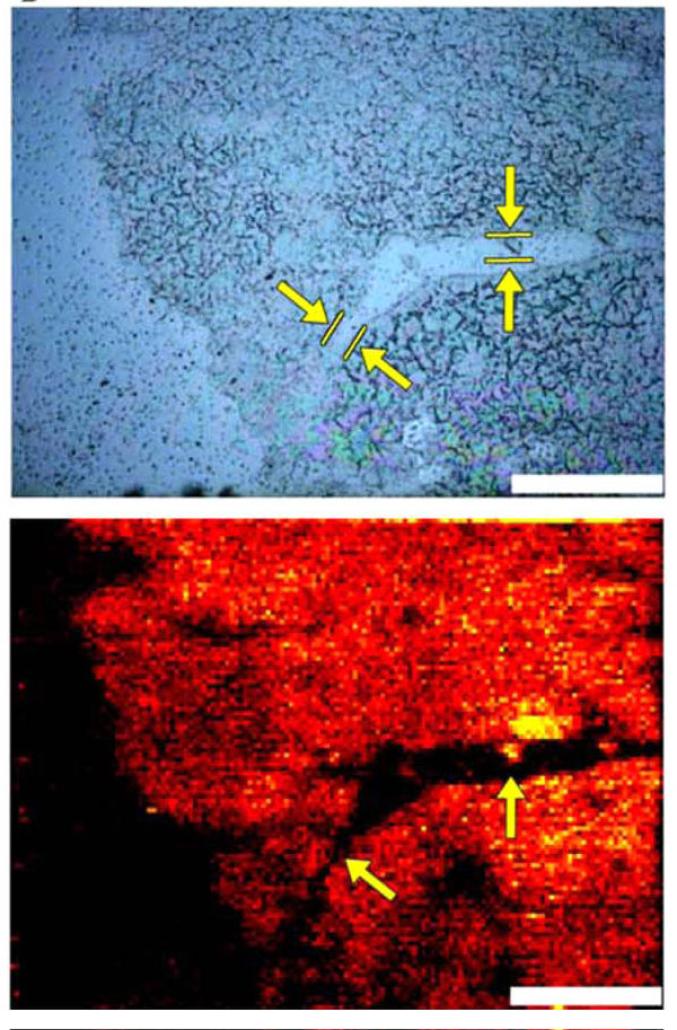
A recent example of ambient laser ablation with post-ionization for high lateral resolution MSI is a laser sampling mass spectrometer based on a laser microdissection microscope [107]. A commercial Leica laser inverted microscope microdissection system was used in transmission geometry with a laser ablation spot size of 2 μm in diameter. A 349 nm Nd:YLF laser with a maximum repetition rate of 5 kHz was rastered across the sample at a step size as small as 0.5 μm. The ablated material was captured in a flowing solvent, ionized by electrospray ionization, and mass analyzed in a Q-TOF mass spectrometer. The system was able to detect features as small as 0.6 μm on a polymer sample and could detect cocaine in mouse brain tissue at a lateral resolution of 15 μm (Figure 5).
Figure 5.
Images of a 10 μm-thick mouse brain section from a mouse dosed with cocaine: (top) bright-field and (bottom) mass spectral image of cocaine [107].
5. Near-field Laser Ablation
Near-field optics can be used to localize electromagnetic radiation below the diffraction limit [108, 109]. Ablation can be accomplished in aperture mode with an optical fiber probe that transmits the laser light to a sharp tip or in apertureless mode that uses a sharp conductive tip as an electromagnetic antenna for the laser light [110]. The fiber or conductive tip is positioned within a few nm of the surface using a force-feedback system such as an atomic force microscopy (AFM) [111] or scanning near-field optical microscopy (SNOM) [110]. Material can be desorbed as molecules, ablated as particles, or ions directly formed at the surface. The ablation region is not constrained by the laser wavelength and the optical probe is small and can be placed at the front surface of the sample without blocking the exit of material as would an objective lens.
For MS, ions can be created directly by LDI, immediately post-ionized, or collected and analyzed [35]. In an early report of near-field laser MS, direct LDI was achieved by aperture near-field LDI in vacuum [112]. The scanning head of a near-field microscope was mounted in a vacuum chamber and was used to place an etched uncoated probe approximately 10 nm from the sample surface. The diameter of the probe tip was less than 400 nm and a 337 nm nitrogen laser was used for desorption and ionization. A reported 1 μm lateral resolution in line scan mode was demonstrated for LDI of an acetylcholine sample deposited on a TEM grid.
Direct AP LDI was accomplished under ambient conditions with the ions directed into the mass spectrometer [113]. A 532 nm wavelength and 5 ns pulse width Nd:YAG laser was focused onto an AFM tip for apertureless tip-enhanced ablation. With this configuration, laser ablation craters as small as 50 nm have been observed [114]. For LDI MS, ions were sampled into a 0.8 μm diameter tube that was held 500 μm above and 1 mm away from the tip. A near-field LDI image was recorded at a lateral resolution of 2 μm, which was limited by the mass spectrometer sensitivity rather than the ablation spot size.
