Abstract
Subversion of host cell apoptotic responses is a prominent feature of viral immune evasion strategies to prevent premature clearance of infected cells. Numerous poxviruses encode structural and functional homologs of the Bcl-2 family of proteins, and vaccinia virus harbors antiapoptotic F1L that potently inhibits the mitochondrial apoptotic checkpoint. Recently F1L has been assigned a caspase-9 inhibitory function attributed to an N-terminal α helical region of F1L spanning residues 1–15 (1) preceding the domain-swapped Bcl-2-like domains. Using a reconstituted caspase inhibition assay in yeast we found that unlike AcP35, a well characterized caspase-9 inhibitor from the insect virus Autographa californica multiple nucleopolyhedrovirus, F1L does not prevent caspase-9-mediated yeast cell death. Furthermore, we found that deletion of the F1L N-terminal region does not impede F1L antiapoptotic activity in the context of a viral infection. Solution analysis of the F1L N-terminal regions using small angle x-ray scattering indicates that the region of F1L spanning residues 1–50 located N-terminally from the Bcl-2 fold is an intrinsically unstructured region. We conclude that the N terminus of F1L is not involved in apoptosis inhibition and may act as a regulatory element in other signaling pathways in a manner reminiscent of other unstructured regulatory elements commonly found in mammalian prosurvival Bcl-2 members including Bcl-xL and Mcl-1.
Keywords: apoptosis, B-cell lymphoma 2 (Bcl-2) family, poxvirus, protein structure, small angle x-ray scattering (SAXS), caspases, vaccinia virus
Introduction
Programmed cell death or apoptosis is an evolutionarily conserved mechanism (2) to remove damaged, infected, or unwanted cells, and viruses have evolved numerous strategies to prevent premature apoptosis of infected host cells (3). The fate of a cell is substantially determined by the interactions of members of the Bcl-2 family of proteins (4), which are important regulators of intrinsic or mitochondrially mediated apoptosis. The Bcl-2 family of proteins comprises prosurvival and proapoptotic members, which are characterized by the presence of Bcl-2 homology (BH)4 domains (5). Prosurvival Bcl-2 proteins such as Bcl-2, Bcl-xL, and Mcl-1 maintain cell viability until their inactivation by BH3-only proteins such as Bim, Bad, and Puma (6). BH3-only proteins are up-regulated after cellular insults including exposure to cytotoxic drugs or UV light and activate the cellular apoptotic machinery (7, 8). BH3-only proteins only harbor the α helical BH3 domain, which binds to a canonical binding groove on prosurvival Bcl-2 members (9, 10), although recent evidence suggests that they may also bind transiently to an alternative site on multidomain proapoptotic Bcl-2 such as Bax (11) and Bak (12). Up-regulation of BH3-only proteins leads to the activation of the essential proapoptotic proteins Bak and Bax (13), which drive mitochondrial outer membrane permeabilization (14), thus leading to the release of cytochrome c. Cytochrome c together with Apaf-1, the initiator caspase-9, and ATP form the apoptosome (15). Current models suggest that caspase-9 initially contributes to apoptosome formation in its uncleaved and inactive proform (16, 17) and is activated via dimerization at the apoptosome platform that enables autoactivation via proteolysis. Activated caspase-9 then proteolytically activates the downstream effectors caspase-3 and caspase-7 (18), ultimately leading to the destruction of the cell.
The importance of the Bcl-2 family of proteins in apoptosis regulation is reinforced by the observation that numerous viruses encode recognizable sequence homologs of Bcl-2 to subvert premature host cell apoptosis. These include Epstein-Barr virus BHRF1 (19), adenovirus E1B19K (20), Kaposi sarcoma herpesvirus KsBcl-2 (21), fowlpox virus FPV039 (22, 23), and herpesvirus saimiri vBcl-2 (24), and structural studies of some of these confirmed that they adopt a Bcl-2 fold (25–27). However, a number of viral proteins have been identified that shared no discernible sequence identity to known inhibitors of apoptosis. These include myxoma virus M11L (28), cytomegalovirus vMIA (29) and its mouse counterpart m38.5 (30–32), deerpox virus DPV022 (33), sheeppox virus SPPV14 (34), and vaccinia virus F1L (35) and N1L (36). Structural studies of M11L (37, 38) revealed that although it lacks sequence similarity to the Bcl-2 family of proteins it adopts a Bcl-2-like fold and engages BH3 ligands utilizing the canonical BH3 domain binding groove (38). Vaccinia virus N1L was shown to also adopt a Bcl-2-like fold (39, 40), which enabled it to assume dual functionality by mediating intrinsic apoptosis via the canonical Bcl-2 binding groove as well as NF-κB signaling via an additional non-canonical site (41). Similarly, deerpox virus DPV022 was shown to be a Bcl-2-like protein, albeit with a dimeric topology due to a domain swap (42).
Vaccinia virus encodes antiapoptotic F1L, which has been shown to act on the intrinsic pathway of apoptosis (43). F1L is able to engage Bim (44, 45) and Bak (46, 47) and inhibits Bak activation by functionally replacing Mcl-1 during infection (48). Furthermore, F1L is able to inhibit Bax-mediated apoptosis (44), presumably via an indirect mechanism because F1L appears to not engage Bax in the cellular context. Recently, the interaction of F1L with Bim has been shown to be the primary mechanism underlying F1L-mediated inhibition of apoptosis in the context of a live viral infection (49). Although F1L lacks discernible sequence identity to the Bcl-2 family of proteins, the crystal structure of F1L revealed that it adopts a Bcl-2 fold in a domain-swapped dimer configuration (49, 50). In its entirety, F1L from vaccinia virus (MVA) comprises 222 residues of which only residues 57–190 form the canonical Bcl-2-like domain. Although no experimental structural data are available for the N-terminal region of F1L, recent biochemical studies suggested a role in caspase inhibition for the N-terminal region preceding the Bcl-2 fold (51) as well as a role in modulating inflammasome regulation (52). Subsequent molecular modeling proposed the formation of two α helical segments (1) at the extreme N terminus of F1L that engage in an inhibitory substrate complex with caspase-9, thus abrogating caspase-9 activity.
Experimental Procedures
Protein Expression and Purification
The coding sequence corresponding to MVA F1L amino acids 1–202 (UniProt accession number O57173) was cloned into the pETDuet-1 vector. Protein expression was induced in Escherichia coli BL21(DE3) pLysS cells with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 37 °C. The cells were harvested and lysed using 0.2-mm silica beads in a FastPrep instrument (MP Biomedicals) for 4 × 20-s cycles. Cellular debris was removed via centrifugation at 16,000 × g for 20 min and filtered through a 0.22-μm syringe filter. MVA F1L was purified with 2 × 1-ml HiTrap column charged with nickel (GE Healthcare). The protein was further purified via size exclusion chromatography on a HiLoad 16/60 Superdex 75 prep grade column (GE Healthcare) into a final buffer of 25 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm DTT. MVA F1L protein was concentrated to 3.9 mg/ml, flash frozen, and stored at 193 K.
