Abstract
Secretins are versatile outer membrane pores used by many bacteria to secrete proteins, toxins, or filamentous phages; extrude type IV pili (T4P); or take up DNA. Extrusion of T4P and natural transformation of DNA in the thermophilic bacterium Thermus thermophilus requires a unique secretin complex comprising six stacked rings, a membrane-embedded cone structure, and two gates that open and close a central channel. To investigate the role of distinct domains in ring and gate formation, we examined a set of deletion derivatives by cryomicroscopy techniques. Here we report that maintaining the N0 ring in the deletion derivatives led to stable PilQ complexes. Analyses of the variants unraveled that an N-terminal domain comprising a unique βββαβ fold is essential for the formation of gate 2. Furthermore, we identified four βαββα domains essential for the formation of the N2 to N5 rings. Mutant studies revealed that deletion of individual ring domains significantly reduces piliation. The N1, N2, N4, and N5 deletion mutants were significantly impaired in T4P-mediated twitching motility, whereas the motility of the N3 mutant was comparable with that of wild-type cells. This indicates that the deletion of the N3 ring leads to increased pilus dynamics, thereby compensating for the reduced number of pili of the N3 mutant. All mutants exhibit a wild-type natural transformation phenotype, leading to the conclusion that DNA uptake is independent of functional T4P.
Keywords: EM, protein domain, protein folding, structure/function, type IV pili, Thermus thermophilus, electron cryotomography, natural transformation, ring and gate forming domains, secretin
Introduction
Secretins in the outer membrane of Gram-negative bacteria are key components for the assembly of type IV pili (T4P)2 machineries or type II and type III secretion systems (T2SS and T3SS, respectively) and are required for the export of filamentous phages (1–4). Secretin pores can determine bacterial pathogenicity by their role in T4P extrusion, which is crucial for bacterial adherence and motility on solid surfaces, termed twitching motility (5–7). Natural transformation systems in bacteria, mediating the uptake of free DNA, are also dependent on secretin proteins (8, 9). The latter contribute significantly to the acquisition and distribution of antibiotic resistance, which is an emerging threat to human health (10).
A wide range of secretins have been discovered in different bacteria. Some of them have been subject to biochemical and structural characterization, revealing significantly different architectures. Distinct features of secretin complexes are proposed because of their unique functions and corresponding interaction partners and substrates (1, 2, 11–15). Important structural information has been derived from electron cryomicroscopy of isolated GspD, the T2SS secretin from Vibrio cholerae. GspD forms a 12-fold symmetrical cylinder with a diameter of 155 Å and a length of 200 Å (16, 17). The conserved β sheet-rich C-terminal domains of GspD form a channel through the outer membrane that is closed by an extracellular and a periplasmic gate (2, 17). The periplasmic N-terminal domains N0, N1, N2, and N3 were mapped to the density of the wall of the periplasmic chamber observed by single-particle EM (SP-EM). This revealed that the N3 domain occupies a constriction site that was suggested to be important for the initiation of conformational changes during protein secretion (16).
Recently, we reported the first in situ structure of an entire T4P machinery by electron cryotomography (cryo-ET) using the thermophilic bacterium Thermus thermophilus HB27 as a model organism (18). The central membrane-embedded part of the structure is formed by the secretin PilQ, which plays a dual role in T4P extrusion and natural transformation (9, 18–20).
The secretin complex is suggested to guide the DNA through the outer membrane and through the periplasmic space (20). DNA transport requires dynamics of a pseudopilus comprising different pilins (21–23). This pseudopilus is suggested to be powered by a zinc-containing polymerization ATPase, PilF, which also powers T4P extrusion (24–27). In addition to PilF, several unique membrane-associated proteins, such as PilC, PilM, PilN, and PilO, are suggested to be implicated in polymerization of the pseudopilus (9, 23, 28–30). Interestingly, T4P-mediated twitching motility requires two retraction ATPases, designated PilT1 and PilT2, that are dispensable for natural transformation (24).
PilQ was found to undergo substantial conformational changes between the closed and the open, pilus-extruding state (18). SP-EM analyses of purified PilQ complexes revealed that the structure comprises six stacked rings (N0-N5) and a cone structure (20), consistent with the in situ data of the entire T4P machinery (18). Structural analyses of PilQ complexes formed by different PilQ variants led to the identification of an unusual ααβαββα fold as the N0 ring-forming domain. Production of the truncated PilQ variant in a pilQ-negative background led to a mutant that was non-piliated and non-motile but still naturally transformable (19), leading to the conclusion that pilus structures are not required for natural transformation. However, identification of other ring- and gate-forming domains was not possible because of instability of the PilQ variants. Moreover, the assignment of specific functions to distinct ring structures in secretins has not been reported so far. Here we found that maintaining the N0 ring domain in PilQ deletion derivatives leads to stable PilQ complexes, which we used to perform a thorough structure/function analysis that unraveled one of the gate-forming and four ring-forming domains. We further demonstrated that the individual rings play separate roles in T4P extrusion and T4P dynamics but are dispensable for natural transformation. These results demonstrate that individual ring structures within the secretin complex exhibit distinct functions during T4P extrusion.
Results
The Four Conserved βαββα Folds and the Preceding βββαβ domain Are Not Essential for PilQ Complex Formation
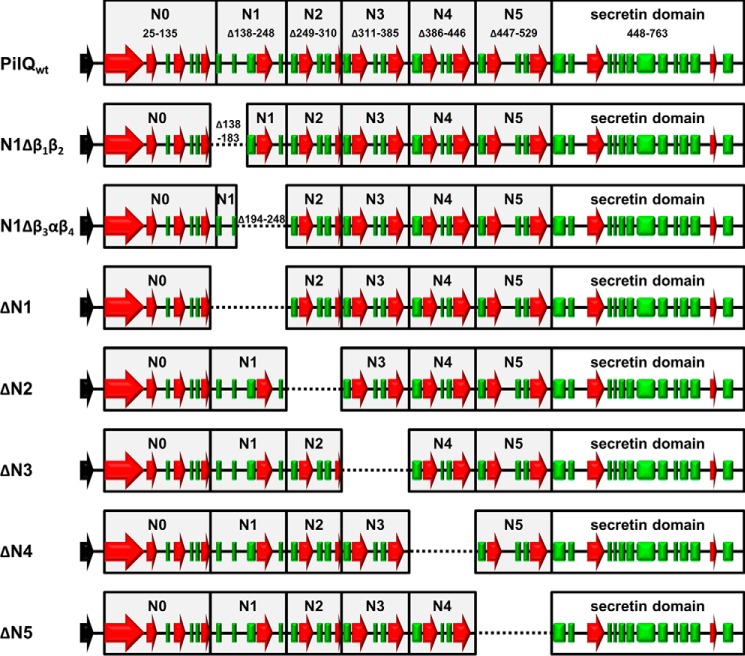
Secondary structure analyses revealed that there are four contiguous βαββα folds (N2 to N5) in the secretin PilQ from T. thermophilus, suggested to form ring-building motifs (20). To analyze the role of these motifs in ring formation, we generated a set of internal PilQ deletion variants (ΔN2, ΔN3, ΔN4, and ΔN5) that were produced in a T. thermophilus pilQ deletion background (Fig. 1). Western blotting analyses of membrane fractions revealed that all variants assembled into SDS-stable PilQ complexes (Fig. 2), demonstrating that deletion of individual βαββα domains does not abolish complex formation.
