Abstract
Effective recognition of viral infection and subsequent triggering of antiviral innate immune responses are essential for the host antiviral defense, which is tightly regulated by multiple regulators, including microRNAs. Previous reports have shown that some microRNAs are induced during virus infection and participate in the regulation of the innate antiviral response. However, whether the type I IFN response is regulated by miR-223 is still unknown. Here, we reported that vesicular stomatitis virus (VSV) infection induced significant up-regulation of miR-223 in murine macrophages. We observed that miR-223 overexpression up-regulated type I IFN expression levels in VSV-infected macrophages. We also demonstrated that miR-223 directly targets FOXO3 to regulate the type I IFN production. Furthermore, type I IFN, which is triggered by VSV infection, is responsible for the up-regulation of miR-223, thus forming a positive regulatory loop for type I IFN production. Our results uncovered a novel mechanism of miR-223-mediated regulation of type I IFN production in the antiviral innate immunity for the first time.
Keywords: antiviral agent, innate immunity, interferon, interferon regulatory factor (IRF), microRNA (miRNA), FOXO3, IRF7, type I interferon, microRNA-223
Introduction
Host innate immune response is the first line of defense following viral infection, which recognizes viral components and produces proinflammatory cytokines and interferons (1–8). The IFN family of antiviral cytokines, especially type I IFN, plays pivotal roles in the host antiviral innate immune response. They are known to inhibit viral replication and mediate protection against viral infection (1–4). Type I IFN production during virus infection should be tightly controlled by multiple intracellular regulators to initiate an appropriate immune response for eliminating the invading pathogens and preventing the development of immunopathological conditions; principal among these is the family of interferon regulatory factors (9–12). Among the interferon regulatory factors, IRF3 and IRF7 are crucial in the host response to virus infection, inducing the transcription of the type I IFN genes, including the Ifn-β gene as well as a number of Ifn-α genes (13–16). It has been reported that IRF7 is regulated at three different levels, including the transcriptional, post-transcriptional, and post-translational levels. Transcription of the Irf7 gene is induced by viral infection or type I IFN stimulation (17), although the stability of IRF7 is regulated by FOXO3 (18).
MicroRNAs (miRNAs)4 are small, non-coding, regulatory RNAs that range from 18 to 22 nucleotides in length. They are mainly encoded by gene introns but are also sometimes encoded by dedicated genes. The miRNAs regulate the expression of specific target proteins by either inhibiting translation or degrading the corresponding mRNA (19, 20). They regulate between one- and two-thirds of the human genome and participate in most of the cell's main functions (including growth, proliferation, differentiation, signal transduction, apoptosis, metabolism, and aging) (21). It has been reported that, with respect to virus infection, miRNAs constitute a critical additional control mechanism in the regulation of both cellular and viral gene expression. In fact, some miRNAs may target cellular genes involved in the virus replication control (22), and miRNAs targeting viral open reading frames may cause degradation of viral mRNAs to exhibit a direct antiviral effect (23). Previous reports have shown that some miRNAs, such as miR-146a and miR-155, are induced during virus infection and participate in the regulation of the innate antiviral response (24–26). What's more, it has also been reported that miR-466l can directly target the 3′-UTR sequences of IFN-α mRNA species, thus inhibiting IFN-α production and enhancing viral replication (27). However, whether the type I IFN response is regulated by other miRNAs is still unknown.
miR-223 is in the first cadre of miRNAs discovered to be highly expressed in myeloid cells of the bone marrow, where it is mainly expressed in the myeloid, granulocytic, and monocytic compartments, and miR-223 has a key role in the development and homeostasis of the immune system (28, 29). To date, miR-223 has been demonstrated to be involved in many types of cancers, inflammatory diseases, autoimmune diseases, and other pathological processes (19). However, there are few reports about the function of miR-223 in the regulation of type I IFN signaling and subsequent antiviral innate immunity. Our initial observation of significant up-regulation of miR-223 in murine macrophages upon VSV infection prompted us to study the potential effect of miR-223 in antiviral innate immunity. We observed that miR-223 overexpression up-regulated type I IFN expression levels in VSV-infected macrophages. We also demonstrated that miR-223 directly targeted FOXO3 to regulate the type I IFN production. Furthermore, type I IFN, which is triggered by VSV infection, is responsible for the up-regulation of miR-223, thus forming a positive regulatory loop for type I IFN production. Our results uncovered an essential mechanism of miR-223-mediated regulation of type I IFN production in the antiviral innate immunity.
Results
VSV-triggered Type I IFN Induces the Increase in miR-223 Expression in Macrophages
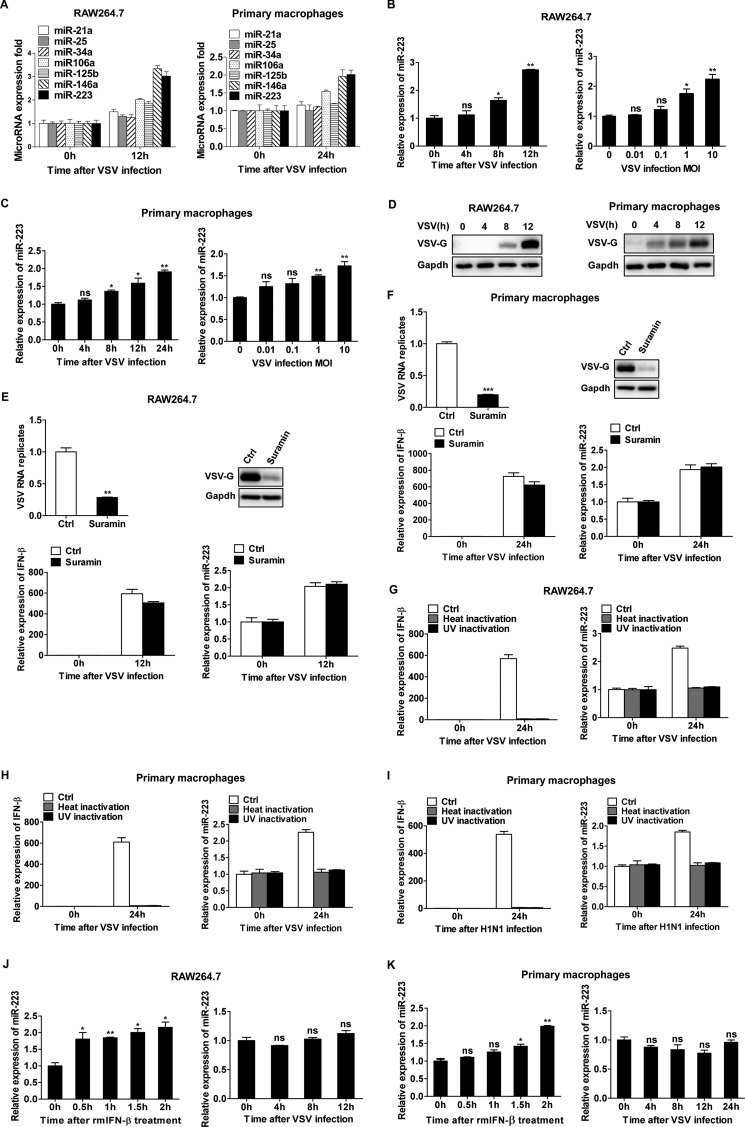
Previous reports have shown that some miRNAs, such as miR-146a and miR-155, are induced during virus infection and participate in the regulation of the innate antiviral response (24–26). To identify other microRNAs that are potentially involved in the regulation of the innate antiviral response, we selected several miRNAs (miR-21, -25, -34a, -106a, -125b, -146a, and -223) that are enriched in myeloid cells, and we analyzed their expression in VSV-infected RAW264.7 cells and primary macrophages via qPCR (Fig. 1A). We observed that one of those miRNAs, miR-223, was significantly increased upon VSV infection. We also found that miR-146a was up-regulated by VSV challenge, which is consistent with a previous report (26). In RAW264.7 cells, miR-223 was significantly increased at 8 h post-infection and was rapidly up-regulated during the next 4 h, reaching the maximum at 12 h, and miR-223 expression was induced by VSV in a dose-dependent manner (Fig. 1B). In primary peritoneal macrophages, the up-regulation for miR-223 by VSV infection was also observed, but the induction extent was weaker than that in RAW264.7 cells (Fig. 1C). The up-regulation of miR-223 upon VSV infection suggests that miR-223 may function as a regulator of VSV-associated signaling events in macrophages. We next investigated the underlying mechanism by which miR-223 was induced. First, we observed that VSV infection increased VSV-G protein level in macrophages (Fig. 1D), so we considered whether viral RNA or viral protein was responsible for the up-regulation of miR-223. It was reported that suramin could selectively inhibit the RNA-dependent RNA polymerase of virus, viral RNA synthesis, and viral protein synthesis (30, 31). So we treated the infected macrophages with suramin, and we found the viral RNA and viral protein were significantly down-regulated; however, miR-223 up-regulation was still induced along with type I IFN production (Fig. 1, E and F). Furthermore, we observed that heat- or UV-inactivated VSV and influenza viruses were no longer able to induce miR-223 up-regulation compared with infectious virus, due to the impaired type I IFN production (Fig. 1, G–I), indicating that miR-223 expression might be regulated by virus-triggered type I IFN signaling. It was reported previously that miR-223 could be induced by type I IFN in macrophages (32), and we further investigated the effect of type I IFN on VSV-induced miR-223 expression. The results showed that miR-223 was significantly induced after rmIFN-β treatment in macrophages (Fig. 1, J and K). On the contrary, after blocking type I IFN receptors with special antibody, the effect of VSV infection-induced miR-223 up-regulation was counteracted (Fig. 1, J and K). Taken together, these results suggest that VSV infection up-regulates miR-223 expression in macrophages mainly through type I IFN-dependent pathway.
