Abstract
Bone's remarkable mechanical properties are a result of its hierarchical structure. The mineralized collagen fibrils, made up of collagen fibrils and crystal platelets, are bone's building blocks at an ultrastructural level. The organization of bone's ultrastructure with respect to the orientation and arrangement of mineralized collagen fibrils has been the matter of numerous studies based on a variety of imaging techniques in the past decades. These techniques either exploit physical principles, such as polarization, diffraction or scattering to examine bone ultrastructure orientation and arrangement, or directly image the fibrils at the sub-micrometre scale. They make use of diverse probes such as visible light, X-rays and electrons at different scales, from centimetres down to nanometres. They allow imaging of bone sections or surfaces in two dimensions or investigating bone tissue truly in three dimensions, in vivo or ex vivo, and sometimes in combination with in situ mechanical experiments. The purpose of this review is to summarize and discuss this broad range of imaging techniques and the different modalities of their use, in order to discuss their advantages and limitations for the assessment of bone ultrastructure organization with respect to the orientation and arrangement of mineralized collagen fibrils.
Keywords: bone ultrastructure organization, ultrastructure orientation and arrangement, mineralized collagen fibrils, collagen orientation, crystal orientation, bone imaging
1. Introduction
Bone is a material of remarkable mechanical properties that are optimized through evolutionary processes and functional adaptation during the lifetime to meet the basic mechanical needs of supporting the human body, transmitting forces for locomotion and protecting vital organs. In order to achieve these mechanical properties, human bone has developed a complicated, composite structure (figure 1). At a macroscopic level, the organ bone is composed of two osseous tissue types: cortical and trabecular bone. These differ at a microstructural level, with cortical bone being composed of osteons or Haversian systems, whereas trabecular rods and plates form the trabecular or cancellous bone compartment. Both cortical and trabecular bone are typically made up of lamellae, which are mostly composed of mineralized collagen fibril bundles or fibres [8,9], a few to several micrometres in diameter. At the ultrastructural level, mineralized collagen fibrils with diameters of the order of approximately 100 nm are the building blocks of bone [8]. It is possible that these do not form bundles or fibres, but have a disordered organization instead [9]. The mineralized collagen fibrils are formed by the combination of collagen fibrils [10] and hydroxyapatite (HA) mineral crystals [11]. The crystals appear in the form of platelets [12], approximately 3 × 25 × 50 nm in size, although significant variations in platelet size have been reported, based on experiments using atomic force microscopy (AFM) [13], transmission electron microscopy (TEM) [14] and X-ray scattering [15] or diffraction [16]. Platelets are formed by hexagonal crystal unit cells, with dimensions a = b = 9.4 Å and c = 6.8 Å [17]. The crystals are either intra- or extra-fibrillar [18], where intra-fibrillar crystals are associated with the gap regions of the collagen fibril [19], while extra-fibrillar crystals are found in the space surrounding the fibrils [20]. It is worth noting that the collagen–mineral interaction is a topic of intense interest and study [21,22]. Further, it has been shown that the c-plane of the unit cells coincides with the direction of crystal platelets [23], and with the direction of the fibrils [24]. This means that, for investigating the orientation of the bone ultrastructure, one can study the orientation of each of the four structural elements of the ultrastructure: (i) the unit crystal, (ii) the crystal platelet, (iii) the collagen fibril, and (iv) the fibril bundle or fibre (if the fibrils have been organized in bundles or fibres).
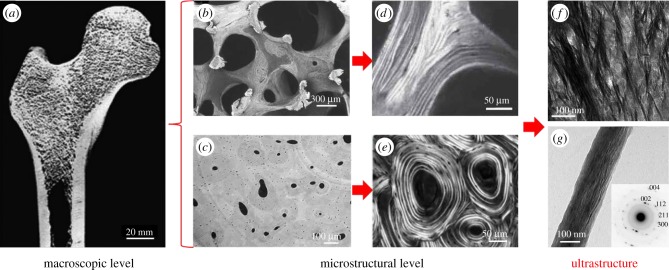
Figure 1.
Bone at different hierarchical levels. At a macroscopic level, bone consists of the cortical and the trabecular bone compartments. On a microstructural level, the trabecular network and the Haversian system are observed, which are typically formed by bone lamellae. At a lower hierarchical level, bone ultrastructure comprises mineralized collagen fibrils, which are arranged randomly or in bundles. This review presents the methods that enable investigations of the organization of the ultrastructure. (a) Human femur cut in half and imaged using scanning electron microscopy (SEM). (Image from [1] with kind permission of the Royal Society of Chemistry.) (b) Trabecular network imaged using SEM in backscattered electron mode. (Image from [2] with kind permission of Humana Press, Inc.) (c) Haversian system imaged using synchrotron radiation-based computed tomography (SR-CT). (Image from [3] with kind permission of SPIE.) (d) Lamellar structure of trabecular bone imaged using polarized second harmonic generation (pSHG) imaging. (Image from [4] with kind permission of the Materials Research Society.) (e) Lamellar structure of cortical bone imaged using circularly polarized light microscopy. (Image from [5] with kind permission of John Wiley and Sons, Inc.) (f) Mineralized collagen fibril bundles imaged using transmission electron microscopy (TEM). (Image from [6] with kind permission of PLoS.) (g) Single mineralized collagen fibril and diffraction pattern (inset) showing the orientation of unit crystal cells imaged using TEM and electron diffraction, respectively. (Image from [7] with kind permission of ACS Publications.) (Online version in colour.)
There exist several factors that are being intensely studied concerning bone's ultrastructure organization [21,25–29]. Among them, the significant contribution of the orientation and arrangement of bone's ultrastructure to its mechanical properties has long been suggested [30–33] and experimentally investigated [34–41]. In addition, several studies conducted at different structural scales have shown that ultrastructure orientation and arrangement are among the best predictors of mechanical properties such as bone strength or elastic modulus [42–45]. Many approaches have been proposed in previous years to investigate the three-dimensional (3D) orientation of at least one of the four structural elements of bone ultrastructure mentioned above, including methods based on visible light, X-rays, electrons or magnetic fields, with some of them providing very promising results. This review intends to present an overview of these approaches and recent progress in their development, in terms of their suitability for the assessment of bone ultrastructure organization, with a specific emphasis on ultrastructure orientation and arrangement. Namely, for each technique we (i) explain its underlying physical principles, (ii) present how the method is applied to study ultrastructure organization, and (iii) critically present the advantages and limitations of those methods in assessing 3D organization of the mineralized collagen fibrils.
It should be mentioned that an overarching limitation of all studies are the artefacts introduced by sample preparation steps, including sample sectioning, decalcification, dehydration or embedding [46]. Depending on the protocol used, these procedures might alter to a greater or lesser extent the tissue structure and, therefore, limit the quantitative aspect of result interpretation. However, quantitative studies of the effects of these factors in relation to the ultrastructure orientation and arrangement are missing. Hence, this review does not include the effects of sample preparation on the final outcome. Finally, this review does not include techniques such as magnetic resonance imaging (MRI) [47], electron backscatter diffraction [48], microwave method [49], small-angle light scattering [50], elastic scattering spectroscopy [51] or ultrasonic methods [52], which have been shown to be able to provide information on the orientation and arrangement of ultrastructure in bone or other tissues, but have not contributed extensively to the assessment of bone ultrastructure organization.
2. Techniques to assess the organization of bone ultrastructure
2.1. Technique categorization
The techniques to assess the organization of the mineralized collagen fibrils or the bone ultrastructure can be divided into two categories, as follows.
The first category represents methods, which can be used to examine directly and specifically the orientation of the structural elements of the bone ultrastructure (fibril bundles/fibres, collagen fibrils, mineral platelets or unit crystals) without providing an image of them, by using polarization, scattering or diffraction of the probe. In this category, only MRI makes use of another physical phenomenon, the orientation-dependent magnetic relaxation. We denote this category of techniques as ‘orientation-specific techniques’.
The second category encompasses methods that provide direct images of bone ultrastructural elements, which we refer to as ‘imaging techniques’, where orientation-specific information can be derived from the images (as a by-product). These methods exhibit spatial resolutions that enable imaging of the ultrastructure of bone, where the ultrastructural elements (mineralized collagen fibrils or fibril bundles) can be visually identified. Quantification of the orientation and arrangement of the ultrastructure is performed by image post-processing of the acquired images, either through specialized orientation-sensitive algorithms [53–55] or, most commonly, through two-dimensional (2D) or 3D Fourier transform (FT) [56], which allow the orientation and degree of orientation (DO) to be derived [57–60]. It should be noted that the indirect assessment of the organization of mineralized collagen fibrils by imaging techniques can lead to artefacts, which are discussed in the introduction of the respective subsection.
Moreover, another distinction between the various techniques is adopted in this review, through the different probes used for the techniques. (Visible) light is the more conventional probe used for many decades in the assessment of the orientation of mineralized collagen fibrils. As its wavelength exceeds the mineral crystal sizes, methods using light as a probe are limited in examining the collagen fibrils or fibril bundles. X-rays and electrons have been used more recently, and can give access to significantly higher spatial resolutions than (visible) light, which allows crystal platelets, unit crystals and also features of the fibrils, such as the typical approximately 67 nm collagen D-spacing, to be probed. The only method that uses a different probe is AFM, which exploits the mechanical interaction of the sample with a sharp tip.
2.2. Orientation-specific techniques
The orientation-specific techniques and their characteristics are presented in table 1. In the following, these techniques are discussed in detail.
Table 1.
Orientation-specific techniques for the assessment of the organization of bone ultrastructure. FOV, field of view; HA, hydroxyapatite.
| technique | spatial resolutiona | feature probed | sample format | FOV dimension | quantitative 3D orientation | main limitation(s) | additional information |
|---|---|---|---|---|---|---|---|
| PLM | 250 nm | fibril + fibril bundle | section | cm | ✗ | sections (destructive) equipment for 3D studies custom-made |
tissue image (in brightfield mode) |
| PRS | 1 µm | collagen + HA mineral | surface | mm | ✗ | limited resolution expensive time-consumingb tissue surface only |
tissue composition tissue quality |
| pFTIR | 2 µm | collagen + HA mineral | section | mm | ✗ | time-consuming sections (destructive) |
tissue composition tissue quality |
| pSHG | 150 nm | fibril + fibril bundle | surface/section | cm | ✗ | expensive low tissue penetration |
SHG image (in SHG mode) |
| SAXS/WAXS | 200 nm synchrotron 100 µm laboratory-based | fibril + HA platelet/unit crystal | sectionc | mm | ✓ | time-consuming sections (destructive)c limited access to synchrotron facilities |
size and shape of ultrastructure features tissue composition |
| X-ray scattering tomography | 200 nm synchrotron 100 µm laboratory-based | fibril + HA platelet/unit crystal | volume | mm | ✓ | high dose limited access to synchrotron facilities long post-processing time |
size and shape of ultrastructure
features tissue composition |
| electron diffraction | 1 nm | unit crystal | section | μm | ✗ | expensive sample preparation time-consuming sections (destructive) limited FOV |
crystal structure very high-resolution tissue images (in TEM mode) |
aSpatial resolution values reflect the current resolution limits of each technique.
bExcept if coherent Raman scattering is used [61], which in exchange leads to significantly higher costs.
cSections not needed when spatially resolved information is not required.
2.2.1. Light-based techniques
2.2.1.1. Polarized light microscopy
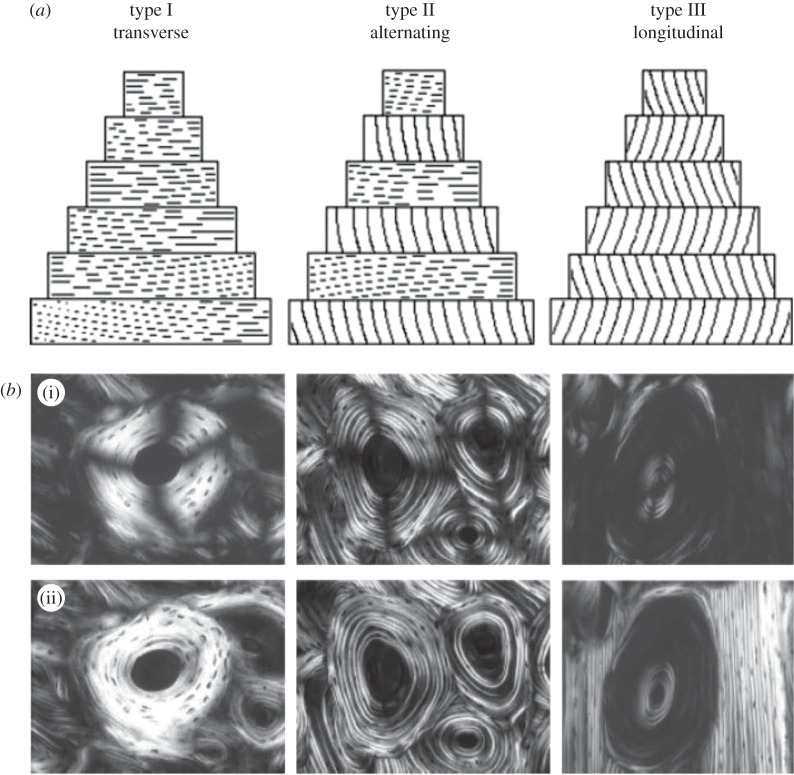
Polarized light microscopy (PLM) has been used since the early nineteenth century to study collagen structure in different biological tissues, exploiting the positive intrinsic and form birefringence of collagen [5,38,62]. Especially for bone, use of PLM has been mostly driven by the early observation of the alternating bright and dark appearance of the lamellae in the osteons (figure 2). The two commonly used forms of PLM are circular PLM and linear PLM (figure 2).
Figure 2.
Determining bone ultrastructure organization using polarized light microscopy (PLM). (a) Sketch of the orientation of the fibrils in the osteon, for the three different osteon types (transverse, alternating and longitudinal). (b) Linear PLM (i) and circular PLM (ii) images of the three types of osteons. The linear PLM images exhibit the ‘Maltese cross’ artefact, because of the polarizer–analyser set-up, leading to a ±90° ambiguity in the orientation of the fibrils in the plane of the section. (Images from [5] with kind permission of Wiley-Liss.)
In circular PLM, image brightness depends on the out-of-plane orientation of the mineralized collagen fibres in the structure and on their DO, with fibres perpendicular to the light path and most highly oriented fibres leading to the highest detected intensities [5,38,63]. In linear PLM, the image brightness also reflects the in-plane orientation of the collagen fibres relative to the polarizer [64], at the plane perpendicular to the light path. In order to retrieve the in-plane orientation, either the sample or the polarizer–analyser system has to be rotated and the results need to be fitted to a sinusoidal curve [64,65]. However, the in-plane results have an ambiguity of ±90°, which is inherent in polarizer–analyser systems (see linear PLM images in figure 2). This ambiguity can be removed with the introduction of a quarter-wavelength plate in the imaging system [66], which is, however, not standard in linear PLM microscopes. Other factors that influence the local image intensity are the section thickness and its optical transparency, the uniformity of the illumination and the initial light intensity [5]. These factors should be well controlled when performing PLM experiments [67]. In addition, local image brightness is susceptible to changes depending on the collagen content/density [64,68] (i.e. the mineral-to-matrix ratio), which is not uniform throughout a bone section, and thus complicates quantification of the 3D orientation of the mineralized collagen fibres from PLM images [64,68]. In general, there is a lack of standardization in the analysis of polarized light images, mainly due to the challenging technical demands and the complex theory of polarized light and birefringence, which can lead to incorrect interpretations of PLM outcomes [62,64].
On the other hand, the applications of PLM in bone research throughout the years have been numerous, provided important insights into the ultrastructural organization of bone [69–72] and allowed structure–function relationships to be investigated [34,35,73–75]. Because of its wide use since the early twentieth century and its relatively low cost compared with most other methods, PLM has been the method of reference for almost all other developed methods investigating the ultrastructural organization [63,76–78]. While other methods, such as X-ray scattering, are increasingly being used as reference methods to quantify bone ultrastructure orientation, shape and size, newer PLM techniques are being developed for quantitative assessment of the 3D orientation of mineralized collagen fibres, by combining the sensitivity of linear PLM for the in-plane orientation and the out-of-plane sensitivity of circular PLM [64,68,79]. The spatial resolution of PLM is in the range of approximately 250 nm and—as typical for optical microscopy systems—is limited by the diffraction limit of visible light at approximately 150 nm (super-resolution microscopy techniques have not been employed in such applications).
2.2.1.2. Polarized Raman spectroscopy
Raman spectroscopy is based on the Raman effect, where incoming photons scatter inelastically on the probed molecule and experience an energy shift. When many photons from the incoming laser interact with the probed material, the outcome is a spectrum of different energy shifts, depending on the material molecules. If one uses a polarized laser, the direction of collagen fibrils affects the Raman signal [80] (figure 3). More specifically, the polarized laser affects some of the peaks in the energy spectrum, such as the amide I and the v1 phosphate peak [67]. This offers a way to investigate the orientation of the collagen fibrils [67,83]. Given that the amide I peak characterizes the organic and the v1 phosphate peak the inorganic part of the ultrastructure, polarized Raman spectroscopy (PRS) can be used to derive information on the collagen and the minerals independently [84,85]. Once the peak positions of the spectra are identified and the heights of the peaks or the areas under the peak are calculated, the data analysis required to extract the 3D orientation of the ultrastructure is similar to that of PLM, as the data have to be fitted to a sinusoidal curve [86]. However, as in PLM, a quantitative 3D analysis is not possible to date.
Figure 3.
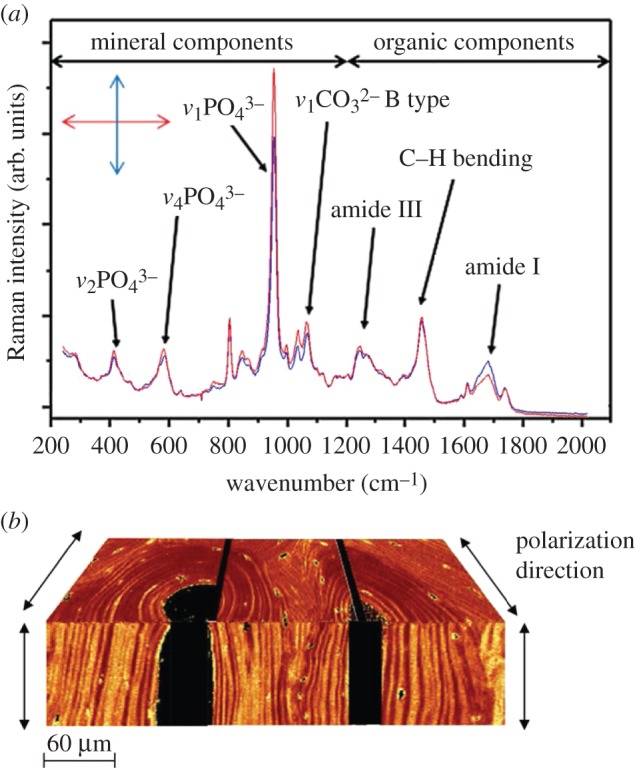
Determining bone ultrastructure organization with Raman spectroscopy. (a) Two Raman spectra of human vertebral trabecular bone embedded in polymethyl methacrylate, where the most important peaks are identified. Spectra were acquired under orthogonal laser polarization directions (double-headed arrow inset). Analysis of the differences in the heights or areas under peaks, such as the v1 phosphate peak (approx. 960 cm−1) or the amide I peak (approx. 1650 cm−1), can provide the ultrastructure orientation. (Image from [67] with kind permission of PLoS, peak assignments according to [81].) (b) A composite of 2D images based on polarized Raman spectral analysis, resulting in a 3D representation of two orthogonal planes of an osteonal structure of human cortical bone. The colour intensity corresponds to the v1 phosphate to amide I ratio, with a spatial resolution of 1–2 mm. (Image from [82] with kind permission of Elsevier.) (Online version in colour.)
In contrast with linear PLM, Raman spectroscopy offers the advantage that the orientation in the plane perpendicular to the light path can be deduced unambiguously as there is no analyser in the experimental set-up. In addition, Raman spectroscopy is performed in reflection mode, meaning that it can be used to analyse a sample without the need to section it, even in vivo [87], and can reach the bone under the skin [88,89]. On the other hand, Raman experiments are much more time-consuming (acquisition of one spectrum typically needs tens of seconds, except if coherent Raman scattering is employed [61]), and offer a lower spatial resolution of approximately 1 µm compared with that of PLM (approx. 250 nm). Despite that, advances in instrumentation have enabled high-resolution, position-resolved analyses of bone ultrastructure orientation [81,82]. This is often combined with composition analysis [90,91], which is an inherent capability of Raman spectroscopes to provide properties that determine different bone quality [92] and other clinically relevant [88,93] properties. Because of the attention Raman spectroscopy has been gaining as an in vivo imaging modality [88,89,94,95], and the advances that have been made in recent years in the tools to characterize mineralized collagen fibril orientation, Raman spectroscopy/imaging can be expected to become a common tool to characterize bone ultrastructure organization in the near future.