FIGURE 1.
Conserved folds and localization of deletions in PilQ. The predicted ring forming domains (N0-N5) are indicated in boxes. The α-helical domains are indicated by red arrows, and β sheets are shown as green boxes.
FIGURE 2.
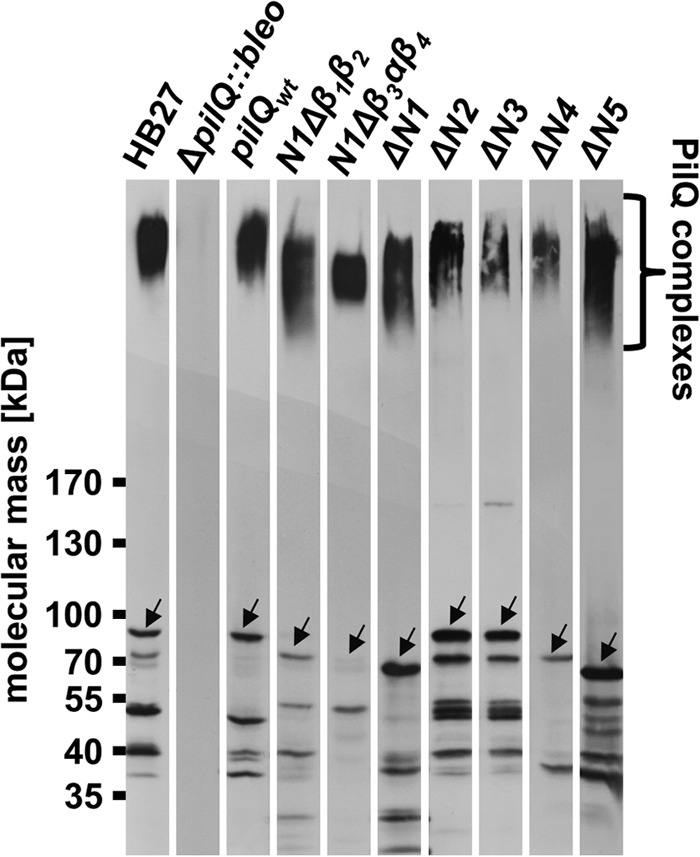
Detection of truncated PilQ complexes. Total membranes (50 μg of protein) of the T. thermophilus HB27 wild type and membranes of pilQwt, ΔpilQ::bleo, N1Δβ1β2, N1β3αβ4, ΔN1, ΔN2, ΔN3, ΔN4, and ΔN5 mutants were separated by 3–12% SDS-PAGE and subjected to Western blotting analysis using polyclonal PilQ antibodies (1:10,000 dilution). The full-length PilQ monomers are indicated by arrows.
The first βαββα fold (N2 domain) is preceded by an unusual βββαβ fold (N1 domain) (Fig. 1). To elucidate the role of this domain in ring formation, we generated a PilQ deletion derivative devoid of either the entire N1 domain (Δβ1β2β3αβ4) or N1 subdomains (N1Δβ1β2 or N1Δβ3αβ4) (Fig. 1). These deletion derivatives also assembled into macromolecular PilQ complexes (Fig. 2), providing clear evidence that the N1 domain is also dispensable for complex assembly.
Identification of the Ring-forming Domains
To unequivocally identify the ring-forming domains in PilQ, His-tagged deletion derivatives were produced in T. thermophilus and purified by affinity and anion exchange chromatography. SDS-PAGE analyses led to the detection of high molecular weight PilQ complexes (Fig. 3). Different size PilQ complexes and varying amounts of monomers were detected for most of the purified deletion derivatives (Fig. 3). The highest amount of monomer accumulation was detected in preparations of PilQ deletion derivatives devoid of the N1 domain and the N1Δβ1β2 subdomain. This leads to the conclusion that, in particular, the N1β1β2 plays a major role in PilQ complex stability.
FIGURE 3.
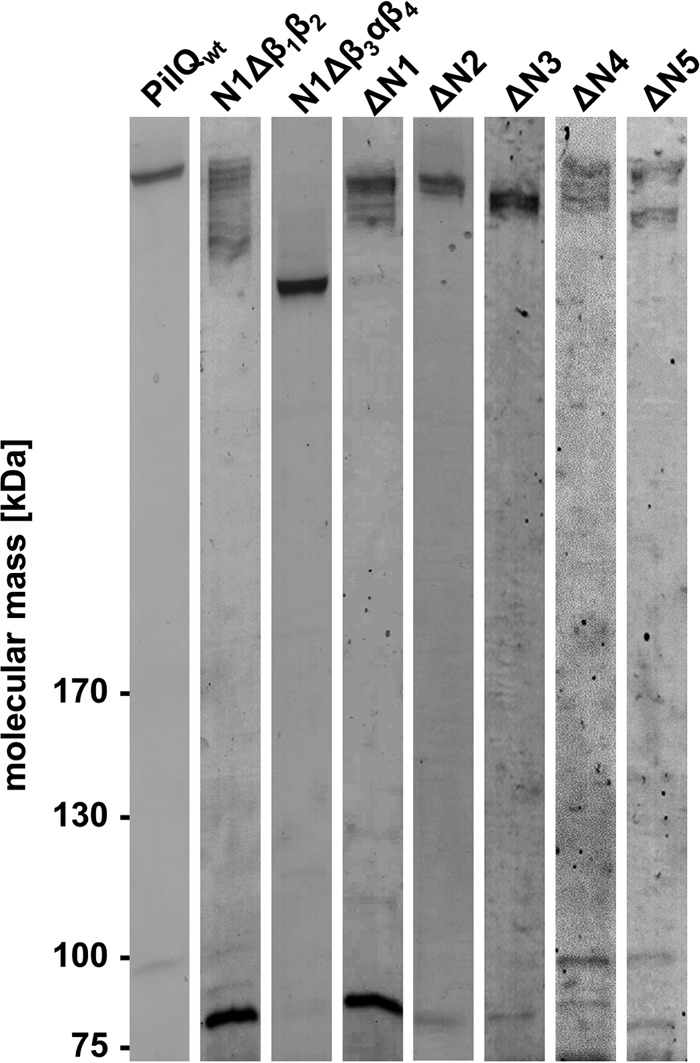
Analysis of PilQ complexes formed by PilQ deletion derivatives. Purified PilQ complexes were separated by 3–12% SDS-PAGE, and proteins were stained using Instant Blue. Each lane contained 2 μg of purified protein.