Small Angle X-ray Scattering Experiments
Synchrotron x-ray scattering data were collected from 50-μl samples of thawed MVA F1L(1–202) protein (including an N-terminal purification tag with the sequence MGSSHHHHHHSQDP) in capillary tubes at the SAXS/WAXS beamline of the Australian Synchrotron using a Pilatus-1M detector. The scattering was measured with exposure times of 1 s at 12 keV at a temperature of ∼286 K with protein concentrations ranging from 3.6 to 0.12 mg/ml in 25 mm HEPES, pH 7.5, NaCl 150 mm, 5 mm DTT. The sample to detector distance was 1575 mm, and the momentum transfer range was 0.01 < s < 0.5 Å−1 at 12 keV. Constant water scattering was determined by subtracting the scattering of an empty capillary from the scattering of a capillary filled with water. Normalization was achieved via integrating the beam stop. To control for radiation damage, the samples were measured in a 1.5-mm quartz capillary and flowed past the beam while measuring 18 × 1 s on samples and blanks (gel filtration buffer and water).
Envelope Modeling
The scattering images were integrated, averaged, and calibrated against water using software specific to the beamline (53). The radius of gyration (Rg) and the forward scattering I(0) were determined by Guinier approximation using PRIMUS from ATSAS (54, 55). Rigid body modeling was performed on the processed data using BUNCH (56) and CORAL (54). Both models were generated using a known crystal structure of MVA F1L (Protein Data Bank code 4d2m) lacking 50 amino acids at the N terminus and 15 amino acids at the C terminus. P2 symmetry was imposed to generate a dimer, and the scattering curve of the 1.80 mg/ml data was used. 14 residues were added to the sequence of F1L corresponding to the pDuet purification tag.
The MVA F1L coordinate file was prepared for rigid body modeling using Coot (57) (removing side chain alternative conformations and renumbering amino acids within the sequence) and MASSHA (fixing monomer orientation to generate a proper dimer upon P2 symmetry) (58).
Yeast Colony Assays
Saccharomyces cerevisiae W303α cells were transformed with either pGALL(LEU2), pGALL(LEU2)-HA-Bax, or pGALL(LEU2)-His6-Bak or co-transformed with pGALL(HIS3)-Apaf-1(1–530), pGALL(LEU2)-caspase-9, and pGALL(URA3)-caspase-3 or the corresponding empty vectors. Yeast bearing these plasmids were then transformed with either pGALL(TRP1), pGALL(TRP1)Bcl-xL, pGALL(TRP1)-VACV(COP)F1L, or pGALL(TRP1)-AcP35. The pGALL(TRP1) and pGALL(LEU2) vectors place genes under the control of a galactose-inducible promoter. Cells were spotted as 5-fold serial dilutions onto medium containing 2% (w/v) galactose (inducing), which induces protein expression, or 2% (w/v) glucose (repressing), which prevents protein expression, as described previously (59). Plates were incubated for 48 (glucose) or 72 h (galactose) at 30 °C and then photographed.
Cell Lines
HEK293T and HeLa cells, both obtained from the American Type Culture Collection (ATCC), were maintained at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen), 200 μm l-glutamine (Invitrogen), 50 units of penicillin (Invitrogen)/ml, and 50 μg of streptomycin (Invitrogen)/ml.
Plasmid Construction
pGALL(TRP1)-VACV(COP)F1L was generated by subcloning from synthetic cDNA encoding for full-length wild-type VACV(COP)F1L (GenScript) using available BamHI and EcoRI sites. pGALL(LEU2), pGALL(LEU2)-caspase-9, pGALL(HIS3)-Apaf-1(1–530), pGALL(URA3)-caspase-3, pGALL(TRP1), pGALL(TRP1)Bcl-xL, and pGALL(TRP1)-AcP35 have been described previously (60–62). pGALL(LEU2)-HA-Bax was kindly provided by Jamie Fletcher. FLAG-F1L(43–226), FLAG-F1L(50–226), and FLAG-F1L(60–226) were amplified by polymerase chain reaction (PCR) using codon-optimized pcDNA3-FLAG-F1L as a template and Pwo (Pyrococcus woesei) polymerase (Roche Applied Science). The forward primers used, all containing a BamHI restriction site, were 5′-GGATCCATGGACTACAAAGACGATGACGACAAGGAGAACATGGTGTACCGGTTC-3′ for FLAG-F1L(43–226), 5′-GGATCCATGGACTACAAAGACGATGACGACAAGGACAAGTCTACCAATATCCTG-3′ for FLAG-F1L(50–226), and 5′-GGATCCATGGACTACAAAGACGATGACGACAAGAGCACCGAGCGGGACCACGTG-3′ for FLAG-F1L(60–226). The reverse primer used for all three constructs was 5′-GAATTCTCAGCCGATCATGTACTTCAG-3′ containing an EcoRI restriction site. The three PCR products were subcloned into the shuttle vector pGemT (Promega) followed by an additional subcloning step into the final destination vector pcDNA3 (Invitrogen).
Transient Transfection
Transfection of HEK293T and HeLa cells (1 × 106) seeded in 6-cm cell culture dishes (Corning Inc.) was done using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Unless otherwise stated, cells were transfected with 2 μg of pcDNA3-FLAG-A6L, pcDNA3-FLAG-BakΔBH3, pcDNA3-FLAG-F1L, pcDNA3-FLAG-F1L(43–226), pcDNA3-FLAG-F1L(50–226), or pcDNA3-FLAG-F1L(60–226); 1 μg of pcDNA3-HA-Bak; or 0.5 μg of pEGFP-C3. Transfected cells were supplemented with 20% FBS (DMEM, 20% FBS, 200 μm l-glutamine) 2 h post-transfection and maintained at 37 °C and 5% CO2.
Whole Cell Lysates
To determine the expression levels of the F1L N-terminal truncation mutants, HEK293T cells were mock transfected or transiently transfected with pcDNA3-FLAG-BakΔBH3, pcDNA3-FLAG-F1L, pcDNA3-FLAG-F1L(43–226), pcDNA3-FLAG-F1L(50–226), or pcDNA3-FLAG-F1L(60–226). Following an 18-h transfection period, cells were washed with phosphate-buffered saline (PBS) and suspended in 150 μl of SDS loading buffer containing 0.06 m Tris, pH 6.8 (Invitrogen), 2% SDS (Fischer Scientific), 32% glycerol (Anachemia), 0.05 m β-mercaptoethanol (Bioshop), and 0.005% bromphenol blue (Bio-Rad). Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Confocal Microscopy
To determine the subcellular localization of the F1L N-terminal truncation mutants, HeLa cells were seeded onto 18-mm coverslips (Fisher Scientific) in 3.5-cm-diameter culture dishes (Corning Inc.). After 24 h, 5 × 105 cells were transfected with pcDNA3-FLAG-A6L, pcDNA3-FLAG-F1L, pcDNA3-FLAG-F1L(43–226), pcDNA3-FLAG-F1L(50–226), or pcDNA3-FLAG-F1L(60–226). After a 12-h transfection period, the cells were fixed in 4% paraformaldehyde (Sigma-Aldrich), permeabilized in 1% Nonidet P-40 (Sigma-Aldrich), and blocked in 30% goat serum (Invitrogen). Cells were then stained with polyclonal rabbit anti-FLAG M2 antibody (Sigma-Aldrich) at a dilution of 1:200 and monoclonal mouse anti-cytochrome c antibody (BD Pharmingen) at a dilution of 1:150. Signals were amplified with Alexa Fluor 488-conjugated donkey anti-mouse antibody (Invitrogen) and Alexa Fluor 546-conjugated donkey anti-rabbit antibody (Invitrogen), both at a dilution of 1:400. Coverslips were mounted using mounting solution containing DAPI stain and visualized with a Zeiss Axiovert laser scanning microscope.
