Abstract
Recent studies identified the adaptor protein Ajuba as a positive regulator of Yes-associated protein (YAP) oncogenic activity through inhibiting large tumor suppressor (Lats1/2) core kinases of the Hippo pathway, a signaling pathway that plays important roles in cancer. In this study, we define a novel mechanism for phospho-regulation of Ajuba in mitosis and its biological significance in cancer. We found that Ajuba is phosphorylated in vitro and in vivo by cyclin-dependent kinase 1 (CDK1) at Ser119 and Ser175 during the G2/M phase of the cell cycle. Mitotic phosphorylation of Ajuba controls the expression of multiple cell cycle regulators; however, it does not affect Hippo signaling activity, nor does it induce epithelial-mesenchymal transition. We further showed that mitotic phosphorylation of Ajuba is sufficient to promote cell proliferation and anchorage-independent growth in vitro and tumorigenesis in vivo. Collectively, our discoveries reveal a previously unrecognized mechanism for Ajuba regulation in mitosis and its role in tumorigenesis.
Keywords: cell proliferation, cyclin-dependent kinase (CDK), Hippo pathway, mitosis, phosphorylation, signal transduction
Introduction
Genetic screens in Drosophila have discovered the Hippo pathway (1) and extensive studies have demonstrated important roles for Hippo signaling in tissue homeostasis, stem cell function, and cancer biology (2–5). Protein kinases Mst1/2 (together with the adaptor protein WW45) and Lats1/2 (with the regulatory subunit Mob1) form the core complexes in the Hippo pathway and these proteins regulate each other through phosphorylation. This core kinase signaling subsequently phosphorylates and inactivates the downstream effectors, oncoproteins YAP and TAZ, by sequestering them in the cytoplasm and promoting ubiquitination-dependent degradation (5). During past years, many regulators and input signals have been identified that influence Hippo-YAP signaling activity, such as the cell polarity and adherens junctions proteins, mechanical force, actin cytoskeleton (6–8), hypoxia (9), energy stress (10, 11), and mitosis/cytokinesis stress (12–15). The downstream effectors YAP/TAZ also cross-talk with, or function as, mediators of many other signaling pathways, such as the G-protein coupled receptor, Wnt/β-catenin, TGF-β/SMAD, EGF, Notch, Hedgehog, and KRas/MAPK pathways (16).
A previous study identified Drosophila jub (orthologous to Ajuba proteins in mammals) as a negative regulator of the Hippo pathway (17). Djub promotes Yki (Drosophila ortholog of YAP/TAZ) activation through interacting with, and inhibiting, Warts (Drosophila ortholog of Lats1/2) kinase, and this function/mechanism appears to be conserved in mammalian cells (17). Subsequent studies revealed that Ajuba functions as an adaptor protein that links EGFR-MAPK signaling to the Hippo pathway in both Drosophila and mammals (18). Furthermore, Djub/Ajuba are also required for JNK-mediated activation of Yki/YAP, implying a conserved link between JNK signaling and Hippo pathways (19). Interestingly, cytoskeletal tension modulates organ growth through Yki in a Djub-dependent manner in Drosophila, although such a link in mammalian cells has not been identified (20).
Ajuba family proteins, including Ajuba, LIM-domain containing protein 1 (LIMD1), and Wilms tumor 1 interacting protein (WTIP), are adaptor/scaffold proteins with three LIM domains at their C termini. The LIM domains interact with other proteins in various subcellular locations to exert the biological functions of Ajuba proteins (21). The Ajuba family is involved in many cellular processes such as cell-cell adhesion, gene transcription, cell proliferation, cell migration, and mitosis/cytokinesis (21). Interestingly, several studies also suggest that Ajuba family proteins function as potential tumor suppressors or oncoproteins (22–26). Some reports indicate that Ajuba is a critical member of the mitotic machinery. For example, Ajuba activates Aurora-A kinase to recruit the CDK1-cyclin B complex to centrosomes, and it contributes to mitotic entry (27). Similarly, Ajuba associates with Lats2 at centrosomes during mitosis and regulates the integrity of the spindle apparatus (28). Ajuba is also a microtubule-associated protein and plays a role in metaphase-anaphase transition through interactions with Aurora-B and BubR1 at kinetochores (29). Collectively, these studies suggest an important role of Ajuba in mitosis, and indicate that Ajuba may exert its oncogenic or tumor suppressive function via dysregulation of mitosis. Ajuba has been observed to be phosphorylated by Aurora-A (27) and Lats2 (28) in mitosis; however, the phosphorylation site(s) and its biological function have remained elusive.
We have recently investigated how the Hippo pathway (core members and their regulators) is regulated in mitosis. We have shown that KIBRA (an upstream regulator of the Hippo pathway) (30, 31), YAP (12, 13), and TAZ (15) are phosphorylated by mitotic kinases. Importantly, mitotic phosphorylation of YAP/TAZ is critical for proper mitotic progression and for their oncogenic activity in cancer cells (12, 13, 15). These studies prompted us to further examine whether other components or regulators of the Hippo pathway are regulated by phosphorylation during mitosis. In this study, we found that many of the Hippo members/regulators, including Ajuba, are indeed phosphorylated during antimitotic drug-induced G2/M phase arrest. We characterized the phospho-regulation of Ajuba in mitosis, and identified CDK1 as a major kinase for mitotic phosphorylation of Ajuba. We further examined the functional significance of the phosphorylation and found that mitotic phosphorylation promotes the oncogenic activity of Ajuba independently of the Hippo pathway, suggesting a novel mechanism that regulates Ajuba in cancer cells.