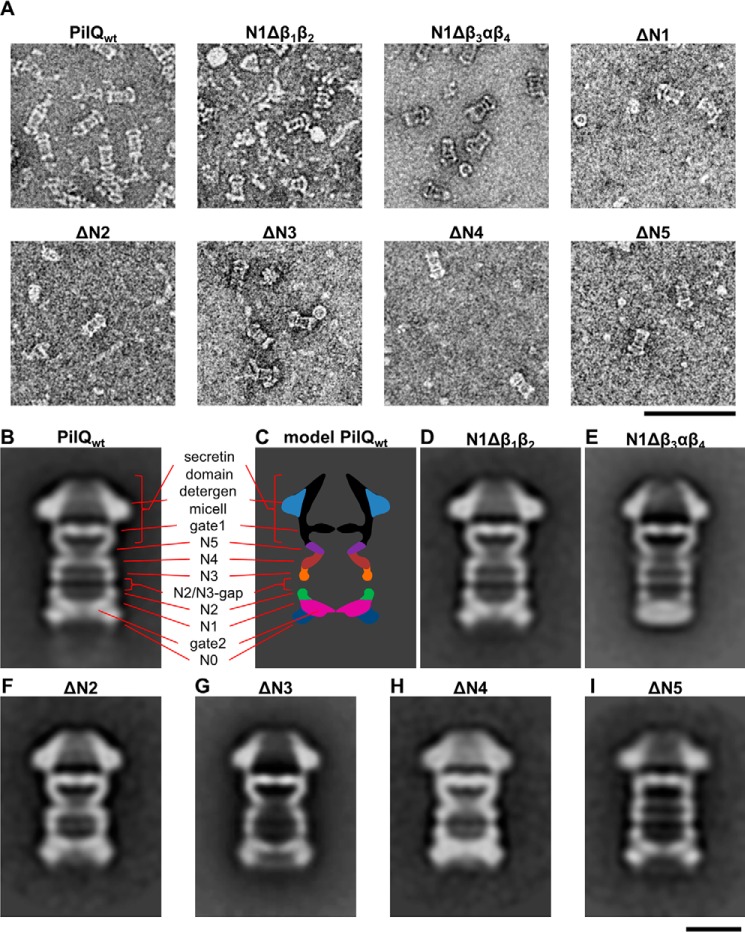
SP-EM studies of PilQ showed six rings and a cone domain that were also seen in situ by cryo-ET (18–20). To analyze the effect of the domain deletions on the structure of PilQ, we analyzed the purified deletion derivatives by SP-EM (Fig. 4A). In all cases, with the exception of the ΔN1 mutant, the amount of protein complex was sufficient for SP-EM analysis (Fig. 4A). The overall shape of the complex was comparable with the in situ cryo-ET structure of the PilQ complex. Deletion of the first subdomain of N1 (N1Δβ1β2) did not result in an obvious change in the structure compared with the wild type (Fig. 4D). In contrast, deletion of the second part of N1 (N1Δβ3αβ4) changed the structure dramatically (Fig. 4E). The periplasmic end of the complex, corresponding to the N0, N1, and N2 rings, was significantly narrower. The N0 ring, which is the widest ring in the wild-type complex (Fig. 4, B and C), was no longer visible. In the wild-type structure, a second dense domain was observed close to the N0 ring that extends into the center of the complex, forming gate 2 (Fig. 4, B and C). This gate is still clearly present in PilQ complexes devoid of the partial deletions in the N1 domain. The N1 ring is followed by two further rings (N2 and N3) that are similar in size and are separated by a gap of ∼12 Å, designated as the N2/N3 gap (Fig. 4, B and C). The ΔN2 and ΔN3 variants were structurally similar, with one ring and the N2/N3 gap missing (Fig. 4, F and G). Deletion of the βαββα domain downstream of the N3 domain led to loss of the N4 ring, confirming its position in the complex (Fig. 4, C and H). The C-terminal part of all variants as well as the wild type formed a trapezoid (“cone structure”) that shows a strong central density (Fig. 4B). Below the cone is a “cup” structure with a shape characteristic of the secretin domain. Based on its position, the lower part of the cone is likely to be gate 1, as identified by cryo-ET and subtomogram averaging (Fig. 4, B and C) (18). In all ring variants, the secretin domain and gate 1 were clearly visible (Fig. 4, B–I), suggesting that a specific motif in the secretin domain is responsible for forming this gate. The narrow ring beneath gate 1 in the cup is missing in the ΔN5 mutant complex (Fig. 4I), indicating that it is formed by the βαββα domain in N5 (Fig. 4I).
FIGURE 4.
SP-EM analysis of the PilQ wild-type complex and the PilQ mutant complexes. A, purified PilQ complexes were negatively stained with 2% ammonium molybdate and analyzed by EM. Images were taken at high defocus (−1.5–2.5 μm) to give an overview of the samples. In all cases, the purifications yielded protein complexes that did not aggregate. However, the number and shape of the complexes assembled from the deletion derivatives varied considerably. Scale bar = 100 nm. B and D—I, reference-free class averages of a dataset of 9896 (PilQwt), 6648 (N1Δβ1β2), 7109 (N1Δβ3αβ4), 1568 (ΔN2), 5000 (ΔN3), 2028 (ΔN4), and 1974 (ΔN5) particle images revealed PilQ complex formation. Representative class averages are shown for each PilQ complex. The number of particles images per class for PilQwt was 726, for N1Δβ1β2 949, for N1Δβ3αβ4 726, for ΔN2 341, for ΔN3 900, for ΔN4 and ΔN5 439, respectively. C, a model of the PilQwt complex is shown. Scale bar = 10 nm.