Immunoprecipitation
To detect the interaction between the N-terminal truncation mutants of F1L with Bak, HEK293T cells were co-transfected with pcDNA3-HA-BAK along with pcDNA3-FLAG-A6L, pcDNA3-FLAG-F1L, pcDNA3-FLAG-F1L(43–226), pcDNA3-FLAG-F1L(50–226), or pcDNA3-FLAG-F1L(60–226). After 18 h, transfected cells were lysed for 1.5 h in 2% CHAPS lysis buffer containing 2% (w/v) CHAPS (Sigma-Aldrich), 150 mm NaCl, 50 mm Tris, pH 8.0 (Invitrogen), and EDTA-free proteinase inhibitor (Roche Applied Science). FLAG-tagged constructs in the cell lysates were immunoprecipitated with monoclonal mouse anti-FLAG M2 antibody (Sigma-Aldrich) (1:4000 dilution) for 2 h followed by precipitation of the immune complexes with lysis buffer-equilibrated protein G-Sepharose beads (GE Healthcare) for 1 h. Beads were washed three times in 2% CHAPS lysis buffer and resuspended in 50 μl of SDS gel loading buffer. Lysate samples were acetone-precipitated and suspended in 50 μl of SDS gel loading buffer. The proteins were analyzed by loading 40% of each sample on SDS-polyacrylamide gels and blotted for FLAG and Bak.
Apoptosis Assay
To determine the ability of the F1L N-terminal truncation mutants to protect against apoptosis, HeLa cells were co-transfected with pcDNA3-FLAG-A6L, pcDNA3-FLAG-F1L, pcDNA3-FLAG-F1L(43–226), pcDNA3-FLAG-F1L(50–226), or pcDNA3-FLAG-F1L(60–226) along with pEGFP-C3 at a ratio of 4:1 (FLAG:EGFP) that served as a marker of transfection. After an 18-h transfection period, cells were treated with 10 ng/ml tumor necrosis factor α (TNFα) (Roche Applied Science) along with 5 μg/ml cycloheximide to induce apoptosis. Cells were then stained with 0.2 μm tetramethylrhodamine ethyl ester (Molecular Probes) for 30 min. The cells were subsequently washed twice with PBS supplemented with 1% FBS and analyzed by flow cytometry. Flow cytometric analysis was performed on a BD Biosciences FACScan with EGFP fluorescence measured through the FL-1 channel equipped with a 489-nm filter (band pass, 42 nm) and tetramethylrhodamine ethyl ester fluorescence measured through the FL-2 channel equipped with a 585-nm filter (band pass, 42 nm). Data were acquired on 20,000 cells/sample with fluorescence signals at logarithmic gain. Standard deviations were generated from three independent experiments. To assess the expression of the FLAG-tagged constructs in the presence of EGFP, whole cell lysates were harvested after an 18-h transfection period and analyzed by SDS-PAGE and immunoblotting.
SDS-PAGE and Immunoblotting
Cellular lysates were analyzed using SDS-PAGE. Lysates were suspended in SDS loading buffer, boiled for 10 min, and run on 15% polyacrylamide gels. Proteins were then transferred to a polyvinylidene fluoride membrane (GE Healthcare) using a semidry transfer apparatus (Tyler Research Instruments) for 2 h at 420 mA. Membranes were blocked in 5% skim milk powder in TBST (Tris-buffered saline with 0.1% Tween) overnight at 4 °C. The membranes were probed with monoclonal mouse anti-FLAG M2 antibody (1:5000) to detect FLAG-tagged constructs or polyclonal rabbit anti-Bak N terminus antibody (1:500) (Upstate) to detect Bak. Horseradish peroxidase-conjugated secondary donkey anti-mouse or donkey anti-rabbit antibody (Jackson ImmunoResearch Laboratories) was then used at a concentration of 1:25,000. Proteins were visualized by chemiluminescence after treatment with ECL reagent (GE Healthcare).
Results
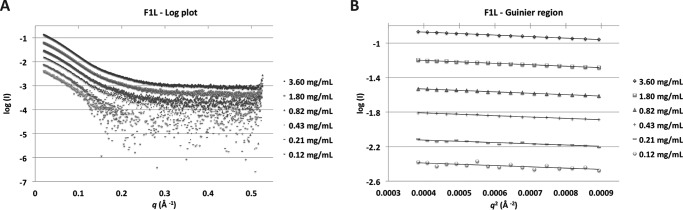
To understand the function of the N-terminal region of F1L preceding the Bcl-2 fold, we investigated the solution structure of F1L with an intact N terminus using small angle x-ray scattering (Table 1). Recombinant MVA F1L(1–202) was measured at six concentrations, ranging from 0.12 to 3.6 mg/ml. The scattering curve profile is conserved throughout the concentration range tested with the exception of the highest concentration in which interparticle interference is observed (Fig. 1A and Table 2). The scattering conforms to a straight line in the low q region on a Guinier plot (Fig. 1B), and the calculated radius of gyration does not vary significantly with the measured concentration range, suggesting an absence of significant concentration effects from the second highest concentration (Table 2).
TABLE 1.
Data collection and scattering-derived parameters
| Data collection parameters | |
| Instrument | SAXS/WAXS beamline Australian Synchrotron |
| Beam geometry (μm) | 80 × 200 |
| Wavelength (keV) | 12 |
| q range (Å−1) | 0.025–0.500 |
| Exposure time (s) | 1 (per frame; 18 frames) |
| Concentration range (mg ml−1) | 0.12–3.60 |
| Temperature (K) | 293 |
| Structural parametersa | |
| I(0) (cm−1) (from Guinier) | 0.073 ± 0.000 |
| Rg (Å) (from Guinier) | 34.10 ± 0.243 |
| Molecular mass determinationa | |
| Partial specific volume (cm3 g−1)b | 0.728 |
| Contrast ( Δρ × 1010 cm−2)b | 3.021 |
| Mr (from I(0)) | 51,679 |
| Calculated monomeric Mr from sequence | 25,290 |
| Software used | |
| Primary data reduction | SAXS/WAXS beamline software |
| Data processing | PRIMUS |
| Rigid body modelling | CORAL, BUNCH |
| Three-dimensional graphics representation | PyMOL |
| Graphics representation | Excel, SASPLOT |
a Reported for 1.80 mg ml−1.
b Determined with MULCh (72).
FIGURE 1.
F1L(1–202) SAXS analysis and oligomeric state. A, log plot of SAXS raw data. Concentrations are in descending order, commencing at 3.60 mg/ml followed by 1.80, 0.82, 0.43, 0.21, and 0.12 mg/ml. B, Guinier plots of SAXS data. Concentrations are as in A.
TABLE 2.
