Abstract
Sirtuins are NAD+-dependent lysine deacylases, regulating a variety of cellular processes. The nuclear Sirt1, the cytosolic Sirt2, and the mitochondrial Sirt3 are robust deacetylases, whereas the other sirtuins have preferences for longer acyl chains. Most previous studies investigated sirtuin-catalyzed deacylation on peptide substrates only. We used the genetic code expansion concept to produce natively folded, site-specific, and lysine-acetylated Sirt1–3 substrate proteins, namely Ras-related nuclear, p53, PEPCK1, superoxide dismutase, cyclophilin D, and Hsp10, and analyzed the deacetylation reaction. Some acetylated proteins such as Ras-related nuclear, p53, and Hsp10 were robustly deacetylated by Sirt1–3. However, other reported sirtuin substrate proteins such as cyclophilin D, superoxide dismutase, and PEPCK1 were not deacetylated. Using a structural and functional approach, we describe the ability of Sirt1–3 to deacetylate two adjacent acetylated lysine residues. The dynamics of this process have implications for the lifetime of acetyl modifications on di-lysine acetylation sites and thus constitute a new mechanism for the regulation of proteins by acetylation. Our studies support that, besides the primary sequence context, the protein structure is a major determinant of sirtuin substrate specificity.
Keywords: acetylation, GTPase, p53, sirtuin, synthetic biology
Introduction
Lysine acetylation has been known for a long time to occur on histones, where it is crucial for the regulation of gene expression (1–3). Progress in mass spectrometry in the last decade enabled the identification and relative quantification of thousands of lysine acetylation sites in all cellular compartments (3–6). These studies revealed that lysine acetylation is highly dynamic, conserved from bacteria to man, and is present in proteins covering all essential cellular functions (6–11). Although a rigorous functional investigation of most of these sites is lacking, data from a proteomic study suggest that lysine acetylation occurs predominantly in structured regions in contrast to phosphorylation (3). Furthermore, several reports suggested compartment-specific consensus sequences for lysine acetylation (3, 9, 12).
Because of the high level of sensitivity of today's mass spectrometers, one of the biggest challenges in lysine acetylation research is to distinguish biologically relevant from irrelevant sites among the thousands of sites identified (13–16). Measures for the biological significance of a particular lysine acetylation site are its regulation by lysine acetyltransferases (KATs),3 lysine deacetylases (KDACs) and, unless it results in a gain-of-function or dominant-negative effect, its accumulation to sufficiently high stoichiometries (13–16). In contrast to phosphorylation, which mostly occurs on serine/threonine or tyrosine residues and where hundreds of kinases and phosphatases are known, the number of genes encoding for KATs and KDACs is limited (17–26). To date, 22 proteins with KAT activity have been identified (17–19, 27, 28). These enzymes use acetyl-CoA as an acetyl donor molecule and transfer the acetyl moiety to the ϵ-amino group of lysine side chains (27). Because of its low sequence conservation, their identification is difficult, and a recent report suggests that there might be more KATs, termed orphan-KATs, encoded in the human genome (18). For KATs, it has been proposed that their incorporation into multiprotein complexes regulates their substrate specificity.
KDACs can be subdivided into two major groups, the classical KDACs, showing a Zn2+-dependent catalytic mechanism, and the sirtuins (derived from yeast Sir2, silent information regulator 2), using NAD+ as a cofactor for catalysis (28–32). The human genome encodes for 11 classical deacetylases (classes I, II, and IV) and for seven sirtuins. Although classical KDACs exist in multiprotein complexes that determine their substrate specificity, the determinants of sirtuin substrate specificity are less well understood (28, 30, 33–36). Sirtuins were discovered in the 1980s in a genetic screen for genes that silence the mating type loci in Saccharomyces cerevisiae (37, 38). Later, it was found that Sir2 regulates aging and life span in yeast and that it has KDAC activity (39, 40). The role of sirtuins to mediate the effect of dietary restriction on life span was shown later for diverse organisms such as Drosophila, Caenorhabditis elegans, and even for mice (41–43). Sirtuins are involved in many aspects of cellular regulation, and their dysfunction can have drastic consequences on cellular function promoting the organism's aging process and the development of severe diseases such as cancer and neurodegenerative diseases (44–52). Because of the dependence on the levels of acetyl-CoA as activated acyl donor molecules and NAD+ as a cofactor for sirtuin-mediated deacetylation, the acetylation status of the proteome is tightly connected to the cellular metabolic state (50, 53, 54).
Sirtuins are present in all cellular compartments, with Sirt1, -6, and -7 localized predominantly in the nucleus, Sirt2 in the cytosol, and Sirt3, -4, and -5 in the mitochondrial matrix. For Sirt5, recent reports suggest that besides its mitochondrial localization it is also active in the cytosol (55). Importantly, of the seven sirtuins encoded in the human genome, only Sirt1, -2, and -3 have a robust deacetylase activity. The other sirtuins have preferences for longer acyl chains (56, 57).
Much research is focused on the development of novel strategies to activate or inhibit sirtuin function (58, 59). Several small molecule compounds were developed to achieve this goal (22, 30, 58, 60–65). However, it is still often difficult to design inhibitors that specifically target one sirtuin without affecting others. Therefore, another promising strategy is to design mechanistic inhibitors by combining substrate-based peptides with acetyl-lysine analogues such as trifluoroacetyl-lysine and thioacetyl-lysine, which show a markedly reduced rate of deacetylation (32, 66–68). We and others show that some sirtuins present a remarkable level of substrate specificity for certain acetylation sites (14, 15, 69). However, whether structural features or the primary sequence is the main determinant of specificity remains an unresolved question. A major drawback of nearly all the functional and structural investigations performed so far is that they used peptide substrates to draw conclusions about specificities and putative consensus sequences for sirtuin-catalyzed deacetylation. However, how the specificity of sirtuin-catalyzed deacetylation is determined in natively folded substrate proteins has hardly been investigated thus far. One study, in which deacetylation by sirtuins was analyzed with chemically acetylated protein, suggests that structural features do in fact play an important role for substrate recognition (70). Here, we present the first data on sirtuin specificity in a context of site-specifically acetylated full-length substrate proteins. We discovered that Sirt1–3 are able to deacetylate two neighboring acetylated lysine side chains and that structural components are major determinants of sirtuin substrate specificity. These results suggest that data derived from peptide-based experiments should be cross-checked for their validity in the natively folded context.
Experimental Procedures
Expression and Purification of Proteins
Purification of site-specifically acetylated proteins was performed as described (14). Sirt1(225–664), Sirt2(50–356), Ran, Hsp10, CypD(43–207), MnSOD(25–222), and PEPCK1 were purified as His6-tagged fusion proteins from pRSF-Duet-1 or the described modified version thereof for site-specific incorporation of acetyl-l-lysine (pRSF-Duet-1-MbtRNACUA-MbPylRS) (14). Sirt3(118–399) and Sirt2(50–356) (without a His tag for crystallization) were expressed as GST fusion proteins also as described from pGEX4T5/TEV (based on pGEX-4T1, GE Healthcare). p53 was also expressed as a GST fusion protein using a pRSF-Duet-1-MbtRNACUA-MbPylRS-derived vector, which had its dual multiple cloning site replaced by the GST open reading frame and multiple cloning site of the described pGEX-4T5/TEV vector (14). In our hands, GST-p53 fusion protein was not efficiently cleaved by TEV protease when still bound to the column. Thus, the GST-p53 was first eluted from the column, digested with TEV in solution overnight, and subsequently concentrated in a 30-kDa MWCO Centricon for a final S200 size exclusion chromatography. p53 eluted early after the size exclusion peak.
Antibodies
The following antibodies (ABs) from Abcam were used (catalogue number, dilution): acetyl-lysine (ab21623, 1:1500); His tag (ab18184, 1:2000); and HRP-coupled secondary ABs against rabbit (ab6721, 1:10,000) and mouse (ab6728, 1:10,000). The ABs against Sirt3 (sc49744, 1:400) and p53 (sc-99, 1:1000) were purchased from Santa Cruz Biotechnology, Inc. For the anti-Ran AcK37 AB, two rabbits were immunized with an acetyl-TGEFE(AcK)KYVAT-(C)-peptide. A 72-day-bleed yielding the optimal Ran AcK37/Ran-WT signal ratio was further purified by negative adsorption with the non-acetylated peptide. The flow-through was subjected to positive adsorption using the acetylated peptide. All these steps were carried out by Thermo Fisher custom antibody services. The resulting Ran AcK37 AB was used at a dilution of 1:65.
Isothermal Titration Calorimetry (ITC)
ITCs were carried out on MicroCal ITC200 or Auto-ITC200 instruments (GE Healthcare) based on Ref. 71. All measurements were done in buffer A (100 mm NaCl, 50 mm Tris-Cl, pH 7.4, 5 mm MgCl2, 2 mm β-mercaptoethanol). For the measurements with protein, 10 μm nicotinamide was included in the buffer. Parameters were as follows: 20 °C, 2 μl at 4-s injections, 1000 rpm stirring speed, target differential power value of 6. The isotherms were fitted with the built-in one-site binding model of the Origin MicroCal extension software.
Crystallization
Sirt2(50–356) was crystallized in sitting drop plates (150-nl drop size) at a concentration of 10 mg/ml in buffer A and supplemented with a 1.2-fold molar excess of the RanTFAcK37 13-mer peptide in 0.1 m HEPES, pH 7.5, 2.0 m NH4SO4. Crystals grew after 3 days at 20 °C and were washed twice in mother liquor, which was supplemented with 15 and 30% d-glucose, 10% glycerol as cryoprotectant, respectively, prior to storage in liquid N2. The crystals belonged to the space group P6122 with two heterodimers per asymmetric unit. The native dataset was collected at the Swiss Light Source in Villigen, Switzerland, with the X06DA/PX3 beamline at a wavelength of 1.0 Å and 100 K using a Dectris PILATUS 2 M detector. The oscillation range was 0.1°, and 1200 frames were collected. The program iMOSFLM was used for indexing and integration (72, 73). AIMLESS was used for scaling (74). Initial phases were determined with the program Phaser (as part of the suite Phenix-dec-1893) (75) and by using the Sirt2(43–370)·S2iL5 structure as search model (PDB code 4L3O) (76). The program Coot 0.7.1 was used to build a model into the 2Fo − Fc and Fo − Fc electron density maps in iterative rounds of refinement, which were carried out with REFMAC5 (77–79). Quality assessment of the structure model was done with Molprobity (80). Figures of structures were prepared with PyMOL version 1.7.2.0 (81). Data collection and refinement statistics are given in Table 1. Rwork was calculated as follows: Rwork = Σ| Fo − Fc|/Σ Fo, with Fo and Fc as the observed and calculated structure factor amplitudes, respectively. Rfree is calculated as Rwork using the test set reflections only.
TABLE 1.
Data collection and refinement statistics (molecular replacement)
The structure has been deposited at the Protein Data Bank (code 5FYQ).
| Sirt2(50–356)·Ran(31–43) TfAcK37 | |
|---|---|
| Data collection | |
| Space group | P6122 |
| Cell dimensions | |
| a, b, c (Å) | 114.95, 114.95, 206.48 |
| α, β, γ (°) | 90.0, 90.0, 120.0 |
| Resolution (Å)a | 56.61–3.00 (3.18–3.00) |
| Observed reflections | 187,174 (31766) |
| Unique reflections | 16,888 (2655) |
| Rsym or Rmergeb (%) | 18.0 (72.1) |
| Rmeasc (%) | 19.7 (78.6) |
| I/σI | 13.7 (3.9) |
| CC1/2d | 0.994 (0.840) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 11.1 (12.0) |
| Refinement | |
| Resolution (Å) | 56.61 (3.0) |
| No. of used reflections | 15,978 |
| Rwork/Rfreee | 23.16/27.11 |
| No. of non-hydrogen atoms | |
| Total | 4606 |
| Protein | 4417 |
| SO42−/Zn2+ | 10/2 |
| Water | 177 |
| B-factors (Å2) | |
| Protein | 49.05 |
| SO42− | 72.23 |
| Zn2+ | 93.43 |
| Water | 51.94 |
| Average B-factors (Å2) | |
| Main chain | 48.75 |
| Side chain | 49.35 |
| All atoms | 49.23 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.537 |
| Ramachandran plotf (%) | |
| Favored | 95.4 |
| Allowed | 4.4 |
| Outliers | 0.2 |
a Values for the highest resolution shell are shown in parentheses.
b Rsym = ΣΣ|I(hkl; j) − 〈I(hkl)〉|/ΣΣ〈I(hkl)〉, with I(hkl; j) being the jth measurement of the intensity of the unique reflection (hkl), and 〈I(hkl)〉 being the mean overall symmetry-related measurements (107).
c Rmeas = ΣΣ[n(hkl)/(n(hkl − 1)]1/2|I(hkl; j) − 〈I(hkl)〉|/ΣΣ〈I(hkl)〉 (107).
d CC1/2 correlation coefficient from (108).
e Rwork = Σ |Fo − Fc|/Σ Fo, where Fo and Fc are the observed and calculated structure factor amplitudes. Rfree is calculated similarly to Rwork using random 5% of working set of reflections (109).
f MolProbity is from Ref. 110.
