Abstract
This paper focuses on the ability of a folate-decorated triblock copolymer to deliver a targeted dose of siRNA in order to overcome chemotherapy resistance which can commonly cause complications in ovarian cancer patients. The micelleplexes that are formed upon electrostatic interaction with siRNA are used to deliver siRNA in a targeted manner to SKOV-3 ovarian cancer cells that overexpress folate receptor-α (FRα). The triblock copolymer consists of polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) (PEI-g-PCL-b-PEG-Fol). In this work, polymers of different molecular weights of PEG, as well as different grafting degrees of the (g-PCL-b-PEG-Fol) chains to PEI, were analyzed to optimize targeted siRNA delivery. The polymers, their micelleplexes, and the in vitro performance of the latter were characterized by nuclear magnetic resonance, dynamic light scattering, transmission electron microscopy, flow cytrometry, western blot, confocal microscopy, and in luciferase assays. The different PEI-g-PCL-b-PEG-Fol conjugates showed suitable sizes below 260 nm, especially at N/P 5, which also allowed for full siRNA condensation. Furthermore, flow cytometry and Western blot analysis demonstrated that our best polymer was able to effectively deliver siRNA and that siRNA delivery resulted in efficient protein knockdown of toll-like receptor 4 (TLR4). Consequently, TLR4 knock down within SKOV-3 cells resensitized them toward paclitaxel (PTX) treatment, and apoptotic events increased. This study demonstrates that PEI-g-PCL-b-PEG-Fol conjugates are a reliable delivery system for siRNA and are able to mediate therapeutic protein knockdown within ovarian cancer cells. Additionally, this study provides further evidence to link TLR4 levels to chemotherapy resistance.
Graphical abstract
INTRODUCTION
While healthy tissues generally do not express an abundance of folate receptor-α (FRα), several cancers have been found to significantly overexpress FRα. Most notable, in approximately 85–90% of ovarian cancers, there is an overexpression of FRα with an increasing expression as the histological grade of the cancer increases.1–4 Outside of a full oophorectomy for early stage patients, treatments for late stage ovarian cancer includes radiation and a combination of platinum and taxane chemotherapeutic agents. Often times, late stage ovarian cancer patients experience a reoccurrence of their disease where resistance to first line treatment is commonly seen.5 To overcome challenges seen within the clinic, such as chemotherapy resistance, relapse of the disease, and off target toxicity, we are taking advantage of the overly expressed FRα commonly observed in ovarian cancer patients by using folate receptor targeted nanoparticles. Targeted nanoparticle delivery, formulated and designed specifically for enhanced tumor targeting and uptake tackling chemoresistance could therefore become a novel approach for treating refractory ovarian cancer.
A new theory to treating cancer eventually within the clinic is the use of nanoparticles to deliver a targeted payload to the tumor, while decreasing uptake of the drug inside healthy tissues. Both Doxil and Abraxane are nanopartcles that are currently being used within the clinic to treat cancer.6,7 However, both of these nanoparticle formulations solely rely on the enhanced permeation and retention effect (EPR effect) to passively target the tumor by means of extravasation out of the tumor’s leaky blood vasculature.8,9 A targeted delivery, such as demonstrated within this paper, can be achieved by attaching a targeting ligand to the surface of the nanoparticle to increase its interaction with the cell.10–13 The folate receptor is an excellent receptor to target due to its nature of receptor–ligand interaction. FRα is an internalizing transmembrane receptor which will endocytose once folic acid, its ligand, binds, and the receptor–ligand complex is internalized. The ligand, and anything that is conjugated to it, is subsequently deposited into the cytoplasm, while the receptor is recycled back to the cell surface.14–16 This provides a selective gate to deliver chemotherapeutics, but even macromolecules such as therapeutic RNA (siRNA), into the cytoplasm of the cell where they can take effect.14 In order to overcome the hurdles commonly seen with ovarian cancer treatment, such as relapse and resistance, a wide variety of combinational therapies that include siRNA are currently being studied.17–23 However, our own approach incorporates targeted delivery of siRNA to ovarian cancer cells for therapeutic knock down of specific oncogenes that give rise to chemotherapy resistance, such as toll-like recepter 4 (TLR4).24–27 We hypothesize that knock down of these proteins resensitizes ovarian cancer cells toward first-line chemotherapeutic agents. Our results show that folate decorated nanoparticles can effectively deliver siRNA into the cancer cells and achieve a drastic and sustained knockdown of TLR4. Our approach of using a triblock copolymer that consists of polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol), or folate coupled PEI-g-PCL-b-PEG-Fol, overcomes typical obstacles of siRNA delivery, such as rapid clearance and degradation in circulation. PEI-g-PCL-b-PEG polymers have been shown to form stable micelles with siRNA that exhibit enhanced circulation time, and folate coupled PEI-g-PCL-b-PEG-Fol conjugates have been reported to transfect receptor overexpressing cells in a targeted manner.28–35 Within the polymer, PEI electrostatically condenses and shields the siRNA from degradation by nucleases, while the conjugated folic acid ligand on the particle surface gives specificity toward cells that overexpress FR. In addition, the PCL block increases the hydrophobic content of the nanoparticle, which forms the inner core of the micelle where paclitaxel (PTX) can be encapsulated for combination therapy with the same particle.28 Lastly, the addition of PEG increases the biocompatibility and acts as a stealth mechanism to avoid macrophage detection of the nanoparticles.32 Collectively, these four components are hypothesized to effectively encapsulate their payload and yield a targeted delivery to the cancer cells of interest.
Altogether, our strategy within this project is to create an effective, targeted siRNA therapy to meet the following goals: (1) develop a biocompatible folate-decorated nanoparticle, which can deliver siRNA in a targeted fashion to ovarian cancer cells that overexpress FRα; (2) achieve a targeted tumor uptake and specificity; (3) accomplish increased pharmacokinetic parameters such as bioavailability and prolonged circulation; and (4) overcome the barrier of chemotherapy resistance. In response to the aforementioned, we hypothesize that by effectively delivering siRNA against TLR4 with our folate-conjugated triblock copolymer to ovarian cancer cells, we can achieve a targeted therapeutic effect in FR-overexpressing cells, decrease off-target toxicity, and overcome chemotherapy resistance in combination with PTX.
4. EXPERIMENTAL SECTION
Materials
Heterobifunctional poly(ethylene glycol) (HO-PEG-COOH, 3.5 and 5 kDa), as well as monofunctional (CH3-PEG-COOH, 5 kDa) was purchased from JenKem Technologies (Plano, TX, U.S.A.). Hyperbranched polyethylenimine (hyPEI, 25k Da) was obtained from BASF (Ludwigshafen, Germany). All other reagents for synthesis were obtained from Sigma-Aldrich (St. Louis, MO, U.S.A.) and used without further modification. Dicer substrate double-stranded siRNA (DsiRNA) targeting the enhanced green fluorescent protein gene (EGFP siRNA, 25/27), siRNA for toll-like receptor 4 (TLR4), and a scrambled nonspecific control (siNegCon) DsiRNA as well as Alexa Fluor-488 and TYE-563 labeled siRNA were purchased from Integrated DNA Technologies (IDT, Coralville, IA, U.S.A.). Dulbecco’s Modified Eagle’s Medium (10×) without folic acid, Dulbecco’s phosphate buffered saline (PBS), heat-inactivated fetal bovine serum (FBS), d-(+)-glucose, sodium bicarbonate, sodium pyruvate, 2-mercaptoethanol, dimethyl sulfoxide (DMSO, ≥ 99.7%), ethylenediaminetetraacetic acid (EDTA, 99.4–100.06%), trypan blue (0.4%, sterile filtered), and luciferin were purchased from Sigma-Aldrich (St. Louis, MO, U.S.A.).
Synthesis of Triblock Copolymers and Characterization
The overall reaction scheme, adjusted from Liu et al.,36 can be found in the Supporting Information, Scheme 1. The triblock copolymers were synthesized by a six step reaction process consisting of coupling an azido functionalized folic acid (molecule A) with a heterobifunctional acrylate-PCL-b-PEG-alkyne (molecule B) via click chemistry reaction. This was followed by coupling the previous product of acrylate-PCL-b-PEG-Fol (molecule C) with hy-PEI (25 kDa), as previously described.36 A total of six different conjugates were synthesized consisting of two different PEG lengths (3.5 kDa or 5 kDa), varying grafting densities of PCL-b-PEG-Fol (10 or 30 µmol per 10 µmol of PEI), as well as one null folate conjugate (Table 1). Compounds synthesized were characterized by 1HNMR, UV spectroscopy, and a folate composition assay.
Table 1.