Results
Ajuba Family Proteins Are Phosphorylated during Antimitotic Drug-induced G2/M Arrest
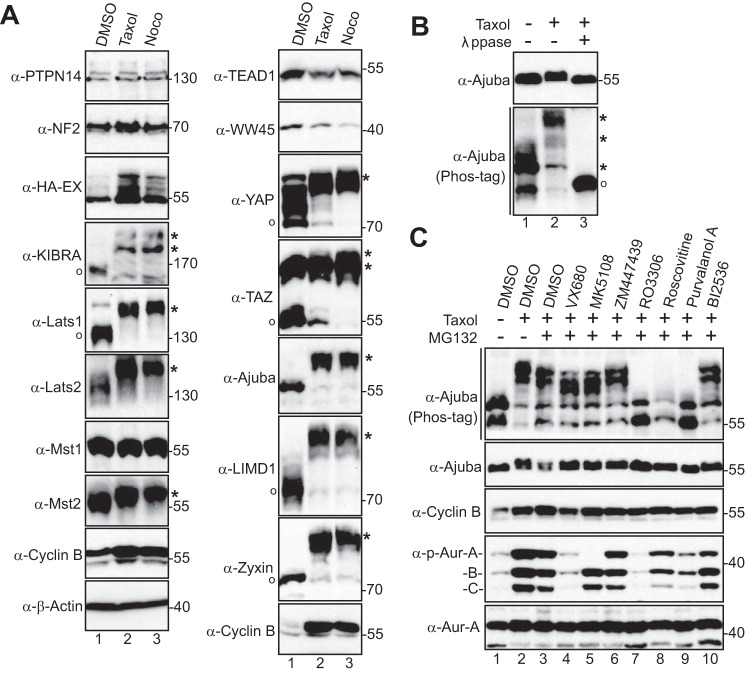
To further explore whether members of the Hippo pathway are regulated by phosphorylation during mitosis, we examined the phosphorylation status of the Hippo pathway proteins during G2/M arrest induced by Taxol or Nocodazole. As shown in Fig. 1A, consistent with previous reports, there was a dramatic up-shift of Lats1 and Lats2 mobility (due to mitotic phosphorylation) (32) during Taxol or Nocodazole treatment (Fig. 1A). As expected, the mobility of KIBRA, YAP, and TAZ were all significantly retarded due to phosphorylation during G2/M arrest (Fig. 1A) (12, 15, 30, 31). Taxol or Nocodazole treatment did not cause any evident change in the mobility/phosphorylation for PTPN14, NF2, or EX (which are all upstream regulators of the Hippo-YAP pathway), for WW45 or TEAD1 (Fig. 1A). Interestingly, Mst2, but not Mst1, was phosphorylated during G2/M arrest (Fig. 1A). One of the most prominent changes we observed was the striking mobility up-shift of the Ajuba and Zyxin family proteins including Ajuba, LIMD1, and Zyxin (Fig. 1A). In this study, we have chosen to focus on Ajuba, and so we further investigated its phosphorylation status. λ-Phosphatase treatment completely converted all slow-migrating bands to fast-migrating bands, confirming that the mobility shift of Ajuba during G2/M arrest is caused by phosphorylation (Fig. 1B).
FIGURE 1.
CDK1-dependent phosphorylation of Ajuba during G2/M arrest. A, HeLa cells were treated with DMSO, Taxol (100 nm for 16 h), or Nocodazole (Noco, 100 ng/ml for 16 h). Total cell lysates were probed with the indicated antibodies against Hippo components on Phos-tag SDS-polyacrylamide gels (see ”Experimental Procedures“). O and * mark the non-phosphorylated and phosphorylated proteins, respectively. B, HeLa cells were treated with Taxol as indicated and cell lysates were further treated with (+) or without (−) λ-phosphatase (ppase). Total cell lysates were probed with anti-Ajuba antibody. C, HeLa cells were treated with Taxol together with or without various kinase inhibitors as indicated. VX680 (2 μm), MK5108 (10 μm), ZM447439 (1 μm), RO3306 (5 μm), Roscovitine (30 μm), Purvalanol A (10 μm), and BI2536 (100 nm) were used. Inhibitors were added (with MG132 to prevent cyclin B from degradation and cells from exiting from mitosis) 1–2 h before harvesting the cells. Total cell lysates were subjected to Western blotting with the indicated antibodies.
Identification of the Corresponding Kinase for Ajuba Phosphorylation
Next, we used various kinase inhibitors to identify the candidate kinase for Ajuba phosphorylation. In contrast to the findings in a previous study (27), our data demonstrated that inhibition of Aurora-A (with MK5108) or Aurora-A, -B, and -C (with VX680) kinases only mildly reduced Ajuba phosphorylation (Fig. 1C). Addition of BI2536 (an inhibitor for mitotic kinase Plk1) had no effect on the Ajuba mobility shift/phosphorylation (Fig. 1C). Interestingly, treatments with RO3306 (CDK1 inhibitor), Roscovitine (inhibits CDK1/2/5), or Purvalanol A (CDK1/2/5 inhibitor) almost completely reverted the mobility shift/phosphorylation (Fig. 1C, lanes 7–9). CDK1 is a well known mitotic kinase. These data suggest that CDK1 is likely the corresponding kinase for Ajuba phosphorylation induced by Taxol or Nocodazole treatments. Inhibition of MEK-ERK kinases (with U0126), p38 (with SB203580), GSK-3β (with SB216763), mechanistic target of rapamycin (with rapamycin), PI-3K (with LY294002), or Akt (MK2206) failed to alter the phosphorylation of Ajuba during G2/M arrest (data not shown).
