Abstract
At sites of chronic inflammation, epithelial cells are exposed to high levels of reactive oxygen species and undergo cancer-associated DNA methylation changes, suggesting that inflammation may initiate epigenetic alterations. Previously, we demonstrated that oxidative damage causes epigenetic silencing proteins to become part of a large complex that is localized to GC-rich regions of the genome, including promoter CpG islands that are epigenetically silenced in cancer. However, whether these proteins were recruited directly to damaged DNA or during the DNA repair process was unknown. Here we demonstrate that the mismatch repair protein heterodimer MSH2-MSH6 participates in the oxidative damage-induced recruitment of DNA methyltransferase 1 (DNMT1) to chromatin. Hydrogen peroxide treatment induces the interaction of MSH2-MSH6 with DNMT1, suggesting that the recruitment is through a protein–protein interaction. Importantly, the reduction in transcription for genes with CpG island-containing promoters caused by oxidative damage is abrogated by knockdown of MSH6 and/or DNMT1. Our findings provide evidence that the role of DNMT1 at sites of oxidative damage is to reduce transcription, potentially preventing transcription from interfering with the repair process. This study uniquely brings together several factors that are known to contribute to colon cancer, namely inflammation, mismatch repair proteins, and epigenetic changes.
Keywords: mismatch repair, epigenetics, oxidative damage, DNA repair, transcription, DNMT1, SIRT1
Introduction
Histones, a major component of chromatin, can be posttranslationally modified affecting chromatin compaction and gene expression. Gene expression can also be affected by DNA methylation, with hypermethylation of CpG islands causing transcriptional repression of the associated genes. Transcription, DNA replication and DNA repair all occur on chromatin and these processes utilize various histone modifying and chromatin remodeling enzymes in addition to incorporating histone variants (Groth et al., 2007). Most of the time DNA methylation and histone modification changes that occur during DNA repair are likely transient, being restored back to normal through active DNA demethylation (potentially through the ten-eleven translocation (TET) family of DNA demethylases) (Kohli and Zhang, 2013) or histone modification as repair is completed. However, when cells are repeatedly exposed to DNA damage and repair some chromatin modifications or DNA methylation changes may persist heritably affecting transcription and resulting in epigenetic alterations.
Individual tumors have hundreds to thousands of genes aberrantly silenced by promoter CpG island hypermethylation, including tumor suppressor genes whose silencing plays a direct role in carcinogenesis (Lao and Grady, 2011). Inflammation and the associated increase in reactive oxygen species (ROS) also play a key role in the initiation and progression of a majority of human epithelial cancers (Niwa and Ushijima, 2010). While a strong association between chronic inflammation and aberrant epigenetic silencing of tumor suppressor genes in many cancers is well established, the mechanism by which inflammation initiates cancer-specific epigenetic changes remains to be determined (Niwa and Ushijima, 2010).
DNA is methylated by the DNA methyltransferase (DNMT) family of enzymes. DNMT1 DNA methylates the daughter strand of hemi-methylated DNA during DNA replication and is involved in maintaining epigenetic silencing in cancer cells (Leonhardt et al., 1992; Rhee et al., 2002). While the exact mechanism is unclear, the DNMTs also play a role in maintaining genomic stability, potentially through their participation in DNA repair. DNMT1 along with chromatin modifiers, including the histone deacetylase sirtuin 1 (SIRT1) and the polycomb group protein Enhancer of Zeste Homolog 2 (EZH2), are recruited to double-strand breaks (DSBs) and sites of oxidative damage in an S-phase independent manner (Mortusewicz et al., 2005; O'Hagan et al., 2008, 2011; Ha et al., 2010). While it has been demonstrated that DNMT1 is recruited to DSBs through its interaction with proliferating cell nuclear antigen (PCNA), it is unclear how DNMT1 and other epigenetic silencing proteins are recruited to sites of oxidative damage (Mortusewicz et al., 2005; Ha et al., 2010). Localization of repressive proteins may play a role in locally inhibiting transcription at sites of DNA damage to prevent transcription from interfering with DNA repair (Soria et al., 2012; O'Hagan, 2014).
Exposure to ROS causes oxidative damage of proteins, lipids and DNA. Oxidative DNA damage can be in the form of base damage, single strand breaks, and DSBs. The base excision repair pathway (BER) repairs most base damage, including the most common oxidative lesion 7,8-dihydro-8-oxo-deoxyguanosine (8-oxodG) (Lindahl, 1982; Neeley and Essigmann, 2006). Canonical mismatch repair (MMR) is a S-phase dependent process that repairs single nucleotide mismatches and small insertion and deletion loops created during DNA replication (Zlatanou et al., 2011). The heterodimers responsible for recognizing mismatches are MSH2-MSH6 (MutSα) and MSH2-MSH3 (MutSβ). Subsequently, MutLα (MLH1 and PMS2 heterodimer) is recruited to participate in the excision step of repair. Impaired MMR causes alterations in the length of short, repetitive DNA sequences resulting in microsatellite instability (MSI). A link between MMR proteins and oxidative lesions was first hypothesized because basal and induced 8-oxodG levels are higher in cells lacking MSH2 than their MSH2 expressing counterparts (DeWeese et al., 1998; Colussi et al., 2002). Recently, it was demonstrated that the human MSH2-MSH6 heterodimer acts with ubiquitylated PCNA and DNA polymerase eta to repair clustered oxidative DNA lesions in an S-phase independent fashion (Zlatanou et al., 2011).
Herein we establish that the MMR proteins, MSH2 and MSH6, are involved in the recruitment of DNMT1 to sites of oxidative DNA damage likely through a protein–protein interaction. Furthermore, we determine for the first time that the oxidative damage-induced reduction in nascent expression of target genes with CpG island-containing promoters is dependent on MSH2-MSH6 and DNMT1. The connection between oxidative damage, MSH2-MSH6, DNMT1, and the reduction in transcription of target genes provides a potential mechanism by which some of these genes become susceptible to aberrant epigenetic silencing during carcinogenesis.
