Abstract
We analyzed the cellular and molecular mechanisms governing the adhesive and migratory behavior of enteric neural crest cells (ENCCs) during their collective migration within the developing mouse gut. We aimed to decipher the role of the complement anaphylatoxin C3a during this process, because this well-known immune system attractant has been implicated in cephalic NCC co-attraction, a process controlling directional migration.
We used the conditional Ht-PA-cre transgenic mouse model allowing a specific ablation of the N-cadherin gene and the expression of a fluorescent reporter in migratory ENCCs without affecting the central nervous system. We performed time-lapse videomicroscopy of ENCCs from control and N-cad-herin mutant gut explants cultured on fibronectin (FN) and micropatterned FN-stripes with C3a or C3aR antagonist, and studied cell migration behavior with the use of triangulation analysis to quantify cell dispersion. We performed ex vivo gut cultures with or without C3aR antagonist to determine the effect on ENCC behavior. Confocal microscopy was used to analyze the cell-matrix adhesion properties. We provide the first demonstration of the localization of the complement anaphylatoxin C3a and its receptor on ENCCs during their migration in the embryonic gut. C3aR receptor inhibition alters ENCC adhesion and migration, perturbing directionality and increasing cell dispersion both in vitro and ex vivo. N-cad-herin-null ENCCs do not respond to C3a co-attraction. These findings indicate that C3a regulates cell migration in a N-cadherin-dependent process. Our results shed light on the role of C3a in regulating collective and directional cell migration, and in ganglia network organization during enteric nervous system ontogenesis. The detection of an immune system chemokine in ENCCs during ENS development may also shed light on new mechanisms for gastrointestinal disorders.
Keywords: Enteric nervous system, Neural crest cells, Co-attraction, Complement anaphylatoxin C3a, N-cadherin, Migration, Adhesion
1. Introduction
The enteric nervous system (ENS) controls the motor and secretory functions of the gastrointestinal tract. It consists of enteric neurons and glial cells, most of which are produced from the vagal enteric neural crest cells (ENCCs), which invade the foregut mesenchyme of mice at E9.0–9.5, subsequently migrating rostrocaudally within the gut mesenchyme. At early stages of development, the gut is a straight tubular structure that increases in length and undergoes morphological changes and regionalization during its colonization by ENCCs. During the gut colonization process, these cells proliferate, differentiate and progress caudally in the midgut. They pause at the cecum and then continue to migrate until they reach the distal hindgut (Druckenbrod and Epstein, 2007, 2005; Young et al., 2004; Lake and Heuckeroth, 2013; Xu et al., 2014). In addition, a transmesenteric subpopulation of ENCCs migrates from the midgut towards the hindgut without passing through the cecum (Nishiyama et al., 2012).
During gut colonization, the ENCCs migrate collectively, establishing transient cell-cell contacts and migrating as chains of cells, with the lead cells following unpredictable local trajectories (Druckenbrod and Epstein, 2005; Young et al., 2014). The extending strands of cells form nodes and diverge to form new nodes, suggesting that attraction-repulsion behavior occurs during network formation (Druckenbrod and Epstein, 2007). ENCC behavior is driven mostly by migration and proliferation. Some ENCCs advance caudally whereas others populate individual regions by “directional dispersion”, thereby ensure that enteric neurons are present along the entire length of the gut; the ENCCs may therefore be considered to colonize the gut while migrating (Young et al., 2014). ENCC proliferation is observed throughout the developing gut and at the front of cell migration whose progression is probably driven by the ENCC-free space ahead (Newgreen et al., 2013).
In addition to the frontal expansion other mechanisms were shown to control collective NCC behavior including the response to gradients in all NCC populations, the “follow-the leader” in trunk and cranial NCCs, and the combined action of co-attraction and contact inhibition of locomotion (CIL) in cranial NCCs (Kulesa and McLennan, 2015). CIL was discovered by Abercrombie and Heaysman (Abercrombie and Heaysman, 1953, 1954), and is described as a repulsive interaction causing a change of migration toward the opposite direction upon contact with an adjacent cell. The cranial and trunk NCCs exhibit a collective migration after emerging from the neural tube. They retain their cohesiveness and the same neighbors for a long period mostly via transient and dynamics contacts, and migrate between locations in closely associated groups, as streams or sheets. In chick, cranial NCCs migrate as stream of cells and adopt a follow-the-leader behavior (Wynn et al., 2013) regulated by biased cell-cell contact and respond to environmental cues. In Xenopus the regulation of cranial NCCs involves two mechanisms, a CIL where repulsive local intercellular interaction between cells redirects their migration and chemotaxis towards a NCC-secreted chemoattractant (co-attraction) (Theveneau et al., 2010; Theveneau and Mayor, 2011; Woods et al., 2014). Mutual chemoattraction and repulsion between cells act as complementary mechanisms with opposite effects (Carmona-Fontaine et al., 2011). N-cadherin has been implicated in the CIL of cranial NCCs (Theveneau et al., 2010), a process regulated by Rho GTPases and Wnt signaling (Carmona-Fontaine et al., 2008). The Complement anaphylatoxin C3a was identified as the essential co-attractant factor acting in concert with CIL by counteracting cranial NCC dispersion, thereby orchestrating their collective and directional migration in Xenopus (Carmona-Fontaine et al., 2011).
Complement is known to play a central role in the immune system, in which it acts as a rapid and efficient surveillance system, eliminating cell debris and infectious microbes, orchestrating immune responses and maintaining homeostasis (Ricklin and Lambris, 2013; Shinjyo et al., 2009). C3a has chemotactic properties in the immune system. It also binds neural progenitor cells in a specific and reversible manner, stimulating their differentiation into neurons, and it modulates the SDF-1a-induced differentiation and migration of these cells (Shinjyo et al., 2009). The Complement system is activated via three independent pathways converging on the cleavage of C3 into two fragments, C3b and C3a. The C3a fragment is a small anaphylatoxin peptide that binds to the receptor C3aR. In Xenopus cranial NCCs, C3a is localized in the NC territory at the premigratory and migratory stages. It is colocalized with C3aR only at the migratory stage, and these two molecules together regulate the collective migration of these cells (Carmona-Fontaine et al., 2011). The Complement pathway is highly conserved and plays several unexpected roles in animal development, highlighting its importance for the control of cell behavior (Leslie and Mayor, 2013).
ENCCs also require transient cell-cell adhesion for forward progression in the gut wall, because isolated cells migrate more slowly than high-density groups of cells (Young et al., 2004). Isolated cells exhibit an unbiased random walk behavior, while nonsolitary cells or cells in chains are more directional and progress caudally (Young et al., 2014). ENCCs may therefore be considered to be a population of cells migrating collectively, however in contrast to the cranial NCCs they do not retain their neighbors. Several adhesion receptors have been implicated in the colonization of the developing gut by ENCCs (Anderson et al., 2006; Breau et al., 2006). Cooperation between N-cadherin and beta1 integrins has been shown to control the balance between cell-cell and cellextracellular matrix adhesion in ENCCs, to regulate the migration of these cells and to be required for the correct ontogenesis of the ENS (Breau et al., 2009; Watanabe et al., 2013; Broders-Bondon et al., 2012). The interactions of ENCCs with their surroundings may also contribute to the response of cells to cues from the gut environment (Wynn et al., 2013).
In this study, we investigated whether ENCCs used a C3a-based and N-cadherin dependent mechanism to regulate their collective behavior. We show here that ENCCs express the complement C3aR and that the anaphylatoxin C3a is present in gut and in cultured gut explants. We also found that C3a/C3aR inhibitors disturbed the migratory chains and neuronal network organization during the ENCC colonization of the embryonic gut, their cohesiveness, co-attraction, and directionality of migration, through an N-cadherin-dependent process.
2. Materials and methods
2.1. Mouse maintenance and genotyping
Mutant mice were crossed as previously described (Broders-Bondon et al., 2012). Ht-PA-Cre;Ncadneo/+ breeding males were crossed with Ncadfl/fl;beta1fl/fl;R26RYFPfl/fl females. Two genotypes were obtained in the progeny: Ht-PA-Cre;Ncadneo/fl;beta1+/fl;R26R YFP+/fl (referred to as Ncad−/− or the Ncad mutant), and Ht-PA-Cre;Ncad+ /fl;beta1+/fl;R26R YFPwt/fl (referred to as controls). ENCCs from Ncad mutants are referred to as Ncad−/− ENCCs. This crossing strategy allows the expression of the YFP reporter protein in all the ENCCs.
The protocol was approved by the Committee on the Ethics of Animal Experiments of the Institut Curie (National registration number: #118). Genotyping was performed with primers synthesized by Eurogentec (Belgium); the primers for Ht-PA-Cre mice have been described elsewhere (Pietri et al., 2003). The sense and antisense primers used for N-cadherin neo amplification have been described elsewhere (Broders-Bondon et al., 2012).