Post-ionization of the near-field ablated material allows ionization to be performed independently of desorption. Near-field laser desorption and electron ionization post-ionization was accomplished using a SNOM apparatus and quadrupole mass spectrometer. A 355 nm Nd:YAG laser was coupled into an optical fiber-pulled SNOM tip and the pulsed laser light was directed at the target surface. The laser-ablated material was sampled into a 20 cm-long and 20 μm-ID stainless steel capillary that was positioned 5 μm from the tip. Nitrogen gas was detected from the laser-induced decomposition of bis(phenyl-N,N-diethyltriazene) from 200 nm near-field ablation spots. A similar apparatus used a 2 kHz repetition rate 349 nm Nd:YLF coupled into a SNOM tip with laser-desorbed neutrals sampled into an ion trap mass spectrometer with a 250 μm-ID capillary [115]. Tips with a 500 to 800 nm diameter aperture produced craters as small as 200 nm in diameter. Spatially resolved images of anthracene at 5 μm lateral resolution were reported. It was found that much of the ejected material was redeposited as particulate [116] and the overall efficiency of ion production was 10−4 [117].
Collection of near-field ablated material allows off-line MS analysis of biomolecules extracted from the removed particulate. An atomic force microscope and a gold coated silicon tip with a 60 nm diameter has been used for apertureless near-field laser ablation for off-line MALDI MS [118, 119]. A pulsed laser was set to a distance of 15 nm from the surface in tapping mode and the tip was irradiated with a mildly focused laser beam. Wavelengths of 355, 532, and 1064 nm from a Nd:YAG laser have been used for near-field ablation. Craters approximately 1 μm in diameter are produced. The ablated material is captured on a metal wire that can be analyzed directly in the mass spectrometer or the compounds extracted into a matrix for MALDI MS. The 355 nm laser was used to ablate insulin and bradykinin standards for MALDI MS with only limited fragmentation observed for the insulin [118]. Proteins detected using the 532 nm laser irradiation showed no fragmentation [119].
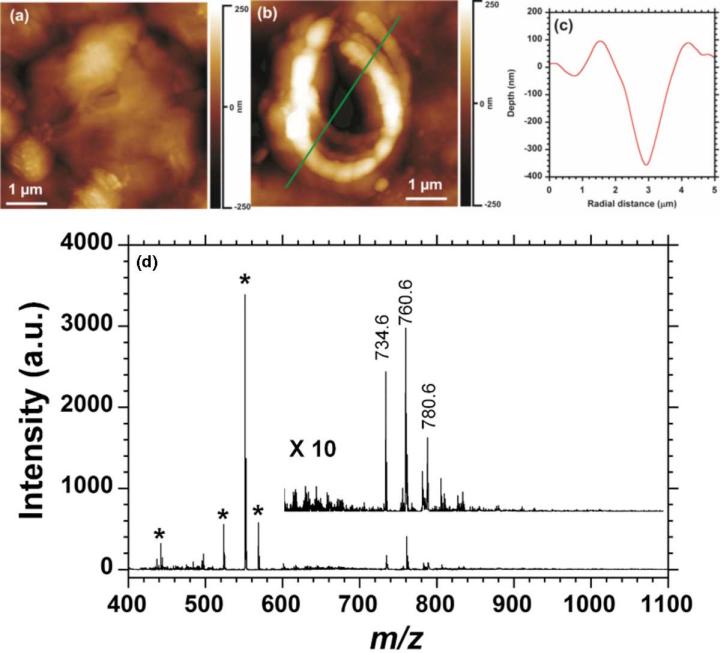
The 1064 nm wavelength was used for tissue ablation with the rationale that the near-IR absorption window of the tissue will be minimally disruptive. Rat brain tissue sections with a thickness of 10 μm were placed on the AFM stage and observed from the AFM video camera for the selection of an area of interest. Figure 6a shows the AFM height image of the sample region scanned prior to ablation. The ablation was performed by placing the AFM probe 15 nm above the surface of the rat brain tissue and directing the laser onto the AFM probe for 3 seconds with a repetition rate of 10 Hz at a laser fluence of 12 kJ/m2. The resulting crater is shown in Figure 6b and the depth profile of the crater along the cursor line indicated in Figure 6b is shown in Figure 6c. The average depth of the ablation craters from three replicates under the conditions used for Figure 6b was 400 ± 30 nm and the average ablation spot area was 10 μm2.
Figure 6.
Atomic force microscopy (AFM) near-field laser ablation crater on rat brain tissue (a) before and (b) after ablation, (c) depth profile and (d) MALDI mass spectrum of the collected material with phosphatidylcholine ions at m/z 734.6 [PC(32:0) + H]+ , 760.6 [PC(34:1) + H]+ , and 788.6 [PC(36:1) + H]+ shown.