Results
The oxidative damage-induced increase in affinity of DNMT1 for chromatin is independent of PCNA
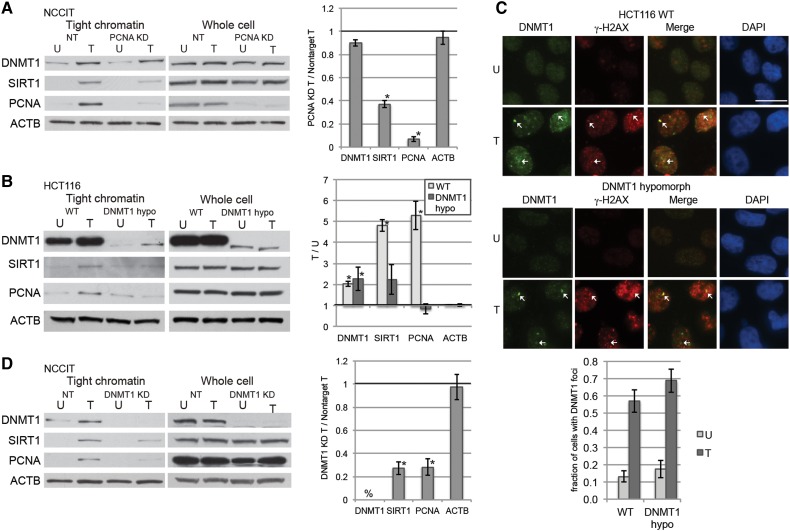
DNMT1 is recruited to replication foci and DSBs in part through its interaction with PCNA (Chuang et al., 1997; Mortusewicz et al., 2005; Ha et al., 2010). Therefore, we initially hypothesized that DNMT1 is recruited to sites of oxidative DNA damage through its interaction with PCNA. Previously, by extracting cell pellets with increasing concentrations of salt we demonstrated that DNMT1 was loosely bound to chromatin in untreated cells and became more tightly bound to chromatin after oxidative damage (O'Hagan et al., 2011). Here we used a related assay using a single high concentration salt buffer wash designed to examine the levels of protein tightly bound to chromatin. Knockdown of PCNA in NCCIT embryonic carcinoma cells did not affect the H2O2-induced increase in affinity of DNMT1 for chromatin, although the levels of SIRT1 bound tightly to chromatin were reduced by PCNA knockdown (Figure 1A). The total cellular levels of DNMT1 and SIRT1 were not affected by PCNA knockdown (Figure 1A). Treatment of cells with H2O2 for 30 min reduced cell survival as expected, but even at 2 mM H2O2 a significant fraction of cells are viable 24 h post-exposure (Supplementary Figure S1A).
Figure 1.
Oxidative damage-induced recruitment of DNMT1 to chromatin is not dependent on PCNA. (A) NCCIT cells were transfected with nontarget siRNAs (NT) or PCNA siRNA. After 72 h, they were untreated (U) or treated with 1 mM H2O2 for 30 min (T). Data are displayed as mean ± SEM of the ratio of the indicated protein levels in PCNA-knockdown H2O2-treated cells over nontarget H2O2-treated cells (n = 2). *P < 0.05. (B) HCT116 parental (WT) or DNMT1 hypomorphic (DNMT1 hypo) cells were untreated (U) or treated with 4 mM H2O2 for 30 min (T). Data are displayed as mean ± SEM of the ratio of the indicated protein levels in H2O2-treated cells over untreated cells for the given cell line (n = 3). *P < 0.05. (C) HCT116 parental (WT) or DNMT1 hypomorphic (DNMT1 hypo) cells were untreated (U) or treated with 4 mM H2O2 for 30 min (T). Immunofluorescence analysis was performed after using a preextraction buffer. Arrows indicate foci where DNMT1 and γ-H2AX colocalize. Data presented are mean ± SEM of the percentage of cells with DNMT1 foci in their nuclei from three biological replicates with at least 20 cells counted for each replicate. Scale bar, 5 μm. (D) NCCIT cells were infected with nontarget shRNAs (NT) or DNMT1 shRNA. After 96 h, they were untreated (U) or treated with 1 mM H2O2 for 30 min (T). Data are displayed as mean ± SEM of the ratio of the indicated protein levels in DNMT1-knockdown H2O2-treated cells over nontarget H2O2-treated cells (n = 3). *P < 0.05. %, DNMT1 is not detectable by western blot.
To further confirm these results, we treated HCT116 parental cells, a colorectal cancer cell line, or HCT116 DNMT1 hypomorph cells, which only express low levels of a truncated form of DNMT1 that lacks the PCNA binding domain, with H2O2 (Egger et al., 2006). In the parental and DNMT1 hypomorph cells, DNMT1 was more tightly bound to chromatin after H2O2 treatment than in the respective untreated cells (Figure 1B). Notably, the H2O2-induced increase in binding of SIRT1 and PCNA to chromatin was less in the DNMT1 hypomorph cells than the parental cells, even though their total cellular protein levels were not different (Figure 1B). Previously, it had been demonstrated that DNMT1 forms foci after treatment with H2O2 (O'Hagan et al., 2011), and here we demonstrated that hypomorphic DNMT1 still formed oxidative damage-induced nuclear foci that colocalize with γ-H2AX (Figure 1C). Therefore, we knocked down DNMT1 and found a reduced amount of SIRT1, as shown previously (O'Hagan et al., 2011), as well as PCNA tightly bound to chromatin after H2O2 treatment (Figure 1D).
Previously it had been demonstrated that modulating levels of the DNA glycosylase for 8-oxodG, OGG1, moderately affect the binding of DNMT1 to chromatin after oxidative damage (O'Hagan et al., 2011). However, treating cells with methyl methanesulfonate (MMS), an agent that induces DNA damage mainly repaired by BER (Yan et al., 2014), did not cause a change in binding of DNMT1, SIRT1, or PCNA to chromatin (Supplementary Figure S1B), suggesting that activation of the BER pathway is not sufficient to cause the increase in affinity of DNMT1 and SIRT1 for chromatin that is caused by oxidative damage.
The mismatch repair proteins MSH2 and MSH6 participate in the oxidative damage-induced increase in affinity of DNMT1 for chromatin
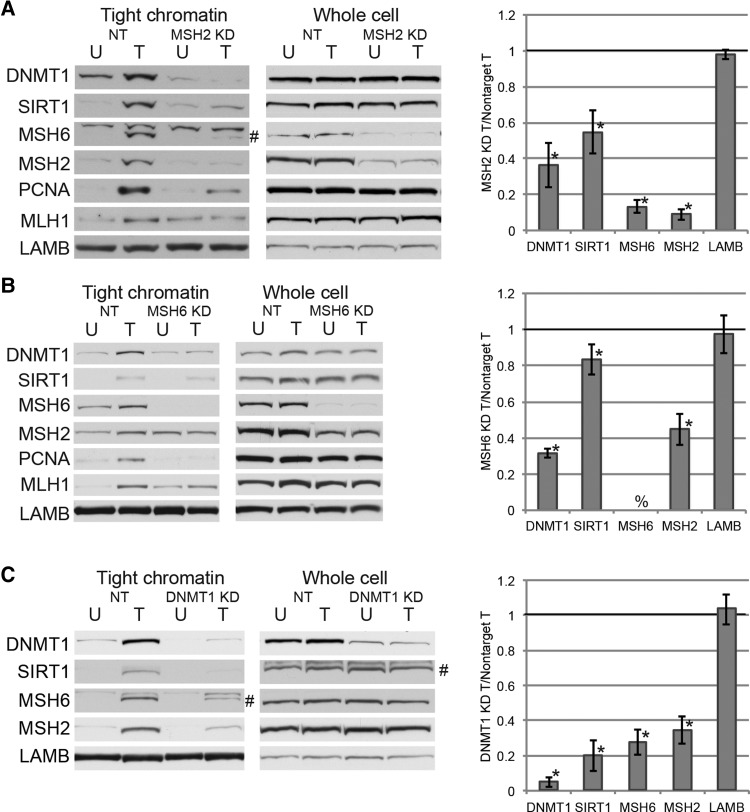
Since the MSH2-MSH6 heterodimer is involved in the repair of clustered oxidative lesions in a manner that involves ubiquitylation of PCNA, we hypothesized that MSH2 and/or MSH6 may be involved in the recruitment of DNMT1 to chromatin after oxidative damage (Zlatanou et al., 2011). H2O2 treatment of NCCIT cells increased the tightness of binding of MSH2, MSH6, and MLH1 to chromatin (Figure 2A). Therefore, we analyzed the binding of DNMT1 and SIRT1 to chromatin following knockdown of MSH2 or MSH6. Knockdown of MSH2 reduced the H2O2-induced increase in affinity of DNMT1, SIRT1, MLH1, and PCNA for chromatin, without affecting the total levels of these proteins (Figure 2A). There was a decrease in the total protein levels as well as the oxidative damage-induced increase in affinity for chromatin of MSH2 and MSH6 in the MSH2 knockdown cells, consistent with MSH6 protein stability being dependent on the presence of MSH2 (Edelbrock et al., 2013). Knockdown of MSH6 also reduced the H2O2-induced increase in the affinity of DNMT1, SIRT1, MLH1, and PCNA for chromatin without affecting total protein levels of these proteins (Figure 2B).