CDK1 Phosphorylates Ajuba in Vitro
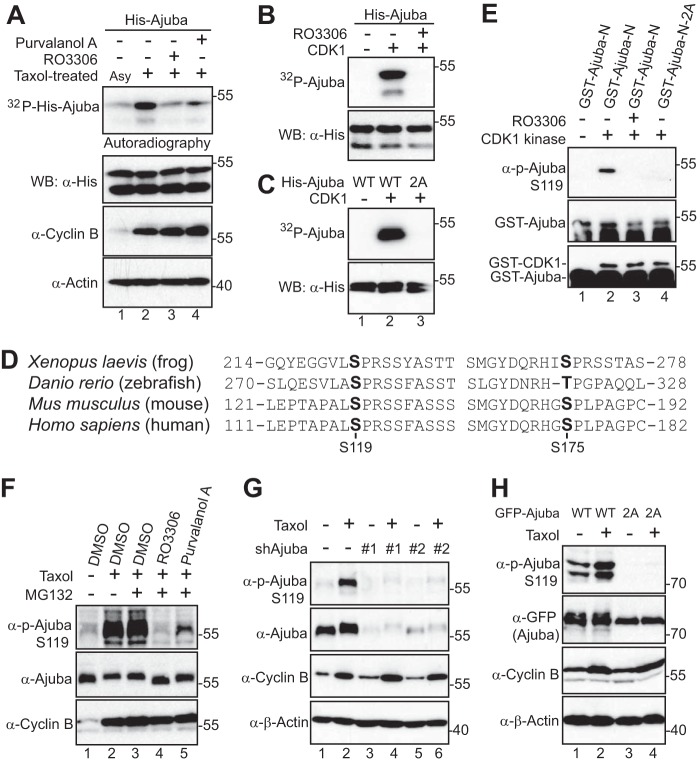
To determine whether CDK1 kinase can directly phosphorylate Ajuba, we performed in vitro kinase assays with His-tagged Ajuba proteins as substrates. Fig. 2A shows that Taxol-treated mitotic lysates robustly phosphorylated Ajuba and that addition of RO3306 or Purvalanol A greatly reduced phosphorylation of His-Ajuba (Fig. 2A). As expected, purified CDK1-cyclin B kinase complex phosphorylated His-Ajuba proteins in vitro (Fig. 2B). These results indicate that CDK1 directly phosphorylates Ajuba in vitro.
FIGURE 2.
Ajuba is phosphorylated by CDK1 in vitro and in cells. A, in vitro kinase assays using HeLa cell lysates to phosphorylate recombinant His-Ajuba. Asy, asynchronized; Tax, Taxol-treated. Total cell lysates were probed with cyclin B and β-actin antibodies. RO3306 (5 μm) or Purvalanol A (10 μm) was used to inhibit CDK1 kinase activity. B, in vitro kinase assays with purified CDK1/cyclin B complex. RO3306 (5 μm) was used to inhibit CDK1 kinase activity. C, in vitro kinase assays with purified CDK1/cyclin B complex. 2A, S119A/S175A. D, conservation of the mitotic phosphorylation sites of Ajuba. E, in vitro kinase assays were done as in B except anti-phospho-Ajuba Ser119 antibodies were used. F, HeLa cells were treated with Taxol together with or without various kinase inhibitors as indicated. Inhibitors were added (with MG132 to prevent cyclin B from degradation and cells from exiting from mitosis) 1 h before harvesting the cells. Total cell lysates were subjected to Western blotting with the indicated antibodies. G, RKO colon cancer cells expressing Tet-control shRNA or Tet-shRNA Ajuba (#1 and #2) in the presence of doxycycline (1 μg/ml for 2 days) were treated with (+) or without (−) Taxol and total cell lysates were subjected to Western blotting with the indicated antibodies. H, HEK293T cells were transfected with GFP-Ajuba or GFP-Ajuba mutants. At 32 h post-transfection, the cells were treated with Taxol for 16 h. The immunoprecipitates (with GFP antibodies) were probed with anti-phospho-Ajuba and subsequent anti-GFP antibodies. Total cell lysates before immunoprecipitation were also included (cyclin B and β-actin).
CDK1-Cyclin B Complex Phosphorylates Ajuba at Ser119 in Vitro and in Cells
CDK1 phosphorylates substrates at a minimal proline-directed consensus sequence (33). Ajuba contains a total of 6 (S/T)P motifs (Thr30, Ser119, Ser137, Ser175, Ser196, and Ser237). Interestingly, two of them (Ser119 and Ser175) were identified as mitotic phosphorylation sites by previous phospho-proteomic studies (34) and mutating these two sites to alanine abolished the 32P incorporation in His-Ajuba, suggesting that Ser119 and Ser175 are the main CDK1 sites in Ajuba in vitro (Fig. 2C). Ser119 and Ser175 are highly conserved in vertebrates (Fig. 2D). Therefore, these two sites were chosen for further study.
