Abstract
Antibodies somatically mutate to attain high affinity in germinal centers (GCs). There, competition between B cell clones and among somatic mutants of each clone drives an increase in average affinity across the population. The extent to which higher-affinity cells eliminating competitors restricts clonal diversity is unknown. By combining multiphoton microscopy and sequencing, we show that tens to hundreds of distinct B cell clones seed each GC, and that GCs lose clonal diversity at widely disparate rates. Furthermore, efficient affinity maturation can occur in the absence of homogenizing selection, ensuring that many clones can mature in parallel within the same GC. Our findings have implications for development of vaccines in which antibodies with non-immunodominant specificities must be elicited, as is the case for HIV-1 and influenza.
The average affinity of specific antibodies increases dramatically over the course of an immune response (1, 2). This phenomenon is known as affinity maturation, and is the result of a Darwinian process that alternates stochastic somatic hypermutation (SHM) of immunoglobulin (Ig) genes with the selection and clonal expansion of B cells that have acquired affinity-enhancing mutations (2). Successive iterations of this cycle drive an increase in the overall affinity of antibodies over time, which is essential for their effectiveness in curtailing and preventing infection.
Affinity maturation takes place in germinal centers (GCs), defined microanatomical clusters containing up to a few thousand B cells that emerge in multiple copies within secondary lymphoid organs upon infection or immunization (3–7). Each GC is traditionally viewed as arising from a handful of independent “founder clones,” many of which are lost as affinity-based competition leads to progressive focusing of the repertoire on the most successful progeny (which we refer to as “homogenizing selection”) (8). Though it is inducible under experimental settings (9), the extent to which homogenizing selection takes place during normal immunization or infection has been difficult to determine, mostly due to technical limitations in the methods used to measure clonal diversity within individual GCs (10–14). Here, we report the development of two imaging-based approaches that expand our ability to study GC clonal composition in response to any antigen. Our findings highlight the heterogeneity of clonal evolution pathways in GCs.
Measuring clonal diversity in GCs using multiphoton microscopy
As an approach to estimating GC clonal diversity by imaging, we made use of a “brainbow” allele for multicolor fate mapping to permanently tag individual B cells and their progeny with different combinations of fluorescent proteins (15). This method, as implemented in the Rosa26Confetti allele (16), relies on stochastic Cre-mediated recombination to commit cells to expression of one of four possible fluorescent proteins, generating ten different color combinations when two alleles are recombined in homozygous mice (fig. S1A–B). Because recombination stops upon cessation of Cre activity, selective proliferation of an individual GC B cell would lead to the appearance of clusters of daughter cells of the same color.
We generated Rosa26Confetti/Confetti mice carrying the Mx1-Cre transgene, which triggers Confetti recombination spontaneously during early life (17). Multiphoton imaging of popliteal lymph nodes (pLN) in naïve Mx1-Confetti mice showed that, in the absence of immunization, cells of different colors are intermixed, as expected from the polyclonal nature and migratory behavior of naïve lymphocytes (Fig. 1A, movie S1). In contrast, subcutaneous immunization with alum-adsorbed chicken gamma globulin (CGG) induced the appearance in the draining lymph node (LN) of clusters of cells of the same color, suggestive of monoclonal expansion (Fig. 1B, movies S2 and S3). Such clusters could also be found in mesenteric LNs (Fig. 1C) and even occasionally in Peyer’s patches (fig. S1C) of unimmunized mice, where GCs form spontaneously in response to intestinal microbiota. Thus, clonal expansions within GCs can be readily detected by multicolor fate mapping, even in cases where the driving antigen is unknown.
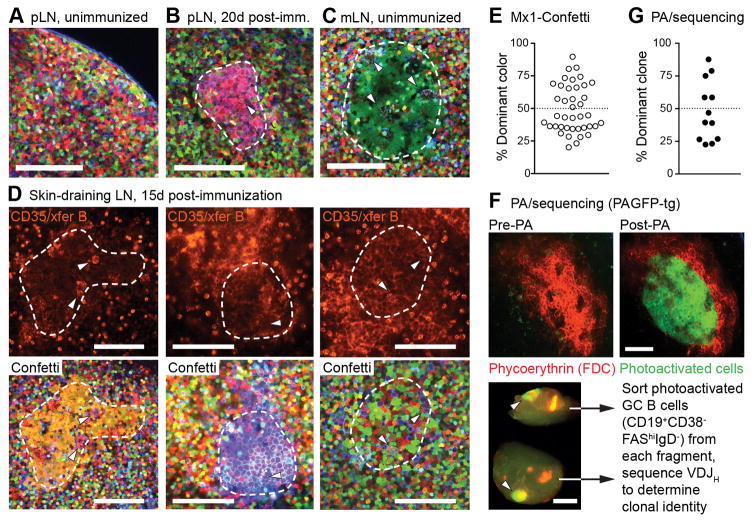
Figure 1. Visualizing clonal expansions in GCs using a brainbow allele.