Identification of the Gate 2-forming Domain
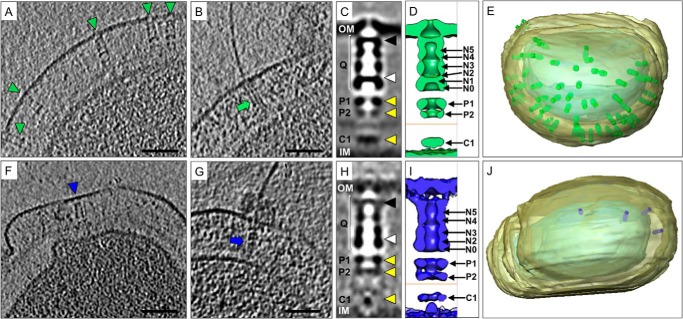
Because of the poor stability of the ΔN1 mutant, it was not suitable for SP-EM analysis. To investigate the effect of the N1 mutation, the intact T4P machinery composed of PilQ and accessory proteins was analyzed in situ by cryo-ET and subtomogram averaging. The assembled machinery in the closed state (without pili) was identified in whole cells (Fig. 5F) and detected exclusively at the cell poles, in agreement with previous data (18). However, the number of complexes was severely reduced compared with wild-type cells (Fig. 5, A, E, F, and J), probably as a result of reduced protein stability. Closed-state complexes were selected, aligned, and averaged to determine the three-dimensional structure of the ΔN1 mutant in its native membrane-bound state. Comparison with the wild-type complex (Fig. 5, C and D) shows that the N1 domain is responsible for the formation of gate 2 (Fig. 5, H and I). The outer membrane-embedded secretin domain was poorly resolved in the average, most likely because of the inherent flexibility of the complex and the reduced number of particles available. Removal of gate 2 did not affect the ability of PilQ to assemble with accessory proteins (designated P1, P2, and C1) into the entire T4P machinery (Fig. 5, C, D, H, and I). In rare cases, cells lacking gate 2 in PilQ were also found to assemble pili (Fig. 5G). However, the shape of the PilQ complex was abnormal compared with the wild type (compare Fig. 5, B and C), and it was associated with extracellular material, possibly from cell leakage (Fig. 5B). Conventional Fourier shell correlation, applying the 0.5 FSC criterion, are presented in Fig. 6.
FIGURE 5.
Subtomogram averaging of the in situ T4P machinery from wild-type and pilQΔN1 mutant cells. A–J, representative data from wild-type cells (A–E) and from the ΔN1 mutant (F–J). Tomographic slices of T. thermophilus wild-type cells (A and B) and ΔN1 mutant cells (F and G) show the T4P machinery in the closed (A and F, blue and green arrowheads) and open, pilus-assembled (B and G, blue and green arrows) state. Scale bars = 50 nm. Subtomogram averaging reveals the three-dimensional structure of the wild-type (C) and ΔN1 (H) closed-state complexes. PilQ (Q) is indicated with white brackets, the position of gate 1 with a black arrowhead, gate 2 with a white arrowhead, and the positions of accessory proteins P1, P2, and C1 with yellow arrowheads. The outer membrane (OM) and inner membrane (IM) are also shown. D and I, the corresponding averages are shown as surface representations. The position of the peptidoglycan layer is indicated with an orange line, and the positions of the ring domains and accessory proteins are also shown. E and J, docking the subtomogram averages back into the corresponding tomographic volumes reveals the number and distribution of T4P complexes in whole cells.
FIGURE 6.
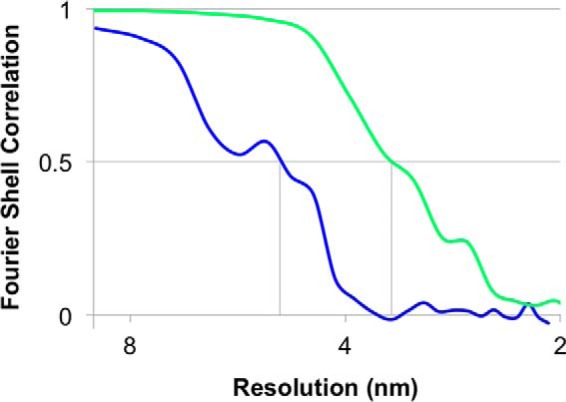
FSC curves for subtomogram averages. Resolution estimates were based on conventional FSC measurements and the 0.5 criterion in IMOD. Calculations for the closed state of the ΔN1 mutant (blue, 50 Å) is shown with the wild-type complex (green, 35 Å) for comparison. Data for the wild-type complex have been shown previously (18).
The Ring-forming Domains Are Important for Pilus Extrusion
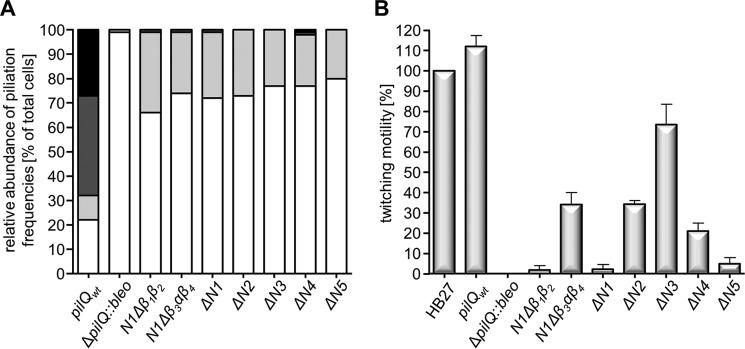
To analyze the role of the ring-forming domains in pilus biogenesis, the degree of piliation in different pilQ deletion mutants was analyzed in whole T. thermophilus cells by EM. Seventy-eight percent of wild-type cells were piliated with 6 ± 3 pili/cell, whereas only 1% of the ΔpilQ::bleo mutant cells were piliated with one pilus at the cell surface, mostly on disrupted cells (Fig. 7A), in agreement with previous data (19). All PilQ variants could extrude pili; however, the percentage of piliated cells decreased significantly to 20–34% (Fig. 7A). Moreover, the number of pili per cell decreased dramatically (Fig. 7A). In the wild-type cells, only 10% showed less than three pili, whereas 41% exhibited four to seven pili and 27% more than seven. In contrast, the piliated mutant cells extruded only one to three pili (Fig. 7A).
FIGURE 7.
Evaluation of the piliation and twitching motility data. A, piliation of the pilQ mutants. In total, 300–350 cells/strain were analyzed, and this value was set to 100%. The percentage of non-piliated cells is shown in white, cells exhibiting one to three pili are colored light gray, cells exhibiting four to seven pili are shown in dark gray, and cells exhibiting more than eight pili are shown in black. B, the twitching motility of the pilQ mutants. The diameter of the twitching zones of wild-type T. thermophilus HB27 was set to 100%.
Twitching Motility of the pilQ Mutants
To test the role of the N1-N5 domains and the N1 subdomains (N1Δβ1β2 and N1Δβ3αβ4) in T4P-mediated twitching motility, we assessed the ability of cells to twitch. Both wild-type cells and the ΔpilQ::bleo mutant complemented with wild-type pilQ were able to move on solid surfaces with twitching zones of ∼3.2 and ∼3.5 cm, respectively (positive controls) (Fig. 7B). The non-complemented ΔpilQ::bleo mutant (negative control) did not show twitching at all (Fig. 7B), in agreement with our previous findings (19). Depending on the specific pilQ mutation, the cells exhibited various changes in their motility. The ΔN3 mutant cells were as mobile as the wild type (Student's t test (95%)) (Fig. 7B). The clear reduction in piliation but wild-type levels of twitching suggests that the remaining pilus structures must exhibit increased dynamics, thereby compensating for their reduced numbers. An alternative explanation would be that only a fraction of pili present on wild-type cells contributes to twitching motility.