We have generated phospho-specific antibodies against Ser119 and Ser175. In vitro kinase assays demonstrated that CDK1 readily phosphorylates Ajuba at Ser119 (Fig. 2E). Very weak signal was detected when the phospho-Ser175 antibody was used under these conditions (data not shown). Addition of RO3306 or mutating Ser119 to alanine abolished the phosphorylation (Fig. 2E). These data suggest that CDK1 phosphorylates Ajuba at Ser119 in vitro. Taxol treatment significantly increased the phosphorylation of Ser119 on endogenous Ajuba (Fig. 2F). Using inhibitors for CDK1 kinase, we demonstrated that phosphorylation of Ajuba Ser119 is CDK1 kinase dependent (Fig. 2F). The signal of Ajuba Ser119 during Taxol treatment was significantly reduced in Ajuba knockdown cells, confirming the specificity of the phospho-Ser119 antibody (Fig. 2G). Taxol treatment also increased the phosphorylation of Ser119 on transfected Ajuba, and the signal was abolished by mutating Ser119 to alanine (Fig. 2H), suggesting that these antibodies specifically recognize phosphorylated Ajuba. Taken together, these observations indicate that Ajuba is phosphorylated at Ser119 by CDK1 in cells during antimitotic drug-induced G2/M arrest.
CDK1/Cyclin B Mediates Ajuba Phosphorylation at Ser119 and Ser175 in Cells
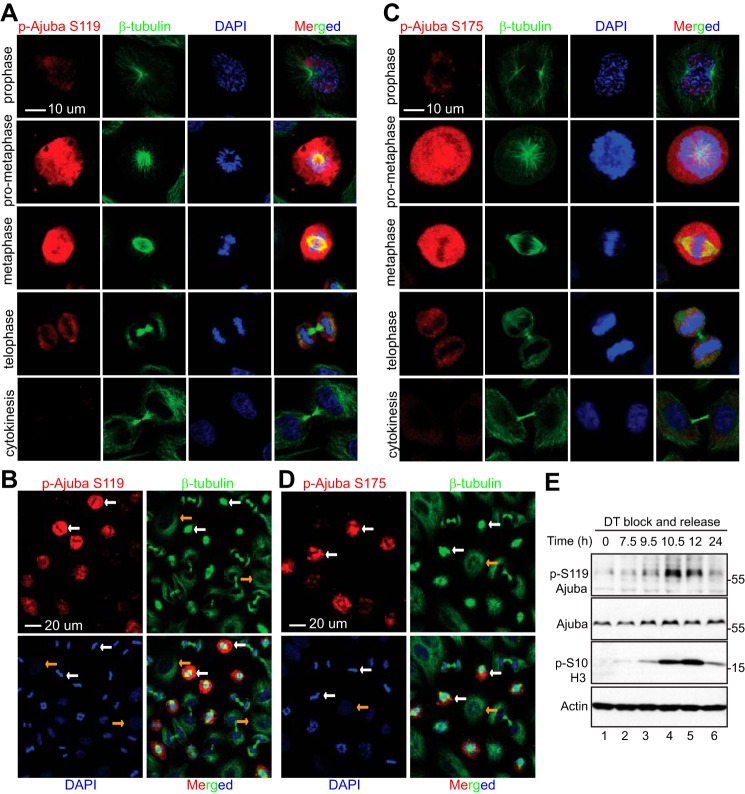
We next performed immunofluoresence microscopy with these phospho-specific antibodies. Both antibodies against Ser119 and Ser175 detected strong signals in Nocodazole-arrested prometaphase cells (Fig. 3, A–C, white arrows). The signal was very low or not detectable in interphase cells (Fig. 3, A–C, yellow arrows). The specificity of the antibodies was further confirmed by peptide blocking assays. Phosphopeptide, but not control non-phosphopeptide, incubation completely blocked the signal, suggesting that these antibodies specifically recognize Ajuba only when it is phosphorylated (Fig. 3, A and B). Again, addition of RO3306 or Purvalanol A largely diminished the signals detected by p-Ajuba Ser119 and Ser175 antibodies in Nocodazole-treated prometaphase cells, further indicating that the phosphorylation is CDK1 dependent (Fig. 3, A and B, low panels).
FIGURE 3.
CDK1 mediates the phosphorylation of Ajuba at Ser119 and Ser175 during G2/M phase arrest. A, HeLa cells were treated with Nocodazole for 8 h and then fixed. Before the cells were stained with phospho-specific antibody against Ser119 of Ajuba, the cells were preincubated with PBS (no peptide control), or non-phosphorylated (control) peptide, or the phosphorylated peptide used for immunizing rabbits. CDK1 inhibitors RO3306 (5 μm) or Purvalanol A (10 μm) together with MG132 (25 μm) were added 2 h before the cells were fixed (bottom two rows). B, experiments were done similarly as in A with phospho-specific antibody against Ser175 of Ajuba. C, HeLa cells were treated and stained with phospho-specific antibodies as in A and B. A ×63 oil objective lens was used to view fewer cells in a field. P-H3 S10 was used as a mitotic marker. White and yellow arrows mark some of the prometaphase cells and the interphase cells, respectively.