Mass Spectrometry
Mass spectrometry was performed as described previously but Glu-C was used instead of trypsin for the digest, and MaxQuant search parameters were changed accordingly (14).
Deacetylation Assays
Deacetylase assays were done in buffer A supplemented with 1 mm NAD+ (AppliChem) at 23 °C. Where indicated, deacetylation assays for PEPCK1 were performed in a different buffer (see below). For the single turnover deacetylation experiment (Fig. 1, G and H), NAD+ concentrations were as indicated. Proteins were used at 12 μm unless otherwise stated and deacetylase concentrations as indicated. Samples taken at the indicated time points were heated for 5 min at 95 °C to stop the reaction. Samples were separated by SDS-PAGE, blotted on PVDF membranes, blocked with 5% skim milk (in PBS/Tween 20), incubated with the primary AB overnight at 4 °C, and incubated with the secondary HRP-coupled AB for 1 h at room temperature. Detection of HRP was carried out with ECL solution (Carl Roth) and a Vilber Fusion Xpress or by exposure to x-ray film. The ImageJ Gel Analyzer tool was used for densitometric quantification.
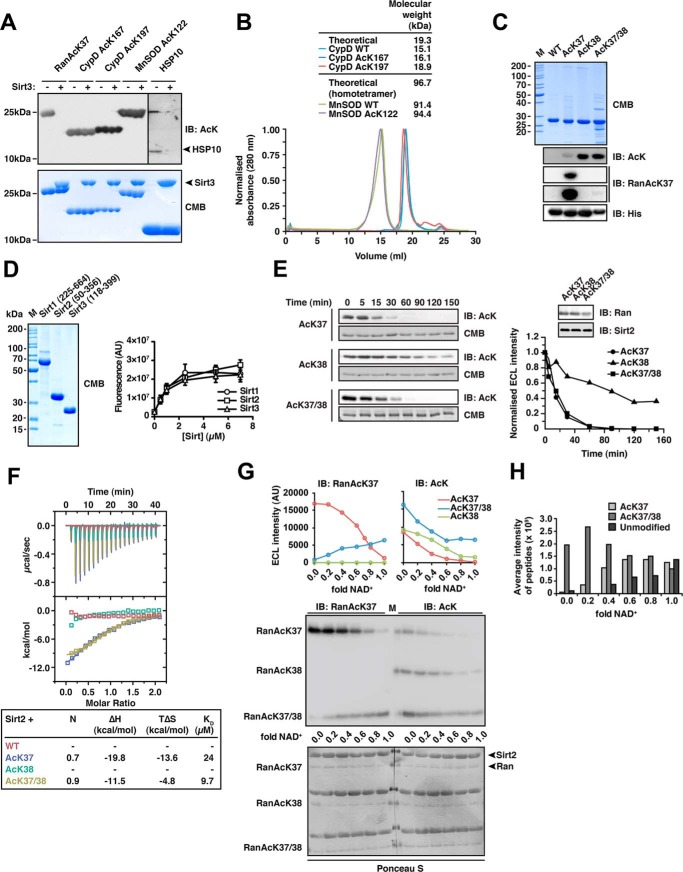
FIGURE 1.
Sirt2 can deacetylate di-acetylated Ran. A, deacetylation of selected substrate proteins by Sirt3. We prepared site-specifically acetylated Ran AcK37, CypD AcK167, and AcK197, MnSOD AcK122, and Hsp10 AcK56 using the genetic code expansion concept and analyzed its deacetylation by Sirt3 by immunoblotting using an anti-acetyl-lysine antibody (anti-AcK-AB). Sirt3 was used in equimolar ratio with the respective protein substrate (40 μm), except for Hsp10 (40 μm Sirt3 and 80 μm Hsp10). Coomassie Brilliant (CMB) blue staining is shown as loading control. B, analytical size exclusion chromatography of CypD and MnSOD proteins used in this study (Superdex 200 10/300). To assess the quality of the non-acetylated and site-specifically lysine-acetylated CypD (WT, AcK167, and AcK197) and MnSOD (WT and MnSOD AcK122) proteins used here, we performed analytical SEC experiments. All CypD proteins eluted as apparent monomer from the SEC column. MnSOD behaved as apparent tetramer on SEC, as reported earlier. For both, MnSOD and CypD, all acetylated proteins behaved as the non-acetylated proteins showing that lysine acetylation did not affect protein folding or oligomerization. C, Ran was lysine-acetylated at Lys-38 and at Lys-37/Lys-38 using the genetic code expansion concept. Coomassie Brilliant Blue staining of an SDS-PAGE with 5 μg of protein shows the final purity. Anti-AcK-AB was used to show the successful incorporation of acetyl-l-lysine into Ran. The anti-AcK-AB detects acetyl-l-lysine with different sensitivity depending on the sequence context. Detection with anti-His6-AB serves as a loading control. An antibody raised against a Ran AcK37-derived 11-mer peptide (see “Experimental Procedures”) is specific for Ran AcK37, showing only a very weak signal for RanAcK37/38 after long exposure to x-ray film and does not detect Ran AcK38. D, final purity of the Sirt1, Sirt2, and Sirt3 enzymes used in this study. 5 μg of each sirtuin was separated by SDS-PAGE and the gel Coomassie Brilliant Blue-stained to assess the final purity. The enzymatic activities were tested with a fluor-de-lys assay. All enzymes are active and show a similar activity. Error bars represent the standard error of the mean for three independent measurements. E, kinetics of Ran AcK37, AcK38, and AcK37/38 deacetylation by Sirt2. 12 μm site-specifically lysine-acetylated Ran protein was incubated with 0.14 μm Sirt2 for the indicated times. The level of Ran acetylation was assessed by immunoblotting using a specific anti-AcK-AB (left). Ran AcK37 and di-acetylated Ran AcK37/38 were deacetylated with similar kinetics, whereas Ran AcK38 shows a strongly reduced Sirt2-catalyzed deacetylation rate. The SDS-polyacrylamide gels corresponding to the Western blots were stained with Coomassie Brilliant Blue and serve as a loading control. The reactions were quantified densitometrically using ImageJ software (right). F, Ran AcK37 and AcK37/38 bind to Sirt2 as shown by isothermal titration calorimetry. 45 μm Ran-WT, Ran AcK37, AcK38, or AcK37/38 protein was titrated with 450 μm Sirt2 (50–356, non-His6-tagged). Ran AcK37 and RanAcK37/38 bind with affinities of 24 and 9.7 μm, respectively. Both reactions are solely driven by the reaction enthalpy (Ran AcK37 ΔH, −19.8 kcal/mol; TΔS, −13.6 kcal/mol; Ran AcK37/38 ΔH,−11.5 kcal/mol; TΔS, −4.8 kcal/mol). No binding heat was observed under these conditions for Ran-WT and Ran AcK38. G, deacetylation of Ran AcK38 by Sirt2 is favored over AcK37 in di-acetylated Ran AcK37/38.12 μm Ran AcK37. AcK38 and AcK37/38 were used in Sirt2-mediated deacetylation reactions with increasing amounts of the essential cofactor NAD+ (Sirt2 concentration, 24 μm, molar ratio Ran AcK/NAD+, 0–1.0). Using the anti-Ran AcK37 AB, we observed a linear decrease of acetylated Ran AcK37. In contrast, the signal increased for di-acetylated AcK37/38 as a function of the NAD+ concentration. This suggests that AcK38 in the context of Ran AcK37/38 is deacetylated preferentially by Sirt2. As expected, AcK38 shows no signal using the anti-Ran AcK37 AB. Using the pan-anti-AcK-AB, all acetylated Ran proteins showed a linear decrease in the acetylation level. However, although AcK37 and AcK38 were nearly completely deacetylated at a molar Ran AcK/Sirt2 ratio of 1:1, this was not the case for di-acetylated AcK37/38R. Detection with an anti-Ran-AB was used as a loading control. Densitometric analyses of the signals obtained with the anti-AcK-AB and anti-Ran AcK37 are shown at the top. H, mass spectrometric analysis of the experiment is shown in F. The reactions were analyzed by digest with Glu-C followed by ESI-MS/MS. Without NAD+, there is only di-acetylated Ran present. With increasing NAD+ concentrations, the average intensity of peptides corresponding to di-acetylated Ran decreases and that of the non-acetylated Ran increases. The amount of Ran AcK37 peptide increases with increasing NAD+ concentrations suggesting that Ran AcK38 is deacetylated first. IB, immunoblot.
Activity Assay for Sirtuins
Activity assays for purified Sirts were performed in buffer B (25 mm Tris, pH 8.0, 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2, 1 mm NAD+) using a fluorogenic substrate (SRP0308, Sigma) at 5 μm and the indicated amounts of enzyme. The reaction volume was 50 μl, and reactions were set up in triplicate in black 96-well plates. After incubation at 37 °C for 30 min, the reaction was stopped with 5 μl of developer solution (5 μg/ml trypsin, 100 mm nicotinamide) and subsequently incubated for 30 min at room temperature. Fluorescence readout was done on a Beckman Paradigm (wavelength, excitation 350 nm; emission 450 nm).
PEPCK1 Activity Assay
PEPCK1 activity was measured essentially as described (82). The assay was performed with an LS55 fluorescence spectrometer (PerkinElmer Life Sciences) and Quartz SUPRASIL cuvettes (10-mm light path, Hellma Analytics) in buffer containing 100 mm HEPES-NaOH, pH 7.2, 100 mm KHCO3, 3 mm phosphoenolpyruvate, 2 mm GDP, 2 mm MgCl2, 0.2 mm MnCl2, 10 mm DTT, 0.2 mm NADH, and 2 units/ml of malate dehydrogenase (Amresco, Ultra Pure). PEPCK1 was added at a final concentration of 25 nm, and the fluorescence drop was followed over 60 min (excitation wavelength of 345 nm and a readout wavelength of 470 nm).
Analytical Size Exclusion Chromatography (SEC)
Analytical SEC was performed using a Superdex 200 10/300 GL column (GE Healthcare) with a flow rate of 0.5 ml/min. A calibration curve was generated using a gel filtration low and high molecular weight kit (GE Healthcare) according to the manufacturer's instructions. SECs for PEPCK1 were performed in 100 mm HEPES-NaOH, pH 7.2, 100 mm KHCO3, 2 mm MgCl2, 0.2 mm MnCl2, 10 mm DTT. A solution of 100 μl at 3 mg/ml was loaded onto the column. Fractions corresponding to the correct molecular weight were concentrated using a 30-kDa MWCO Centricon and subsequently used for the activity assay and, where indicated, also for deacetylation assays (buffer was supplemented with 1 mm NAD+ for the deacetylation experiment). SECs for CypD and MnSOD were performed in buffer A.
Analysis of Potency of 2′/3′-O-Acetyl-ADP-ribose to Lysine-acetylated MnSOD und PEPCK1
To test whether 2′/3′-O-acetyl-ADP-ribose (Santa Cruz Biotechnology, Inc.; sc-481663) can non-enzymatically lysine-acetylate PEPCK1 or MnSOD, we incubated recombinantly expressed MnSOD and PEPCK1 (both at a concentration of 0.6 μm) with 2 mm 2′/3′-O-acetyl-ADP-ribose in vitro (2 h, 23 °C) and analyzed the reactions by immunoblotting using an anti-AcK antibody. The buffer for PEPCK1 was as described above for the SEC runs, additionally containing 1 mm NAD+.