The N1Δβ3αβ4, ΔN2, and ΔN4 mutants were impaired in twitching motility and showed only 34%, 34%, and 21% of wild-type motility, respectively (Fig. 7B). An even stronger effect was exerted by deletion of the N5 domain. These mutants exhibited only 5% of wild-type motility (Fig. 7B). Moreover, the N1Δβ1β2 and ΔN1 mutant were completely defective in twitching (Fig. 7B). These data suggest that mutations in close proximity to either the N or C terminus of PilQ have a strong effect on T4P-mediated twitching motility and that the N5 domain and the N-terminal β1β2 fold in the N1 subdomain are absolutely essential.
The Ring-forming Domains Are Not Required for Natural Transformation
The pilQ mutants had transformation frequencies of 1 × 10−3 ± 3.4 × 10−4 to 2.1 × 10−3 ± 8.2 × 10−4 transformants/living count, which are comparable with those of the pilQ mutant complemented with the pilQ wild-type gene (1.9 × 10−3 ± 9.4 × 10−4). The negative control, a T. thermophilus ΔpilQ::bleo mutant, was not naturally transformable at all. The complete defect of the N1Δβ1β2 and the ΔN1 mutants in twitching motility together with their wild-type transformation phenotype led to the conclusion that pilus-mediated twitching motility is not required for natural transformation (19, 25). This corresponds to our recent finding that pilus structures on the cell surface are not essential for natural transformation (19, 25)
Discussion
PilQ in its monomeric state is only ∼83 kDa, and 12–14 copies assemble to a remarkably long structure spanning the outer membrane and half of the periplasm (18, 20). The complex has distinct structural features, such as rings and gates, but the domains in the monomer responsible for these functions were so far elusive. Here we provide the first insights into the topology of this macromolecular complex. Variants of PilQ described in this manuscript allowed us to identify the ring- and gate-forming motifs based on SP-EM and in situ by cryo-ET. In a previous study, we identified an unusual N0 ring-forming domain (ααβαββα fold) in PilQ (19). However, all previous attempts to identify further ring-forming domains (N1 to N5) by nested deletions beyond the N0 domain have failed because mutants did not assemble PilQ complexes (19). Here we show that, in the presence of the N0 domain, internal deletion derivatives of PilQ can still assemble into complexes (Fig. 2). This indicates that none of the rings are individually essential for complex assembly, which subsequently enables further detailed molecular analyses.
All PilQ variants were unstable, as indicated by multiple bands on SDS-PAGE. Complexes generated from ΔN1 and N1Δβ1β2 deletion derivatives were least stable, indicating that the β1β2 subdomain of N1 plays a major role in PilQ complex stabilization. Cryo-ET of the in situ T4P machinery in the ΔN1 mutant revealed that the complexes were devoid of gate 2. This suggests that the unusual βββαβ fold in the N1 domain plays a dual role in ring and gate formation. Gate 2 might act as a seal to prevent leakage of periplasmic components into the secretin complex. This is supported by the observation of an in situ “leaky” phenotype (Fig. 5G). The β1β2β3αβ4 fold is likely highly dynamic, supporting our earlier data demonstrating major conformational changes in the region of gate 2 (18). Conformational changes in gate 2, triggered by interactions with growing T4P, are suggested to allow extrusion of T4P.
SP-EM analysis of a set of PilQ deletion derivatives revealed that four distinct βαββα domains form the rings N2 to N5. These βαββα folds have also been detected in other ring-forming domains of secretin family proteins (31, 32) and are similar to nuclear ribonucleoprotein complex, K protein homology (KH) domains. KH domains consist of a three-stranded β sheet packed against two or three α helices and are involved in DNA binding (31, 32). A role of PilQ in DNA binding is also supported by the finding that a pilQ mutant was defect in DNA binding and uptake (23). Therefore, it is tempting to speculate that the βαββα domains in PilQ might be involved in DNA binding. However, the typical GXXG motif required for DNA interactions, which is present in the KH domain of the secretin HofQ and in eukaryotic proteins such as the neuronal splicing factor Nova-1 (32, 33), is missing in PilQ.
The diameters of the PilQ rings differ significantly (∼10 nm at N5 to ∼16 nm at N0) in the closed state, as observed by both cryo-ET and SP-EM (18–20). These structural differences may be due to different mutual orientations of the domains, as seen for T2SS and T3SS (2). The invagination in ring N5 may form a stabilizer for the pilus within the central channel of PilQ.
Several T2SS and T3SS secretins have been studied by EM to date, revealing a number of common features. These include a cylindrical arrangement of 12–15 subunits per ring and a large periplasmic vestibule comprising two to four rings, closed at the periplasmic end by one gate (2). The structure of the T. thermophilus secretin complex differs significantly from all other known secretin complexes by its composition of six stacked rings. Moreover, in T. thermophilus, the PilQ channel was found to be sealed by a unique second N-terminal gate located in the N1 ring (18). These features may be a result of adaptation to the increased thickness of the cell envelope, which might be due to adaptation to the extreme environment in which T. thermophilus thrives.
The ΔN1 domain deletion or the N1 subdomain deletion N1Δβ1β2 abolished twitching motility, whereas deletion of the N1β3αβ4 subdomain of the N1 fold had only a minor effect but changed the structure of the PilQ complex dramatically. The structural changes observed in this complex formed by the N1β3αβ4 variant might be the result of a fusion of the N0 ring to the remaining N1 domain. This suggests that the interaction between the N1 β1β2 subdomain and the T4P is important in determining pilus dynamics. We hypothesize that the first two β sheets of the N1 ring might act as a lever controlling the opening and closing of the gate, analogous to VirB of the T4SS of Agrobacterium tumefaciens (34).
In conclusion, we find that secretins share many common features, such as cup-forming domains, but also significant architectural differences, such as the number and shape of the rings and the number and localization of gates. The structure/function analyses of the PilQ mutants described here have helped to unravel the structural role of distinct domains (Fig. 8). These data provide a first insight into the function of distinct substructures of a secretin complex playing a dual role in T4P dynamics and DNA uptake. Our findings are an important milestone in the elucidation of the mechanism of secretin-mediated T4P extrusion and pilus dynamics.
FIGURE 8.