Figure 2.
Knockdown of MSH2 or MSH6 reduces the recruitment of DNMT1 to damaged chromatin. (A) NCCIT cells were infected with nontarget shRNAs (NT) or MSH2 shRNA. After 96 h, they were untreated (U) or treated with 1 mM H2O2 for 30 min (T). Data are displayed as mean ± SEM of the ratio of the indicated protein levels in MSH2-knockdown H2O2-treated cells over nontarget H2O2-treated cells (n = 3). *P < 0.05. # indicates MSH6 band. Upper band is non-specific. (B) NCCIT cells were infected with nontarget shRNAs (NT) or MSH6 shRNA. Data are displayed as mean ± SEM of the ratio of the indicated protein levels in MSH6-knockdown H2O2-treated cells over nontarget H2O2-treated cells (n = 3). *P < 0.05 by one-tail t-test. %, MSH6 is not detectable by western blot. (C) NCCIT cells were infected with nontarget shRNAs (NT) or DNMT1 shRNA. Data are displayed as mean ± SEM of the ratio of the indicated protein levels in DNMT1-knockdown H2O2-treated cells over nontarget H2O2-treated cells. *P < 0.05. # indicates correct band. Upper band is non-specific.
To determine whether or not MLH1, a MMR protein involved in later steps of the canonical MMR process, affects the recruitment of DNMT1 or SIRT1 to chromatin after oxidative damage HCT116 cells, which lack MLH1 and MSH3 expression, or HCT116 + chr3 cells, which have restored MLH1 expression (Koi et al., 1994), were treated with H2O2. Levels of DNMT1 and SIRT1 in the tight chromatin fraction after treatment were similar in both cell lines (Supplementary Figure S2A), suggesting that MLH1 levels do not affect the H2O2-induced increase in affinity of DNMT1 and SIRT1 for chromatin.
To examine whether DNMT1 levels affect recruitment of MSH2 or MSH6 to chromatin, DNMT1 was knocked down prior to treatment with H2O2. Knockdown of DNMT1 reduced the H2O2-induced increase in binding of SIRT1 to chromatin as shown previously (O'Hagan et al., 2011) as well as MSH6 and MSH2 without affecting total cellular levels (Figure 2C).
Ionizing radiation (IR) also induces oxidative DNA damage in cells along with DSBs, yet previously we had demonstrated that IR does not induce an increase in affinity of DNMT1 and SIRT1 for chromatin (O'Hagan et al., 2011). Interestingly, IR also does not induce an increase in affinity of MSH2 or MSH6 for chromatin further linking the recruitment of DNMT1 and SIRT1 to chromatin to MSH2/6 (Supplementary Figure S2B). Treatment with the alkylating agent MNNG has been demonstrated to cause an S-phase independent increase in binding of MMR proteins and PCNA to chromatin (Schroering and Williams, 2008). To determine whether the increase in the affinity of DNMT1 and SIRT1 for chromatin will occur after another type of damage that is recognized by the MMR proteins, we treated cells with MNNG and isolated the proteins tightly bound to chromatin. After MNNG treatment, there was a modest increase in the tightness of binding of MSH6 and MLH1 to chromatin as compared with untreated cells, but no change in DNMT1 or SIRT1 binding, unlike in H2O2-treated cells where all of these proteins became more tightly bound to chromatin (Supplementary Figure S2C).
MSH6 expression restores the recruitment of DNMT1 to chromatin after oxidative damage
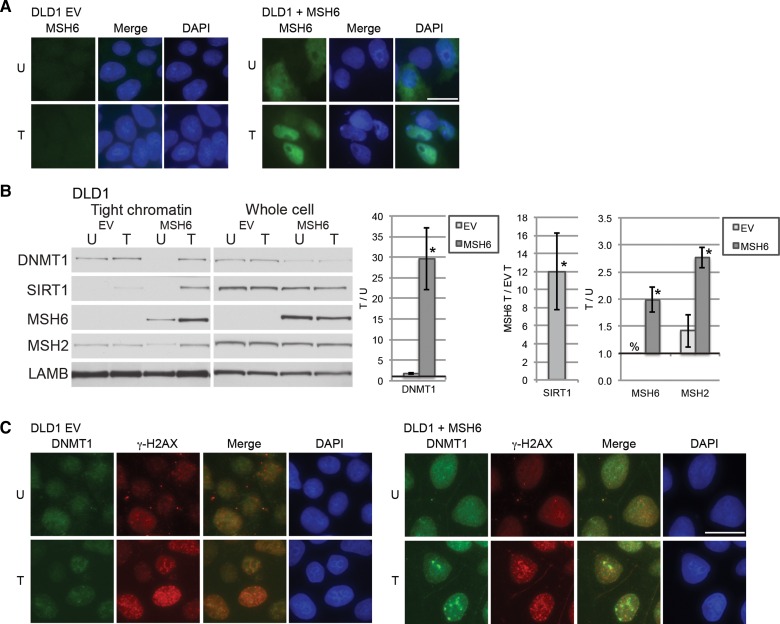
In untreated cells, MSH2 and MSH6 are mostly cytoplasmic, but become nuclear in response to DNA damage (Christmann and Kaina, 2000). Nuclear localization signals are present in the N-terminal region of human MSH6 but not in human MSH2 (Gassman et al., 2011). It is believed that in humans MSH2 and MSH6 heterodimerize in the cytosol because nuclear levels of MSH2 are decreased in cells lacking MSH6 (Christmann and Kaina, 2000). Additionally, it was demonstrated that MSH2, MSH6, and MSH3 are recruited to sites of laser irradiation, and that the recruitment of MSH2 requires either MSH6 or MSH3 (Hong et al., 2008). In an attempt to examine whether MSH2 alone or both MSH2 and MSH6 are needed for the increase in affinity of DNMT1 for chromatin after oxidative damage, two additional colon cancer cell lines were treated with H2O2 with and without restoration of expression of the MMR proteins. LOVO cells lack expression of MSH2, MSH6, and MSH3. Restoration of MSH2 expression in LOVO cells was not sufficient to restore the H2O2-induced increase in affinity of DNMT1 or MSH2 for chromatin (Supplementary Figure S3). LOVO cells did not survive reexpression of MSH6 with or without MSH2 overexpression (data not shown). DLD1 cells express MSH2 and MSH3, but lack MSH6 expression (Umar et al., 1997). Transiently overexpressed MSH6 in these cells was localized throughout the cell in untreated cells and detected in the nucleus after H2O2 treatment (Figure 3A). We found that in DLD1 cells expressing MSH6, there is an increase in the tight chromatin levels of MSH2 (as well as MSH6) after H2O2 treatment compared with untreated MSH6 expressing cells, but not in control empty vector (EV) cells (Figure 3B), suggesting that the response of MSH2 to oxidative damage is dependent on MSH6. Furthermore, H2O2 treatment induced a greater increase in the tightness of binding of DNMT1 to chromatin in DLD1 cells overexpressing MSH6 than in EV cells when compared with their respective untreated cells. SIRT1 was not detectable in the untreated cells, but there was more SIRT1 in the tight chromatin faction of the MSH6 expressing cells than the EV cells. Lower levels of DNMT1 in the untreated tight chromatin fraction of MSH6 expressing cells when compared with the EV cells are likely explained by the observation that total cellular levels of DNMT1 are decreased in the cells expressing MSH6.