2.2.1.3. Polarized Fourier transform infrared spectroscopy
Raman and infrared (IR) spectroscopy are two vibrational spectroscopy methods that can detect specific chemical bonds in a sample [96]. Their difference lies in the fact that IR spectroscopy detects the absorption of photons by the sample for a range of IR frequencies, as opposed to the energy shift due to Raman scattering for a single frequency. Fourier transform IR (FTIR) is the most commonly applied type of IR spectroscopy, because of its higher speed, accuracy and signal-to-noise ratio compared with conventional (dispersive) IR techniques [97]. Similar to Raman spectroscopy, the orientation and arrangement of mineralized collagen fibres can be investigated through the use of a polarized laser [77,98]. However, IR spectroscopy detects asymmetric rather than symmetric vibrational modes (stretches) [99]. Consequently, the technique cannot be used to image aqueous samples, as opposed to Raman spectroscopy. IR spectroscopy is usually conducted in transmission mode, and thus requires more extensive (and destructive) sample preparation steps. On the other hand, the equipment for IR spectroscopy is significantly less expensive, making its application more common than Raman spectroscopy. Although FTIR has been routinely used to examine the composition of bone [100], relatively few studies have investigated the ultrastructural organization of bone [101,102] and neighbouring tissues such as the ligament-to-bone insertion [103] or cartilage [77,104]. This is due to the relatively recent idea of using polarized light for different sample or polarization rotation angles, which allows collagen fibril orientation to be examined in FTIR [77,98]. It should be noted that, as for PLM and Raman spectroscopy, FTIR is an inherently 2D technique, and cannot provide quantitative 3D orientation information. Its spatial resolution is somewhat lower than that of Raman spectroscopy [92], in the range of a few to several micrometres.
2.2.1.4. Polarized second harmonic generation imaging
Second harmonic generation (SHG) imaging [105,106] is a relatively new technique that has gained a lot of attention during the past two decades, partly because it can be realized using existing multi-photon microscopy instrumentation (SGH and multi-photon microscopy are described in more detail in §2.3). SHG exhibits high specificity and, thus, good image contrast for collagen [107,108], which makes it an ideal imaging method for all collagenous tissues including bone (figure 4). Direct investigation of the orientation of the fibrils can take place with the help of a polarizer–analyser couple [106,110]. Compared with PLM, pSHG offers higher image contrast for collagen fibrils. Another advantage of pSHG is the capability to penetrate tissue. However, this is limited to less than approximately 50 µm in the case of dense tissues such as bone [4,111], as the signal is compromised with increasing tissue depth [112,113]. On this account, there have been only a few studies published for bone using pSHG [109,114], and these are restricted to regions close to the bone surface, where similar results can be achieved using PLM. On the other hand, SHG is a popular technology that has gained a lot of attention more recently, and it is regularly being used for other softer tissues such as tendon [108], cartilage [112] or intervertebral disc [110]. However, similarly to PLM, SHG cannot be employed to quantitatively determine the collagen density and, hence, to derive the DO of the fibrils, but provides the in-plane [115] and/or out-of-plane [116] orientation in a semi-quantitative way [117].
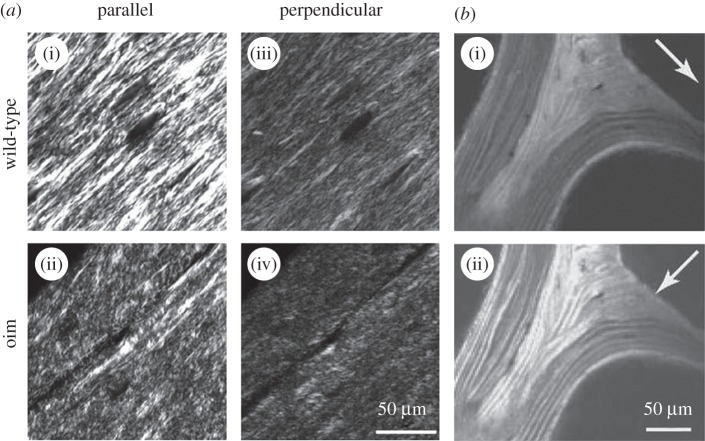
Figure 4.
Determining bone ultrastructure organization using polarized second harmonic generation (pSHG). (a) Comparison of organized wild-type (i) and disorganized osteogenesis imperfecta (oim) (ii) bone ultrastructure from 5 µm thick sections of demineralized femoral mouse bones. The images in (a(i),(ii); parallel) are taken with a polarizer angle difference of 90° with respect to the images in (a(iii),(iv); perpendicular). (Images from [109] with kind permission of SPIE.) (b) pSHG of human vertebral trabecular bone in transmission mode. The white arrows indicate the polarization direction of the incident laser beam. Mineralized collagen fibril bundles/fibres arranged in lamellae are clearly visible when aligned with the laser polarization direction (b(ii)). (Image from [4] with kind permission of Cambridge Journals.)
2.2.2. X-ray-based techniques
2.2.2.1. Small-angle/wide-angle X-ray scattering
Small-angle X-ray scattering (SAXS) and wide-angle X-ray scattering (WAXS), also referred to as small-angle and wide-angle X-ray diffraction (SAXD and WAXD), are phenomena occurring when incoming X-rays are scattered by a sample, at smaller or larger angles, respectively. SAXS and WAXS exploit differences in electron density distributions of the different materials within the sample; X-ray photons interact with ordered and periodic systems such as collagen and mineral crystals, resulting in scattered X-ray waves that interfere constructively or destructively to create the corresponding intensity patterns on the detector, depending on the size and the spatial distribution of the scatterers [118]. SAXS and WAXS can both be used for the analysis of bone ultrastructure [119]: WAXS provides information from scatterers with dimensions in the sub-nanometre range, which are typical for crystallites and spacings between crystal lattice planes. By contrast, SAXS can be employed to retrieve information from tissue features of the order of 1–100 nm, from both collagen [120] and mineral crystal platelets [121,122], thus providing information on both organic and inorganic phases of bone [123]. When used in combination, SAXS and WAXS can simultaneously provide information on the unit crystals, crystal platelets and collagen fibrils in bone [123]. The area detectors used to record diffraction patterns in SAXS and WAXS provide information on tissue anisotropy based on the anisotropic scattering [124]. SAXS especially has been regularly employed in the past few decades to investigate collagen fibril orientation in many collagen-rich tissues [125]. For bone, studies have been undertaken to investigate ultrastructure organization in animal [126,127] and in human bone tissue [63,128,129], in cortical [63,122,130] and trabecular bone [76,101], and in the bone–cartilage interface [131–133], looking at the influence of age [134], disease [135,136], drug use [137–139], fracture healing [140,141] or genetic modifications [142,143].
The high brilliance of synchrotron radiation (SR) facilities and recent advances in fast-readout and low-noise detectors have enabled fast acquisition of X-ray scattering patterns, which have allowed spatially resolved investigations of bone tissue through scanning small-angle X-ray scattering (sSAXS) [76,144]. In typical sSAXS protocols thin sections are used to obtain information from discrete probed tissue volumes. Common practice is to match the thickness of the sections with the size of the X-ray beam, so that the probed volume is cubic. This practice also helps to avoid averaging information over extended sample volumes, where ultrastructural orientations may vary significantly. However, the use of thin sections for spatially resolved investigations is a destructive method. Typical spatial resolutions of sSAXS are in the range of tens of micrometres, but can reach the sub-micrometre level. However, higher spatial resolutions usually also need thinner tissue sections (to ensure cubic probed volumes), which restricts the field of view (FOV).
SAXS diffraction patterns provide 2D orientation information only, which is merely a projection of the 3D orientation information of the ultrastructure [145]. Recently, there have been efforts towards deriving the 3D ultrastructure orientation from SAXS data (3D SAXS), by probing the sample under different rotation angles [45,141,146]. In a new method, called 3D sSAXS [145], the 3D ultrastructure orientation has been derived quantitatively in a spatially resolved manner for small bone trabecular volumes [67] (figure 5). These results underline the potential of SAXS and WAXS for studying bone ultrastructure orientation and arrangement, with its main limitation being the difficulty in accessing the special synchrotron facilities required for such investigations. Another limitation is the need for thin sections, when spatially resolved information is needed, making the method destructive. Compared with PLM, SAXS and WAXS offer better capabilities to characterize the organization of mineralized and non-mineralized collagen fibrils in a quantitative fashion, as the DO of the fibrils can be normalized by the transmission information that is being simultaneously recorded [147]. Finally, it should be noted that SAXS and WAXS can be combined with in situ mechanical testing [148,149] to investigate load transfer mechanisms in normal [150], diseased [151] or treated [152] bone, broadening their range of applications and providing insight into bone structure–function relationships.
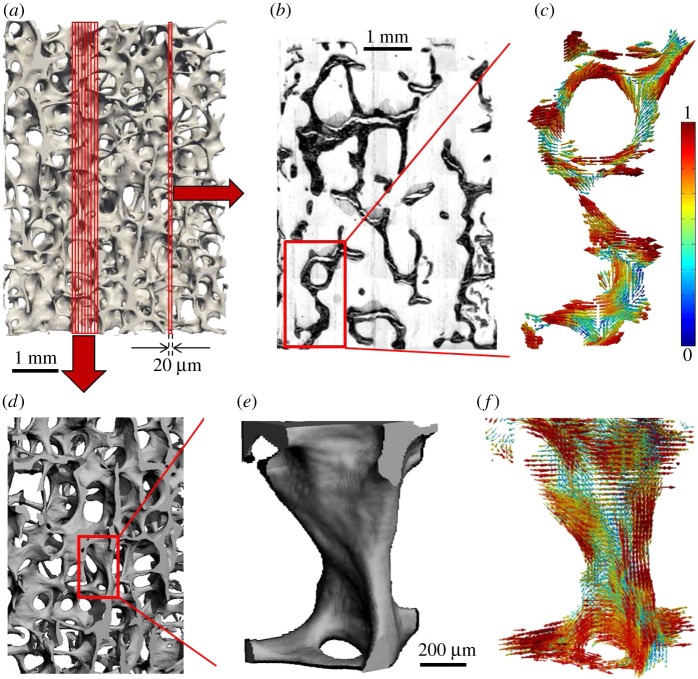
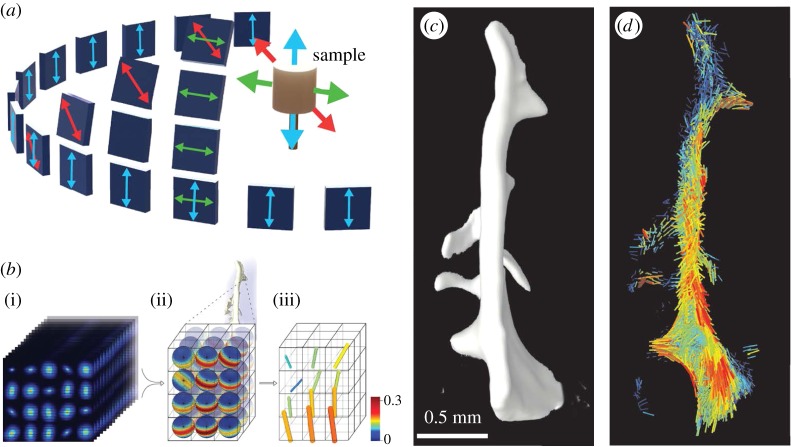
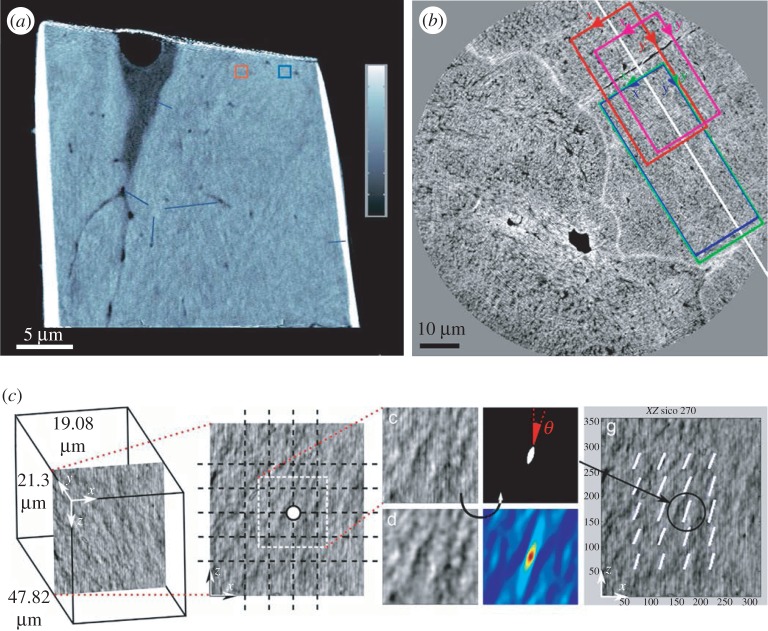
Figure 5.
Determining bone ultrastructure organization with 3D scanning small-angle X-ray scattering (3D sSAXS). (a) Trabecular bone volume, which includes the trabeculae of interest, imaged with micro-computed tomography. (b) Thin section cut out of the volume in (a). The region of interest (rectangle) is scanned with sSAXS for different rotation angles. (c) Local 3D orientation for every bone sub-volume, based on the analysis of the diffraction patterns of each sub-volume for the different rotation angles. The level of the degree of orientation (DO) is denoted by the length of the vector, as well as by the colour of the colourmap (in the online version). (d) Many consecutive thin sections stacked together. In each section, the region of interest contained in the red rectangle is scanned. (e) The trabecular structure under investigation. (f) Reconstruction of the 3D orientation map for each sub-volume of the trabecula. The level of the DO can be interpreted by the length of the vector and the colourmap in (c) (in the online version). (Images from [145] with kind permission of Elsevier.) (Online version in colour.)
2.2.2.2. X-ray scattering/diffraction tensor tomography
X-ray scattering/diffraction tomography has been developed and employed in previous decades to tomographically reconstruct SAXS or WAXS information for a sample volume [153]. Briefly, reconstruction techniques used in X-ray absorption tomography have been applied to tomographically reconstruct information from specific q-ranges in the diffraction patterns. This can be used to distinguish different materials [154], tissues [155], tissue features or composition [156,157] within a sample. However, such approaches assume isotropic azimuthal scattering, ignoring the anisotropy in the diffraction patterns. Approaches that take into account the structural anisotropy have also been proposed [158–160], where diffraction information is reconstructed for different azimuthal angles. These studies provide tissue anisotropy information in a tomographic way. However, they do not provide 3D orientation information, as they do not account for the fact that the orientation information in the diffraction pattern is merely a projection of the 3D orientation, which changes with sample rotation [145].
Concerning quantitative ultrastructure organization analysis, three techniques were developed very recently to investigate 3D ultrastructure orientation in a tomographic way, based on the phenomenon of X-ray scattering: X-ray tensor tomography [161], six-dimensional SAXS tomography (6D SAXS tomography) [162] and small-angle scattering tensor tomography (SAS tensor tomography) [163]. The three techniques can retrieve the ultrastructure organization of a volume of material, such as bone, without having to section the sample. These techniques have evolved in different ways: X-ray tensor tomography has evolved from X-ray dark-field imaging using a grating interferometer [164], where ultra-small X-ray scattering is exploited [165]. The intensity modulations due to the rotation of the third grating [166,167] or of the sample [168,169] reveal the 2D orientation of the ultrastructure, and have been used to retrieve 2D ultrastructure organization of bone [170] or dentin [166]. By rotating the sample around two axes, and using an iterative reconstruction algorithm, it is possible to retrieve the 3D ultrastructure orientation [161]. Applications to bone ultrastructure are expected to follow. SAS tensor tomography and 6D SAXS tomography, on the other hand, have evolved from SAXS, combining the concepts of SAXS tomography [171] and 3D sSAXS [145], while adding a sample rotation around a second axis. For the reconstruction of ultrastructure orientation, 6D SAXS tomography employs a finite number of virtual tomographic axes (figure 6a): for each axis direction, only the ultrastructure orientations that are parallel to the axis are reconstructed. The use of 6D SAXS tomography has very recently been employed to evaluate the ultrastructure orientation in bone dentin [162]. On the other hand, SAS tensor tomography uses an iterative tensor tomography algorithm based on spherical harmonics for the reconstruction of the ultrastructure orientation (figure 6b). Known internal sample symmetries—such as the rotational symmetry in mineralized collagen fibrils—can be exploited to reduce post-processing time. SAS tensor tomography was very recently applied to successfully retrieve the ultrastructure organization of a bone trabecula [163] (figure 6c,d). All three techniques are non-destructive, and open new paths towards ultrastructure organization investigations of whole sample volumes. It should be noted that this comes at the cost of higher X-ray dose, and long post-processing times needed to handle the vast amount of acquired data [172]. In addition, X-ray tensor tomography is currently limited to the assessment of only the 3D orientation and not the DO, as it cannot quantify the amount of scatterers in each voxel. The spatial resolution of these techniques is similar to the SAXS techniques, i.e. in the range of tens of micrometres in synchrotron facilities and hundreds of micrometres for laboratory-based systems.
Figure 6.
Determining bone ultrastructure organization with six-dimensional SAXS tomography (6D SAXS tomography) and small-angle scattering tensor tomography (SAS tensor tomography). (a) The virtual-tomography-axis technique in 6D SAXS tomography, where, for each virtual sample axis, the corresponding projections (arrows with matching colour) are used to reconstruct the ultrastructure orientation. (Image from [162] with kind permission of Nature Publishing Group.) (b) The iterative spherical harmonics technique in SAS tensor tomography. The thousands of SAXS patterns corresponding to the same voxel under different angles (i) are fitted to a spherical harmonics equation (represented by a single sphere in (ii)) that represents the local orientation and arrangement of the mineralized collagen fibrils (iii). (c) Computed tomography reconstruction based on the recorded transmitted intensity. (d) Reconstruction of the orientation and arrangement information for a bone trabecula based on the iterative spherical harmonics algorithm. (Images from [163] with kind permission of Nature Publishing Group.) (Online version in colour.)
2.2.3. Electron-based techniques
2.2.3.1. Electron transmission diffraction
Electron transmission diffraction pattern detection can be an additional feature in TEM set-ups [173] (described below). It is used to provide information on the orientation of mineral crystals [23,24,174], derived from the diffraction pattern of the electrons that interact with the crystal lattice planes (figure 7). However, extensive preparation protocols for TEM that typically include fixation, dehydration, drying, enhancing feature contrast, preparing small samples for subsequent cutting with an ultramicrotome and the following handling of very small specimens have restricted the use of electron diffraction for quantifying bone ultrastructure organization to a handful of studies over the past decades [6,7,175–178] (figure 7), examining either the crystal arrangement in single platelets or fibrils or a limited number of points within a TEM section. TEM gives access to very small features at the nanometre scale (e.g. single platelets), which are inaccessible with other techniques, and enables the analysis of the 3D orientation of mineral platelets [179,180]. Nevertheless, the restricted FOV (of the order of a few micrometres) and elaborate sample preparation procedures are major limiting factors for electron transmission diffraction to become widely used in the study of the organization of bone ultrastructure.
Figure 7.
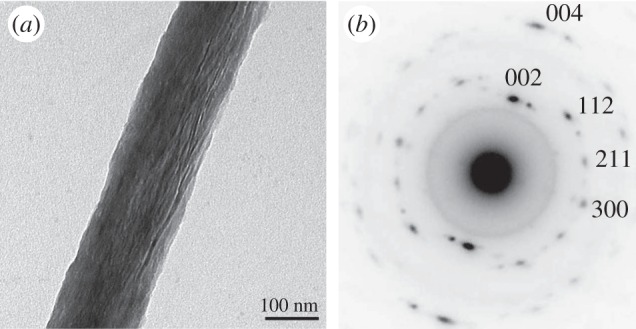
Determining bone ultrastructure organization with electron transmission diffraction. (a) Single mineralized collagen fibril. Data assessed by TEM. (b) A diffraction pattern from an area of the fibril, with the c-axis of the crystals (strong black dots) being aligned with the direction of the fibril. (Images from [7] with kind permission of American Chemical Society Publications.)