The ablated material was captured and analyzed by MALDI-TOF MS according to the previously reported protocol [118]. The ablated material was extracted from the capture wire by immersing the tip in 2 μL of a saturated solution of -cyano-4-hydroxycinnamic acid matrix in a 50:50 (v/v) solution of acetonitrile and 0.1% aqueous trifluoroacetic acid. The solution was then deposited on a MALDI target for analysis and mass spectra were acquired from an average of 500 laser shots. The resulting mass spectrum is shown in Figure 6a. In this spectrum the most intense peaks in the m/z range between 400 and 600 result from matrix ions (indicated with asterisks). A magnified view of the spectrum is shown as an inset in the m/z range between 600 and 1100 where phospholipids are typically detected [120, 121]. No peaks were observed above m/z 1100 for any of the tissue samples. The most intense peaks in Figure 6 between m/z 600 and 1100 correspond to phosphatidylcholines (PCs). In this spectrum, prominent ions at m/z 734.6 [PC(32:0) + H] + , 760.6 [PC(34:1) + H]+, and 788.6 [PC(36:1) + H]+ are labeled.
6. Conclusions
To summarize the current state of the art: laser-based MSI can be performed for peptides and proteins with a lateral resolution of 5 μm and small molecules at 1 μm lateral resolution. Reflection geometry MALDI has been used to image proteins greater than 20 kDa in mass at 5 μm lateral resolution [76] and transmission mode MALDI has been used to image proteins > 10 kDa with a 1 μm spot and 2.5 μm raster step [62]. AP MALDI has been used to image peptides up to 2500 Da at 5 μm lateral resolution using a high mass resolution and accuracy Orbitrap mass spectrometer [105]. AP laser ablation and liquid sample collection was used to detect a small molecule, cocaine, in rat brain tissue at 1 μm lateral resolution [107] and near-field ablation and capture was used to sample (but not image) lipids from rat brain tissue using 1 μm ablation spots.
The recent development of high lateral resolution commercial MALDI MSI mass spectrometers with 5 μm laser spot size points to high-resolution MSI of large biomolecules in tissue becoming routine [18, 62]. Even with a small laser spot size, there remain challenges for biomolecule imaging in developing protocols for matrix addition without sample migration, achieving adequate sensitivity for large molecules at small laser spot sizes [4], and developing methods for compound identification and quantification [122, 123]. Dry matrix addition methods are promising [67-69], but must be improved for broad applicability to large biomolecules. Quantification in MALDI MSI can be accomplished using stable isotope standards [124-126] or parallel liquid chromatography tandem MS of biomolecules extracted from the bulk tissue [127-130]. Tandem MS can also be used in compound identification [125, 126]. Laser microdissection [131-135], laser ablation sample transfer [136, 137], or manual dissection [123, 138] can be used for selection of regions for MS analysis. Improving the speed and spatial resolution of these methods is necessary for them to be compatible with high-resolution MALDI MSI.
Commercial MALDI MSI is approaching the point where single cell MS can be accomplished. However, continued technology development is needed. Transmission geometry MSI has advantages over reflection geometry in lateral resolution, but the requirement for thin tissue sections is a drawback. Mass spectrometers that incorporate both imaging modalities could be useful.
Further, microscope mode imaging will become more applicable with faster electronics, but requires an expensive specialized instrument. However, it has the potential to be a useful option on a SIMS instrument with microscope mode ion optics. Ambient laser MS is a new area for MSI that is rapidly growing with many innovative ionization and sampling methods. A number of these methods use infrared lasers that suffer a disadvantage in lateral resolution due to their wavelength. Nonetheless, better optical configurations such as transmission geometry and high numerical aperture objectives can bring IR laser imaging performance closer to high-resolution MSI. Near-field ablation is a novel approach for very small scale imaging and sampling. At the sub-micrometer scale, ionization and sampling efficiency are paramount issues and targeted approaches such as antibody labeling may be needed for them to achieve their full potential.
Table 1.
List of abbreviations
| Abbreviation | Term |
|---|---|
| SNOM | Scanning near-field optical microscopy |
| AFM | Atomic force microscopy |
| DESI | Desorption electrospray ionization |
| FTICR | Fourier transform ion cyclotron resonance |
| LAMMA | Laser microprobe mass analyzer |
| LDI | Laser desorption/ionization |
| MALDI | Matrix-assisted laser desorption/ionization |
| MS | Mass spectrometry |
| SIMS | Secondary ion mass spectrometry |
| TOF | Time-of-flight |
Highlights.
Mass spectrometry imaging using laser desorption and ablation is reviewed.
Lateral resolution of 10 μm for single cell imaging.
Laser microprobe, MALDI, ambient and near-field imaging.
High numerical aperuture reflective and through-hole objectives.
Oversampling, beam shaping, transmission geometry and microscope mode imaging.