Results
Sirtuin-catalyzed Deacetylation of Natively Folded Lysine-acetylated Substrate Proteins
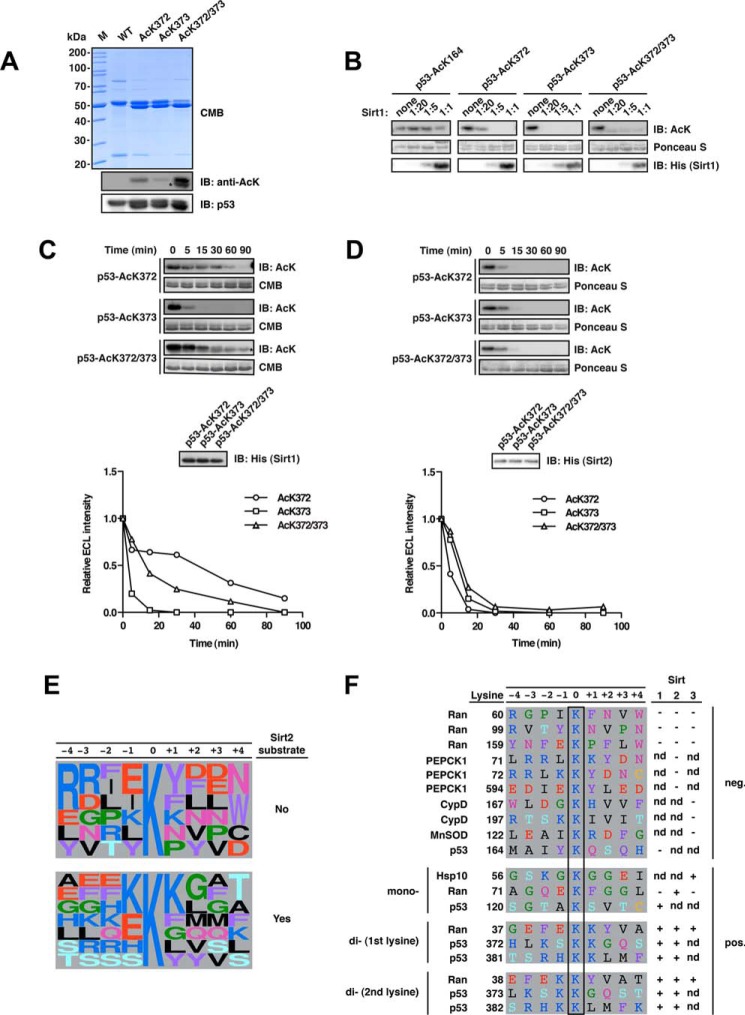
A synthetically evolved, orthogonal acetyl-lysyl-tRNA synthetase/tRNACUA pair from Methanosarcina barkeri expressed in Escherichia coli was used to site-specifically incorporate N-(ϵ)-acetyl-l-lysine (AcK) into sirtuin substrate proteins. To this end, we selected reported substrate proteins of Sirt1–3, showing a robust lysine deacetylase activity (Sirt1, p53; Sirt2, Ran, PEPCK1; Sirt3, CypD, MnSOD, Hsp10) (83–90). Notably, for the natively folded mitochondrial proteins MnSOD acetylated at Lys-122 and CypD acetylated at Lys-167 (mouse, Lys-166) and Lys-197 (mouse, Lys-196), we could not detect any deacetylation by Sirt3 even using molar enzyme/substrate concentrations of 1:1, although these were reported Sirt3 substrates. To exclude that the failure to detect sirtuin-catalyzed deacetylation of CypD and MnSOD was due to the protein quality being compromised, we performed analytical SEC experiments. As expected, acetylated CypD eluted at a volume corresponding to a monomer of ∼16.1 and 18.9 kDa for AcK167 and AcK197, respectively, (theoretical size, 19.3 kDa). MnSOD eluted at a volume consistent with its known homotetrameric form (theoretical size of the tetramer, 96.7 kDa; Fig. 1B). For Hsp10, we observed a robust AcK56 deacetylation catalyzed by Sirt3, although the signal we obtained using the anti-AcK antibody was very weak (Fig. 1A).
To investigate how and whether the structural context, besides its primary sequence, affects the specificity of sirtuin-catalyzed deacetylation, we used acetylated Ran as a model system. We have previously shown that Ran acetylated at Lys-37 (Ran AcK37) is most efficiently deacetylated by recombinant Sirt2 but also by Sirt1 and -3 (14). We tested whether Sirt2 is also able to deacetylate the adjacent AcK38 of natively folded Ran, which is located in a similar structural environment as AcK37 and was also identified as being acetylated in human (3). Moreover, we speculated that acetylation of both lysines might interfere with the deacetylation by Sirt2 at this di-acetyl-lysine site. Ran was thus purified in three different site-specifically acetylated forms as follows: Ran AcK37, AcK38, and Ran AcK37/38 (Fig. 1C). To test the deacetylation of these proteins in vitro, Sirt2(50–356) was purified from E. coli, and its activity was confirmed using a fluor-de-lys assay (Fig. 1D). We observed that the deacetylation of AcK38R (superscript R indicates Ran) was much slower compared with AcK37R. Surprisingly, when Ran was acetylated at both Lys-37 and Lys-38 (AcK37/38R), deacetylation occurred at a rate comparable with that observed for AcK37R (Fig. 1E). Consistently, affinity measurement of Sirt2 to full-length Ran AcK37 and Ran AcK37/38 by ITC resulted in similar KD values of 24 and 9.7 μm, whereas binding was neither detected for Ran AcK38 nor for non-acetylated Ran under the assay conditions (Fig. 1F).
Sirt2 Di-deacetylates Ran AcK37/38
Given the slow rate of AcK38R deacetylation, we speculated that di-deacetylation of Ran occurs sequentially with AcK38R being deacetylated first. We tested this hypothesis by incubation of all three acetylated versions of Ran, namely AcK37R, AcK38R, and AcK37/38R, with a 2-fold molar excess of Sirt2 and increasing NAD+ concentrations up to a molar NAD+/Ran ratio of 1:1. Because enzymatic activity of sirtuins is strictly dependent on the cofactor NAD+, we expected to observe only partial deacetylation of each substrate when NAD+ is present in submolar amounts. To discriminate between different acetylation sites on a Western blot, we used a Ran AcK37-specific antibody (anti-Ran AcK37 AB). This antibody shows only a weak signal for di-acetylated Ran AcK37/38 and does not recognize Ran-WT (WT is wild type) or Ran AcK38 (Fig. 1G). With this anti-Ran AcK37 AB, an increase in the signals for Ran AcK37/38 was observed toward higher concentrations of NAD+ suggesting that Ran AcK37 accumulated under these conditions. As expected, analyzing the reactions using a pan-anti-AcK AB, we observed a decrease in signal intensity for all three Ran AcKs with increasing NAD+ concentrations. The same applies to Ran AcK37 detected with the anti-Ran AcK37 AB (Fig. 1G). To confirm these data, we performed an identification and quantification of the peptides after deacetylation of Ran AcK37/38 by Sirt2 by mass spectrometry. Consistent with the immunoblotting data, for the Sirt2-catalyzed deacetylation of Ran AcK37/38, we observed increasing peptide intensities for Ran AcK37 toward higher NAD+ concentrations, after Glu-C digest followed by LC-MS/MS analysis. Moreover, the amounts of unmodified peptide increased with higher NAD+ concentrations, although peptides corresponding to Ran AcK38 were either not identified or only in trace amounts (Fig. 1H). Taken together, these data suggest that Sirt2 first deacetylates Ran AcK37/38 at AcK38 and subsequently at AcK37 and that this di-deacetylation reaction occurs with a significantly faster rate than mono-deacetylation of Ran AcK38.
Sirt2 Binds to 13-mer Ran Peptides Encompassing AcK37, AcK38, and AcK37/38
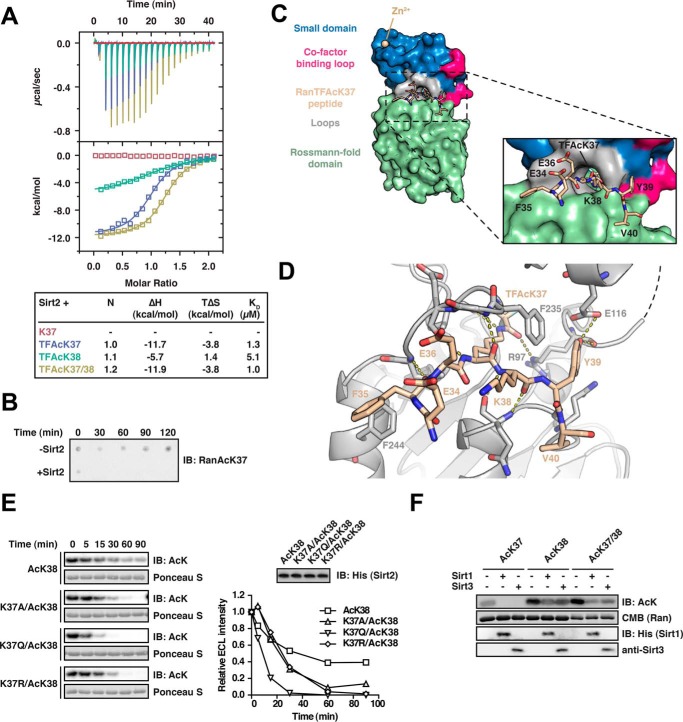
To further structurally characterize the interaction of (di-)acetylated Ran and Sirt2, we initiated crystallization trials with Sirt2 and full-length Ran AcK37, AcK38, and AcK37/38. However, because our attempts to crystallize the Ran protein in complex with Sirt2 failed, we decided to use Ran-derived 13-mer peptides containing the acetyl-lysine analogue Nϵ-trifluoroacetyl-l-lysine (TFAcK, peptide sequence 31LTGEFEKKYVATL43) to stabilize the complex (68). Using ITC, we first confirmed that the three TFAcK-13-mer Ran peptides (Ran(31–43)-TFAcK37, Ran(31–43)-TFAcK38, and Ran(31–43)-TFAcK37/38) behaved similar to the full-length protein regarding the binding to Sirt2. We obtained an ∼5-fold weaker affinity and less favorable reaction enthalpy for the Ran TFAcK38–13-mer (KD, 5.0 μm; ΔH, −5.7 kcal/mol) compared with TFAcK37- and TFAcK37/38–13-mers (KD, 1.3 μm; ΔH, −11.7 kcal/mol; and KD, 1.0 μm; ΔH, −11.9 kcal/mol; Fig. 2A). This suggests that the amino acid sequence surrounding residues Lys-37R and Lys-38R affects the binding specificity of Sirt2 toward Ran and that our structural data would thus provide insights into the molecular events leading to deacetylation of both sites. Interestingly, in a recent microarray screen for sirtuin targets using immobilized acetylated peptides, the 13-mer for Ran AcK37 was not found to be deacetylated by Sirt2 (69). However, when we analyzed its deacetylation in solution, we found that the 13-mer not only binds but that it is also efficiently deacetylated as also observed for the full-length Ran protein (Fig. 2B). Thus, this particular peptide was most likely not properly accessible upon immobilization on the microarray surface, which, however, was apparently not the case for other peptides (as determined by Rauh et al. (69)).
FIGURE 2.
Characterization of the Ran-Sirt2 interaction using trifluoroacetyl-lysine peptides and Ran mutants. A, Ran TFAcK37, TFAcK38, and TFAcK37/38 13-mer peptides bind to Sirt2 in the micromolar range as shown by isothermal titration calorimetry. 30 μm Sirt2 (50–356, non-His6-tagged) was titrated with 300 μm trifluorolysine-acetylated Ran 13-mer peptide (Ran amino acids 31–43). The reactions are all driven by a favorable reaction enthalpy, ΔH, whereas the reaction of the TFAcK38 peptide is the only one driven by both favorable enthalpy and favorable entropy, TΔS. This shows that the TFAcK38 peptide uses a distinct mechanism of binding. B, Ran 13-mer AcK37 peptide is deacetylated by Sirt2. 133 μm of Ran AcK37 13-mer peptide (Ran amino acids 31–43) was spotted on a nitrocellulose membrane. Addition of 0.5 μm Sirt2 leads to a complete deacetylation already after 30 min, compared with the non-enzyme control (−Sirt2). The specific anti-Ran AcK37 AB was used for detection by immunoblotting. C, crystal structure of Sirt2 (amino acids 50–356) in complex with the TFAcK37 13-mer Ran peptide (amino acids 31–43). The structure was solved at a resolution of 3 Å. Left, overall Sirt2·Ran(31–43) TFAcK37 structure. Sirt2 is shown as a surface representation. Blue, small Zn2+-binding domain; red, cofactor binding loop; green, Rossmann-fold domain; beige, Ran(31–43) TFAcK37 peptide. Right, close-up of the Ran(31–43) TFAcK37 peptide and Sirt2. Three charged residues Glu-34R, Glu-36R, and the Lys-38R are solvent-exposed. The hydrophobic tunnel leading to the Sirt2 active site is too narrow to simultaneously adopt two acetylated lysine residues. The TFAcK37 penetrates into a hydrophobic tunnel formed by the small subdomain and the Rossmann-fold domain. D, two aromatic residues Phe-35R and Tyr-39R of the Ran(31–43) 13-mer TFAcK37 peptide act as anchor points through the formation of stacking interactions with the aromatic residues in Phe-244 and Phe-235 of Sirt2, respectively. Furthermore, the peptide forms several main chain hydrogen bonds with Sirt2, highlighted with the dotted yellow lines, and the hydroxyl group of Tyr-39R forms a hydrogen bond with the main chain carbonyl-oxygen of Arg-97 of Sirt2. The peptide adopts a crescent- and w-shaped conformation. E, effects of Ran mutations on the kinetics of Sirt2-mediated di-deacetylation of Ran AcK38. To assess whether electrostatic or steric properties of AcK37R increase the deacetylation rate of AcK38R, we created Lys-37R mutant proteins in the Ran AcK38 background. The non-mutated mono-acetylated Ran AcK38 shows the slowest Sirt2-catalyzed deacetylation of the proteins compared. All mutant variants (RanK37A/AcK38, K37R/AcK38, and K37Q/AcK38) show an accelerated Sirt2-catalyzed deacetylation compared with the mono-acetylated Ran AcK38 protein (12 μm Ran and 0.06 μm Sirt2). This shows that both electrostatic and steric properties of AcK37R contribute to the observed accelerated Sirt2-mediated deacetylation of AcK38R. F, Sirt1 and Sirt3 are able to di-deacetylate Ran AcK37/38. Ran AcK37, Ran AcK38, and di-acetylated Ran AcK37/38 were incubated with Sirt1 or Sirt3, and the acetylation level was assessed by immunoblotting (IB) using an anti-AcK antibody (12 μm Ran was incubated with 0.8 μm of the indicated Sirtuin for 30 min). Ran AcK38 shows a relatively strong remaining anti-AcK signal compared with the di-acetylated AcK37/38R, suggesting that, as observed for Sirt2, the presence of AcK37R stimulates the deacetylation of AcK38R by Sirt1 and Sirt3. Coomassie (CMB) staining is shown as loading control for Ran. Sirtuins were detected with the indicated antibodies.