2.3. Imaging techniques
The imaging techniques presented here provide direct images of bone ultrastructural elements, where orientation-specific information can be derived from the images (as a by-product). These ultrastructural elements include mineralized collagen fibril bundles (for light-based techniques), mineralized collagen fibrils (for X-ray-based techniques) or fibril features such as the approximately 67 nm D-spacing and crystal platelets (for electron-based techniques). By applying image post-processing steps to the gathered image data (e.g. FT) and with the goal to derive orientation-specific information, the organization of the ultrastructure can be analysed [53,55–59].
The imaging techniques typically have a considerably lower FOV than the orientation-specific techniques employing the same probe (presented earlier). The reason for the FOVs being different is that the ultrastructural elements need to be visually identified, which relies on high spatial resolutions, which in turn limit the FOV, and, consequently, the size of the area/volume that can be investigated. One way to overcome this limitation and to extend the effective FOV is through imaging adjacent sample areas [181].
Another consequence of the image-based approaches is the need to discretize the image data in order to derive the ultrastructure orientation via orientation-specific algorithms, which usually include a FT of a 2D or 3D dataset (figure 8). However, most algorithms are not specific for the fibrils, but they rather average the information from all features in the image. For the case of bone, this can be features of the lacuno-canalicular network, lamellar boundaries, cement lines, bone–canal or bone–marrow interfaces and possibly image artefacts. Despite this, the imaging techniques presented here offer the advantage of providing visual information of bone tissue at the scale of the structural elements, which also enables observations concerning other aspects of the bone tissue, such as the size of the collagen fibrils or fibres, the lacuno-canalicular network or the lamellar structure. Such information can then also be used to create ultrastructural models that enhance the analytical tools to study bone's hierarchical structure [6,182–184].
Figure 8.
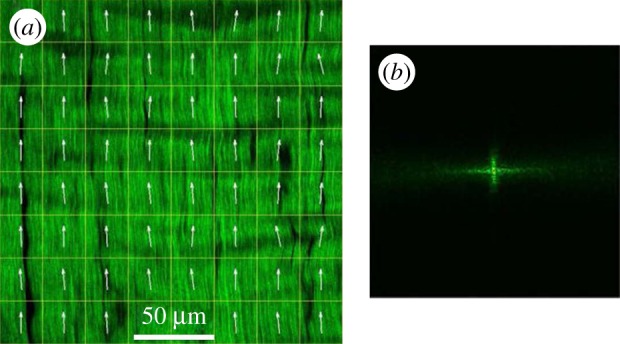
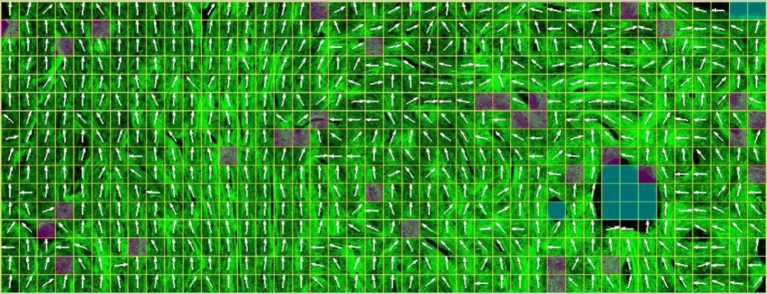
Typical ultrastructure orientation analysis for the presented imaging techniques. (a) Tendon collagen fibres imaged using second harmonic generation microscopy. The image is split into multiple compartments, in each of which the orientation is determined via an orientation-specific image-processing algorithm. (b) FT of a single compartment in (a), to retrieve the 2D orientation of the fibres in this compartment. FT is the most common method to retrieve orientation information from the data in imaging techniques. (a,b) The spatial resolution of imaging techniques, where the orientation-specific information is derived as a by-product, has to be in the sub-micrometre range, so that fibres can be identified, which limits the FOV that can be covered. Also, the necessary compartmentalization (white grid in (a)) reduces the ultrastructure organization analysis resolution (here it is reduced to approx. 25 µm). Moreover, irrelevant structural features (such as the blood vessels that appear black in figure 8a), or possible imaging artefacts, are also taken into account, unless special care is taken to eliminate their influence of the orientation-specific measures. (Image from [59] with kind permission of OSA publishing.) (Online version in colour.)
The most commonly used imaging techniques for assessing the mineralized collagen fibril organization are presented in table 2.
Table 2.
Imaging techniques for the assessment of the organization of bone ultrastructure.
| technique | spatial resolutiona | feature imaged | sample format | FOV dimension | quantitative 3D orientation | main limitation(s) | additional information |
|---|---|---|---|---|---|---|---|
| CLSM | 150 nm | fibril bundle | surface/section | cm | ✗ | cannot resolve single fibrils low depth penetration |
macroscopic image of 3D tissue volume |
| SHG | 100 nm | fibril bundle | surface/section | cm | ✗ | cannot resolve single fibrils low depth penetration expensive |
macroscopic image of collagen in 3D tissue volume |
| SR-CT | 20 nm | fibril bundle | volume | cm | ✗ | cannot resolve single fibrils (up to now) limited access to synchrotron facilities |
mesoscopic 3D tissue image tissue mineral density trabecular architecture lacuno-canalicular network |
| phase-contrast CT | 15 nm | fibril | volume | μm | ✓ | currently at the limit of resolving single fibrils time-consuming limited access to synchrotron facilities |
microscopic 3D tissue image tissue mineral density lacuno-canalicular network |
| TEM | 0.1 nm | fibril + HA platelet | section | μm | ✗ | very extensive sample preparation expensive limited FOV |
nanoscopic 2D tissue image platelet shape and size mineral–collagen interface fibril diameter collagen D-period |
| SEM | 1 nm | fibril | surface | μm | ✗ | extensive sample preparation only tissue surface expensive limited FOV |
nanoscopic 2D tissue image |
| FIB SEM/SBF SEM | 10 nm | fibril | volume | μm | ✓ | extensive sample preparation only tissue surface expensive limited FOV time-consuming |
microscopic 3D tissue image lacuno-canalicular network |
| AFM | 0.1 nm | fibril + HA platelets | surface | μm | ✗ | limited FOV irreproducible results due to possible probe tip damage |
nanoscopic 2D tissue image platelet shape and size mineral–collagen interface fibril diameter collagen D-period mechanical properties |
aSpatial resolution values reflect the current resolution limits of each technique.
2.3.1. Light-based imaging techniques
2.3.1.1. Confocal laser scanning microscopy
Confocal laser scanning microscopy (CLSM) enables imaging ‘inside’ tissues by selectively collecting information from a specific plane (the focal plane) via the use of pinholes in the light path [185]. As different focal planes can be chosen, CLSM enables taking a so-called z-stack of images, which contains information of a volume inside the tissue. It should be noted that this capability is limited in the case of hard tissues such as bone, where CLSM penetration is restricted to a few hundreds of micrometres. The contrast in CLSM images is a result of differences in the refractive index of materials, which is typically small within biological tissues. Consequently, CLSM is often combined with fluorescence microscopy, in order to provide additional biochemical information characterizing the sample. Collagen is well fitted for fluorescence imaging without the need of fluorescent dyes because of its autofluorescence [186]. CLSM has been regularly used to investigate bone and other tissues [187], including the organization of the bone ultrastructure [188–190]. However, bone tissue assessed by CLSM can provide images of the mineralized collagen fibril bundles, which are on the same size scale as its spatial resolution capabilities (approx. 200 nm). Thus, studying the orientation of the bone ultrastructure using CLSM provides qualitative rather than quantitative results. In addition, confocal microscopes have recently lost their charm to multi-photon microscopes to some extent, as the latter offer higher tissue penetration depths and more specific information from the focal plane/point (figure 9) as well as other advantages described in the following.
Figure 9.
Imaging bone with confocal laser scanning microscopy (CLSM). (a) CLSM image of a single lamella from a human femur. The arrow represents a qualitative assessment of the orientation of the collagen fibrils. (Image from [63] with kind permission of Elsevier.) (b,c) Difference in the focusing capabilities of CLSM versus multi-photon microscopy. (Images from [191] with kind permission of Nature Publishing Group.) (b) In the case of CLSM, the laser beam excites molecules outside of the focal plane on its path through the tissue. (c) In multi-photon microscopy, (at least) two photons are combined to specifically excite only the molecules at the focal spot. (Online version in colour.)
2.3.1.2. Second harmonic generation in multi-photon microscopy
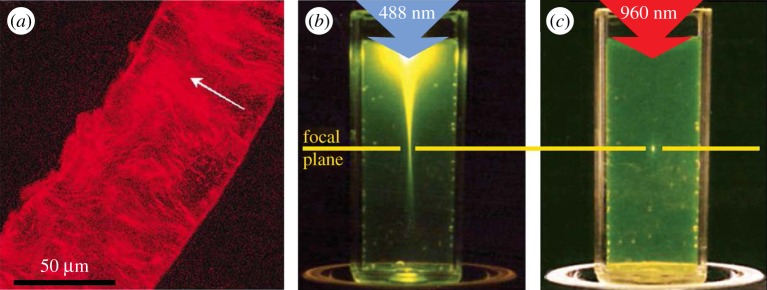
Multi-photon microscopy uses short-pulsed laser to create a high spatial and temporal photon density at a well-defined focal point in the specimen, where two or more lower-energy photons are combined to reach the energy levels necessary for fluorescence excitation [192] (figure 9). The set-up for multi-photon microscopy experiments is very similar to those for CLSM if one removes the pinholes and switches to a very short-pulsed laser [193]. Yet, multi-photon microscopy offers several advantages: (i) higher tissue penetration depth (photons at higher wavelength penetrate deeper into scattering tissues), (ii) significantly greater selectivity of the imaging plane, as photons are combined at, and excite, only one desired point in space, (iii) higher photon yield, due to the lack of the pinholes present in CLSM, which are not needed as the excited point emits fluorescence photons only, (iv) less photodamage (damage to the tissue by harmful radiation), and (v) less photobleaching (progressive destruction of the fluorescence properties of the fluorophore by continuous excitation) [194]. Multi-photon microscopy is routinely used in vivo and is continuously advancing [195]. For the investigation of collagen-rich structures, an extended experimental set-up of multi-photon microscopes is commonly employed, which is called SHG microscopy.
SHG microscopy [105,106] is based on the homonymous phenomenon, where a high photon density beam passing through a strongly birefringent material excites electrons to a virtual state, which results in the emission of photons at an energy twofold the excitation energy. SHG microscopy is thus an appropriate imaging technique for strongly birefringent materials such as collagen, which can be well distinguished from other tissue components [107,196]. It can experimentally be implemented using the same set-up as for multi-photon microscopy, with the addition of a detection filter at half the wavelength of the emission laser. Owing to the great popularity of the technique [191], more dedicated and sophisticated set-ups have been developed that take full advantage of the capabilities of SHG [111]. In recent years, there have been many studies on collagenous tissues using SHG microscopy, including cartilage [197], tendon [59], muscle [198], skin [199], fetal membranes [200] and vessels [201].
At the same time, bone ultrastructure organization has been investigated using SHG imaging in a handful of studies only [45,202–204] (figure 10), which is due to the limited penetration depth of SHG in mineralized tissues (less than 100 µm [45]). Nonetheless, the capability of SHG to provide spatial resolutions down to approximately 30 nm [206], the development of methods to quantify the 3D orientation of collagen fibrils [5], and the advantage to scan deep within the tissue in vivo [121,207,208] or to image in conjunction with mechanical testing [209,210] point towards a wider use of SHG for a quantitative investigation of the bone ultrastructure in the future.
Figure 10.
Imaging bone with second harmonic generation (SHG) microscopy. Collagen fibril bundles from porcine cortical bone (in green in the online version). A grid has been superimposed in order to compartmentalize the picture that enables semi-quantitative assessment of the 2D orientation of the collagen fibrils (white arrows). (Image from [205] with kind permission of Elsevier.) (Online version in colour.)
2.3.2. X-ray-based imaging techniques
2.3.2.1. Absorption-based X-ray imaging
SR-based computed tomography (SR-CT) is a CT-based technique [211] that can reach resolutions in the sub-micrometre range [212]. Compared with conventional laboratory-based micro-computed tomography (µCT) imaging systems, SR-CT can deliver higher resolution images of bone tissue [213] with increased signal-to-noise ratios, mainly due to the high X-ray flux available at (third generation) X-ray synchrotron sources [213]. At the same time, the use of quasi-monochromatic X-ray light in SR-CT imaging prevents beam hardening effects, which are typically present in CT scans from laboratory-based µCT systems that are equipped with a standard (polychromatic) X-ray tube.
Moreover, the parallel X-ray beam set-up for SR-CT imaging is free of cone beam artefacts known from classical µCT systems, where X-rays are emitted in a cone beam fashion [214]. SR-CT allows bone features of sizes similar to the mineralized collagen fibrils or fibres [3,215,216] to be examined in a quantitative way [217,218]. Examples are osteocyte lacunae, which are in the micrometre range, and canaliculi, with dimensions of the order of 100 nm. These bone microporosities can be identified and segmented straightforwardly due to their high contrast compared with the surrounding (mineralized) bone matrix. However, identifying single fibrils requires higher spatial resolutions and contrast, as is shown for SR-CT at sub-micrometre resolution, where a mineralized collagen fibril arrangement is visible [202] (figure 11). The technique is not currently at a stage where bone ultrastructure organization can be studied in a quantitative fashion, yet continuous advancements in SR-CT [212,219] at ever increasing spatial resolutions, currently reaching 20 nm [220], will eventually enable single collagen fibrils to be resolved in three dimensions, and hence quantitative investigations of 3D orientation of mineralized collagen fibrils in bone. Nevertheless, improvements in resolution typically come at the cost of a reduced FOV.
Figure 11.
Imaging bone with synchrotron-based computed tomography (SR-CT). Three images of human femoral bone at a voxel size of 280 nm, where bone ultrastructure organization can be identified, but not quantified. (Image from [202] with kind permission of the American Association of Physicists in Medicine.)
2.3.2.2. Phase-contrast X-ray imaging
Unlike the traditional X-ray absorption techniques, phase-contrast X-ray imaging exploits the phase of the propagation X-ray waves (not their amplitude), which is modulated by interaction with the object when coherent X-rays beams are used, provided explicitly by SR sources.
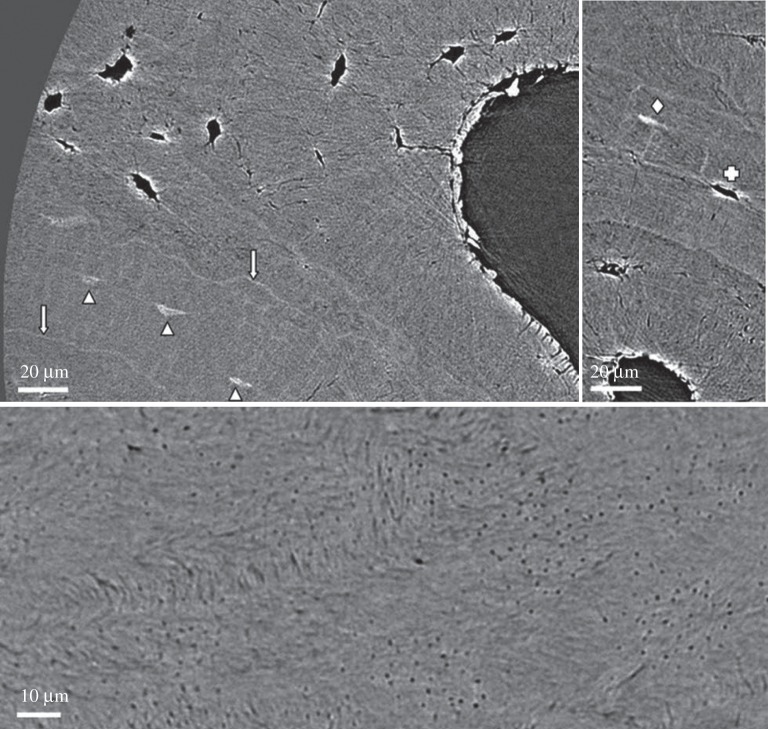
The various phase-contrast techniques offer in many cases higher image contrast at lower X-ray exposure or dose than X-ray absorption techniques, which makes them good candidates for in vivo studies [221–223]. The applications of phase-contrast X-ray imaging techniques are rapidly increasing [222] and specific imaging modalities such as X-ray phase nanotomography [224] and ptychographic (or lensless) CT [225] have been used to image bone with spatial resolutions at the nanometre scale. The first studies on bone ultrastructure organization have been performed on volumes at the micrometre level [226] (figure 12). Considering the capabilities of ptychographic CT for quantitative analysis of materials and tissues [227] at spatial resolutions below 20 nm [228], where single collagen fibrils could be resolved, phase-contrast methods offer the potential for a direct insight into bone ultrastructure organization of single osteons or trabeculae.
Figure 12.
Imaging bone with X-ray phase-contrast techniques. (a) Bone ultrastructure of mouse femur assessed with ptychographic CT at 65 nm isotropic voxel size, where bone microporosities (osteocyte lacunae and canaliculi) are visible. The continuous advances in synchrotron radiation-based imaging techniques led to spatial resolutions below 20 nm, which allows direct assessment of the bone ultrastructure organization. (Image from [225] with kind permission of Nature Publishing Group.) (b) Human femoral bone imaged using X-ray phase nanotomography at 60 nm isotropic voxel size. (c) Compartmentalization of the images via a 3 µm grid and image post-processing results in the assessment of the ultrastructure orientation. ((b,c) Images from [226] with kind permission of Elsevier.) (Online version in colour.)
2.3.3. Electron-based imaging techniques
2.3.3.1. Transmission electron microscopy
TEM has very high spatial resolution capability, which is well in the sub-nanometre range [229]. TEM examines sub-micrometre thin material sections, which are difficult to prepare [230]. The image contrast is imparted through differences in the quantity of transmitted electrons through the sample. The high financial equipment and sample preparation costs, the extensive sample preparation time and expertise required [231] and the restricted FOV (as a consequence of the high spatial resolution) have limited the use of TEM in the study of bone ultrastructure organization.
On the other hand, the available studies using TEM portrayed the bone ultrastructure at a nanoscopic scale, which has led to important arrangement models of the bone ultrastructure, such as the twisted plywood pattern in the osteons proposed by Giraud-Guille [232]. TEM has also allowed local qualitative assessment of the ultrastructure arrangement of healthy and osteoporotic bone [233,234], but in two dimensions only (figure 13). Furthermore, TEM has been applied to make important contributions to what we know about bone structure at very small scales, through investigations of the shape and size of bone crystals [14,235], their spatial relationship and arrangement in relation to the collagen fibrils [175,236] or features of the collagen fibrils such as the approximately 67 nm D-spacing [7] (figure 13). Finally, TEM is also playing an important role in studying crystal arrangement during the bone mineralization process [237].
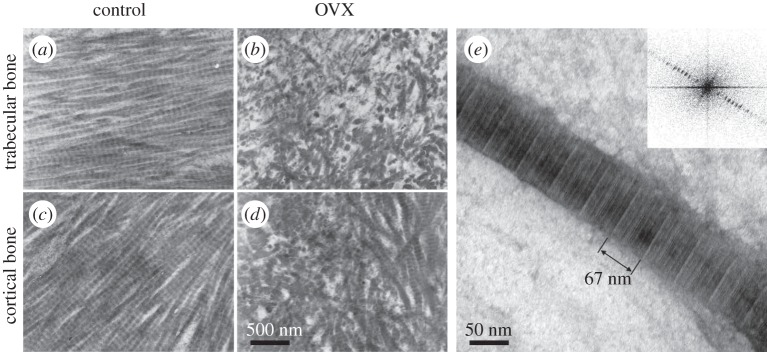
Figure 13.
Imaging bone with transmission electron microscopy (TEM). (a–d) Bone ultrastructure arrangement of decalcified trabecular (a,b) and cortical (c,d) rat tibiae. Collagen arrangement in bones of control animals (a,c) in qualitative comparison with bones from ovariectomized animals (b,d). (Images from [233] with kind permission of Elsevier.) (e) Single mineralized collagen fibril, where the approximately 67 nm D-spacing of collagen is visible and can be measured using Fourier transform of the image (inset). (Image from [7] with kind permission of American Chemical Society Publications.)
2.3.3.2. Scanning electron microscopy
Scanning electron microscopy (SEM) is the most widely used electron microscopy technique. SEM provides nanometre resolution of the sample surface, so there is no need to prepare very thin samples, as for TEM. However, equipment is costly and bone sample preparation procedures labour intensive [2,238]. Secondary electrons allow sample texture and topography to be analysed, and have been used in SEM to determine collagen fibril orientation in different tissues such as ligaments [57], menisci [239], tendons [240,241] and cartilage [242,243], as well as bone [203,209,210]. SEM gives access to mineralization levels and elemental analysis using backscattered electrons and energy-dispersive X-rays, respectively, generated by the interaction of the electron beam with the sample [2,204]. However, only the sample surface is imaged. At the present time, SEM is typically employed in the study of 3D ultrastructure orientation in combination with volume electron microscopy techniques, which give access to volumetric information, such as serial focused ion beam SEM (FIB SEM) and serial block-face SEM (SBF SEM) discussed below.