2.2. Micropatterns
Micropatterns were created with deep UV as previously described (Azioune et al., 2009; Lautenschlager and Piel, 2013), with 13 μm-wide micropatterned lines separated by 40 μm-wide nonadhesive intervals. They were incubated with Alexa 594-conjugated fibronectin (FN) (Molecular Probes, Eugene, OR) and unlabeled FN (Sigma F1141, 10 μg/ml) and were used as substrate for gut explants.
2.3. Antibodies and immunostainings
The C3b/C3c and C3a complement monoclonal antibodies (mAb2/11 and 3/11, respectively) have been described elsewhere (Mastellos et al., 2004). They were used at a dilution of 1/100 and 1/50, respectively. C3aR antibody was obtained from Santa Cruz Technology and used at a dilution of 1/50. C3a agonist peptide (H-CNHITKLREQHRRDHVLGLAR-COOH sequence) was synthesized as previously described (Hoeprich and Hugli, 1986). The selective C3aR antagonist peptide was obtained from Calbiochem (SB290157) and used at concentrations of 100 nM in gut explant cultures and 20 μM for whole gut ex vivo treatment, respectively. Paraffin sections were immunostained as previously described (Pietri et al., 2004), using primary antibodies for the following cell markers: P75NTR receptor (Promega ab8875, 1/250), Sox10 N-20 (Santa Cruz Technology, 1/50), TUJ1 (Eurogentec, 1/1000), β1-integrin (Chemicon 9EG7, 1/100), paxillin (BD Bioscience, 1/100), phosphorylated cortactin (Cell Signaling, 1/50), GFP (Molecular Probes A11122, 1/200), Dapi (Molecular Probes, 1/ 10,000). Alexa Fluor 488- and 647-conjugated secondary antibodies were obtained from Invitrogen. CY3-conjugated secondary antibodies were obtained from Jackson ImmunoResearch. All were used at a dilution of 1/500. Whole-mount immunostaining of the embryonic guts for TUJ1 and YFP were performed as previously described (Stanchina et al., 2006).
2.4. Organotypic cultures, video time-lapse imaging and confocal microscopy
Cultures of E12.5 midgut rings on fibronectin (FN, Sigma F1141, 10 μg/ml) and ex vivo gut cultures were carried out as previously described (Breau et al., 2009).
Briefly, midgut rings were cultured in defined culture medium made of DMEM-F12 (Invitrogen) supplemented with glutamine (Gibco 25030-081, 1/100e), penicillin-streptomycin (Gibco 15140-122, 1/100) and ITS (insulin, transferrin, selenium solution, Invitrogen; 1/100). Phase contrast images were captured as described in the legend of movies. Whole guts were cultured in defined culture medium supplemented with 5% horse serum (Babco). In experiments where samples were treated with C3aR antagonist peptide the controls correspond to samples treated with the solvent (DMSO) at the same dilution. For ex vivo timelapse imaging, gut segments were placed in a drop of growth factor-reduced Matrigel (Becton Dickinson) on the bottom of a microdish in DMEM/F12 (Invitrogen)+ 5% horse serum (Babco). YFP images were captured every 10 min for at least 12 h.
Video-time lapses of ex vivo gut cultures and in vitro gut explant cultures were carried out with a Nikon Eclipse Ti inverted video-microscope equipped with a cooled CDD-camera, at 37 °C, in a humid atmosphere containing 5% CO2.
Image acquisition and analysis were performed on workstations of the Nikon Imaging Centre @ Institut Curie-CNRS" and Institut Mondor de Recherche Biomédicale, U955 INSERM (IMRB). Whole-mount immunostained guts were analyzed with an inverted confocal A1-R Nikon and LSM 510 confocal microscopes.
2.5. Image analysis
Individual ENCCs of ex vivo gut cultures were tracked manually using Metamorph 7.7.3.0 software to determine their mean speed of locomotion as described previously (Breau et al., 2009). Directionality was evaluated by measuring the angle between the rostro-caudal axis of the gut and the straight line separating the initial and final positions of the cell. The rostro-caudal axis was identified at the initial position of the cell by the median axis of the hindgut cylinder portion that is parallel to the gut wall. The caudal and rostral orientation corresponds to 0° and 180°, respectively. Cells migrating caudally toward the right and left side of the hindgut correspond to an orientation at 90° and 270°, respectively.
Individual ENCCs from in vitro cultures on 2D-surfaces and FN-stripes were tracked manually with the Manual Tracking plugin available from http://rsb.info.nih.gov/ij/plugins/track/track.html running under ImageJ/FIJI software. The output was a new stack in which cell centers were represented as dots, and an Excel file containing for each cell trajectory, the positions occupied by the cell over time.
For the analysis of cell dispersion following chemokine treatment, we devised an ImageJ macro determining for each cell, represented as a dot, its two closest neighbors, on the basis of a Delaunay triangulation algorithm, and the areas of the triangles formed, which were proportional to cell dispersion. For the analysis of cell movements on 2D and FN-stripes, we developed a Matlab script for determining, from the Excel file containing the manual tracks: (i) the directionality, the index of which was calculated as the distance between the start and end points (a Euclidean distance) divided by the length of the actual path taken by the cells (a geodesic distance, i.e. the sum of all displacements), (ii) the mean speed, in μm per frame, calculated as the geodesic distance divided by the number of time points, which is not associated with any particular notion of direction; (iii) the exploration speed in the main direction (μm per frame), calculated as the longest axis of the ellipse fitting the convex hull of the trajectory divided by the number of time points, associated with the notion of efficient movement in one direction. The non-parametric Kruskal-Wallis test for multiple comparisons was used for statistical analysis (in Matlab). This analysis workflow is described in Supplementary Fig. 1.
For the quantification of overlapping events between ENCCs, we used the ImageJ tracking plugin. We analyzed migration of 7 adjacent cells during 3 h on gut explant cultures on 2D. For quantification of the influence of C3a on cell adhesion, focal adhesions (FAs) were segmented by manual thresholding. The number, area and Feret's diameter of the FAs were then quantified with the Analyze Particle plugin of ImageJ. Statistical analysis was performed using Student’s t test. For quantification of the influence of the C3aR antagonist on ENCC network during gut colonization, the number of isolated cells and chains that are disconnected from the network (“broken chains”) were counted on Z-stack confocal acquisitions performed on whole-mount immunostained guts. Statistical significance of differences was assessed in two-tailed Mann-Whitney U test.
3. Results
We analyzed the cellular and molecular mechanisms governing the adhesive and migratory behavior of ENCCs during the colonization of the developing gut in mice, focusing on the putative role of C3a and its cooperation with N-cadherin-mediated cell-cell adhesion.
3.1. Complement C3 fragments are specifically present in enteric neural crest cells during early embryonic development
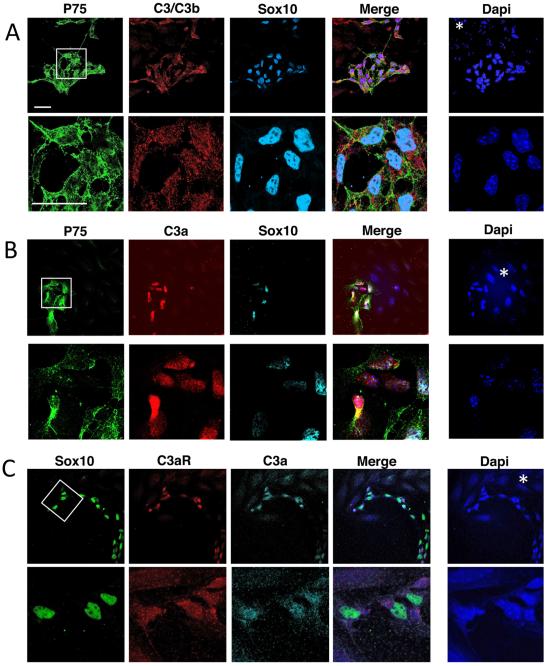
On E12.5 embryonic gut sections, we observed that ENCC progenitors (p75NTR + , YFP + , Sox10 + ) are stained for activated C3b/iC3b (Fig. 1A). At higher magnification (Fig. 1A, bottom panels), p75NTR was clearly located at the membrane, Sox10 was clearly present in the nucleus and C3b/iC3b was detected in the cytoplasm and at the membrane of ENCCs. A staining was also observed in the proximity of ENCCs, associated with cells negative for Sox10 and p75NTR. The mesenchymal, endothelial, epithelial and muscle cells (Dapi + , p75NTR − , Sox10− , Fig. 1A, *) only exhibited a faint staining for C3b/iC3b, suggesting that C3b/iC3b were mainly expressed by ENCCs migrating in the developing intestine. ENCCs were labeled with the anti-C3a antibody (Fig. 1B and Supplementary Fig. 2A), whereas p75NTR − cells were not, except some higher staining observed in few cells located close to the epithelium. Observations at a higher magnification (Fig. 1B, bottom panels) showed C3a to be located mostly in the cytoplasm of ENCCs and in a lesser extent at the membrane.