Ajuba Phosphorylation Occurs during Normal Mitosis
To determine whether phosphorylation of Ajuba occurs during normal mitosis, we collected samples from a double thymidine block and release (35) and performed immunofluoresence staining on cells in different cell-cycle phases. Consistent with Fig. 3, a very weak signal was detected in interphase or cytokinesis cells (Fig. 4, A and B). The p-Ajuba Ser119 signal was increased in prophase and the strongest signals were detected in prometaphase/metaphase cells. The signals were then again weakened during telophase (Fig. 4, A and B). Similar staining patterns were observed with p-Ajuba Ser175 antibody (Fig. 4, C and D). After being released from the double thymidine block, cells enter into mitosis at 10–12 h revealed by increased phospho-H3 Ser10 and the p-Ajuba Ser119 signal was also increased in these cells (Fig. 4E). These results indicate that Ajuba phosphorylation occurs dynamically during normal mitosis.
FIGURE 4.
Ajuba is phosphorylated at Ser119 and Ser175 during unperturbed mitosis. A and B, HeLa cells were synchronized by a double thymidine (DT) block and release method. Cells were stained with antibodies against p-Ajuba Ser119 or β-tubulin, or with DAPI. A ×40 objective lens was used to view various phases of the cells in a field (B). C and D, experiments were done similarly as in A and B with p-Ajuba Ser175 antibodies. White and yellow arrows (in panels B and D) mark the metaphase and interphase cells, respectively. E, HeLa cells were synchronized by a double thymidine block and release method. Total cell lysates were harvested at the indicated time points and subjected to Western blotting analysis.
Mitotic Phosphorylation of Ajuba Impacts Cell Cycle Regulators Without Affecting YAP Activity
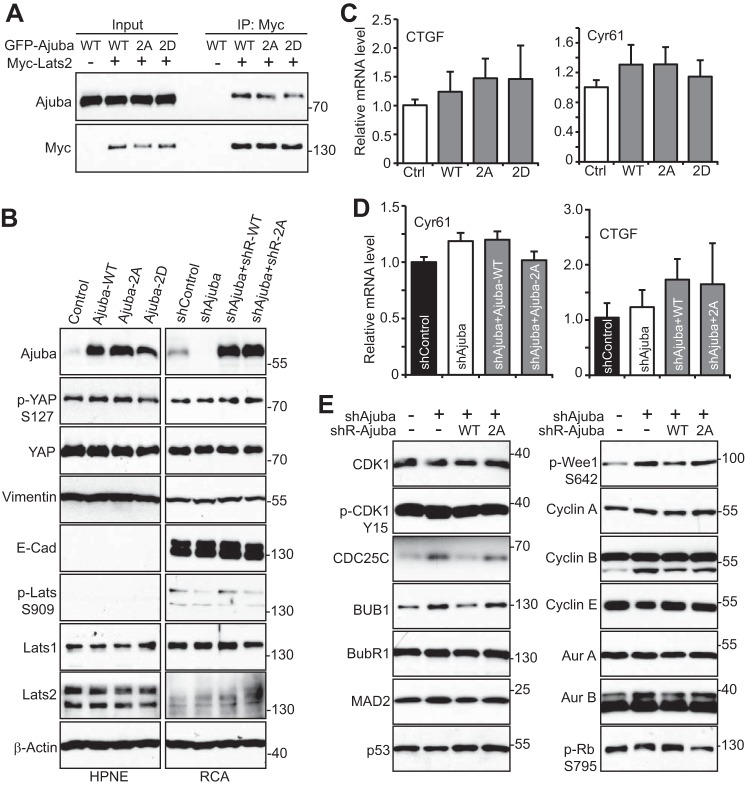
Ajuba was shown to affect the Hippo-YAP signaling activity through interacting with Lats1/2 kinase (17–19). We confirmed that the association between Ajuba and Lats2 was readily detected (Fig. 5A). Non-phosphorylatable (Ajuba-2A, S119A/S175A) or a phosphomimetic (Ajuba-2D, S119D/S175D) mutant has similar binding affinity with Lats2 as wild type Ajuba (Fig. 5A), suggesting that Ajuba phosphorylation does not impact its association with Lats2. YAP Ser127 phosphorylation, Lats activity (revealed by phospho-Ser909), and the levels of YAP and Lats proteins were not significantly altered when Ajuba was overexpressed (in HPNE, immortalized pancreatic epithelial cells) or knocked down (in RCA colon cancer cells) (Fig. 5B). Epithelial-mesenchymal transition is a critical process during development, wound healing, and stem cell behavior, and contributes pathologically to cancer progression and metastasis (36). Several members of the Hippo-YAP signaling regulate epithelial-mesenchymal transition. However, manipulation of Ajuba expression failed to influence the expression of the epithelial-mesenchymal transition markers (Fig. 5B). In line with these observations, the targets expression of YAP was not affected by Ajuba expression in HPNE and RCA cells (Fig. 5, C and D). These results suggest that mitotic phosphorylation of Ajuba does not affect YAP activity and that Ajuba influences Hippo-YAP activity in a context-dependent manner.
FIGURE 5.