Figure 3.
MSH6 recruits DNMT1 to damaged chromatin. (A) DLD1 cells transfected with an empty vector (EV) or a MSH6 plasmid (MSH6) were untreated (U) or treated with 4 mM H2O2 for 30 min and followed by immunofluorescence analysis. Scale bar, 5 μm. (B) Cells were transfected and treated as in A. Data were first normalized to LAMB and then displayed as mean ± SEM of the ratio of the indicated protein levels in H2O2-treated cells over untreated cells for DNMT1, MSH2, or MSH6 in the given cell line (n = 3). SIRT1 data are displayed as the ratio of the SIRT1 levels in MSH6-expressing H2O2-treated cells over EV H2O2-treated cells. *P < 0.05. (C) Cells were transfected and treated as in A. Immunofluorescence analysis was performed after using a preextraction buffer. The data presented are from one representative experiment of three biologic replicates.
Furthermore, oxidative damage-induced foci of DNMT1 were not present in DLD1 cells transfected with an empty vector, but were observed in the DLD1 cells overexpressing MSH6 (Figure 3C). The foci colocalized with γ-H2AX suggesting that DNMT1 was recruited to sites of DNA damage in an MSH6-dependent manner.
DNMT1 interacts with MSH2 and MSH6
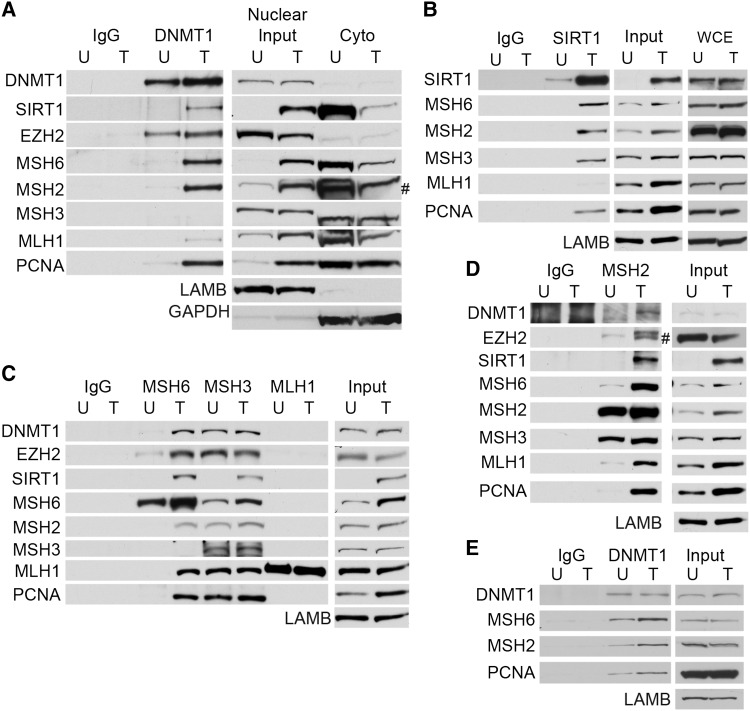
The finding that the oxidative damage-induced increase in affinity of DNMT1 for chromatin is dependent on MSH2-MSH6 suggests that DNMT1 may interact with MSH6 and/or MSH2. Therefore, DNMT1 was co-immunoprecipitated (co-IPed) from the nuclear extracts of untreated or H2O2-treated NCCIT cells. The MMR proteins MSH6, MSH2, MLH1, and PCNA all became more nuclear and less cytoplasmic after treatment (Figure 4A). LAMB and GAPDH are nuclear and cytoplasmic proteins, respectively, and were controls for the nuclear extraction. After H2O2 treatment, DNMT1 interacted with MSH6, MSH2, PCNA and weakly with MLH1, while it did not appear to interact with these proteins in untreated cells or with MSH3 in either untreated or treated cells (Figure 4A). Previously it has been demonstrated that DNMT1 interacts with SIRT1 and the polycomb group member, EZH2, after oxidative damage and we demonstrated similar findings here (O'Hagan et al., 2011). By co-IPing with SIRT1, we found that SIRT1 also interacts with MSH6, MSH2, and PCNA after H2O2 treatment, as well as MSH3 (Figure 4B). Again these proteins became enriched in the nuclear fraction after treatment, whereas the total cellular levels of these proteins did not change. Co-IPs with MSH6 and MSH2 demonstrated that oxidative damage induces the interaction of these proteins with DNMT1, SIRT1, and EZH2 and the repair proteins MLH1 and PCNA (Figure 4C and D). MSH2 interacted with MSH3 but the amount of interaction did not appear to be affected by H2O2 treatment (Figure 4D). Co-IP with MLH1 did not pull down DNMT1, SIRT1, or EZH2, in agreement with the findings that MLH1 did not appear to be needed for the recruitment of DNMT1 to chromatin after oxidative damage (Figure 4C). Interestingly, co-IP with MSH3 pulled down equal levels of DNMT1, EZH2, and MSH2 in untreated and H2O2-treated cells, suggesting that these proteins may interact basally, but that this interaction does not change with damage (Figure 4C). Similarly as to the SIRT1 co-IP, MSH3 interaction with SIRT1 increased after oxidative damage. MSH3 was also found to interact with MSH6 as has been suggested previously (Havugimana et al., 2012). Because in untreated cells MSH2 and MSH6 are mostly cytoplasmic and DNMT1 is mostly nuclear, it is possible that they do not interact because they are in separate cellular compartments. Therefore, we performed co-IPs for DNMT1 after allowing all cellular proteins to be in solution together and demonstrated that DNMT1 can interact with MSH6, MSH2, and PCNA in solution (Figure 4E). However, the interactions increase after H2O2 treatment. Altogether these data indicate that H2O2 treatment induces the interaction of the epigenetic proteins DNMT1, SIRT1, EZH2 with the DNA repair proteins MSH2, MSH6, and PCNA.
Figure 4.