Fig. 1.
Localization of C3 fragments and C3aR in E12.5 gut. (A) Confocal images of midgut sections immunostained with antibodies against C3b/iC3b (red), P75NTR (P75, green), YFP (cytoplasmic staining, green), Sox10 (nuclear staining, cyan) and labeled with Dapi (blue). (B) Confocal images of E12.5 embryo sections at the level of gut and immunostained with antibodies against C3a (red), P75NTR (green) and labeled with Dapi (blue). (C) Confocal images of midgut sections with antibodies against C3aR (red), P75NTR (green), YFP (green). The white boxes in the upper panels indicate the regions shown at higher magnification in the lower panels. The white asterisk and arrows indicate the position of mesenchymal cells and ENCCs, respectively. Scale bars in A represent 30 μm. Scale bar in C represent 50 μm.
In the gut sections stained with anti-C3aR antibody we observed a faint signal in the gut tissue and a higher staining for C3aR in a subset of ENCCs (Fig. 1C, Supplementary Fig. 2B upper panels). Some p75NTR − cells positive for C3aR were found in the mesenchymal layer (Supplementary Fig. 2B, white arrows) located close to ENCCs (Supplementary Fig. 2B, white arrowheads) or close to the epithelium. In the sections stained with control Ig we observed some background staining that does not correspond to ENCC (Supplementary Fig. 2B lower panels and C). The presence of C3b/iC3b, C3a and C3aR in ENCCs was observed on nine gut sections from embryos from three independent crosses.
3.2. Complement C3 fragments are specifically present in enteric neural crest cells in gut explant cultures
We also analyzed the presence of C3a fragments in gut explants cultured in vitro (Fig. 2). In these 2D cultures, ENCCs usually migrate outside of the explant, often before the escape of mesenchymal cells, and the ENCCs are mostly located at the edge of the outgrowth. Fig. 2 shows confocal images of ENCCs at the periphery of the explants. The enteric progenitors displayed staining for C3b/ iC3b, C3a and C3aR (Fig. 2A–C, respectively) whereas mesenchymal cells did not contain these molecules. At higher magnification, the C3b/iC3b fragments were localized principally in the cytoplasm of ENCCs, whereas C3a and C3aR were present in both the cytoplasm and nuclei. C3aR has recently been detected in the nuclei of mesenchymal stem cells, suggesting that it is translocated into the nucleus following stimulation with C3a (DiScipio et al., 2013). A similar process probably occurs in ENCCs. Identical results, with C3b/iC3b, C3a and C3aR specifically detected in ENCCs, were obtained for seven gut explants from embryos from four independent crosses.
Fig. 2.
Detection of C3 fragments and C3aR in E12.5 midgut explant cultures. (A) Confocal images of cultures immunostained with antibodies against C3b/iC3b fragments (red), P75NTR (P75, green), YFP (cytoplasmic staining, green), Sox10 (nuclear staining, cyan) and labeled with Dapi (blue). (B) Confocal images of cultures immunostained with antibodies against C3a (red), YFP (green), Sox10 (cyan) and labeled with Dapi (blue). (C) Confocal images of cultures showing the expression of C3aR (red) by ENCC progenitors (Sox10 + ; nuclear, green). The white boxes in the upper panels indicate the regions shown at higher magnification in the lower panels. The white asterisk shows the position of mesenchymal cells. Scale bars in A represent 30 μm.
3.3. Blockade with a C3aR antagonist affects gut colonization and neuronal network organization
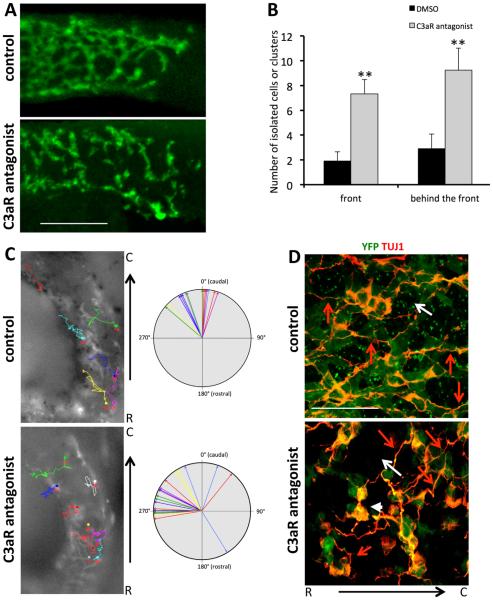
Our results suggest that the anaphylatoxin C3a and its receptor may be involved in the colonization of the developing gut by ENCCs. We investigated whether the C3a anaphylatoxin was required during ENCC migration within the gut wall, by carrying out ex vivo experiments in the presence of C3aR antagonist, which specifically blocks the C3a-C3aR interaction (Ames et al., 2001). We treated E12.5 guts with (n = 6) and without the C3aR antagonist (n = 5) for 16 h. We did not observe a significant delay in the caudal advance of the front in the presence of the C3aR antagonist during the overnight treatment compared to control. We mostly observed an effect on the organization of ENCCs within the gut wall. By contrast to the control conditions, in which ENCCs formed a network of ramified and interconnected chains of cells in the hindgut (Fig. 3A, upper panel), in the presence of the C3aR antagonist, ENCCs were more dispersed and the network more discontinuous with disconnected chains from the network (Fig. 3A, lower panel). This refiects the formation of isolated groups of cells that did not form a continuous network of connected cells resembling that formed by control ENCCs. We quantified the number of isolated ENCCs and disconnected chains from the network (“broken chains”), in a zone covering 200 μm from the migratory front or 200 μm behind this zone. We observed a significant increase in the number of single cells and disconnected chains in C3aR antagonist-treated guts compared to control guts (Fig. 3B).
Fig. 3.
The addition of a C3aR antagonist disturbs gut colonization by ENCCs. (A) Whole-mount anti-GFP immunostaining of E12.5 control (solvent-treated, upper panel) and C3aR antagonist-treated (lower panel) gut taken at the level of the migratory front. ENCCs are visible in green. (B) Quantification of the number of isolated cells, disconnected chains or clusters (broken chains) in control (DMSO-treated, black bars) and C3aR antagonist-treated gut (gray bars) counted in a zone covering 200 μm from the migratory front (front) or 200 μm behind this zone (behind the front). Error bars are SEM, **p < 0.01. (C) Mean directionality of ENCCs tracked in ex vivo gut culture in control condition (in the presence of solvent, DMSO) and in the presence of the C3aR antagonist. Directionality was evaluated by measuring the angle between the rostro-caudal axis of the gut and the straight line separating the initial and final positions of the cell. Each arrow corresponds to individual ENCC. The pictures show individual trajectories of ENCCs (YFP + ) within the proximal hindgut in control condition (n = 16 cells) and in the presence of the C3aR antagonist (n = 23 cells). The tracks overlay the first image in the time series, with the initial positions of the cells indicated by circles. (D) Whole-mount TUJ1 immunostaining (red) of E12.5 gut. ENCCs are visible in green. The ENCC network in C3aR antagonist-treated guts (bottom panel) is disorganized with larger ENCC-free spaces (white arrow) and small groups of non-cohesive cells (white arrowhead) compared to control gut (upper panel). Red arrows point on the orientation of neurites and ENCC network Scale bars in A and D represent 100 and 20 μm, respectively.
We then analyzed the effect of C3aR antagonist on YFP + ENCC dynamics in the mouse proximal hindgut tissue by performing time-lapse video-microscopy on ex vivo whole gut culture. At the migratory front of guts treated with C3aR antagonist, ENCCs only exhibit a slight decrease speed of locomotion (mean speed: 23 ± 4 μm/h; n = 8) compared to control ENCCs (mean speed: 26.7 ± 3 μm/h; n = 10; Student’s t-test, p Value < 0.05). The ENCC directionality of C3aR antagonist-treated guts was perturbed and characterized by more trajectories oriented perpendicularly to the rostro-caudal axis compared to control cells (Fig. 3C). The behavior of ENCCs in the C3aR antagonist-treated gut was similar to that of Ncad−/− ENCCs (Broders-Bondon et al., 2012), which displayed transient adhesion and altered directionality.
In the colonized part of the C3aR antagonist-treated E12.5 mouse gut, we observed a disorganized network of ENCCs (Fig. 3D, n = 3), with empty spaces (white arrow) and small groups of noncohesive cells (white arrowhead). By contrast, the control cells formed a network in which the neurons (TUJ1 + ) and ENCCs (YFP + ) were well connected (Fig. 3D, n = 3). Neurites (red arrows) and ENCC network were mostly oriented parallel to the rostrocaudal axis in the control while they were randomly oriented in C3aR antagonist-treated E12.5 mouse gut. This may refiect in part the impact of C3aR antagonist on the organization of ENCC chains that are disrupted and progression of the ENCCs in the gut wall during the colonization process.