Sirt2 Binds Ran AcK37 Using a Binding Site Flanked by Aromatic Residues
Attempts to crystallize Sirt2 with the Ran-13-mer TFAcK37/38 peptide failed, but we were able to solve the atomic structure of Sirt2 in complex with the Ran TFAcK37–13-mer peptide by molecular replacement to a final resolution of up to 3.0 Å (Table 1). Although we performed the crystallization trials with the 31LTGEFE-TFAcK-KYVATL43 peptide, we did not obtain electron density for the three far N- and C-terminal residues. These residues may thus be flexible in the Sirt2-bound state and most likely do not contribute to Sirt2 substrate recognition. As expected, the peptide is located in the substrate-binding groove formed between the Sirt2 Rossmann-fold domain and the small domain, and the TFAcK37R residue is buried in the active site of the enzyme (Fig. 2C). Consistent with previous reports (91–93), the peptide is bound to Sirt2 through extensive polar interactions between main chain atoms of the peptide and Sirt2 (e.g. between the amide nitrogen of Gln-267Sirt2 and the carbonyl oxygen of Lys-38R or between the amide nitrogen of Leu-239Sirt2 and the carbonyl oxygen of Phe-35R). Interestingly, the two aromatic residues (Phe-35R and Tyr-39R), which are flanking the TFAcK37 residue, are oriented toward aromatic residues Phe-244Sirt2 and Phe-235Sirt2, respectively, and thus likely contribute to peptide binding through stacking interactions. The hydroxyl group of Tyr-39R furthermore forms a hydrogen bond with the main chain carbonyl oxygen of Arg-97Sirt2, which also forms a hydrogen bond to the trifluoroacetyl-lysine side chain. These additional interactions might orient and position AcK37R for catalysis and thus explain the observed faster deacetylation of Ran AcK37 compared with Ran AcK38 (Fig. 2D).
Because it was still an open question how the presence of an acetyl moiety at Ran Lys-37 primes the deacetylation of Ran AcK38 due to the lack of structural data for the di-acetylated variant, we generated Lys-37 mutant versions (K37A/K37Q/K37R) of Ran in the AcK38 background and analyzed these for deacetylation with Sirt2. Interestingly, all three mutants (K37A/AcK38R, K37Q/AcK38R, and K37R/AcK38R) were deacetylated faster than non-mutated Lys-38-acetylated Ran, suggesting that a lysine neighboring AcK38R is the least favorable amino acid for Sirt2 catalysis, at least in this sequence context. Therefore, electrostatic and steric effects at Lys-37R contribute to the increased deacetylation rate of the di-acetylation of Ran AcK37/38.
Ability to Di-deacetylate Two Neighboring Acetyl-lysines Is a Common Feature of Sirt1–3
Next, we tested whether the closely related Sirt1 and Sirt3 are also able to deacetylate two adjacent acetyl-lysine residues. As reported earlier, Ran AcK37 constitutes a substrate not only for Sirt2 but also for Sirt1 and Sirt3 (14). We purified Sirt1(255–664) and Sirt3(118–399) and performed a fluor-de-lys assay, which showed that both enzymes possess a deacetylase activity similar to our recombinantly expressed and purified Sirt2 (Fig. 1D). Importantly, as for Sirt2, Sirt1 and Sirt3 deacetylated Ran AcK38 less efficiently than Ran AcK37/38. This suggests that Sirt1–3 share features of substrate recognition and that di-deacetylation by sirtuins is a common mechanism, at least for Ran AcK37/38 (Fig. 2F).
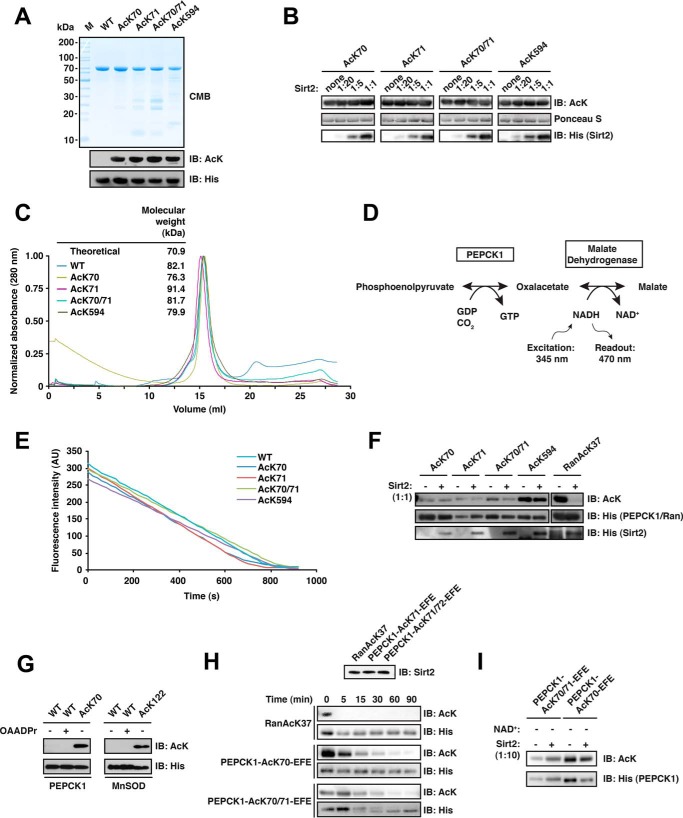
PEPCK1 Can Be Converted into a Substrate of Sirt2 by Mutation of the Sequence N-terminal of Its Di-acetylation Site AcK70/71 to the Corresponding Ran Sequence
In a physiological context, the lifetime of an acetyl modification of a protein may be relatively long due to slow sirtuin-mediated deacetylation at the respective site. The subsequent acetylation of a neighboring lysine residue could release this state by making both sites favored targets of a particular sirtuin. The dynamics of this regulatory mechanism depends on which of two adjacent lysine residues is acetylated first by the respective KAT and which lysine is preferentially deacetylated. We asked whether di-deacetylation constitutes a general mechanism exerted by sirtuins on different substrate proteins. To answer this question, we screened the literature for proteins shown to be di-acetylated at two neighboring lysines and for which deacetylation is mediated by Sirt1, -2, or -3.
For phosphoenolpyruvate carboxykinase 1 (PEPCK1), it has been shown that gluconeogenesis is negatively regulated by acetylation at Lys-70, Lys-71, and Lys-594. Acetylation of PEPCK1 leads to its degradation and is counterbalanced by Sirt2 (88). Interestingly, as for Ran, the sequence of PEPCK1 at the di-deacetylation site is 70KKY72, possibly representing a short motif for di-deacetylation. Evidence for KKY being a high affinity motif at least for Sirt2 also comes from an in vitro selection screen for circular peptidic Sirt2 inhibitors where, in 10 out of 15 cases, clones were isolated that expressed peptides containing the sequence R(I/V)(TFAcK)RY with IC50 values for Sirt2 in the low nanomolar range. Notably, even the linear peptide RI(TFAcK)RY inhibited Sirt2 with an IC50 of 31 nm (94). With arginine having similar physicochemical properties as lysine, we thus speculated that the KKY motif binds strongly to Sirt2 and that it could furthermore be a prerequisite for di-deacetylation.
We purified full-length PEPCK1 AcK70, AcK71, AcK70/71, and for completeness also AcK594 (Fig. 3A), and we tested which of those sites are deacetylated by Sirt2 in vitro. To detect also weak deacetylation reactivity, we incubated all PEPCK1 variants with increasing concentrations of Sirt2 up to an equimolar ratio and an excess of NAD+. Unexpectedly, under the conditions used in our in vitro assay, none of the reported PEPCK1 acetylation sites was deacetylated by Sirt2 (Fig. 3B). To rule out that this was due to PEPCK1 being improperly folded or that protein quality was otherwise compromised, we performed analytical SEC runs with all acetylated PEPCK1 variants in a previously described buffer, which has been optimized for PEPCK1 activity (82). This confirmed that the PEPCK1 preparations were of the correct molecular weight and showed no signs of aggregation (Fig. 3C). We then used the PEPCK1 from the analytical SEC runs to confirm the activity of wild type and the acetylated PEPCK1 preparations in a coupled enzyme assay (Fig. 3, D and E), again as described (82). Subsequently, also the deacetylation assays were repeated with this PEPCK1 material. However, even under these optimized conditions and using a Sirt2/PEPCK1 ratio of 1:1, we did not observe deacetylation for any of the three mono-acetylated PEPCK1 variants nor for the di-acetylated PEPCK AcK70/71 (Fig. 3F). We then sought to rule out the unlikely case that deacetylation does occur but is followed by non-enzymatic acetylation by 2′/3′-O-acetyl-ADP-ribose arising during the reaction, which might then result in a similarly high acetylation signal. To this end, we tested whether incubation of PEPCK1-WT with 2 mm 2′/3′-O-acetyl-ADP-ribose resulted in an acetylation signal with the anti-AcK-AB. As shown in Fig. 3G, this was not the case.
FIGURE 3.
PEPCK1 is not directly deacetylated by Sirt2 in vitro but can be converted into a Sirt2 substrate. A, final purity and quality of PEPCK1-WT and the acetylated PEPCK1 AcK70, AcK71, AcK70/71, and AcK594 proteins. For details on the experimental procedures see Fig. 1A. B, PEPCK1 is not directly deacetylated by Sirt2 at AcK70, AcK71, AcK70/71, or AcK594 in vitro. PEPCK1 was site-specifically lysine-acetylated and used as substrate for Sirt2-catalyzed deacetylation. Deacetylation assay was performed with increasing concentrations of Sirt2 for 2 h at 23 °C. Molar ratios of Sirt2/PEPCK1 are indicated. The concentration of PEPCK1 was 12 μm. Ponceau S staining was used as a loading control, Sirt2 was detected using an anti-His6 antibody. C, analytical size exclusion chromatography of PEPCK1 variants. The molecular mass calculated from the elution volume using a standard curve is indicated together with the theoretical mass of ∼70.9 kDa. All PEPCK1 proteins used in this study behave as the non-acetylated wild type PEPCK1 and elute as monomers from the analytical SEC column (Superdex 200 10/300 GL). D, reaction scheme of a fluorescence-based coupled enzymatic assay to analyze PEPCK1 activity. In the first and rate-limiting reaction step, PEPCK1 catalyzes the conversion of phosphoenolpyruvate to oxaloacetate under the consumption of GDP and CO2. In the second reaction step, oxaloacetate is converted to malate by malate dehydrogenase, which involves the oxidation of NADH to NAD+. The accompanied drop in fluorescence upon oxidation of NADH to NAD+ (excitation, 345 nm; emission, 470 nm) served as an indirect measure of PEPCK1 activity. E, acetylated PEPCK1 is active as determined by a coupled enzyme assay as described in B. The activity of PEPCK1-WT and its acetylated versions (AcK70, AcK71, AcK70/71, and AcK594) was analyzed as described (concentration, 25 nm). All enzymes showed an activity comparable with the wild type, non-acetylated protein. F, PEPCK1 is not deacetylated by Sirt2 in vitro at lower concentrations and in an optimized PEPCK1 buffer. Site-specifically lysine-acetylated PEPCK1 (0.6 μm) was used as a substrate for Sirt2-catalyzed deacetylation (0.6 μm). None of the acetylated PEPCK1 (AcK70, AcK71, AcK70/71, and AcK594) variants was directly deacetylated. Ran AcK37 (12 μm) was used as a positive control, showing complete deacetylation under the assay conditions. Acetylation levels were detected with an anti-AcK AB. An anti-His6 antibody was used detect PEPCK1, Ran, and Sirt2. G, anti-acetyl-lysine immunoreactivity of PEPCK1-WT (0.6 μm) after incubation with 2 mm 2′/3′-O-acetyl-ADP-ribose for 2 h at 23 °C. Immunoblotting using an anti-AcK antibody was used to detect the acetylation level of PEPCK1. An anti-His6 antibody was used to stain for PEPCK1. H, replacing the PEPCK1 sequence 67RRLKKY72 by 67EFEKKY72 (corresponding to the sequence N-terminal to the Ran di-acetylation site) converts PEPCK1 into a Sirt2 substrate. To analyze what are the determinants for Sirt2 activity on PEPCK1, we replaced the sequence preceding the Lys-70 (67RRL69) for the corresponding sequence in Ran AcK37 (34EFE36). This replacement is sufficient to convert PEPCK1 into a Sirt2 substrate. PEPCK1 AcK70 and AcK70/71 67EFE69 mutants (indicated as PEPCK1-EFE) were incubated with 0.06 μm Sirt2 for the indicated time. Ran AcK serves as a positive control and is completely deacetylated already after less than 5 min. PEPCK1 was deacetylated after 90 min. All substrates were used at 0.6 μm. The di-acetylated PEPCK1 AcK70/71 shows a different running behavior than the non-acetylated variant as seen in the respective anti-His6 loading control. I, deacetylation assay with PEPCK1-EFE mutants in the absence of NAD+. The loss of anti-AcK-immunoreactivity of PEPCK1 (0.6 μm) is dependent on the presence of the sirtuin cofactor NAD+, as the signal remains constant after 90 min of incubation with Sirt2 (0.06 μm). CMB, Coomassie Brilliant Blue; IB, immunoblot.