2.3.3.3. Serial focused ion beam SEM and serial block-face SEM
In recent years, different volume electron microscopy techniques have been developed, such as serial focused ion beam SEM (FIB SEM) and serial block-face SEM (SBF SEM) [206]. Both methods follow the idea of imaging a volume slice by slice, by alternating between SEM imaging and removing a very thin section of the sample block at the surface, either with an ion beam for FIB SEM or with an ultramicrotome for SBF SEM. The result is stacks of 2D images that provide a 3D volume of the sample. Given that the result of such a procedure is a micrometre volume with nanometre resolution, the data can be used to study ultrastructure organization of tissues such as muscle, tendon [207] or bone, either in two dimensions [208,244,245] or in three dimensions [9,246] (figure 14). Although only a limited tissue volume can be assessed when using volume electron techniques (the spatial resolution at nanometre levels restricts the FOV to tens of micrometres), and despite the fact that these techniques are destructive, they provide important insights concerning the organization of the bone ultrastructure, such as in Haversian systems [9] or trabeculae [247]. While the spatial resolution of FIB SEM is of the order of a few nanometres, compared with the tens of nanometres for SBF SEM (owing to the finer sectioning or milling capabilities of FIB), both techniques are expected to play an important role in further investigations of bone ultrastructure organization at the nanometre scale. It should be noted that both techniques also allow investigations of bone microporosities, including the lacuno-canalicular network [245]. Still, the extended sample preparation protocols and the extended imaging times involved for serial sectioning electron microscopy consume a lot of resources and time [246].
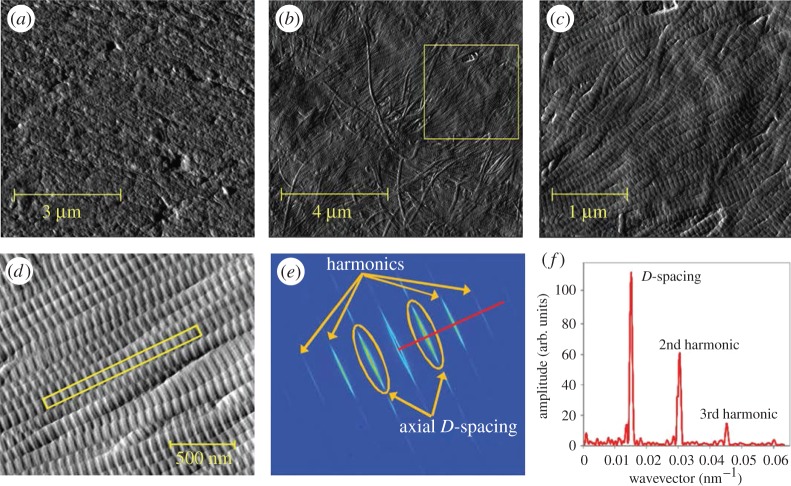
Figure 14.
Imaging bone with serial focused ion beam scanning electron microscopy (FIB SEM). (a–d) Four images out of a 3D stack from demineralized rat tibiae (cortical bone), imaged using serial FIB SEM at a (lateral) pixel size and a slice thickness of about 10 nm. Fast Fourier transform is performed to assess the ultrastructure orientation in two dimensions (insets). (Images from [208] with kind permission of Elsevier.)
2.3.4. Other imaging techniques
2.3.4.1. Atomic force microscopy
AFM [248] is the only technique among the presented ones where the probe mechanically interacts with the sample: a sharp tip mounted on a cantilever is used to probe the sample surface, piezoelectric elements are used to move the sample (or the tip) in the x–y direction and a third element is used to move it in the z-direction. A force-feedback loop has the task of keeping the interaction force between the tip and the sample constant, by acting on the z-piezoelectric element: when the force deviates from the set value, the element moves the sample (or the tip) accordingly, so that the force goes back to the pre-assigned levels. AFM spatial resolution capabilities are impressive and are only comparable with TEM among the techniques considered in this review: it can reach spatial resolutions in the sub-nanometre range, in biological samples and even at room temperature [249]. In addition, AFM can act as a nanoindentation tool to measure mechanical properties of micrometre-scale samples [250,251]. Compared with electron microscopy equipment, AFM is inexpensive and less destructive, while requiring much less copious sample preparation. However, its results are less repeatable, as the probe can be gradually or abruptly blunted by the contact with the sample, or it can pick up a small particle on its tip, events that can cause artefacts in the resulting image. Also, in order to reach the resolution levels needed for imaging single fibrils, the FOV has to be restricted to a few micrometres [252]. Moreover, AFM probes the surface of materials, and can thus provide only 2D information on the organization of the ultrastructure. This is why AFM's numerous applications in bone research [253,254] have been mainly focused on the study of its mechanical properties at a tissue [255–257] and single fibril level [258–260], while very few studies have investigated the organization of the mineralized collagen fibrils [261,262] (figure 15). However, similarly to TEM, AFM has been extensively used to examine bone features at the nanometre scale, such as the size of mineral platelets [13,264], collagen fibril characteristics (e.g. its diameter and D-spacing) [263,265,266] and the spatial relationship between collagen fibrils and mineral platelets [267–269].
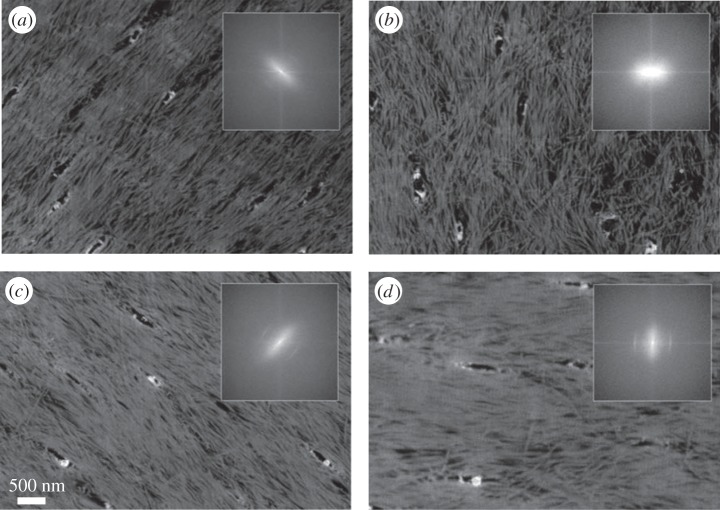
Figure 15.
Imaging bone with atomic force microscopy (AFM). (a–c) Murine cortical bone. (Images from [253] with kind permission of Elsevier.) (a) Polished cortical bone surface, mineralized. (b) Cortical bone surface demineralized using ethylenediaminetetraacetic acid (EDTA). The collagen fibrils are nicely exposed. (c) Collagen fibrils corresponding to the inset area in (b). The characteristic D-spacing as well as their 2D organization are clearly visible. (d–f) D-spacing measurements of collagen fibrils of ovariectomized sheep dermis. (Images from [263] with kind permission of Wiley-VCH Verlag GmbH & Co.) (Online version in colour.)
3. Discussion
The presented techniques that can be used to assess the orientation and arrangement of the mineralized collagen fibrils, the ultrastructural units of bone, were grouped into two categories. For the orientation-specific techniques, the orientation of the collagen fibrils is probed directly based on physical principles such as polarization, diffraction and scattering (see table 1). On the other hand, for the imaging techniques, the images of the ultrastructure must be post-processed using algorithms such as Fourier transform in order to derive quantitative information about the organization of the ultrastructure (table 2).
The presented techniques are numerous, and of different nature, and are used to study the orientation and arrangement of the ultrastructure at different hierarchical levels, in three dimensions or two dimensions only and/or in conjunction with mechanical testing. Some of the techniques have the potential to be applied in vivo, whereas some methods are inherently destructive, and others have the potential to reveal the organization of bone ultrastructure with further advances in technology. Here, we provide a critical assessment of the presented techniques with respect to the above-mentioned aspects, taking into account their capability and potential to provide insight into the organization of bone ultrastructure.
3.1. Assessment at different hierarchical levels and additional information on bone tissue
The presented techniques can provide insight into bone structure and organization at different length scales, probing distinct features at different hierarchical levels (figure 16). In general, orientation-specific techniques provide a larger FOV, as single fibrils or fibril bundles do not need to be resolved individually to provide quantitative information on the 3D organization of the ultrastructure. By contrast, imaging techniques where single collagen fibrils or fibril bundles need to be spatially resolved offer a more limited FOV due to their intrinsic inverse relationship between FOV and spatial resolution. Additionally, the presented techniques provide complementary information, other than orientation and arrangement of mineralized collagen fibrils. Imaging techniques visualize bone structure, probing tissue features apparent at different scales, which allows measures such as ultrastructure feature shapes and sizes and other structural information to be derived, for example about local mineralization, the collagen–mineral interface and the lacuno-canalicular network.
Figure 16.
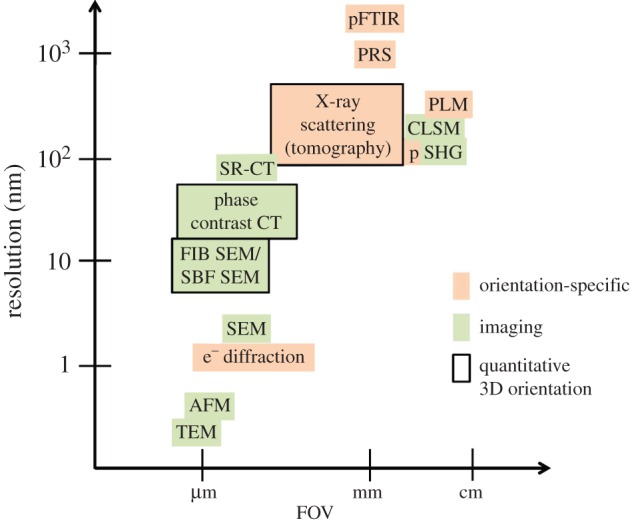
Imaging and orientation-specific techniques for the assessment of bone ultrastructure organization, based on their capabilities in terms of their spatial resolution and the field of view they cover, and their ability to derive the 3D orientation and arrangement of mineralized collagen fibrils in a quantitative way. (Online version in colour.)
More specifically, regarding imaging techniques, AFM and electron microscopy techniques can directly image mineralized collagen fibrils, with a FOV covering areas at a scale of tens of nanometres to a few tens of micrometres [9]. Imaging bone at this scale provides useful information on other structural features, such as the collagen–mineral interface [21], mineral platelet sizes and shapes [13,14] or collagen D-period [7,263] for very high-resolution techniques such as TEM and AFM, and offers data revealing structural details of the lacuno-canalicular network [245] and bone remodelling sites [270] for high-resolution techniques such as SEM. X-ray phase-contrast techniques such as ptychographic CT [225] or X-ray phase nanotomography [226] operate at similar scales to volume SEM, and in addition offer the important advantage of tomographic (i.e. non-destructive) assessment. These techniques can also provide insight into features such as the lacuno-canalicular network and mineralization levels at the nanoscale [225,271]. At higher FOVs, SR-CT offers a tomographic approach, with the limitation that SR-CT cannot resolve single fibrils and is limited to visualization of fibril bundles only. Moreover, SR-CT allows structural features to be studied, such as the intracortical canal network or the lacuno-canalicular network [3,217], microcracks [272], pathological cysts [273] and trabecular bone micro-architecture [274]. Similar to SR-CT, optical microscopy techniques, such as CLSM or SHG, are limited to visualization of fibril bundles only. On the other hand, optical microscopy techniques provide insight into the mesoscopic bone organization.
Orientation-specific techniques can offer additional information on bone tissue. Electron diffraction gives access to information about crystal lattice, shape, size [23] and orientation [180] at spatial resolutions at the nanometre scale, but is expensive, requires extensive sample preparation and provides a very restricted FOV only. When the TEM set-up is used in imaging mode (and not diffraction mode), it allows for very high-resolution images of bone tissue [174]. Using X-ray scattering techniques, such as scanning SAXS and scattering tomographic techniques, one can achieve sub-micrometre resolutions and FOVs at the millimetre level. In addition to crystal or fibril orientation [145,163], X-ray scattering techniques disclose features such as the collagen 67 nm D-period [15,16], the shape and size of HA platelets [126], the load partitioning between different bone phases [149,150] and also information on tissue composition [155,156,159,275]. Polarized spectroscopic techniques such as polarized FTIR (pFTIR) and PRS come along with a comparable low resolution (at the micrometre scale), but they can provide a wealth of information on bone chemical composition and tissue quality, such as mineralization/mineral-to-matrix ratio, crystallinity, collagen cross-linking/maturity and carbonate-to-phosphate ratio [92,100]. Optical microscopy techniques such as pSHG and PLM are also limited in their spatial resolution, which is around 200 nm, but their wider FOV allows studies of macroscopic samples.
3.2. Quantitative assessment of three-dimensional orientation and arrangement of mineralized collagen fibrils
The capability to quantitatively assess the 3D organization of the ultrastructure is key to the understanding of structure–function relationships in bone tissue. As can be seen in tables 1 and 2, four different techniques can provide quantitative measures for the mineralized collagen fibril orientation and arrangement within a tissue volume. First, for the orientation-specific techniques, 3D sSAXS gathers data that can be used to derive the 3D orientation and arrangement of bone tissue ultrastructure for tissue sections with resolutions at the micrometre scale. By stacking these results from consecutive thin sections, one obtains volumetric information for samples at the millimetre scale, as exemplified for complete trabeculae [67,145], that can also provide insight into important ultrastructure–microstructure relationships [67]. X-ray scattering tomography techniques, including 6D SAXS tomography [162] and SAS tensor tomography [163], provide bone ultrastructure organization information in a tomographic, non-destructive way, at the cost of higher X-ray doses and longer data post-processing times. Nevertheless, X-ray scattering tomography techniques seem to open the way to tomographic investigations that provide ultrastructure orientation information in three dimensions. Regarding imaging techniques, volume electron microscopy techniques, such as FIB SEM [208] or SBF SEM [206], provide information by serially removing thin sections and imaging the underlying surface, to reconstruct volumes of tens of micrometres in size. Finally, through phase-contrast techniques, such as ptychographic CT [225] and phase-contrast nanotomography [226], bone tissue samples at overall dimensions of tens of micrometres can be imaged in a tomographic and thus non-destructive way, with spatial resolutions at the nanometre scale. Whereas both volume electron microscopy and phase-contrast techniques use post-processing algorithms, such as FT, to provide the 3D orientation and arrangement of mineralized collagen fibrils at the nanoscale [226,247], phase-contrast approaches have the important advantage of being non-destructive, hence allowing further investigations on the tissue post hoc.
3.3. Combination with in situ mechanical testing
The capability to assess the organization of the ultrastructure in combination with in situ mechanical testing is another important aspect to understand structure–function relationships in bone tissue. Not all presented techniques can be combined with mechanical testing, e.g. techniques that need sample sectioning, such as PLM, FTIR, SHG in transmission mode, TEM (and electron transmission diffraction) or volume electron microscopic techniques (e.g. FIB SEM or SBF SEM). Electron-based techniques relying on an SEM set-up are not ideal for in situ experiments, mainly because of the imaging process taking place under high vacuum and due to the extended sample preparation that alters the mechanical properties of the tissue. Although both requirements have been alleviated with the advent of environmental SEM, which can image samples ‘wet’ and under moderate pressures and has thus enabled in situ experiments of biological samples, reported in situ SEM experiments have been very limited until now for bone tissue, because they currently cannot provide sufficient spatial resolution that would allow assessment of the bone ultrastructure organization. X-ray-based techniques such as SAXS and WAXS have been used in recent years for in situ experiments [149,150,152,276–279]. However, these studies have been carried out for a limited number of discrete points in space within the sample only, and they could not provide quantitative results in terms of the 3D orientation of the ultrastructure. The orientation reconstruction technique of 3D sSAXS [145] could be applied to whole sample measurements in order to provide such 3D information. Also, the X-ray scattering tomography techniques of SAS tensor tomography [163] and 6D SAXS tomography [162] could be used to provide the ultrastructure organization in situ. However, the biggest limitations of X-ray-based techniques for use in conjunction with in situ experiments are the radiation dose and the time needed to acquire the experimental image data, both of which have an important effect on the mechanical properties of the bone sample [280]. On the other hand, light-based techniques do not induce specimen damage—even though long laser residence times can detrimentally affect the sample [281]. PRS would involve acquisition times of tens of seconds for every spot, enough for bone to exhibit its viscoelastic behaviour [282]. Reflection mode SHG (either polarized or not) has been used in combination with in situ experiments of bone [283] and other tissues [200,201]. However, as discussed earlier, SHG in reflection mode can provide qualitative orientation information only, which originates from the sample surface or from sites very close to the surface.
3.4. In vivo assessment
Electron microscopic techniques cannot be applied in vivo due to the sample preparation procedures and imaging conditions needed, which are incompatible with living mammalian cells [284]. Also, in vivo measurements require techniques that can reach bone tissue a few millimetres below the skin surface. X-rays have the inherent ability to penetrate deep into tissues; however, X-ray imaging techniques used to investigate bone ultrastructure organization operate at high resolutions, requiring very small samples and involving significant X-ray doses, which is detrimental for in vivo applications. Local tomography [285,286], where the reconstructed region of interest is smaller than the sample, could be used to significantly reduce the dose, yet at the expense of reconstruction errors [287] such as cupping (radial increase of the grey values towards the edge of the reconstruction circle) or other non-uniform errors over the FOV. Application of SAXS and WAXS in vivo is also very difficult due to the high X-ray dose required for signal detection, because the primary X-ray beam is blocked and the signal is only generated from the scattered X-ray photons, which are several orders of magnitude less than the transmitted ones. However, X-ray phase-contrast methods based on grating-based dark-field imaging [164,288] could be more easily adopted to be used in vivo in animals [221,289] and eventually in humans [290,291], whereas they can also be combined with standard X-ray absorption methods [223,292]. Their use in providing information on ultrastructure organization [161,169], by exploiting ultrastructure orientation-dependent signal modulations [168,293], is expected to rise in the future, as these methods have not been adequately explored to date [169]. In addition, the two recent non-destructive X-ray scattering tomographic methods [162,163] are potential candidates for being applied in vivo, although that would require significant technological advances, mainly in detector technology, in order to reduce the dose deposited in the sample.
Visible light is less harmful to biological tissues and could be used for in vivo investigations of ultrastructure organization. The assessment would however be restricted to superficial areas, due to the (very) low penetration depth of light in hard tissues such as bone. For instance, it has been shown that Raman spectroscopy can be performed on bone transcutaneously [88,89,94,95], whereas application of SHG in vivo in tissues underneath the skin is possible through the use of endomicroscopes [294,295], which can in addition preserve the laser polarization and, thus, also enable pSHG imaging [296,297]. Therefore, the use of PRS and (p)SHG for the in vivo assessment of the ultrastructural organization of bone can be envisaged in the future.
4. Conclusion
Bone's composite nature and hierarchical structure impart its remarkable mechanical properties. At the ultrastructural scale, the mineralized collagen fibrils, with a diameter of approximately 100 nm, are bone's building units. Their organization has been shown to be of importance in determining the mechanical properties at different levels. For this reason, multiple techniques that assess the orientation and arrangement of the mineralized collagen fibrils have been developed.
This article reviewed these different imaging techniques suitable for the assessment of bone ultrastructure organization, and evaluated their ability to determine the orientation and arrangement of the mineralized collagen fibrils at different scales, using different probes and exploiting various different physical phenomena. Their advantages, limitations and most important applications in the study of bone ultrastructure arrangement were presented. Finally, we evaluated the techniques’ capabilities to assess the ultrastructure organization quantitatively and in three dimensions, and in terms of combination with in situ experiments and their suitability for in vivo studies.
It seems that we are currently at a point where both the interest in bone ultrastructure organization is high and the technology potential to assess it is sufficient. As technology is advancing on many fronts (e.g. probe strength and size, lens quality, detector sensitivity) these techniques are going to offer an improved ability to assess bone's ultrastructure organization, and it is very probable that new techniques based on similar physical principles will emerge.
Authors' contributions
M.G. wrote the manuscript, R.M. and P.S. critically revised it. All authors gave final approval for publication.
Competing interests
The authors have no competing interest.
Funding
M.G. was supported by ETH research grant no. ETH-39 11-1.