Summary of the SAXS data and analysis of MVA F1L(1–202) oligomeric state in solution
| Concentration | Rg | Oligomeric state |
|---|---|---|
| mg/ml | Å | |
| 3.60 | 34.80 | 2.20 |
| 1.80 | 34.10 | 2.04 |
| 0.82 | 33.70 | 2.09 |
| 0.43 | 33.10 | 2.12 |
| 0.21 | 31.50 | 2.14 |
| 0.12 | 32.50 | 2.17 |
The calculated molecular mass from I(0) on the absolute scattering scale across the concentration range is ∼53 kDa, corresponding to a dimeric oligomerization state (Table 2). The dimer obtained was expected and previously observed in the known MVA F1L crystal structures (49, 50). Details of the scattering analysis are summarized in Table 1. The experimentally determined Rg for F1L is 34 Å compared with the calculated Rg from the crystal structure of F1L Bcl-2 fold of 20.89 Å.
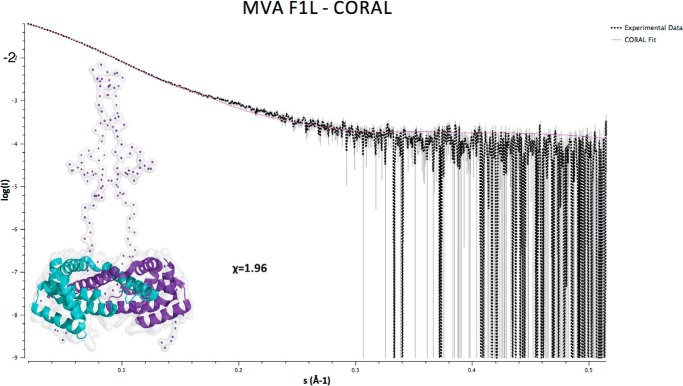
Because partial crystal structures of MVA F1L were available (49, 50), we attempted to model the missing regions of the available structures using a rigid body modeling approach. Modeling was carried out with CORAL (54) and BUNCH (56) using the monomeric F1L structure (residues 51–186; Protein Data Bank code 4d2m) and generating the dimer via imposition of a P2 symmetry. A model for F1L obtained using BUNCH fits the experimental scattering data poorly as indicated by a χ of 2.7 (data not shown). In contrast, a model calculated using CORAL resulted in an improved fit of the scattering curves with a value of χ of 1.96 (Fig. 2). In the model, both F1L N termini protrude away from the Bcl-2 fold in an extended configuration spanning residues 1–50 in addition to the N-terminal hexahistidine tag (Fig. 2). The shape of both F1L N termini in the model suggests an absence of ordered secondary structure, thus rendering F1L residues 1–50 unfolded.
FIGURE 2.
F1L(1–202) CORAL envelope and fit to the scattering data (1.80 mg/ml).
We next examined the ability of F1L to inhibit Apaf-1-activated apoptosis using a model system based on S. cerevisiae. In this assay, expression of a constitutively active form of Apaf-1 together with both caspase-9 and caspase-3 results in yeast death (61), which can be efficiently rescued by the overexpression of caspase inhibitors. Co-expression of full-length VACV(COP)F1L with Apaf-1, caspase-9, and caspase-3 did not protect yeast cells from cell death (Fig. 3A) when compared with the established potent pan-caspase inhibitor AcP35 (63). In contrast, in a complementary yeast-based assay where yeast growth arrest is induced by overexpression of Bak or Bax (64), VACV(COP)F1L was able to rescue yeast growth arrest during Bak overexpression, similar to mammalian Bcl-xL, suggesting that the lack of anticaspase activity of F1L is not due to a lack of expression (Fig. 3B).
FIGURE 3.
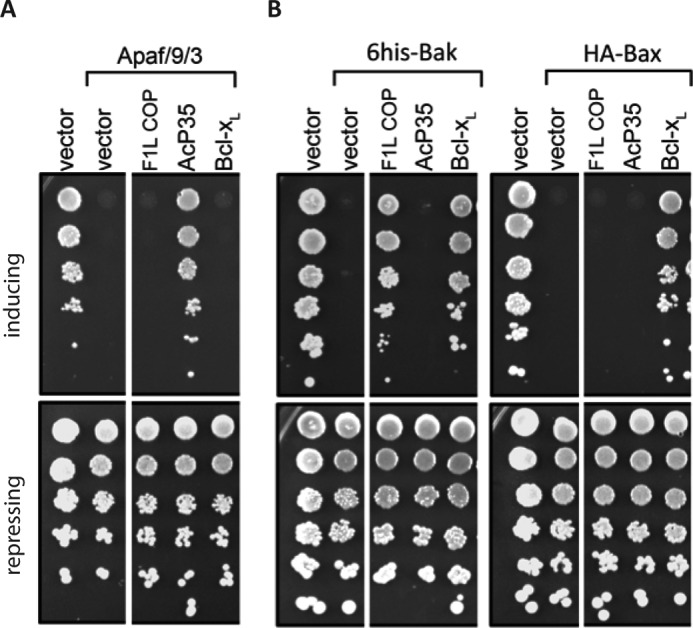
F1L is not able to prevent caspase-mediated yeast cell death. A, reconstitution of the caspase activation pathway (Apaf-1, caspase-9, and caspase-3) in S. cerevisiae. Yeast were co-transformed with constructs encoding Apaf-1, caspase-9, and caspase-3 and the indicated apoptosis regulatory proteins or empty vector, each under the control of an inducible (GAL) promoter. 5-Fold serial dilutions were spotted onto inducing galactose or repressing glucose plates. Colony size indicates growth rate, and colony number reflects cell viability. Each dilution was also spotted onto a control plate (glucose) to verify that equivalent numbers of each transformant were spotted. B, yeast co-transformed with constructs encoding Bax or Bak and the indicated prosurvival proteins, each under the control of an inducible (GAL) promoter, were spotted onto inducing galactose or repressing glucose plates as 5-fold serial dilutions. A and B, images are representative of two independent experiments. White spaces indicate where an irrelevant lane was spliced out of the plate photographs.