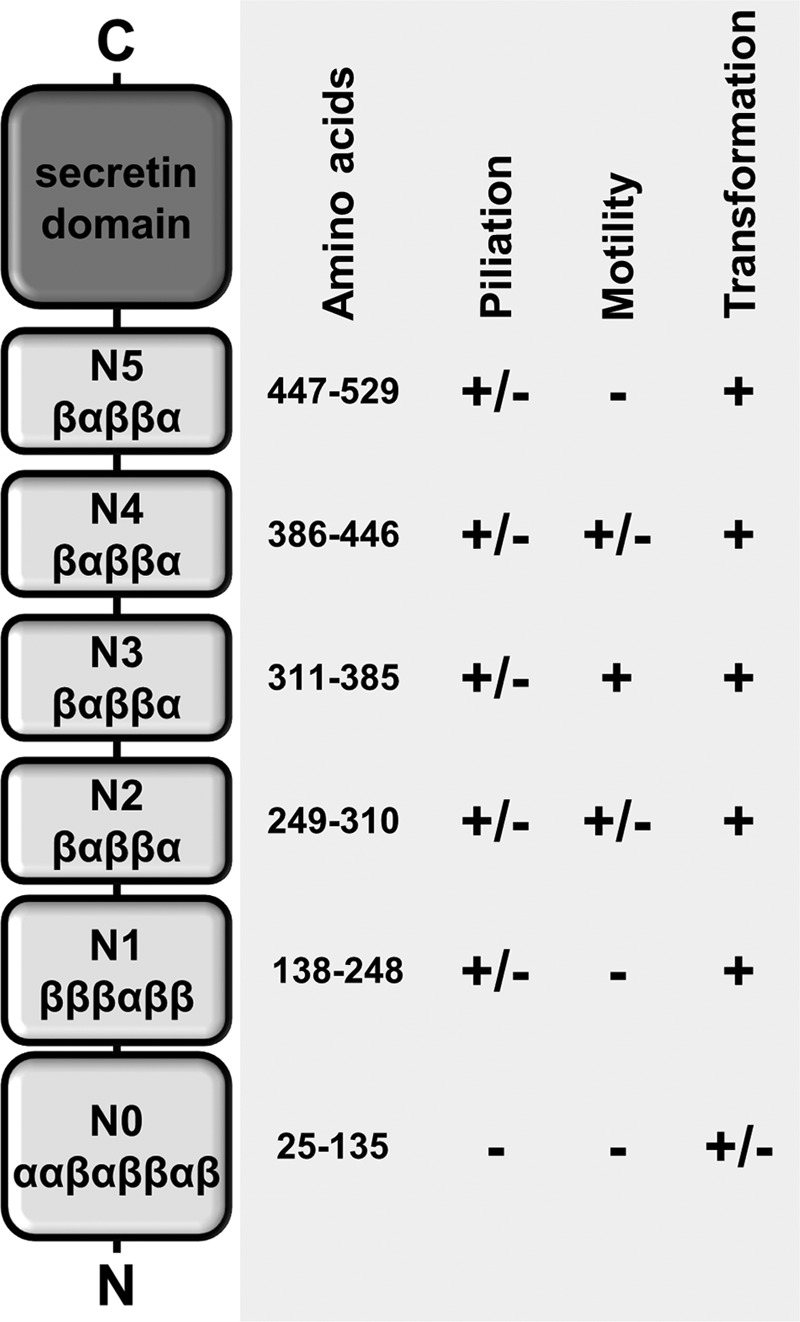
Structural and functional role of distinct domains in the secretin PilQ. +, comparable with the wild-type phenotype; ±, less than the wild type; − not detected.
Experimental Procedures
Organisms and Cultivation
T. thermophilus was grown in TM+ medium (8 g/liter Tryptone, 4 g/liter yeast extract, 3 g/liter NaCl, 0.17 mm CaCl2, 0.6 mm MgCl2, pH 7.5) at 68 °C (35). Kanamycin (20 μg/ml of liquid medium, 40 μg/ml of solid medium) and/or bleomycin (5 μg/ml of liquid medium, 15 μg/ml of solid medium) was added for growth of T. thermophilus mutants. Escherichia coli DH5α was grown in lysogenic broth medium at 37 °C (36). Antibiotics were added (kanamycin 20 μg/ml and/or ampicillin 100 μg/ml) where appropriate.
Site-directed Mutagenesis
For deletion of the N1, N2, N3, N4, or N5 domains or the N1 subdomains (N1Δβ1β2 and N1Δβ3αβ4) of PilQ, the Phusion site-directed mutagenesis (New England Biolabs, Frankfurt, Germany) protocol was used. The E. coli/Thermus shuttle vector pDM12-pilQ-6his (19) was used as template DNA, and primers used are listed in Table S1. Plasmids carrying the mutated pilQ genes were verified by sequencing and transferred into the T. thermophilus pilQ::bleo deletion mutant via electroporation (24). In the various ring deletions, amino acids 138–248 (ΔN1), 249–310 (ΔN2), 311–385 (ΔN3), 386–446 (ΔN4), and 447–529 (ΔN5) were deleted. In the partial deletions of the N1 ring, amino acids 138–183 (N1Δβ1β2) and 194–248 (N1Δβ3αβ4) were deleted.
Membrane Preparation
Membranes were isolated as described recently (37).
Western Blotting Analysis
Membranes from T. thermophilus HB27 wild-type (50 μg of protein) and membranes from the mutants pilQwt, ΔpilQ::bleo, N1Δβ1β2, N1Δβ3αβ4ΔN1, ΔN2, ΔN3, ΔN4, and ΔN5 (50 μg of protein each) were separated by denaturating SDS-PAGE (3–12%). Proteins were transferred onto Protran BA83 membranes (GE Healthcare) and subjected to Western blotting analyses using polyclonal PilQ antibodies (1:10,000 dilution).
Transformation Assays
Transformation frequencies were calculated as transformants per living count as described previously (38). 5 μg of chromosomal DNA of a spontaneously streptomycin-resistant T. thermophilus mutant was used for transformation assays.
Twitching Motility Studies
T. thermophilus strains were grown for 3 days on minimal medium containing 1% BSA. The plates were stained with Coomassie Blue. T. thermophilus cells were detached from the agar, and the twitching zones were defined as the resulting colorless areas. Assays were performed as described previously (24).
Investigation of Piliation Phenotypes by Electron Microscopy
The piliation phenotype of T. thermophilus cells was analyzed by electron microscopy as described recently (25). Cells were transferred to copper grids and shadowed at a unidirectional angle of 25° with platinum-carbon. The amount of pili was counted from 300 randomly selected cells.