References
- 1.Fratzl P, Gupta HS, Paschalis EP, Roschger P. 2004. Structure and mechanical quality of the collagen-mineral nano-composite in bone. J. Mater. Chem. 14, 2115–2123. ( 10.1039/B402005G) [DOI] [Google Scholar]
- 2.Boyde A. 2012. Scanning electron microscopy of bone. In Bone research protocols (eds Helfrich MH, Ralston SH), pp. 365–400. Methods in Molecular Biology 816 Totowa, NJ: Humana Press. [Google Scholar]
- 3.Peyrin F, Dong P, Pacureanu A, Zuluaga M, Olivier C, Langer M, Cloetens P.. 2012. Synchrotron radiation CT from the micro to nanoscale for the investigation of bone tissue. Proc. SPIE 8506, 85060L ( 10.1117/12.939473) [DOI] [Google Scholar]
- 4.Donnelly E, Williams RM, Downs SA, Dickinson ME, Baker SP, van der Meulen MCH. 2006. Quasistatic and dynamic nanomechanical properties of cancellous bone tissue relate to collagen content and organization. J. Mater. Res. 21, 2106–2117. ( 10.1557/jmr.2006.0259) [DOI] [Google Scholar]
- 5.Bromage TG, Goldman HM, McFarlin SC, Warshaw J, Boyde A, Riggs CM. 2003. Circularly polarized light standards for investigations of collagen fiber orientation in bone. Anat. Rec. 274B, 157–168. ( 10.1002/ar.b.10031) [DOI] [PubMed] [Google Scholar]
- 6.McNally EA, Schwarcz HP, Botton GA, Arsenault AL. 2012. A model for the ultrastructure of bone based on electron microscopy of ion-milled sections. PLoS ONE 7, e29258 ( 10.1371/journal.pone.0029258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshpande AS, Beniash E. 2008. Bioinspired synthesis of mineralized collagen fibrils. Crystal Growth Des. 8, 3084–3090. ( 10.1021/cg800252f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiner S, Wagner HD. 1998. The material bone: structure mechanical function relations. Annu. Rev. Mater. Sci. 28, 271–298. ( 10.1146/annurev.matsci.28.1.271) [DOI] [Google Scholar]
- 9.Reznikov N, Shahar R, Weiner S. 2014. Three-dimensional structure of human lamellar bone: the presence of two different materials and new insights into the hierarchical organization. Bone 59, 93–104. ( 10.1016/j.bone.2013.10.023) [DOI] [PubMed] [Google Scholar]
- 10.Wagermaier W, Collagen FP. 2012. Polymer science: a comprehensive reference, vol. 9 (eds Matyjaszewski K, Möller M). Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 11.Beniash E. 2011. Biominerals--hierarchical nanocomposites: the example of bone. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 3, 47–69. ( 10.1002/wnan.105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bocciarelli DS. 1970. Morphology of crystallites in bone. Calcif. Tissue Res. 5, 261 ( 10.1007/BF02017554) [DOI] [PubMed] [Google Scholar]
- 13.Eppell SJ, Tong W, Katz JL, Kuhn L, Glimcher MJ. 2001. Shape and size of isolated bone mineralites measured using atomic force microscopy. J. Orthop. Res. 19, 1027–1034. ( 10.1016/S0736-0266(01)00034-1) [DOI] [PubMed] [Google Scholar]
- 14.Weiner S, Traub W. 1989. Crystal size and organization in bone. Connect. Tissue Res. 21, 259–265. ( 10.3109/03008208909050015) [DOI] [PubMed] [Google Scholar]
- 15.Fratzl P, Schreiber S, Klaushofer K. 1996. Bone mineralization as studied by small-angle X-ray scattering. Connect. Tissue Res. 35, 9–16. ( 10.3109/03008209609005268) [DOI] [PubMed] [Google Scholar]
- 16.Ziv V, Weiner S. 1994. Bone crystal sizes: a comparison of transmission electron microscopic and X-ray diffraction line width broadening techniques. Connect. Tissue Res. 30, 165–175. ( 10.3109/03008209409061969) [DOI] [PubMed] [Google Scholar]
- 17.Posner AS, Perloff A, Diorio AF. 1958. Refinement of the hydroxyapatite structure. Acta Crystallogr. 11, 308–309. ( 10.1107/S0365110X58000815) [DOI] [Google Scholar]
- 18.Katz EP, Li S. 1973. Structure and function of bone collagen fibrils. J. Mol. Biol. 80, 1–15. ( 10.1016/0022-2836(73)90230-1) [DOI] [PubMed] [Google Scholar]
- 19.Weiner S, Traub W. 1986. Organization of hydroxyapatite crystals within collagen fibrils. FEBS Lett. 206, 262–266. ( 10.1016/0014-5793(86)80993-0) [DOI] [PubMed] [Google Scholar]
- 20.Lees S, Prostak K. 1988. The locus of mineral crystallites in bone. Connect. Tissue Res. 18, 41–54. ( 10.3109/03008208809019071) [DOI] [PubMed] [Google Scholar]
- 21.Stock SR. 2015. The mineral–collagen interface in bone. Calcif. Tissue Int. 97, 262–280. ( 10.1007/s00223-015-9984-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair AK, Gautieri A, Chang SW, Buehler MJ. 2013. Molecular mechanics of mineralized collagen fibrils in bone. Nat. Commun. 4, 9 ( 10.1038/ncomms2720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moradianoldak J, Weiner S, Addadi L, Landis WJ, Traub W. 1991. Electron imaging and diffraction study of individual crystals of bone, mineralized tendon and synthetic carbonate apatite. Connect. Tissue Res. 25, 219–228. ( 10.3109/03008209109029158) [DOI] [PubMed] [Google Scholar]
- 24.Traub W, Arad T, Weiner S. 1989. Three-dimensional ordered distribution of crystals in turkey tendon collagen fibers. Proc. Natl Acad. Sci. USA 86, 9822–9826. ( 10.1073/pnas.86.24.9822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garnero P. 2015. The role of collagen organization on the properties of bone. Calcif. Tissue Int. 97, 229–240. ( 10.1007/s00223-015-9996-2) [DOI] [PubMed] [Google Scholar]
- 26.Wagermaier W, Klaushofer K, Fratzl P. 2015. Fragility of bone material controlled by internal interfaces. Calcif. Tissue Int. 97, 201–212. ( 10.1007/s00223-015-9978-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fantner GE, et al. 2006. Hierarchical interconnections in the nano-composite material bone: fibrillar cross-links resist fracture on several length scales. Compos. Sci. Technol. 66, 1205–1211. ( 10.1016/j.compscitech.2005.10.005) [DOI] [Google Scholar]
- 28.Cen L, Liu W, Cui L, Zhang W, Cao Y. 2008. Collagen tissue engineering: development of novel biomaterials and applications. Pediatr. Res. 63, 492–496. ( 10.1203/PDR.0b013e31816c5bc3) [DOI] [PubMed] [Google Scholar]
- 29.Varga P, et al. 2015. Synchrotron X-ray phase nano-tomography-based analysis of the lacunar–canalicular network morphology and its relation to the strains experienced by osteocytes in situ as predicted by case-specific finite element analysis. Biomech. Model Mechanobiol. 14, 267–282. ( 10.1007/s10237-014-0601-9) [DOI] [PubMed] [Google Scholar]
- 30.Gebhardt W. 1905. Functionally important alignments of fine and coarse elements of the bones in vertebrates. II. Special part 1. The construction of Haver's lamella systems and its functional importance. Arch. Entwicklungsmech. Org. 20, 187–322. ( 10.1007/BF02162810) [DOI] [Google Scholar]
- 31.Currey JD. 1969. The relationship between the stiffness and the mineral content of bone. J. Biomech. 2, 477–480. ( 10.1016/0021-9290(69)90023-2) [DOI] [PubMed] [Google Scholar]
- 32.Sasaki N, Ikawa T, Fukuda A. 1991. Orientation of mineral in bovine bone and the anisotropic mechanical properties of plexiform bone. J. Biomech. 24, 57–61. ( 10.1016/0021-9290(91)90326-I) [DOI] [PubMed] [Google Scholar]
- 33.Baumann AP, Deuerling JM, Rudy DJ, Niebur GL, Roeder RK. 2012. The relative influence of apatite crystal orientations and intracortical porosity on the elastic anisotropy of human cortical bone. J. Biomech. 45, 2743–2749. ( 10.1016/j.jbiomech.2012.09.011) [DOI] [PubMed] [Google Scholar]
- 34.Ascenzi A, Bonucci E. 1967. The tensile properties of single osteons. Anat. Rec. 158, 375–386. ( 10.1002/ar.1091580403) [DOI] [PubMed] [Google Scholar]
- 35.Ascenzi A, Bonucci E. 1968. The compressive properties of single osteons. Anat. Rec. 161, 377–392. ( 10.1002/ar.1091610309) [DOI] [PubMed] [Google Scholar]
- 36.Evans FG, Vincentelli R. 1974. Relations of the compressive properties of human cortical bone to histological structure and calcification. J. Biomech. 2, 63 ( 10.1016/0021-9290(69)90042-6) [DOI] [PubMed] [Google Scholar]
- 37.Simkin A, Robin G. 1974. Fracture formation in differing collagen fiber pattern of compact bone. J. Biomech. 7, 183 ( 10.1016/0021-9290(74)90059-1) [DOI] [PubMed] [Google Scholar]
- 38.Boyde A, Riggs CM. 1990. The quantitative study of the orientation of collagen in compact bone slices. Bone 11, 35–39. ( 10.1016/8756-3282(90)90069-B) [DOI] [PubMed] [Google Scholar]
- 39.Nakano T, Kaibara K, Tabata Y, Nagata N, Enomoto S, Marukawa E, Umakoshi Y. 2002. Unique alignment and texture of biological apatite crystallites in typical calcified tissues analyzed by microbeam X-ray diffractometer system. Bone 31, 479–487. ( 10.1016/S8756-3282(02)00850-5) [DOI] [PubMed] [Google Scholar]
- 40.Skedros JG, Dayton MR, Sybrowsky CL, Bloebaum RD, Bachus KN. 2006. The influence of collagen fiber orientation and other histocompositional characteristics on the mechanical properties of equine cortical bone. J. Exp. Biol. 209, 3025–3042. ( 10.1242/jeb.02304) [DOI] [PubMed] [Google Scholar]
- 41.Takano Y, Turner CH, Owan I, Martin RB, Lau ST, Forwood MR, Burr DB. 1999. Elastic anisotropy and collagen orientation of osteonal bone are dependent on the mechanical strain distribution. J. Orthop. Res. 17, 59–66. ( 10.1002/jor.1100170110) [DOI] [PubMed] [Google Scholar]
- 42.Martin RB, Ishida J. 1989. The relative effects of collagen fiber orientation, porosity, density, and mineralization on bone strength. J. Biomech. 22, 419–426. ( 10.1016/0021-9290(89)90202-9) [DOI] [PubMed] [Google Scholar]
- 43.Martin RB, Boardman DL. 1993. The effects of collagen fiber orientation, porosity, density, and mineralization on bovine cortical bone bending properties. J. Biomech. 26, 1047–1054. ( 10.1016/S0021-9290(05)80004-1) [DOI] [PubMed] [Google Scholar]
- 44.Riggs CM, Vaughan LC, Evans GP, Lanyon LE, Boyde A. 1993. Mechanical implications of collagen fibre orientation in cortical bone of the equine radius. Anat. Embryol. (Berl). 187, 239–248. ( 10.1007/bf00195761) [DOI] [PubMed] [Google Scholar]
- 45.Granke M, Gourrier A, Rupin F, Raum K, Peyrin F, Burghammer M, Saïed A, Laugier P. 2013. Microfibril orientation dominates the microelastic properties of human bone tissue at the lamellar length scale. PLoS ONE 8 ( 10.1371/journal.pone.0058043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.An YH, Martin KL. 2003. Handbook of histology methods for bone and cartilage, 6th edn Totowa, NJ: Humana Press Inc. [Google Scholar]
- 47.Szeverenyi NM, Bydder GM. 2011. Dipolar anisotropy fiber imaging in a goat knee meniscus. Magn. Reson. Med. 65, 463–470. ( 10.1002/mrm.22645) [DOI] [PubMed] [Google Scholar]
- 48.Griesshaber E, Goetz AJ, Howard L, Ball A, Ruff S, Schmahl WW. 2012. Crystal architecture of the tooth and jaw bone (pyramid) of the sea urchin Paracentrotus lividus. Bioinspir. Biomim. Nanobiomat. 1, 133–139. ( 10.1680/bbn.11.00018) [DOI] [Google Scholar]
- 49.Osaki S, Tohno S, Tohno Y, Ohuchi K, Takakura Y. 2002. Determination of the orientation of collagen fibers in human bone. Anat. Rec. 266, 103–107. ( 10.1002/ar.10043) [DOI] [PubMed] [Google Scholar]
- 50.Sacks MS, Chuong CJ. 1992. Characterization of collagen fiber architecture in the canine diaphragmatic central tendon. J. Biomech. Eng. Trans. ASME 114, 183–190. ( 10.1115/1.2891370) [DOI] [PubMed] [Google Scholar]
- 51.Kostyuk O, Brown RA. 2004. Novel spectroscopic technique for in situ monitoring of collagen fibril alignment in gels. Biophys. J. 87, 648–655. ( 10.1529/biophysj.103.038976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raum K, Grimal Q, Laugier P, Gerisch A. 2011. Multiscale structure-functional modeling of lamellar bone. Proc. Meet. Acoust. 9, 020005. [Google Scholar]
- 53.Wu J, Rajwa B, Filmer DL, Hoffmann CM, Yuan B, Chiang C, Sturgis J, Robinson JP. 2003. Automated quantification and reconstruction of collagen matrix from 3D confocal datasets. J. Microsc. Oxf. 210, 158–165. ( 10.1046/j.1365-2818.2003.01191.x) [DOI] [PubMed] [Google Scholar]
- 54.Daniels F, ter Haar Romeny B, Rubbens M, van Assen H. 2007. Quantification of collagen orientation in 3D engineered tissue. In Proc. 3rd Kuala Lumpur Int. Conf. on Biomedical Engineering 2006 (Biomed 2006), Kuala Lumpur, Malaysia, 11–14 December 2006 (eds F Ibrahim, N Osman, J Usman, N Kadri), pp. 282–286. Berlin, Germany: Springer.
- 55.Osman OS, Selway JL, Harikumar PE, Stocker CJ, Wargent ET, Cawthorne MA, Jassim S, Langlands K. 2013. A novel method to assess collagen architecture in skin. BMC Bioinformatics 14, 260 ( 10.1186/1471-2105-14-260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russ JC. 2011. The image processing handbook, 6th edn, pp. 345–351. Boca Raton, FL: Taylor & Francis Group, LLC. [Google Scholar]
- 57.Chaudhuri S, Nguyen H, Rangayyan RM, Walsh S, Frank CB. 1987. A Fourier domain directional filtering method for analysis of collagen alignment in ligaments. IEEE Trans. Biomed. Eng. 34, 509–518. ( 10.1109/TBME.1987.325980) [DOI] [PubMed] [Google Scholar]
- 58.van Zuijlen PP, de Vries HJ, Lamme EN, Coppens JE, van Marle J, Kreis RW, Middelkoop E. 2002. Morphometry of dermal collagen orientation by Fourier analysis is superior to multi-observer assessment. J. Pathol. 198, 284–291. ( 10.1002/path.1219) [DOI] [PubMed] [Google Scholar]
- 59.Sivaguru M, Durgam S, Ambekar R, Luedtke D, Fried G, Stewart A, Toussaint KC. 2010. Quantitative analysis of collagen fiber organization in injured tendons using Fourier transform-second harmonic generation imaging. Opt. Express 18, 24 983–24 993. ( 10.1364/OE.18.024983) [DOI] [PubMed] [Google Scholar]
- 60.Lau TY, Ambekar R, Toussaint KC. 2012. Quantification of collagen fiber organization using three-dimensional Fourier transform-second-harmonic generation imaging. Opt. Express. 20, 21 821–21 832. ( 10.1364/OE.20.021821) [DOI] [PubMed] [Google Scholar]
- 61.Suhalim JL, Boik JC, Tromberg BJ, Potma EO. 2012. The need for speed. J. Biophoton. 5, 387–395. ( 10.1002/jbio.201200002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolman M, Kasten FH. 1986. Polarized-light microscopy in the study of the molecular structure of collagen and reticulin. Histochemistry 85, 41–49. ( 10.1007/BF00508652) [DOI] [PubMed] [Google Scholar]
- 63.Ascenzi MG, Ascenzi A, Benvenuti A, Burghammer M, Panzavolta S, Bigi A. 2003. Structural differences between ‘dark’ and ‘bright’ isolated human osteonic lamellae. J. Struct. Biol. 141, 22–33. ( 10.1016/S1047-8477(02)00578-6) [DOI] [PubMed] [Google Scholar]
- 64.Rieppo J, Hallikainen J, Jurvelin JS, Kiviranta I, Helminen HJ, Hyttinen MM. 2008. Practical considerations in the use of polarized light microscopy in the analysis of the collagen network in articular cartilage. Microsc. Res. Tech. 71, 279–287. ( 10.1002/jemt.20551) [DOI] [PubMed] [Google Scholar]
- 65.Bennett JM. 1995. Polarization. In Handbook of optics, vol. 1 (ed. Bass M.), 2nd edn New York, NY: McGraw-Hill. [Google Scholar]
- 66.Glazer AM, Lewis JG, Kaminsky W. 1996. An automatic optical imaging system for birefringent media. Proc. R. Soc. Lond. A 452, 2751–2765. ( 10.1098/rspa.1996.0145) [DOI] [Google Scholar]
- 67.Georgiadis M, et al. In press. Ultrastructure organization of human trabeculae assessed by 3D sSAXS. PLoS ONE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Turnhout MC, Kranenbarg S, van Leeuwen JL. 2009. Modeling optical behavior of birefringent biological tissues for evaluation of quantitative polarized light microscopy. J. Biomed. Opt. 14, 054018 ( 10.1117/1.3241986) [DOI] [PubMed] [Google Scholar]
- 69.Boyde A, Bianco P, Barbos MP, Ascenzi A. 1984. Collagen orientation in compact bone, I. A new method for the determination of the proportion of collagen parallel to the plane of compact bone sections. Metab. Bone Dis. Relat. Res. 5, 299–307. ( 10.1016/0221-8747(84)90017-1) [DOI] [PubMed] [Google Scholar]
- 70.Kalmey JK, Lovejoy CO. 2002. Collagen fiber orientation in the femoral necks of apes and humans: do their histological structures reflect differences in locomotor loading? Bone 31, 327–332. ( 10.1016/S8756-3282(02)00828-1) [DOI] [PubMed] [Google Scholar]
- 71.Goldman HM, Bromage TG, Thomas CDL, Clement JG. 2003. Preferred collagen fiber orientation in the human mid-shaft femur. Anat. Rec. A 272A, 434–445. ( 10.1002/ar.a.10055) [DOI] [PubMed] [Google Scholar]
- 72.Beraudi A, Stea S, Bordini B, Baleani M, Viceconti M. 2010. Osteon classification in human fibular shaft by circularly polarized light. Cells Tissues Organs 191, 260–268. ( 10.1159/000240045) [DOI] [PubMed] [Google Scholar]
- 73.Riggs CM, Lanyon LE, Boyde A. 1993. Functional associations between collagen fibre orientation and locomotor strain direction in cortical bone of the equine radius. Anat. Embryol. (Berl.) 187, 231–238. ( 10.1007/bf00195760) [DOI] [PubMed] [Google Scholar]
- 74.Goldman HM, Thomas CDL, Clement JG, Bromage TG. 2005. Relationships among microstructural properties of bone at the human midshaft femur. J. Anat. 206, 127–139. ( 10.1111/j.1469-7580.2005.00385.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turunen MJ, Saarakkala S, Helminen HJ, Jurvelin JS, Isaksson H. 2012. Age-related changes in organization and content of the collagen matrix in rabbit cortical bone. J. Orthop. Res. 30, 435–442. ( 10.1002/jor.21538) [DOI] [PubMed] [Google Scholar]
- 76.Rinnerthaler S, Roschger P, Jakob HF, Nader A, Klaushofer K, Fratzl P. 1999. Scanning small angle X-ray scattering analysis of human bone sections. Calcif. Tissue Int. 64, 422–429. ( 10.1007/PL00005824) [DOI] [PubMed] [Google Scholar]