Given the lack of Sirt2 activity toward PEPCK1 in vitro, we asked whether this is due to a suboptimal recognition sequence or due to the absence of certain cofactors/interaction partners of Sirt2 that would assist in substrate recognition in vivo. Interestingly, it was possible to convert PEPCK1 into an in vitro substrate of Sirt2. Upon replacement of the sequence N-terminally adjacent of Lys-70/71 in PEPCK1 (67RRL69) by the corresponding Ran sequence (34EFE36), we observed deacetylation of both mono-acetylated PEPCK1 AcK70 and di-acetylated PEPCK1 AcK70/71 in an NAD+-dependent manner (Fig. 3, H and I). Given the strong signal of both individual acetylation sites AcK70 and AcK71 with our anti-AcK-AB, the complete loss of acetylation signal suggests that in the di-acetylated background Sirt2 deacetylates both sites of the PEPCK1-AcK70/71-EFE mutant. The deacetylation rate is however rather slow compared with Ran AcK37, suggesting that additional factors contribute to the efficient recognition of Ran AcK37 by Sirt2. Taken together, these results show that in vitro Sirt2 does not deacetylate PEPCK1 at the reported sites but can be converted into a substrate for di-deacetylation by mutation of three residues N-terminal to the 70KKY72 motif.
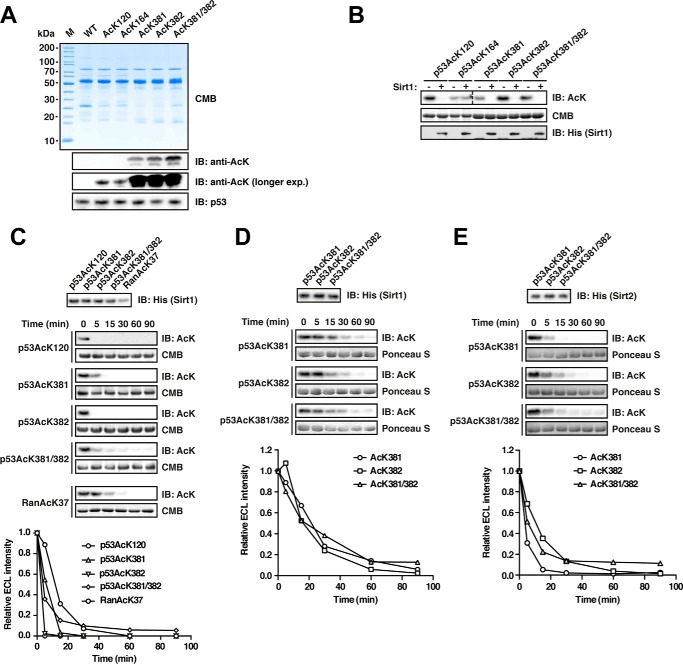
Sirt1 and Sirt2 Deacetylate p53 AcK381, AcK382, and AcK381/382 with Similar Rates
Another well characterized regulatory acetylation occurs at multiple sites of the tumor suppressor p53. Acetylation of p53 is required for its stabilization and for its transactivation leading to apoptosis and/or cell cycle arrest (95). To date, p53 acetylation has been reported for 13 different lysines, most of them located in its C-terminal domain but also in its DNA-binding and tetramerization domains. Interestingly, two lysine pairs are present in the C terminus (Lys-372/373 and Lys-381/382) of p53, all of which have been found to be acetylated (87, 96). HDAC1 and Sirt1 were described as the major p53 deacetylases. HDAC1 is active on the acetylated lysines Lys-320P (superscript P is p53), Lys-373P, and Lys-382P, whereas Sirt1 acts on Lys-382P (97, 98). Evidence also points toward deacetylation of Lys-120P and Lys-164P by Sirt1 (99). The KATs p300/CBP catalyze, inter alia, the acetylation of the lysine pair Lys-381/382P (85). Given our observations regarding the di-deacetylation of Ran by Sirt2, we wondered whether p53 and its deacetylase Sirt1 constitute a similar di-deacetylation system, where p53 can be locked in a mono-acetylated state, and this locked state can be released upon acetylation of the adjacent lysine.
To test this, we purified full-length p53 acetylated at Lys-381, Lys-382, or both using the genetic code expansion concept. For comparison, we also produced the other two described Sirt1-substrate proteins p53 AcK120 and p53 AcK164 (Fig. 4A). When we incubated the different p53-acetylated proteins with recombinant Sirt1, we observed, as expected, that AcK120P and AcK382P were completely deacetylated (after 2 h and at a p53/Sirt1-ratio of 1:20) (Fig. 4B). However, acetylation signals for p53-AcK164 remained almost unchanged suggesting that, at least in vitro, this site is not a direct target of Sirt1. Strikingly, Sirt1 was also able to deacetylate both p53-AcK381 and di-acetylated p53-AcK381/382 (Fig. 4B). To gain insights into the dynamics of the deacetylation reaction, we compared the Sirt1-catalyzed deacetylation of p53 for the positive p53 sites (AcK120, AcK381, and AcK382) and Ran AcK37 in a time course experiment, again with a molar Sirt1/p53-ratio of 1:20. Deacetylation occurred rapidly for all p53 sites, and the rates were comparable with Ran AcK37 (Fig. 4C). We further resolved the deacetylation reaction of the acetylated lysines pair AcK381 and AcK382 at a Sirt1/p53-ratio of 1:200 and found that the mono-acetylated sites and the di-acetylated site are deacetylated at highly comparable rates (Fig. 4D).
FIGURE 4.
Deacetylation of p53 by Sirt1 and Sirt2. A, p53 was site-specifically lysine-acetylated at several sites (AcK120, AcK164, AcK381, AcK382, and AcK381/382). For details on the experimental procedures, see Fig. 1A. An anti-p53 antibody was used to detect p53. B, p53 is deacetylated by Sirt1 at AcK120, AcK381, AcK382, and AcK381/382 but not AcK164 in vitro. Deacetylation assay with site-specifically lysine-acetylated p53 and Sirt1 at a molar Sirt1/p53 ratio of 1:20 (p53, 12 μm; Sirt1, 0.6 μm). The reaction was performed for 2 h at 23 °C, and the acetylation level after Sirt1-catalyzed deacetylation is determined with an anti-AcK antibody. Notably, AcK381, AcK382, and even the di-acetylated AcK381/382 were completely deacetylated under the assay conditions. Coomassie Brilliant Blue (CMB) staining was used as a loading control. An anti-His6 antibody was used to stain for Sirt1. C, time course experiment to assess Sirt1-catalyzed p53-AcK120, -AcK381, -AcK382, and -AcK381/382 deacetylation (p53, 12 μm; Sirt1, 0.6 μm). Ran AcK37 (12 μm) was used as a control. An anti-AcK antibody was used to stain for lysine acetylation, Coomassie Brilliant Blue (CMB), as a loading control. The quantification of the kinetics shown in the right panel was performed using ImageJ. Anti-His6 antibody was used to detect Sirt1. D, time course experiments as for B but with a molar p53/Sirt1 ratio of 1:200 (Sirt1, 0.06 μm) to more sensitively assess the p53-catalyzed deacetylation of p53 AcK381, AcK382, and AcK381/382. As visible in the immunoblottings and the quantifications (lower panel), both mono-acetylated p53 proteins and the di-acetylated protein show highly similar Sirt1-catalyzed deacetylation. E, Sirt2 also deacetylates p53 at AcK381, AcK382, and AcK381/382. Time course experiments were to analyze whether all three sites are also deacetylated by Sirt2 (molar ratio 1:200 as in D). All three acetylated p53 proteins were deacetylated by Sirt2 with similar rates. IB, immunoblot.
This prompted us to test whether the preferential deacetylation of two adjacent acetyl-lysines for Ran AcK37/38 is a unique feature of Sirt2 or rather a matter of the amino acid sequence surrounding the target acetyl-lysine. We thus analyzed the deacetylation of p53 AcK381, AcK382, and AcK381/382 by Sirt2 and found that, like Sirt1, it catalyzes deacetylation of all these sites with very similar rates (Fig. 4E). Thus, in p53, the di-acetylation pair AcK381/382P as well as the mono-acetylated sites AcK381P and AcK383P are targeted with equal preference by both Sirt1 and Sirt2.
Presence of AcK373 in p53 Promotes Deacetylation of AcK372 Only for Sirt1
Less is known about the deacetylation of the second acetylated lysine pair in the C terminus of p53, AcK372/373, although, at least for AcK373P, two studies suggest that this site is also targeted by Sirt1 (100, 101). We tested the di-deacetylation of these acetylated lysine residues. To this end, we prepared site specifically acetylated p53 AcK372, AcK373, and the di-acetylated AcK372/373, all of which showed a double band on an SDS-polyacrylamide gel (Fig. 5A). The smaller species originated from translational termination during the expression using the genetic code expansion concept and thus represents a truncated p53 (86). Because of the similar size compared with full-length p53 and the p53 oligomerization, this truncation product of p53 cannot be removed during the purification process. The same applies to the purifications of the p53-Lys-381/382 but the two bands are not resolved on standard SDS-PAGE.
FIGURE 5.
Deacetylation of p53 at Lys-372 is enhanced upon acetylation of the neighboring Lys-373 for Sirt1 but not Sirt2. A, final purity and quality of p53-WT and acetylated p53 (AcK372, AcK373, and AcK372/372). For details on the experimental procedures see Fig. 1A. Notably, for p53 AcK372, AcK373, AcK381, AcK382, and the di-acetylated proteins, we obtained acetylated truncation products, which we could not remove during purification due to the similar size of the truncation product and the full-length protein and possibly due to oligomerization of the p53 protein. An asterisk denotes the anti-AcK-immunoreactive truncation product. B, p53 deacetylation by Sirt1 at AcK164, AcK372, AcK373, and AcK372/373 was analyzed at increasing molar Sirt1/p53 ratios (p53, 12 μm). The immunoblot with an anti-AcK-antibody shows that, at a molar ratio of Sirt1/p52 of 1:20, the weakest p53 deacetylation by Sirt1 occurs at AcK372, supporting the mechanism of di-deacetylation, by which the presence of AcK373 accelerates deacetylation at AcK372. p53-AcK164 is not deacetylated by Sirt1 in vitro even at a substrate/enzyme ratio of 1:1. C, time course of the Sirt1-catalyzed p53 AcK372, AcK373, and AcK372/373 deacetylation. p53 AcK373 is deacetylated by Sirt1 showing complete deacetylation after 30 min, whereas deacetylation is not completed for AcK372 after 90 min. The di-acetylated p53 AcK372/373 is deacetylated faster than p53 AcK372 (p53, 12 μm; Sirt1, 0.24 μm; molar ratio 1:50). The densitometric quantification was done using ImageJ software. The signal originating from the truncation product remains constant and was subtracted from the signal of the full-length p53 AcK372/373 (the band of the truncated fragment is denoted with an asterisk and runs slightly lower). The acetylation level was assessed using an anti-AcK-antibody. Coomassie Brilliant Blue (CMB) staining was used as a loading control for p53 and an anti-His6 antibody to detect Sirt1. D, time course of Sirt2-catalyzed deacetylation of p53 AcK372, AcK373, and AcK372/373 as described in C but with an enzyme/substrate ratio of 1:200 (0.06 μm Sirt1). All acetylated p53 variants show highly similar deacetylation kinetics with Sirt2. E, frequency plot of sequence context of lysine acetylation sites found in this and our previous study to be substrates or non-substrates of Sirt2 as indicated (14). Representation was created with WebLogo (106). F, sequence alignment of primary sequences surrounding di-acetylation motifs analyzed in this study and in our previous study (14). We discovered that several di-acetylation sites were deacetylated by Sirt1, Sirt2, and/or Sirt3 (Ran AcK37/38, p53 AcK372/373, and p53 AcK381/382). Interestingly, PEPCK1, although also containing a possible di-acetylation motif (AcK70/71) followed by a tyrosine residue, is not deacetylated by Sirt2, suggesting that the structure is an important denominator of Sirt specificity. Notably, the residues 67RRL69 in PEPCK1 preceding Lys-70/Lys-71 form a β-strand as part of an antiparallel β-sheet. It needs further investigation whether the replacement of this sequence by the corresponding Ran sequence 34EFE36 confers Sirt2 activity or whether this is due to structural effects interfering with the formation of this β-sheet, maybe affecting the loops flexibility. IB, immunoblot.