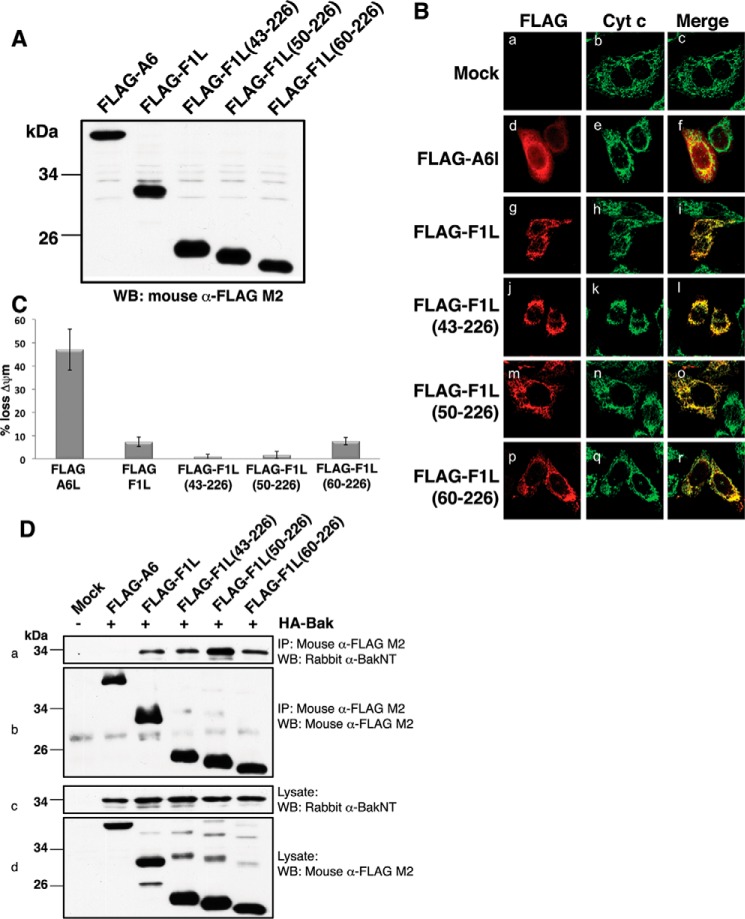
We then sought to define the contribution that the 60 N-terminal residues make to F1L-mediated apoptosis inhibition. We generated FLAG-tagged F1L constructs spanning residues 43–226, 50–226, and 60–226 as well as full-length F1L. All constructs were expressed at comparable levels in HEK293T cells (Fig. 4A), and fluorescence microscopy revealed that all constructs co-localized to mitochondria (Fig. 4B), suggesting that the N-terminal part of F1L does not play a role in determining its subcellular localization. Next we determined the ability of N-terminally truncated VACV(COP)F1L to protect against TNFα-induced apoptosis using flow cytometry. FLAG-F1L(43–226), FLAG-F1L(50–226), FLAG-F1L(60–226), and FLAG-F1L(1–226) inhibited TNFα-induced cell death with comparable potency when compared with a control protein (vaccinia virus A6L), suggesting that deletion of the N-terminal 60 residues has no bearing on F1L-mediated inhibition of apoptosis (Fig. 4C). Furthermore, co-immunoprecipitation experiments revealed that all F1L truncations retained their ability to bind Bak, suggesting that the truncation mutants are folded and active (Fig. 4D).
FIGURE 4.
Functional characterization of F1L truncation mutants. A, VACV(COP)F1L N-terminal truncations expression levels in HeLa cells. B, subcellular localization of VACV(COP)F1L N-terminal truncations in HeLa cells. HeLa cells were transiently transfected with empty vector (panels a–c), FLAG-F1L (panels g–i), FLAG-F1L(43–226) (panels j–l), FLAG-F1L(50–226) (panels m–o), FLAG-F1L(60–226) (panels p–r), or FLAG-A6L (panels d–f) as a control. 12 h post-transfection cells were stained with rabbit anti-FLAG M2 antibody and imaged using a Zeiss Axiovert laser scanning microscope. Mitochondria were visualized by staining for cytochrome (Cyt) c. C, VACV(COP)F1L N-terminal truncations potently protect HeLa cells against TNFα-induced apoptosis. Apoptosis was induced with 10 ng/ml TNFα combined with 5 mg/ml cycloheximide. Apoptosis was assessed by quantifying tetramethylrhodamine ethyl ester fluorescence via flow cytometry, and the percentage of cells that demonstrated a loss of mitochondrial membrane potential (ΔΨm) is given on the y axis. All experiments were performed in triplicate; error bars show S.D. D, F1L truncations efficiently immunoprecipitate Bak. HEK293T cells were co-transfected with pcDNA3-HA-BAK as well as with pcDNA3-FLAG-A6L, pcDNA3-FLAG-F1L, pcDNA3-FLAG-F1L(43–226), pcDNA3-FLAG-F1L(50–226), or pcDNA3-FLAG-F1L(60–226). FLAG-tagged F1L was immunoprecipitated with monoclonal mouse anti-FLAG M2 antibody, Bak was detected using a polyclonal rabbit anti-Bak N terminus (NT) antibody (panel a), and FLAG-F1L constructs were detected using mouse anti-FLAG M2 antibody (panel b). Panels c and d show whole cell lysates probed with polyclonal rabbit anti-Bak N terminus antibody or mouse anti-FLAG M2 antibody as loading controls, respectively. IP, immunoprecipitation; WB, Western blotting.
Discussion
Viruses utilize a range of strategies when subverting premature host cell apoptosis including receptor homologs, inhibitors of apoptosis proteins, Bcl-2 homologs, and direct caspase inhibitors. Although the vast majority of these effector molecules have been shown to fulfill only a single purpose to date, recently emerging evidence is pointing to the potential for multifunctionality (5) as showcased by vaccinia virus N1L. In addition to being an inhibitor of the intrinsic apoptosis pathway (39, 40), N1L also inhibits NF-κB with this dual functionality being mediated via two independent binding sites on N1 (41). N1L dimerization has proven crucial for the NF-κB inhibition, having no effect on apoptosis regulation and BH3-only protein binding.
Similarly, vaccinia virus F1L, an established Bcl-2-like antiapoptotic protein, has been assigned additional functions: the ability to inhibit caspase-9 (51) as well as a role in inflammasome activation (52). According to previous studies, F1L inhibits the recruitment of procaspase-9 to Apaf-1 through its binding to caspase-9 by its N-terminal residues. These results were further supported by data that suggest that F1L N-terminal 15-residue motif is key for caspase-9 inhibition in vitro.
Because these activities were identified in an F1L N-terminal region of unknown structure, we investigated MVA F1L with its intact N terminus using small angle x-ray scattering. Our structural analysis indicates that the MVA F1L N-terminal residues 1–50 prior to the Bcl-2-like domain form an extended unfolded region preceding the Bcl-2 globular fold described previously.
Because this F1L N-terminal domain should in principle be able to access the caspase-9 active site, we next investigated the ability of VACV(COP)F1L to inhibit caspase-9 in a reconstituted caspase-3/caspase-9/Apaf-1 system that efficiently mimics caspase activity in yeast. However, we were unable to observe any inhibition of caspase-9 by VACV(COP)F1L in contrast to the established pan-caspase inhibitor AcP35, a potent caspase inhibitor from the insect virus Autographa californica multiple nucleopolyhedrovirus (63, 65). Unlike VACV(COP)F1L, AcP35 fully prevented yeast death in this system, suggesting that VACV(COP)F1L is not a caspase-9 or indeed a caspase-3/Apaf-1 inhibitor. Furthermore, VACV(COP)F1L was able to rescue yeast from Bak-induced growth arrest in a complementary yeast assay, suggesting that F1L is efficiently expressed in yeast in an active form. Lastly, mutant VACV(COP)F1L that lacked the N-terminal section implicated in caspase-9 inhibition showed no discernible effect on the efficiency and potency of VACV(COP)F1L-mediated inhibition of apoptosis in cellular systems.