(A) Popliteal lymph node (pLN) of an unimmunized Rosa26Confetti/Confetti.Mx1-Cre mouse, imaged by multiphoton microscopy. (B) pLN of a mouse immunized 20 days previously with 10 μg CGG in alum in the hind footpad. The image shows a cluster of similar-colored cells corresponding to a GC (dashed line), as evidenced by the presence of tingible body macrophages (arrowheads). (C) Single-colored GC (dashed line) in a mesenteric lymph node (mLN) from an unimmunized mouse. (D) GCs in draining LN of mice immunized subcutaneously with CGG in alum 15 days prior to imaging. The location of the GC dark zone (dashed line) was determined by injection of labeled antibody to CD35 and surface-labeled naïve B cells (top panels, fluorescence is from Alexa 633 label). For each GC, Confetti colors (bottom panels) were imaged independently and used for quantification. Confetti colors are as shown in fig. S1B. GC identity is confirmed by presence of tingible-body macrophages (arrowheads). Scale bars, 100 μm. Second harmonic generation from collagen fibers is shown in blue. (E) Quantification of data as in (D). Each symbol represents one GC. Graph shows percentage of cells expressing the most abundant color combination. Data pooled from four mice, two independent experiments. (F) Quantifying GC clonality by photoactivation (PA). Photoactivatable-GFP-transgenic mice were immunized in the footpad with 10 μg CGG in alum and imaged 15 days later. FDC networks were labeled with phycoerytrhin immune complexes. Top, images of a single GC within a pLN, prior to and after photoactivation (Scale bar, 100 μm). Bottom, single pLN containing two photoactivated GCs (arrowheads) dissected into two fragments, each of which is separately processed for sorting of PA+ GC B cells and Igh sequencing (Scale bar, 500 μm). (G) Quantification of clonal dominance in multiple GCs. Data obtained as in (F), with clonal identity assigned based on Igh sequence. Each symbol represents one GC, with 2 GCs sequenced per LN (full clonality charts in fig. S2). Given is the percentage of cells belonging to the most abundant clone. Data are from 5 mice, 3 independent experiments.
Quantification of cell colors in GC dark zones at 15 days after immunization (Fig. 1D) revealed a wide range of color dominances (from 20.2% to 89.7%; median, 44.0%), and a relative paucity of predominantly single-colored GCs (6 of 40 (15%) GCs with color dominance > 70%; Fig. 1E). To validate these estimates, we used in-situ photoactivation followed by FACS sorting (9, 18) to obtain single B cells from individual GCs, the clonal identity of which we then determined by Igh mRNA sequencing (Fig. 1F). This approach again showed varying degrees of clonal dominance among individual GCs (3/12 (25%) GCs with dominance >70%; range, 22.5% to 87.5%; median, 43.0%; Fig. 1G and fig. S2). It also revealed the frequent presence of clones that were shared between two individual GCs in the same LN, indicative of synchronous origin (colored slices in fig. S2). We conclude that GCs display variable degrees clonal dominance, even when induced synchronously by immunization. While predominantly monoclonal GCs do exist, these are relatively rare at the time point assayed.
Early GCs are highly diverse
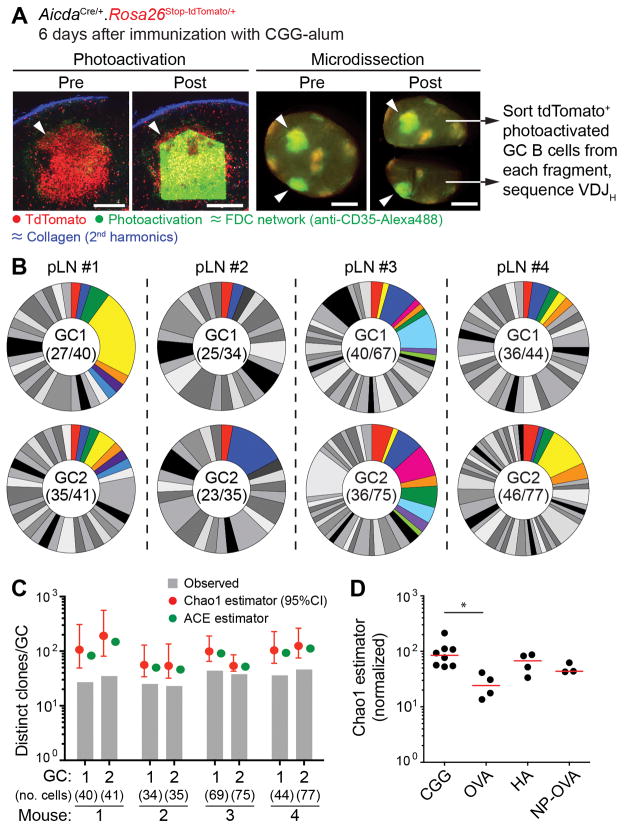
The diversity of clonal dominance levels among mature GCs led us to question the generalizability of reports proposing that GCs are seeded pauciclonally (11, 12, 19). To address this, we generated mice expressing the photoactivatable-GFP transgene along with Cre recombinase driven by the endogenous Aicda locus (AicdaCre) (20), expressed in activated B cells, and a Rosa26lox-stop-lox-tdTomato reporter. At 6 days after immunization with CGG alum, early GCs were identifiable in these mice as clusters of tdTomato+ cells within follicular dendritic cell (FDC) networks. We photoactivated two such clusters per LN, and sorted photoactivated GC B cells from each cluster for Igh sequencing (Fig. 2A and fig. S3A). Early GCs were highly and uniformly polyclonal, with 23 to 46 unique VDJ rearrangements detected when sequencing 34 to 77 single cells per GC (Fig. 2B). Extrapolation of these numbers using the Chao1 and ACE estimators of species richness (21) gave rough estimates of GC clonality that ranged from ~50 to ~200 clones per GC (median Chao1, 102 clones) for the 4 pairs of GCs analyzed (Fig. 2C). Overall mutation and class switching levels were low at this time point (fig. S3B), confirming that these were indeed early GCs. We were again able to identify B cell clones that were shared between two GCs in the same pLN (colored slices, Fig. 2B; mean 15.8% (SD 6.4) of clones found in one GC were also found in its neighbor). Similar experiments using other antigens showed that, although the extent of early GC diversity can vary depending on the immunizing antigen, high diversity is not a unique feature of the response to CGG (Fig. 2D and fig. S3C–E). Diversity was also not the result of non-specific recruitment of B cells to early GCs (fig. S3C and fig S4). We conclude that early GCs are highly diverse, containing tens to hundreds of distinct clones, depending on the antigen used for immunization. The progression from uniform diversity (Fig. 2) to variable degrees of clonal dominance (Fig. 1) in the CGG response implies that individual GCs display different rates of homogenizing selection acting subsequently to this coalescence.