- 77.Bi X, Li G, Doty SB, Camacho NP. 2005. A novel method for determination of collagen orientation in cartilage by Fourier transform infrared imaging spectroscopy (FT-IRIS). Osteoarthritis Cartilage 13, 1050–1058. ( 10.1016/j.joca.2005.07.008) [DOI] [PubMed] [Google Scholar]
- 78.Rubenstein JD, Kim JK, Moravaprotzner I, Stanchev PL, Henkelman RM. 1993. Effects of collagen orientation on MR-imaging characteristics of bovine articular-cartilage. Radiology 188, 219–226. ( 10.1148/radiology.188.1.8511302) [DOI] [PubMed] [Google Scholar]
- 79.Dohmen M, Menzel M, Wiese H, Reckfort J, Hanke F, Pietrzyk U, Zilles K, Amunts K, Axer M. 2015. Understanding fiber mixture by simulation in 3D polarized light imaging. NeuroImage 111, 464–475. ( 10.1016/j.neuroimage.2015.02.020) [DOI] [PubMed] [Google Scholar]
- 80.Makowski AJ, Patil CA, Mahadevan-Jansen A, Nyman JS. 2013. Polarization control of Raman spectroscopy optimizes the assessment of bone tissue. J. Biomed. Opt. 18, 055005 ( 10.1117/1.JBO.18.5.055005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kozielski M, Buchwald T, Szybowicz M, Blaszczak Z, Piotrowski A, Ciesielczyk B. 2011. Determination of composition and structure of spongy bone tissue in human head of femur by Raman spectral mapping. J. Mater. Sci. Mater. Med. 22, 1653–1661. ( 10.1007/s10856-011-4353-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kazanci M, Wagner HD, Manjubala NI, Gupta HS, Paschalis E, Roschger P, Fratzl P. 2007. Raman imaging of two orthogonal planes within cortical bone. Bone 41, 456–461. ( 10.1016/j.bone.2007.04.200) [DOI] [PubMed] [Google Scholar]
- 83.Galvis L, Dunlop JWC, Duda G, Fratzl P, Masic A. 2013. Polarized Raman anisotropic response of collagen in tendon, towards 3D orientation mapping of collagen in tissues. PLoS ONE 8, e63518 ( 10.1371/journal.pone.0063518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raghavan M, Sahar ND, Wilson RH, Mycek M-A, Pleshko N, Kohn DH, Morris MD. 2010. Quantitative polarized Raman spectroscopy in highly turbid bone tissue. J. Biomed. Opt. 15, 037 001–037 007. ( 10.1117/1.3426310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Falgayrac G, Facq S, Leroy G, Cortet B, Penel G. 2010. New method for Raman investigation of the orientation of collagen fibrils and crystallites in the Haversian system of bone. Appl. Spectrosc. 64, 775–780. ( 10.1366/000370210791666255) [DOI] [PubMed] [Google Scholar]
- 86.Rousseau ME, Lefevre T, Beaulieu L, Asakura T, Pezolet M. 2004. Study of protein conformation and orientation in silkworm and spider silk fibers using Raman microspectroscopy. Biomacromolecules 5, 2247–2257. ( 10.1021/bm049717v) [DOI] [PubMed] [Google Scholar]
- 87.Wang H, Lee AMD, Lui H, McLean DI, Zeng H. 2013. A method for accurate in vivo micro-Raman spectroscopic measurements under guidance of advanced microscopy imaging. Sci. Rep. 3, 1890 ( 10.1038/srep01890) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Buckley K, Kerns JG, Gikas PD, Birch HL, Vinton J, Keen R, Parker AW, Matousek P, Goodship AE. 2014. Measurement of abnormal bone composition in vivo using noninvasive Raman spectroscopy. IBMS BoneKEy 11, 602, 602 ( 10.1038/bonekey.2014.97) [DOI] [Google Scholar]
- 89.Matousek P, Draper ERC, Goodship AE, Clark IP, Ronayne KL, Parker AW. 2006. Noninvasive Raman spectroscopy of human tissue in vivo. Appl. Spectrosc. 60, 758–763. ( 10.1366/000370206777886955) [DOI] [PubMed] [Google Scholar]
- 90.Kazanci M, Roschger P, Paschalis EP, Klaushofer K, Fratzl P. 2006. Bone osteonal tissues by Raman spectral mapping: orientation-composition. J. Struct. Biol. 156, 489–496. ( 10.1016/j.jsb.2006.06.011) [DOI] [PubMed] [Google Scholar]
- 91.Schrof S, Varga P, Galvis L, Raum K, Masic A. 2014. 3D Raman mapping of the collagen fibril orientation in human osteonal lamellae. J. Struct. Biol. 187, 266–275. ( 10.1016/j.jsb.2014.07.001) [DOI] [PubMed] [Google Scholar]
- 92.Morris MD. 2010. Raman spectroscopy of bone and cartilage. In Emerging Raman applications and techniques in biomedical and pharmaceutical fields. (eds Matousek P, Morris MD), pp. 347–364. New York, NY: Springer. [Google Scholar]
- 93.Choi S, Jung GB, Kim KS, Lee GJ, Park HK. 2014. Medical applications of atomic force microscopy and Raman spectroscopy. J. Nanosci. Nanotechnol. 14, 71–97. ( 10.1166/jnn.2014.9112) [DOI] [PubMed] [Google Scholar]
- 94.Schulmerich MV, Cole JH, Kreider JM, Esmonde-White F, Dooley KA, Goldstein SA, Morris MD. 2009. Transcutaneous Raman spectroscopy of murine bone in vivo. Appl. Spectrosc. 63, 286–295. ( 10.1366/000370209787599013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Esmonde-White FWL, Morris MD. 2013. Validating in vivo Raman spectroscopy of bone in human subjects. Proc. SPIE 8565, 85656K ( 10.1117/12.2005679) [DOI] [Google Scholar]
- 96.Nakamoto K. 2009. Infrared and Raman spectra of inorganic and coordination compounds, part A, theory and applications in inorganic chemistry, 6th edn New York, NY: Wiley-Interscience. [Google Scholar]
- 97.Faix O. 1992. Fourier transform infrared spectroscopy. In Methods in lignin chemistry (eds Lin S, Dence C), pp. 83–109. Springer Series in Wood Science Berlin, Germany: Springer. [Google Scholar]
- 98.Coats AM, Hukins DWL, Imrie CT, Aspden RM. 2003. Polarization artefacts of an FTIR microscope and the consequences for intensity measurements on anisotropic materials. J. Microsc. Oxf. 211, 63–66. ( 10.1046/j.1365-2818.2003.01198.x) [DOI] [PubMed] [Google Scholar]
- 99.Barth A, Zscherp C. 2002. What vibrations tell us about proteins. Q. Rev. Biophys. 35, 369–430. ( 10.1017/S0033583502003815) [DOI] [PubMed] [Google Scholar]
- 100.Boskey A, Mendelsohn R. 2005. Infrared analysis of bone in health and disease. J. Biomed. Opt. 10, 031102 ( 10.1117/1.1922927) [DOI] [PubMed] [Google Scholar]
- 101.Camacho NP, Rinnerthaler S, Paschalis EP, Mendelsohn R, Boskey AL, Fratzl P. 1999. Complementary information on bone ultrastructure from scanning small angle X-ray scattering and Fourier-transform infrared microspectroscopy. Bone 25, 287–293. ( 10.1016/S8756-3282(99)00165-9) [DOI] [PubMed] [Google Scholar]
- 102.Boskey A, Camacho NP. 2007. FT-IR imaging of native and tissue-engineered bone and cartilage. Biomaterials 28, 2465–2478. ( 10.1016/j.biomaterials.2006.11.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spalazzi JP, Boskey AL, Pleshko N, Lu HH. 2013. Quantitative mapping of matrix content and distribution across the ligament-to-bone insertion. PLoS ONE 8, 16 ( 10.1371/journal.pone.0074349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Camacho NP, West P, Torzilli PA, Mendelsohn R. 2001. FTIR microscopic imaging of collagen and proteoglycan in bovine cartilage. Biopolymers 62, 1–8. ( 10.1002/1097-0282(2001)62:1%3C1::AID-BIP10%3E3.0.CO;2-O) [DOI] [PubMed] [Google Scholar]
- 105.Millard AC, Campagnola PJ, Mohler W, Lewis A, Loew LM. 2003. Second harmonic imaging microscopy. Biophoton. B 361, 47–69. ( 10.1016/S0076-6879(03)61005-0) [DOI] [PubMed] [Google Scholar]
- 106.Campagnola PJ, Loew LM. 2003. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat. Biotechnol. 21, 1356–1360. ( 10.1038/nbt894) [DOI] [PubMed] [Google Scholar]
- 107.Zoumi A, Yeh A, Tromberg BJ. 2002. Imaging cells and extracellular matrix in vivo by using second-harmonic generation and two-photon excited fluorescence. Proc. Natl Acad. Sci. USA 99, 11 014–11 019. ( 10.1073/pnas.172368799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williams RM, Zipfel WR, Webb WW. 2005. Interpreting second-harmonic generation images of collagen I fibrils. Biophys. J. 88, 1377–1386. ( 10.1529/biophysj.104.047308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nadiarnykh O, Plotnikov S, Mohler WA, Kalajzic I, Redford-Badwal D, Campagnola PJ. 2007. Second harmonic generation imaging microscopy studies of osteogenesis imperfecta. J. Biomed. Opt. 12, 9 ( 10.1117/1.2799538) [DOI] [PubMed] [Google Scholar]
- 110.Stoller P, Reiser KM, Celliers PM, Rubenchik AM. 2002. Polarization-modulated second harmonic generation in collagen. Biophys. J. 82, 3330–3342. ( 10.1016/S0006-3495(02)75673-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen XY, Nadiarynkh O, Plotnikov S, Campagnola PJ. 2012. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat. Protoc. 7, 654–669. ( 10.1038/nprot.2012.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mansfield JC, Winlove CP, Moger J, Matcher SJ. 2008. Collagen fiber arrangement in normal and diseased cartilage studied by polarization sensitive nonlinear microscopy. J. Biomed. Opt. 13, 044020 ( 10.1117/1.2950318) [DOI] [PubMed] [Google Scholar]
- 113.Nadiarnykh O, Campagnola PJ. 2009. Retention of polarization signatures in SHG microscopy of scattering tissues through optical clearing. Opt. Express. 17, 5794–5806. ( 10.1364/OE.17.005794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yasui T, Tohno Y, Araki T. 2004. Determination of collagen fiber orientation in human tissue by use of polarization measurement of molecular second-harmonic-generation light. Appl. Opt. 43, 2861–2867. ( 10.1364/AO.43.002861) [DOI] [PubMed] [Google Scholar]
- 115.Ambekar R, Lau TY, Walsh M, Bhargava R, Toussaint KC. 2012. Quantifying collagen structure in breast biopsies using second-harmonic generation imaging. Biomed. Opt. Express. 3, 2021–2035. ( 10.1364/BOE.3.002021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee H, Huttunen MJ, Hsu KJ, Partanen M, Zhuo GY, Kauranen M, Chu S-W. 2013. Chiral imaging of collagen by second-harmonic generation circular dichroism. Biomed. Opt. Express. 4, 909–916. ( 10.1364/BOE.4.000909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hovhannisyan VA, Hu PS, Tan HY, Chen SJ, Dong CY. 2012. Spatial orientation mapping of fibers using polarization-sensitive second harmonic generation microscopy. J. Biophoton. 5, 768–776. ( 10.1002/jbio.201100123) [DOI] [PubMed] [Google Scholar]
- 118.Guinier A, Fournet G. 1955. Small-angle scattering of X-rays. New York, NY: Wiley. [Google Scholar]
- 119.Guagliardi A, Giannini C, Cedola A, Mastrogiacomo M, Ladisa M, Cancedda R. 2009. Toward the X-ray microdiffraction imaging of bone and tissue-engineered bone. Tissue Eng. B 15, 423–442. ( 10.1089/ten.teb.2009.0034) [DOI] [PubMed] [Google Scholar]
- 120.Meek KM, Boote C. 2009. The use of X-ray scattering techniques to quantify the orientation and distribution of collagen in the corneal stroma. Prog. Retin. Eye Res. 28, 369–392. ( 10.1016/j.preteyeres.2009.06.005) [DOI] [PubMed] [Google Scholar]
- 121.White SW, Hulmes DJS, Miller A, Timmins PA. 1977. Collagen-mineral axial relationship in calcified turkey leg tendon by X-ray and neutron diffraction. Nature 266, 421–425. ( 10.1038/266421a0) [DOI] [PubMed] [Google Scholar]
- 122.Ascenzi A, Bigi A, Koch MHJ, Ripamonti A, Roveri N. 1985. A low-angle X-ray diffraction analysis of osteonic inorganic phase using synchrotron radiation. Calcif. Tissue Int. 37, 659–664. ( 10.1007/BF02554926) [DOI] [PubMed] [Google Scholar]
- 123.Pabisch S, Wagermaier W, Zander T, Li C, Fratzl P. 2013. Imaging the nanostructure of bone and dentin through small- and wide-angle X-ray scattering. Methods Enzymol. 532, 391–413. ( 10.1016/B978-0-12-416617-2.00018-7) [DOI] [PubMed] [Google Scholar]
- 124.Paris O. 2008. From diffraction to imaging: new avenues in studying hierarchical biological tissues with x-ray microbeams. Biointerphases 3, FB16–FB26. ( 10.1116/1.2955443) [DOI] [PubMed] [Google Scholar]
- 125.Fratzl P. 2008. Collagen: structure and mechanics. New York, NY: Springer. [Google Scholar]
- 126.Fratzl P, Groschner M, Vogl G, Plenk H, Eschberger J, Fratzl-Zelman N, Koller K, Klaushofer K. 1992. Mineral crystals in calcified tissues: a comparative study by SAXS. J. Bone Miner. Res. 7, 329–334. ( 10.1002/jbmr.5650070313) [DOI] [PubMed] [Google Scholar]
- 127.Fratzl P, Schreiber S, Boyde A. 1996. Characterization of bone mineral crystals in horse radius by small-angle X-ray scattering. Calcif. Tissue Int. 58, 341–346. ( 10.1007/BF02509383) [DOI] [PubMed] [Google Scholar]
- 128.Ascenzi A, Benvenuti A, Bigi A, Foresti E, Koch MHJ, Mango F, Ripamonti A, Roveri N. 1998. X-ray diffraction on cyclically loaded osteons. Calcif. Tissue Int. 62, 266–273. ( 10.1007/s002239900428) [DOI] [PubMed] [Google Scholar]
- 129.Wagermaier W, Gupta HS, Gourrier A, Paris O, Roschger P, Burghammer M, Riekel C, Fratzl P. 2007. Scanning texture analysis of lamellar bone using microbeam synchrotron X-ray radiation. J. Appl. Crystallogr. 40, 115–120. ( 10.1107/S0021889806044888) [DOI] [Google Scholar]
- 130.Wagermaier W, Gupta HS, Gourrier A, Burghammer M, Roschger P, Fratzl P. 2006. Spiral twisting of fiber orientation inside bone lamellae. Biointerphases 1, 1–5. ( 10.1116/1.2178386) [DOI] [PubMed] [Google Scholar]
- 131.Zizak I, Paris O, Roschger P, Bernstorff S, Amenitsch H, Klaushofer K, Fratzl P. 2000. Investigation of bone and cartilage by synchrotron scanning-SAXS and -WAXD with micrometer spatial resolution. J. Appl. Crystallogr. 33, 820–823. ( 10.1107/S0021889800001321) [DOI] [Google Scholar]
- 132.Zizak I, Roschger P, Paris O, Misof BM, Berzlanovich A, Bernstorff S, Amenitsch H, Klaushofer K, Fratzl P. 2003. Characteristics of mineral particles in the human bone/cartilage interface. J. Struct. Biol. 141, 208–217. ( 10.1016/S1047-8477(02)00635-4) [DOI] [PubMed] [Google Scholar]
- 133.Kaabar W, Gundogdu O, Laklouk A, Bunk O, Pfeiffer F, Farquharson MJ, Bradley DA. 2010. mu-PIXE and SAXS studies at the bone-cartilage interface. Appl. Radiat. Isot. 68, 730–734. ( 10.1016/j.apradiso.2009.09.038) [DOI] [PubMed] [Google Scholar]
- 134.Fratzl P, Fratzl-Zelman N, Klaushofer K, Vogl G, Koller K. 1991. Nucleation and growth of mineral crystals in bone studied by small-angle X-ray scattering. Calcif. Tissue Int. 48, 407–413. ( 10.1007/BF02556454) [DOI] [PubMed] [Google Scholar]
- 135.Giannini C, Siliqi D, Bunk O, Beraudi A, Ladisa M, Altamura D, Stea S, Baruffaldi F. 2012. Correlative light and scanning X-ray scattering microscopy of healthy and pathologic human bone sections. Sci. Rep. 2, 435 ( 10.1038/srep00435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fratzl P, Paris O, Klaushofer K, Landis WJ. 1996. Bone mineralization in an osteogenesis imperfecta mouse model studied by small-angle X-ray scattering. J. Clin. Invest. 97, 396–402. ( 10.1172/JCI118428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fratzl P, Roschger P, Eschberger J, Abendroth B, Klaushofer K. 1994. Abnormal bone mineralization after fluoride treatment in osteoporosis: a small-angle X-ray-scattering study. J. Bone Miner. Res. 9, 1541–1549. ( 10.1002/jbmr.5650091006) [DOI] [PubMed] [Google Scholar]
- 138.Roschger P, Rinnerthaler S, Yates J, Rodan GA, Fratzl P, Klaushofer K. 2001. Alendronate increases degree and uniformity of mineralization in cancellous bone and decreases the porosity in cortical bone of osteoporotic women. Bone 29, 185–191. ( 10.1016/S8756-3282(01)00485-9) [DOI] [PubMed] [Google Scholar]
- 139.Bunger MH, et al. 2010. Strontium and bone nanostructure in normal and ovariectomized rats investigated by scanning small-angle x-ray scattering. Calcif. Tissue Int. 86, 294–306. ( 10.1007/s00223-010-9341-8) [DOI] [PubMed] [Google Scholar]
- 140.Liu YF, Manjubala I, Schell H, Epari DR, Roschger P, Duda GN, Fratzl P. 2010. Size and habit of mineral particles in bone and mineralized callus during bone healing in sheep. J. Bone Miner. Res. 25, 2029–2038. ( 10.1002/jbmr.84) [DOI] [PubMed] [Google Scholar]
- 141.Yifei L, Manjubala I, Roschger P, Schell H, Duda GN, Fratzl P. 2010. Mineral crystal alignment in mineralized fracture callus determined by 3D small-angle X-ray scattering. J. Phys. Conf. Ser. 247, 012031 ( 10.1088/1742-6596/247/1/012031) [DOI] [Google Scholar]
- 142.Misof BM, et al. 2003. Targeted overexpression of vitamin D receptor in osteoblasts increases calcium concentration without affecting structural properties of bone mineral crystals. Calcif. Tissue Int. 73, 251–257. ( 10.1007/s00223-002-2139-6) [DOI] [PubMed] [Google Scholar]
- 143.Roschger P, Matsuo K, Misof BM, Tesch W, Jochum W, Wagner EF, Fratzl P, Klaushofer K. 2004. Normal mineralization and nanostructure of sclerotic bone in mice overexpressing Fra-1. Bone 34, 776–782. ( 10.1016/j.bone.2004.01.004) [DOI] [PubMed] [Google Scholar]
- 144.Deyhle H, Bunk O, Muller B. 2011. Nanostructure of healthy and caries-affected human teeth. Nanomed. Nanotechnol. Biol. Med. 7, 694–701. ( 10.1016/j.nano.2011.09.005) [DOI] [PubMed] [Google Scholar]
- 145.Georgiadis M, Guizar-Sicairos M, Zwahlen A, Trüssel AJ, Bunk O, Müller R, Schneider P. 2015. 3D scanning SAXS: a novel method for the assessment of bone ultrastructure orientation. Bone 71, 42–52. ( 10.1016/j.bone.2014.10.002) [DOI] [PubMed] [Google Scholar]
- 146.Seidel R, Gourrier A, Kerschnitzki M, Burghammer M, Fratzl P, Gupta HS, Wagermaier W. 2012. Synchrotron 3D SAXS analysis of bone nanostructure. Bioinspir. Biomim. Nanobiomat. 1, 123–132. ( 10.1680/bbn.11.00014) [DOI] [Google Scholar]
- 147.Bunk O, Bech M, Jensen TH, Feidenhans'l R, Binderup T, Menzel A, Pfeiffer F. 2009. Multimodal X-ray scatter imaging. N. J. Phys. 11, 123016 ( 10.1088/1367-2630/11/12/123016) [DOI] [Google Scholar]
- 148.Karunaratne A, Terrill NJ, Gupta HS. 2013. Synchrotron X-ray nanomechanical imaging of mineralized fiber composites. Methods Enzymol. 532, 415–473. ( 10.1016/B978-0-12-416617-2.00019-9) [DOI] [PubMed] [Google Scholar]
- 149.Almer JD, Stock SR. 2005. Internal strains and stresses measured in cortical bone via high-energy X-ray diffraction. J. Struct. Biol. 152, 14–27. ( 10.1016/j.jsb.2005.08.003) [DOI] [PubMed] [Google Scholar]