FIGURE 1.
VSV-triggered type I IFN induces miR-223 expression in macrophages. A, RAW264.7 cells were infected with or without VSV at m.o.i. 0.1 for 12 h. Mouse peritoneal macrophages were infected with or without VSV at m.o.i. 10 for 24 h. The expressions of different miRNAs were measured by qPCR and normalized to the expression of U6 in each sample. B, RAW264.7 cells were infected with or without VSV at m.o.i. 0.1 for indicated times or at indicated m.o.i. for 12 h, and the expression of miR-223 was measured by qPCR and normalized to the expression of U6 in each sample. C, mouse peritoneal macrophages were infected with or without VSV at m.o.i. 10 for indicated times or at indicated m.o.i. for 24 h, and the expression of miR-223 was measured. D, RAW264.7 cells were infected with VSV at m.o.i. 0.1, and mouse peritoneal macrophages were infected VSV at m.o.i. 10. Total protein levels of VSV-G in lysates were detected by immunoblot at the indicated time. E, RAW264.7 were treated with suramin (200 μm) after infection with VSV at m.o.i. 0.1 for 1 h and then infected with VSV for 12 h; the expression of VSV RNA replicates, IFN-β and miR-223, were measured by qPCR, and total protein levels of VSV-G in lysates were detected by immunoblot. F, mouse peritoneal macrophages were treated with suramin (200 μm) after infection with VSV at m.o.i. 10 for 1 h and then infected with VSV for 24 h; the expression of VSV RNA replicates, IFN-β and miR-223, were measured by qPCR, and total protein levels of VSV-G in lysates were detected by immunoblot. G, RAW264.7 cells were infected with infectious VSV, heat- or UV-inactivated VSV at m.o.i. 0.1 for 12 h; IFN-β and miR-223 were measured by qPCR. H, mouse peritoneal macrophages were infected with infectious VSV, heat- or UV-inactivated VSV at m.o.i. 10 for 24 h; IFN-β and miR-223 were measured by qPCR. I, mouse peritoneal macrophages were infected with infectious H1N1, heat- or UV-inactivated H1N1 at m.o.i. 10 for 24 h; IFN-β and miR-223 were measured by qPCR. J, RAW264.7 macrophages were treated with rmIFN-β (25 ng/ml) for indicated times or pretreated with anti-mouse IFNAR1 antibody (10 μg/ml) for 2 h, then infected with VSV at m.o.i. 0.1 for indicated times, and the expression of miR-223 was measured. K, mouse peritoneal macrophages were treated as in E, infected with VSV at m.o.i. 10 for indicated times, and miR-223 expression was measured. Data are the mean ± S.D. (n = 3) of one representative experiment. Similar results were obtained in three independent experiments. ***, p < 0.1; **, p < 0.01; *, p < 0.05; ns, not significant; Ctrl, control.
miR-223 Positively Regulates VSV-triggered Type I IFN Production in Macrophages
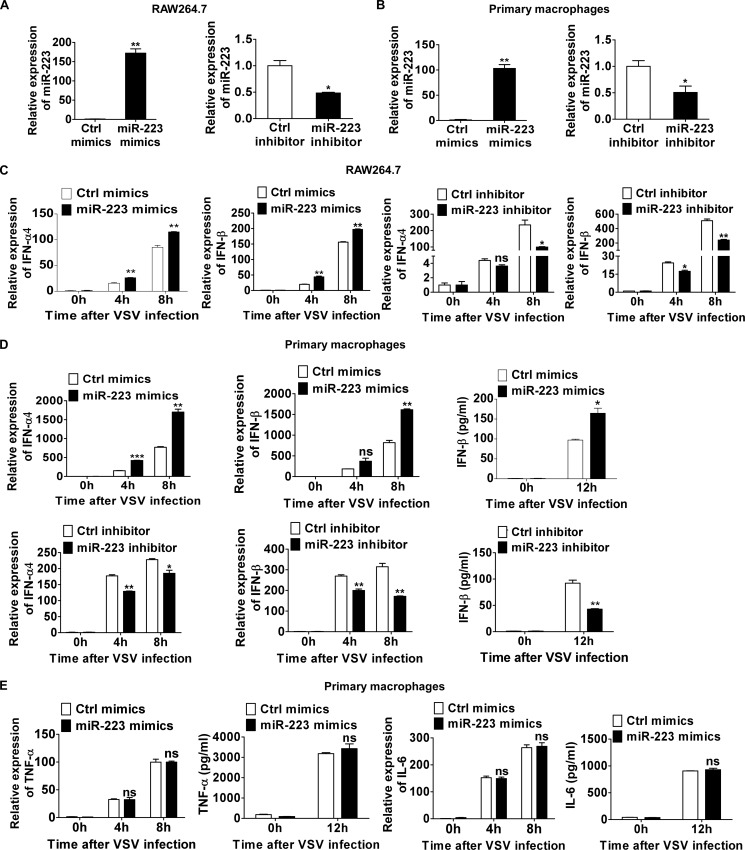
To further identify whether VSV-induced miR-223 expression could affect VSV-triggered response in macrophages, we investigated the role of miR-223 in type I IFN production after VSV challenge. As shown in Fig. 2, A and B, transfection of miR-223 mimics increased miR-223 expression in macrophages, whereas miR-223 inhibitors decreased its expression level. miR-223 overexpression promoted VSV-triggered IFN-β production at both the mRNA and protein levels (Fig. 2, C and D), whereas inhibition of miR-223 inhibited VSV-triggered IFN-β production (Fig. 2, C and D). Positive regulation of VSV-triggered IFN-α4 mRNA production by miR-223 was also observed, which was similar to positive regulation of IFN-β by miR-223 (Fig. 2, C and D). However, we found no significant differences in mRNA or protein levels of TNF-α and IL-6 (Fig. 2E). Taken together, these results demonstrate that miR-223 positively regulates VSV-triggered type I IFN production but does not significantly affect TNF-α and IL-6 production in macrophages.