All Six Conjugates Synthesized with the Proposed Scheme
| name | PEG chain size (kDa) |
PCL chain size (kDa) |
hy-PEI (kDa) |
feed ratio of PCL-b-PEG(-folate) (µmol) |
statistical (final grafting degree) |
folate (mol per mg polymer) |
|---|---|---|---|---|---|---|
| 3.5k 10 µmol | 3.5 | 1000 | 25 | 10 | 5 | 2.4 × 10−06 |
| 3.5k 30 µmol | 3.5 | 1000 | 25 | 30 | 5.5 | 3.1 × 10−06 |
| 5k 10 µmol | 5 | 1000 | 25 | 10 | 5 | 1.2 × 10−05 |
| 5k 30 µmol | 5 | 1000 | 25 | 30 | 4.7 | 8.9 × 10−06 |
| mixed conjugate |
3.5 and 5 | 1000 | 25 | 10 | 0.5 | 3.9 × 10−05 |
| null folate | 5 | 1000 | 25 | 10 | 2.5 | 0 |
Folate Composition Assay
A UV spectroscopy assay used for determining the folic acid concentration within each sample. Each sample was read in triplicates on a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT, U.S.A.). Folic acid standards were dissolved in DMSO at a concentration of 2, 1, 0.5, 0.25, 0.125, and 0 mg/mL. Conjugates were weighed out and dissolved in water. A 100 µL aliquot of each sample was added to a 96 transparent well plate and read at 360 nm. Blank values of DMSO and water were used to eliminate any background signal. Results were analyzed by Graphpad Prism 5.0 and are displayed as mean values.
Cell Culture
SKOV-3 cells are a human ovarian cancer cell line and were obtained from ATTC (LG Promochem, Wesel, Germany). The SKOV-3/LUC cell line was transfected to stably express the reporter gene luciferase as described before.37 SKOV-3 ovarian cancer cells were cultured in folate free DMEM cell culture medium (Sigma-Aldrich) supplemented with 0.584 g/L of l-glutamine, 3.7 g/L sodium bicarbonate, 10% fetal bovine serum (Thermo Scientific Hyclone), and 1% penicillin/streptomycin. Cells were grown in 75 cm2 cell culture flasks (Thermo Scientific) at 37 °C and 5% CO2, and passaged every 2–3 days when they had reached confluency.
A549 cells are a human adenocarcinoma alveolar based lung cancer cell line and were obtained from ATTC (LG Promochem, Wesel, Germany). A549 lung adenocarcinoma cells were cultured in DMEM cell culture medium (Sigma-Aldrich) and supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were grown in 75 cm2 cell culture flasks at 37 °C and 5% CO2 and passaged twice a week when they reached confluency.
Preparation of PEI-g-PCL-b-PEG-Fol Micelleplexes
Each polymer was dissolved in water to yield a 1 mg/mL concentration based upon the PEI 25 kDa content. Once dissolved, samples were filtered through a 0.22 µm filter for sterilization. In order to prepare the micelleplexes, a specific ratio between the amine groups found within the polymer (N) and the phosphate groups of the siRNA (P) was chosen. The N/P ratios were calculated based upon the formula seen below:
In the equation listed above, m refers to the mass of the polymer needed, n refers to the amount of siRNA used per well, 52 represents the amount of phosphate groups within one 25/27 nucleotide siRNA molecule, the MW represents the molecular weight of the protonable unit found within the polymer (43.1 g/mol for hy-PEI), and the N/P is the desired ratio between amine groups on the polymer and phosphate groups on the siRNA.
SYBR Gold and Heparin Assays
SYBR gold assay were used to assess the ability of each conjugate to successfully condense siRNA at varying N/P ratios (0–20). SYBR Gold is a fluorescent dye that intercalates with uncomplexed double-stranded nucleic acids and experiences a more than 1000-fold fluorescence enhancement upon intercalation. However, once siRNA is condensed within the micelleplex, the dye can no longer intercalate and exhibits very weak fluorescence. All conjugates were tested against hy-PEI (25 kDa) as positive control. At varying N/P ratios (0–20), 50 µL of polymer dilution and 50 µL of 1 µM EGFP siRNA was added to each well in a total of 100 µL of a 5% glucose solution. Once mixed, solutions were incubated for 20 min at room temperature. Afterward, 30 µL of a 4× SYBR Gold solution (Life Technologies, Carlsbad, CA, U.S.A.) was added to each well and incubated in the dark for 10 min at room temperature. Samples were measured in triplicates for fluorescence at 495 (excitation) and 537 nm (emission) on a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT, U.S.A.). Samples were normalized based on the following criteria. The fluorescence level of 100% free siRNA was calculated based on the fluorescence of free siRNA, noncondensed, with SYBR gold dye. The fluorescence of zero percent free siRNA was calculated with SYBR gold dye in glucose solution only at the absence of siRNA. Results were analyzed by Graphpad Prism 5.0 and are displayed as mean values and standard deviation.
Similarly, heparin assays were used to determine the stability of the micelleplexes, the conjugates formed at a physiologically relevant pH (7.4), as well as at a lower pH, to resemble the late endosome (4.5). The lowest N/P ratios that showed full condensation for each polymer (N/P 5) were used for testing the stability against increasing amounts of heparin (0–1.0 IU). The samples at pH 7.4 were made in 5% glucose solution, while the samples measured at pH 4.5 were made in sodium acetate buffer. Samples were prepared as described for the SYBR gold assay with the exception of adding 10 µL of heparin solution at various concentrations (0–1 international unit (IU) per well). Samples were incubated for varying time points (30 min-4 h) at room temperature. Afterward, fluorescence was measured in triplicates at 495 nm (excitation) and 537 nm (emission) on a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT, U.S.A.). Results of the heparin assays were analyzed as described for the SYBR gold assays.
Hydrodynamic Diameter and Zeta (ζ) Potential Measurements
Measurements of the hydrodynamic diameters of micelleplexes were performed by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments Inc., Malvern, U.K.). Micelleplexes were made as described above and measured at N/P 5, 6, and 7, complexing 40 pmol of scrambled siRNA. Samples were diluted with 5% glucose to a total volume of 75 µL within a disposable cuvette. Each sample was read in triplicates with each run consisting of 15 scans. Results are represented as average size (nm) ± standard deviation. The samples were then diluted with 5% glucose to 800 µL and transferred to disposable capillary cells, and ζ-potential measurements were taken. ζ-Potential measurements were read in triplicates by laser doppler anemometry (LDA), with each run consisting of 30 scans. Results are shown in mV ± standard deviation.
Transmission Electron Microscopy (TEM)
Transmission electron microscopy was used in order to assess the size and morphology of the micelleplexes after siRNA condensation. For TEM analysis, micelleplexes were made as described above in a total volume of 20 µL of 5% glucose containing 20 pmol of siRNA. Samples were added dropwise to a copper-coated grid, air-dried, and imaged with a transmission electron microscope (JEOL 2010 TEM, JEOL, Tokyo, Japan).
Cellular Uptake of Micelleplexes by Flow Cytometry
In 24-well plates (Corning Incorporated, Corning, NY), 60000 SKOV-3 cells were seeded and incubated overnight at 37 °C and 5% CO2. In each experiment, freshly made micelleplexes containing 50 pmol of AF488 siRNA at varying N/P ratios were added per well. Negative controls consisted of blank/untreated cells, and cells treated with free siRNA. Positive control cells were transfected with Lipofectamine 2000 (Life Technologies, Carlsbad, CA, U.S.A.) following standard protocol. Unless otherwise stated, cells were transfected for 4 h in 37 °C and 5% CO2 with 50 µL of micelleplexe solution containing 50 pmol siRNA within a total volume of 500 µL of serum containing cell culture media. In order to quench any extracellular fluorescence, the cells were incubated with 100 µL of 0.4% Trypan Blue (Fisher Scientific). Results were compared between cells treated with and without Trypan Blue in order to gain insight on each polymer’s uptake profile. Cells were then washed twice with 1× PBS + 2 mM EDTA, trypsinized and spun down at 350 g for 5 min. After centrifugation, the supernatant was decanted, and the cells were washed twice with 1× PBS + 2 mM EDTA. Samples were analyzed via flow cytometry (Applied Biosystems Attune Acoustic Focusing Cytometer), and the median fluorescence intensity (MFI) was collected and recorded. Samples were run in triplicates, with each sample consisting of a minimum of 10000 viable cells. The siRNA was excited at 488 nm, and emission detected using a 530/30 band-pass filter set. Analysis and presentation of the data was performed by GraphPad Prism 5.0 software calculating mean values and standard deviation.
Monensin Assay
To determine the extent of siRNA being trapped within the endosome, a monensin assay was utilized using flow cytometry. In 24-well plates, 60000 SKOV-3 cells were seeded and incubated overnight at 37 °C and 5% CO2. Freshly made micelleplexes containing 50 pmol of AF488 siRNA were added per well. Negative controls consisted of blank/untreated cells. Cells were transfected for 24 h in 37 °C and 5% CO2 with 50 µL of micelleplexe solution containing 50 pmol siRNA within a total volume of 500 µL of serum containing cell culture media. In order to quench any extracellular fluorescence, certain cells were incubated with 100 µL of 0.4% Trypan Blue, while others were treated with monensin. Results were compared between cells treated with and without Trypan Blue and monensin in order to gain insight on each polymer’s uptake profile. Cells were then washed twice with 1× PBS + 2 mM EDTA, trypsinized, and spun down at 350 g for 5 min. After centrifugation, the supernatant was decanted, and the cells were washed once with 1× PBS + 2 mM EDTA and incubated at 4 °C for 30 min with 50 µM monensin. Afterward, cells were washed once with 1× PBS + 2 mM EDTA and were analyzed via flow cytometry; the median fluorescence intensity (MFI) was collected and recorded for each sample. Samples were run in triplicates, with each sample consisting of a minimum of 10000 viable cells. The siRNA was excited at 488 nm, and emission detected using a 530/30 band-pass filter set. Analysis and presentation of the data was performed by GraphPad Prism 5.0 software calculating mean values and standard deviation.