3.4. ENCC co-attraction is regulated by C3a and requires N-cadherin
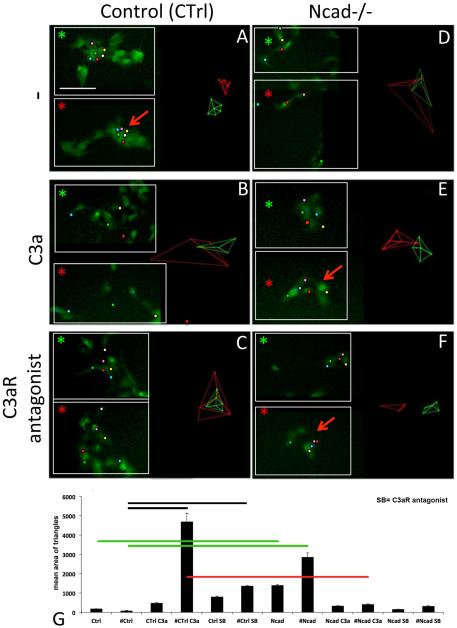
In vitro, while migrating ENCCs interact together via transient cell-cell interaction, they exchange their neighbors within a cluster that is not a strongly cohesive group of cells. An additional parameter to transient adhesion should be necessary for migratory ENCCs to stay close together in these conditions. We investigated the contribution of C3a-C3aR to the maintenance of the ENCC cluster and thus the attraction between ENCCs during their migration, by carrying out gut explant cultures in the presence of C3aR agonist or C3a. We analyzed the dynamic behavior of small groups of adjacent ENCCs in response to the factors added. We studied both control ENCCs (Ctrl, Fig. 4A–C) and N-cadherin-null ENCCs (Ncad−/− ; Fig. 4D–F) treated with C3a (B, E; n = 18) and C3aR antagonist (C, F; n = 15), comparing these cells with untreated cells (A, D; n = 18). Time-lapse video microscopy was performed, with one image taken every five minutes, before and immediately after the addition of factors, over a period of 3.5 h. We followed the dynamic behavior of groups of adjacent cells migrating out of gut explants (Fig. 4); small inserts in each panel show the positions of the tracked cells at t = 0 (green asterisk) and t = 3.5 h (red asterisk). Groups of five or six adjacent ENCCs were tracked manually and cell dispersion was quantified with the Delaunay triangulation algorithm (see supplementary Fig. 1), as previously described (Carmona-Fontaine et al., 2011). This algorithm determines the two closest neighbors for each cell, and determines the areas of the triangle formed by the space between these cells, providing an indication of cell dispersion. Panels A–F of Fig. 4, show the Delaunay triangles corresponding to analyses over the two time periods: the first 25 min (green triangles), and all time points from 75 min to 3.5 h (red triangles).
Fig. 4.
Dynamic analysis of ENCCs in gut explants cultured in vitro on a 2D FN-coated substrate. (A–C) Behavior of control ENCCs (Ctrl, YFP + , green) in culture medium (A), and with C3a (B) or the C3aR antagonist (C). In ENCC clusters, the tracked cells are highlighted with colored dots at two time points: t = 5 minutes (top inset, green asterisk) and t = 3.5 h (bottom inset, red asterisk). The Delaunay triangles obtained are shown on the left part of the panels. (D–F) Behavior of Ncad−/− ENCCs in culture medium (D), and with C3a (E) or the C3aR antagonist (F). (G) Quantitative analysis of the Delaunay triangulation results. The mean areas of Delaunay triangles are combined for each set of conditions into two groups. One group corresponds to the first 25 min of time-lapse video microscopy for untreated control (Ctrl) and Ncad −/− ENCCs (Ncad), for C3a-treated control (Ctrl C3a) and Ncad −/− ENCCs (Ncad C3a), and for control (Ctrl SB) and Ncad −/− ENCCs (Ncad SB) treated with C3aR antagonist (SB). The second group corresponds to all time points after 75 min of time-lapse video microscopy for untreated control (#Ctrl) and Ncad −/− ENCCs (#Ncad), for C3a-treated control (#Ctrl C3a) and Ncad −/− ENCCs (#Ncad C3a), and for control (#Ctrl SB) and Ncad −/− ENCCs (#Ncad SB) treated with C3aR antagonist (SB). The black, green and red lines indicate significant differences (p < 0.01) between the corresponding conditions. The errors bars are SEM. Scale bar in A represents 30 μm.
The untreated control (Ctrl) ENCCs remained close to their neighbors (Fig. 4A, similar areas for the green and red triangles), whereas treatment with soluble C3a (Fig. 4B) or C3aR antagonist (Fig. 4C) significantly increased cell dispersion, resulting in larger triangle areas (compare the green and red triangles in Fig. 4B and C). Thus, exogenous C3a stimulates ENCCs and favors their dispersion from the neighbors. By contrast, C3aR antagonist inhibits C3aR and impedes the maintenance of ENCC clusters, thereby promoting their dispersion. This indicates that C3a/C3aR is required for ENCCs to keep their neighbors close to them and suggests that an attractive effect counteracts dispersion.
Ncad−/− ENCCs displayed weaker cell-cell adhesion (Fig. 4D) than control ENCCs (Fig. 4A), but they nevertheless retained some capacity for intercellular adhesion, mediated by the other adhesion receptors on their surface as previously described (Broders-Bondon et al., 2012). Ncad−/− ENCCs located in close proximity to each other dispersed over time. The addition of exogenous soluble C3a and C3aR antagonist to Ncad−/− cells did not promote dispersion (Fig. 4E and F, red arrow). Thus, N-cadherin is implicated in the response of ENCCs to C3a.
After 75 min, control ENCCs (#Ctrl) displayed significantly lower levels of dispersion (p value < 0.01) than ENCCs treated with C3a (#Ctrl C3a) and C3aR antagonist (#Ctrl, SB) (Fig. 4G, black lines), and Ncad−/− ENCCs (#Ncad) (Fig. 4G, green lines). Furthermore, the dispersion response of Ncad−/− ENCCs to C3a (# Ncad C3a) was significantly weaker (p value < 0.01) than that of control ENCCs (#Ctrl C3a) (Fig. 4G, red lines), and not significantly different from that of untreated Ncad−/− cells (#Ncad). Our results indicate that C3a/C3aR-mediated co-attraction is involved in and required to maintain the cohesiveness between ENCCs, a process that requires N-cadherin function.
3.5. The ENCC behavior is regulated by C3a- and N-cadherin-mediated responses on 2D surfaces
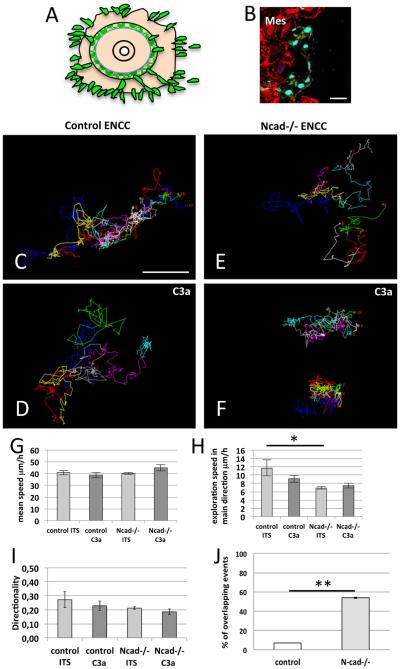
The effect of C3a on the migratory behavior of ENCCs was analyzed by time-lapse imaging overnight, on surfaces (2D, with a homogeneous FN coating), with one image taken every five minutes beginning just before and continuing for six hours after the addition of C3a. On 2D surfaces, ENCCs escaped from the explant with mesenchymal cells (Fig. 5A and B) but are often observed at the periphery of the outgrowth. ENCCs were able to move in any direction (Fig. 5C–F). Control ENCCs had more dispersed trajectories and explored a larger territory in the presence of C3a than untreated cells (Fig. 5C, n = 11; D, n = 13). The trajectory of neighboring ENCCs with time (Fig. 5C) appeared more intricate in control conditions than in the presence of C3a. In contrast, Ncad−/− ENCCs had less dispersed trajectories in the presence of C3a than in its absence (Fig. 5E, n = 13; F, n = 12). Thus, control and C3a-treated Ncad−/− ENCCs on the one hand, and Ncad−/− and C3a-treated control ENCCs on the other, had similar behavior.
Fig. 5.