FIGURE 2.
miR-223 positively regulates VSV-triggered type I IFN production. A, 0.5 ml of 2 × 105 RAW264.7 cells were transfected with control (Ctrl) mimics or miR-223 mimics (left), control inhibitors or miR-223 inhibitors (right) as indicated at a final concentration of 20 nm. After 48 h, miR-223 expression was measured. B, 0.5 ml of 2 × 105 mouse peritoneal macrophages were transfected as described in A, and after 48 h miR-223 expression was measured. C, 0.5 ml of 2 × 105 RAW264.7 cells were transfected as described in A. After 48 h, cells were infected by VSV at m.o.i. 0.1 for indicated times. IFN-α4 (left) and IFN-β (right) mRNA expression were measured by qPCR and normalized to the expression of β-actin in each sample. D, 0.5 ml of 2 × 105 mouse peritoneal macrophages were transfected as described in A. After 48 h, cells were infected by VSV at m.o.i. 10 for indicated times. IFN-α4 and IFN-β mRNA expression were measured by qPCR. IFN-β in supernatants was measured by ELISA. E, 0.5 ml of 2 × 105 peritoneal macrophages were transfected with control mimics or miR-223. After 48 h, cells were infected by VSV at m.o.i. 10 for indicated times. TNF-α and IL-6 mRNA expressions were measured by qPCR. TNF-α and IL-6 in supernatants were measured by ELISA. Data are the mean ± S.D. (n = 3) of one representative experiment. Similar results were obtained in three independent experiments. ***, p < 0.1; **, p < 0.01; *, p < 0.05; ns, not significant.
miR-223 Feedback Attenuates the Viral Replication in Macrophages
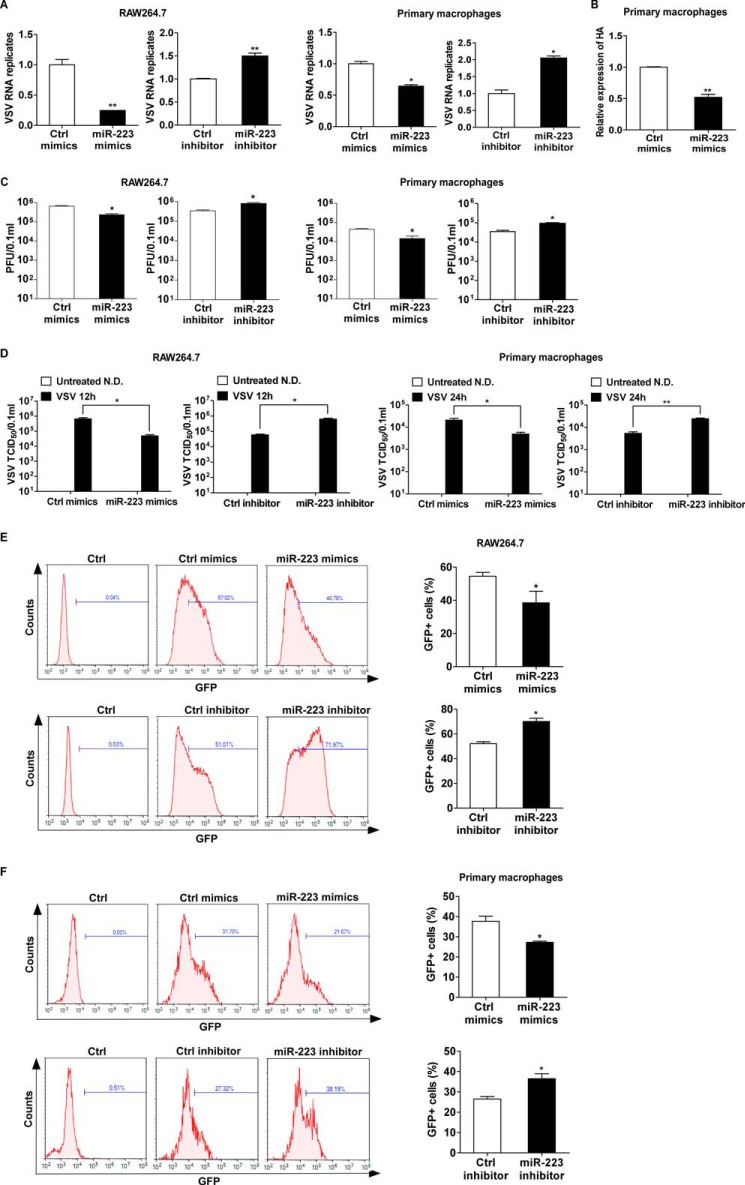
To investigate the biological significance of up-regulated miR-223 during viral infection, we further examined the effect of miR-223 on VSV replication in macrophages. By measuring intracellular VSV RNA replicates of the infected macrophages, we found that overexpression of miR-223 suppressed VSV replication, whereas inhibition of induced miR-223 facilitated VSV replication (Fig. 3A). Furthermore, we also observed that miR-223-induced viral resistance is not only against VSV infection but also the infection of influenza A H1N1 (Fig. 3B). Consistent with the results of intracellular VSV RNA replicates, VSV plaque assay and 50% tissue culture infectious dose (TCID50) assay in cultural supernatants also showed that VSV replication was inhibited by miR-223 overexpression and increased by miR-223 inhibition (Fig. 3, C and D). To further verify the effect of miR-223 on VSV replication, we challenged macrophages with a VSV strain expressing GFP (VSV-GFP). The VSV genome was modified by inserting the GFP transgene into the viral genome, and thus the presence of GFP in cells indicates VSV infection and replication (33). We quantified VSV-GFP expression by the percentage of VSV-GFP-positive cells, which should reflect the degree of viral infection across the population. As shown in Fig. 3, E and F, overexpression of miR-223 reduced the degree of VSV-GFP infection, whereas inhibition of induced miR-223 increased the degree of VSV-GFP infection. Thus, we conclude that induced miR-223 expression, as a positive feedback, attenuates the viral replication in virus-infected macrophages.