Confocal Scanning Laser Microscopy
SKOV-3 cells were seeded in a Permanox 8 chamber slide (Nunc, Fisher Scientific, Waltham, MA, U.S.A.) at a density of 25000 cells in a total volume of 300 µL and allowed to incubate overnight in 37 °C and 5% CO2. Micelleplexes were made as described above using 40 pmol of labeled siRNA. After incubating the cells with the micelleplexes for 4, 12, or 24 h, the supernatant was decanted. Following, the cells were washed with 300 µL of PBS for 2–3 min each while shaking. Afterward, cells were fixed with 4% paraformaldehyde solution in PBS for 20 min at room temperature. This solution was then discarded, and cells were washed twice with 300 µL of PBS for 2–3 min each while shaking. The nucleus was stained with DAPI at a concentration of 175 ng/mL (Life Technologies, Carlsbad, CA, U.S.A.) for 20 min at room temperature while shaking. Cells were then washed twice with 300 µL of PBS. The chambers were then removed, the slides were blotted to remove any excess wash solution with a Kimwipe, and 1 drop of Fluorsave (CalBiochem, San Diegeo, CA, U.S.A.) was added per coverslip. The coverslips were mounted and let sit for 1–2 h in the dark. For excitation of TYE-563, an excitation wavelength of 570 nm was used while emission was detected with a spectral detector at 590 nm. DAPI staining was excited with a UV laser that had an excitation wavelength of 364 nm, and emission was detected at 385 nm. Images were recorded using a Zeiss LSM 780 confocal microscope and overlaid with brightfield light to gain information about cellular structures.
Protein Knockdown by Western Blot and Luciferase Assay
SKOV-3 and A549 cells were seeded in 6 well plates with a seeding density of 300000 cells per well and allowed to attach overnight at 37 °C and 5% CO2. For the assessment of TLR4 expression, SKOV-3 and A549 cells were harvested after 24 h of incubation. For transfection of SKOV-3 cells, micelleplexes were formed as previously described containing 100 pmol of TLR4 targeted siRNA. Cells were incubated with micelleplexes for 48 and 72 h to assess protein knockdown. Afterward, they were washed twice with ice cold PBS followed by a 2 min incubation in 100 µL of RIPA lysis buffer. The lysis solution was collected, transferred to a conical tube, and pipetted up and down to ensure complete cell lysis. Cells were incubated on ice for 30 min, followed by sonication and centrifugation at 15000 g for 20 min at 4 °C. Once centrifuged, the pellet was discarded and samples were analyzed via a Pierce BCA Protein Assay Kit (Life Technologies, Carlsbad, CA, U.S.A.) to determine the protein concentration.
The samples were prepared for loading the 10% polyacrylamide gel by denaturing 30 µg of protein by adding 1× final concentration loading buffer, β-mercaptoethanol, and by diluting them to 35 µL with RIPA buffer. Samples were added to a dry heat bath (at 95 °C) for 5 min before loading onto the gel. Once loaded, samples were run at 110 V for approximately 1–2 h at room temperature. The gel was then transferred to a membrane by running at 0.4 Å current for 1 h at room temperature. In order to keep the box cold, it was placed on ice. The membrane was then blocked in 5% milk in phosphate buffered saline containing Tween-20 (PBST) for 1 h at room temperature, followed by overnight incubation with 1:1000 diluted primary anti-TLR4 antibody 76B357.1 (abcam, Cambridge, MA, U.S.A.) at 4 °C. On the next day, the membrane was washed three times for 10 min each with PBST, followed by incubation with a secondary antibody goat antimouse IgG-HRP SC-2005 diluted 1:10,000 (Santa Cruz Biotechnology, Dallas, Texas, U.S.A.). Samples were incubated at room temperature for 1 h. This was last followed by another set of three washes of PBST for 10 min each. Afterward, the membrane was imaged using an ImageQuant LAS4000. Membranes were also probed for β-actin with a mouse monoclonal antibody 3700P (Cell Signaling Technology, Beverly, Massachusetts, U.S.A.) in order to test for proper loading controls.
Protein knockdown was also measured with a luciferase knockdown experiment. SKOV-3/LUC cells which have a NF-KB binding site on the CMV luciferase promoter were used in this experiment. In 24-well plates, 60000 SKOV-3/LUC cells were seeded per well and incubated overnight at 37 °C and 5% CO2. Micelleplexes were made as described previously with 50 pmol of TLR4 targeted or scrambled siRNA. Cells were incubated at 37 °C and 5% CO2 with the micelleplexes for 24 h before adding 1000 nM paclitaxel to each well for an incubation period of 48 h. After 48 h, cells were washed twice with 200 µL of PBS and treated with 300 µL of lysis buffer (Cell Culture Lysis Reagent, CCLR, Promega, Madison, WI, U.S.A.) per well. Each well was scraped with a pipet to effectively dislodge cell debris on the bottom of the well. The plate was then rocked for 5 min at room temperature. Cell lystates were transferred to conical tubes and set on ice. Each tube was vortexed for 10–15 s and then centrifuged at 12000 g for 2 min at 4 °C. The supernatant was collected, and 20 µL of each sample was added to a white 96-well plate to be analyzed for luminescence using a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT, U.S.A.). Each well was injected with 100 µL of luciferase assay reagent containing 10 mM luciferin (Sigma-Aldrich, St. Louis, MO, U.S.A.) by the plate reader immediately before the measurement. Samples were measured in triplicates and analyzed using GraphPad Prism 5.0 software.
MTT Assays
SKOV-3 cells were seeded at a density of 10000 cells per well in 200 µL of medium within a 96-well plate and incubated overnight at 37 °C and 5% CO2. Lipofectamine and each conjugate were diluted to varying concentrations from 0 to 16 µg mL−1 and added to the SKOV-3 cells and incubated for 24 h at 37 °C and 5% CO2. Upon 24 h of incubation, 20 µL of a sterile 5 mg/mL MTT solution was added to the cells and incubated for 4 h at 37 °C and 5% CO2. The media was then removed and 200 µL of DMSO was added to each well for 10 min. The plates absorbance was read at 540 nm using a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT, U.S.A.). The percentage of viable cells was calculated by the ratio of absorbance of treated cells compared with untreated cells. Samples were measured in triplicates and analyzed using GraphPad Prism 5.0 software.
For assessment of resensitization of SKOV-3 cells toward PTX, SKOV-3 cells were transfected with the micelleplex containing 780 nmol TLR4 siRNA within a T-75 flask for 24 h at 37 °C and 5% CO2, day 0. On day 1, 6000 SKOV-3 cells were seeded per well within a 96-well plate for the MTT assay in 200 µL of media. Subsequently, on day 2, the cells were treated with PTX at concentrations ranging from 0 to 1000 µM for 48 h in 200 µL of media. Upon 48 h incubation, on day 5, 20 µL of a sterile 5 mg/mL MTT solution was added to the cells in serum-free media and incubated for 4 h at 37 °C and 5% CO2. The media was then removed and 200 µL of DMSO was added to each well for 10 min. The absorbance of each well was read at 540 nm using a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT, U.S.A.). The data was analyzed with GraphPad Prism 5.0 software for IC50 values.
Annexin Assays
SKOV-3 cells were seeded at 60000 cells per well in 24-well plates and incubated overnight at 37 °C and 5% CO2. Micelleplexes were made as described previously with 50 pmol of TLR4 targeted or scrambled siRNA. Cells were incubated at 37 °C and 5% CO2 with the micelleplexes for 24 h before adding paclitaxel (0–1000 nM). Cells were incubated for another 48 h. Afterward, cells were washed with PBS while keeping each supernatant. Cells were trypsinized for 3–5 min, fresh media was added to each well, and the supernatant was added to each tube. Samples were centrifuged at 350 g for 10 min, and the supernatants were decanted. According to the Alexa Fluor 488 Annexin V/Dead Cell Apoptosis Kit, 1× Annexin Binding Buffer was prepared consisting of 10 mM HEPES/NaOH, 140 mM NaCl, and 2.5 mM CaCl2 at pH 7.4. Cells were resuspended in 100 µL of 1× Annexin Binding Buffer followed by the addition of 5 µL of Alexa Flour 488 annexin V and 1 µL of 100 µg/mL of Propidium Iodine (PI). Cells were incubated at room temperature for 15 min and then diluted with 400 µL of 1× binding buffer. Samples were kept on ice and analyzed by flow cytometry at excitation of 488 nm and emission at 530 nm. Samples were measured in triplicates and analyzed using GraphPad Prism 5.0 software.
Statistics
All statistical analyses within this paper were performed in triplicates. Results are given as mean values ± standard deviation (SD). GraphPad Prism 5.0 software was utilized to address significance by means of either a one or two way ANOVA.