7. Acknowledgements
This work was supported by the National Science Foundation (Grant CHE-1152106) and National Institutes of Health Grant Number R21DA035504.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chughtai K, Heeren RM. Chem. Rev. 2010;110:3237–3277. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Hove ERA, Smith DF, Heeren RM. J. Chromatogr. A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Heeren RMA. Int. J. Mass Spectrom. 2015;377:672–680. [Google Scholar]
- 4.Spengler B. Anal. Chem. 2015;87:64–82. doi: 10.1021/ac504543v. [DOI] [PubMed] [Google Scholar]
- 5.Boxer SG, Kraft ML, Weber PK. Annu. Rev. Biophys. 2009;38:53–74. doi: 10.1146/annurev.biophys.050708.133634. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher JS, Vickerman JC. Anal. Chem. 2013;85:610–639. doi: 10.1021/ac303088m. [DOI] [PubMed] [Google Scholar]
- 7.Gessel MM, Norris JL, Caprioli RM. J. Proteomics. 2014:1–12. doi: 10.1016/j.jprot.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norris JL, Caprioli RM. Chem. Rev. 2013;113:2309–2342. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C, Dill AL, Eberlin LS, Cooks RG, Ifa DR. Mass Spectrom. Rev. 2013;32:218–243. doi: 10.1002/mas.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding X, Duan Y. Mass Spectrom. Rev. 2013 doi: 10.1002/mas.21415. [DOI] [PubMed] [Google Scholar]
- 11.Castaing R, Slodzian G. J. Microscopie. 1962;1:395–410. [Google Scholar]
- 12.Benninghoven A. Angew. Chem. Int. Ed. 1994;33:1023–1043. [Google Scholar]
- 13.Winograd N. Anal. Chem. 2005;77:142 A–149 A. [Google Scholar]
- 14.Bich C, Touboul D, Brunelle A. Mass Spectrom. Rev. 2013 doi: 10.1002/mas.21399. [DOI] [PubMed] [Google Scholar]
- 15.Hillenkamp F, Karas M. Int. J. Mass Spectrom. 2000;200:71–77. [Google Scholar]
- 16.Verbueken AH, Bruynseels FJ, Van Grieken RE. Biomed. Mass Spectrom. 1985;12:438–463. doi: 10.1002/bms.1200120903. [DOI] [PubMed] [Google Scholar]
- 17.Caprioli RM, Farmer T, Gile J. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 18.Ogrinc Potoc nik N, Porta T, Becker M, Heeren RMA, Ellis SR. Rapid Commun. Mass Spectrom. 2015;29:2195–2203. doi: 10.1002/rcm.7379. [DOI] [PubMed] [Google Scholar]
- 19.Laiko VV, Baldwin MA, Burlingame AL. Anal. Chem. 2000;72:652–657. doi: 10.1021/ac990998k. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Shrestha B, Vertes A. Anal. Chem. 2007;79:523–532. doi: 10.1021/ac061577n. [DOI] [PubMed] [Google Scholar]
- 21.Brady JJ, Judge EJ, Levis RJ. Rapid Commun. Mass Spectrom. 2009;23:3151–3157. doi: 10.1002/rcm.4226. [DOI] [PubMed] [Google Scholar]
- 22.Trimpin S. J. Am. Soc. Mass Spectrom. 2015;27:4–21. doi: 10.1007/s13361-015-1253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cramer R, Pirkl A, Hillenkamp F, Dreisewerd K. Angew. Chem. Int. Ed. 2013;52:2364–2367. doi: 10.1002/anie.201208628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryumin P, Brown J, Morris M, Cramer R. Methods. 2016:1–10. doi: 10.1016/j.ymeth.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Nemes P, Vertes A. Meth. Mol. Biol. 2010;656:159–171. doi: 10.1007/978-1-60761-746-4_9. [DOI] [PubMed] [Google Scholar]
- 26.Galhena AS, Harris GA, Nyadong L, Murray KK, Fernandez FM. Anal. Chem. 2010;82:2178–2181. doi: 10.1021/ac902905v. [DOI] [PubMed] [Google Scholar]
- 27.Passarelli MK, Ewing AG. Curr. Opin. Chem. Biol. 2013;17:854–859. doi: 10.1016/j.cbpa.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubakhin SS, Lanni EJ, Sweedler JV. Curr. Opin. Biotechnol. 2013;24:95–104. doi: 10.1016/j.copbio.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoppe PS, Coutu DL, Schroeder T. Nat. Cell Biol. 2014;16:919–927. doi: 10.1038/ncb3042. [DOI] [PubMed] [Google Scholar]