Analysis of the migratory behavior of ENCCs on 2D substrates. (A) Diagram of the organotypic culture of gut explants with ENCCs escaping the explant and moving on 2D-FN-coated substrates (ENCCs, green; other cells of the gut in light orange). (B) Immunofluorescence staining of a gut explant culture after 18 h for YFP (green) and paxillin (red), with Sox10 (cyan), showing ENCCs (Sox10 + ) and mesenchymal cells (highly positive for paxilline staining, Mes). (C–E) Representative tracks followed by the ENCCs on FN coated-surfaces for control (C, D) and Ncad−/− ENCCs (E, F) in culture medium without (C, E) or with C3a (D, F). (G–I) Quantification of mean speed (G), exploration speed (H) and directionality (I). (J) Quantification of overlapping/crossing events between control (white bar) and Ncad −/− (gray bar) ENCCs. Error bars are SEM, *p < 0.05. Scale bars in B and C represent 30 and 50 μm, respectively.
We observed that Ncad−/− cells entering into contact with other ENCCs displayed protrusions extending toward the top of contacting cells (overlaps) and had the tendency to cross them. We quantified the number of overlapping/crossing events between control (n = 30) and Ncad−/− (n = 31) ENCCs on 2D surface (Fig. 5J) by analyzing the behavior of groups of 7 adjacent cells from 3 distinct experiments. Movies 1 and 2 in the supplementary materials illustrate the behavior of a group of control and Ncad−/− ENCCs, respectively. We detected that a small % of moving ENCCs were able to overlap or cross their neighbors upon contact. In contrast, we observed a strong increase in these events for Ncad−/− cells (p value = 0.01). This indicates that control and Ncad−/− ENCCs exhibit a different migratory behavior upon intercellular adhesion. This could resemble a CIL behavior, which is abolished in Ncad−/− cells.
We next analyzed the effect of cell-cell contact on in vitro ENCC behavior by time-lapse imaging at a higher magnification, with images taken at a much higher frequency, once every 10 s. Movie 3 in the supplementary materials illustrates the intercellular adhesion-mediated changes in the direction of control ENCC migration characteristic of the contact-dependent repolarization of cells causing them to move away from other cells, in a process of CIL. In this movie, we can see an ENCC located on the left migrating towards another ENCC located on the right. Upon contact, the cells retract and the cell on the left repolarizes in the opposite direction and moves away. Thus, in addition to a process of co-attraction between ENCCs controlled by C3a-C3aR-dependent mechanisms, these cells can respond to transient contact with each other, through a CIL mechanism.
Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2016.03.022.
3.6. The ENCC behavior is regulated by C3a- and N-cadherin-mediated responses on FN-stripes
In situations in which mesenchymal cells are located beyond the ENCCs, this might affect ENCC behavior. We investigated this aspect with a micropatterning strategy, making it possible to control cell shape and behavior, including migration (Thery et al., 2006; Thery and Bornens, 2006; Thery and Piel, 2009). Gut explants were deposited on substrates micropatterned with 13 μm wide straight FN-stripes (Fig. 6A and B). The choice of dimension for these lines was based on the size of adherent ENCCs, to prevent the strong effects on cell motility observed with narrower stripes (Doyle et al., 2009). The first cell to escape from the explant at a FN-stripe was the first to migrate on it; it can be an ENCC or a mesenchymal cell. Because ENCCs are often found escaping first the explant as group of cells we often observed several ENCCs migrating along the stripes but more rarely a mixture of ENCCs and mesenchymal cells. This made it possible to carry out a more detailed analysis of ENCC migration behavior, as previously reported for cranial NCCs (Scarpa et al., 2013). The migratory behavior of control and Ncad−/− ENCCs observed on cell tracking indicated that the cells moved as expected on the basis of the spatial restrictions imposed by the patterns (Fig. 6C–F), as shown by comparisons with 2D conditions. Control ENCCs had longer trajectories on FN-stripes in the presence of C3a (Fig. 6D, n = 17) than in its absence (Fig. 6C, n = 13). More back and forth behaviors on stripes were observed for control ENCCs than for C3a-treated cells indicating a tendency for ENCCs to move toward other ENCCs. In contrast, Ncad−/− ENCCs had more dispersed trajectories in the absence of C3a (Fig. 6E, n = 7) than in its presence C3a (Fig. 6F, n = 8). The cells therefore migrated to similar extents in 2D and FN-stripes cultures. Similar patterns of ENCC migration were observed for the cells of embryos from two independent crosses.
Fig. 6.
Analysis of the migratory behavior of ENCCs on FN stripes. (A) Diagram of gut explant cultured on micro-patterned-FN stripes (13-μm FN-stripes /40-μm non-adhesive stripes; ENCCs, green; other cells of the gut, light orange). (B) Immunostaining of gut cultures for Sox10 and P75 (green), and cortactin (red), with Dapi labeling (blue), on Alexa594-coupled stripes of FN (cyan). The ENCCs migrate on the stripes (merge). (C–F) Representative tracks followed by ENCCs on FN stripes for control (C, D) and Ncad − / − ENCCs (E, F) without (C, E) or with C3a (D, F). (G–I) Quantification of mean speed (G), exploration speed (H) and directionality (I) in culture medium (ITS) without or with C3a. Error bars are SEM, *** p < 0.005. Scale bars in B and C represent 30 and 50 μm, respectively.
3.7. The migratory parameters are differentially affected on 2D surfaces and FN-stripes
We decided to analyze ENCC behavior more precisely, quantifying it with other parameters in a mathematical analysis, as described in the Materials and Methods. We analyzed the tracks for mean speed, exploration speed and directionality. Mean speed, regardless of direction, did not differ significantly between ENCCs with and without C3a treatment or between control and Ncad−/− cells on 2D surface and FN-stripes (Figs. 5G and 6G). The exploration speed was used as an indicator of the efficiency of movement in the principal direction of migration. Exploration speed was significantly lower in Ncad−/− ENCCs, which moved more randomly towards the main direction than control ENCCs on FN-stripes. We observed differences between 2D and FN-stripes conditions (Figs. 5H and 6H), with a lower exploration speed for control C3a-treated ENCCs in 2D (p < 0.05) but no difference in exploration speed in the presence and absence of treatment on FN-stripes. Finally, we analyzed the directionality index, which was calculated by dividing the straight-line distance between the start and end points by the length of the actual trajectory. The values of this index can vary between 1 (directional: shortest straight line) and close to 0 (non-directional: trajectories frequently changing direction and covering long distances). The directionality and mean speed values of ENCCs were slightly lower on FN-stripes than on 2D substrates, due to the constraints imposed by the stripes, forcing the cells to migrate forward or to change direction and migrate backward (Scarpa et al., 2013), whereas they can move in any direction on 2D substrates. Control ENCCs treated with C3a had a higher directionality than untreated ENCCs (p < 0.005) on FN lines, but no such difference was observed on 2D substrates. Thus, the restricted migration of C3a-treated cells on FN lines made their directionality predictable, highlighting the effect of this exogenous chemoattractant on the migratory behavior of ENCCs. By contrast, Ncad−/− ENCCs behaved similarly in the presence and absence of C3a, confirming that mutant ENCCs do not respond to C3a.
3.8. C3a modulates the size of focal adhesion sites
Gut explants cultured for 24 h on FN-coated surfaces were examined by labeling with antibodies against Sox10 (enteric progenitors), paxillin (maker of focal adhesions, FA) and activated beta1 integrin (open conformation of the receptor) to identify cell-FN adhesion sites and with Dapi (Fig. 7). The area and Feret's diameter of FA were quantified. Feret's diameter is the longest axis of the FA. The area and length of paxillin-positive FAs were significantly lower for ENCCs treated with C3a or C3aR antagonist than for untreated cells (Fig. 7A–C, p < 0.0001). Those of FAs positive for activated beta1 integrins were significantly lower for ENCCs treated with C3a than for untreated cells (Fig. 7D–E, p < 0.0001), indicating that C3a/C3aR stimulation modulates the cell-matrix adhesion properties of ENCCs. By contrast, Ncad −/− ENCCs had smaller FAs (activated beta1 + , p < 0.01) than control cells and the size of their FAs was not modified by C3a treatment (Fig. 7D and E), indicating that N-cadherin depletion modulates the cell-matrix adhesion properties of ENCCs and that this molecule is required for the C3a-dependent modulation of ENCC adhesions.
Fig. 7.