FIGURE 3.
miR-223 feedback attenuates the viral replication. A, RAW264.7 cells transfected with miR-223 mimics or inhibitors were infected by VSV at m.o.i. 0.1 for 1 h, then washed, and fresh medium was then added; after 12 h, VSV intracellular VSV RNA replicates were quantified using qRT-PCR and normalized to that of β-actin in each sample. Mouse peritoneal macrophages transfected with miR-223 mimics or inhibitors were infected by VSV at m.o.i. 10 for 1 h, then washed, and fresh medium was then added; after 24 h, intracellular VSV RNA replicates were quantified using qPCR and normalized to that of β-actin in each sample. B, mouse peritoneal macrophages transfected with miR-223 mimics were infected by H1N1 at m.o.i. 10 for 1 h, then washed, and fresh medium was then added; after 24 h, intracellular HA mRNA was quantified using quantitative RT-PCR. C and D, RAW264.7 and peritoneal macrophages were treated as in A, and VSV Plaque assay and TCID50 assay in cultural supernatants were done on Vero cells (N.D., not detected). E and F, RAW264.7 cells transfected with miR-223 mimics or inhibitors were infected by VSV-GFP at m.o.i. 0.1 for 1 h, then washed, and fresh medium was then added; after 12 h, VSV-GFP infection was quantified using flow cytometry. Mouse peritoneal macrophages transfected with miR-223 mimics or inhibitors were infected by VSV-GFP at m.o.i. 10 for 1 h, then washed, and fresh medium was then added; after 24 h, VSV-GFP infection was quantified using flow cytometry. Data are the mean ± S.D. (n = 3) of one representative experiment. Similar results were obtained in three independent experiments. **, p < 0.01; *, p < 0.05; Ctrl, control.
miR-223 Enhances IRF7 Expression by Targeting FOXO3
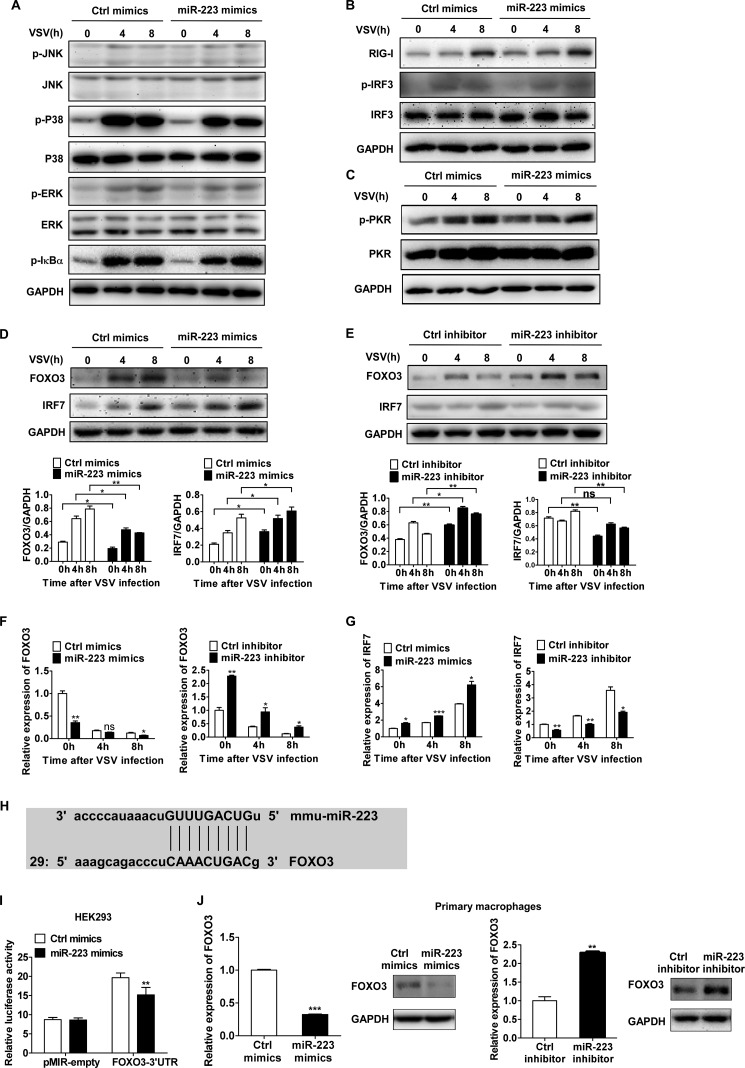
We next investigated the effect of miR-223 on RNA virus-activated downstream signal pathways in macrophages. We found that miR-223 overexpression did not significantly increase phosphorylation of p38, ERK, JNK, and IκBα after VSV challenge (Fig. 4A). Moreover, we did not observe a significant difference in the expression of RIG-I and phosphorylation of IRF3 (Fig. 4B). The phosphorylation of the dsRNA-dependent protein kinase, a primary regulator of antiviral activities (34), did not significantly increase after miR-223 overexpression (Fig. 4C). Interestingly, however, we observed that IRF7 was increased at both the protein and mRNA levels by miR-223 overexpression but decreased by miR-223 inhibition (Fig. 4, D and E).
FIGURE 4.
miR-223 enhances IRF7 expression by targeting FOXO3. A–C, 0.5 ml of 2 × 105 mouse peritoneal macrophages were transfected with control (Ctrl) mimics or miR-223; after 48 h, cells were infected by VSV at m.o.i. 10. Phosphorylated (p-) or total proteins in lysates were detected by immunoblot at the indicated times. D and E, 0.5 ml of 2 × 105 mouse peritoneal macrophages were transfected with miR-223 mimics or inhibitors; after 48 h, cells were infected by VSV at m.o.i. 10. Total protein levels of FOXO3 and IRF7 in lysates were detected by immunoblot at the indicated times. Band intensity was calculated using ImageJ software, and ratios of FOXO3/GAPDH or IRF7/GAPDH were determined. F and G, FOXO3 mRNA (F) and IRF7 mRNA expression (G) were measured by qPCR and normalized to the expression of β-actin in each sample. H, alignment of miR223 and its target sites in 3′-UTR of FOXO3 is shown. I, HEK293 cells were cotransfected with pMIR-REPORTTM FOXO3–3′-UTR luciferase reporter plasmid or empty pMIR-REPORTTM and pTK-RL plasmid, together with miR-223 mimics. After 24 h, firefly luciferase activity was measured and normalized by Renilla luciferase activity. J, 0.5 ml of 2 × 105 mouse peritoneal macrophages were transfected with miR-223 mimics or inhibitors; after 48 h, FOXO3 mRNA and protein levels were detected by qPCR and immunoblot. Data are the mean ± S.D. (n = 3) of one representative experiment. Similar results were obtained in three independent experiments. ***, p < 0.1; **, p < 0.01; *, p < 0.05; ns, not significant.
We then further investigated the possible targets of miR-223 that could modulate the expression of IRF7. Because it has been reported previously that the stability of IRF7 is regulated by FOXO3 (18), we next examined whether FOXO3 is a potential target of miR-223. This presumption was confirmed by the observation that FOXO3 was decreased at both the protein and mRNA levels by miR-223 overexpression but was increased by miR-223 inhibition (Fig. 4, F and G). Then we combined two algorithms, Miranda and PicTar, to search for the genes with putative miR-223 targeting sites in their 3′-UTRs, and we found that FOXO3 with one evolutionarily conserved seed region for miR-223 binding in the 3′-UTR was predicted to be a potential target (Fig. 4H). To certify the possibility that FOXO3 was regulated posttranscriptionally by miR-223, we constructed reporter plasmids by cloning the mouse FOXO3 3′-UTR into the pMIR-REPORTTM luciferase vector. By cotransfection of the reporter plasmids and internal control pRL-TK-Renilla luciferase plasmids with miR-223 mimics in HEK293 cells, we observed that miR-223 mimics markedly decreased the luciferase level (Fig. 4I). Furthermore, transfection of miR-223 mimics decreased FOXO3 expression in macrophages at both the protein and mRNA levels, whereas miR-223 inhibition increased FOXO3 (Fig. 4J), suggesting that FOXO3 expression could be inhibited by miR-223 via both translational inhibition and mRNA degradation. Together, the results show that miR-223 enhances IRF7 expression by targeting FOXO3, and mouse FOXO3 is a new target of miR-223.