DNMT1 and MSH2-MSH6 interact after oxidative damage. (A) NCCIT cells were untreated (U) or treated with 2 mM H2O2 for 30 min. Nuclear extracts were prepared and co-immunoprecipitations were performed with control IgG or anti-DNMT1 antibodies. # indicates that lower band is MSH2. (B) Cells were treated as in A and co-immunoprecipitations were performed with control IgG or anti-SIRT1 antibodies. (C) Cells were treated as in A and co-immunoprecipitations were performed with control IgG, anti-MSH6, anti-MSH3, and anti-MLH1 antibodies. (D) Cells were treated as in A and co-immunoprecipitations were performed with control IgG or anti-MSH2 antibodies. # indicates that lower band is EZH2. (E) Cells were treated as in A and co-immunoprecipitations were performed after allowing all cellular proteins to be in solution together with control IgG and anti-DNMT1 antibodies.
We hypothesized that DNMT1's methyltransferase activity does not affect the H2O2-induced increase in affinity of DNMT1 for chromatin and/or DNMT1's interaction with MSH2/6. Here, by using HCT116 DNMT1 hypomorph cells that stably express WT or catalytically inactive DNMT1 (Clements et al., 2012), we demonstrated that while cells expressing WT DNMT1 had higher DNMT activity, H2O2 treatment induced a similar increase in affinity for chromatin of both forms of DNMT1 (Supplementary Figure S4A and B). Additionally, both WT and catalytically inactive DNMT1 interacted with MSH2 and MSH6 after H2O2 treatment (Supplementary Figure S4C).
Knockdown of MSH6 and/or DNMT1 affects the H2O2-induced reduction in nascent transcription
Previously, it has been demonstrated that oxidative damage induces the enrichment of γ-H2AX, DNMT1, SIRT1, and EZH2 at promoter CpG islands (O'Hagan et al., 2011). This enrichment likely occurs because these islands are GC-rich and since guanine is the most easily oxidized base, the CpG islands undergo the most DNA damage after exposure to H2O2 (O'Hagan et al., 2011). Because it is likely that several oxidative lesions are occurring simultaneously in CpG islands and clustered sites of oxidative damage were demonstrated to be repaired by an MSH2-MSH6 dependent pathway (Zlatanou et al., 2011), we hypothesized that the oxidative damage at CpG islands is being repaired by MSH2 and MSH6. The model proposed here suggests that MSH6 with MSH2 recruits DNMT1 and the other epigenetic proteins to sites of oxidative damage (Figure 5D).
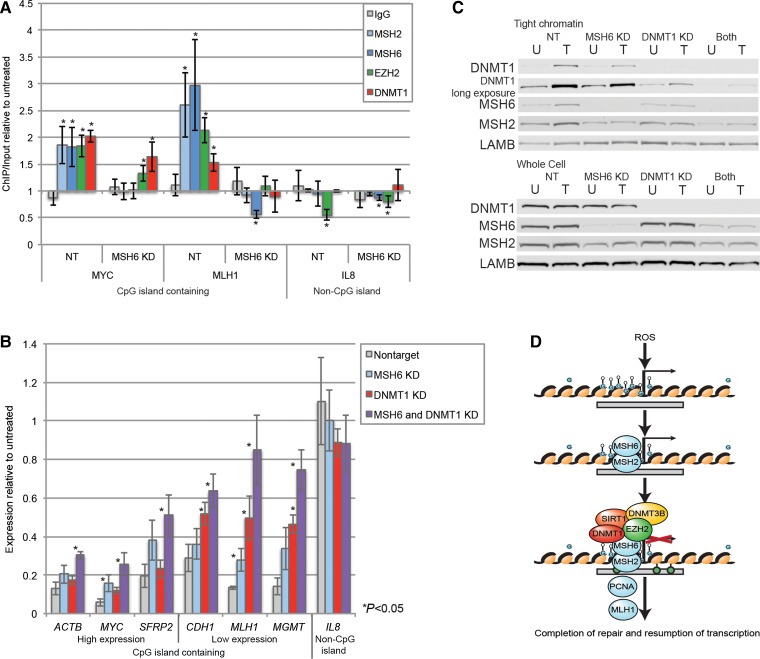
Figure 5.
MSH6 and DNMT1 participate in reducing transcription of promoter CpG island-containing genes following oxidative damage. (A) NCCIT cells were infected with nontarget or MSH6 shRNAs. After 96 h, cells were untreated or treated with 2 mM H2O2 for 30 min followed by local ChIP for control IgG, MSH2, MSH6, EZH2, and DNMT1 and analyzed by qPCR. Data are presented as mean ± SEM of treated ChIP/input values normalized to untreated ChIP/Input values (n = 3-5). *P < 0.05. (B) NCCIT cells were infected with nontarget, MSH6, DNMT1, or both MSH6 and DNMT1 shRNAs. After 96 h, cells were untreated (U) or treated with 0.5 mM H2O2 for 30 min (T), and nascent RNA was labeled concurrently. qPCR data are presented as mean ± SEM of the treated over the untreated values (n = 3). *P < 0.05. (C) Tight chromatin and total cellular (Whole cell) protein was isolated from a portion of the cells from B. Tight chromatin is the remaining protein in the chromatin pellet after extraction with 0.45 M NaCl buffer. The data presented are from one representative experiment of two biologic replicates. (D) A working model for the recruitment of epigenetic silencing proteins to chromatin after oxidative damage. When cells are exposed to reactive oxygen species (ROS), clustered oxidative damage occurs at promoter CpG islands, which by definition have a high GC-content. The MSH2-MSH6 heterodimer is recruited to sites of damage and by protein–protein interactions recruits epigenetic silencing proteins, including DNMT1, SIRT1, and EZH2, to promoter CpG islands. This recruitment results in the transient reduction of transcription while repair is taking place. Once repair is completed, the transcription is resumed.
Therefore, we performed chromatin immunoprecipitation (ChIP) for MSH2, MSH6, DNMT1, and EZH2 in nontarget and MSH6 knockdown cells to determine whether reduction in MSH6 levels will affect the recruitment of the epigenetic proteins to promoter CpG islands. We found that DNMT1, EZH2, MSH2, and MSH6 were all significantly enriched in the promoter CpG islands of genes (MYC and MLH1) after H2O2 treatment in nontarget knockdown cells, but not in a non-CpG island-containing gene promoter (IL8) (Figure 5A). Knockdown of MSH6 reduced the H2O2-induced enrichment of both the MMR proteins MSH2 and MSH6 as well as the epigenetic proteins DNMT1 and EZH2 in the promoter CpG islands (MYC and MLH1). Interestingly, the non-CpG island containing promoter of IL8 had a decrease in enrichment of EZH2 after H2O2 treatment as compared with untreated cells in both the nontarget and MSH6 knockdown cells. These results suggest that MSH2-MSH6 indeed contribute to the localization of the epigenetic silencing proteins to CpG islands after oxidative damage.
It has been hypothesized that epigenetic silencing proteins are recruited to sites of DNA damage in part to inhibit transcription so that it does not interfere with the repair process (Soria et al., 2012). Through an unknown mechanism, exposure of cells to H2O2 or alkylating agents causes a significant inhibition of global mRNA synthesis that is dependent on MMR proteins but is independent of S-phase (Yanamadala and Ljungman, 2003). If DNMT1 is involved in repression of transcription after oxidative damage, we hypothesized that knockdown of either DNMT1 or MSH6 will negate the H2O2-induced reduction of transcription. Control cells treated with H2O2 had lower levels of expression of promoter CpG island-containing genes (ACTB, MYC, CDH1, MLH1, MGMT), but not of a non-CpG island-containing gene (IL8) (Figure 5B). MSH6 or DNMT1 knockdown cells had significantly less of a H2O2-induced decrease in nascent expression than nontarget shRNA-treated cells for one or four out of the six CpG island-containing genes examined, respectively. Combining knockdown of MSH6 and DNMT1 further decreased the effect of H2O2 treatment on transcription resulting in significant dampening of the nascent transcription blockade after H2O2 treatment as compared with nontarget knockdown cells for all six of the CpG island-containing genes examined (Figure 5B). Knockdown of DNMT1 and/or MSH6 did not significantly affect the basal nascent expression levels of the genes examined (data not shown).