2. RESULTS AND DISCUSSION
Synthesis of PEI-PCL-b-PEG-Fol Conjugates
The first step in this project was to synthesize several triblock copolymers and to characterize them via several criteria. Two different molecular weight blocks of heterobifunctional PEG chains were used in order to start the synthesis, namely 3.5 kDa and 5 kDa. Both were heterobifunctional with a hydroxyl and carboxylic acid group at either end. Two different groups of copolymers were synthesized with either molecular weight of PEG. Additionally, the grafting degree of PCL-b-PEG-Fol was varied in terms of two different molar ratios per fixed amount of PEI, namely, equimolar or 3-fold molar excess. One copolymer was synthesized to bear 10 µmol of the grafted PCL-b-PEG-folate chains per 10 µmol of PEI, and the other one was grafted with 30 µmol per 10 µmol of PEI. Furthermore, we synthesized a PEI-g-PCL-b-PEG null folate polymer which had a monofunctional mPEG chain and no folic acid attached, and a mixed conjugate which had both 3.5 kDa and 5 kDa PEG blocks in the PCL-b-PEG-folate chains. Interestingly, the feed ratio of PCL-b-PEG-folate chains did not have a significant influence on the polymers’ final structure. Both equimolar ratios and 3-fold excess yielded in copolymers with statistically about 5 PCL-b-PEG-folate chains per molecule PEI. It is possible that, especially in the reactions of equimolar ratios, a large amount of unreacted PEI or PEI with just one PCL-b-PEG-folate chain were present but were lost during the purification of the polymers by ultrafiltration with a 30 kDa MWCO membrane. A folate concentration assay was performed on each of the five conjugates which were made with folic acid attached to the PCL–PEG chain. The average amount of folate was 1.313 × 10−5 mol folate/mg of polymer. This was similar, and in some cases higher, than what has been previously reported in the literature.36 Table 1 describes each polymer’s composition along with its corresponding and designated name. Diagrams of these polymers can be seen in Scheme 1. Furthermore, conjugates were successfully characterized by 1H NMR (D2O) as described before,36,38,39 and the spectra were as follows: δ (ppm) = 8.6, 8.0, 7.6, 6.8 peaks characterizing the folate terminus; strong singlet peak at 3.6 (OCH2CH2O); and a broad but weak peak 1.6–1.2 (COCH2CH2CH2CH2CH2O; Supporting Information, Spectra 1).
Scheme 1. Schematic Representation of All Synthesized Folate Decorated ConjugatesA.
ANumbers 1–6 are all screened for siRNA condensation, as shown in Figure 1. Red depicts PCL, while blue and yellow signify PEG and folic acid, respectively.
siRNA Condensation Ability and Retention
As explained above, our approach utilizes a PEI block to electrostatically condense and protect the siRNA from nuclease degradation in vitro and in vivo. Therefore, we tested whether the triblock copolymers were still able to effectively condense the siRNA compared to unmodified, hyperbranched hy-PEI as described previously.40 In order to do this, SYBR gold assays were carried out with each polymer at varying polymer/siRNA (N/P) ratios ranging from 0 to 20. Figure 1A shows that the grafting of PCL-PEG-Fol did not decrease the copolymers’ ability to condense the siRNA. In order to mimimize toxic side effects that could result from an excess of free polymer in the micelleplex suspension, the ideal N/P ratio was defined as the lowest one which fully condenses the siRNA. The SYBR Gold assays showed the optimal N/P ratios of all conjugates to be between 5 and 7. Therefore, these N/P ratios were continuously used for all subsequent experiments.
Figure 1.
(A–C) SYBR gold assays for each conjugate in comparison to PEI. The conjugates’ ability to electrostatically condense siRNA was analyzed from N/P 0–20 (A). Heparin assays at pH 7.4 mimic the pH during in vivo circulation. Each conjugate was tested at N/P 5 and incubated in the presence of heparin for 30 min (B). Heparin assays at pH 4.5 mimic the late endosome. Each conjugate was tested at N/P 5 and incubated in the presence of heparin for 30 min (C).
Additionally, the appropriate retention and release of siRNA are critical for effective delivery. Therefore, heparin assays were utilized in order to mimic release of the siRNA during circulation (pH 7.4) and in the late endosome (pH 4.5).31,41 Heparin is a polyanion and in the presence of the micelleplexes can be used to mimic competition for the electrostatic binding with the polymer. It is known that the presence of polyanions in serum can cause premature release of the siRNA from merely electrostatiacally self-assembled polyplexes.42 Each conjugate was tested at N/P 5 in the presence of increasing amounts of heparin (0–1.0 IU). These data, seen in Figure 1B, demonstrate that the copolymers only released 3–12% of the total siRNA in the presence of the competing polyanion heparin at pH 7.4, whereas PEI polyplexes released up to 20% siRNA in the concentration range of heparin tested here. Conversely, micelleplexes need to have good releases profiles at low pH values in order to be released from the polymer to achieve optimal protein knockdown. Based on our hypothesis that our micelleplexes are internalized within the cell by means of folate receptor mediated endocytosis, the micelleplexes have to escape out of an endocytic vesicle to deposit the siRNA into the cytoplasm. Thus, the same heparin assay was run with similar conditions as before, except at pH 4.5 to mimic the pH of the late endosome. Figure 1C demonstrates that all micelleplexes were able to successfully release more than 95% of the siRNA, while unmodified PEI only released about 50%. Collectively, these data indicate that all six copolymers would not only be stable in circulation (see release at pH 7.4, Figure 1B), but could efficiently release siRNA into the cytoplasm upon being endocytosed (see release at pH 4.5, Figure 1C). Release of siRNA is into the cytoplasm is hypothesized to occur due to the proton sponge effect which results in bursting of the endosome and subsequent siRNA deposition into the cytoplasm. Evidence for siRNA deposition and action within the cytoplasm is reflected in the protein knockdown data that is discussed later in this paper. Similar assays were performed to assess siRNA release over time while in the presence of heparin. It was found that after 30 min, the release profiles did not change, but were comparable to the curves in Figure 1C (data not shown).
Characterizing Nanoparticle Morphology, Hydrodynamic Diameter, and Zeta Potential
Two important characteristics for nanoparticle delivery are their sizes and zeta potentials. An effective carrier should form nanoparticles within the nanometer scale with a slightly positive zeta potential to increase the likelihood of cell binding and consecutive endocytosis.23,29,43–46 Maintaining small sizes of nanoparticles is important for permeation out of the blood vessels and into the tumor as well as avoiding detection from the host’s natural defense mechanisms, such as macrophages. In the lung, an optimal size to avoid macrophage detection and subsequent endocytosis is 260 nm.45,47 However, optimal sizes of nanoparticles that are administered intravenously should also be below 260 nm, if not smaller, in order to avoid macrophage detection and phagocytosis, along with other side effects.43,44,46 Here, micelleplex formulations with 50 pmol of siRNA at N/P 5, 6, and 7 were characterized by dynamic light scattering (DLS). As seen in Figure 2A, at N/P 5, the hydrodynamic diameters were all below 150 nm, and slightly increased as the N/P ratio increased. At the higher N/P ratios (6 and 7), the hydrodynamic diameters were either at or below 260 nm. The sizes determined here with DLS measurement are smaller than other folic acid chitosan low molecular weight PEI delivery systems reported in literature (220–250 nm).48 Conversely, other previously published folic acid targeted delivery systems utilizing platinum based nanoparticles have much smaller hydrodynamic diameters.49 Collectively, this suggests that our micelleplexes have adequate sizes to evade the host immune system and easily permeate out of the blood vessel and into the tumor interstitium and correspond adequately with what is published.
Figure 2.
Characterizing nanoparticle morphology, size, and zeta potential. Each micelleplex’s hydrodynamic diameter was tested at N/P 5, 6, and 7 with 50 pmol of siRNA (A). Each micelleplex formulation was then diluted to measure zeta potentials via LDA (B). TEM images of 5k 30 µmol (C) and mixed conjugate (D) are shown.
Furthermore, the zeta potentials of the micelleplexes are important for delivery as the cell membrane carries a slight negative charge. If the micelleplexes are negatively charged, they can be electrostatically repelled by the cell membrane, and the siRNA uptake would suffer. Conversely, with a slight positive charge to the micelleplex, attraction between the cell membrane and micelleplexes is expected which helps promote binding to the cell membrane and nonspecific, adsorptive uptake. However, a micelleplex that is strongly positively charged can be toxic to the cells, which is a common problem with polycation delivery vectors for siRNA. Ideally, this toxicity should be avoided. In case of targeted nanoparticles, a strong positive zeta potential is not necessary, since uptake is promoted by receptor-mediated endocytosis. A slight positive charge, however, can be helpful to orient the particles in close proximity of the cell membrane and receptor. In our experiments, the corresponding zeta potentials for every micelleplex at every N/P ratio were all positive, and below +20 mV. Although there has not been a reported threshold for cationic polymers zeta potential in correlation to toxicity, generally, the less cationic nanoparticles are, the fewer cytotoxic effects will be seen within the cell. Most N/P formulations for each micelleplex were around +8–12 mV, corroborating other nanoparticle formulations, which are published.50 Figure 2B represents the average zeta potential for each micelleplex containing siRNA. Cytotoxicity studies were carried out with an MTT assay comparing lipofectamine, a known toxic transfection reagent, against our conjugates. The MTT assay was carried out at concentrations at which lipofectamine was used for transfections. However, our conjugates are used at much lower concentrations and additionally exhibit higher IC50 values, thus, suggesting that there is little toxicity concerns when transfecting with our folate decorated micelleplexes (Supporting Information, Figure 1). This has been seen in literature with similar PEI–PEG/siRNA delivery systems.37 Based on the data acquired from zeta potential measurements, our micelleplexes should be able to have a slight attraction to the cell membrane and not be inherently toxic.