- 30.Khoo BL, Chaudhuri PK, Ramalingam N, Tan DSW, Lim CT, Warkiani ME. Int. J. Cancer. 2016 doi: 10.1002/ijc.30006. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 31.Milo R. BioEssays. 2013;35:1050–1055. doi: 10.1002/bies.201300066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller BO, Li L. J. Am. Soc. Mass Spectrom. 2001;12:1055–1063. [Google Scholar]
- 34.Marginean I, Tang K, Smith RD, Kelly RT. J. Am. Soc. Mass Spectrom. 2013;25:30–36. doi: 10.1007/s13361-013-0749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang Z, Yin Z, Yang H, Xiao Y, Hang W, Li J. Trends Anal. Chem. 2016;75:24–34. [Google Scholar]
- 36.Luxembourg S, McDonnell L, Mize T, Heeren RM, Proteome Res J. 2005;4:671–673. doi: 10.1021/pr049762+. [DOI] [PubMed] [Google Scholar]
- 37.Soltwisch J, Göritz G, Jungmann JH, Kiss A, Smith DF, Ellis SR, Heeren RMA. Anal. Chem. 2014;86:321–325. doi: 10.1021/ac403421v. [DOI] [PubMed] [Google Scholar]
- 38.Becker J, Matusch A, Wu B. Anal. Chim. Acta. 2014;835:1–18. doi: 10.1016/j.aca.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 39.Di Palma S, Bodenmiller B. Curr Opin Biotechnol. 2015;31:122–129. doi: 10.1016/j.copbio.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Hare DJ, New EJ, de Jonge MD, McColl G. Chem. Soc. Rev. 2015 doi: 10.1039/c5cs00055f. [DOI] [PubMed] [Google Scholar]
- 41.Cotter RJ. Anal. Chim. Acta. 1987;195:45–59. [Google Scholar]
- 42.Conzemius RJ, Capellen JM. Int. J. Mass Spectrom. Ion Phys. 1980;34:197–271. [Google Scholar]
- 43.Fenner NC. Rev. Sci. Instrum. 1966;37:1068. [Google Scholar]
- 44.Hillenkamp F, Unsöld E, Kaufmann R, Nitsche R. Nature. 1975;256:119–120. doi: 10.1038/256119a0. [DOI] [PubMed] [Google Scholar]
- 45.Hillenkamp F, Unsold E, Kaufmann R, Nitsche R. Appl. Phys. 1975;8:341–348. [Google Scholar]
- 46.Böhm R, Kapr T, Schmitt HU, Albrecht J. J. Anal. Appl. Pyrol. 1985;8:448–461. [Google Scholar]
- 47.Holm R, Kaempf G, Kirchner D, Heinen HJ. Anal. Chem. 1984;56:690–692. [Google Scholar]
- 48.Vogt H, Heinen HJ, Meier S, Wechsung R. Fresenius Z. Anal. Chem. 1981;308:195–200. [Google Scholar]
- 49.Wilk ZA, Hercules DM. Anal. Chem. 1987;59:1819–1825. [Google Scholar]
- 50.Van Vaeck L, Struyf H, Van Roy W, Adams F. Mass Spectrom. Rev. 1994;13:189–208. [Google Scholar]
- 51.Brenna JT, Creasy WR, McBain W, Soria C. Rev. Sci. Instrum. 1988;59:873. [Google Scholar]
- 52.Pelletier M, Kreier G, Muller JF, Weil D, Johnston M. Rapid Commun. Mass Spectrom. 1988;2:146–150. [Google Scholar]
- 53.Struyf H, Van Roy W, Van Vaeck L, Van Griecken R. Anal. Chim. Acta. 1993;283:139–151. [Google Scholar]
- 54.Van Vaeck L, Van Roy W, Struyf H, Adams F, Caravatti P. Rapid Commun. Mass Spectrom. 1993;7:323–331. [Google Scholar]
- 55.Erdös P. JOSA. 1959;49:877–883. [Google Scholar]
- 56.Becker S. Inorganic Mass Spectrometry: Principles and Applications. John Wiley; 2008. [Google Scholar]
- 57.Dingle T, Griffiths BW, Ruckman JC. Vacuum. 1981 [Google Scholar]
- 58.Ruckman JC, Davey AR, Clarke NS. Vacuum. 1984;34:911–924. [Google Scholar]
- 59.Behm J, Hemminger J, Lykke KR. Anal. Chem. 1996;68:713–719. doi: 10.1021/ac9510999. [DOI] [PubMed] [Google Scholar]
- 60.Savina MR, Lykke KR. Anal. Chem. 1997;69:3741–3746. [Google Scholar]