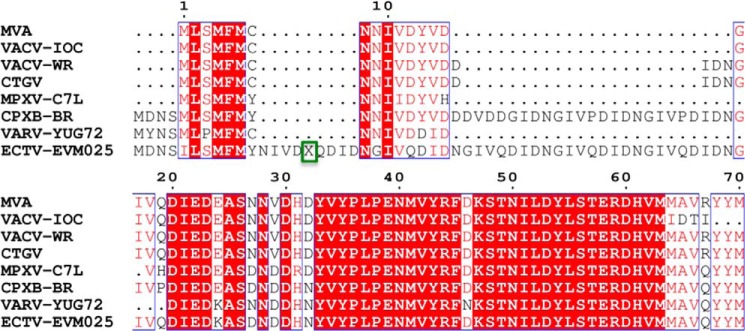
Mammalian prosurvival Bcl-2 family members have been shown to harbor intrinsically disordered regions (IDRs) in addition to a folded Bcl-2 domain that have been assigned important regulatory functions. IDRs in the Bcl-2 proteins frequently bear multiple regulation sites such as those for phosphorylation, deamidation, and ubiquitination (66). Although the presence of IDRs is a recurring feature of the Bcl-2 family, their location within the protein varies. Bcl-2 and Bcl-xL both contain large IDRs of ∼50 residues as insertions between the α1 and α2 helices, whereas in other family members such insertions are substantially shorter. In contrast, the loop in Mcl-1 connecting α1 and α2 is structured with an extended IDR of low complexity spanning 160 residues N-terminally found prior to the Bcl-2-like fold (67). In Boo (68) and Bcl-B (69), an IDR insertion has been identified that connects the α5 and α6 helices. Viral Bcl-2 proteins appear to be largely free of IDRs (5). In BHRF1, the loop connecting α1 and α2 is unstructured in solution (26) but adopts a short helix in the crystal structure (27). All other structures of viral Bcl-2 proteins indicate a highly compact architecture with predominantly short loops connecting the α helical secondary structure elements. We now show that vaccinia virus F1L is a notable exception to this general observation because it harbors a 60-residue unfolded region N-terminal to the Bcl-2 fold. The N-terminal 60 residues in F1L do not display substantial sequence variations among F1L ORFs from a range of vaccinia viruses or related poxviruses, suggesting that the majority if not all N-terminal regions in the various F1L ORFs are unstructured (Fig. 5).
FIGURE 5.
Sequence alignment of F1L protein N termini from different orthopoxviruses. Shown is sequence alignment of the N-terminal 70 residues of MVA F1L (UniProt accession number O57173), vaccinia virus strain IOC (VACV-IOC) (UniProt accession number: A5HDI4), vaccinia virus strain Western Reserve (VACV-WR) (UniProt accession number P24356), Cantagalo virus (CTGV) (UniProt accession number A5HDH9), monkeypox virus C7L (MPXV-C7L) (UniProt accession number Q8V547), cowpox virus strain Brighton Red (CPXB-BR) (UniProt accession number Q8QN17), variola virus strain Yugoslavia 1972 V72-164 (VARV-YUG72) (UniProt accession number Q0N5C2), and ectromelia virus EVM025 (ECTV-EVM025) (UniProt accession number Q8JLH9). Conserved regions are boxed in red. X highlighted with a green box represents the following sequence: NGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIVQDIDNGIV.
A number of F1L homologs have been identified within Poxviridae including ectromelia virus EVM025, monkeypox virus C7L, and variola virus F1L; however, only EVM025 and variola virus F1L have been studied in any detail. Both EVM025 and variola virus F1L were shown to be apoptosis inhibitors with EVM025 inhibiting Bak directly, whereas Bax-mediated apoptosis was inhibited via the sequestration of Bim (70). In contrast, variola virus F1L was only able to inhibit Bak-mediated apoptosis and did not show any ability to engage Bim (71). Both EVM025 and variola virus F1L harbor long N-terminal extensions prior to their Bcl-2 fold; however, neither has been functionally characterized.
We have shown that the N-terminal region prior to the Bcl-2 fold in vaccinia virus F1L adopts an extended, unstructured configuration. Furthermore, biochemical and cellular assays suggest that this region is unable to functionally inhibit caspase-9. The lack of impact of deletion of the extended N-terminal region of F1L on its ability to inhibit apoptosis suggests that any capability of this region to inhibit caspases is vestigial and only plays a minor role in modulating apoptosis.
Author Contributions
S. C. and B. M. designed, performed, and analyzed the experiments shown in Figs. 1 and 2 and contributed to writing the manuscript. R-L. B. and S. C. designed, performed, and analyzed the experiments shown in Fig. 4. D. P-E. and C. J. H. designed, performed, and analyzed the experiments shown in Fig. 3. M. B. designed experiments in Fig. 4, conceived the project, and contributed to writing the manuscript. M. K. designed experiments in Figs. 1 and 2, conceived the project, and wrote the manuscript. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Nathan Cowieson for helpful discussion and staff at the SAXS/WAXS beamline at the Australian Synchrotron for help with x-ray data collection.
This work was supported in whole or part by Australian Research Council Grant FT130101349 and National Health and Medical Research Council Project Grant APP1007918. The authors declare that they have no conflicts of interest with the contents of this article.
Small angle x-ray scattering data were deposited at the Small Angle Scattering Biological Data Bank (SASDB) under accession code SASDBV4.
- BH
- Bcl-2 homology
- MVA
- modified vaccinia Ankara
- SAXS
- small angle x-ray scattering
- WAXS
- wide angle x-ray scattering
- EGFP
- enhanced GFP
- Rg
- radius of gyration
- VACV(COP)
- vaccinia virus strain Copenhagen
- IDR
- intrinsically disordered region.
References
- 1.Yu E., Zhai D., Jin C., Gerlic M., Reed J. C., and Liddington R. (2011) Structural determinants of caspase-9 inhibition by the vaccinia virus protein, F1L. J. Biol. Chem. 286, 30748–30758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaux D. L., Haecker G., and Strasser A. (1994) An evolutionary perspective on apoptosis. Cell 76, 777–779 [DOI] [PubMed] [Google Scholar]
- 3.Galluzzi L., Brenner C., Morselli E., Touat Z., and Kroemer G. (2008) Viral control of mitochondrial apoptosis. PLoS Pathog. 4, e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youle R. J., and Strasser A. (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 5.Kvansakul M., and Hinds M. G. (2015) The Bcl-2 family: structures, interactions and targets for drug discovery. Apoptosis 20, 136–150 [DOI] [PubMed] [Google Scholar]
- 6.Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., and Huang D. C. (2005) Differential targeting of pro-survival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 7.Giam M., Huang D. C., and Bouillet P. (2008) BH3-only proteins and their roles in programmed cell death. Oncogene 27, Suppl. 1, S128–S136 [DOI] [PubMed] [Google Scholar]
- 8.Kvansakul M., and Hinds M. G. (2014) The structural biology of BH3-only proteins. Methods Enzymol. 544, 49–74 [DOI] [PubMed] [Google Scholar]
- 9.Liu X., Dai S., Zhu Y., Marrack P., and Kappler J. W. (2003) The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity 19, 341–352 [DOI] [PubMed] [Google Scholar]
- 10.Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., Thompson C. B., and Fesik S. W. (1997) Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275, 983–986 [DOI] [PubMed] [Google Scholar]