Additionally, transmission electron microscopy (TEM) was used to validate our findings obtained with dynamic light scattering in terms of micelleplex sizes. TEM imaging serves to learn more about the size but also the morphology of our micelleplexes. As presented in Figure 2C,D, the sizes determined by TEM corroborate extremely well with DLS measurements. In addition, the micelle formation of the inner core and outer corona is clearly shown. Concluding these results, all conjugates formed micelleplexes with the siRNA with adequate sizes (100–200 nm) and zeta potentials (+8–12 mV). Based on these data, our micelleplexes contain optimal characteristics to condense siRNA and allow for interaction with the cellular membrane.
Assessing siRNA Uptake and Targeted Delivery
After the appropriate characterizations were performed to assess our micelleplexes’ size, zeta potential, and siRNA release/retention profiles, flow cytometry was utilized to perform siRNA delivery studies. For all siRNA uptake flow cytometry experiments, unless otherwise stated, Alexa fluor 488 labeled siRNA was used. In order to identify which conjugate worked the best, all folate-decorated micelleplexes were tested against one another as well as compared with PEI and lipofectamine. As seen in Figure 3A, comparable results to PEI were obtained, while slightly less transfection efficiency was seen in comparison to lipofectamine. Although there seemed to be no statistical difference in the initial screen between all conjugates, we did notice a trend that the 10 µmol conjugates performed better than their 30 µmol counterparts. Due to this, the three most promising conjugates appeared to be PEI grafted with 10 µmol of the chains containing 3.5 kDa PEG (3.5k 10 µmol), PEI grafted with 10 µmol of the chains containing 5 kDa PEG (5k 10 µmol), and the mixed conjugate. With this small selection, we determined the optimal N/P ratio for each conjugate’s siRNA delivery. Due to the smaller sizes obtained at N/P 5 and no difference in uptake with higher N/P ratios, siRNA was formulated with the copolymers mentioned at N/P 5, which may also reduce any possible toxicity seen with excess polymer at increasing N/P ratios (data not shown). Once delivery conditions were optimal, folate-decorated conjugates were compared against a null folate conjugate to assess targeted FRα-mediated uptake. First, a time course analysis was carried out comparing the three best folate decorated conjugates and the null–folate conjugate seen in Figure 3B. Each micelleplex was transfected and allowed to incubate for 24, 48, 72, and 96 h before harvesting cells for flow cytometry. At each time point, every folate-decorated conjugate outperformed the null folate polymer in SKOV-3 FRα positive cells. This led us to believe that there are two possible mechanisms for uptake of our micelleplexes. As one possibility, non-receptor-mediated, but charge-mediated adsorptive endocytosis across the cellular membrane could occur due to sedimentation of the micelleplexes over a period of time and might be promoted by their amphiphilic properties. Second, the folate-decorated micellepexes are able to be endocytosed utilizing the folate-receptor-mediated endocytosis pathway. With this in mind, we hypothesized that our folate-decorated micelleplexes can take advantage of both mechanisms, thus, resulting in a more significant uptake over time.
Figure 3.
siRNA uptake studies using flow cytometry. Uptake study across all folate decorated conjugates using Alexa Fluor 488 for 4 h (A). Time course uptake study with the three most promising conjugates against folate null conjugate (B). Uptake in SKOV-3 cells incubated for 2 or 4 h at 37 °C versus 4 °C (C). Uptake study with and without an excess of free folic acid to determine competitive inhibition of binding (D). Uptake study with trypan blue and monensin treatment to assess localization of siRNA (E); ***p < 0.05, ****p < 0.01.
The next step was to test whether the uptake that was demonstrated in Figure 3B was folate receptor driven. To answer this question, two experiments were performed. First, we analyzed the micelleplexes’ siRNA delivery at 37 and 4 °C. When cells are incubated at 4 °C, the lower temperature inhibits active uptake such as FRα-mediated endocytosis, but leaves receptor-mediated binding or charge-mediated binding to the cell still an available option for cell-associated fluorescence. Figure 3C shows that significant inhibition of active uptake occurs for SKOV-3 cells incubated with folate-decorated micelleplexes at 4 °C in comparison to 37 °C. Conversely, the folate null micelleplexes showed little decrease between the two conditions, thus, suggesting no receptor-mediated but possibly adsorptive endocytosis was inhibited for this conjugate. Most interestingly, the uptake efficacy of the targeted formulation (3.5k 10 µmol) was significantly higher than that of the nontargeted micelleplexes (Null folate) if incubated at 37 °C. However, if incubated at 4 °C, the uptake of both formulations was comparable. Additionally, competitive inhibition of receptor-mediated uptake was analyzed after preincubating SKOV-3 cells with an excess of the receptor substrate, free folic acid (FF). The recycling rates of FRα vary depending on the tissue and the tumor cell line. However, on average, the in vivo recycling rate is just under 5.7 h, and recycling of a receptor previously blocked with free folic acid renders the former available for binding folate-conjugated nanoparticles.51 Therefore, our studies were performed at early time points with or without preincubation of free folic acid to get a better picture of whether the targeted micelleplexes are binding with FRα.1,51 Figure 3D demonstrates that when excess free folic acid was added (20 µg), there is no significant drop in siRNA uptake for neither the folate-decorated micelleplex nor the nonfolate micelleplex. Excess folic acid concentrations were used based on previous studies with similar concentrations and findings.11,52,53 Benoit et al. did not observe any competitive inhibition in the presence of 10 µg/mL of free folic acid.11 Conversely, at higher concentrations, above 1 mM, uptake was inhibited as shown by Arima et al.52 Tied into the fact that a nonspecific inhibition was seen across the board with our folate-decorated micelleplexes as well as the nondecorated ones at concentrations above 1 mM of free folic acid per well (data not shown), we have seen a different result of competitive uptake than Arima et al. The lack of competition with low amounts of free folic acid could be due to the fact that our micelleplexes are multivalent and, thus, have a stronger binding avidity to the receptor when compared to the affinity of a monovalent folic acid to the receptor. Therefore, it is expected that the folate-decorated micelleplexes will easily outcompete folic acid for the folate receptors binding sites. At higher concentrations of free folic acid, uptake can be inhibited but the observed inhibition is not necessarily an inhibition of receptor-mediated endocytosis. Taken together, these results imply that uptake of these micelleplexes by means of diffusion does not explain the difference of targeted versus nontargeted formulations, while the folate-decorated micelleplexes are able to utilize FRα-mediated uptake for siRNA delivery if (1) energy-dependent endocytosis is possible and (2) the receptor is not blocked by a free competing ligand. These data were further reinforced by confocal microscopy showing significantly more siRNA deposition within the cell by the folate-decorated nanoparticles. As seen in Figure 4, all folate-decorated nanoparticles (C–E) were able to effectively delivery siRNA more efficiently than their null folate counterpart (F). At first glance, PEI seemed to deliver a significantly larger amount of siRNA to the cells. However, the fluorescence may arise from PEI complexes merely bound to the surface of the cell due to the strong electrostatic interaction and not effectively being endocytosed into the cell. When analyzed with flow cytometry and using trypan blue to quench the extracellular fluorescence, the MFI for PEI drops significantly in comparison to targeted micelleplexes, indicating that a higher percentage of polyplexes is not internalized, but only cell-bound. Additionally, PEI/siRNA polyplexes that are taken up into an endosome may not release siRNA as efficiently into the cytoplasm as the micelleplexes do due to the very strong binding of the polymer to siRNA (Figure 1C). Taken together, these results appear different from the flow cytometry data in Figure 3A. Due to CLSM being a different type of measurement revealing information on spatial siRNA deposition in the cell, it is not surprising that there is a discrepancy seen here. This could also be attributed to self-quenching of the fluorescently labeled siRNA molecules when in close proximity to other fluorphores which could be stronger for the Tye-563 dye than observed with Alexa Fluor 488.54 However, our CLSM data strengthens our previous observations that folate decorated micelleplexes can utilize a receptor-mediated mode of uptake, while the null folate micelleplexes cannot, thus, resulting in greater siRNA delivery ability. To determine if the micelleplexes are taken up by endocytosis into acidic vesicles, a monensin assay was performed. Fluorescence was measured after transfection without any treatment for total fluorescence, measured after treatment with trypan blue to quench any extracellular fluorescence, as well as measured after treatment with monensin to quench any fluorescence located within acidic vesicles. With this technique, we are able to elucidate where in the cell the fluorescent signal originates from. As shown in Figure 3E, the majority of the fluorescence is intracellular for both folate-targeted and nontargeted micelleplexes. At 24 h, the overall uptake was higher for targeted versus nontargeted micelleplexes, which is in line with Figure 3B. Interestingly, a higher extent of targeted particles was trafficked into acidic vesicles compared to nontargeted ones. This observation can be explained by the intracellular trafficking of the folate receptor, which can either be recycled at the early endosome stage or ripen to a late endosome. Most importantly, however, the targeted micelleplexes are shown to be more efficient in terms of delivery to the cytoplasm, which is the site of action for siRNA. These results are also in line with our observations based on confocal microscopy which, especially in Figure 4D, show siRNA in the cytoplasm, rather than in vesicles. The percentage of particles that is trapped in vesicles, however, may not result in a bright signal in the CLSM images, and therefore explain the discrepancy with the flow cytometric results.