Mitotic phosphorylation controls the expression of cell cycle regulators, but does not affect the Hippo-YAP activity. A, HEK293T cells were transfected with various DNA plasmids as indicated. The immunoprecipitates (with Myc antibodies) were probed with anti-Ajuba and subsequent anti-Myc antibodies. Total cell lysates before immunoprecipitation were also included (Input). B, total cell lysates from various HPNE and RCA cell lines as indicated were probed with the indicated antibodies. HPNE cells were stably transduced with vector, Ajuba, Ajuba-2A, or Ajuba-2D. Tet-On-inducible Ajuba-knockdown cell lines expressing shRNA-resistant Ajuba or Ajuba-2A in RCA colon cancer cells were also established (see ”Experimental Procedures“). 2A, S119A/S175A; 2D, S119D/S175D. C and D, quantitative RT-PCR for CTGF and Cyr61 in cell lines established in B. E, total cell lysates were harvested from RCA cell lines established in B and were subjected to Western blotting analysis with various cell cycle regulators.
We further determined whether Ajuba/mitotic phosphorylation affects cell cycle regulators. Interestingly, several such genes' expression (including CDC25C, BUB1 and phosphorylated Wee1) were increased upon Ajuba knockdown in RCA cells (Fig. 5E). Moreover, re-expression of wild type Ajuba, but not the Ajuba-2A mutant, rescued the phenotype (Fig. 5E). These observations suggest that Ajuba and its phosphorylation may have a role in cell cycle progression through regulating the expression of cell cycle regulators.
Mitotic Phosphorylation of Ajuba Is Required for Cell Proliferation and Anchorage-independent Growth
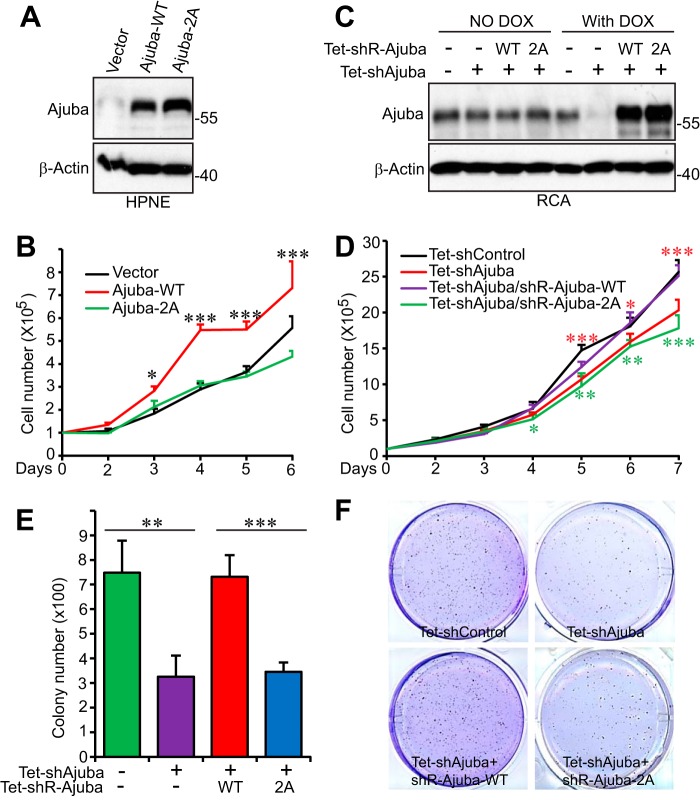
Next we asked what is the biological significance of mitotic phosphorylation of Ajuba. To address this question, we first established HPNE cell lines stably expressing Ajuba or non-phosphorylatable Ajuba mutant (Ajuba-2A) (Fig. 6A). Interestingly, overexpression of Ajuba significantly increased cell proliferation when compared with control cells. However, cells expressing Ajuba-2A proliferated at a rate similar to that of control cells, suggesting that mitotic phosphorylation of Ajuba promotes cell proliferation (Fig. 6B). Ectopic expression of Ajuba (wild type or 2A or 2D) was not sufficient to stimulate anchorage-independent growth in soft agar in HPNE cells (data not shown).
FIGURE 6.
Mitotic phosphorylation of Ajuba is required for cell proliferation and anchorage-independent growth. A, HPNE cells stably expressing vector, Ajuba, or Ajuba-2A were established, and expression of Ajuba and Ajunba-2A were confirmed by Western blotting. 2A, S119A/S175A. B, cell proliferation assays with transduced HPNE cells established in A. Data were expressed as the mean ± S.D. of three independent experiments. ***, p < 0.001; *, p < 0.05 (Ajuba-WT versus Ajuba-2A) (t test). C, establishment of Tet-On-inducible Ajuba-knockdown cell lines expressing shRNA-resistant Ajuba or Ajuba-2A in RCA colon cancer cells (see ”Experimental Procedures“). Cells were kept on Tet-approved FBS and doxycycline was added (1 μg/ml) to the cells 2 days prior to the experiments. D, cell proliferation assays in RCA cells established in C in the presence of doxycycline (DOX). Data were expressed as the mean ± S.D. of three independent experiments. Red asterisks mark the comparisons between shControl and shAjuba. Green asterisks indicate the comparisons between Tet-Ajuba-WT and Tet-Ajuba-2A. ***, p < 0.001; **, p < 0.01; *, p < 0.05 (t test). E and F, colony assays in soft agar to assess anchorage-independent growth of RCA cells established in C in the presence of doxycycline. Data were expressed as the mean ± S.D. of three repeats (E) and representative images were shown (F). ***, p < 0.001; **, p < 0.01 (t test).