- 61.Kaletaş B.a.K., Van Der Wiel IM, Stauber J, Dekker LJ, Güzel C.k., Kros JM, Luider TM, Heeren RM. Proteomics. 2009;9:2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 62.Zavalin A, Yang J, Hayden K, Vestal M, Caprioli RM. Anal. Bioanal. Chem. 2015;407:2337–2342. doi: 10.1007/s00216-015-8532-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Römpp A, Spengler B. Histochem. Cell Biol. 2013;139:759–783. doi: 10.1007/s00418-013-1097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prentice BM, Chumbley CW, Caprioli RM. J. Mass Spectrom. 2015;50:703–710. doi: 10.1002/jms.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Passarelli MK, Wang J, Mohammadi AS, Trouillon R, Gilmore I, Ewing AG. Anal. Chem. 2014;86:9473–9480. doi: 10.1021/ac501228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poetsch A, Schlüsener D, Florizone C, Eltis L, Menzel C, Rögner M, Steinert K, Roth U. J Biomol Tech. 2008;19:129–138. [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas A, Charbonneau JL, Fournaise E, Chaurand P. Anal. Chem. 2012;84:2048–2054. doi: 10.1021/ac2033547. [DOI] [PubMed] [Google Scholar]
- 68.Yang J, Caprioli RM. Anal. Chem. 2011;83:5728–5734. doi: 10.1021/ac200998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dufresne M, Thomas A, Breault-Turcot J, Masson J-F, Chaurand P. Anal. Chem. 2013;85:3318–3324. doi: 10.1021/ac3037415. [DOI] [PubMed] [Google Scholar]
- 70.Jurchen JC, Rubakhin SS, Sweedler JV. J. Am. Soc. Mass Spectrom. 2005;16:1654–1659. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 71.Thiery-Lavenant G, Zavalin AI, Caprioli RM. J. Am. Soc. Mass Spectrom. 2013;24:609–614. doi: 10.1007/s13361-012-0563-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spraggins JM, Caprioli RM. J. Am. Soc. Mass Spectrom. 2011;22:1022–1031. doi: 10.1007/s13361-011-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siegman AE. Bhowmik A, editor. OE/LASE'93: Optics, Electro-Optics, & Laser Applications in Science& Engineering, SPIE1993, pp. :2–12. [Google Scholar]
- 74.Dreisewerd K, Schuerenberg M, Karas M, Hillenkamp F. Int. J. Mass. Spectrom. and Ion Processes. 1995;141:127–148. [Google Scholar]
- 75.Qiao H, Spicer V, Ens W. 2008;22:2779–2790. doi: 10.1002/rcm.3675. [DOI] [PubMed] [Google Scholar]
- 76.Zavalin A, Yang J, Haase A, Holle A, Caprioli R. J. Am. Soc. Mass Spectrom. 2014;25:1–4. doi: 10.1007/s13361-014-0872-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kettling H, Vens-Cappell S, Soltwisch J, Pirkl A, Haier J, Müthing J, Dreisewerd K. Anal. Chem. 2014;86:7798–7805. doi: 10.1021/ac5017248. [DOI] [PubMed] [Google Scholar]
- 78.Musapelo T, Murray KK. J. Am. Soc. Mass Spectrom. 2013;24:1108–1115. doi: 10.1007/s13361-013-0631-z. [DOI] [PubMed] [Google Scholar]
- 79.Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM. J. Mass Spectrom. 2012;47:1473–1481. doi: 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- 80.Spengler B, Hubert M. J. Am. Soc. Mass Spectrom. 2002;13:735–748. doi: 10.1016/S1044-0305(02)00376-8. [DOI] [PubMed] [Google Scholar]
- 81.Bouschen W, Schulz O, Eikel D, Spengler B. 2010;24:355–364. doi: 10.1002/rcm.4401. [DOI] [PubMed] [Google Scholar]
- 82.Chaurand P, Schriver KE, Caprioli RM. J. Mass Spectrom. 2007;42:476–489. doi: 10.1002/jms.1180. [DOI] [PubMed] [Google Scholar]
- 83.Luxembourg SL, Mize TH, McDonnell LA, Heeren RMA. Anal. Chem. 2004;76:5339–5344. doi: 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- 84.Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Science. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 85.Huang M, Yuan C, Cheng S, Cho Y, Shiea J. Annu Rev Anal Chem. 2010;3:43–65. doi: 10.1146/annurev.anchem.111808.073702. [DOI] [PubMed] [Google Scholar]
- 86.Hsu C-C, Dorrestein PC. Curr Opin Biotechnol. 2014;31C:24–34. doi: 10.1016/j.copbio.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dill AL, Eberlin LS, Ifa DR, Cooks RG. Chem. Comm. 2011;47:2741–2746. doi: 10.1039/c0cc03518a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cooks RG, Ouyang Z, Takats Z, Wiseman J. Science. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 89.Van Berkel GJ, Pasilis SP, Ovchinnikova O. J. Mass Spectrom. 2008;43:1161–1180. doi: 10.1002/jms.1440. [DOI] [PubMed] [Google Scholar]