Figure 4.
siRNA uptake studies using confocal laser scanning microscopy. Uptake study across the three most promising folate decorated conjugates compared to hy-PEI and null folate for 4 h. Cell nuclei are stained with DAPI. In order, images are of blank cells (A), PEI treated (B), 3.5k 10 µmol (C), 5k 10 µmol (D), mixed conjugate (E), and null folate (F).
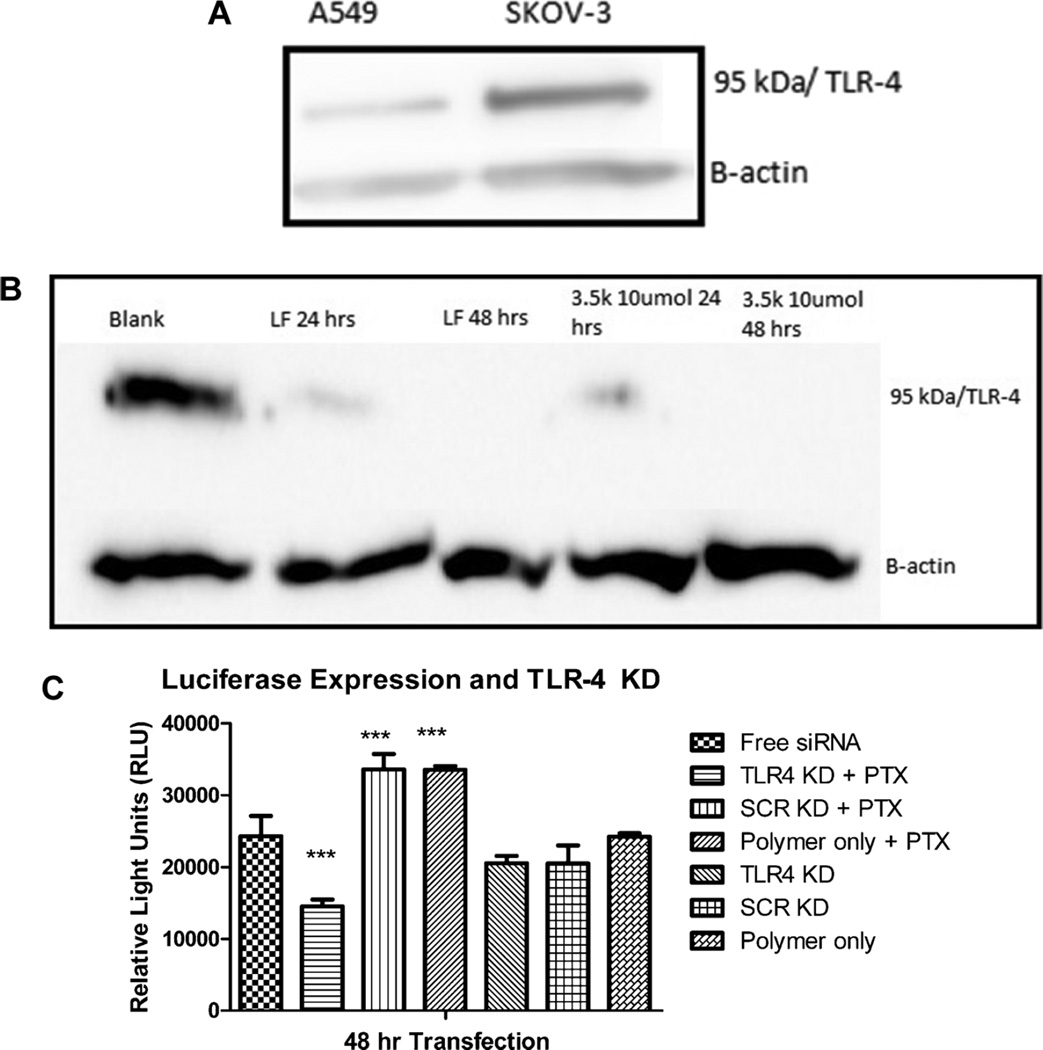
Protein Knockdown In Vitro and Resensitization toward Paclitaxel Treatment
Ultimately, the main goal of siRNA delivery is to be able to achieve protein knockdown. Furthermore, our objective was to mediate protein knockdown in a targeted manner to achieve paclitaxel resensitization. For preliminary experiments, we chose SKOV-3 cells that are PTX resistant and FRα overexpressing, in comparison to a PTX sensitive cell line, here A549, which have only a basal FRα expression. The TLR4 expression in both cell lines was assess via Western Blot analysis and found to be about 4-fold increased in SKOV-3 cells as compared to A549, seen in Figure 5A. Previous literature studying ovarian cancer suggests that a rise in TLR4 expression leads to increased chemo-resistance.24–26,55 Subsequently, we transfected SKOV-3 cells with siRNA against TLR4 using the 3.5k 10 µmol conjugate and lipofectamine as a positive control and determined the gene knockdown knockdown after 48 and 72 h via Western blot analysis. Figure 5B shows significant knockdown of TLR4 at 48 h and knockout at 72 h for both lipofectamine and the 3.5k 10 µmol conjugate.
Figure 5.
(A) Western Blot analysis of TLR4 levels within SKOV-3 and A549 cells. (B) Western Blot analysis of TLR4 knockdown at 24 and 48 h using lipofectamine and 3.5k 10 µmol. (C) Luciferase assay in SKOV-3/LUC cells assessing TLR4 knockdown on luciferase expression 48 h post transfection when treated with 1000 µM of PTX.
To further elucidate the impact of TLR4 knockdown, we used a SKOV-3 luciferase cell line with luciferase expression controlled by a CMV promoter with a NF-KB binding site. In this cell culture model, NF-KB activation results in enhanced luciferase activity. Combined with the fact that PTX treatment causes NF-KB activation downstream through activating the TLR4 pathway, this model allows determining the effects of TLR4 knockdown on chemosensitivity of SKOV-3 cells toward PTX through measuring luciferase expression.55 Figure 5C shows that the luciferase expression of SKOV-3/LUC cells is clearly upregulated upon treatment with PTX, unless TLR4 is down regulated upon TLR4 knockdown by the 3.5k 10 µmol micelleplex. In this case, a dramatic decrease in luciferase expression is observed compared to cells not treated with PTX. However, cells treated with PTX upon transfection with a scrambled siRNA or polymer only showed an increase of luciferase expression, thus demonstrating no knockdown of TLR4 occurred. Interestingly, TLR4 knockdown did not affect the basal luciferase expression of SKOV-3/LUC cells when this expression was not triggered by treatment with PTX. Thus, Figure 5 demonstrates three processes: (1) if SKOV-3 cells are treated with PTX, NF-KB is activated and pro-survival genes may be activated, (2) if TLR4 is knocked down, NF-KB activation is inhibited, and (3) NF-KB is not necessary for basal luciferase expression but strongly triggers the latter. Taken together, Figure 5 demonstrates that the 3.5k 10 µmol conjugate is able to successfully knockdown TLR4 protein with a therapeutically relevant effect on PTX treatment.
Lastly, cells were transfected with the 3.5k 10 µmol conjugate containing TLR4 siRNA followed by treatment with PTX in order to assess SKOV-3 resensitization to PTX. MTT cell viability assays confirmed that TLR4 knockdown results in a decrease in IC50 value for PTX in comparison to cells not transfected with micelleplexes containing TLR4 siRNA (Figure 6A). Their corresponding IC50 values were 9.34 and 21.72 nM, respectively. In addition, cells were analyzed via flow cytometry to assess the percentage of apoptotic cells between different treatments. Figure 6A,B shows that pretreatment with 3.5k 10 µmol micelleplexes containing TLR4 siRNA caused a resensitization of SKOV-3 cells at varying concentrations of PTX. This effect was most drastic at higher PTX concentrations. At 1000 nM, only 27% and 23% of cells were apoptotic or dead if the cells underwent no pretreatment or if they were transfected with 3.5k 10 µmol conjugate containing scrambled siRNA, respectively. Conversely, when TLR4 specific siRNA was used, more than double the cells stained positive for annexin V binding to phosphatidyl serine on the cellular surface, a marker of apoptosis. Collectively, SKOV-3 cells that were pretreated with 3.5k 10 µmol micelleplexes containing TLR4 siRNA before PTX treatment resulted in a significant increase in cell death. A loss in TLR4 activity has been cited in literature render SKOV-3 cells sensitive to PTX treatment. Szajnik et. al showed that by using siRNA to knockdown TLR4 there was a 2–3-fold increase in cell death upon PTX treatment.25 Other groups using shRNA to stably knock down TLR4 have displayed similar resensitization, a 3-fold increase in caspase activity, upon PTX treatment.24 Our results are in line with these published results; however, our approach utilizes a targeted micelleplex delivery system to focus the TLR4 knockdown to cells that overexpress FRα.
Figure 6.
(A) MTT assay of SKOV-3 cells after TLR4 knockdown at 48 h. (B) Annexin flow cytometry stain: Flow cytometry analysis of cell death via Annexin V staining for apoptosis.