We further determined the impact of mitotic phosphorylation of Ajuba in cancer cells. We established RCA cell lines in which the endogenous Ajuba was replaced with shRNA-resistant Ajuba or Ajuba-2A in a Tet-inducible manner (Fig. 6C). Without doxycycline induction, these cell lines express similar levels of endogenous Ajuba proteins (Fig. 6C, left 4 lanes) and no proliferation or other differences were detected among these cells. Addition of doxycycline to the cell culture medium induced endogenous Ajuba knockdown and expression of shRNA-resistant Ajuba or its non-phosphorylatable mutant (Fig. 6C, right 4 lanes). Consistent with the Ajuba overexpression results in HPNE cells, knockdown of Ajuba in RCA cells decreased proliferation, and importantly, expression of wild type Ajuba, but not the non-phosphorylatable mutant Ajuba-2A, completely rescued the cell proliferation defects (Fig. 6D). Furthermore, Ajuba knockdown also significantly decreased anchorage-independent growth in soft agar, and again, re-expression of Ajuba-2A failed to rescue the defects, whereas wild type Ajuba did (Fig. 6, E and F). These data suggest that mitotic phosphorylation is essential for Ajuba to promote cell proliferation and anchorage-independent growth.
Mitotic Phosphorylation of Ajuba Is Required for Tumorigenesis
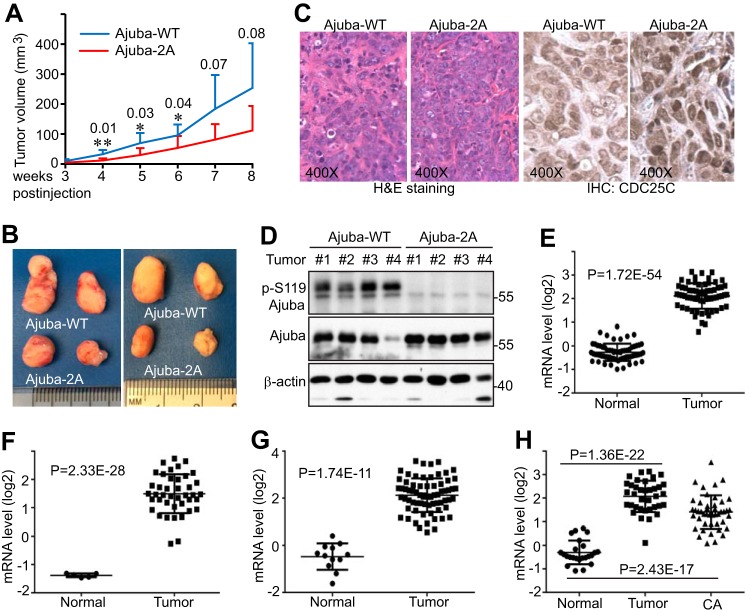
We next evaluated the influence of Ajuba and its mitotic phosphorylation on tumor growth in animals. RCA cells in which the endogenous Ajuba was replaced with shRNA-resistant wild type Ajuba or Ajuba-2A (Fig. 6C) were subcutaneously inoculated into immunodeficient mice. Interestingly, tumors from mice harboring Ajuba-2A-expressing cells tended to be smaller when compared with those from mice injected with Ajuba-expressing cells (Fig. 7, A and B). Histopathological examination revealed no significant differences among these tumors (Fig. 7C). Consistent with Fig. 5E, CDC25C expression was higher in Ajuba-2A-expressing tumors than Ajuba-WT tumors (Fig. 7C). Western blotting analysis confirmed the phosphorylation status of Ser119, and verified that Ajuba (wild type or 2A) expression levels were similar in most of these tumors (Fig. 7D). These results support the hypothesis that mitotic phosphorylation is essential for Ajuba-promoted tumor growth in vivo.
FIGURE 7.
Ajuba phosphorylation is essential for tumorigenesis in mice. A, tumor growth curve. RCA cells expressing Tet-shRNA Ajuba and shRNA-resistant wild type Ajuba or Ajuba-2A were subcutaneously inoculated into athymic nude mice (Ajuba-WT on the left flank and Ajuba-2A on the right flank) and the mice were kept on doxycycline (0.5 mg/ml) in their drinking water throughout the experiment. Two of 10 mice did not form visible tumors (both left and right sides) and were excluded from the analysis. Therefore, the tumor volume at each point was the average of 8 tumors. The p values are also shown. **, p < 0.01; *, p < 0.05 (t test). B, the largest four tumors in each group were excised and photographed at the end point. C, hematoxylin and eosin (H&E) and CDC25C IHC staining in tumors shown in B. D, Western blotting analysis with tumor samples from B. E–H, mRNA levels of Ajuba in normal colon and colon tumors from public data sets. Data were mined from Oncomine.org. The original studies were as follows: Refs. 49 (E), 50 (F), 51 (G), and 52 (H). Tumors, colorectal carcinoma; CA, colorectal adenocarcinoma.