Effects of C3a/C3aR on the FAs of ENCCs. (A) Immunofluorescence staining of ENCCs cultured without (–), with C3a, C3aR antagonist or a mixture of C3a + C3aR antagonist, for paxillin (red), YFP (green), Sox10 (cyan), and with Dapi labeling (blue). The small white boxes in the upper panels indicate the regions shown the paxillin staining at higher magnification in the left inserts. (B, C) Quantification of paxillin-positive adhesion sites (FAs): the graphs show the mean area (B) and mean Feret's diameter (C)( ± SEM). n > 40 cells; n > 300 sites and n > 50 cells; n > 500 sites from control cells with and without C3a treatment; n > 20 cells; n > 79 sites and n > 24 cells; n > 400 sites from control cells with C3aR antagonist and C3a + C3aR antagonist, respectively. Two independent experiments were carried out for this analysis. (D, E) Quantification of activated beta1 integrin-positive adhesion sites: the graphs show the mean area (E) and mean Feret's diameter (F) ( ± SEM). n > 40 cells; n > 300 sites and n > 50 cells; n > 500 sites from control cells with and without C3a treatment. n > 20 cells; n > 260 sites and n > 15 cells; n > 220 sites from Ncad −/− cells without and with C3a treatment, respectively. Two independent experiments were carried out for this analysis. Error bars are SEM, ****; p < 0.0001. **, p < 0.01). Scale bar in A represents 10 μm.
4. Discussion
4.1. Chemokine and colonization of the gut by ENCCs
In the fetal gut, ENCCs progress through the gut mesenchyme, from which they receive environmental signals. GDNF and endothelin-3 (EDN-3) are two of the main factors expressed by the gut mesenchyme. Their downstream signaling cascades regulate ENCC behavior (Goto et al., 2013) and are critical for the survival, proliferation and maintenance of the pool of ENS progenitors, which are important parameters regulating ENCC migration and gut colonization, and their differentiation (Hearn et al., 1998; Nagy and Goldstein, 2006; Hotta et al., 2010; Barlow et al., 2003; Simpson et al., 2007). GDNF is increasingly expressed in the developing gut mesenchyme at the stage of the colonization by ENCCs. It is enriched in the cecum and proximal midgut (Young et al., 2001), and functions as a chemoattractant molecule for ENCC migration in vitro and in vivo (Young et al., 2001; Natarajan et al., 2002). EDN-3 is enriched in the cecum and colon (Leibl et al., 1999). Perturbation of EDN-3/EDNRB signaling impaired gut colonization (Nagy and Goldstein, 2006; Barlow et al., 2003) and decreased their speed of migration in the gut tissue without affecting their directionality (Young et al., 2014). Our study shows that, in addition to these two factors, the Complement anaphylatoxin C3a is expressed in the developing gut by ENCCs (Fig. 1 and supplementary Fig. 2). The perturbation of C3a/C3aR interaction alters the organization of non-solitary ENCCs at the migratory front and behind by disrupting migratory chains and increasing the number of isolated cells with consequence in their speed of locomotion and directionality (Fig. 3). This illustrates a role for the C3a/C3aR signaling during the ENS ontogenesis.
Recent data suggest that individual ENCCs cannot sense the polarity of the gut and are unable to detect chemotactic gradients along it, as revealed by their unbiased random walk behavior (Young et al., 2014). In the case of solitary cells located at a distance from the main ENCC population their random walk behavior may not the signature of a lack of a co-attraction but the result of other effects. Their behavior may depend on other parameters counteracting the co-attraction between ENCCs. Mesenchymal cells surround these isolated ENCCs and this may impede the diffusion of co-attraction signal. Alternatively, solitary cells at the front may respond to another cue, which compete with the coattraction favoring cell dispersion. However, these hypotheses remain to be further investigated.
Xu and colleagues summarize views reconciling contradiction between the self-organizing ability of ENCCs and the roles of the gut mesenchyme-derived factors, because this situation favors the generation of local gradients (Xu et al., 2014). These local gradients may involve chemokines, small secreted proteins that diffuse locally and GDNF has been identified as a good candidate for directing migration (Young et al., 2001). It has been proposed that ENCCs produce GDNF gradients themselves by consuming it locally, thus favoring enteric progenitor migration toward higher levels of untapped GDNF in gut region beyond the migratory front. These stimuli could act in synergy with the C3a-dependent coattraction between ENCCs to regulate their collective behavior during ENS ontogenesis.
4.2. C3a and co-attraction between ENCCs
We observed that ENCCs express C3a and its receptor C3aR (Figs. 1, 2 and supplementary Fig. 2). We did not use heparin-acrylic beads soaked in purified factor to test C3a-dependent attraction process, as described for in vitro chemotaxis assays on clusters of cephalic NCCs (Carmona-Fontaine et al., 2011), because of the cell heterogeneity and topology of gut explants, and the timing of ENCC escape from the explant. We found that purification steps (flow cytometry) isolating ENCCs from the surrounding gut tissue before their culture, in the defined medium we used in the present study, altered the migratory properties of these cells (Broders-Bondon, unpublished data). Instead, we used in vitro techniques based on use of the C3a anaphylatoxin or C3aR antagonist, added to the culture medium in organotypic culture, and devised several approaches to quantifying ENCC dispersion and directionality in the absence or presence of specific factors. While ENCCs are highly migratory they are rarely dispersed and rather tend to stay close to their neighbors forming groups of cells.
We showed that these cells can response to C3a (Figs. 4–6). We expect that in ENCC clusters a significant level of C3a is achieved locally that sustains the ability of cells to stay in close proximity. An external C3a source disrupts the paracrine effect of C3a on surrounding cells and favors cell dispersion, due to the competition between exogenous C3a and the endogenous C3a secreted by ENCCs. This indicates that C3a is responsible for the ability of ENCCs to keep their neighbors close to them and thus to be “attractive” to avoid cell dispersion. If C3aR antagonist perturbs C3a/ C3aR signal the ENCC clusters disperse. Untreated ENCCs migrating in chains on FN stripes exhibit more back and forth behaviors (Fig. 6) compared to C3a-treated cells, which display higher directionality revealing the ability of ENCCs to move toward other ENCCs. Thus C3a/C3aR signal has a short-range effect by maintaining ENCCs together in a cluster contributing to an attractive effect of the ENCCs on neighbors. A C3a-C3aR mediated co-attraction between ENCCs regulates their migratory behavior.
Control ENCC clusters can disperse in response to exogenous C3a or C3a antagonist whereas cells lacking N-cadherin cannot, suggesting that C3a could regulate cell cluster maintenance and migration through a N-cadherin-dependent process.
4.3. Intercellular interactions, contact inhibition of migration and co-attraction in NCCs
The collective migration of cells is a complex process driven by their intrinsic properties, interaction with other cells and response to extrinsic cues. Intercellular contacts between ENCCs favor their directionality during gut colonization (Young et al., 2014). N-cad-herin depletion in ENCC alters their directionality during gut colonization (Broders-Bondon et al., 2012) and provokes a transient delay in the colonization of the developing gut. During gut colonization ENCC chains are often seen in close association with growing axons (Hao and Young, 2009; Landman et al., 2011) and this type of interaction is retained in N-cadherin-depleted ENCCs (Broders-Bondon et al., 2012). Landman and colleagues (Landman et al., 2011) used cellular automata simulations to model and analyze several hypotheses including those of the contribution of axon-dependent and -independent formation of ENCC chains during gut colonization. They identified important parameters governing ENCC patterns. In the second hypothesis it was taken in account the fact that ENCCs prefer to visit a site that has been previously visited by ENCCs, called reinforced random walk, possibly involving mechanisms such as ENCCs locally depositing attractive cues, locally depleting repulsive cues or altering the extracellular matrix (Landman et al., 2011). A C3a-dependent co-attraction could contribute to such mechanism.
Desai et al. (2013) have demonstrated that discrete differences in cell repolarization speed at the <<tail>> and <<head>> part of cell upon CIL contribute to the formation of train of cells from an initial population of isolated cells migrating on 2D-stripes. It would be interesting to probe whether co-attraction can improve or affect the occurrence of this behavior.
Wynn et al. (2013) have analyzed the role of intercellular contact-mediated guidance, including those mediated through filopodia, and cell interaction with extrinsic signals in dictating the “follow-the-leader” mode of cranial NCC migration both using transplants and mathematical simulations. They demonstrated that the persistence of cell-cell contact is an important parameter but alone it does not explain the migration in chain, which required the contribution of additional parameter including cell interaction with the environment (extracellular matrix). ENCCs establish transient contacts but did not retain their neighbors. In this case an additional parameter keeping cells close together may help in regulating their collective behavior and we propose that co-attraction mediated by C3a could contribute to regulate this process, in addition to N-cadherin mediated transient contacts.