Figure 2. Clonal diversity in early GCs.
(A) Photoactivation of single early GC clusters. photo activatable-GFP-transgenic. AicdaCre. Rosa26lox-stop-lox-tdTomato mice were immunized in the footpad with 10 μg CGG in alum and imaged 6 days later. FDC networks were marked by injection of labeled antibody to CD35. Left panels show images of a single tdTomato+ cluster (arrowheads) within a pLN prior to and after photoactivation (Scale bar, 200 μm). Right panels show dissection of a single pLN with two photoactivated GCs (arrowheads) into two fragments, each of which is separately processed for cell sorting (Scale bar, 500 μm). (B) Pie charts showing clonal diversity in early GCs. Each slice represents one clone. Colored slices represent clones that were found in both GCs (upper and lower pie charts) from the same pLN. Numbers in the center of each chart are (number of clones observed/total number of cells sequenced). Clonal identity was assigned based on Igh sequence. Pairs are from 4 different mice in 3 independent experiments. (C) Estimation of total clonal richness in individual GCs using the Chao1 and ACE estimators. Graphs show observed clonal richness (from panel (B)), and total richness according to the indicated estimator. (D) Estimated clonal richness (Chao1) in GCs elicited by various antigens. Mice were immunized with 10 μg of the indicated antigen, and imaged/photoactivated as in (A). Each symbol represents one GC, bar indicates median. For comparison purposes, estimates are normalized by interpolation to the size of the smallest sample (34 cells). Note that numbers for CGG GCs in this panel differ from those in panel (C) due to normalization. Further details in Fig. S3. * p < 0.05, Kruskall-Wallis test with Dunn’s post-test. All other comparisons were not significant.
Extent of homogenizing selection in individual GCs
Clonal dominance in GCs may arise by parallel selection of multiple members of a clone present in the early GC or by strong expansion of single SHM variants over cells of the same and of different clones. To investigate these dynamics, we generated mice in which recombination of Rosa26Confetti was driven by the tamoxifen-inducible AicdaCreERT2 allele, specifically expressed in activated B cells (22). We thus delayed recombination until after GC formation, allowing different members of the same clone to express different color combinations, each giving origin to an independent early GC “lineage.” In this system, GCs dominated by the descendants of a single SHM variant would be identifiable as having “resolved” to a single dominant color (Fig. 3A).
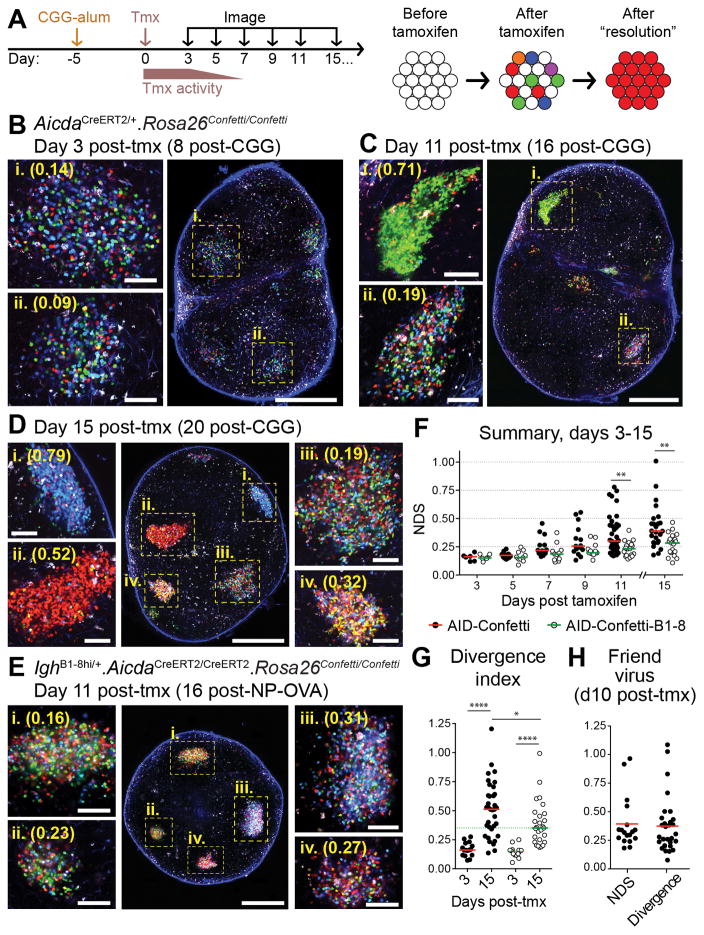
Figure 3. Kinetics of color dominance in individual GCs.
(A) Graphic representation of the experimental protocol. AID-Confetti mice were immunized in the footpad with 10 μg CGG in alum, treated with tamoxifen (tmx) 5 days later, and imaged at the indicated time points. Tamoxifen triggers recombination of one or both Confetti alleles in individual GC B cells, independently of clonal origin. (B-E) Whole lymph node (large panel; scale bar, 500 μm) and higher-magnification images (side panels; scale bar, 100 μm) showing GCs at different times after tmx administration. Cell colors as in fig. S1B. Second harmonic generation from collagen fibers is shown in blue. (B-D), AID-Confetti mice imaged at the time points indicated. (E) WT recipients of 1–2 × 104 adoptively-transferred AID-Confetti-B1-8 Igλ+ B cells, immunized with NP-OVA as in (A) and imaged 11 days post-tmx. Higher-magnification panels show independently-acquired images of the GCs indicated in the overview panel. (F) Quantification of data as shown in (B-E). NDS, normalized dominance score. Bars represent the median. (G) Divergence index for AID-Confetti and AID-Confetti-B1-8 mice at days 3 and 15 post-tmx. Bars represent the median. Green dotted line placed at the median of the day 15 AID-Confetti-B1-8 data for reference. (H) Quantification of GC selection in AID-Confetti mice infected with Friend Virus as detailed in fig. S7. Graph shows NDS and divergence index at day 10 post-tmx (day 30 post-infection). Bars represent the median. For panels (F-H), each symbol represents one GC. Data are pooled from 2–6 replicate experiments. For NDS quantification, we exclude GCs with density of fluorescent cells below 0.4 cells/100 μm2 (equivalent to approximately 40% of cells having recombined a Confetti allele; see Supplementary Text). * p<0.02; ** p < 0.01; **** p < 0.0001, Mann-Whitney U test.