Purification of PilQ
T. thermophilus ΔpilQ::bleo mutants carrying pDM12-pilQN1Δβ1β2, pDM12-pilQN1Δβ3αβ4, pDM12-pilQNΔ1, pDM12-pilQΔN2, pDM12-pilQΔN3, pDM12-pilQΔN4, or pDM12-pilQΔN5, respectively, were grown in TM+ medium in a 10-liter fermenter to stationary growth phase (8 liters/min aeration, 600 rpm stirring). Cells were harvested, and 30 g of cell pellet was resuspended in buffer (30 mm Tris/HCl and 200 mm NaCl (pH 7.5)). Cell disruption was carried out with a French press (3 × 1000 psi) (Thermo Fisher Scientific, Schwerte, Germany). Membranes were prepared by ultracentrifugation, and PilQ complexes were solubilized using 3% dodecyl maltoside (DDM) for 30 min at room temperature. Samples were centrifuged (100,000 × g, 4 °C, 45 min), and the supernatants diluted to a final concentration of 1% DDM and subsequently incubated with 5 ml of TALON® metal affinity resin (Takara Bio Europe (Clontech Laboratories, Inc., St-Germain-en-Laye, France) for 45 min at 4 °C. The column was washed with buffer containing 0.05% DDM and 12.5 mm imidazole. Elution was carried out with buffer containing 150 mm imidazole and 0.05% DDM. The elution fraction was diluted 1:1 with buffer without NaCl (0.05% DDM) and subjected to anion exchange (Q-Sepharose, GE Healthcare). The PilQ-containing fractions were detected by Western blotting and Coomassie staining (InstantBlue, Expedeon, Cambridge, UK), and the elution fractions containing PilQ-complexes were concentrated (VivaSpin, 50,000 MWCO PES, Sartorius Stedim Biotech GmbH, Göttingen, Germany).
Electron Microscopy and Image Processing
PilQ complexes were diluted to a final concentration of 0.02 mg/ml and negatively stained with 2% (w/v) ammonium molybdate. 3 μl of protein sample was applied onto freshly glow-discharged carbon-coated copper grids. The sample was reduced to a thin film by blotting, and the staining solution was then immediately applied to the grid and blotted off from the same side. The negative stain process was repeated three times. Electron micrographs were collected using a FEI Tecnai G2 Spirit operated at 120 kV at a nominal magnification of ×28,000. Images were collected on a Gatan 4k × 4k charge-coupled device camera with a pixel size of 4.2 Å with a defocus range of −0.2 to −1.0 μm. Particles were selected using the boxer module in EMAN (39). Image processing and reference-free two-dimensional classification were performed with Xmipp 3.1 using the classification algorithm CL2D (40) and with RELION (41).
Electron Cryotomography and Subtomogram Averaging
Cryo-ET and subtomogram averaging were performed as described previously (18), with the following exceptions. Tomograms were collected at tilt steps of 2.5° and 7-μm defocus using a JEM-3200FSC microscope (JEOL, Tokyo, Japan) equipped with a field emission gun operating at 300 keV and an in-column energy filter operated with a slit width of 40 eV. Tomograms were collected with a K2 Summit direct electron detection camera (Gatan) at a nominal magnification of ×10,000 corresponding to a pixel size of 3.9 Å. The total dose per tomogram was ∼140e−/Å2. For subtomogram averaging, coordinates corresponding to the outer membrane and inner membrane domains of the T4P machinery were marked manually in IMOD from three-times binned tomograms and averaged using PEET (42). The final average was obtained from 432 particles (C12 symmetry applied) using a mask drawn around PilQ. The resolution estimate is ∼50 Å using conventional “even/odd” FSC, applying the 0.5 FSC criterion (Fig. 6). A mask was drawn around the protein to exclude the membrane from this estimate. Surface views were drawn with UCSF Chimera (43). For comparisons with the wild-type complex, data were used that have been published previously (18) and is now displayed in a different way.
Author Contributions
R. S. designed and performed functional analyses, cloning, complex purifications, and electron microscopy analyses and drafted the manuscript. E. D. performed the electron microscopy studies. V. A. M. G. conducted the electron cryotomography studies. I. R. helped with protein purification. M. D. helped with generation of deletion derivatives and protein purification. J. V. performed the data analysis of the electron microscopy studies. B. A. supervised the study and revised the manuscript.
Supplementary Material
Acknowledgments
We thank Bernd Ludwig (Goethe University, Frankfurt, Germany) for plasmid pDM12. We also thank Werner Kühlbrandt for support; Deryck Mills, Simone Prinz, and Friederike Joos for help with electron microscopy; and Juan Castillo-Hernández and Özkan Yildiz for computer support.
This work was supported by Deutsche Forschungsgemeinschaft Grant AV 9/6-1. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
The subtomogram average was uploaded to the EMDataBank with ID code EMDB-8224. The data corresponding to the closed state of the wild-type complex can be found under EMDB-3021 and EMDB-3022.
- T4P
- type IV pili
- T2SS
- type II secretion system
- T3SS
- type III secretion system
- SP-EM
- single-particle EM
- cryo-ET
- electron cryotomography
- DDM
- dodecyl maltoside
- KH
- nuclear ribonucleoprotein complex, K protein homology.
References
- 1.Costa T. R., Felisberto-Rodrigues C., Meir A., Prevost M. S., Redzej A., Trokter M., and Waksman G. (2015) Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359 [DOI] [PubMed] [Google Scholar]
- 2.Korotkov K. V., Gonen T., and Hol W. G. (2011) Secretins: dynamic channels for protein transport across membranes. Trends Biochem. Sci. 36, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koster M., Bitter W., and Tommassen J. (2000) Protein secretion mechanisms in Gram-negative bacteria. Int. J. Med. Microbiol. 290, 325–331 [DOI] [PubMed] [Google Scholar]
- 4.Tosi T., Estrozi L. F., Job V., Guilvout I., Pugsley A. P., Schoehn G., and Dessen A. (2014) Structural similarity of secretins from type II and type III secretion systems. Structure 22, 1348–1355 [DOI] [PubMed] [Google Scholar]
- 5.Burrows L. L. (2012) Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66, 493–520 [DOI] [PubMed] [Google Scholar]
- 6.Nudleman E., and Kaiser D. (2004) Pulling together with type IV pili. J. Mol. Microbiol. Biotechnol. 7, 52–62 [DOI] [PubMed] [Google Scholar]
- 7.Berry J. L., and Pelicic V. (2015) Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss army knives. FEMS Microbiol. Rev. 39, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krüger N.-J., and Stingl K. (2011) Two steps away from novelty: principles of bacterial DNA uptake. Mol. Microbiol. 80, 860–867 [DOI] [PubMed] [Google Scholar]
- 9.Friedrich A., Prust C., Hartsch T., Henne A., and Averhoff B. (2002) Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 68, 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roca I., Akova M., Baquero F., Carlet J., Cavaleri M., Coenen S., Cohen J., Findlay D., Gyssens I., Heure O. E., Kahlmeter G., Kruse H., Laxminarayan R., Liebana E., Lopez-Cerero L., et al. (2015) The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 6, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karuppiah V., Berry J. L., and Derrick J. P. (2011) Outer membrane translocons: structural insights into channel formation. Trends Microbiol. 19, 40–48 [DOI] [PubMed] [Google Scholar]