Antiviral Function of miR-223 Is Mainly through Targeting FOXO3
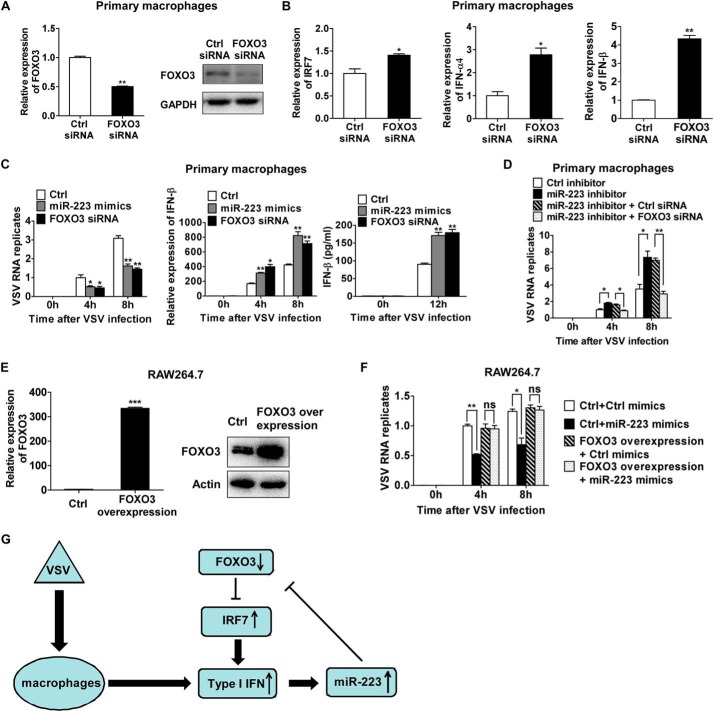
To demonstrate the role of targeting FOXO3 in the antiviral function of miR-223, we examined the effects of knockdown of FOXO3 on VSV-triggered type I IFN production. It has been shown previously that FOXO3 functions as a negative regulator of basal IRF7 transcription, thus suppressing the IRF7-dependent antiviral response (18). Consistent with this, we have observed that knockdown of FOXO3 promoted IRF7 expression and markedly increased VSV-triggered type I IFN production in primary mouse peritoneal macrophages (Fig. 5, A and B). These data suggest that positive regulation of VSV-triggered type I IFN production by miR-223 could be mediated by inhibition of its target FOXO3.
FIGURE 5.
Antiviral function of miR-223 is mainly through targeting FOXO3. A and B, murine peritoneal macrophages were transfected with control siRNA and FOXO3 siRNA as indicated. After 48 h, FOXO3, IRF7, IFN-α4, and IFN-β mRNA expressions and FOXO3 protein levels were detected. C, murine peritoneal macrophages were transfected with control (Ctrl), miR-223 mimics, and FOXO3 siRNA, respectively, as indicated. After 48 h, macrophages were infected by VSV at m.o.i. 10 for 1 h and washed; intracellular VSV RNA replicates at indicated time points were quantified using qRT-PCR, and the value at 4-h time point in control cells served as 1-fold control. The relative IFN-β mRNA expression at indicated time points was detected by qRT-PCR, and IFN-β in supernatants was measured by ELISA. D, murine peritoneal macrophages were transfected with control inhibitor, miR-223 inhibitor, miR-223 inhibitors combined with control siRNA, and miR-223 inhibitors combined with FOXO3 siRNA, respectively, as indicated. After 48 h, macrophages were infected by VSV at m.o.i. 10 for 1 h and washed; intracellular VSV RNA replicates at indicated time points were quantified using qRT-PCR, and the value at 4 h in cells transfected with control inhibitors served as 1-fold control. E, RAW264.7 cells were transfected with FOXO3 overexpression plasmid. After 48 h, FOXO3 mRNA and protein levels were detected by qRT-PCR (left) and immunoblot (right). F, RAW264.7cells were transfected with control (Ctrl) mimics combined with control plasmid, miR-223 mimics combined with control plasmid, or control mimics combined with FOXO3 overexpression plasmid, and miR-223 mimics combined with FOXO3 overexpression plasmid. After 48 h, macrophages were infected by VSV at m.o.i. 10 for 1 h and washed; intracellular VSV RNA replicates at indicated time points were quantified using qRT-PCR, and the level at 4-h time point in cells transfected with control plasmid and control mimics served as 1-fold control. G, proposed positive regulatory loop of type I IFN/miR-223/FOXO3/IFR7 pathway in regulating antiviral innate immunity. Data are the mean ± S.D. (n = 3) of one representative experiment. Similar results were obtained in three independent experiments. ***, p < 0.1; **, p < 0.01; *, p < 0.05; ns, not significant.
Then, to further demonstrate the role of targeting FOXO3 in the antiviral function of miR-223, we carried out experiments of FOXO3 knockdown and overexpression. In murine peritoneal macrophages, FOXO3 knockdown attenuated VSV replication, which resembled the effect of miR-223 overexpression (Fig. 5C). The VSV replication increased by miR-223 inhibition was rescued by FOXO3 knockdown (Fig. 5D). These results suggest that FOXO3 knockdown phenocopied the antiviral effect of induced miR-223 and counteracted the effect of miR-223 inhibition.
Furthermore, we confirmed that the FOXO3-overexpressed cell clone of RAW264.7 macrophages, which expressed FOXO3 mRNA without its 3′-UTR sequence, had significantly elevated FOXO3 expression at both the mRNA and protein levels (Fig. 5E). Because the FOXO3-overexpressed RAW264.7 cell clones transcribed FOXO3 mRNA without its 3′-UTR, miR-223 should no longer target FOXO3 expression in these cells. We also did not find that miR-223 overexpression could influence VSV proliferation in FOXO3-overexpressed RAW264.7 cells, as documented by measuring intracellular VSV RNA replicates (Fig. 5F). Therefore, we concluded that induced miR-223 upon viral infection exhibits its antiviral function mainly through suppressing endogenous FOXO3 expression and subsequently promoting type I IFN signaling.
Discussion
miR-223 is highly expressed in myeloid cells of the bone marrow (28). It is expressed specifically in cells of the granulocytic lineage and its expression changes during maturation, becoming incrementally higher as granulocytes mature (35). To date, miR-223 has been demonstrated to be involved in the regulation of a broad range of important cellular processes (including cell cycle regulation) and the invasiveness of many different cell types, hematopoietic differentiation, and immune cell function (19). Specifically, it was reported that miR-223 played a role in the pathogenesis of certain infectious diseases, such as influenza and chronic hepatitis B virus infection, and an antiviral effect for miR-223 was also observed in severe acute respiratory syndrome (SARS)-coronavirus-infected bronchoalveolar stem cells and in freshly isolated HIV-1-infected monocytes (36–40). Furthermore, miR-223 was found to inhibit dengue virus 2 (DENV2) replication in human endothelium-like EAhy926 cells by targeting microtubule-destabilizing protein stathmin 1 (STMN1) (41). However, less is known about the detailed roles of miR-223 in the regulation of the innate antiviral response.
In this study, we report a positive regulatory loop for type I IFN production. First, we found that VSV infection could induce miR-223 up-regulation in a murine macrophage cell line and in murine primary macrophages, which was accompanied by a significant up-regulation of type I IFN. The most plausible hypothesis is that type I IFN can induce miR-223 up-regulation after VSV infection. Consistent with this, we observed that miR-223 was significantly induced after rmIFN-β treatment in macrophages. On the contrary, after blocking type I IFN receptors with special antibody, the effect of VSV infection-induced miR-223 up-regulation was counteracted. Second, we have proved that miR-223 positively regulates VSV-triggered type I IFN production and inhibits VSV replication. Third, we have demonstrated that miR-223 enhances IRF7 expression by targeting FOXO3 and then promotes the type I IFN production. Therefore, we present a new model in which type I IFN, miR-223, FOXO3, and IRF7 form a positive regulatory loop for type I IFN production (Fig. 5G).