4.4. N-cadherin adhesion and C3a/C3aR signaling
Ncad −/− ENCCs exhibited different response to C3a and C3aR antagonist than control cells on gut culture explants (Figs. 4–6). One explanation could be that Ncad −/− cells exhibit a perturbed 98 F. Broders-Bondon et al. / Developmental Biology 414 (2016) 85–99 C3aR signaling or a defective C3aR expression. We have detected very faint staining of ENCCs for C3aR in gut section of Ncad−/− conditional mutant gut (data not shown). However, the staining for the receptor is already faint in control ENCCs and it was not possible to determine if these differences are significant or not. Another possibility is that C3a/C3aR signaling has an effect on cell adhesion properties. We detected small changes in ENCC focal adhesions in the presence of C3a (Fig. 7). It could not be excluded that C3a produces some changes in intercellular adhesion by modifying adhesion half-life or cadherin function. It was shown that C3aR morpholino-treated cells did not have altered N-cad-herin based adhesion (Carmona-Fontaine et al., 2011) but the effect of cranial NCC stimulation by C3a on this process has not been tested. C3a alters E-cadherin expression in epithelial cells (Tang et al., 2009). If such mechanism occurs for N-cadherin, the stimulation by C3a should not change the Ncad −/− ENCC behavior. It would be interesting to assess the C3a impact on ENCC cadherin-based adhesion.
Cranial NCC collective behavior involves a co-attraction process combined to a N-cadherin dependent CIL. We observed that when an ENCC is moving toward another ENCC and came into contact it repolarizes and migrates away, in vitro, (Supplementary movie 3). The analysis of collisions between ENCCs migrating in opposite direction is very difficult to observed in vivo but a CIL behavior can be occasionally observed ex vivo (Young et al., 2014). in vitro timelapse imaging of groups of cells revealed that migrating ENCCs rarely cross their neighbors while more than 50% of Ncad−/− ENCCs can extent protrusions on their neighbors and exhibit a crossing behavior (Fig. 5J). Crossing the neighbors reveals that cells do not display a CIL. The presence of N-cadherin in control ENCCs decrease the crossing behavior indicating that their locomotion is modified upon contact and may reveal of potential for CIL. The combined effects of co-attraction and contact dependent regulation of locomotion could contribute in ENCC collective migration during the colonization of the developing gut. Our results on ENCCs are in agreement with previous studies on cranial NCCs where N-cadherin anti-sense morphant induces wide cell overlapping (Theveneau et al., 2010). CIL of cranial NCCs requires N-cadherin function. A similar N-cadherin dependent regulation of locomotion or CIL process could dictate ENCC collective behavior at least in a subset of cells but this requires further detailed investigation.
4.5. Cell-matrix adhesion during collective ENCC migration
Cell adhesion molecules mediate interactions between ENCCs and between ENCCs and the extracellular matrix along which they migrate during gut colonization. ENCCs express a large repertoire of cell surface receptors of the integrin family, controlling adhesion to the extracellular matrix (Breau et al., 2009; McKeown et al., 2013), and cadherins, controlling intercellular adhesion (Breau et al., 2006; Broders-Bondon et al., 2012; Gaidar et al., 1998). Cellcell and cell-matrix interactions cooperate to ensure the efficient migration of ENCCs and the correct organization of the ENS. ENCCs express N-cadherin strongly, and cadherin-6 and -11 to a lesser extent (Breau et al., 2006). They express a repertoire of integrins, including α4β1, α5β1, α6β1, αvβ1, αvβ3 and αvβ5 (Breau and Dufour, 2009). β1-integrins mediate the interaction of ENCCs with FN, whereas αV-integrins tend to be involved principally in adhesion to vitronectin substrates. Once the integrins have been activated, they recruit structural and signaling proteins that contribute to the maturation of the nascent adhesion towards FA and regulate cell migration. The size and dynamics of FAs are important parameters controlling cell migration. ENCCs are migratory cells producing small focal adhesions compared to the gut mesenchymal cells (Broders-Bondon F. and Gazquez E., data not shown). in vitro, the treatment of ENCCs with C3a reduces FA size and favors cell dispersion. The effect of C3a on ENCC FA size may reflect a change in FA maturation or dynamics. Our results indicate that C3a regulates the adhesion properties of ENCCs, which are probably linked to changes in cell polarity and dispersion during the co-attraction process. C3a was unable to modulate β1-in-tegrin-mediated adhesions in N-cadherin-depleted ENCCs suggesting that these mutant cells may have an impaired response to C3a signaling but this hypothesis remains to be investigated.
5. Conclusions
Co-attraction regulated by the C3a anaphylatoxin and its receptor contributes to ENS ontogenesis. Ncad−/− ENCCs do not respond to the co-attraction effect, which suggests that crosstalk between N-cadherin-mediated adhesion and C3a/C3aR signaling may control the interactions between ENCCs during their collective migration in the developing gut (Fig. 8).
Fig. 8.

Proposed additional mechanisms involved in the collective migration of ENCCs during gut colonization. In addition to other mechanisms, the C3a/C3aR-dependent co-attraction and the N-cadherin dependent regulation of ENCC dispersion upon intercellular contact could contribute to regulate ENCC behavior.
Supplementary Material
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique, the Institut Curie and the Fondation ARC pour la recherche contre le cancer (Grants 4864) and National Institutes of Health Grant AI068730, and by funding from the European Community Seventh Framework Program under grant agreement number 602699 (DIREKT). EG was supported by a PhD fellowship from the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche. We thank Nicolas Carpi and Emmanuel Theriac for advice and discussions on micro-patterns and microfiuidics, Jean-Loup Duband for helpful discussions. Lucie Sengmanivong and Ludovic Leconte from the Nikon Imaging Centre @Institut Curie-CNRS. We thank the PICT-IBiSA Lhomond Imaging facility of Institut Curie and Vincent Fraisier, François Waharte for their help with video and confocal microscopy imaging. We thank the imaging facility of the Institut Mondor de Recherche Biomédicale (IMRB), U955 INSERM and Xavier Decrouy for his help with confocal imaging. We thank Isabelle Grandjean, Sonia Jannet and Stéphanie Boissel, from the mouse facility of Institut Curie, and Rachid Souktani, Marjorie Collery and Damien Fois from the mouse facility of IMRB.
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2016.03.022.
References
- Abercrombie M, Heaysman JE. Observations on the social behaviour of cells in tissue culture. I. Speed of movement of chick heart fibroblasts in relation to their mutual contacts. Exp. Cell Res. 1953;5(1):111–131. doi: 10.1016/0014-4827(53)90098-6. [DOI] [PubMed] [Google Scholar]
- Abercrombie M, Heaysman JE. Observations on the social behaviour of cells in tissue culture. II. Monolayering of fibroblasts. Exp. Cell Res. 1954;6(2):293–306. doi: 10.1016/0014-4827(54)90176-7. [DOI] [PubMed] [Google Scholar]
- Ames RS, et al. Identification of a selective nonpeptide antagonist of the anaphylatoxin C3a receptor that demonstrates antiinflammatory activity in animal models. J. Immunol. 2001;166(10):6341–6348. doi: 10.4049/jimmunol.166.10.6341. [DOI] [PubMed] [Google Scholar]
- Anderson RB, et al. The cell adhesion molecule L1 Is required for chain migration of neural crest cells in the developing mouse gut. Gastroenterology. 2006;130(4):1221–1232. doi: 10.1053/j.gastro.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Azioune A, et al. Simple and rapid process for single cell micro-patterning. Lab Chip. 2009;9(11):1640–1642. doi: 10.1039/b821581m. [DOI] [PubMed] [Google Scholar]
- Barlow A, de Graaff E, Pachnis V. Enteric nervous system progenitors are coordinately controlled by the G protein-coupled receptor EDNRB and the receptor tyrosine kinase RET. Neuron. 2003;40(5):905–916. doi: 10.1016/s0896-6273(03)00730-x. [DOI] [PubMed] [Google Scholar]
- Breau MA, et al. Lack of beta1 integrins in enteric neural crest cells leads to a Hirschsprung-like phenotype. Development. 2006;133(9):1725–1734. doi: 10.1242/dev.02346. [DOI] [PubMed] [Google Scholar]
- Breau MA, et al. Beta1 integrins are required for the invasion of the caecum and proximal hindgut by enteric neural crest cells. Development. 2009;136(16):2791–2801. doi: 10.1242/dev.031419. [DOI] [PubMed] [Google Scholar]
- Breau MA, Dufour S. Biological development of the enteric nervous system. In: Núñez RN, López-Alonso M, editors. Hirschsprung’s Disease: Diagnosis and Treatment. Congenital Disorders – Laboratory and Clinical Research Series; Nova, New York: 2009. pp. 15–44. [Google Scholar]