- 12.Bouley J., Condemine G., and Shevchik V. E. (2001) The PDZ domain of OutC and the N-terminal region of OutD determine the secretion specificity of the type II out pathway of Erwinia chrysanthemi. J. Mol. Biol. 308, 205–219 [DOI] [PubMed] [Google Scholar]
- 13.Guilvout I., Hardie K. R., Sauvonnet N., and Pugsley A. P. (1999) Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J. Bacteriol. 181, 7212–7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowal J., Chami M., Ringler P., Müller S. A., Kudryashev M., Castaño-Díez D., Amstutz M., Cornelis G. R., Stahlberg H., and Engel A. (2013) Structure of the dodecameric Yersinia enterocolitica secretin YscC and its trypsin-resistant core. Structure 21, 2152–2161 [DOI] [PubMed] [Google Scholar]
- 15.Kudryashev M., Stenta M., Schmelz S., Amstutz M., Wiesand U., Castaño-Díez D., Degiacomi M. T., Münnich S., Bleck C. K., Kowal J., Diepold A., Heinz D. W., Dal Peraro M., Cornelis G. R., and Stahlberg H. (2013) In situ structural analysis of the Yersinia enterocolitica injectisome. eLife 2, e00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korotkov K. V., Johnson T. L., Jobling M. G., Pruneda J., Pardon E., Héroux A., Turley S., Steyaert J., Holmes R. K., Sandkvist M., and Hol W. G. (2011) Structural and functional studies on the interaction of GspC and GspD in the type II secretion system. PLoS Pathog. 7, e1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichow S. L., Korotkov K. V., Hol W. G., and Gonen T. (2010) Structure of the cholera toxin secretion channel in its closed state. Nat. Struct. Mol. Biol. 17, 1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gold V. A., Salzer R., Averhoff B., and Kühlbrandt W. (2015) Structure of a type IV pilus machinery in the open and closed state. eLife 4, 10.7554/eLife.07380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burkhardt J., Vonck J., Langer J. D., Salzer R., and Averhoff B. (2012) Unusual N-terminal ααβαββα fold of PilQ from Thermus thermophilus mediates ring formation and is essential for piliation. J. Biol. Chem. 287, 8484–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkhardt J., Vonck J., and Averhoff B. (2011) Structure and function of PilQ, a secretin of the DNA transporter from the thermophilic bacterium Thermus thermophilus HB27. J. Biol. Chem. 286, 9977–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedrich A., Hartsch T., and Averhoff B. (2001) Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl. Environ. Microbiol. 67, 3140–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedrich A., Rumszauer J., Henne A., and Averhoff B. (2003) Pilin-like proteins in the extremely thermophilic bacterium Thermus thermophilus HB27: implication in competence for natural transformation and links to type IV pilus biogenesis. Appl. Environ. Microbiol. 69, 3695–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarzenlander C., Haase W., and Averhoff B. (2009) The role of single subunits of the DNA transport machinery of Thermus thermophilus HB27 in DNA binding and transport. Environ. Microbiol. 11, 801–808 [DOI] [PubMed] [Google Scholar]
- 24.Salzer R., Joos F., and Averhoff B. (2014) Type IV pilus biogenesis, twitching motility, and DNA uptake in Thermus thermophilus: discrete roles of antagonistic ATPases PilF, PilT1, and PilT2. Appl. Environ. Microbiol. 80, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salzer R., Herzberg M., Nies D. H., Joos F., Rathmann B., Thielmann Y., and Averhoff B. (2014) Zinc and ATP binding of the hexameric AAA-ATPase PilF from T. thermophilus: role in complex stability, piliation, adhesion, twitching motility and natural transformation. J. Biol. Chem. 289, 30343–30354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salzer R., Herzberg M., Nies D. H., Biuković G., Grüber G., Müller V., and Averhoff B. (2013) The DNA uptake ATPase PilF of Thermus thermophilus: a reexamination of the zinc content. Extremophiles 17, 697–698 [DOI] [PubMed] [Google Scholar]
- 27.Rose I., Biuković G., Aderhold P., Müller V., Grüber G., and Averhoff B. (2011) Identification and characterization of a unique, zinc-containing transport ATPase essential for natural transformation in Thermus thermophilus HB27. Extremophiles 15, 191–202 [DOI] [PubMed] [Google Scholar]
- 28.Rumszauer J., Schwarzenlander C., and Averhoff B. (2006) Identification, subcellular localization, and functional interactions of PilMNOWQ and PilA4 involved in transformation competency and pilus biogenesis in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 273, 3261–3272 [DOI] [PubMed] [Google Scholar]
- 29.Karuppiah V., Collins R. F., Thistlethwaite A., Gao Y., and Derrick J. P. (2013) Structure and assembly of an inner membrane platform for initiation of type IV pilus biogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, E4638–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karuppiah V., Hassan D., Saleem M., and Derrick J. P. (2010) Structure and oligomerization of the PilC type IV pilus biogenesis protein from Thermus thermophilus. Proteins 78, 2049–2057 [DOI] [PubMed] [Google Scholar]
- 31.Korotkov K. V., Pardon E., Steyaert J., and Hol W. G. (2009) Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure 17, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarry M., Jääskeläinen M., Paino A., Tuominen H., Ihalin R., and Högbom M. (2011) The extra-membranous domains of the competence protein HofQ show DNA binding, flexibility and a shared fold with type I KH domains. J. Mol. Biol. 409, 642–653 [DOI] [PubMed] [Google Scholar]
- 33.Teplova M., Malinina L., Darnell J. C., Song J., Lu M., Abagyan R., Musunuru K., Teplov A., Burley S. K., Darnell R. B., and Patel D. J. (2011) Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1. Structure 19, 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zechner E. L., Lang S., and Schildbach J. F. (2012) Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 367, 1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oshima T., and Imahori K. (1971) Isolation of an extreme thermophile and thermostability of its transfer ribonucleic acid and ribosomes. J. Gen. Appl. Microbiol. 17, 513–517 [Google Scholar]
- 36.Bertani G. (1951) Studies on lysogenesis. J. Bacteriol. 62, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salzer R., Kern T., Joos F., and Averhoff B. (2016) The Thermus thermophilus comEA/comEC operon is associated with DNA binding and regulation of the DNA translocator and type IV pili. Environ. Microbiol. 18, 65–74 [DOI] [PubMed] [Google Scholar]
- 38.Salzer R., Joos F., and Averhoff B. (2015) Different effects of MglA and MglB on pilus-mediated functions and natural competence in Thermus thermophilus. Extremophiles 19, 261–267 [DOI] [PubMed] [Google Scholar]
- 39.Ludtke S. J., Baldwin P. R., and Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 40.Sorzano C. O., Bilbao-Castro J. R., Shkolnisky Y., Alcorlo M., Melero R., Caffarena-Fernández G., Li M., Xu G., Marabini R., and Carazo J. M. (2010) A clustering approach to multireference alignment of single-particle projections in electron microscopy. J. Struct. Biol. 171, 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres S. H. (2012) RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicastro D., Schwartz C., Pierson J., Gaudette R., Porter M. E., and McIntosh J. R. (2006) The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948 [DOI] [PubMed] [Google Scholar]
- 43.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.