- 90.Shrestha B, Vertes A. Anal. Chem. 2009;81:8265–8271. doi: 10.1021/ac901525g. [DOI] [PubMed] [Google Scholar]
- 91.Shelley JT, Ray SJ, Hieftje GM. Anal. Chem. 2008;80:8308–8313. doi: 10.1021/ac801594u. [DOI] [PubMed] [Google Scholar]
- 92.Gray AL. The Analyst. 1985;110:551–556. [Google Scholar]
- 93.Russo RE, Mao X, Liu H, Gonzalez J, Mao S. Talanta. 2002 doi: 10.1016/s0039-9140(02)00053-x. [DOI] [PubMed] [Google Scholar]
- 94.Russo RE, Mao X, Gonzalez JJ, Zorba V, Yoo J. Anal. Chem. 2013;85:6162–6177. doi: 10.1021/ac4005327. [DOI] [PubMed] [Google Scholar]
- 95.Coon JJ, McHale KJ, Harrison WW. Rapid Commun. Mass Spectrom. 2002;16:681–685. doi: 10.1002/rcm.626. [DOI] [PubMed] [Google Scholar]
- 96.Nyadong L, Galhena AS, Fernandez FM. Anal. Chem. 2009;81:7788–7794. doi: 10.1021/ac9014098. [DOI] [PubMed] [Google Scholar]
- 97.Vaikkinen A, Shrestha B, Kauppila TJ, Vertes A, Kostiainen R. Anal. Chem. 2012;84:1630–1636. doi: 10.1021/ac202905y. [DOI] [PubMed] [Google Scholar]
- 98.Shiea J, Huang M-Z, Hsu H-J, Lee C-Y, Yuan C-H, Beech I, Sunner J. Rapid Commun. Mass Spectrom. 2005;19:3701–3704. doi: 10.1002/rcm.2243. [DOI] [PubMed] [Google Scholar]
- 99.Sampson J, Hawkridge AM, Muddiman DC. J. Am. Soc. Mass Spectrom. 2006;17:1712–1716. doi: 10.1016/j.jasms.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 100.Nemes P, Vertes A. Methods Mol. Biol. 2010;656:159–171. doi: 10.1007/978-1-60761-746-4_9. [DOI] [PubMed] [Google Scholar]
- 101.Laiko VV, Taranenko NI, Berkout VD, Yakshin MA, Prasad CR, Lee HS, Doroshenko VM. J. Am. Soc. Mass Spectrom. 2002;13:354–361. doi: 10.1016/s1044-0305(02)00341-0. [DOI] [PubMed] [Google Scholar]
- 102.Coello Y, Jones AD, Gunaratne TC, Dantus M. Anal. Chem. 2010;82:2753–2758. doi: 10.1021/ac9026466. [DOI] [PubMed] [Google Scholar]
- 103.Creaser CS, Ratcliffe L. Curr. Anal. Chem. 2006;2006;29-15(2):9–15. [Google Scholar]
- 104.Koestler M, Kirsch D, Hester A, Leisner A, Guenther S, Spengler B. Rapid Commun. Mass Spectrom. 2008;22:3275–3285. doi: 10.1002/rcm.3733. [DOI] [PubMed] [Google Scholar]
- 105.Guenther S, Römpp A, Kummer W, Spengler B. Int. J. Mass Spectrom. 2011;305:228–237. [Google Scholar]
- 106.Khalil SM, Römpp A, Pretzel J, Becker K, Spengler B. Anal. Chem. 2015;87:11309–11316. doi: 10.1021/acs.analchem.5b02781. [DOI] [PubMed] [Google Scholar]
- 107.Cahill JF, Kertesz V, Van Berkel GJ. Anal. Chem. 2015;87:11113–11121. doi: 10.1021/acs.analchem.5b03293. [DOI] [PubMed] [Google Scholar]
- 108.Novotny L. Prog. Optics. 2007;50:137–184. [Google Scholar]
- 109.Cleveland D, Michel RG. Appl. Spectrosc. Rev. 2008;43:93–110. [Google Scholar]
- 110.Novotny L, Stranick SJ. Annu. Rev. Phys. Chem. 2006;57:303–331. doi: 10.1146/annurev.physchem.56.092503.141236. [DOI] [PubMed] [Google Scholar]
- 111.Jalili N, Laxminarayana K. Mechatronics. 2004;14:907–945. [Google Scholar]
- 112.Kossakovski DA, O'Connor SD, Widmer M, Baldeschwieler JD, Beauchamp JL. Ultramicrosc. 1998 [Google Scholar]
- 113.Bradshaw JA, Ovchinnikova OS, Meyer KA, Goeringer DE. Rapid Commun. Mass. Spectrom. 2009;23:3781–3786. doi: 10.1002/rcm.4313. [DOI] [PubMed] [Google Scholar]
- 114.Meyer KA, Ovchinnikova OS, Ng K, Goeringer DE. Rev. Sci. Instrum. 2008;79:123710. doi: 10.1063/1.3053200. [DOI] [PubMed] [Google Scholar]
- 115.Schmitz TA, Gamez G, Setz PD, Zhu L, Zenobi R. Anal. Chem. 2008;80:6537–6544. doi: 10.1021/ac8005044. [DOI] [PubMed] [Google Scholar]