FOXO3 is a member of Forkhead box transcription factor O subfamily. FOXO3 together with other members, such as FOXO1, FOXO4, etc., participates in cell function regulations related to cell cycle arrest, induction of apoptosis, and oxidative and cellular stress (42–44). In general, FOXO3 is known to inhibit cell cycle progression and to promote cell death, so it has been thought that FOXO3 can be an important target to inhibit cancer cell progression (44). FOXO3 has also been shown to play a role in immunity and inflammation (45–47). For the involvement of FOXO3 in the regulatory loop, a previous report presented that FOXO3 inhibited IRF7 transcription and negatively regulated innate immune response (18). Consistent with this, we have observed that knockdown of FOXO3 promoted IRF7 expression and markedly increased VSV-triggered type I IFN production in primary mouse peritoneal macrophages. Furthermore, FOXO3 knockdown attenuated VSV replication, which resembled the effect of miR-223 overexpression. So, we concluded that induced up-regulation of miR-223 upon viral infection exhibited its antiviral function mainly through suppressing endogenous FOXO3 expression and subsequently promoting type I IFN signaling.
miRNAs have been thought to target multiple mRNAs, named targetomes, to regulate gene expression. A single miRNA might tune protein synthesis from thousands of genes by direct or indirect effects (48). In this study, mouse FOXO3 was proved to be a novel target of miR-223 and contributed to the feedback-positive regulatory function of miR-223 in the regulation of type I IFN production. It is probable that we are far from unveiling the last target of miR-223, and some of the potential targets might also regulate type I IFN production. This presumption may raise interesting future work to reveal the entire functions of miR-223 in innate immune response.
Experimental Procedures
Mice and Reagents
BALB/c mice (6–8 weeks old) were purchased from SIPPR-BK Experimental Animal Ltd. Co. (Shanghai, China). All experiments and animal care were performed according to protocols approved by the Zhejiang University Institutional Animal Care and Use Committee. VSV and VSV-GFP were gifts from Prof. Wang Xiaojian (Zhejiang University, Hangzhou, China). Influenza A (H1N1) virus was kindly provided by Prof. Yang Yu (Zhejiang University, Hangzhou, China). miR-223 mimics and control mimics, miR-223 inhibitors, and control inhibitor, FOXO3 small interfering RNA (siRNA), and control siRNA were from GenePharma (Shanghai, China). Abs specific to β-actin, HRP-coupled secondary Abs, and suramin sodium were from Santa Cruz Biotechnology (Santa Cruz, CA). Abs specific to ERK, JNK, p38, IRF3, RIG-I, FOXO3, phosphorylated ERK, phosphorylated JNK, phosphorylated p38, phosphorylated IRF3, and phosphorylated IκBα were from Cell Signaling Technology (Danvers, MA). Abs specific to IRF7, VSV-G, PKR, and phosphorylated PKR were from Abcam (Cambridge, MA). Ab specific to GAPDH was from Beyotime (Haimen, China). Recombinant mouse IFN-β (rmIFN-β) was from Sino Biological (Beijing, China). Anti-mouse IFNAR1 Ab was from eBioscience (San Diego).
Cell Culture and Transfection
HEK293 cell line was obtained from American Type Culture Collection (Manassas, VA) and cultured as described previously (49). RAW264.7 cell line was obtained from American Type Culture Collection and cultured in DMEM containing 10% FBS. Murine primary macrophages were obtained and cultured as described previously (50). 1 × 104 cells were seeded into 96-well plates and incubated overnight. JetPRIME transfection reagents (Polyplus-transfection, Illkirch, France) were used for the cotransfection of plasmids and RNAs, according to the manufacturer's instructions. 0.5 ml of growth medium containing 2 × 105 cells was seeded into each well of 24-well plates, incubated overnight, and then transfected with RNAs using INTERFERin (Polyplus-transfection), according to the manufacturer's instructions.
Detection of Cytokine Production
Into each well of 24-well plates, 0.5 ml of growth medium containing 2 × 105 cells was seeded, incubated overnight, and then transfected as described above. After 48 h, the cells were infected with VSV for the indicated time periods. The concentrations of IFN-β in culture supernatants were measured with ELISA kits (BioLegend) according to the manufacturer's protocols. TNF-α and IL-6 in the supernatants were measured with ELISA kits (eBioscience) according to the manufacturer's protocols.
Plasmid Construction
FOXO3 mRNA without its 3′-UTR sequence was cloned from the cDNA of RAW264.7 cells and then inserted into pcDNA3.1(+) vector to generate FOXO3-overexpression plasmid. Primers for plasmid construction were 5′-CGCGGATCCATGGCAGAGGCACCAGCC-3′ (forward) and 5′-CCGGAATTCTCAGCCTGGTACCCAGCT-3′ (reverse).
3′-UTR Luciferase Reporter Assays
The wild-type mouse FOXO3 3′-UTR luciferase reporter vectors were constructed by amplifying the mouse FOXO3 mRNA 3′-UTR and cloning it into the pMIR-REPORTTM luciferase vector. HEK293 cells described above were cotransfected with 80 ng of luciferase reporter plasmid, 40 ng of thymidine kinase promoter-Renilla luciferase reporter plasmid, and the indicated miRNA mimics or controls (final concentration, 30 nm). After 24 h, luciferase activities were measured using the Dual-LuciferaseTM reporter assay system (Promega), according to the manufacturer's instructions.
RNA Isolation and Real Time Quantitative PCR (qPCR)
Total RNA was extracted with TRIzol (Invitrogen). Real time quantitative PCR, using SYBR Green detection chemistry, was performed on a 7500 real time PCR system (Applied Biosystems) as we previously described (51). For miRNA analysis, the RT primers used were as follows: miR-21a, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACAT-3′; miR-25, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCAATTG-3′; miR-34a, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAACCA-3′; miR-106a, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCTG-3′; miR-125b, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACAAG-3′; miR-146a, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACCCAT-3′; and miR-223, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGGGGT-3′. The qPCR primers used were as follows: miR-21a, 5′-GCCTAGCTTATCAGACTGA-3′ (forward); miR-25, 5′-ATTAGGCGGAGACTTGGGC-3′ (forward); miR-34a, 5′-GCCTGGCAGTGTCTTAGCT-3′ (forward); miR-106a, 5′-GCCCAAAGTGCTAACAGTG-3′ (forward); miR-125b, 5′-GCCTCCCTGAGACCCTAAC-3′(forward); miR-146a, 5′-GCCTGAGAACTGAATTCCA-3′(forward); miR-223, 5′-TGGCTGTCAGTTTGTCAAAT-3′ (forward), and the universal reverse primer, 5′-GTGTCGTGGAGTCGGCAA-3′. U6 small nuclear RNA was quantified using its reverse primers for reverse-transcriptase reaction and its forward and reverse primers for qPCR, which were 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). The relative expression level of miRNA was normalized by U6 expression. For mouse β-actin, IFN-β, and IFN-α4 mRNA analysis, the qPCR primers were described previously (49, 50). For VSV Indiana serotype, the qPCR primers were 5′-ACGGCGTACTTCCAGATGG-3′ (forward) and 5′-CTCGGTTCAAGATCCAGGT-3′ (reverse). For HA of H1N1, the qPCR primers were 5′-AGTTCAAGTCGGAAATAGCAAT-3′ (forward) and 5′-ATACCAGATCCAGCATTTCTTTC-3′ (forward). For mouse FOXO3, the qPCR primers were 5′-CTGGGGGAACCTGTCCTATG-3′ (forward) and 5′-TCATTCTGAACGCGCATGAAG-3′ (reverse). For mouse IRF7, the qPCR primers were 5′-GAGACTGGCTATTGGGGGAG-3′ (forward) and 5′-GACCGAAATGCTTCCAGGG-3′ (reverse). For mouse TNF-α, the qPCR primers were 5′-AAGCCTGTAGCCCACGTCGTA-3′ (forward) and 5′-GGCACCACTAGTTGGTTGTCTTTG-3′ (reverse). For mouse IL-6, the primers were 5′-TAGTCCTTCCTACCCCAATTTCC-3′ (forward) and 5′-TTGGTCCTTAGCCACTCCTTC-3′ (reverse). Data were normalized by the level of β-actin expression in each sample described above.