The reduction in total cellular protein levels of MSH6, MSH2, and DNMT1 was similar in the single knockdowns as compared with the double knockdown (Figure 5C). However, the combined DNMT1 and MSH6 knockdown uniquely diminished the affinity of DNMT1 for chromatin after H2O2 treatment. Thus there is a correlation between the level of DNMT1 in the chromatin and the decrease in nascent expression after H2O2 treatment. The tight chromatin levels of MSH6 and MSH2 in all the target knockdown cells after H2O2 treatment were lower than the nontarget cells, and the double knockdown did not reduce these levels any further than in the MSH6 knockdown cells.
Discussion
In the present study we link the recruitment of DNMT1 to chromatin after oxidative damage to MMR proteins. Though other groups have demonstrated that PCNA plays a role in recruitment of DNMT1 to DSBs (Ha et al., 2010), we found that PCNA is not required for the recruitment of DNMT1 to chromatin after H2O2 treatment. It is important to note that IR, which induces DSBs as well as oxidative DNA damage, does not cause a change in affinity of MSH2, MSH6, DNMT1, or SIRT1 for chromatin in our assay. Future studies are required to determine why MSH2-MSH6 becomes more tightly bound to chromatin after to H2O2, but not IR, even though both treatments induce oxidative DNA damage. Additional work is also needed to demonstrate that the actual oxidative DNA damage is required for the binding of MSH2-MSH6 to chromatin as we hypothesize. In combination, these findings suggest that after H2O2 exposure MSH2-MSH6 first binds to damaged chromatin and then recruits DNMT1 followed by PCNA to sites of oxidative DNA damage (Figure 5D).
Because MSH2 and MSH6 act as a heterodimer, it is difficult to tease apart their specific roles in the recruitment of DNMT1 to damaged chromatin. Knockdown of either protein reduces the protein levels of the other and we did not have success overexpressing MSH6 alone in cells lacking both MSH2 and MSH6 (HEC59 and LOVO cells, data not shown). Our findings also suggest that MSH2 without MSH6 is not sufficient for the recruitment of DNMT1 to damaged chromatin: we do not observe H2O2-induced tightening of DNMT1 to chromatin in LOVO cells that express MSH2 but not MSH6. Furthermore, because in human cells MSH6 is required for the nuclear translocation of MSH2 in response to damage, it is possible that DNMT1 is recruited to damaged chromatin through MSH2, but that MSH2 first needs to be made nuclear through its interaction with MSH6 (Christmann and Kaina, 2000). We found that MSH2-MSH6 interacts with DNMT1 in total cellular protein lysates from untreated cells, but that this interaction is increased in lysates from treated cells. These findings suggest that physical location plays a partial role in the interaction of DNMT1 with MSH2-MSH6, but that there are likely additional mechanisms that further increase these interactions, possibly including H2O2-induced protein modification, protein–protein crosslinks and/or DNA damage. We did rule out MLH1 as a major player in the recruitment of DNMT1 to damaged chromatin (Supplementary Figure S2A). While MLH1 participates in canonical MMR and perhaps repair of oxidative damage (Pena-Diaz et al., 2012), it likely interacts with MSH2 and MSH6 at a later step in the repair pathway. MSH3 did appear to basally interact with DNMT1 and EZH2 in untreated cells, but this interaction does not change after H2O2 treatment. Additionally, DLD1 parental cells express MSH2 and MSH3, but we do not see the H2O2-induced increase in affinity of DNMT1 for chromatin until we express MSH6 in these cells, indicating that MSH3 is not involved or only minimally involved in the recruitment of DNMT1 to damaged chromatin. The interaction of MSH3 with SIRT1 increases after oxidative damage, which potentially explains why in parental DLD1 cells there is still some increase in SIRT1 binding to chromatin after H2O2 treatment.
Previously DNMT1-deficient cells have been shown to be resistant to drugs that alkylate DNA and have increased slippage rates at microsatellite repeats, which are phenomena normally associated with defects in MMR (Guo et al., 2004; Kim et al., 2004; Wang and James Shen, 2004). One group found that reduction of DNMT1 levels causes a reduction in total levels of MMR components (Loughery et al., 2011). However, we did not see a decrease in total cellular levels of MLH1, MSH2, MSH6, or MSH3 protein in DNMT1 knockdown cells (Figure 2C). In the work presented here, knockdown of DNMT1 reduced the levels of MMR proteins that are tightly bound to chromatin after oxidative damage, suggesting that DNMT1 plays a role in the recruitment and/or the stability of MMR proteins at sites of oxidative DNA damage. Whether DNMT1 interacts with or stabilizes MSH2-MSH6 at sites of canonical MMR remains to be determined.
We hypothesize that DNMT1 acts as a scaffold and brings SIRT1, EZH2, and DNMT3B to sites of oxidative damage where they inhibit transcription through a chromatin-based mechanism, not through DNA methylation. In previous studies, we have demonstrated that the H2O2-induced recruitment of these epigenetic proteins to CpG island-containing promoters results in the formation of more repressive chromatin through histone mark changes at promoter CpG islands of high and low expression genes and increases in DNA methylation at promoter CpG islands of low expression genes (O'Hagan et al., 2011). We also found that knockdown of DNMT1 reduces the amount of SIRT1 bound to chromatin after H2O2 treatment. Furthermore, DNMT1 knockdown abrogates the H2O2-induced reduction in transcription of CpG island-containing genes. Knockdown of DNMT1 and MSH6 together further releases the transcription blockade. Moreover, in other systems catalytically inactive DNMT1 has been shown to be able to silence the transcription of genes by acting as a scaffold for chromatin remodelers (Clements et al., 2012). Because catalytically inactive DNMT1 is recruited to chromatin and interacts with MSH2/6 after H2O2 in a manner similar to WT DNMT1, the DNA methylation activity of DNMT1 is likely not required for the localization of DNMT1 to sites of oxidative DNA damage. Additionally, H2O2-induced recruitment of epigenetic proteins and transcriptional repression occurs at high expression genes that do not have increases in DNA methylation. Therefore, DNA methylation is likely not required for repair of oxidative lesions. The mechanism and relevance of increased DNA methylation at low expression genes needs to be further studied.