Administration of tamoxifen to Rosa26Confetti/Confetti.AicdaCreERT2/+ (AID-Confetti) mice at 5 days after CGG immunization triggered recombination, often of a single Confetti allele, in ~40–50% of GC B cells (fig. S5A–B) by the end of the 3 to 5-day period during which tamoxifen is active (23). Recombination thus ends near the onset of T cell-driven selection, at the beginning of the second week of the response (24) (Fig. 3A). GCs analyzed 3 days after tamoxifen treatment (8 days post-immunization) displayed roughly equal proportions of CFP+, YFP+ and RFP+ cells and a lower proportion of GFP+ and doubly-recombined cells (Fig. 3B), as expected from the original description of the Confetti allele (16). Only residual if any GCs containing recombined cells could be detected in unimmunized or alum-only control mice (fig. S5C).
Color distribution changed over time, with progressive emergence of GCs dominated by a single color/combination (Fig. 3B–G). To quantify color dominance, we calculated a normalized dominance score (NDS) for each GC, which approximates (25) the fraction of cells belonging to a single lineage originating at the time of recombination (fig. S5D). Kinetic analysis was extended to 15 days after tamoxifen treatment (20 days after immunization), near the end of the CGG-alum-induced GC response in most LNs. Median dominance increased monotonically, from 0.16 at day 3 to 0.39 at day 15 post-tamoxifen. GCs with NDS of 0.5 or more (corresponding to ~50% of all GC cells displaying the same color combination) appeared as early as day 9 post-tamoxifen, and a single color could account for close to 80% of GC B cells at day 11 and virtually all cells in a GC four days later (Fig. 3F). However, progression among GCs varied markedly, as dominance in the lowest-scoring GCs at days 11 and 15 after tamoxifen remained close to day 3 levels. Evaluation of color distribution in exceptional LNs in which GCs could still be found at very late time points showed that low-dominance GCs were present up to the very end of the response (fig. S6A–C). Therefore, GC selection does not inevitably lead to high color dominance within the lifetime of the response to CGG-alum.
To determine the extent to which progression of color dominance depends on affinity-based competition, we generated an adoptive transfer model in which wild-type hosts received B cells from AicdaCreERT2/CreERT2. Rosa26Confetti/Confetti. IghB1-8hi/+ (AID-Confetti-B1-8) donors (in which Igλ+ B cells have uniformly high affinity to the NP hapten and do not undergo affinity maturation due to lack of a functional Aicda allele). NP-OVA-induced GCs in recipient mice approached high color dominance only rarely and with much delayed kinetics (Fig. 3E–F and fig. S6D). Therefore, differences in affinity between competing B cells likely contribute towards the rapid rise in dominance observed in a fraction of GCs. To quantify the effect of neutral competition—defined as the change in lineage abundances over time due to stochastic factors, in the absence of differences in affinity—in AID-Confetti-B1-8 GCs, we calculated a divergence index, which computes the difference between the expected and observed proportions of all 10 colors in each GC (fig. S5E), and is thus more sensitive to small changes in color abundance than the NDS. This metric showed marked divergence from baseline distribution in this population (Fig. 3G and fig. S6D), highlighting the effect on GC selection of factors unrelated to affinity. Moreover, the least divergent GCs in “wild-type” AID-Confetti mice remained relatively close to baseline color distribution (at day 15 post-tamoxifen, 25.7% of AID-Confetti GCs scored below the median of the AID-Confetti-B1-8 distribution; Fig. 3G). We conclude that individual GCs are highly heterogeneous with respect to selection: while a fraction of these structures become heavily dominated by cells of one color in a matter of days—suggesting strong expansion of the descendants of a single SHM variant arising at or after the onset of GC selection—others deviate from baseline color distribution at levels that do not exceed those attained by neutral competition.
We extended our findings to a different antigenic system by measuring clonal dominance in GCs elicited by infection with Friend retrovirus (FV) (26). We treated infected AID-Confetti mice with tamoxifen at 20 days post-infection (an early time point in the delayed GC response to FV) and imaged spleen slices 10 days later (fig. S7A–B). A wide range of NDS and divergence scores were also observed in FV-induced GCs (Fig. 3H and fig. S7C). Therefore, heterogeneity in the outcome of selection is a common property of GCs induced by model antigens and by viral infection, and thus likely represents an intrinsic property of GC evolution.
Homogeneous GCs are the product of “clonal bursts”
To investigate the underlying clonal structure of the variation in homogenizing selection rates among GCs, we determined the Igh gene sequences of fluorescent B cells isolated from GCs with different degrees of color dominance. To achieve this, we dissected pLNs from immunized AID-Confetti mice into fragments containing single GCs using vibratome sectioning guided by multiphoton microscopy (Fig. 4A and fig. S8). From each LN, we sorted cells from one high-dominance GC and from a neighboring low-dominance GC for Ig sequencing.