RNA Interference
The FOXO3-specific siRNA was v5′-GGCGGCTCTTGGTGGTCTCTT-3′ (sense) and 5′-GGCGGCUCUUGGUGGUCUCUU-3′ (antisense). The control siRNA sequences were 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense). siRNA duplexes were transfected into RAW264.7 cell or primary murine peritoneal macrophages at a final concentration of 30 nm.
Immunoblotting
The cells were washed twice with cold PBS and lysed with cell lysis buffer (Cell Signaling Technology) supplemented with protease inhibitor mixture (Beyotime). Protein concentrations of the cell lysis extracts were measured with BCA assay (Pierce) and equalized with the extraction reagent. Equal amounts of the extracts were loaded and subjected to SDS-polyacrylamide gel, transferred into PVDF membrane, and blotted as we described previously (52).
VSV Virus Production and Infection
Wild-type VSV (Indiana serotype) and VSV-GFP were propagated in Vero cells after infection at m.o.i. = 0.01. 48 h later, supernatant was collected after centrifugation and then filtered through a 0.45-μm filter (Millipore) and stored at −80 °C. Virus infections were performed in the absence of serum for 1 h and then replaced with fully supplemented growth medium.
VSV Yield Qualification
Macrophages were transfected and infected by VSV as indicated. 0.1 ml of the cultural supernatants were serially diluted on the monolayer of Vero cells, which were obtained from ATCC, and 1 × 104 cells were seeded into 96-well plates 1 day before measurement. The TCID50 was measured after 3 days. Plaque assays were done on Vero cells in 12-well plates at 2 × 105 cells per well for VSV. Supernatants from infected cells were serially diluted and infected on Vero cells for 1 h. The cells were then covered with growth medium containing 0.6% low-melting point agarose. Plaques stained with 0.5% crystal violet (m/v) in 20% ethanol (v/v) were counted after 16 h for VSV. For indicated experiments, cells were mock-infected or infected with VSV-GFP under similar conditions, and GFP expression was analyzed by FACSCaliburTM flow cytometry.
Virus Inactivation
For heat inactivation, the viruses were incubated for 30 min at 56 °C. For UV inactivation, the viruses were placed in a 12-well plate on ice and exposed to UV light for 45 min.
Statistical Analysis
All experiments were repeated three times. Statistical significance was determined by Student's t test, with p < 0.05 considered to be statistically significant.
Author Contributions
Q. W., Y. L., and L. C. designed the research, L. C., Y. S., L. H., X. W., L. L., F. D., and Y. L. performed the experiments and analyzed the data. L. C., Y. L., and Q. W. wrote the paper.
Acknowledgments
We thank Prof. Xiaojian Wang for kindly providing VSV and VSV-GFP. We are grateful for the technical assistance of Ting Pan.
This work was supported by National Natural Science Foundation of China Grants 81230074 and 81571524, National Key Basic Research Program of China Grant 2013CB530502, and Zhejiang Provincial Program for Cultivation of High Level Innovative Health Talents and for Innovative Research Team in Zhejiang Province Grant 2010R50046. The authors declare that they have no conflicts of interest with the contents of this article.
- miRNA
- microRNA
- VSV
- vesicular stomatitis virus
- IRF
- interferon regulatory factor
- rmIFN-β
- recombinant mouse IFN-β
- qPCR
- real time quantitative
- m.o.i.
- multiplicity of infection
- Ab
- antibody.
References
- 1.Stifter S. A., and Feng C. G. (2015) Interfering with immunity: detrimental role of type I IFNs during infection. J. Immunol. 194, 2455–2465 [DOI] [PubMed] [Google Scholar]
- 2.Killip M. J., Fodor E., and Randall R. E. (2015) Influenza virus activation of the interferon system. Virus Res. 209, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann H. H., Schneider W. M., and Rice C. M. (2015) Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36, 124–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W., Han C., Xie B., Hu X., Yu Q., Shi L., Wang Q., Li D., Wang J., Zheng P., Liu Y., and Cao X. (2013) Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 152, 467–478 [DOI] [PubMed] [Google Scholar]
- 5.Barbalat R., Ewald S. E., Mouchess M. L., and Barton G. M. (2011) Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 29, 185–214 [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi O., and Akira S. (2008) MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 20, 17–22 [DOI] [PubMed] [Google Scholar]
- 7.Beutler B., Eidenschenk C., Crozat K., Imler J. L., Takeuchi O., Hoffmann J. A., and Akira S. (2007) Genetic analysis of resistance to viral infection. Nat. Rev. Immunol. 7, 753–766 [DOI] [PubMed] [Google Scholar]
- 8.Akira S., Uematsu S., and Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell. 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 9.Herdy B., Jaramillo M., Svitkin Y. V., Rosenfeld A. B., Kobayashi M., Walsh D., Alain T., Sean P., Robichaud N., Topisirovic I., Furic L., Dowling R. J., Sylvestre A., Rong L., Colina R., et al. (2012) Translational control of the activation of transcription factor NF-κB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat. Immunol. 13, 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J., Li Y., Zhu L., Liu D., Songyang Z., Wang H. Y., and Wang R. F. (2012) NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat. Immunol. 13, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiba S., Baghdadi M., Akiba H., Yoshiyama H., Kinoshita I., Dosaka-Akita H., Fujioka Y., Ohba Y., Gorman J. V., Colgan J. D., Hirashima M., Uede T., Takaoka A., Yagita H., and Jinushi M. (2012) Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat. Immunol. 13, 832–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura T., Yanai H., Savitsky D., and Taniguchi T. (2008) The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26, 535–584 [DOI] [PubMed] [Google Scholar]
- 13.Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., and Taniguchi T. (2005) IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777 [DOI] [PubMed] [Google Scholar]
- 14.Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., and Taniguchi T. (2000) Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13, 539–548 [DOI] [PubMed] [Google Scholar]
- 15.Sato M., Hata N., Asagiri M., Nakaya T., Taniguchi T., and Tanaka N. (1998) Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441, 106–110 [DOI] [PubMed] [Google Scholar]
- 16.Marié I., Durbin J. E., and Levy D. E. (1998) Differential viral induction of distinct interferon-α genes by positive feedback through interferon regulatory factor-7. EMBO J. 17, 6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu R., Au W. C., Yeow W. S., Hageman N., and Pitha P. M. (2000) Regulation of the promoter activity of interferon regulatory factor-7 gene. Activation by interferon and silencing by hypermethylation. J. Biol. Chem. 275, 31805–31812 [DOI] [PubMed] [Google Scholar]
- 18.Litvak V., Ratushny A. V., Lampano A. E., Schmitz F., Huang A. C., Raman A., Rust A. G., Bergthaler A., Aitchison J. D., and Aderem A. (2012) A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature 490, 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taïbi F., Metzinger-Le Meuth V., Massy Z. A., and Metzinger L. (2014) miR-223: an inflammatory oncomiR enters the cardiovascular field. Biochim. Biophys. Acta 1842, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 20.Lewis B. P., Burge C. B., and Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 21.Kloosterman W. P., and Plasterk R. H. (2006) The diverse functions of microRNAs in animal development and disease. Dev. Cell 11, 441–450 [DOI] [PubMed] [Google Scholar]