Interestingly, when we tested the acetylated p53 variants of AcK372, AcK373, and AcK372/373 for deacetylation by Sirt1 at enzyme/substrate ratios from 1:20 to 1:1, we found that all three acetylated proteins are deacetylated. In this experiment, we also confirmed that p53-AcK164 is not deacetylated by Sirt1 even with the highest amount of Sirt1. p53-AcK372 showed a remaining signal at a ratio of 1:20, suggesting that this site is a suboptimal target for Sirt1 and shows slower deacetylation kinetics compared with AcK373P and the di-acetylated protein AcK372/373P (Fig. 5B). Consistent with this observation, p53-AcK372 deacetylation was slower than both the mono-acetylated p53-AcK373 and the di-acetylated p53-AcK372/373 in a separate time course experiment with an enzyme/ratio of 1:50 (Fig. 5C). Thus, the p53 di-acetylation site AcK372/373 behaves similarly to that of Ran (AcK37/38) described above, in that di-deacetylation is favored over mono-acetylation for one of the sites. Notably, in case of p53, it is the first acetylated lysine, AcK372P, that is only weakly deacetylated by Sirt1, and it is the presence of the second acetylated lysine (AcK373) that accelerates its deacetylation. In contrast, for Ran, it is the second AcK38, which is only slowly deacetylated by Sirt2 in the absence of the first acetylated lysine AcK37. Strikingly, however, for p53 this differential recognition of the p53-K372/373 site was only observed for Sirt1 and not for Sirt2, which did not show any preference for AcK372P, AcK373P, or AcK372/373P when resolved at an enzyme/substrate of 1:200 (Fig. 5D).
Taken together, our data suggest that in principle Sirt1, -2, and -3 are able to deacetylate two adjacent lysine residues. As observed for both enzymes with Ran and p53, the presence of one acetylated lysine can accelerate the deacetylation of the neighboring acetylated lysine side chain (which itself is only slowly deacetylated in its mono-acetylated state) by a yet unknown mechanism. However, because this is not the case for all di-acetylated lysines, the dynamics of di-deacetylation are dependent on the sequence context and furthermore seem to reflect the substrate recognition differences between sirtuins.
Discussion
To our knowledge, we present here the first site-specific study on the substrate specificity of sirtuins in a full-length protein context as opposed to acetylated peptides. Using the Genetic Code Expansion Concept allowed us to test individual acetylation sites in natively folded proteins. We selected previously reported substrate proteins of Sirt1, Sirt2, and Sirt3 as the three sirtuins with robust lysine deacetylase activity (Sirt1, p53; Sirt2, Ran, PEPCK1; Sirt3, CypD, MnSOD, Hsp10). Importantly, for most of these candidate proteins, the substrate deacetylase relationship was established based on sirtuin knockdown/knock-out and overexpression studies, in which the acetylation status of the protein was analyzed qualitatively in response to these perturbations. With our direct in vitro deacetylation assays, we were unable to reconstitute the Sirt3-catalyzed deacetylation of CypD at two reported sites, namely AcK167 and AcK197 (corresponding to AcK155 in CypA), as well as MnSOD (AcK122) and the Sirt2-catalyzed deacetylation of PEPCK1 (AcK70, AcK71, and AcK594). In case of p53, except for AcK164, we could confirm all previously identified Sirt1 target sites. Our failure to confirm the deacetylases for selected candidate proteins suggests that either there are no direct substrates for the respective sirtuins or, alternatively, that additional factors are required in vivo to stimulate or enable the deacetylation at these sites, such as scaffolding proteins or post-translational modifications.
Based on our previous work, we decided to further analyze the deacetylation of Ran AcK37 and Ran AcK38, for which we identified Sirt1–3 as deacetylases (14). As these two sites are directly next to each other and thus found in a very similar structural environment, the switch I of Ran, we reasoned that this constellation is a good model to investigate how much the structural context interferes with Sirt2-catalyzed deacetylation at these sites. The striking difference in the deacetylation rate of the two sites, AcK37R and AcK38R, may be interpreted as a support for a “sequence-centric” view of sirtuin substrate specificity. This is also reflected by the fact that the Ran AcK38 peptide shows a 5-fold weaker binding toward Sirt2 compared with the Ran AcK37 peptide. Our structural data show that the aromatic residues Phe-35R and Tyr-39R preceding and following the acetylation sites (31LTGEFEKKYVATL43) are important residues for substrate recognition and binding by Sirt2. This is supported by an earlier study on peptidic Sirt2 inhibitors, which demonstrated that high affinity peptides in many cases contained a tyrosine in the +2 position relative to the acetyl moiety (94). At first glance, the importance of the sequence context was further supported by the fact that replacement of the three N-terminal residues next to the PEPCK1-Lys-70/71 acetylation sites by the corresponding sequence in Ran, including F35R, was sufficient to render it a direct Sirt2 substrate. Previous studies, in which peptides have been used to find sequence dependences of substrate recognition, show that, although classical consensus motifs cannot be found for sirtuins, the sequence context clearly plays a role, albeit in a less intuitive contextual fashion (69, 102, 103). The characteristics of the sequences surrounding substrate and non-substrate sites tested here and in our previous study seem to agree with this context-dependent substrate recognition without a clear consensus (Fig. 5, E and F) (14).
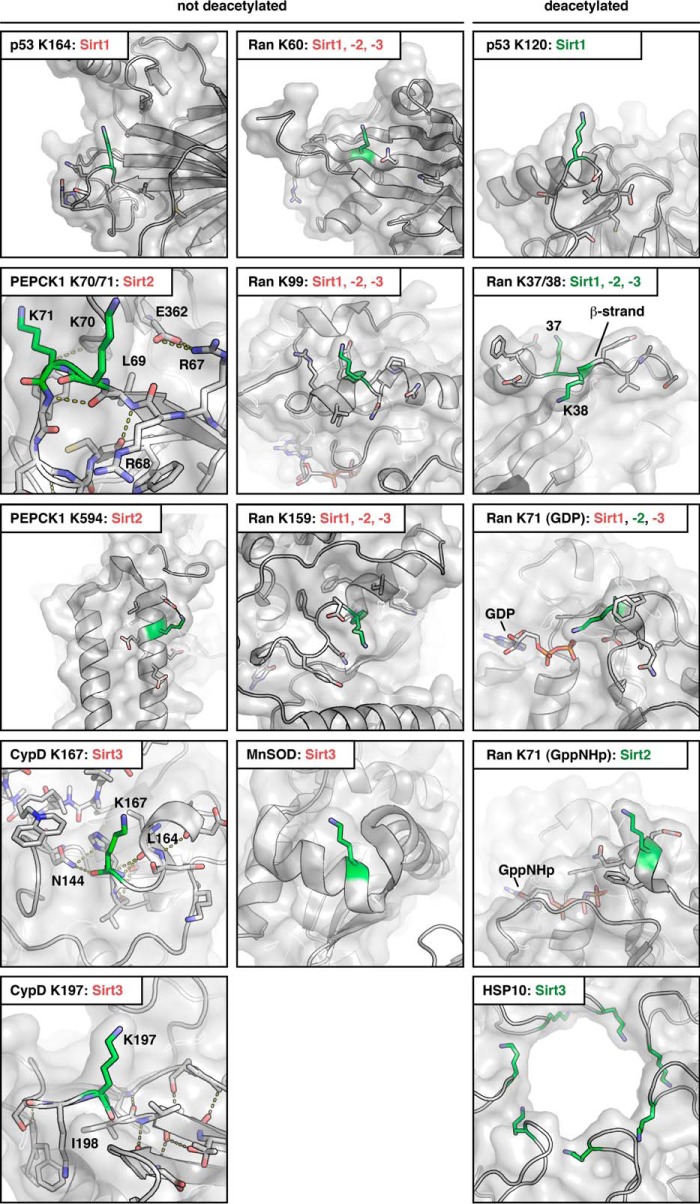
The sequence-centric view is challenged by other aspects of the present and our previous study on Ran acetylation (14). Based on the available structures of the candidate protein substrates in the non-acetylated form, all of the sites found not to be deacetylated by sirtuins are integral parts of α-helices and β-strands, in close proximity to those secondary structure elements or in apparently restrained loop regions as observed for CypD AcK167, CypD AcK197, MnSOD AcK122, and PEPCK1 AcK70/71. Mutation of the N-terminal PEPCK1 sequence 67RRL69 to the corresponding sequence in Ran 34EFE36 might release this restrained conformation, possibly by resolving a salt bridge between Arg-67 and Glu-362 of PEPCK1, which in turn renders PEPCK1 a Sirt2 substrate (Fig. 6). A special case in this regard is the residue p53 Lys-164, which, in addition to its flanking N-terminal residues being part of a β-strand, appears to be inaccessible by Sirt1 due to the localization of an α-helix of the p53 tetramerization domain. In contrast, the acetylation sites efficiently deacetylated by sirtuins are, without exception, located in regions with no profound secondary structure or in loop regions of the substrate proteins (e.g. the C terminus of p53, switch I in Ran). In the hexameric Hsp10, which forms the lid of the Hsp10-Hsp60 chaperonin system, the acetylated Lys-56 is located in a highly accessible and flexible loop at the opening pore (104). For Ran in its inactive GDP-bound conformation, Lys-38 is located on a small β-strand, thereby maybe restricting the access to the sirtuin active site if acetylated. This restraint might be released upon acetylation of the neighboring Lys-37R making AcK38R accessible for deacetylation by sirtuins (Fig. 6). However, the primary sequence affects the interplay of Sirt2 and Ran AcK38 as our ITC data using acetylated Ran peptides show that the AcK38R peptide binds to Sirt2 with a 5-fold reduced affinity compared with the AcK37R-peptide.
FIGURE 6.
Localization of sirtuin substrate and non-substrate lysine acetylation sites in known crystal structures of Ran·GDP (PDB code 1BYU), Ran·GppNHp (PDB code 1IBR), MnSOD (PDB code 1PL4), CypD (PDB code 5A0E), Hsp10 (PDB code 4PJ1), PEPCK1 (PDB code 1NHX), and p53 (PDB code 2OCJ). All non-substrate lysines are located within or directly next to secondary structure elements, as observed in MnSOD Lys-122, or are located in conformationally restrained loops as observed in CypD Lys-167 and Lys-197 and PEPCK1 Lys-70/ Lys-71. CypD Lys-167 is located directly C-terminally of an α-helix. CypD Lys-197 is located directly N-terminally of a β-strand, which restricts its conformational freedom. The main chain nitrogen of Lys-197 makes a hydrogen bond to a bridging water molecule. The side chain of Ile-198, directly adjacent to Lys-197 is part of a hydrophobic core conformationally stabilizing this region. As a direct measure of its conformational rigidity, the loops encompassing Lys-167 and Lys-197 in CypD are well defined in the crystal structure, which is furthermore reflected in the B-factors for the amide nitrogen, the C-α and the carbonyl C-atom, forming the lysine's backbone (these are in the range of 10.59–12.72 Å2 for Lys-167 and 12.45–13.41 Å2 for Lys-197; PDB code 5A0E). These low B-factors suggest that these residues have a low level of flexibility. PEPCK1 Lys-70 and Lys-71 are located within a loop region. However, the residues 67RRL69 preceding these lysines form a small β-strand, which is part of an anti-parallel β-sheet. Furthermore, the loop flexibility is restrained by several side chain and main chain interactions (side chain, Glu-362 with Arg-67, Lys-71 with Asp-365 and Glu-375; main chain, Gly-366 with Lys-71, Leu-77 with Arg-67). This restraint of the loop's conformation could explain why AcK70 and AcK71 are not Sirt2 substrates. Mutation of 67RRL69 in PEPCK1 to the corresponding residues in Ran (giving rise to 67EFE69) makes PEPCK1 a Sirt2 substrate, most likely at least in part by releasing the loop's restrained conformation. However, alteration in the primary sequence might also affect the deacetylation. In p53, Lys-164 lies directly C-terminal to a β-strand; Lys-120 is located within a flexible loop. For the C-terminal lysines Lys-372, Lys-373, Lys-381, and Lys-382, there is no structural information available, as this domain is intrinsically disordered. In Ran, Lys-60 is located within a β-strand and Lys-99 and Lys-159 are within an α-helix. None of the sirtuins tested in our previous study deacetylated any of these three lysine acetylation sites of Ran. In contrast, Lys-37, Lys-38, and Lys-71 are potent sirtuin substrate sites. Although AcK71 is specifically deacetylated only by Sirt2, AcK37 is deacetylated by Sirt1, Sirt2, and Sirt3. AcK38 is deacetylated weakly by Sirt1, -2, and -3 but the presence of AcK37 increases the potency of AcK38 to act as a Sirt1, -2, and -3 substrate. Lys-37 and Lys-38 in Ran are located within the switch I loop. Lys-38R is part of a small β-sheet, restraining the structural flexibility. Acetylation of the neighboring Lys-37 might release these restraints making AcK38 a better sirtuin substrate. Lys-71 is part of the switch II loop, which is structurally more restricted in the active state (here: GppNHp loaded). However, we observed, that AcK71 is a better substrate for Sirt2 if it is present in its GppNHp-loaded state suggesting that this conformation is highly suitable for catalysis. Sirtuin labeling is in red if the respective sirtuin is not active and is in green if a sirtuin is active in deacetylating a protein at the respective site. In the panels depicted in the right column, all substrate proteins and sites are shown that are deacetylated by sirtuins. In the left two columns all sites that were not deacetylated by Sirt1–3 are presented.