- 11.Gavathiotis E., Suzuki M., Davis M. L., Pitter K., Bird G. H., Katz S. G., Tu H. C., Kim H., Cheng E. H., Tjandra N., and Walensky L. D. (2008) BAX activation is initiated at a novel interaction site. Nature 455, 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leshchiner E. S., Braun C. R., Bird G. H., and Walensky L. D. (2013) Direct activation of full-length proapoptotic BAK. Proc. Natl. Acad. Sci. U.S.A. 110, E986–E995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindsten T., Ross A. J., King A., Zong W. X., Rathmell J. C., Shiels H. A., Ulrich E., Waymire K. G., Mahar P., Frauwirth K., Chen Y., Wei M., Eng V. M., Adelman D. M., Simon M. C., et al. (2000) The combined functions of proapoptotic Bcl-2 family members Bak and Bax are essential for normal development of multiple tissues. Mol. Cell 6, 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green D. R., and Kroemer G. (2004) The pathophysiology of mitochondrial cell death. Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 15.Yuan S., Yu X., Topf M., Ludtke S. J., Wang X., and Akey C. W. (2010) Structure of an apoptosome-procaspase-9 CARD complex. Structure 18, 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reubold T. F., and Eschenburg S. (2012) A molecular view on signal transduction by the apoptosome. Cell. Signal. 24, 1420–1425 [DOI] [PubMed] [Google Scholar]
- 17.Riedl S. J., and Salvesen G. S. (2007) The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 8, 405–413 [DOI] [PubMed] [Google Scholar]
- 18.Pop C., and Salvesen G. S. (2009) Human caspases: activation, specificity, and regulation. J. Biol. Chem. 284, 21777–21781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchini A., Tomkinson B., Cohen J. I., and Kieff E. (1991) BHRF1, the Epstein-Barr virus gene with homology to Bc12, is dispensable for B-lymphocyte transformation and virus replication. J. Virol. 65, 5991–6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White E., Sabbatini P., Debbas M., Wold W. S., Kusher D. I., and Gooding L. R. (1992) The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor α. Mol. Cell. Biol. 12, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng E. H., Nicholas J., Bellows D. S., Hayward G. S., Guo H. G., Reitz M. S., and Hardwick J. M. (1997) A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc. Natl. Acad. Sci. U.S.A. 94, 690–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banadyga L., Gerig J., Stewart T., and Barry M. (2007) Fowlpox virus encodes a Bcl-2 homologue that protects cells from apoptotic death through interaction with the proapoptotic protein Bak. J. Virol. 81, 11032–11045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banadyga L., Veugelers K., Campbell S., and Barry M. (2009) The fowlpox virus BCL-2 homologue, FPV039, interacts with activated Bax and a discrete subset of BH3-only proteins to inhibit apoptosis. J. Virol. 83, 7085–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nava V. E., Cheng E. H., Veliuona M., Zou S., Clem R. J., Mayer M. L., and Hardwick J. M. (1997) Herpesvirus saimiri encodes a functional homolog of the human bcl-2 oncogene. J. Virol. 71, 4118–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Q., Petros A. M., Virgin H. W., Fesik S. W., and Olejniczak E. T. (2002) Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus. Proc. Natl. Acad. Sci. U.S.A. 99, 3428–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Q., Petros A. M., Virgin H. W., Fesik S. W., and Olejniczak E. T. (2003) Solution structure of the BHRF1 protein from Epstein-Barr virus, a homolog of human Bcl-2. J. Mol. Biol. 332, 1123–1130 [DOI] [PubMed] [Google Scholar]
- 27.Kvansakul M., Wei A. H., Fletcher J. I., Willis S. N., Chen L., Roberts A. W., Huang D. C., and Colman P. M. (2010) Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1. PLoS Pathog. 6, e1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham K. A., Opgenorth A., Upton C., and McFadden G. (1992) Myxoma virus M11L ORF encodes a protein for which cell surface localization is critical in manifestation of viral virulence. Virology 191, 112–124 [DOI] [PubMed] [Google Scholar]
- 29.Goldmacher V. S., Bartle L. M., Skaletskaya A., Dionne C. A., Kedersha N. L., Vater C. A., Han J.-W., Lutz R. J., Watanabe S., Cahir McFarland E. D., Kieff E. D., Mocarski E. S., and Chittenden T. (1999) A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to bcl-2. Proc. Natl. Acad. Sci. U.S.A. 96, 12536–12541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jurak I., Schumacher U., Simic H., Voigt S., and Brune W. (2008) Murine cytomegalovirus m38.5 protein inhibits Bax-mediated cell death. J. Virol. 82, 4812–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manzur M., Fleming P., Huang D. C., Degli-Esposti M. A., and Andoniou C. E. (2009) Virally mediated inhibition of Bax in leukocytes promotes dissemination of murine cytomegalovirus. Cell Death Differ. 16, 312–320 [DOI] [PubMed] [Google Scholar]
- 32.Fleming P., Kvansakul M., Voigt V., Kile B. T., Kluck R. M., Huang D. C., Degli-Esposti M. A., and Andoniou C. E. (2013) MCMV-mediated inhibition of the pro-apoptotic Bak protein is required for optimal in vivo replication. PLoS Pathog. 9, e1003192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banadyga L., Lam S. C., Okamoto T., Kvansakul M., Huang D. C., and Barry M. (2011) Deerpox virus encodes an inhibitor of apoptosis that regulates Bak and Bax. J. Virol. 85, 1922–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamoto T., Campbell S., Mehta N., Thibault J., Colman P. M., Barry M., Huang D. C., and Kvansakul M. (2012) Sheeppox virus SPPV14 encodes a Bcl-2-like cell death inhibitor that counters a distinct set of mammalian pro-apoptotic proteins. J. Virol. 86, 11501–11511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wasilenko S. T., Stewart T. L., Meyers A. F., and Barry M. (2003) Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc. Natl. Acad. Sci. U.S.A. 100, 14345–14350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartlett N., Symons J. A., Tscharke D. C., and Smith G. L. (2002) The vaccinia virus N1L protein is an intracellular homodimer that promotes virulence. J. Gen. Virol. 83, 1965–1976 [DOI] [PubMed] [Google Scholar]
- 37.Douglas A. E., Corbett K. D., Berger J. M., McFadden G., and Handel T. M. (2007) Structure of M11L: a myxoma virus structural homolog of the apoptosis inhibitor, Bcl-2. Protein Sci. 16, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kvansakul M., van Delft M. F., Lee E. F., Gulbis J. M., Fairlie W. D., Huang D. C., and Colman P. M. (2007) A structural viral mimic of prosurvival bcl-2: a pivotal role for sequestering proapoptotic bax and bak. Mol. Cell 25, 933–942 [DOI] [PubMed] [Google Scholar]
- 39.Aoyagi M., Zhai D., Jin C., Aleshin A. E., Stec B., Reed J. C., and Liddington R. C. (2007) Vaccinia virus N1L protein resembles a B cell lymphoma-2 (Bcl-2) family protein. Protein Sci. 16, 118–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooray S., Bahar M. W., Abrescia N. G., McVey C. E., Bartlett N. W., Chen R. A., Stuart D. I., Grimes J. M., and Smith G. L. (2007) Functional and structural studies of the vaccinia virus virulence factor N1 reveal a Bcl-2-like anti-apoptotic protein. J. Gen. Virol. 88, 1656–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maluquer de Motes C., Cooray S., Ren H., Almeida G. M., McGourty K., Bahar M. W., Stuart D. I., Grimes J. M., Graham S. C., and Smith G. L. (2011) Inhibition of apoptosis and NF-κB activation by vaccinia protein N1 occur via distinct binding surfaces and make different contributions to virulence. PLoS Pathog. 7, e1002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burton D. R., Caria S., Marshall B., Barry M., and Kvansakul M. (2015) Structural basis of deerpox virus-mediated inhibition of apoptosis. Acta Crystallogr. D Biol. Crystallogr. 71, 1593–1603 [DOI] [PubMed] [Google Scholar]