- 22.Lecellier C. H., Dunoyer P., Arar K., Lehmann-Che J., Eyquem S., Himber C., Saïb A., and Voinnet O. (2005) A cellular microRNA mediates antiviral defense in human cells. Science 308, 557–560 [DOI] [PubMed] [Google Scholar]
- 23.Fiorucci G., Chiantore M. V., Mangino G., and Romeo G. (2015) MicroRNAs in virus-induced tumorigenesis and IFN system. Cytokine Growth Factor Rev. 26, 183–194 [DOI] [PubMed] [Google Scholar]
- 24.O'Neill L. A., Sheedy F. J., and McCoy C. E. (2011) MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 11, 163–175 [DOI] [PubMed] [Google Scholar]
- 25.Wang P., Hou J., Lin L., Wang C., Liu X., Li D., Ma F., Wang Z., and Cao X. (2010) Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 185, 6226–6233 [DOI] [PubMed] [Google Scholar]
- 26.Hou J., Wang P., Lin L., Liu X., Ma F., An H., Wang Z., and Cao X. (2009) MicroRNA-146a feedback inhibits RIG-I-dependent type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 183, 2150–2158 [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Fan X., He X., Sun H., Zou Z., Yuan H., Xu H., Wang C., and Shi X. (2012) MicroRNA-466l inhibits antiviral innate immune response by targeting interferon-α. Cell. Mol. Immunol. 9, 497–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C. Z., Li L., Lodish H. F., and Bartel D. P. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86 [DOI] [PubMed] [Google Scholar]
- 29.Lim L. P., Glasner M. E., Yekta S., Burge C. B., and Bartel D. P. (2003) Vertebrate microRNA genes. Science 299, 1540. [DOI] [PubMed] [Google Scholar]
- 30.Mastrangelo E., Pezzullo M., Tarantino D., Petazzi R., Germani F., Kramer D., Robel I., Rohayem J., Bolognesi M., and Milani M. (2012) Structure-based inhibition of norovirus RNA-dependent RNA polymerases. J. Mol. Biol. 419, 198–210 [DOI] [PubMed] [Google Scholar]
- 31.Albulescu I. C., van Hoolwerff M., Wolters L. A., Bottaro E., Nastruzzi C., Yang S. C., Tsay S.-C., Hwu J. R., Snijder E. J., and van Hemert M. J. (2015) Suramin inhibits chikungunya virus replication through multiple mechanisms. Antiviral Res. 121, 39–46 [DOI] [PubMed] [Google Scholar]
- 32.Cobos Jiménez V., Booiman T., de Taeye S. W., van Dort K. A., Rits M. A., Hamann J., and Kootstra N. A. (2012) Differential expression of HIV-1 interfering factors in monocyte-derived macrophages stimulated with polarizing cytokines or interferons. Sci. Rep. 2, 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez M., Porosnicu M., Markovic D., and Barber G. N. (2002) Genetically engineered vesicular stomatitis virus in gene therapy: application for treatment of malignant disease. J. Virol. 76, 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran J. M., Moxley M. A., Buller R. M., and Corbett J. A. (2005) Encephalomyocarditis virus induces PKR-independent mitogen-activated protein kinase activation in macrophages. J. Virol. 79, 10226–10236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnnidis J. B., Harris M. H., Wheeler R. T., Stehling-Sun S., Lam M. H., Kirak O., Brummelkamp T. R., Fleming M. D., and Camargo F. D. (2008) Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451, 1125–1129 [DOI] [PubMed] [Google Scholar]
- 36.Choi E. J., Kim H. B., Baek Y. H., Kim E. H., Pascua P. N., Park S. J., Kwon H. I., Lim G. J., Kim S., Kim Y. I., and Choi Y. K. (2014) Differential microRNA expression following infection with a mouse-adapted, highly virulent avian H5N2 virus. BMC Microbiol. 14, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J., Wu C., Che X., Wang L., Yu D., Zhang T., Huang L., Li H., Tan W., Wang C., and Lin D. (2011) Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 50, 136–142 [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Chan E. Y., Li J., Ni C., Peng X., Rosenzweig E., Tumpey T. M., and Katze M. G. (2010) MicroRNA expression and virulence in pandemic influenza virus-infected mice. J. Virol. 84, 3023–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Ye L., Hou W., Zhou Y., Wang Y. J., Metzger D. S., and Ho W. Z. (2009) Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood 113, 671–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mallick B., Ghosh Z., and Chakrabarti J. (2009) MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS ONE 4, e7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu N., Gao N., Fan D., Wei J., Zhang J., and An J. (2014) miR-223 inhibits dengue virus replication by negatively regulating the microtubule-destabilizing protein STMN1 in EAhy926 cells. Microbes Infect. 16, 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salih D. A., and Brunet A. (2008) FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr. Opin. Cell Biol. 20, 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho K. K., Myatt S. S., and Lam E. W. (2008) Many forks in the path: cycling with FoxO. Oncogene 27, 2300–2311 [DOI] [PubMed] [Google Scholar]
- 44.Dansen T. B., and Burgering B. M. (2008) Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 18, 421–429 [DOI] [PubMed] [Google Scholar]
- 45.Dejean A. S., Beisner D. R., Ch'en I. L., Kerdiles Y. M., Babour A., Arden K. C., Castrillon D. H., DePinho R. A., and Hedrick S. M. (2009) Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat. Immunol. 10, 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riou C., Yassine-Diab B., Van grevenynghe J., Somogyi R., Greller L. D., Gagnon D., Gimmig S., Wilkinson P., Shi Y., Cameron M. J., Campos-Gonzalez R., Balderas R. S., Kelvin D., Sekaly R. P., and Haddad E. K. (2007) Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells. J. Exp. Med. 204, 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luron L., Saliba D., Blazek K., Lanfrancotti A., and Udalova I. A. (2012) FOXO3 as a new IKK-ϵ-controlled check-point of regulation of IFN-β expression. Eur. J. Immunol. 42, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 48.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., and Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 49.Liu X., Yao M., Li N., Wang C., Zheng Y., and Cao X. (2008) CaMKII promotes TLR-triggered proinflammatory cytokine and type I interferon production by directly binding and activating TAK1 and IRF3 in macrophages. Blood 112, 4961–4970 [DOI] [PubMed] [Google Scholar]
- 50.An H., Zhao W., Hou J., Zhang Y., Xie Y., Zheng Y., Xu H., Qian C., Zhou J., Yu Y., Liu S., Feng G., and Cao X. (2006) SHP-2 phosphatase negatively regulates the TRIF adaptor protein-dependent type I interferon and proinflammatory cytokine production. Immunity 25, 919–928 [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Chen Q., Song Y., Lai L., Wang J., Yu H., Cao X., and Wang Q. (2011) MicroRNA-98 negatively regulates IL-10 production and endotoxin tolerance in macrophages after LPS stimulation. FEBS Lett. 585, 1963–1968 [DOI] [PubMed] [Google Scholar]
- 52.Wang Q. Q., Li H., Oliver T., Glogauer M., Guo J., and He Y. W. (2008) Integrin β1 regulates phagosome maturation in macrophages through Rac expression. J. Immunol. 180, 2419–2428 [DOI] [PubMed] [Google Scholar]