Our results suggest that certain structural prerequisites have to be met for efficient substrate recognition by sirtuins. An interesting case is the acetylation site Lys-71 of Ran, which is located in the switch II loop and for which we previously reported that in vitro it is specifically targeted only by Sirt2 (14). Because this site is deacetylated faster if Ran is in its active conformation, in which this loop adopts a more rigid conformation compared with the inactive GDP-bound state, in which the switch II loop is more flexible, it seems that in this case substrate recognition is promoted by certain structural elements. This indicates that sirtuins may as well be able to deacetylate sites located in structured or rigid regions of a protein.
The ability of Sirt1–3 to deacetylate two neighboring acetyl-lysines has further interesting implications. Over the course of this study, we have characterized in detail four such lysine pairs, one for Ran (Lys-37/38), one in PEPCK1 (Lys-70/71), and two for p53 (Lys-372/373 and Lys-381/382). We observed that some di-acetylation sites are targeted by sirtuins, regardless of which combination of mono-acetylation or di-acetylation is present. As an example, p53 AcK381, AcK382, and AcK381/382 were all deacetylated equally well by Sirt1 and Sirt2. This might reflect that a low selection pressure prevailed during evolution to restrict the deacetylation of those sites to one enzyme or, alternatively, that efficient removal of any possible combination of acetyl modifications at these sites posed a selective advantage. The deacetylation may as well be regulated by other mechanisms, such as the subcellular localization or the expression patterns of the deacetylases and the substrate protein in these cases.
For Ran AcK37, AcK38, and AcK37/38, we found that Sirt2 deacetylates AcK38R less efficiently than AcK37R. The presence of AcK37R can strongly accelerate the deacetylation of AcK38R. This suggests a new mechanism to increase lifetime lysine-acetyl modifications, without losing the possibility to switch off the acetylated state. We also confirmed that this mechanism is not restricted to Sirt2, as shown for Sirt1 and the p53 di-acetyl-lysine pair AcK372/373. Notably, for this pair it was the presence of an acetyl modification at the second lysine (Lys-373P) that accelerated the deacetylation at the first acetylated lysine (Lys-372P). Importantly, we discovered that Sirt2 deacetylates p53 with similar rates at these sites, showing that differences exist between Sirt1 and Sirt2 regarding substrate recognition.
Acetylation of p53 plays an important role in the activation of its tumor-suppressive functions. An understanding of the dynamics of their deacetylation is thus clinically relevant. As noted above, it was not clear how the simultaneous acetyl modification of adjacent lysine residues would interfere with deacetylation by Sirt1. Whether this differential deacetylation has a biologically relevant impact on the lifetime of the activated state of p53 or other substrate proteins is an interesting question for future studies. It should be noted that acetylation of two neighboring lysines is currently known for 978 di-lysine sites on 771 proteins in human (according to current entries on Phosphosite.org) (105), although it is not clear how many of these potential di-acetylation sites occur simultaneously or in a mutually exclusive manner. Di-acetylation may thus be a common strategy throughout the human proteome, including histones. In this regard, it is not clear whether KATs are able to transfer acetyl moieties to neighboring lysine side chains. Many bromodomains are able to bind to two acetylated lysines. However, whether they can bind to directly adjacent acetylated lysines has not been investigated so far.
Taken together, our results reveal that substrate recognition by sirtuins is not only affected by the primary sequence but to a large extent also by the protein structure. Thus, in future studies the results obtained using acetylated peptides for deacetylation by sirtuin deacetylases should be integrated with additional data from natively folded proteins. The Genetic Code Expansion Concept enabled the site-specific study of previously reported sirtuin deacetylation sites, which in turn led to the discovery of the ability sirtuins to deacetylate two adjacent acetyl-lysines. This has important implications for the regulation of proteins by acetylation as it allows us to prolong the lifetime of an acetylation event and to remove it by an additional acetylation of a neighboring lysine. Although the exact molecular mechanism needs to be further investigated, our structural and biochemical data suggest that sirtuins cannot accommodate two acetylated lysines simultaneously in their active site but that instead di-deacetylation occurs sequentially. Given the increasing number of acetylation sites identified through advanced high throughput mass spectrometry, we believe that it will become more important to discriminate sites that are of high biological relevance from those that may be regarded as background noise. This task will involve the determination of absolute stoichiometries of acetylation sites and the development of reliable prediction tools for their regulation by lysine acetyltransferases and KDACs. Our data on the substrate recognition of sirtuins are thus an important step to ultimately gain a deeper understanding of the regulation of the acetylome.
Author Contributions
All authors designed experiments and analyzed data. P. K. and M. L. wrote the manuscript. M. S. and U. B. took x-ray dataset. I. N. synthesized the peptides. P. K., S. d. B., L. S., A. E., N. K., and L. B. performed experiments.
Acknowledgments
We thank the beamline groups at the Swiss Light Source, Paul-Scherrer-Institute, Villigen/Switzerland whose outstanding efforts have made these experiments possible. We thank Astrid Wilbrand-Hennes and Rene Grandjean (CECAD proteomics facility) for support in mass spectrometry and Hendrik Nolte for critical and helpful discussions of the mass spectrometry data. We are grateful to Prof. Dr. Bernhard Schermer for the p53 construct.
This work was supported in part by Emmy Noether Programme of the German Research Foundation (Deutsche Forschungsgemeinschaft) Grant LA2984-1/1 and by European Community's Seventh Framework Programme FP7/2007-2013b by Grant 283570 (BioStruct-X). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5FYQ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- KAT
- lysine acetyltransferase
- KDAC
- lysine deacetylase
- Ran
- Ras-related nuclear
- CypD
- cyclophilin D
- MnSOD
- superoxide dismutase, mitochondrial
- AcK
- acetyl-l-lysine
- Sir
- sirtuin
- ITC
- isothermal titration calorimetry
- AB
- antibody
- PDB
- Protein Data Bank
- TEV
- tobacco etch virus
- SEC
- size exclusion chromatography
- GppNHp
- guanosine 5′-[β,γ-imido]triphosphate
- TFAcK
- Nϵ-trifluoroacetyl-l-lysine.
References
- 1.Allfrey V. G., Faulkner R., and Mirsky A. E. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 51, 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hebbes T. R., Thorne A. W., and Crane-Robinson C. (1988) A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7, 1395–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 4.Beli P., Lukashchuk N., Wagner S. A., Weinert B. T., Olsen J. V., Baskcomb L., Mann M., Jackson S. P., and Choudhary C. (2012) Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 46, 212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choudhary C., Weinert B. T., Nishida Y., Verdin E., and Mann M. (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550 [DOI] [PubMed] [Google Scholar]
- 6.Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., and Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 7.Weinert B. T., Wagner S. A., Horn H., Henriksen P., Liu W. R., Olsen J. V., Jensen L. J., and Choudhary C. (2011) Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci. Signal. 4, ra48. [DOI] [PubMed] [Google Scholar]
- 8.Weinert B. T., Iesmantavicius V., Moustafa T., Schölz C., Wagner S. A., Magnes C., Zechner R., and Choudhary C. (2014) Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 10, 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lundby A., Lage K., Weinert B. T., Bekker-Jensen D. B., Secher A., Skovgaard T., Kelstrup C. D., Dmytriyev A., Choudhary C., Lundby C., and Olsen J. V. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriksen P., Wagner S. A., Weinert B. T., Sharma S., Bacinskaja G., Rehman M., Juffer A. H., Walther T. C., Lisby M., and Choudhary C. (2012) Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteomics 11, 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C. F., Grishin N. V., and Zhao Y. (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnad F., Ren S., Choudhary C., Cox J., and Mann M. (2010) Predicting post-translational lysine acetylation using support vector machines. Bioinformatics 26, 1666–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knyphausen P., Kuhlmann N., de Boor S., and Lammers M. (2015) Lysine acetylation as a fundamental regulator of Ran function: implications for signaling of proteins of the Ras-superfamily. Small GTPases 6, 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Boor S., Knyphausen P., Kuhlmann N., Wroblowski S., Brenig J., Scislowski L., Baldus L., Nolte H., Krüger M., and Lammers M. (2015) Small GTP-binding protein Ran is regulated by posttranslational lysine acetylation. Proc. Natl. Acad. Sci. U.S.A. 112, E3679–E3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhlmann N., Wroblowski S., Knyphausen P., de Boor S., Brenig J., Zienert A. Y., Meyer-Teschendorf K., Praefcke G. J., Nolte H., Krüger M., Schacherl M., Baumann U., James L. C., Chin J. W., and Lammers M. (2016) Structural and mechanistic insights into the regulation of the fundamental Rho-regulator RhoGDIα by lysine acetylation. J. Biol. Chem. 291, 5484–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhlmann N., Wroblowski S., Scislowski L., and Lammers M. (2016) RhoGDIα acetylation at K127 and K141 affects binding toward nonprenylated RhoA. Biochemistry 55, 304–312 [DOI] [PubMed] [Google Scholar]
- 17.Albaugh B. N., Arnold K. M., and Denu J. M. (2011) KAT(ching) metabolism by the tail: insight into the links between lysine acetyltransferases and metabolism. Chembiochem 12, 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montgomery D. C., Sorum A. W., and Meier J. L. (2014) Chemoproteomic profiling of lysine acetyltransferases highlights an expanded landscape of catalytic acetylation. J. Am. Chem. Soc. 136, 8669–8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth S. Y., Denu J. M., and Allis C. D. (2001) Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120 [DOI] [PubMed] [Google Scholar]
- 20.Haberland M., Montgomery R. L., and Olson E. N. (2009) The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falkenberg K. J., and Johnstone R. W. (2014) Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 13, 673–691 [DOI] [PubMed] [Google Scholar]
- 22.Yuan H., and Marmorstein R. (2012) Structural basis for sirtuin activity and inhibition. J. Biol. Chem. 287, 42428–42435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 24.Tonks N. K. (2006) Protein-tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7, 833–846 [DOI] [PubMed] [Google Scholar]
- 25.Goldenson B., and Crispino J. D. (2015) The aurora kinases in cell cycle and leukemia. Oncogene 34, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor S. S., and Kornev A. P. (2011) Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 36, 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berndsen C. E., and Denu J. M. (2008) Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr. Opin. Struct. Biol. 18, 682–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marmorstein R., and Zhou M. M. (2014) Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feldman J. L., Dittenhafer-Reed K. E., and Denu J. M. (2012) Sirtuin catalysis and regulation. J. Biol. Chem. 287, 42419–42427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dittenhafer-Reed K. E., Feldman J. L., and Denu J. M. (2011) Catalysis and mechanistic insights into sirtuin activation. Chembiochem 12, 281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith B. C., and Denu J. M. (2006) Sirtuins caught in the act. Structure 14, 1207–1208 [DOI] [PubMed] [Google Scholar]
- 32.Smith B. C., Hallows W. C., and Denu J. M. (2008) Mechanisms and molecular probes of sirtuins. Chem. Biol. 15, 1002–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischle W., Dequiedt F., Hendzel M. J., Guenther M. G., Lazar M. A., Voelter W., and Verdin E. (2002) Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45–57 [DOI] [PubMed] [Google Scholar]
- 34.Kelly R. D., and Cowley S. M. (2013) The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem. Soc. Trans. 41, 741–749 [DOI] [PubMed] [Google Scholar]