- 150.Almer JD, Stock SR. 2007. Micromechanical response of mineral and collagen phases in bone. J. Struct. Biol. 157, 365–370. ( 10.1016/j.jsb.2006.09.001) [DOI] [PubMed] [Google Scholar]
- 151.Karunaratne A, Davis GR, Hiller J, Esapa CT, Terrill NJ, Brown SDM, Cox RD, Thakker RV, Gupta HS. 2012. Hypophosphatemic rickets is associated with disruption of mineral orientation at the nanoscale in the flat scapula bones of rachitic mice with development. Bone 51, 553–562. ( 10.1016/j.bone.2012.04.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gallant MA, et al. 2014. Bone cell-independent benefits of raloxifene on the skeleton: a novel mechanism for improving bone material properties. Bone 61, 191–200. ( 10.1016/j.bone.2014.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Birkbak ME, Leemreize H, Frolich S, Stock SR, Birkedal H. 2015. Diffraction scattering computed tomography: a window into the structures of complex nanomaterials. Nanoscale 7, 18 402–18 410. ( 10.1039/C5NR04385A) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Harding G, Kosanetzky J. 1985. Elastic scatter computed tomography. Phys. Med. Biol. 30, 183 ( 10.1088/0031-9155/30/2/008) [DOI] [PubMed] [Google Scholar]
- 155.Kleuker U, Suortti P, Weyrich W, Spanne P. 1998. Feasibility study of x-ray diffraction computed tomography for medical imaging. Phys. Med. Biol. 43, 2911 ( 10.1088/0031-9155/43/10/017) [DOI] [PubMed] [Google Scholar]
- 156.Jensen TH, Bech M, Bunk O, Menzel A, Bouchet A, Le Duc G, Feidenhans'l R, Pfeiffer F. 2011. Molecular X-ray computed tomography of myelin in a rat brain. NeuroImage 57, 124–129. ( 10.1016/j.neuroimage.2011.04.013) [DOI] [PubMed] [Google Scholar]
- 157.Leemreize H, Almer JD, Stock SR, Birkedal H. 2013. Three-dimensional distribution of polymorphs and magnesium in a calcified underwater attachment system by diffraction tomography. J. R. Soc. Interface 10, 20130319 ( 10.1098/rsif.2013.0319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stock SR, de Carlo F, Almer JD. 2008. High energy X-ray scattering tomography applied to bone. J. Struct. Biol. 161, 144–150. ( 10.1016/j.jsb.2007.10.001) [DOI] [PubMed] [Google Scholar]
- 159.Deymier-Black AC, Veis A, Cai Z, Stock SR. 2014. Crystallographic texture and elemental composition mapped in bovine root dentin at the 200 nm level. Scanning 36, 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gürsoy D, Biçer T, Almer JD, Kettimuthu R, Stock SR, De Carlo F. 2015. Maximum a posteriori estimation of crystallographic phases in X-ray diffraction tomography. Phil. Trans. R. Soc. A 373, 20140392 ( 10.1098/rsta.2014.0392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Malecki A, et al. 2014. X-ray tensor tomography. Europhys. Lett. 105, 6 ( 10.1209/0295-5075/105/38002) [DOI] [Google Scholar]
- 162.Schaff F, Bech M, Zaslansky P, Jud C, Liebi M, Guizar-Sicairos M, Pfeiffer F. 2015. Six-dimensional real and reciprocal space small-angle X-ray scattering tomography. Nature 527, 353–356. ( 10.1038/nature16060) [DOI] [PubMed] [Google Scholar]
- 163.Liebi M, Georgiadis M, Menzel A, Schneider P, Kohlbrecher J, Bunk O, Guizar-Sicairos M. 2015. Nanostructure surveys of macroscopic specimens by small-angle scattering tensor tomography. Nature 527, 349–352. ( 10.1038/nature16056) [DOI] [PubMed] [Google Scholar]
- 164.Pfeiffer F, Bech M, Bunk O, Kraft P, Eikenberry EF, Bronnimann Ch, Grünzweig C, David C. 2008. Hard-X-ray dark-field imaging using a grating interferometer. Nat. Mater. 7, 134–137. ( 10.1038/nmat2096) [DOI] [PubMed] [Google Scholar]
- 165.Yashiro W, Terui Y, Kawabata K, Momose A. 2010. On the origin of visibility contrast in x-ray Talbot interferometry. Opt. Express. 18, 16 890–16 901. ( 10.1364/OE.18.016890) [DOI] [PubMed] [Google Scholar]
- 166.Jensen TH, et al. 2010. Directional x-ray dark-field imaging of strongly ordered systems. Phys. Rev. B 82, 214103 ( 10.1103/PhysRevB.82.214103) [DOI] [Google Scholar]
- 167.Jensen TH, Bech M, Bunk O, Donath T, David C, Feidenhans'l R, Pfeiffer F. 2010. Directional X-ray dark-field imaging. Phys. Med. Biol. 55, 3317 ( 10.1088/0031-9155/55/12/004) [DOI] [PubMed] [Google Scholar]
- 168.Bayer F, Zabler S, Brendel C, Pelzer G, Rieger J, Ritter A, Weber T, Michel T, Anton G. 2013. Projection angle dependence in grating-based X-ray dark-field imaging of ordered structures. Opt. Express. 21, 19 922–19 933. ( 10.1364/OE.21.019922) [DOI] [PubMed] [Google Scholar]
- 169.Bayer FL, Hu S, Maier A, Weber T, Anton G, Michel T, Riess CP. 2014. Reconstruction of scalar and vectorial components in X-ray dark-field tomography. Proc. Natl Acad. Sci. USA 111, 12 699–12 704. ( 10.1073/pnas.1321080111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Potdevin G, et al. 2012. X-ray vector radiography for bone micro-architecture diagnostics. Phys. Med. Biol. 57, 3451–3461. ( 10.1088/0031-9155/57/11/3451) [DOI] [PubMed] [Google Scholar]
- 171.Jensen TH, Bech M, Bunk O, Thomsen M, Menzel A, Bouchet A, Le Duc G, Feidenhans'l R, Pfeiffer F. 2011. Brain tumor imaging using small-angle X-ray scattering tomography. Phys. Med. Biol. 56, 1717–1726. ( 10.1088/0031-9155/56/6/012) [DOI] [PubMed] [Google Scholar]
- 172.Fratzl P. 2015. Imaging techniques: extra dimension for bone analysis. Nature 527, 308–309. ( 10.1038/527308a) [DOI] [PubMed] [Google Scholar]
- 173.Qu S-X, Lu X, Leng Y. 2007. TEM study of bone and scaffold materials. In Advanced bioimaging technologies in assessment of the quality of bone and scaffold materials (eds Qin L, Genant H, Griffith J, Leung K), pp. 373–392. Berlin, Germany: Springer. [Google Scholar]
- 174.Liu Y, Kim Y-K, Dai L, Li N, Khan SO, Pashley DH, Tay FR. 2011. Hierarchical and non-hierarchical mineralisation of collagen. Biomaterials 32, 1291–1300. ( 10.1016/j.biomaterials.2010.10.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Weiner S, Arad T, Traub W. 1991. Crystal organization in rat bone lamellae. FEBS Lett. 285, 49–54. ( 10.1016/0014-5793(91)80722-F) [DOI] [PubMed] [Google Scholar]
- 176.Ziv V, Wagner HD, Weiner S. 1996. Microstructure-microhardness relations in parallel-fibered and lamellar bone. Bone 18, 417–428. ( 10.1016/8756-3282(96)00049-X) [DOI] [PubMed] [Google Scholar]
- 177.Olszta MJ, Cheng XG, Jee SS, Kumar R, Kim YY, Kaufman MJ, Douglas EP, Gower LB. 2007. Bone structure and formation: a new perspective. Mater. Sci. Eng. R Rep. 58, 77–116. ( 10.1016/j.mser.2007.05.001) [DOI] [Google Scholar]
- 178.Landis WJ, Glimcher MJ. 1978. Electron diffraction and electron probe microanalysis of the mineral phase of bone tissue prepared by anhydrous techniques. J. Ultrastruct. Res. 63, 188–223. ( 10.1016/S0022-5320(78)80074-4) [DOI] [PubMed] [Google Scholar]
- 179.Vainshtein BK. 1964. The geometrical theory of electron diffraction patterns. In Structure analysis by electron diffraction (ed. Vainshtein BK.), pp. 27–113. Oxford, UK: Pergamon. [Google Scholar]
- 180.Kolb U, Gorelik T, Kubel C, Otten MT, Hubert D. 2007. Towards automated diffraction tomography: part I—data acquisition. Ultramicroscopy 107, 507–513. ( 10.1016/j.ultramic.2006.10.007) [DOI] [PubMed] [Google Scholar]
- 181.Mokso R, Quaroni L, Marone F, Irvine S, Vila-Comamala J, Blanke A, Stampanoni M. 2012. X-ray mosaic nanotomography of large microorganisms. J. Struct. Biol. 177, 233–238. ( 10.1016/j.jsb.2011.12.014) [DOI] [PubMed] [Google Scholar]
- 182.Alexander B, Daulton TL, Genin GM, Lipner J, Pasteris JD, Wopenka B, Thomopoulos S. 2012. The nanometre-scale physiology of bone: steric modelling and scanning transmission electron microscopy of collagen-mineral structure. J. R. Soc. Interface 9, 1774–1786. ( 10.1098/rsif.2011.0880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li YP, Aparicio C. 2013. Discerning the subfibrillar structure of mineralized collagen fibrils: a model for the ultrastructure of bone. PLoS ONE 8, 12 ( 10.1371/journal.pone.0076782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jager I, Fratzl P. 2000. Mineralized collagen fibrils: a mechanical model with a staggered arrangement of mineral particles. Biophys. J. 79, 1737–1746. ( 10.1016/S0006-3495(00)76426-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Paddock SW. 2000. Principles and practices of laser scanning confocal microscopy. Mol. Biotechnol. 16, 127–149. ( 10.1385/MB:16:2:127) [DOI] [PubMed] [Google Scholar]
- 186.Monici M. 2005. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol. Annu. Rev. 11, 227–256. ( 10.1016/S1387-2656(05)11007-2) [DOI] [PubMed] [Google Scholar]
- 187.Jones CW, Smolinski D, Keogh A, Kirk TB, Zheng MH. 2005. Confocal laser scanning microscopy in orthopaedic research. Prog. Histochem. Cytochem. 40, 1–71. ( 10.1016/j.proghi.2005.02.001) [DOI] [PubMed] [Google Scholar]
- 188.Ascenzi MG, Lomovtsev A. 2006. Collagen orientation patterns in human secondary osteons, quantified in the radial direction by confocal microscopy. J. Struct. Biol. 153, 14–30. ( 10.1016/j.jsb.2005.08.007) [DOI] [PubMed] [Google Scholar]
- 189.Ascenzi MG, Gill J, Lomovtsev A. 2008. Orientation of collagen at the osteocyte lacunae in human secondary osteons. J. Biomech. 41, 3426–3435. ( 10.1016/j.jbiomech.2008.09.010) [DOI] [PubMed] [Google Scholar]
- 190.Ascenzi MG, et al. 2012. Parathyroid hormone treatment improves the cortical bone microstructure by improving the distribution of type I collagen in postmenopausal women with osteoporosis. J. Bone Miner. Res. 27, 702–712. ( 10.1002/jbmr.1497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zipfel WR, Williams RM, Webb WW. 2003. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 21, 1369–1377. ( 10.1038/nbt899) [DOI] [PubMed] [Google Scholar]
- 192.Denk W, Strickler JH, Webb WW. 1990. 2-photon laser scanning fluorescence microscopy. Science 248, 73–76. ( 10.1126/science.2321027) [DOI] [PubMed] [Google Scholar]
- 193.Denk W, Piston D, Webb W. 1995. Two-photon molecular excitation in laser-scanning microscopy. In Handbook of biological confocal microscopy (ed. Pawley J.), pp. 445–458. New York, NY: Springer. [Google Scholar]
- 194.Denk W, Svoboda K. 1997. Photon upmanship: why multiphoton imaging is more than a gimmick. Neuron 18, 351–357. ( 10.1016/S0896-6273(00)81237-4) [DOI] [PubMed] [Google Scholar]
- 195.Hoover EE, Squier JA. 2013. Advances in multiphoton microscopy technology. Nat. Photon. 7, 93–101. ( 10.1038/nphoton.2012.361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, Webb WW. 2003. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc. Natl Acad. Sci. USA 100, 7075–7080. ( 10.1073/pnas.0832308100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhu X, Tang Y, Chen J, Xiong S, Zhuo S, Chen J. 2013. Monitoring wound healing of elastic cartilage using multiphoton microscopy. Osteoarthritis Cartilage 21, 1799–1806. ( 10.1016/j.joca.2013.08.016) [DOI] [PubMed] [Google Scholar]
- 198.Plotnikov SV, Millard AC, Campagnola PJ, Mohler WA. 2006. Characterization of the myosin-based source for second-harmonic generation from muscle sarcomeres. Biophys. J. 90, 693–703. ( 10.1529/biophysj.105.071555) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wu S, Li H, Yang H, Zhang X, Li Z, Xu S. 2011. Quantitative analysis on collagen morphology in aging skin based on multiphoton microscopy. J. Biomed. Opt. 16, 040502 ( 10.1117/1.3565439) [DOI] [PubMed] [Google Scholar]
- 200.Mauri A, Perrini M, Mateos JM, Maake C, Ochsenbein-Koelble N, Zimmermann R, Ehrbar M, Mazza E. 2013. Second harmonic generation microscopy of fetal membranes under deformation: normal and altered morphology. Placenta 34, 1020–1026. ( 10.1016/j.placenta.2013.09.002) [DOI] [PubMed] [Google Scholar]
- 201.Chen H, Slipchenko MN, Liu Y, Zhao XF, Cheng JX, Lanir Y, Kassab GS. 2013. Biaxial deformation of collagen and elastin fibers in coronary adventitia. J. Appl. Physiol. 115, 1683–1693. ( 10.1152/japplphysiol.00601.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pacureanu A, Langer M, Boller E, Tafforeau P, Peyrin F. 2012. Nanoscale imaging of the bone cell network with synchrotron X-ray tomography: optimization of acquisition setup. Med. Phys. 39, 2229–2238. ( 10.1118/1.3697525) [DOI] [PubMed] [Google Scholar]
- 203.Shah FA, Zanghellini E, Matic A, Thomsen P, Palmquist A. 2016. The orientation of nanoscale apatite platelets in relation to osteoblastic-osteocyte lacunae on trabecular bone surface. Calcif. Tissue Int. 98, 193–205. ( 10.1007/s00223-015-0072-8) [DOI] [PubMed] [Google Scholar]
- 204.Zhou W, Apkarian R, Wang Z, Joy D. 2007. Fundamentals of scanning electron microscopy (SEM). In Scanning microscopy for nanotechnology (eds Zhou W, Wang Z), pp. 1–40. New York, NY: Springer. [Google Scholar]
- 205.Ambekar R, Chittenden M, Jasiuk I, Toussaint KC. 2012. Quantitative second-harmonic generation microscopy for imaging porcine cortical bone: comparison to SEM and its potential to investigate age-related changes. Bone 50, 643–650. ( 10.1016/j.bone.2011.11.013) [DOI] [PubMed] [Google Scholar]
- 206.Peddie CJ, Collinson LM. 2014. Exploring the third dimension: volume electron microscopy comes of age. Micron 61, 9–19. ( 10.1016/j.micron.2014.01.009) [DOI] [PubMed] [Google Scholar]
- 207.Pingel J, et al. 2014. 3-D ultrastructure and collagen composition of healthy and overloaded human tendon: evidence of tenocyte and matrix buckling. J. Anat. 224, 548–555. ( 10.1111/joa.12164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Reznikov N, Almany-Magal R, Shahar R, Weiner S. 2013. Three-dimensional imaging of collagen fibril organization in rat circumferential lamellar bone using a dual beam electron microscope reveals ordered and disordered sub-lamellar structures. Bone 52, 676–683. ( 10.1016/j.bone.2012.10.034) [DOI] [PubMed] [Google Scholar]
- 209.Suda K, Abe K, Kaneda K. 1999. Changes in the orientation of collagen fibers on the superficial layer of the mouse tibial bone after denervation: scanning electron microscopic observations. Arch. Histol. Cytol. 62, 231–235. ( 10.1679/aohc.62.231) [DOI] [PubMed] [Google Scholar]
- 210.Boyde A, Hobdell MH. 1968. Scanning electron microscopy of lamellar bone. Z. Zellforsch. Mikrosk. Anat. 93, 213–231. ( 10.1007/BF00336690) [DOI] [PubMed] [Google Scholar]
- 211.Kinney JH, Nichols MC. 1992. X-ray tomographic microscopy (XTM) using synchrotron radiation. Annu. Rev. Mater. Sci. 22, 121–152. ( 10.1146/annurev.ms.22.080192.001005) [DOI] [Google Scholar]
- 212.Weitkamp T, et al. 2010. Status and evolution of the ESRF beamline ID19. AIP Conf. Proc. 1221, 33–38. ( 10.1063/1.3399253) [DOI] [Google Scholar]
- 213.Cattaneo PM, Dalstra M, Beckmann F, Donath T, Melsen B. 2004. Comparison of conventional and synchrotron-radiation-based microtomography of bone around dental implants. Proc. SPIE 5535, 757 ( 10.1117/12.559630) [DOI] [Google Scholar]
- 214.Peter Z, Peyrin F. 2011. Synchrotron radiation micro-CT imaging of bone tissue. In Theory and applications of CT imaging and analysis (ed. Homma N.), pp. 233–254. Rijeka, Croatia: InTech. See http://www.intechopen.com/books/theory-and-applications-of-ct-imaging-andanalysis/synchrotron-radiation-micro-ct-imaging-of-bone-tissue. [Google Scholar]
- 215.Koester KJ, Ager JW III, Ritchie RO. 2008. The true toughness of human cortical bone measured with realistically short cracks. Nat. Mater. 7, 672–677. ( 10.1038/nmat2221) [DOI] [PubMed] [Google Scholar]
- 216.Peyrin F, Dong P, Pacureanu A, Langer M. 2014. Micro- and nano-CT for the study of bone ultrastructure. Curr. Osteoporos. Rep. 12, 465–474. ( 10.1007/s11914-014-0233-0) [DOI] [PubMed] [Google Scholar]
- 217.Dong P, Pacureanu A, Zuluaga MA, Olivier C, Grimal Q, Peyrin F. 2014. Quantification of the 3D morphology of the bone cell network from synchrotron micro-CT images. Image Anal. Stereol. 33, 10 ( 10.5566/ias.v33.p157-166) [DOI] [Google Scholar]
- 218.Schneider P, Stauber M, Voide R, Stampanoni M, Donahue LR, Mueller R. 2007. Ultrastructural properties in cortical bone vary greatly in two inbred strains of mice as assessed by synchrotron light based micro- and nano-CT. J. Bone Miner. Res. 22, 1557–1570. ( 10.1359/jbmr.070703) [DOI] [PubMed] [Google Scholar]
- 219.Martin T, Koch A. 2006. Recent developments in X-ray imaging with micrometer spatial resolution. J. Synchrot. Radiat. 13, 180–194. ( 10.1107/S0909049506000550) [DOI] [PubMed] [Google Scholar]
- 220.Vila-Comamala J, Pan Y, Lombardo JJ, Harris WM, Chiu WKS, David C, David C, Wang Y. 2012. Zone-doubled Fresnel zone plates for high-resolution hard X-ray full-field transmission microscopy. J. Synchrot. Radiat. 19, 705–709. ( 10.1107/S0909049512029640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bech M, Tapfer A, Velroyen A, Yaroshenko A, Pauwels B, Hostens J, Bruyndonckx P, Sasov A, Pfeiffer F. 2013. In-vivo dark-field and phase-contrast x-ray imaging. Sci. Rep. 3, 3209 ( 10.1038/srep03209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bravin A, Coan P, Suortti P. 2013. X-ray phase-contrast imaging, from pre-clinical applications towards clinics. Phys. Med. Biol. 58, R1–R35. ( 10.1088/0031-9155/58/1/R1) [DOI] [PubMed] [Google Scholar]
- 223.Wang ZT, Hauser N, Singer G, Trippel M, Kubik-Huch RA, Schneider CW, Stampanoni M. 2014. Non-invasive classification of microcalcifications with phase-contrast X-ray mammography. Nat. Commun. 5, 9 ( 10.1038/ncomms4797) [DOI] [PubMed] [Google Scholar]