We hypothesize that the reduction in transcription at sites of DNA damage prevents transcription from interfering with the DNA repair process. Recruitment of repressive factors may also be associated with DNA damage response signaling as has been demonstrated for other repair pathways (Soria et al., 2012). In a majority of cells the DNA damage is likely repaired and the chromatin, DNA methylation and transcription are restored back to normal, but it is possible that at sites of chronic inflammation, such transient recruitment of epigenetic silencing proteins may occasionally result in the stable epigenetic silencing of promoters. In this regard, it has been demonstrated that recruitment of epigenetic proteins including DNMT1, DNMT3B, SIRT1, and EZH2, to an induced DSB can induce epigenetic silencing of the associated promoter in a small percentage of cells, suggesting that damage-induced recruitment of epigenetic silencing proteins can lead to stable epigenetic silencing (O'Hagan et al., 2008). Interestingly, in this model system transcriptional silencing was initially mediated by repressive chromatin, with dense DNA methylation only occurring with cell passage suggesting that repressive chromatin formation after oxidative damage may lead to DNA methylation with further passaging under conditions where silencing the given gene is favorable for the cell. Further work is needed to determine the duration of the transient effect on transcription after oxidative damage, to understand what if any benefit it gives to repair of the damaged chromatin, and to establish whether oxidative damage can result in similar silencing events as were demonstrated for a DSB.
There is a known association between defects in MMR and hereditary and sporadic colon cancer, with sporadic colon cancer often being associated with loss of expression of MLH1 due to DNA methylation of its promoter (Jass, 2007). In these cancers, methylation of MLH1 is associated with MSI, a high number of CpG islands with aberrant DNA hypermethylation (CIMP), and mutations in RAS or BRAF (Jass, 2007). However, the findings presented here suggest that loss of MSH2-MSH6 could potentially result in less promoter CpG island DNA methylation. Interestingly, The Cancer Genome Atlas (TCGA) colon cancer study found that 20% of hypermutated sporadic colon tumors examined were unmethylated for MLH1 and did not have a CIMP phenotype, but had mutations in MSH2, MSH6, and/or Rad18, a ubiquitin ligase involved in repair of oxidative damage by MSH2-MSH6, in addition to mutations in the proofreading domain of POLE (the catalytic subunit of DNA polymerase epsilon) (Muzny et al. , 2012; Palles et al., 2013). These findings suggest that loss of MSH2-MSH6 may be associated with fewer DNA methylation changes than that occur with loss of MLH1.
In conclusion, our results link repair of oxidative damage by MSH2-MSH6 to transcriptional inhibition by DNMT1 at the sites of DNA damage. While many questions remain as to how these processes may result in abnormal stable epigenetic silencing that contributes to cancer formation, this work brings together many known factors involved in colon cancer including inflammation, MMR proteins, and DNA methylation.
Materials and methods
Cell culture and treatments
NCCIT, LOVO, and DLD1 cells were obtained from American Tissue Type Culture Collection and maintained at 37°C and 5% CO2 in RPMI-1640, McCoy's5A, and DMEM-F12, respectively. HCT116 DNMT1 hypomorph cells were maintained as described previously (Rhee et al., 2002). For H2O2 exposure 30% H2O2 (Sigma) was diluted in PBS immediately before adding it to the media and cells were collected 30 min later. Because the cell lines used had different sensitivities to H2O2, the concentration of H2O2 used for each cell line for the tight chromatin assay was based on the H2O2 concentration for that cell line that induced similar levels of nuclear accumulation of SIRT1 to NCCIT cells treated with 1 mM H2O2 (data not shown).
siRNA and shRNA knockdown and MMR protein expression
For siRNA knockdown of PCNA cells were transfected with PCNA or nontarget siRNA at 72 h before collection following the manufacturer's protocol (Dharmacon). For shRNA knockdown of DNMT1, MSH2, and MSH6, cells were transduced with the indicated lentiviral particles 96H before collection following the manufacturer's protocol (Sigma). MSH2 (DNASU, HsCD00434282) was expressed using lentiviral particles following the manufacturer's protocol (The RNAi Consortium (TRC) Broad Institute). For MSH6 expression, cells were transfected with a MSH6 plasmid (TrueClone, NM_000179) using Lipofectamine 3000 (Invitrogen) following the manufacturer's protocol.
Tight chromatin and whole cell isolation
Cells were treated with 4 mM (HCT116, LOVO, DLD1) or 1 mM H2O2 (NCCIT) and pellets were sequentially washed in CEBN buffer [10 mM HEPES pH 7.8, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.2% NP-40, 1× protease inhibitor cocktail (Thermo Scientific), 1× phosphatase Inhibitor cocktail (Sigma), N-ethylmaleimide (Sigma)], CEB buffer (CEBN buffer without NP-40), soluble nuclear buffer (3 mM EDTA, 0.2 mM EGTA, inhibitors), and 0.45 M NaCl buffer (50 mM Tris pH 8.0, 0.05% NP40, 0.45 M NaCl, inhibitors). The remaining pellet was lysed in 4% SDS buffer using a Qiashredder (Qiagen) and referred to as tight chromatin. Band densitometry data for western blots were collected using ImageJ software. Whole cell extracts were prepared from 1/10 of the pellet collected after treatment before beginning the tight chromatin isolation.
Immunofluorescence
Cells grown on coverslips coated with poly-l-lysine were treated with 4 mM H2O2 and were or were not preextracted as indicated using 0.5% Triton-X 100 in 10 mM HEPES (pH 7.4), 2 mM MgCl2, 100 mM KCl, 1 mM EDTA, and then fixed in 4% paraformaldehyde, permeabilized in PBS + 0.5% Triton-X 100, blocked in PBST (PBS with 0.1% Tween-20) containing 1% BSA, and incubated with the antibodies indicated in the Supplementary material. DAPI was used to stain nuclei.
Co-immunoprecipitation
Cells were treated with 2 mM H2O2. Nuclear extraction was performed using CEBN and CEB buffers. The nuclear pellet or the whole cell pellet was resuspended in modified RIPA buffer (50 mM Tris pH 7.5, 100 mM NaCl, 3 mM EDTA, 0.5% NP40, 50 mM NaF), sonicated for three cycles of 30 sec on/30 sec off using a BioruptorPico sonicator (Diagenode), rotated at 4°C for 1 h with 60 mM spermine and 20 mM spermidine to release chromatin bound proteins, sonicated for two cycles of 30 sec on/30 sec off, and cleared by high-speed centrifugation. Lysates were rotated with antibody for 4 h at 4°C. Protein A/G-magnetic beads (Pierce) were added and the samples were rotated O/N at 4°C. The beads were washed six times with TNE + buffer (50 mM Tris pH7.5, 150 mM NaCl, 5 mM EDTA, 0.5% NP40, 0.5% TritonX-100, 50 mM NaF, inhibitors) for 10 min at 4°C. Complexes were eluted off the beads in loading buffer at 65°C for 15 min.
Chromatin immunoprecipitation
Cells untreated or treated with 2 mM H2O2 were crosslinked using 1% formaldehyde. Nuclear extraction was performed using CEBN and CEB buffers. Nuclear pellets were sonicated in sonication buffer containing 1% SDS for 3 sets of 10 cycles of 30 sec on/30 sec off using a BioruptorPico sonicator. Sonicated chromatin was diluted to a final SDS concentration of 0.1% and aliquots were rotated with antibody O/N at 4°C. Protein A/G Magnetic beads (Invitrogen) were added and the samples were rotated for 3 h at 4°C. Beads were washed for 10 min at RT with each of the following buffers twice: low salt, high salt, and LiCl (Millipore). Eluted DNA was analyzed by qPCR using primers indicated in Supplementary Table S1.