- 35.Yang T., Jian W., Luo Y., Fu X., Noguchi C., Bungert J., Huang S., and Qiu Y. (2012) Acetylation of histone deacetylase 1 regulates NuRD corepressor complex activity. J. Biol. Chem. 287, 40279–40291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S. H., Vickers E., Brehm A., Kouzarides T., and Sharrocks A. D. (2001) Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 21, 2802–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rine J., and Herskowitz I. (1987) Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivy J. M., Klar A. J., and Hicks J. B. (1986) Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol. Cell. Biol. 6, 688–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaeberlein M., McVey M., and Guarente L. (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imai S., Armstrong C. M., Kaeberlein M., and Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 41.Baur J. A., Chen D., Chini E. N., Chua K., Cohen H. Y., de Cabo R., Deng C., Dimmeler S., Gius D., Guarente L. P., Helfand S. L., Imai S., Itoh H., Kadowaki T., Koya D., et al. (2010) Dietary restriction: standing up for sirtuins. Science 329, 1012–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bordone L., Cohen D., Robinson A., Motta M. C., van Veen E., Czopik A., Steele A. D., Crowe H., Marmor S., Luo J., Gu W., and Guarente L. (2007) SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell 6, 759–767 [DOI] [PubMed] [Google Scholar]
- 43.Guarente L. (2012) Sirtuins and calorie restriction. Nat. Rev. Mol. Cell Biol. 13, 207. [DOI] [PubMed] [Google Scholar]
- 44.Haigis M. C., and Sinclair D. A. (2010) Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson G., and Maes M. (2014) Neurodegeneration in Parkinson's disease: interactions of oxidative stress, tryptophan catabolites and depression with mitochondria and sirtuins. Mol. Neurobiol. 49, 771–783 [DOI] [PubMed] [Google Scholar]
- 46.Braidy N., Jayasena T., Poljak A., and Sachdev P. S. (2012) Sirtuins in cognitive ageing and Alzheimer's disease. Curr. Opin. Psychiatry 25, 226–230 [DOI] [PubMed] [Google Scholar]
- 47.Chakraborty C., and Doss C. G. (2013) Sirtuins family–recent development as a drug target for aging, metabolism, and age related diseases. Curr. Drug Targets 14, 666–675 [DOI] [PubMed] [Google Scholar]
- 48.Denu J. M., and Gottesfeld J. M. (2012) Minireview series on sirtuins: from biochemistry to health and disease. J. Biol. Chem. 287, 42417–42418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donadini A., Rosano C., Felli L., and Ponassi M. (2013) Human sirtuins: an overview of an emerging drug target in age-related diseases and cancer. Curr. Drug Targets 14, 653–661 [DOI] [PubMed] [Google Scholar]
- 50.Imai S., and Guarente L. (2014) NAD+ and sirtuins in aging and disease. Trends Cell Biol. 24, 464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Outeiro T. F., Kontopoulos E., Altmann S. M., Kufareva I., Strathearn K. E., Amore A. M., Volk C. B., Maxwell M. M., Rochet J. C., McLean P. J., Young A. B., Abagyan R., Feany M. B., Hyman B. T., and Kazantsev A. G. (2007) Sirtuin 2 inhibitors rescue α-synuclein-mediated toxicity in models of Parkinson's disease. Science 317, 516–519 [DOI] [PubMed] [Google Scholar]
- 52.Chalkiadaki A., and Guarente L. (2015) The multifaceted functions of sirtuins in cancer. Nat. Rev. Cancer 15, 608–624 [DOI] [PubMed] [Google Scholar]
- 53.Chang H. C., and Guarente L. (2014) SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 25, 138–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakagawa T., and Guarente L. (2014) SnapShot: sirtuins, NAD, and aging. Cell Metab. 20, 192–192 [DOI] [PubMed] [Google Scholar]
- 55.Nishida Y., Rardin M. J., Carrico C., He W., Sahu A. K., Gut P., Najjar R., Fitch M., Hellerstein M., Gibson B. W., and Verdin E. (2015) SIRT5 Regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell 59, 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feldman J. L., Dittenhafer-Reed K. E., Kudo N., Thelen J. N., Ito A., Yoshida M., and Denu J. M. (2015) Kinetic and structural basis for acyl-group selectivity and NAD+ dependence in sirtuin-catalyzed deacylation. Biochemistry 54, 3037–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feldman J. L., Baeza J., and Denu J. M. (2013) Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 288, 31350–31356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sinclair D. A., and Guarente L. (2014) Small-molecule allosteric activators of sirtuins. Annu. Rev. Pharmacol. Toxicol. 54, 363–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milne J. C., and Denu J. M. (2008) The Sirtuin family: therapeutic targets to treat diseases of aging. Curr. Opin Chem. Biol. 12, 11–17 [DOI] [PubMed] [Google Scholar]
- 60.Winnik S., Auwerx J., Sinclair D. A., and Matter C. M. (2015) Protective effects of sirtuins in cardiovascular diseases: from bench to bedside. Eur. Heart J. 36, 3404–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gertz M., Nguyen G. T., Fischer F., Suenkel B., Schlicker C., Fränzel B., Tomaschewski J., Aladini F., Becker C., Wolters D., and Steegborn C. (2012) A molecular mechanism for direct sirtuin activation by resveratrol. PLoS ONE 7, e49761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imai S. (2010) A possibility of nutriceuticals as an anti-aging intervention: activation of sirtuins by promoting mammalian NAD biosynthesis. Pharmacol. Res. 62, 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dan L., Klimenkova O., Klimiankou M., Klusman J. H., van den Heuvel-Eibrink M. M., Reinhardt D., Welte K., and Skokowa J. (2012) The role of sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica 97, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Green K. N., Steffan J. S., Martinez-Coria H., Sun X., Schreiber S. S., Thompson L. M., and LaFerla F. M. (2008) Nicotinamide restores cognition in Alzheimer's disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phospho-τ. J. Neurosci. 28, 11500–11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen G. T., Schaefer S., Gertz M., Weyand M., and Steegborn C. (2013) Structures of human sirtuin 3 complexes with ADP-ribose and with carba-NAD+ and SRT1720: binding details and inhibition mechanism. Acta Crystallogr. D Biol. Crystallogr. 69, 1423–1432 [DOI] [PubMed] [Google Scholar]
- 66.Hirsch B. M., and Zheng W. (2011) Sirtuin mechanism and inhibition: explored with N(ϵ)-acetyl-lysine analogs. Mol. Biosyst. 7, 16–28 [DOI] [PubMed] [Google Scholar]
- 67.Suzuki T., Asaba T., Imai E., Tsumoto H., Nakagawa H., and Miyata N. (2009) Identification of a cell-active non-peptide sirtuin inhibitor containing N-thioacetyl lysine. Bioorg. Med. Chem. Lett. 19, 5670–5672 [DOI] [PubMed] [Google Scholar]
- 68.Smith B. C., and Denu J. M. (2007) Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. J. Biol. Chem. 282, 37256–37265 [DOI] [PubMed] [Google Scholar]
- 69.Rauh D., Fischer F., Gertz M., Lakshminarasimhan M., Bergbrede T., Aladini F., Kambach C., Becker C. F., Zerweck J., Schutkowski M., and Steegborn C. (2013) An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat. Commun. 4, 2327. [DOI] [PubMed] [Google Scholar]
- 70.Khan A. N., and Lewis P. N. (2005) Unstructured conformations are a substrate requirement for the Sir2 family of NAD-dependent protein deacetylases. J. Biol. Chem. 280, 36073–36078 [DOI] [PubMed] [Google Scholar]
- 71.Wiseman T., Williston S., Brandts J. F., and Lin L. N. (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 72.Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leslie A. G., and Powell H. R. (2007) Processing diffraction data with MOSFLM. Evolving Methods for Macromolecular Crystallography 245, 41–51 [Google Scholar]
- 74.Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamagata K., Goto Y., Nishimasu H., Morimoto J., Ishitani R., Dohmae N., Takeda N., Nagai R., Komuro I., Suga H., and Nureki O. (2014) Structural basis for potent inhibition of SIRT2 deacetylase by a macrocyclic peptide inducing dynamic structural change. Structure 22, 345–352 [DOI] [PubMed] [Google Scholar]
- 77.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B. 3rd., Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DeLano W. L. (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA [Google Scholar]
- 82.Case C. L., and Mukhopadhyay B. (2007) Kinetic characterization of recombinant human cytosolic phosphoenolpyruvate carboxykinase with and without a His10-tag. Biochim. Biophys. Acta 1770, 1576–1584 [DOI] [PubMed] [Google Scholar]
- 83.Tao R., Coleman M. C., Pennington J. D., Ozden O., Park S. H., Jiang H., Kim H. S., Flynn C. R., Hill S., Hayes McDonald W., Olivier A. K., Spitz D. R., and Gius D. (2010) Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell 40, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hafner A. V., Dai J., Gomes A. P., Xiao C. Y., Palmeira C. M., Rosenzweig A., and Sinclair D. A. (2010) Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging 2, 914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gu W., and Roeder R. G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 [DOI] [PubMed] [Google Scholar]
- 86.Arbely E., Natan E., Brandt T., Allen M. D., Veprintsev D. B., Robinson C. V., Chin J. W., Joerger A. C., and Fersht A. R. (2011) Acetylation of lysine 120 of p53 endows DNA-binding specificity at effective physiological salt concentration. Proc. Natl. Acad. Sci. U.S.A. 108, 8251–8256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reed S. M., and Quelle D. E. (2014) p53 acetylation: regulation and consequences. Cancers 7, 30–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang W., Wang S., Xiao M., Lin Y., Zhou L., Lei Q., Xiong Y., Guan K. L., and Zhao S. (2011) Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell 43, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu Z., Chen Y., Aponte A. M., Battaglia V., Gucek M., and Sack M. N. (2015) Prolonged fasting identifies heat shock protein 10 as a Sirtuin 3 substrate: elucidating a new mechanism linking mitochondrial protein acetylation to fatty acid oxidation enzyme folding and function. J. Biol. Chem. 290, 2466–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simic Z., Weiwad M., Schierhorn A., Steegborn C., and Schutkowski M. (2015) The varϵ-amino group of protein lysine residues is highly susceptible to nonenzymatic acylation by several physiological Acyl-CoA thioesters. Chembiochem 16, 2337–2347 [DOI] [PubMed] [Google Scholar]
- 91.Zhao K., Chai X., and Marmorstein R. (2003) Structure of the yeast Hst2 protein deacetylase in ternary complex with 2′-O-acetyl ADP ribose and histone peptide. Structure 11, 1403–1411 [DOI] [PubMed] [Google Scholar]
- 92.Avalos J. L., Bever K. M., and Wolberger C. (2005) Mechanism of sirtuin inhibition by nicotinamide: altering the NAD+ cosubstrate specificity of a Sir2 enzyme. Mol. Cell 17, 855–868 [DOI] [PubMed] [Google Scholar]
- 93.Avalos J. L., Celic I., Muhammad S., Cosgrove M. S., Boeke J. D., and Wolberger C. (2002) Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell 10, 523–535 [DOI] [PubMed] [Google Scholar]
- 94.Morimoto J., Hayashi Y., and Suga H. (2012) Discovery of macrocyclic peptides armed with a mechanism-based warhead: isoform-selective inhibition of human deacetylase SIRT2. Angew Chem. Int. Ed Engl. 51, 3423–3427 [DOI] [PubMed] [Google Scholar]
- 95.Tang Y., Zhao W., Chen Y., Zhao Y., and Gu W. (2008) Acetylation is indispensable for p53 activation. Cell 133, 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brooks C. L., and Gu W. (2011) The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2, 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., and Weinberg R. A. (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 98.Ito A., Kawaguchi Y., Lai C. H., Kovacs J. J., Higashimoto Y., Appella E., and Yao T. P. (2002) MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21, 6236–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Z. N., Chung S. K., Xu Z., and Xu Y. (2014) Oct4 maintains the pluripotency of human embryonic stem cells by inactivating p53 through Sirt1-mediated deacetylation. Stem Cells 32, 157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Frazzi R., Valli R., Tamagnini I., Casali B., Latruffe N., and Merli F. (2013) Resveratrol-mediated apoptosis of Hodgkin lymphoma cells involves SIRT1 inhibition and FOXO3a hyperacetylation. Int. J. Cancer 132, 1013–1021 [DOI] [PubMed] [Google Scholar]
- 101.Knights C. D., Catania J., Di Giovanni S., Muratoglu S., Perez R., Swartzbeck A., Quong A. A., Zhang X., Beerman T., Pestell R. G., and Avantaggiati M. L. (2006) Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J. Cell Biol. 173, 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gurard-Levin Z. A., Kilian K. A., Kim J., Bähr K., and Mrksich M. (2010) Peptide arrays identify isoform-selective substrates for profiling endogenous lysine deacetylase activity. ACS Chem. Biol. 5, 863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith B. C., Settles B., Hallows W. C., Craven M. W., and Denu J. M. (2011) SIRT3 substrate specificity determined by peptide arrays and machine learning. ACS Chem. Biol. 6, 146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Landry S. J., Steede N. K., and Maskos K. (1997) Temperature dependence of backbone dynamics in loops of human mitochondrial heat shock protein 10. Biochemistry 36, 10975–10986 [DOI] [PubMed] [Google Scholar]
- 105.Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Diederichs K., and Karplus P. A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]
- 108.Diederichs K., and Karplus P. A. (2013) Better models by discarding data? Acta Crystallogr. D Biol. Crystallogr. 69, 1215–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brünger A. T. (1997) Free R value: cross-validation in crystallography. Methods Enzymol. 277, 366–396 [DOI] [PubMed] [Google Scholar]
- 110.Chen V. B., Arendall W. B. 3rd., Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]