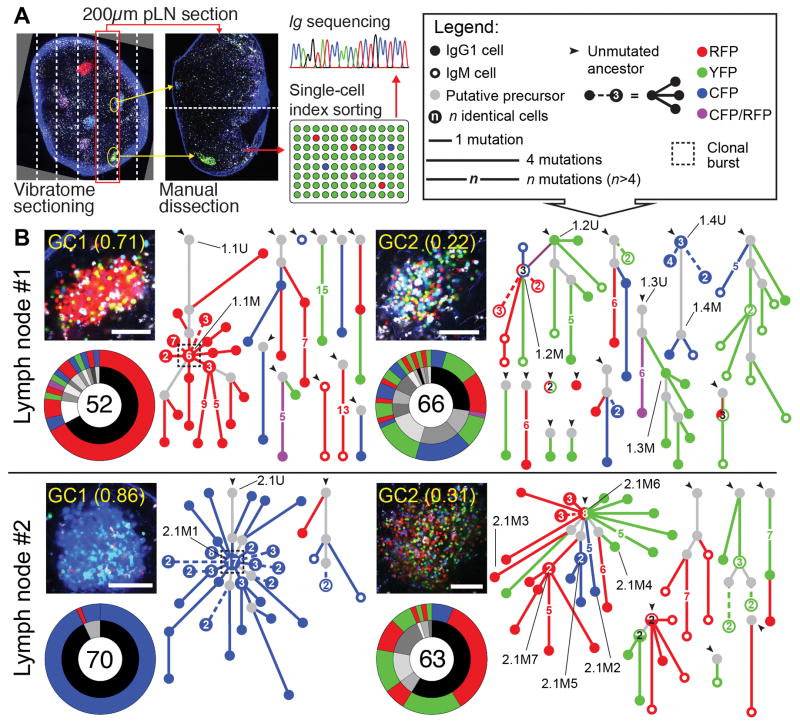
Figure 4. Clonal relationships among cells obtained from GCs with high or low color dominance.
(A) Method used to obtain Ig sequences. (B) Igh sequence relationship among B cells from 2 pairs of individual GCs from 2 pLNs of different mice, obtained 10 days after tamoxifen administration (15 days post-immunization), as described in Fig. 3. Each panel contains: top left, multiphoton image (scale bar, 100 μm, cell colors as in fig. S1B, second harmonic generation from collagen fibers is shown in blue); bottom left, clonal distribution pie-chart (with clones represented in grayscale in the inner ring and Confetti colors in the outer ring; number of cells sequenced is indicated in the center); and right, trees representing the phylogeny of Ig heavy-chain V-segment sequences within each clone (symbols according to the legend in the top right corner). Dashed lines within phylogenies indicate multiple variants distanced the same number of mutations from the originating node. IDs of variants for which affinity was measured in Fig. 5 are indicated by black lines. For each LN, GC1 and GC2 were considered as displaying high and low color dominance, respectively
We obtained Igh sequences from 52 to 74 single cells per GC from 3 pairs of pLN GCs harvested 10 days after tamoxifen treatment (15 days after immunization with CGG-alum) (Fig. 4B and fig. S9A). Comparison of SHM levels between high- and low-dominance GCs from the same LN (fig. S9B) and between Aicda-sufficient and haploinsufficient GCs (fig. S9C) indicated that heterogeneity was not the consequence of asynchronous GC formation or of substantially impaired AID activity. Most cells of the dominant color in the 3 high-dominance GCs (Fig. 4B, and fig. S9A, left panels) derived from a single expanded clone. In all 3 GCs, dominance could be readily attributed to the selective expansion of a single SHM variant, 3 to 5 VH mutations (4 to 9 total mutations) distanced from the unmutated ancestor (UA) (dashed boxes in Fig. 4B and fig. S9A). Thus, high-dominance GCs are predominantly monoclonal, and appear to result from “clonal bursts,” in which a single SHM variant is heavily expanded over a short period of time, leading to extensive loss of clonal diversity concomitant with broad diversification of the expanded variant. On the other hand, low-dominance GCs (Fig. 4B and fig. S9A, right panels) were more clonally diverse than their high-dominance neighbors. Low color dominance is unlikely to be the result of failure to identify a dominant nonfluorescent lineage, since fluorescent and nonfluorescent B cells obtained from two independently-sequenced low-dominance GCs were largely clonally related (fig. S10). Although low-dominance GCs consisted predominantly of small independent expansions, one of three GCs sequenced (LN2/GC2) contained a larger clone (accounting for 59% of all recombined cells), within which multiple colors were represented. This expanded clone carried heavy and light chain V(D)J rearrangements identical to the dominant clone in the single-colored GC sequenced from the same LN (LN2/GC1), indicative of a common cell of origin. In contrast to the clonal bursts of single-colored GCs, however, this multicolored expansion branched off from the Igh UA itself, and developed in parallel along several distinct lineages. Thus, expansion of this clone began in the pre/early GC period, prior to the end of the tamoxifen pulse, and subsequent competition in the mature GC failed to focus on a single dominant SHM variant. Together, these data indicate that, while clonal dominance in GCs can arise by parallel expansion of members of the same clone, loss of diversity is greatest in GCs in which strong clonal bursts rapidly expand and diversify single SHM variants.