- Broders-Bondon F, et al. N-cadherin and beta1-integrins cooperate during the development of the enteric nervous system. Dev. Biol. 2012;364(2):178–191. doi: 10.1016/j.ydbio.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C, et al. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456(7224):957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C, et al. Complement fragment C3a controls mutual cell attraction during collective cell migration. Dev. Cell. 2011;21(6):1026–1037. doi: 10.1016/j.devcel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai RA, et al. Contact inhibition of locomotion probabilities drive solitary versus collective cell migration. J. R. Soc. Interface. 2013;10(88):20130717. doi: 10.1098/rsif.2013.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiScipio RG, et al. Complement C3a signaling mediates production of angiogenic factors in mesenchymal stem cells. J. Biomed. Sci. Eng. 2013;06(08):1–13. [Google Scholar]
- Doyle AD, et al. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 2009;184(4):481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckenbrod NR, Epstein ML. The pattern of neural crest advance in the cecum and colon. Dev. Biol. 2005;287(1):125–133. doi: 10.1016/j.ydbio.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Druckenbrod NR, Epstein ML. Behavior of enteric neural crest-derived cells varies with respect to the migratory wavefront. Dev. Dyn. 2007;236(1):84–92. doi: 10.1002/dvdy.20974. [DOI] [PubMed] [Google Scholar]
- Gaidar YA, et al. Distribution of N-cadherin and NCAM in neurons and endocrine cells of the human embryonic and fetal gastroenteropancreatic system. Acta Histochem. 1998;100(1):83–97. doi: 10.1016/S0065-1281(98)80008-1. [DOI] [PubMed] [Google Scholar]
- Goto A, et al. GDNF and endothelin 3 regulate migration of enteric neural crest-derived cells via protein kinase A and Rac1. J. Neurosci. 2013;33(11):4901–4912. doi: 10.1523/JNEUROSCI.4828-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao MM, Young HM. Development of enteric neuron diversity. J. Cell. Mol. Med. 2009;13(7):1193–1210. doi: 10.1111/j.1582-4934.2009.00813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn CJ, Murphy M, Newgreen D. GDNF and ET-3 differentially modulate the numbers of avian enteric neural crest cells and enteric neurons in vitro. Dev. Biol. 1998;197(1):93–105. doi: 10.1006/dbio.1998.8876. [DOI] [PubMed] [Google Scholar]
- Hoeprich PD, Jr., Hugli TE. Helical conformation at the carboxy-terminal portion of human C3a is required for full activity. Biochemistry. 1986;25(8):1945–1950. doi: 10.1021/bi00356a017. [DOI] [PubMed] [Google Scholar]
- Hotta R, et al. Effects of tissue age, presence of neurones and endothelin-3 on the ability of enteric neurone precursors to colonize recipient gut: implications for cell-based therapies. Neurogastroenterol. Motil. 2010;22(3):331–e86. doi: 10.1111/j.1365-2982.2009.01411.x. [DOI] [PubMed] [Google Scholar]
- Kulesa PM, McLennan R. Neural crest migration: trailblazing ahead. F1000prime Rep. 2015;7:02. doi: 10.12703/P7-02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JI, Heuckeroth RO. Enteric nervous system development: migration, differentiation, and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;305(1):G1–24. doi: 10.1152/ajpgi.00452.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman KA, et al. Building stable chains with motile agents: Insights into the morphology of enteric neural crest cell migration. J. Theor. Biol. 2011;276(1):250–268. doi: 10.1016/j.jtbi.2011.01.043. [DOI] [PubMed] [Google Scholar]
- Lautenschlager F, Piel M. Microfabricated devices for cell biology: all for one and one for all. Curr. Opin. Cell Biol. 2013;25(1):116–124. doi: 10.1016/j.ceb.2012.10.017. [DOI] [PubMed] [Google Scholar]
- Leibl MA, et al. Expression of endothelin 3 by mesenchymal cells of embryonic mouse caecum. Gut. 1999;44(2):246–252. doi: 10.1136/gut.44.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie JD, Mayor R. Complement in animal development: unexpected roles of a highly conserved pathway. Semin. Immunol. 2013;25(1):39–46. doi: 10.1016/j.smim.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastellos D, et al. Novel monoclonal antibodies against mouse C3 interfering with complement activation: description of fine specificity and applications to various immunoassays. Mol. Immunol. 2004;40(16):1213–1221. doi: 10.1016/j.molimm.2003.10.019. [DOI] [PubMed] [Google Scholar]
- McKeown SJ, Wallace AS, Anderson RB. Expression and function of cell adhesion molecules during neural crest migration. Dev. Biol. 2013;373(2):244–257. doi: 10.1016/j.ydbio.2012.10.028. [DOI] [PubMed] [Google Scholar]
- Nagy N, Goldstein AM. Endothelin-3 regulates neural crest cell proliferation and differentiation in the hindgut enteric nervous system. Dev. Biol. 2006;293(1):203–217. doi: 10.1016/j.ydbio.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Natarajan D, et al. Requirement of signalling by receptor tyrosine kinase RET for the directed migration of enteric nervous system progenitor cells during mammalian embryogenesis. Development. 2002;129(22):5151–5160. doi: 10.1242/dev.129.22.5151. [DOI] [PubMed] [Google Scholar]
- Newgreen DF, et al. Simple rules for a “simple” nervous system? Molecular and biomathematical approaches to enteric nervous system formation and malformation. Dev. Biol. 2013;382(1):305–319. doi: 10.1016/j.ydbio.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama C, et al. Trans-mesenteric neural crest cells are the principal source of the colonic enteric nervous system. Nat. Neurosci. 2012;15(9):1211–1218. doi: 10.1038/nn.3184. [DOI] [PubMed] [Google Scholar]
- Pietri T, et al. The human tissue plasminogen activator-Cre mouse: a new tool for targeting specifically neural crest cells and their derivatives in vivo. Dev. Biol. 2003;259(1):176–187. doi: 10.1016/s0012-1606(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Pietri T, et al. Conditional beta1-integrin gene deletion in neural crest cells causes severe developmental alterations of the peripheral nervous system. Development. 2004;131(16):3871–3883. doi: 10.1242/dev.01264. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Lambris JD. Complement in immune and inflammatory disorders: therapeutic interventions. J. Immunol. 2013;190(8):3839–3847. doi: 10.4049/jimmunol.1203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa E, et al. A novel method to study contact inhibition of locomotion using micropatterned substrates. Biol. Open. 2013;2(9):901–906. doi: 10.1242/bio.20135504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinjyo N, et al. Complement-derived anaphylatoxin C3a regulates in vitro differentiation and migration of neural progenitor cells. Stem Cells. 2009;27(11):2824–2832. doi: 10.1002/stem.225. [DOI] [PubMed] [Google Scholar]
- Simpson MJ, et al. Cell proliferation drives neural crest cell invasion of the intestine. Dev. Biol. 2007;302(2):553–568. doi: 10.1016/j.ydbio.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Stanchina L, et al. Interactions between Sox10, Edn3 and Ednrb during enteric nervous system and melanocyte development. Dev. Biol. 2006;295(1):232–249. doi: 10.1016/j.ydbio.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Tang Z, et al. C3a mediates epithelial-to-mesenchymal transition in proteinuric nephropathy. J. Am. Soc. Nephrol. 2009;20(3):593–603. doi: 10.1681/ASN.2008040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery M, et al. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA. 2006;103(52):19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery M, Bornens M. Cell shape and cell division. Curr. Opin. Cell Biol. 2006;18(6):648–657. doi: 10.1016/j.ceb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Thery M, Piel M. Adhesive micropatterns for cells: a microcontact printing protocol. Cold Spring Harb. Protoc. 2009;(7) doi: 10.1101/pdb.prot5255. pdb prot5255. [DOI] [PubMed] [Google Scholar]
- Theveneau E, et al. Collective chemotaxis requires contact-dependent cell polarity. Dev. Cell. 2010;19(1):39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Can mesenchymal cells undergo collective cell migration? The case of the neural crest. Cell Adhes. Migr. 2011;5(6):490–498. doi: 10.4161/cam.5.6.18623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, et al. Sox10 and Itgb1 interaction in enteric neural crest cell migration. Dev. Biol. 2013;379(1):92–106. doi: 10.1016/j.ydbio.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Woods ML, et al. Directional collective cell migration emerges as a property of cell interactions. PLoS One. 2014;9(9):e104969. doi: 10.1371/journal.pone.0104969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn ML, et al. Follow-the-leader cell migration requires biased cell-cell contact and local microenvironmental signals. Phys. Biol. 2013;10(3):035003. doi: 10.1088/1478-3975/10/3/035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Heanue T, Pachnis V. Travelling within the fetal gut: simple rules for an arduous journey. BMC Biol. 2014;12:50. doi: 10.1186/s12915-014-0050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young HM, et al. GDNF is a chemoattractant for enteric neural cells. Dev. Biol. 2001;229(2):503–516. doi: 10.1006/dbio.2000.0100. [DOI] [PubMed] [Google Scholar]
- Young HM, et al. Dynamics of neural crest-derived cell migration in the embryonic mouse gut. Dev. Biol. 2004;270(2):455–473. doi: 10.1016/j.ydbio.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Young HM, et al. Colonizing while migrating: how do individual enteric neural crest cells behave? BMC Biol. 2014;12:23. doi: 10.1186/1741-7007-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.