CONCLUSION
The overexpression of FRα in ovarian cancer cells offers the ability to specifically target and deliver siRNA in a Trojan-horse like mechanism explicitly to these cancer cells. Utilizing polymer based siRNA delivery systems for therapeutic purposes offers a very wide range of possibilities due to the modularity of this approach. Theoretically, as long as a sequence is available for a protein’s mRNA, complementary siRNA sequences can be made against that protein to abrogate its expression. The delivery system discussed in this paper has the opportunity and ability to target cancer cells through FRα overexpression for example found in ovarian cancer cells. Furthermore, targeted micelleplexes, as seen here, have the ability to provide beneficial therapy to patients while decreasing off target toxicity which is commonly seen with most chemotherapy treatments. Our triblock copolymer, consisting of PEI-g-PCL-b-PEG-Fol has shown impressive ability to condense and protect siRNA along with favorable release profiles at acidic pH values as found in late endosomal vesicles. Furthermore, several physical techniques such as DLS, and TEM were utilized to show adequate sizes (100–200 nm), zeta potentials (0–30 mV), and a core–corona structure. Utilizing fluorescently-labeled siRNA and flow cytometry, transfection conditions were optimized, and the micelleplexes were demonstrated to utilize FR-mediated endocytosis for cellular uptake. Protein knockdown with 3.5k 10 µmol micelleplexes was analyzed by Western blots and luciferase assays. Both demonstrated efficient protein knockdown of TLR4. Upon knockdown of TLR4, a resensitization occurred for SKOV-3 cells to PTX treatment and a significant increase in apoptotic cells were detected with flow cytometry. This approach displays similar therapeutic effect with published literature but utilizes a targeted delivery mechanism. Collectively, these findings based on cell culture models suggest the feasibility of targeted gene knockdown of TLR4 and resensitization of PTX resistant cells toward PTX therapy. Currently, in vivo targeting and therapeutic efficacy are being tested.
Supplementary Material
Acknowledgments
This work was supported by the Wayne State Start-Up and NanoIncubator Grant to Olivia Merkel as well as the Ruth L. Kirschstein National Research Award T32-CA009531 fellowship to S.K.J. The NIH Center Grant P30CA22453 supporting the Wayne State Microscopy, Imaging and Cytometry Resources (MICR), and the SOM summer research fellowship to V.L. are gratefully acknowledged. We are grateful to Dr. Anna Moszczynska and Bryan Killinger (Department of Pharmaceutical Sciences, Wayne State University) for generous use of lab equipment and technical support.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- MTT assay of lipofectamine against folate decorated micelleplexes on SKOV-3 cells; Concentration ranges were from 0 to 16 µg/mL and transfected for 24 h (PDF).
The authors declare no competing financial interest.
REFERENCES
- 1.Markert S, Lassmann S, Gabriel B, Klar M, Werner M, Gitsch G, Kratz F, Hasenburg A. Alpha-folate receptor expression in epithelial ovarian carcinoma and non-neoplastic ovarian tissue. Anticancer Res. 2008;28(6A):3567–3572. [PubMed] [Google Scholar]
- 2.Parker N, Turk MJ, Westrick E, Lewis JD, Low PS, Leamon CP. Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay. Anal. Biochem. 2005;338(2):284–293. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 3.Ross JF, Chaudhuri PK, Ratnam M. Differential regulation of folate receptor isoforms in normal and malignant tissues in vivo and in established cell lines. Physiologic and clinical implications. Cancer. 1994;73(9):2432–2443. doi: 10.1002/1097-0142(19940501)73:9<2432::aid-cncr2820730929>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 4.NIH National Cancer Institute. NIH Publication 06-1561. Bethesda, MD: National Cancer Institute; 2006. What you need to know about Ovarian Cancer. [Google Scholar]
- 5.Institute, N. C. Recurrent or Persistent Ovarian Epithelial, Fallopian Tube, and Primary Peritoneal Cancer Treatment. http://www.easterncarolinawomens.com/womens-health/hw-view.php?DOCHWID=ncicdr0000062963. [Google Scholar]
- 6.Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, Barry ST, Gabizon A, Grodzinski P, Blakey DC. Challenges and key considerations of the enhanced permeability and retention effect for nanomedicine drug delivery in oncology. Cancer Res. 2013;73(8):2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ledermann JA, Kristeleit RS. Optimal treatment for relapsing ovarian cancer. Ann. Oncol. 2010;21(Suppl 7):vii218–vii222. doi: 10.1093/annonc/mdq377. [DOI] [PubMed] [Google Scholar]
- 8.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007;2(12):751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 9.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J. Controlled Release. 2000;65(1–2):271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 10.Leamon CP. Folate-targeted drug strategies for the treatment of cancer. Curr. Opin. Investig. Drugs. 2008;9(12):1277–1286. [PubMed] [Google Scholar]
- 11.Benoit DS, Srinivasan S, Shubin AD, Stayton PS. Synthesis of folate-functionalized RAFT polymers for targeted siRNA delivery. Biomacromolecules. 2011;12(7):2708–2714. doi: 10.1021/bm200485b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Li K, Pan J, Liu B, Feng SS. Folic acid conjugated nanoparticles of mixed lipid monolayer shell and biodegradable polymer core for targeted delivery of Docetaxel. Biomaterials. 2010;31(2):330–338. doi: 10.1016/j.biomaterials.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 13.Morris RT, Joyrich RN, Naumann RW, Shah NP, Maurer AH, Strauss HW, Uszler JM, Symanowski JT, Ellis PR, Harb WA. Phase II study of treatment of advanced ovarian cancer with folate-receptor-targeted therapeutic (vintafolide) and companion SPECT-based imaging agent (99mTc-etarfolatide) Ann. Oncol. 2014;25(4):852–858. doi: 10.1093/annonc/mdu024. [DOI] [PubMed] [Google Scholar]
- 14.Vlahov IR, Leamon CP. Engineering folate-drug conjugates to target cancer: from chemistry to clinic. Bioconjugate Chem. 2012;23(7):1357–1369. doi: 10.1021/bc2005522. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Li S, Shi Y, Chuan X, Li J, Zhong T, Zhang H, Dai W, He B, Zhang Q. The development of site-specific drug delivery nanocarriers based on receptor mediation. J. Controlled Release. 2014;193:139–153. doi: 10.1016/j.jconrel.2014.05.028. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, Low PS. Folate-mediated delivery of macromolecular anticancertherapeutic agents. Adv. Drug Delivery Rev. 2002;54:675–693. doi: 10.1016/s0169-409x(02)00042-x. [DOI] [PubMed] [Google Scholar]
- 17.Huang YH, Bao Y, Peng W, Goldberg M, Love K, Bumcrot DA, Cole G, Langer R, Anderson DG, Sawicki JA. Claudin-3 gene silencing with siRNA suppresses ovarian tumor growth and metastasis. Proc. Natl. Acad. Sci. U. S. A. 2009;106(9):3426–3430. doi: 10.1073/pnas.0813348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shahzad MM, Lu C, Lee JW, Stone RL, Mitra R, Mangala LS, Lu Y, Baggerly KA, Danes CG, Nick AM, Halder J, Kim HS, Vivas-Mejia P, Landen CN, Lopez-Berestein G, Coleman RL, Sood AK. Dual targeting of EphA2 and FAK in ovarian carcinoma. Cancer Biol. Ther. 2009;8(11):1027–1034. doi: 10.4161/cbt.8.11.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Gu W, Chen J, Chen W, Xu ZP. Co-delivery of siRNAs and anti-cancer drugs using layered double hydroxide nanoparticles. Biomaterials. 2014;35(10):3331–3339. doi: 10.1016/j.biomaterials.2013.12.095. [DOI] [PubMed] [Google Scholar]
- 20.Shen J, Yin Q, Chen L, Zhang Z, Li Y. Co-delivery of paclitaxel and survivin shRNA by pluronic P85-PEI/TPGS complex nanoparticles to overcome drug resistance in lung cancer. Biomaterials. 2012;33(33):8613–8624. doi: 10.1016/j.biomaterials.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh SK, Yigit MV, Uchida M, Ross AW, Barteneva N, Moore A, Medarova Z. Sequence-dependent combination therapy with doxorubicin and a survivin-specific small interfering RNA nanodrug demonstrates efficacy in models of adenocarcinoma. Int. J. Cancer. 2014;134(7):1758–1766. doi: 10.1002/ijc.28499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sourbier C. Ovarian cancer: emerging molecular-targeted therapies. Biologics. 2012;6:147–154. doi: 10.2147/BTT.S24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg MS, Xing D, Ren Y, Orsulic S, Bhatia SN, Sharp PA. Nanoparticle-mediated delivery of siRNA targeting Parp1 extends survival of mice bearing tumors derived from Brca1-deficient ovarian cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108(2):745–750. doi: 10.1073/pnas.1016538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang AC, Su QB, Wu FX, Zhang XL, Liu PS. Role of TLR4 for paclitaxel chemotherapy in human epithelial ovarian cancer cells. Eur. J. Clin. Invest. 2009;39(2):157–164. doi: 10.1111/j.1365-2362.2008.02070.x. [DOI] [PubMed] [Google Scholar]