- 116.Zhu L, Gamez G, Schmitz T, Krumeich F, Zenobi R. Anal. Bioanal. Chem. 2010;396:163–172. doi: 10.1007/s00216-009-2919-1. [DOI] [PubMed] [Google Scholar]
- 117.Zhu L, Stadler J, Schmitz TA, Krumeich F, Zenobi R. J. Phys. Chem. 2011;C(115):1006–1013. [Google Scholar]
- 118.Ghorai S, Seneviratne CA, Murray KK. J. Am. Soc. Mass Spectrom. 2015;26:63–70. doi: 10.1007/s13361-014-1005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Murray KK, Ghorai S, Seneviratne CA. MRS Proc. 2015;1754:mrsf14–1754-pp1708-1704. [Google Scholar]
- 120.Jackson SN, Wang H-YJ, Woods AS. J. Am. Soc. Mass Spectrom. 2005;16:2052–2056. doi: 10.1016/j.jasms.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 121.Park S-G, Murray KK. Anal. Chem. 2012;84:3240–3245. doi: 10.1021/ac3006704. [DOI] [PubMed] [Google Scholar]
- 122.Hattori K, Kajimura M, Hishiki T, Nakanishi T, Kubo A, Nagahata Y, Ohmura M, Yachie-Kinoshita A, Matsuura T, Morikawa T, Nakamura T, Setou M, Suematsu M. Antioxid. Redox Signal. 2010;13:1157–1167. doi: 10.1089/ars.2010.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Quanico J, Franck J, Gimeno JP, Sabbagh R, Salzet M, Day R, Fournier I. Chem. Com. 2015;51:4564–4567. doi: 10.1039/c4cc08331h. [DOI] [PubMed] [Google Scholar]
- 124.Nilsson A, Fehniger TE, Gustavsson L, Andersson M, Kenne K, Marko-Varga G, Andren PE. PLoS One. 2010;5:e11411–11418. doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pirman DA, Yost RA. Anal. Chem. 2011;83:8575–8581. doi: 10.1021/ac201949b. [DOI] [PubMed] [Google Scholar]
- 126.Pirman DA, Reich RF, Kiss A, Heeren RMA, Yost RA. Anal. Chem. 2013;85:1081–1089. doi: 10.1021/ac302960j. [DOI] [PubMed] [Google Scholar]
- 127.Sun N, Ly A, Meding S, Witting M, Hauck SM, Ueffing M, Schmitt-Kopplin P, Aichler M, Walch A. Proteomics. 2014;14:913–923. doi: 10.1002/pmic.201300407. [DOI] [PubMed] [Google Scholar]
- 128.Cole LM, Bluff JE, Carolan VA, Paley MN, Tozer GM, Clench MR. Proteomics. 2014;14:890–903. doi: 10.1002/pmic.201300429. [DOI] [PubMed] [Google Scholar]
- 129.Berisha A, Dold S, Guenther S, Desbenoit N, Takáts Z, Spengler B, Römpp A. Rapid Commun. Mass Spectrom. 2014;28:1779–1791. doi: 10.1002/rcm.6960. [DOI] [PubMed] [Google Scholar]
- 130.Prideaux B, Elnaggar MS, Zimmerman M, Wiseman JM, Li X, Dartois V. Int. J. Mass Spectrom. 2014:1–10. doi: 10.1016/j.ijms.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Johann DJ, Rodriguez-Canales J, Mukherjee S, Prieto DA, Hanson JC, Emmert-Buck M, Blonder J. J. Proteome Res. 2009;8:2310–2318. doi: 10.1021/pr8009403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.He J, Zhu J, Liu Y, Wu J, Nie S, Heth JA, Muraszko KM, Fan X, Lubman DM. Electrophoresis. 2013;34:1627–1636. doi: 10.1002/elps.201200566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhu J, Nie S, Wu J, Lubman DM. J. Proteomics Res. 2013;12:2791–2804. doi: 10.1021/pr400139c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hebbar S, Schulz WD, Sauer U, Schwudke D. Anal. Chem. 2014;86:5345–5352. doi: 10.1021/ac500276r. [DOI] [PubMed] [Google Scholar]
- 135.Sethi S, Vrana JA, Theis JD, Leung N, Sethi A, Nasr SH, Fervenza FC, Cornell LD, Fidler ME, Dogan A. Kidney Int. 2012;82:226–234. doi: 10.1038/ki.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Donnarumma F, Murray KK. J. Mass Spectrom. 2015 doi: 10.1002/jms.3744. in press. [DOI] [PubMed] [Google Scholar]
- 137.Donnarumma F, Cao F, Murray KK. J. Am. Soc. Mass Spectrom. 2016 doi: 10.1007/s13361-015-1249-0. 10.1007/s13361-015-1249-0. in press. [DOI] [PubMed] [Google Scholar]
- 138.Franck J, Quanico J, Wisztorski M, Day R, Salzet M, Fournier I. Anal. Chem. 2013;85:8127–8134. doi: 10.1021/ac4009397. [DOI] [PubMed] [Google Scholar]