- 224.Langer M, Pacureanu A, Suhonen H, Grimal Q, Cloetens P, Peyrin F. 2012. X-ray phase nanotomography resolves the 3D human bone ultrastructure. PLoS ONE 7, e35691 ( 10.1371/journal.pone.0035691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Dierolf M, Menzel A, Thibault P, Schneider P, Kewish CM, Wepf R, Bunk O, Pfeiffer F. 2010. Ptychographic X-ray computed tomography at the nanoscale. Nature 467, 436–439. ( 10.1038/nature09419) [DOI] [PubMed] [Google Scholar]
- 226.Varga P, Pacureanu A, Langer M, Suhonen H, Hesse B, Grimal Q, Cloetens P, Raum K, Peyrin F. 2013. Investigation of the three-dimensional orientation of mineralized collagen fibrils in human lamellar bone using synchrotron X-ray phase nano-tomography. Acta Biomater. 9, 8118–8127. ( 10.1016/j.actbio.2013.05.015) [DOI] [PubMed] [Google Scholar]
- 227.Diaz A, Trtik P, Guizar-Sicairos M, Menzel A, Thibault P, Bunk O. 2012. Quantitative X-ray phase nanotomography. Phys. Rev. B. 85, 020104(R) ( 10.1103/PhysRevB.85.020104) [DOI] [Google Scholar]
- 228.Holler M, et al. 2014. X-ray ptychographic computed tomography at 16 nm isotropic 3D resolution. Sci Rep. 4, 3857 ( 10.1038/srep03857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kisielowski C, et al. 2008. Detection of single atoms and buried defects in three dimensions by aberration-corrected electron microscope with 0.5-angstrom information limit. Microsc. Microanal. 14, 469–477. ( 10.1017/S1431927608080902) [DOI] [PubMed] [Google Scholar]
- 230.Grandfield K, Engqvist H. 2012. Focused ion beam in the study of biomaterials and biological matter. Adv. Mater. Sci. Eng. 6, 841961 ( 10.1155/2012/841961) [DOI] [Google Scholar]
- 231.Graham L, Orenstein JM. 2007. Processing tissue and cells for transmission electron microscopy in diagnostic pathology and research. Nat. Protocol. 2, 2439–2450. ( 10.1038/nprot.2007.304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Giraud-Guille MM. 1988. Twisted plywood architecture of collagen fibrils in human compact bone osteons. Calcif. Tissue Int. 42, 167–180. ( 10.1007/BF02556330) [DOI] [PubMed] [Google Scholar]
- 233.Kafantari H, Kounadi E, Fatourous M, Milonakis M, Tzaphlidou M. 2000. Structural alterations in rat skin and bone collagen fibrils induced by ovariectomy. Bone 26, 349–353. ( 10.1016/S8756-3282(99)00279-3) [DOI] [PubMed] [Google Scholar]
- 234.Rubin MA, Jasiuk I. 2005. The TEM characterization of the lamellar structure of osteoporotic human trabecular bone. Micron 36, 653–664. ( 10.1016/j.micron.2005.07.010) [DOI] [PubMed] [Google Scholar]
- 235.Rubin MA, Jasiuk L, Taylor J, Rubin J, Ganey T, Apkarian RP. 2003. TEM analysis of the nanostructure of normal and osteoporotic human trabecular bone. Bone 33, 270–282. ( 10.1016/S8756-3282(03)00194-7) [DOI] [PubMed] [Google Scholar]
- 236.Su X, Sun K, Cui FZ, Landis WJ. 2003. Organization of apatite crystals in human woven bone. Bone 32, 150–162. ( 10.1016/S8756-3282(02)00945-6) [DOI] [PubMed] [Google Scholar]
- 237.Li DS, Nielsen MH, Lee JRI, Frandsen C, Banfield JF, De Yoreo JJ. 2012. Direction-specific interactions control crystal growth by oriented attachment. Science 336, 1014–1018. ( 10.1126/science.1219643) [DOI] [PubMed] [Google Scholar]
- 238.Boyde A, Jones SJ. 1996. Scanning electron microscopy of bone: instrument, specimen, and issues. Microsc. Res. Tech. 33, 92–120. ( 10.1002/(SICI)1097-0029(19960201)33:2%3C92::AID-JEMT2%3E3.0.CO;2-0) [DOI] [PubMed] [Google Scholar]
- 239.Petersen W, Tillmann B. 1998. Collagenous fibril texture of the human knee joint menisci. Anat. Embryol. Berl. 197, 317–324. ( 10.1007/s004290050141) [DOI] [PubMed] [Google Scholar]
- 240.Sasaki K, Yamamoto N, Kiyosawa T, Sekido M. 2012. The role of collagen arrangement change during tendon healing demonstrated by scanning electron microscopy. J. Electron. Microsc. 61, 327–634. ( 10.1093/jmicro/dfs057) [DOI] [PubMed] [Google Scholar]
- 241.Franchi M, Trire A, Quaranta M, Orsini E, Ottani V. 2007. Collagen structure of tendon relates to function. Sci. World J. 7, 404–420. ( 10.1100/tsw.2007.92) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kaab MJ, Gwynn IA, Notzli HP. 1998. Collagen fibre arrangement in the tibial plateau articular cartilage of man and other mammalian species. J. Anat. 193, 23–34. ( 10.1046/j.1469-7580.1998.19310023.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Clark JM. 1991. Variation of collagen fiber alignment in a joint surface—a scanning electron-microscope study of the tibial plateau in dog, rabbit, and man. J. Orthop. Res. 9, 246–257. ( 10.1002/jor.1100090213) [DOI] [PubMed] [Google Scholar]
- 244.Faingold A, Cohen SR, Reznikov N, Wagner HD. 2013. Osteonal lamellae elementary units: lamellar microstructure, curvature and mechanical properties. Acta Biomater. 9, 5956–5962. ( 10.1016/j.actbio.2012.11.032) [DOI] [PubMed] [Google Scholar]
- 245.Schneider P, Meier M, Wepf R, Mueller R. 2011. Serial FIB/SEM imaging for quantitative 3D assessment of the osteocyte lacuno-canalicular network. Bone 49, 304–311. ( 10.1016/j.bone.2011.04.005) [DOI] [PubMed] [Google Scholar]
- 246.Starborg T, Kalson NS, Lu Y, Mironov A, Cootes TF, Holmes DF, Kadler KE. 2013. Using transmission electron microscopy and 3View to determine collagen fibril size and three-dimensional organization. Nat. Protoc. 8, 1433–1448. ( 10.1038/nprot.2013.086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Reznikov N, Chase H, Brumfeld V, Shahar R, Weiner S. 2015. The 3D structure of the collagen fibril network in human trabecular bone: relation to trabecular organization. Bone 71, 189–195. ( 10.1016/j.bone.2014.10.017) [DOI] [PubMed] [Google Scholar]
- 248.Binnig G, Quate CF, Gerber C. 1986. Atomic force microscope. Phys. Rev. Lett. 56, 930–933. ( 10.1103/PhysRevLett.56.930) [DOI] [PubMed] [Google Scholar]
- 249.Iwata K, Yamazaki S, Mutombo P, Hapala P, Ondracek M, Jelinek P, Sugimoto Y. 2015. Chemical structure imaging of a single molecule by atomic force microscopy at room temperature. Nat. Commun. 6, 7766 ( 10.1038/ncomms8766) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Burnham NA, Colton RJ. 1989. Measuring the nanomechanical properties and surface forces of materials using an atomic force microscope. J. Vac. Sci. Technol. A 7, 2906–2913. ( 10.1116/1.576168) [DOI] [Google Scholar]
- 251.Stolz M, et al. 2009. Early detection of aging cartilage and osteoarthritis in mice and patient samples using atomic force microscopy. Nat. Nanotechnol. 4, 186–192. ( 10.1038/nnano.2008.410) [DOI] [PubMed] [Google Scholar]
- 252.Kotova SL, Timashev PS, Guller AE, Shekhter AB, Misurkin PI, Bagratashvili VN, Solovieva AB. 2015. Collagen structure deterioration in the skin of patients with pelvic organ prolapse determined by atomic force microscopy. Microsc. Microanal. 21, 324–333. ( 10.1017/S1431927615000148) [DOI] [PubMed] [Google Scholar]
- 253.Wallace JM. 2012. Applications of atomic force microscopy for the assessment of nanoscale morphological and mechanical properties of bone. Bone 50, 420–427. ( 10.1016/j.bone.2011.11.008) [DOI] [PubMed] [Google Scholar]
- 254.Thurner PJ. 2009. Atomic force microscopy and indentation force measurement of bone. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 1, 624–649. ( 10.1002/wnan.56) [DOI] [PubMed] [Google Scholar]
- 255.Hengsberger S, Kulik A, Zysset P. 2001. A combined atomic force microscopy and nanoindentation technique to investigate the elastic properties of bone structural units. Eur. Cells Mater. 1, 12–17. [DOI] [PubMed] [Google Scholar]
- 256.Tai K, Dao M, Suresh S, Palazoglu A, Ortiz C. 2007. Nanoscale heterogeneity promotes energy dissipation in bone. Nat. Mater. 6, 454–462. ( 10.1038/nmat1911) [DOI] [PubMed] [Google Scholar]
- 257.Yao H, Dao M, Carnelli D, Tai K, Ortiz C. 2011. Size-dependent heterogeneity benefits the mechanical performance of bone. J. Mech. Phys. Solids 59, 64–74. ( 10.1016/j.jmps.2010.09.012) [DOI] [Google Scholar]
- 258.Andriotis OG, Manuyakorn W, Zekonyte J, Katsamenis OL, Fabri S, Howarth PH, Davies DE, Thurner PJ. 2014. Nanomechanical assessment of human and murine collagen fibrils via atomic force microscopy cantilever-based nanoindentation. J. Mech. Behav. Biomed. Mater. 39, 9–26. ( 10.1016/j.jmbbm.2014.06.015) [DOI] [PubMed] [Google Scholar]
- 259.Thompson JB, Kindt JH, Drake B, Hansma HG, Morse DE, Hansma PK. 2001. Bone indentation recovery time correlates with bond reforming time. Nature 414, 773–776. ( 10.1038/414773a) [DOI] [PubMed] [Google Scholar]
- 260.Wenger MPE, Bozec L, Horton MA, Mesquida P. 2007. Mechanical properties of collagen fibrils. Biophys. J. 93, 1255–1263. ( 10.1529/biophysj.106.103192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Thurner PJ, et al. 2007. Imaging of bone ultrastructure using atomic force microscopy. In Modern research and educational topics in microscopy (eds Méndez-Vilas AD, Díaz J), pp. 37–48. Extremadura, Spain: Formatex. [Google Scholar]
- 262.Thalhammer S, Heckl WM, Zink A, Nerlich AG. 2001. Atomic force microscopy for high resolution imaging of collagen fibrils—a new technique to investigate collagen structure in historic bone tissues. J. Archaeol. Sci. 28, 1061–1068. ( 10.1006/jasc.2000.0644) [DOI] [Google Scholar]
- 263.Erickson B, Fang M, Wallace JM, Orr BG, Les CM, Banaszak Holl MM. 2013. Nanoscale structure of type I collagen fibrils: quantitative measurement of D-spacing. Biotechnol. J. 8, 117–126. ( 10.1002/biot.201200174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tong W, Glimcher MJ, Katz JL, Kuhn L, Eppell SJ. 2003. Size and shape of mineralites in young bovine bone measured by atomic force microscopy. Calcif. Tissue Int. 72, 592–598. ( 10.1007/s00223-002-1077-7) [DOI] [PubMed] [Google Scholar]
- 265.Habelitz S, Balooch M, Marshall SJ, Balooch G, Marshall GW Jr. 2002. In situ atomic force microscopy of partially demineralized human dentin collagen fibrils. J. Struct. Biol. 138, 227–236. ( 10.1016/S1047-8477(02)00029-1) [DOI] [PubMed] [Google Scholar]
- 266.Wallace JM, Erickson B, Les CM, Orr BG, Banaszak Holl MM. 2010. Distribution of type I collagen morphologies in bone: relation to estrogen depletion. Bone 46, 1349–1354. ( 10.1016/j.bone.2009.11.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Tao J, Battle KC, Pan H, Salter EA, Chien Y-C, Wierzbicki A, De Yoreo JJ. 2015. Energetic basis for the molecular-scale organization of bone. Proc. Natl Acad. Sci. USA 112, 326–331. ( 10.1073/pnas.1404481112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sasaki N, Tagami A, Goto T, Taniguchi M, Nakata M, Hikichi K. 2002. Atomic force microscopic studies on the structure of bovine femoral cortical bone at the collagen fibril-mineral level. J. Mater. Sci. Mater. Med. 13, 333–337. ( 10.1023/A:1014079421895) [DOI] [PubMed] [Google Scholar]
- 269.Hassenkam T, Fantner GE, Cutroni JA, Weaver JC, Morse DE, Hansma PK. 2004. High-resolution AFM imaging of intact and fractured trabecular bone. Bone 35, 4–10. ( 10.1016/j.bone.2004.02.024) [DOI] [PubMed] [Google Scholar]
- 270.Mosekilde L. 1990. Consequences of the remodeling process for vertebral trabecular bone-structure—a scanning electron-microscopy study (uncoupling of unloaded structures). Bone Miner. 10, 13–35. ( 10.1016/0169-6009(90)90046-I) [DOI] [PubMed] [Google Scholar]
- 271.Hesse B, et al. 2015. Canalicular network morphology is the major determinant of the spatial distribution of mass density in human bone tissue: evidence by means of synchrotron radiation phase-contrast nano-CT. J. Bone Miner. Res. 30, 346–356. ( 10.1002/jbmr.2324) [DOI] [PubMed] [Google Scholar]
- 272.Larrue A, Rattner A, Peter ZA, Olivier C, Laroche N, Vico L, Peyrin F. 2011. Synchrotron radiation micro-CT at the micrometer scale for the analysis of the three-dimensional morphology of microcracks in human trabecular bone. PLoS ONE 6, e21297 ( 10.1371/journal.pone.0021297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Chiba K, Nango N, Kubota S, Okazaki N, Taguchi K, Osaki M, Ito M. 2012. Relationship between microstructure and degree of mineralization in subchondral bone of osteoarthritis: a synchrotron radiation microCT study. J. Bone Miner. Res. 27, 1511–1517. ( 10.1002/jbmr.1609) [DOI] [PubMed] [Google Scholar]
- 274.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Mueller R. 2010. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 25, 1468–1486. ( 10.1002/jbmr.141) [DOI] [PubMed] [Google Scholar]
- 275.Stock SR, Veis A, Telser A, Cai Z. 2011. Near tubule and intertubular bovine dentin mapped at the 250 nm level. J. Struct. Biol. 176, 203–211. ( 10.1016/j.jsb.2011.07.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Karunaratne A, Boyde A, Esapa CT, Hiller J, Terrill NJ, Brown SDM, Cox RD, Thakker RV, Gupta HS. 2013. Symmetrically reduced stiffness and increased extensibility in compression and tension at the mineralized fibrillar level in rachitic bone. Bone 52, 689–698. ( 10.1016/j.bone.2012.10.029) [DOI] [PubMed] [Google Scholar]
- 277.Singhal A, Yuan F, Stock SR, Almer JD, Brinson LC, Dunand DC. 2013. Evolution of phase strains during tensile loading of bovine cortical bone. Adv. Eng. Mater. 15, 238–249. ( 10.1002/adem.201200204) [DOI] [Google Scholar]
- 278.Deymier-Black AC, Yuan F, Singhal A, Almer JD, Brinson LC, Dunand DC. 2012. Evolution of load transfer between hydroxyapatite and collagen during creep deformation of bone. Acta Biomater. 8, 253–261. ( 10.1016/j.actbio.2011.08.014) [DOI] [PubMed] [Google Scholar]
- 279.Hoo RP, Fratzl P, Daniels JE, Dunlop JWC, Honkimaeki V, Hoffman M. 2011. Cooperation of length scales and orientations in the deformation of bovine bone. Acta Biomater. 7, 2943–2951. ( 10.1016/j.actbio.2011.02.017) [DOI] [PubMed] [Google Scholar]
- 280.Barth HD, Zimmermann EA, Schaible E, Tang SY, Alliston T, Ritchie RO. 2011. Characterization of the effects of x-ray irradiation on the hierarchical structure and mechanical properties of human cortical bone. Biomaterials 32, 8892–8904. ( 10.1016/j.biomaterials.2011.08.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Niemz MH. 2004. Medical applications of lasers. Laser-tissue interactions, pp. 151–247. Berlin, Germany: Springer. [Google Scholar]
- 282.Lakes RS, Katz JL, Sternstein SS. 1979. Viscoelastic properties of wet cortical bone—I. Torsional and biaxial studies. J. Biomech. 12, 657 ( 10.1016/0021-9290(79)90016-2) [DOI] [PubMed] [Google Scholar]
- 283.Wentzell S, Nesbitt RS, Macione J, Kotha S. 2013. Measuring strain using digital image correlation of second harmonic generation images. J. Biomech. 46, 2032–2038. ( 10.1016/j.jbiomech.2013.06.003) [DOI] [PubMed] [Google Scholar]
- 284.Kirk SE, Skepper JN, Donald AM. 2009. Application of environmental scanning electron microscopy to determine biological surface structure. J. Microsc. 233, 205–224. ( 10.1111/j.1365-2818.2009.03111.x) [DOI] [PubMed] [Google Scholar]
- 285.Xiao X, De Carlo F, Stock SR. 2008. X-ray zoom in tomography of calcified tissue. Proc. SPIE 7078, 707810 ( 10.1117/12.796638) [DOI] [Google Scholar]
- 286.Guizar-Sicairos M, Boon JJ, Mader K, Diaz A, Menzel A, Bunk O. 2015. Quantitative interior X-ray nanotomography by a hybrid imaging technique. Optica 2, 259–266. ( 10.1364/OPTICA.2.000259) [DOI] [Google Scholar]
- 287.Xiao X, De Carlo F, Stock S. 2007. Practical error estimation in zoom-in and truncated tomography reconstructions. Rev. Sci. Instrum. 78, 063705 ( 10.1063/1.2744224) [DOI] [PubMed] [Google Scholar]
- 288.Kagias M, Wang Z, Villanueva-Perez P, Jefimovs K, Stampanoni M. 2016. 2D-Omnidirectional hard-X-ray scattering sensitivity in a single shot. Phys. Rev. Lett. 116, 093902 ( 10.1103/PhysRevLett.116.093902) [DOI] [PubMed] [Google Scholar]
- 289.Velroyen A, et al. 2015. Grating-based X-ray dark-field computed tomography of living mice. EBioMedicine 2, 1500–1506. ( 10.1016/j.ebiom.2015.08.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Momose A, et al. 2014. X-ray phase imaging: from synchrotron to hospital. Phil. Trans. R. Soc. A 372, 20130023 ( 10.1098/rsta.2013.0023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Tapfer A, et al. 2012. Experimental results from a preclinical X-ray phase-contrast CT scanner. Proc. Natl Acad. Sci. USA 109, 15 691–15 696. ( 10.1073/pnas.1207503109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Thuring T, Guggenberger R, Alkadhi H, Hodler J, Vich M, Wang Z, David C, Stampanoni M. 2013. Human hand radiography using X-ray differential phase contrast combined with dark-field imaging. Skelet. Radiol. 42, 827–835. ( 10.1007/s00256-013-1606-7) [DOI] [PubMed] [Google Scholar]
- 293.Revol V, Kottler C, Kaufmann R, Neels A, Dommann A. 2012. Orientation-selective X-ray dark field imaging of ordered systems. J. Appl. Phys. 112, 114903 ( 10.1063/1.4768525) [DOI] [Google Scholar]
- 294.Gu M, Bao H, Kang H. 2014. Fibre-optical microendoscopy. J. Microsc. 254, 13–18. ( 10.1111/jmi.12119) [DOI] [PubMed] [Google Scholar]
- 295.Lefort C, Hamzeh H, Louradour F, Pain F, Haidar DA. 2014. Characterization, comparison, and choice of a commercial double-clad fiber for nonlinear endomicroscopy. J. Biomed. Opt. 19, 8 ( 10.1117/1.JBO.19.7.076005) [DOI] [PubMed] [Google Scholar]
- 296.Bao HC, Boussioutas A, Jeremy R, Russell S, Gu M. 2010. Second harmonic generation imaging via nonlinear endomicroscopy. Opt. Express. 18, 1255–1260. ( 10.1364/OE.18.001255) [DOI] [PubMed] [Google Scholar]
- 297.Fu L, Gu M. 2008. Polarization anisotropy in fiber-optic second harmonic generation microscopy. Opt. Express. 16, 5000–5006. ( 10.1364/OE.16.005000) [DOI] [PubMed] [Google Scholar]