- 43.Stewart T. L., Wasilenko S. T., and Barry M. (2005) Vaccinia virus F1L protein is a tail-anchored protein that functions at the mitochondria to inhibit apoptosis. J. Virol. 79, 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor J. M., Quilty D., Banadyga L., and Barry M. (2006) The vaccinia virus protein F1L interacts with Bim and inhibits activation of the pro-apoptotic protein Bax. J. Biol. Chem. 281, 39728–39739 [DOI] [PubMed] [Google Scholar]
- 45.Fischer S. F., Ludwig H., Holzapfel J., Kvansakul M., Chen L., Huang D. C., Sutter G., Knese M., and Häcker G. (2006) Modified vaccinia virus Ankara protein F1L is a novel BH3-domain binding protein and acts together with the early viral protein E3L to block virus-associated apoptosis. Cell Death Differ. 13, 109–118 [DOI] [PubMed] [Google Scholar]
- 46.Wasilenko S. T., Banadyga L., Bond D., and Barry M. (2005) The vaccinia virus F1L protein interacts with the proapoptotic protein Bak and inhibits Bak activation. J. Virol. 79, 14031–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Postigo A., Cross J. R., Downward J., and Way M. (2006) Interaction of F1L with the BH3 domain of Bak is responsible for inhibiting vaccinia-induced apoptosis. Cell Death Differ. 13, 1651–1662 [DOI] [PubMed] [Google Scholar]
- 48.Campbell S., Hazes B., Kvansakul M., Colman P., and Barry M. (2010) Vaccinia virus F1L interacts with Bak using highly divergent Bcl-2 homology domains and replaces the function of Mcl-1. J. Biol. Chem. 285, 4695–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell S., Thibault J., Mehta N., Colman P. M., Barry M., and Kvansakul M. (2014) Structural insight into BH3 domain binding of vaccinia virus antiapoptotic F1L. J. Virol. 88, 8667–8677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kvansakul M., Yang H., Fairlie W. D., Czabotar P. E., Fischer S. F., Perugini M. A., Huang D. C., and Colman P. M. (2008) Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands. Cell Death Differ. 15, 1564–1571 [DOI] [PubMed] [Google Scholar]
- 51.Zhai D., Yu E., Jin C., Welsh K., Shiau C. W., Chen L., Salvesen G. S., Liddington R., and Reed J. C. (2010) Vaccinia virus protein F1L is a caspase-9 inhibitor. J. Biol. Chem. 285, 5569–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerlic M., Faustin B., Postigo A., Yu E. C., Proell M., Gombosuren N., Krajewska M., Flynn R., Croft M., Way M., Satterthwait A., Liddington R. C., Salek-Ardakani S., Matsuzawa S., and Reed J. C. (2013) Vaccinia virus F1L protein promotes virulence by inhibiting inflammasome activation. Proc. Natl. Acad. Sci. U.S.A. 110, 7808–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirby N. M., and Cowieson N. P. (2014) Time-resolved studies of dynamic biomolecules using small angle x-ray scattering. Curr. Opin. Struct. Biol. 28, 41–46 [DOI] [PubMed] [Google Scholar]
- 54.Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 56.Petoukhov M. V., and Svergun D. I. (2005) Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J 89, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 58.Konarev P. V., Petoukhov M. V., and Svergun D. I. (2001) MASSHA—a graphics system for rigid-body modelling of macromolecular complexes against solution scattering data. J. Appl. Crystallogr. 34, 527–532 [Google Scholar]
- 59.Jabbour A. M., Puryer M. A., Yu J. Y., Lithgow T., Riffkin C. D., Ashley D. M., Vaux D. L., Ekert P. G., and Hawkins C. J. (2006) Human Bcl-2 cannot directly inhibit the Caenorhabditis elegans Apaf-1 homologue CED-4, but can interact with EGL-1. J. Cell Sci. 119, 2572–2582 [DOI] [PubMed] [Google Scholar]
- 60.Hawkins C. J., Wang S. L., and Hay B. A. (1999) A cloning method to identify caspases and their regulators in yeast: identification of Drosophila IAP1 as an inhibitor of the Drosophila caspase DCP-1. Proc. Natl. Acad. Sci. U.S.A. 96, 2885–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hawkins C. J., Silke J., Verhagen A. M., Foster R., Ekert P. G., and Ashley D. M. (2001) Analysis of candidate antagonists of IAP-mediated caspase inhibition using yeast reconstituted with the mammalian Apaf-1-activated apoptosis mechanism. Apoptosis 6, 331–338 [DOI] [PubMed] [Google Scholar]
- 62.Beaumont T. E., Shekhar T. M., Kaur L., Pantaki-Eimany D., Kvansakul M., and Hawkins C. J. (2013) Yeast techniques for modeling drugs targeting Bcl-2 and caspase family members. Cell Death Dis. 4, e619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brand I. L., Civciristov S., Taylor N. L., Talbo G. H., Pantaki-Eimany D., Levina V., Clem R. J., Perugini M. A., Kvansakul M., and Hawkins C. J. (2012) Caspase inhibitors of the P35 family are more active when purified from yeast than bacteria. PLoS One 7, e39248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jurgensmeier J. M., Krajewski S., Armstrong R. C., Wilson G. M., Oltersdorf T., Fritz L. C., Reed J. C., and Ottilie S. (1997) Bax- and Bak-induced cell death in the fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell 8, 325–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brand I. L., Green M. M., Civciristov S., Pantaki-Eimany D., George C., Gort T. R., Huang N., Clem R. J., and Hawkins C. J. (2011) Functional and biochemical characterization of the baculovirus caspase inhibitor MaviP35. Cell Death Dis. 2, e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rautureau G. J., Day C. L., and Hinds M. G. (2010) Intrinsically disordered proteins in bcl-2 regulated apoptosis. Int. J. Mol. Sci. 11, 1808–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Day C. L., Chen L., Richardson S. J., Harrison P. J., Huang D. C., and Hinds M. G. (2005) Solution structure of prosurvival Mcl-1 and characterization of its binding by proapoptotic BH3-only ligands. J. Biol. Chem. 280, 4738–4744 [DOI] [PubMed] [Google Scholar]
- 68.Rautureau G. J., Day C. L., and Hinds M. G. (2010) The structure of Boo/Diva reveals a divergent Bcl-2 protein. Proteins 78, 2181–2186 [DOI] [PubMed] [Google Scholar]
- 69.Rautureau G. J., Yabal M., Yang H., Huang D. C., Kvansakul M., and Hinds M. G. (2012) The restricted binding repertoire of Bcl-B leaves Bim as the universal BH3-only prosurvival Bcl-2 protein antagonist. Cell Death Dis. 3, e443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mehta N., Taylor J., Quilty D., and Barry M. (2015) Ectromelia virus encodes an anti-apoptotic protein that regulates cell death. Virology 475, 74–87 [DOI] [PubMed] [Google Scholar]
- 71.Marshall B., Puthalakath H., Caria S., Chugh S., Doerflinger M., Colman P. M., and Kvansakul M. (2015) Variola virus F1L is a Bcl-2-like protein that unlike its vaccinia virus counterpart inhibits apoptosis independent of Bim. Cell Death Dis. 6, e1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whitten A. E., Cai S., and Trewhella J. (2008) MULCh: modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J. Appl. Crystallogr. 41, 222–226 [Google Scholar]