Nascent transcription
Nascent transcription assays were performed using the Click-iT Nascent RNA Capture Kit (Invitrogen). Cells were labeled with ethynyl uridine for 30 min concurrently with the 0.5 mM H2O2 treatment if indicated. cDNA was analyzed by qPCR using primers indicated in Supplementary Table S1.
Genes are called high expression or low expression based on their number of reads from a NCCIT mRNA-sequencing GEO dataset (GSM644996) (Jung et al., 2012).
Statistical analysis
Band densitometry, local ChIP and nascent transcription data are presented as the mean ± standard error (SEM). These data are evaluated by one-tail t-test and considered statistically significant with a P-value < 0.05.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported by the National Institute of Environmental Health Sciences (RO1ES023183 to H.M.O'H. and RO1ES011858 to S.B.B.).
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We thank Drs T.A. Kunkel and A.B. Clark from NIEHS, Research Triangle Park, NC, USA for the generous gift of the HCT116 parental and +chr3 cells. We thank the Indiana University Light Microscopy Imaging Center and the Indiana University Radiological Safety Office for their assistance.
References
- Christmann M., Kaina B. (2000). Nuclear translocation of mismatch repair proteins MSH2 and MSH6 as a response of cells to alkylating agents. J. Biol. Chem. 275, 36256–36262. [DOI] [PubMed] [Google Scholar]
- Chuang L.S.H., Ian H.I., Koh T.W. et al. (1997). Human DNA (cytosine-5) methyltransferase PCNA complex as a target for p21(WAF1). Science 277, 1996–2000. [DOI] [PubMed] [Google Scholar]
- Clements E.G., Mohammad H.P., Leadem B.R. et al. (2012). Dnmt1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 40, 4334–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi C., Parlanti E., Degan P. et al. (2002). The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr. Biol. 12, 912–918. [DOI] [PubMed] [Google Scholar]
- DeWeese T.L., Shipman J.M., Larrier N.A. et al. (1998). Mouse embryonic stem cells carrying one or two defective Msh2 alleles respond abnormally to oxidative stress inflicted by low-level radiation. Proc. Natl Acad. Sci. USA 95, 11915–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelbrock M.A., Kaliyaperumal S., Williams K.J. (2013). Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat. Res. 743–744, 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G., Jeong S., Escobar S.G. et al. (2006). Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl Acad. Sci. USA 103, 14080–14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassman N.R., Clodfelter J.E., McCauley A.K. et al. (2011). Cooperative nuclear localization sequences lend a novel role to the N-terminal region of MSH6. PLoS One 6, e17907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A., Rocha W., Verreault A. et al. (2007). Chromatin challenges during DNA replication and repair. Cell 128, 721–733. [DOI] [PubMed] [Google Scholar]
- Guo G., Wang W., Bradley A. (2004). Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature 429, 891–895. [DOI] [PubMed] [Google Scholar]
- Ha K., Lee G.E., Palii S.S. et al. (2010). Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery. Hum. Mol. Genet. 20, 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havugimana P.C., Hart G.T., Nepusz T. et al. (2012). A census of human soluble protein complexes. Cell 150, 1068–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Jiang J., Hashiguchi K. et al. (2008). Recruitment of mismatch repair proteins to the site of DNA damage in human cells. J. Cell Sci. 121, 3146–3154. [DOI] [PubMed] [Google Scholar]
- Jass J.R. (2007). Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 50, 113–130. [DOI] [PubMed] [Google Scholar]
- Jung I., Kim S.K., Kim M. et al. (2012). H2B monoubiquitylation is a 5′-enriched active transcription mark and correlates with exon-intron structure in human cells. Genome Res. 22, 1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Trinh B.N., Long T.I. et al. (2004). Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 32, 5742–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli R.M., Zhang Y. (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koi M., Umar A., Chauhan D.P. et al. (1994). Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N’-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 54, 4308–4312. [PubMed] [Google Scholar]
- Lao V.V., Grady W.M. (2011). Epigenetics and colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 8, 686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H., Page A.W., Weier H.U. et al. (1992). A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71, 865–873. [DOI] [PubMed] [Google Scholar]
- Lindahl T. (1982). DNA repair enzymes. Annu. Rev. Biochem. 51, 61–87. [DOI] [PubMed] [Google Scholar]
- Loughery J.E., Dunne P.D., O'Neill K.M. et al. (2011). DNMT1 deficiency triggers mismatch repair defects in human cells through depletion of repair protein levels in a process involving the DNA damage response. Hum. Mol. Genet. 20, 3241–3255. [DOI] [PubMed] [Google Scholar]
- Mortusewicz O., Schermelleh L., Walter J. et al. (2005). Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl Acad. Sci. USA 102, 8905–8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzny D.M., Chang K., Dinh H.H. et al. (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeley W.L., Essigmann J.M. (2006). Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 19, 491–505. [DOI] [PubMed] [Google Scholar]
- Niwa T., Ushijima T. (2010). Induction of epigenetic alterations by chronic inflammation and its significance on carcinogenesis. Adv. Genet. 71, 41–56. [DOI] [PubMed] [Google Scholar]
- O'Hagan H.M. (2014). Chromatin modifications during repair of environmental exposure-induced DNA damage: A potential mechanism for stable epigenetic alterations. Environ. Mol. Mutagen. 55, 278–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan H.M., Mohammad H.P., Baylin S.B. (2008). Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 4, e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan H.M., Wang W., Sen S. et al. (2011). Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG islands. Cancer Cell 20, 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palles C., Cazier J.-B., Howarth K.M. et al. (2013). Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 45, 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Diaz J., Bregenhorn S., Ghodgaonkar M. et al. (2012). Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell 47, 669–680. [DOI] [PubMed] [Google Scholar]
- Rhee I., Bachman K.E., Park B.H. et al. (2002). DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416, 552–556. [DOI] [PubMed] [Google Scholar]
- Schroering A.G., Williams K.J. (2008). Rapid induction of chromatin-associated DNA mismatch repair proteins after MNNG treatment. DNA Repair 7, 951–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G., Polo S.E., Almouzni G. (2012). Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol. Cell 46, 722–734. [DOI] [PubMed] [Google Scholar]
- Umar A., Koi M., Risinger J.I. et al. (1997). Correction of hypermutability, N-methyl-N’-nitro-N-nitrosoguanidine resistance, and defective DNA mismatch repair by introducing chromosome 2 into human tumor cells with mutations in MSH2 and MSH6. Cancer Res. 57, 3949–3955. [PubMed] [Google Scholar]
- Wang K.Y., James Shen C.K. (2004). DNA methyltransferase Dnmt1 and mismatch repair. Oncogene 23, 7898–7902. [DOI] [PubMed] [Google Scholar]
- Yan S., Sorrell M., Berman Z. (2014). Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell. Mol. Life Sci. 71, 3951–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanamadala S., Ljungman M. (2003). Potential role of MLH1 in the induction of p53 and apoptosis by blocking transcription on damaged DNA templates. Mol. Cancer Res. 1, 747–754. [PubMed] [Google Scholar]
- Zlatanou A., Despras E., Braz-Petta T. et al. (2011). The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol. Cell 43, 649–662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.