Affinity maturation in the absence of homogenizing selection
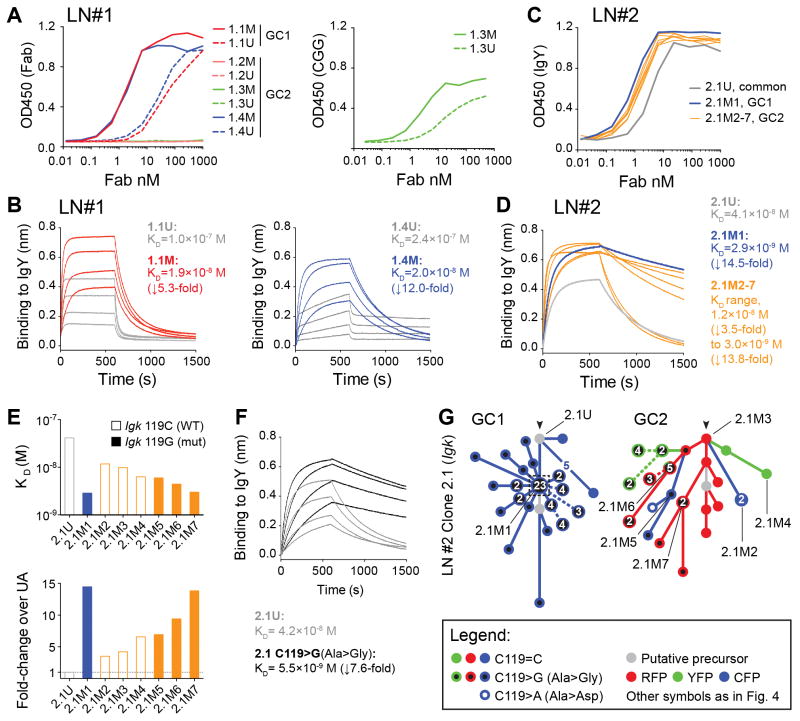
To investigate the relationship between clonal bursting and affinity maturation, we measured the affinity of recombinant Fab fragments derived from B cells originating from high- and low-dominance GCs (Fig. 4B), as well as from their deduced or sequenced UAs. Fab affinities varied markedly between clones, from undetectable to low nanomolar KD (Fig. 5). As expected, the two Fabs obtained from SHM variants that underwent clonal bursts had higher affinity for the immunizing antigen than their respective UAs (5.3-fold increase (1.0×10−7 to 1.9×10−8) for clone 1.1 and 14.5-fold increase (4.1×10−8 to 2.9×10−9) for clone 2.1, Fig. 5). However, affinity maturation was also evident in Fabs cloned from low-dominance GCs. Of the three clones obtained from LN1/GC2, SHM variants of two clones (1.3 and 1.4) showed increased affinity over their UAs, whereas the affinity of the third clone (1.2) remained below detection level (Fig. 5). Clone 1.4 showed a gain in affinity of 12-fold (2.4×10−7 to 2.0×10−8), notably larger than the 5.4-fold change observed for clone 1.1 (associated with a clonal burst in the neighboring single-colored GC). We also determined the affinity of six additional variants of clone 2.1 from LN2/GC2 (which shared its UA with the dominant clone of LN2/GC1, and are thus directly comparable between GCs). Gains in affinity were evident in all of these variants, ranging from 3.5 to 13.8 fold (KD 1.2×10−8 and 3.0×10−9, respectively) (Fig. 5C–E). Although the highest of these was close in magnitude to the 14.5-fold change seen in variant 2.1M1 (associated with the clonal burst in LN2/GC1), the increase in affinity among GC2 Fabs was in general lower (median increase of 6.8-fold). Thus, affinity maturation can occur in the absence of clonal bursts, although limited data from the two LN2 GCs suggests that selection of high-affinity mutants in these cases may not be as efficient.
Figure 5. Affinity maturation in GCs with high or low color dominance.
Affinity measurements for reconstructed Fabs derived from B cell clones/variants indicated in Fig. 4B. (A) Binding of Fabs cloned from LN1 to IgY (right) or CGG (left), measured by ELISA. (B) Bio-layer interferometry for Fabs cloned from LN1 binding to IgY. (C) and (D) as in (A) and (B), respectively, but using Fabs cloned from LN#2. (E) Affinity for IgY among variants of clone 2.1 (Fig. 4B) from LN2/GC1 (blue) and LN2/GC2 (orange), shown as KD (top) and fold-change over UA (bottom). The unmutated ancestor (2.1U) is shown in gray. Open bars have the WT nucleotide (C) in the 119 position, closed bars have the C119>G (Ala>Gly) mutation. (F) Affinity of Fabs reconstructed from clone 2.1, either unmutated (gray lines) or with replacement of a single Igk nucleotide (C119>G (Ala>Gly), black lines). (C) Igk sequence relationships among B cells from clone 2.1 from LN2/GC1 (left) or LN2/GC2 (right). Symbols according to the legend below the figure and in Fig. 4B. Cloned Fab IDs are indicated by black lines. Note that, although not all cells yielded both Igk and Igh sequences, clonal relationships are drawn from all available data, and therefore exact correspondence between the trees in panel (G) and in Fig. 4B is not expected. Bio-layer interferometry was performed with Fabs at 20, 40, 80, and 160 nM. Panel (D) shows only the 160 nM measurement. Reported affinities are the average of two measurements fitted globally at the 20–160 nM range.
Finally, detailed analysis of the SHM trajectory of variant 2.1M1 showed that this burst was associated with a somatic mutation in Igk (C119>G, Ala>Gly) that alone increased the Fab affinity by 7.6-fold (Eig. 5E-G). This mutation was also found to have occurred independently at least twice in LN2/GC2, but without the accompanying burst. Thus, identical mutations occurring within the same clone may still have distinct outcomes when in different GCs, again suggesting a role for stochastic factors in GC selection.
Conclusions
Using a combination of imaging methods and single-cell sequencing, we show that (i) early GCs can be highly diverse, containing up to hundreds of distinct B cell clones; (ii) individual GCs approach homogeneity at different rates, leading to variable loss of diversity; (iii) homogeneous GCs are the product of “clonal bursts” that lead to rapid expansion and further diversification of SHM variants with improved affinity; and (iv) affinity maturation can take place in the presence or absence of such bursts, and thus does not necessarily require radical loss of clonal diversity.