- 25.Szajnik M, Szczepanski MJ, Czystowska M, Elishaev E, Mandapathil M, Nowak-Markwitz E, Spaczynski M, Whiteside TL. TLR4 signaling induced by lipopolysaccharide or paclitaxel regulates tumor survival and chemoresistance in ovarian cancer. Oncogene. 2009;28(49):4353–4363. doi: 10.1038/onc.2009.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly MG, Alvero AB, Chen R, Silasi DA, Abrahams VM, Chan S, Visintin I, Rutherford T, Mor G. TLR-4 signaling promotes tumor growth and paclitaxel chemoresistance in ovarian cancer. Cancer Res. 2006;66(7):3859–3868. doi: 10.1158/0008-5472.CAN-05-3948. [DOI] [PubMed] [Google Scholar]
- 27.Yin Q, Shen J, Chen L, Zhang Z, Gu W, Li Y. Overcoming multidrug resistance by co-delivery of Mdr-1 and survivin-targeting RNA with reduction-responsible cationic poly(beta-amino esters) Biomaterials. 2012;33(27):6495–6506. doi: 10.1016/j.biomaterials.2012.05.039. [DOI] [PubMed] [Google Scholar]
- 28.Endres T, Zheng M, Kilic A, Turowska A, Beck-Broichsitter M, Renz H, Merkel OM, Kissel T. Amphiphilic biodegradable PEG-PCL-PEI triblock copolymers for FRET-capable in vitro and in vivo delivery of siRNA and quantum dots. Mol. Pharmaceutics. 2014;11(4):1273–1281. doi: 10.1021/mp400744a. [DOI] [PubMed] [Google Scholar]
- 29.Zheng M, Liu Y, Samsonova O, Endres T, Merkel O, Kissel T. Amphiphilic and biodegradable hy-PEI-g-PCL-b-PEG copolymers efficiently mediate transgene expression depending on their graft density. Int. J. Pharm. 2012;427(1):80–87. doi: 10.1016/j.ijpharm.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 30.Yu Liu JN, Steele T, Merkel O, Kissel T. A new synthesis method and degradation of hyper-branched polyethylenimine grafted polycaprolactone block mono-methoxyl poly (ethylene glycol) copolymers (hy-PEI-g-PCL-b-mPEG) as potential DNA delivery vectors. Polymer. 2009;50(16):3895–3904. [Google Scholar]
- 31.Zheng M, Librizzi D, Kilic A, Liu Y, Renz H, Merkel OM, Kissel T. Enhancing in vivo circulation and siRNA delivery with biodegradable polyethylenimine-graft-polycaprolactone-block-poly-(ethylene glycol) copolymers. Biomaterials. 2012;33(27):6551–6558. doi: 10.1016/j.biomaterials.2012.05.055. [DOI] [PubMed] [Google Scholar]
- 32.Akhtar S. Non-viral cancer gene therapy: beyond delivery. Gene Ther. 2006;13(9):739–740. doi: 10.1038/sj.gt.3302692. [DOI] [PubMed] [Google Scholar]
- 33.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Delivery Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juliano R, Bauman J, Kang H, Ming X. Biological barriers to therapy with antisense and siRNA oligonucleotides. Mol. Pharmaceutics. 2009;6(3):686–695. doi: 10.1021/mp900093r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkel OM, Kissel T. Nonviral pulmonary delivery of siRNA. Acc. Chem. Res. 2012;45(7):961–970. doi: 10.1021/ar200110p. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Zheng M, Renette T, Kissel T. Modular Synthesis of Folate Conjugated Ternary Copolymers: Polyethylenimine-graft-Polycaprolactone-block-Poly(ethylene glycol)-Folae for Targeted Gene Delivery. Bioconjugate Chem. 2012;23:1211–1220. doi: 10.1021/bc300025d. [DOI] [PubMed] [Google Scholar]
- 37.Merkel OM, Beyerle A, Beckmann BM, Zheng M, Hartmann RK, Stoger T, Kissel TH. Polymer-related off-target effects in non-viral siRNA delivery. Biomaterials. 2011;32(9):2388–2398. doi: 10.1016/j.biomaterials.2010.11.081. [DOI] [PubMed] [Google Scholar]
- 38.Storey RF, Sherman JW. Kinetics and Mechanism of the Stannous Octoate-Catalyzed Bulk Polymerization of e-Caprolactone. Macromolecules. 2002;35:1504–1512. [Google Scholar]
- 39.Hans M, Gasteier P, Keul H, Moeller M. Ring-Opening Polymerization of e-Caprolactone by Means of Mono-and Multifunctional Initiators: Comparison of Chemical and Enzymatic Catalysis. Macromolecules. 2002;39:3184–3193. [Google Scholar]
- 40.Merkel OM, Beyerle A, Librizzi D, Pfestroff A, Behr TM, Sproat B, Barth PJ, Kissel T. Nonviral siRNA delivery to the lung: investigation of PEG-PEI polyplexes and their in vivo performance. Mol. Pharmaceutics. 2009;6(4):1246–1260. doi: 10.1021/mp900107v. [DOI] [PubMed] [Google Scholar]
- 41.Elsayed M, Corrand V, Kolhatkar V, Xie Y, Kim NH, Kolhatkar R, Merkel OM. Influence of oligospermines architecture on their suitability for siRNA delivery. Biomacromolecules. 2014;15(4):1299–1310. doi: 10.1021/bm401849d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Buyens K, Sanders NN, De Smedt SC, Behe M, Kissel T. Stability of siRNA polyplexes from poly(ethylenimine) and poly(ethylenimine)-g-poly-(ethylene glycol) under in vivo conditions: effects on pharmacokinetics and biodistribution measured by Fluorescence Fluctuation Spectroscopy and Single Photon Emission Computed Tomography (SPECT) imaging. J. Controlled Release. 2009;138(2):148–159. doi: 10.1016/j.jconrel.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 43.He C, Hu Y, Yin L, Tang C, Yin C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials. 2010;31(13):3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 44.Storm G, Belliot SO, Daemen T, Lasic DD. Lasic, Surface modification of nanoparticles to oppose uptake by the mononuclear phagocyte system. Adv. Drug Delivery Rev. 1995;17:31. [Google Scholar]
- 45.Sons JW. Antibody-Mediated Drug Delivery Systems: Concepts, Technology, and Applications. Hoboken, NJ: John Wiley and Sons, Inc.; 2012. [Google Scholar]
- 46.Jiang WK, Kim BYS, Rutka JT, Chan WCW. Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 47.Lauweryns JM, Baert JH. Alveolar clearance and the role of the pulmonary lymphatics. Am. Rev. Respir. Dis. 1977;115(4):625–683. doi: 10.1164/arrd.1977.115.4.625. [DOI] [PubMed] [Google Scholar]
- 48.Li JM, Wang YY, Zhang W, Su H, Ji LN, Mao ZW. Low-weight polyethylenimine cross-linked 2-hydroxypopyl-beta-cyclodextrin and folic acid as an efficient and nontoxic siRNA carrier for gene silencing and tumor inhibition by VEGF siRNA. Int. J. Nanomed. 2013;8:2101–2117. doi: 10.2147/IJN.S42440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teow Y, Valiyaveettil S. Active targeting of cancer cells using folic acid-conjugated platinum nanoparticles. Nanoscale. 2010;2(12):2607–2613. doi: 10.1039/c0nr00204f. [DOI] [PubMed] [Google Scholar]
- 50.Yu B, Tang C, Yin C. Enhanced antitumor efficacy of folate modified amphiphilic nanoparticles through co-delivery of chemotherapeutic drugs and genes. Biomaterials. 2014;35(24):6369–6378. doi: 10.1016/j.biomaterials.2014.04.095. [DOI] [PubMed] [Google Scholar]
- 51.Paulos CM, Reddy JA, Leamon CP, Turk MJ, Low PS. Ligand binding and kinetics of folate receptor recycling in vivo: impact on receptor-mediated drug delivery. Mol. Pharmacol. 2004;66(6):1406–1414. doi: 10.1124/mol.104.003723. [DOI] [PubMed] [Google Scholar]
- 52.Arima H, Yoshimatsu A, Ikeda H, Ohyama A, Motoyama K, Higashi T, Tsuchiya A, Niidome T, Katayama Y, Hattori K, Takeuchi T. Folate-PEG-appended dendrimer conjugate with alpha-cyclodextrin as a novel cancer cell-selective siRNA delivery carrier. Mol. Pharmaceutics. 2012;9(9):2591–2604. doi: 10.1021/mp300188f. [DOI] [PubMed] [Google Scholar]
- 53.Fernandes JC, Qiu X, Winnik FM, Benderdour M, Zhang X, Dai K, Shi Q. Low molecular weight chitosan conjugated with folate for siRNA delivery in vitro: optimization studies. Int. J. Nanomed. 2012;7:5833–5845. doi: 10.2147/IJN.S35567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Rompaey E, Engelborghs Y, Sanders N, De Smedt SC, Demeester J. Interactions between oligonucleotides and cationic polymers investigated by fluorescence correlation spectroscopy. Pharm. Res. 2001;18(7):928–936. doi: 10.1023/a:1010975908915. [DOI] [PubMed] [Google Scholar]
- 55.Byrd-Leifer CA, Block EF, Takeda K, Akira S, Ding A. The role of MyD88 and TLR4 in the LPS-mimetic activity of Taxol. Eur. J. Immunol. 2001;31(8):2448–2457. doi: 10.1002/1521-4141(200108)31:8<2448::aid-immu2448>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.