We speculate that affinity maturation arises through a balance between slow but steady elimination of lower affinity clones and variants, punctuated by sporadic clonal bursts of various magnitudes that generate a wealth of SHM variants of a clone, at the expense of clonal diversity. This balance would ensure that the polyclonality of the GC response is maintained, at the same time as very successful clones can be heavily diversified and exported to effector or memory fates, generating the range of GC outcomes observed by us and others (14, 27). Although it cannot be ruled out that differences in outcomes between GCs may be related to factors such as initial clonal composition or timing of the appearance of key mutations, the finding that GCs can deviate substantially from baseline color distribution even when all cells have equal affinity and cannot undergo SHM (Fig. 3F–G and fig. S6D) suggests that stochastic factors unrelated to affinity can strongly influence GC selection. A system in which B cell selection is driven by dynamic encounters with a limiting number of T helper cells would be especially sensitive to such effects (9, 27). The explosive B cell proliferation that can be driven by such encounters would provide a mechanistic basis for the clonal burst phenomenon (28, 29).
The finding that early GCs can contain a large diversity of clones is in contrast to the widely held view that these structures form pauciclonally (10–12, 30). Although this discrepancy may be partly attributable to methodological issues (31), Kuppers et al. (13), reported higher clonal diversity in one of two GCs originating from different reactive human lymph nodes, and speculated that pauciclonal GCs were likely the product of extensive selection acting on originally polyclonal early GCs. Our current data strongly support this view.
Our findings have implications for the design of vaccines against highly variable pathogens such as influenza and HIV, where broadly-protective antibodies targeting conserved, non-immunodominant epitopes must be elicited (32, 33). Understanding whether GC competition restricts the appearance of such clones, and how clonal bursts may promote the somatic diversification required for broad neutralization to emerge may prove key to the successful generation of broadly-protective antibodies by vaccination.
Supplementary Material
Acknowledgments
We thank M. Carroll, A. Schmidt and S. Harrison (Harvard Medical School), H. Wardemann (German Cancer Research Center), and K. Hasenkrug (NIAID) for essential reagents, and Robert K. Colwell (U. Connecticut) and Sebastian Binder (Helmholtz Centre for Infection Research) for help with statistical analysis. We are especially thankful to the late Herman N. Eisen (1918–2014) for the lengthy discussions of affinity maturation on which the present study is based. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. Antibody sequences are also available from the GenBank database, accession numbers KU613419 to KU615568. This work was supported by NIH grant 5DP5OD012146 (GDV), with additional support from Human Frontier Science Program Grant (RGP0033/2015) (GDV and MMH), German Federal Ministry of Education and Research within the Measures for the Establishment of Systems Medicine, eMed project SYSIMIT, FKZ: 01ZX1308B (MMH), Swiss National Science Foundation Postdoc Mobility Fellowship and Cancer Research Institute Irvington Postdoctoral Fellowship (GP), and Norwegian Research Council FRIPRO mobility grant (JTJ). The Octet RED96 Bio-Layer Interferometry System was supported by NIH grant S10 OD016326 to the MIT Biophysical Instrumentation Facility.
Footnotes
Materials and Methods
References and Notes
- 1.Eisen HN, Siskind GW. Variations in Affinities of Antibodies During the Immune Response. Biochemistry. 1964 Jul;3:996. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- 2.Eisen HN. Affinity enhancement of antibodies: how low-affinity antibodies produced early in immune responses are followed by high-affinity antibodies later and in memory B-cell responses. Cancer Immunol Res. 2014 May;2:381. doi: 10.1158/2326-6066.CIR-14-0029. [DOI] [PubMed] [Google Scholar]
- 3.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 4.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996 Jun 27;381:751. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 5.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007 Aug;27:190. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarlinton DM. Evolution in miniature: selection, survival and distribution of antigen reactive cells in the germinal centre. Immunol Cell Biol. 2008 Feb;86:133. doi: 10.1038/sj.icb.7100148. [DOI] [PubMed] [Google Scholar]
- 7.Victora GD, Nussenzweig MC. Germinal centers. Annual review of immunology. 2012;30:429. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 8.Victora GD, Mesin L. Clonal and cellular dynamics in germinal centers. Current opinion in immunology. 2014 Jun;28:90. doi: 10.1016/j.coi.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Victora GD, et al. Germinal Center Dynamics Revealed by Multiphoton Microscopy with a Photoactivatable Fluorescent Reporter. Cell. 2010 Nov 12;143:592. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacob J, Kassir R, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. I. The architecture and dynamics of responding cell populations. J Exp Med. 1991 May 1;173:1165. doi: 10.1084/jem.173.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroese FG, Wubbena AS, Seijen HG, Nieuwenhuis P. Germinal centers develop oligoclonally. Eur J Immunol. 1987 Jul;17:1069. doi: 10.1002/eji.1830170726. [DOI] [PubMed] [Google Scholar]
- 12.Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol. 1991 Dec;21:2951. doi: 10.1002/eji.1830211209. [DOI] [PubMed] [Google Scholar]
- 13.Kuppers R, Zhao M, Hansmann ML, Rajewsky K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. Embo J. 1993 Dec 15;12:4955. doi: 10.1002/j.1460-2075.1993.tb06189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziegner M, Steinhauser G, Berek C. Development of antibody diversity in single germinal centers: selective expansion of high-affinity variants. European journal of immunology. 1994 Oct;24:2393. doi: 10.1002/eji.1830241020. [DOI] [PubMed] [Google Scholar]
- 15.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007 Nov 1;450:56. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 16.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010 Oct 1;143:134. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 17.note1, See Supplementary Materials and Methods.
- 18.Shulman Z, et al. T follicular helper cell dynamics in germinal centers. Science. 2013 Aug 9;341:673. doi: 10.1126/science.1241680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob J, Przylepa J, Miller C, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J Exp Med. 1993 Oct 1;178:1293. doi: 10.1084/jem.178.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008 Dec 12;135:1028. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotelli NJ, Colwell RK. In: Biological Diversity: Frontiers in Measurement and Assessment. Magurran AE, McGill BJ, editors. Oxford University Press; Oxford: 2010. pp. 39–54. [Google Scholar]
- 22.Dogan I, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009 Dec;10:1292. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 23.Jarjour M, et al. Fate mapping reveals origin and dynamics of lymph node follicular dendritic cells. The Journal of experimental medicine. 2014 Jun 2;211:1109. doi: 10.1084/jem.20132409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominguez-Sola D, et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nature immunology. 2012 Nov;13:1083. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.note2, See Supplementary Materials and Methods for mathematical validation of the NDS.
- 26.Nair S, et al. Distinct roles of CD4+ T cell subpopulations in retroviral immunity: lessons from the Friend virus mouse model. Retrovirology. 2011;8:76. doi: 10.1186/1742-4690-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radmacher MD, Kelsoe G, Kepler TB. Predicted and inferred waiting times for key mutations in the germinal centre reaction: evidence for stochasticity in selection. Immunology and cell biology. 1998 Aug;76:373. doi: 10.1046/j.1440-1711.1998.00753.x. [DOI] [PubMed] [Google Scholar]
- 28.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014 May 29;509:637. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gitlin AD, et al. T cell help controls the speed of the cell cycle in germinal center B cells. Science. 2015 Jul 16; doi: 10.1126/science.aac4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob J, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. II. A common clonal origin for periarteriolar lymphoid sheath-associated foci and germinal centers. J Exp Med. 1992 Sep 1;176:679. doi: 10.1084/jem.176.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faro J, Or-Guil M. How oligoclonal are germinal centers? A new method for estimating clonal diversity from immunohistological sections. BMC Bioinformatics. 2013;14(Suppl 6):S8. doi: 10.1186/1471-2105-14-S6-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Victora GD, Wilson PC. Germinal Center Selection and the Antibody Response to Influenza. Cell. 2015 Oct 22;163:545. doi: 10.1016/j.cell.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein F, et al. Antibodies in HIV-1 vaccine development and therapy. Science. 2013 Sep 13;341:1199. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010 Jan;13:133. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995 Sep 8;269:1427. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 36.Mason DY, Jones M, Goodnow CC. Development and follicular localization of tolerant B lymphocytes in lysozyme/anti-lysozyme IgM/IgD transgenic mice. International immunology. 1992 Feb;4:163. doi: 10.1093/intimm/4.2.163. [DOI] [PubMed] [Google Scholar]
- 37.Shih TA, Roederer M, Nussenzweig MC. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat Immunol. 2002 Apr;3:399. doi: 10.1038/ni776. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt AG, et al. Viral receptor-binding site antibodies with diverse germline origins. Cell. 2015 May 21;161:1026. doi: 10.1016/j.cell.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halemano K, et al. Humoral immunity in the Friend retrovirus infection model. Immunologic research. 2013 Mar;55:249. doi: 10.1007/s12026-012-8370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dietze KK, et al. Combining regulatory T cell depletion and inhibitory receptor blockade improves reactivation of exhausted virus-specific CD8+ T cells and efficiently reduces chronic retroviral loads. PLoS pathogens. 2013;9:e1003798. doi: 10.1371/journal.ppat.1003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Browne EP. Toll-like receptor 7 inhibits early acute retroviral infection through rapid lymphocyte responses. Journal of virology. 2013 Jul;87:7357. doi: 10.1128/JVI.00788-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009 Apr 17;324:392. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007 Jan 26;315:528. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 44.Shulman Z, et al. T follicular helper cell dynamics in germinal centers. Science. 2013 Aug 9;341:673. doi: 10.1126/science.1241680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trombetta JJ, et al. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr Protoc Mol Biol. 2014;107:4 22 1. doi: 10.1002/0471142727.mb0422s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiller T, Busse CE, Wardemann H. Cloning and expression of murine Ig genes from single B cells. J Immunol Methods. 2009 Oct 31;350:183. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic acids research. 2009 Jan;37:D1006. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Retter I, Althaus HH, Munch R, Muller W. VBASE2, an integrative V gene database. Nucleic acids research. 2005 Jan 1;33:D671. doi: 10.1093/nar/gki088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasqual G, Angelini A, Victora GD. Triggering positive selection of germinal center B cells by antigen targeting to DEC-205. Methods in molecular biology. 2015;1291:125. doi: 10.1007/978-1-4939-2498-1_10. [DOI] [PubMed] [Google Scholar]
- 50.Chao A. Nonparametric-Estimation of the Number of Classes in a Population. Scand J Stat. 1984;11:265. [Google Scholar]
- 51.Colwell RK, et al. Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. J Plant Ecol-Uk. 2012 Mar;5:3. [Google Scholar]
- 52.Meyer-Hermann M, et al. A theory of germinal center B cell selection, division, and exit. Cell reports. 2012 Jul 26;2:162. doi: 10.1016/j.celrep.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 53.Meyer-Hermann M. Overcoming the dichotomy of quantity and quality in antibody responses. Journal of immunology. 2014 Dec 1;193:5414. doi: 10.4049/jimmunol.1401828. [DOI] [PubMed] [Google Scholar]
- 54.Gu H, Tarlinton D, Muller W, Rajewsky K, Forster I. Most Peripheral B-Cells in Mice Are Ligand Selected. Journal of Experimental Medicine. 1991 Jun 1;173:1357. doi: 10.1084/jem.173.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boersch-Supan ME, Agarwal S, White-Scharf ME, Imanishi-Kari T. Heavy chain variable region. Multiple gene segments encode anti-4-(hydroxy-3-nitro-phenyl)acetyl idiotypic antibodies. The Journal of experimental medicine. 1985 Jun 1;161:1272. doi: 10.1084/jem.161.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.