A previous report showed that Ajuba was up-regulated in colon cancer cell lines and tumors (22). We further analyzed the expression of Ajuba in published data and confirmed that the mRNA levels of Ajuba were significantly increased in colon tumors compared with normal colon (Fig. 7, E–H). Together, these observations indicate that Ajuba functions as a tumor-promoting regulator in colon cancer in a mitotic phosphorylation-dependent manner.
Discussion
Ajuba family proteins (Ajuba, LIMD1, and WTIP) play roles in various cellular processes, and one of the most studied areas is the role of Ajuba protein in mitosis. Ajuba is required for mitotic entry in coordination with Aurora-A kinase and is co-localized at centrosomes with Aurora-A, CDK1/cyclin B (27), and Lats2 (28) during G2/mitosis. Interestingly, Ajuba protein became phosphorylated in mitotic cells; however, there are differing reports regarding the kinase that contributes to this mitotic phosphorylation. Hirota et al. (27) showed that Aurora-A directly phosphorylated Ajuba in vitro but did not investigate whether Ajuba phosphorylation is Aurora-A dependent in cells. Another report suggested that Lats2 contributed to Ajuba phosphorylation during mitosis (28). The current study provided evidence that CDK1 is the major kinase responsible for Ajuba phosphorylation and that CDK1 phosphorylates Ajuba in vitro and in cells during mitosis, adding a new layer of regulation for Ajuba during mitosis. Our data do not exclude the possibility that Aurora-A and Lats2 kinases can phosphorylate Ajuba in cells as well. Future studies are needed to further define the mitotic phosphorylation (phosphorylation sites and their biological function) of Ajuba by Aurora-A and/or Lats2. Of note, several large scale proteomic studies have identified Ser119 and Ser175 as mitotic phosphorylation sites and both sites fit the CDK1-phosphorylation consensus sequence (34).
In Drosophila, Djub promotes cell proliferation and inhibits apoptosis by regulating Hippo-Yki activity (17). Consistent with these observations, our data further confirm that Ajuba is a positive regulator for cell proliferation and anchorage-independent growth in pancreatic and colon cancer cells (Fig. 6). Furthermore, Ajuba also promotes migration and invasion in colon cancer cells (22). Interestingly, whereas these studies clearly showed that Ajuba promotes cell proliferation, Ajuba was shown to suppress malignant mesothelioma cell proliferation (23), suggesting a cell type-specific role of Ajuba in cancer cells. In line with a role of Ajuba in cancer, recent large scale genomic studies found that the Ajuba gene is mutated in 7% of esophageal squamous cell carcinomas (24, 25) and Ajuba is overexpressed in colon cancer patients (22). Our current study further demonstrates that mitotic phosphorylation of Ajuba by CDK1 is critical for its biological function, suggesting that there is a link between the role of Ajuba in cancer and its mitotic regulation and that Ajuba may exert its role in cancer through deregulation in mitosis. Together, these studies suggest that Ajuba may play a role in tumorigenesis, although further confirmation will require genetic animal models. Ajuba is not essential for embryo development and Ajuba knock-out mice have no obvious phenotypes (37). These observations suggest that Ajuba may function as a fine regulator in tumorigenesis and needs an additional allele product to promote/inhibit tumor cell growth. However, knock-out of the Ajuba allele has not been combined with any other oncogenes or tumor suppressors including ones in the Hippo-YAP pathway. In addition, since there is functional redundancy and overlapping expression within the Ajuba family proteins (21), clearly defining the biological role of Ajuba in tumorigenesis may be even more challenging.
Mitotic aberrations cause genomic/chromosome instability, which is characteristic of human malignancy (38). Several reports showed that the Hippo pathway plays important roles in maintaining normal mitosis and suggest a mechanism through which the Hippo tumor suppressor pathway exerts its function. For example, loss of core tumor suppressors in the Hippo pathway (including Lats2, Mst1/2, Mob1, and WW45) leads to severe defects in multiple mitotic processes (39–41). Accordingly, we recently reported that overexpression of active YAP (12, 13) or TAZ (15) is sufficient to trigger mitotic defects, including centrosome amplification, spindle disorganization, chromosome misalignment, and subsequent aneuploidy. Interestingly, we also found that several Hippo core members (Lats1, Lats2, and Mst2) (Fig. 1) or their upstream regulator (KIBRA) (30, 31) or downstream effectors (YAP and TAZ) (12, 13, 15) are phosphorylated during mitosis. Importantly, mitotic phosphorylation is critical for their oncogenic or tumor suppressive functions (12, 13, 15). These observations suggest that in addition to their expression levels, the phosphorylation status of these proteins must also be finely controlled, adding another layer of regulation for Hippo-YAP activity during tumorigenesis. Such studies may provide additional insights into the underlying mechanisms of Hippo-YAP signaling in cancer. Thus, we extended our studies to other Hippo regulators and we found that the Ajuba/Zyxin family proteins (Ajuba, LIMD1, and Zyxin) are also phosphorylated during antimitotic drug-induced mitotic arrest (Fig. 1). Zxyin was previously shown to be phosphorylated and played a role in mitosis; however, the phosphorylation sites, corresponding kinase, and their functional significance remain elusive (42). Although these proteins are structurally and functionally related, sites analogous to Ajuba Ser119 and Ser175 do not exist on LIMD1 and Zyxin. Additionally, the role of LIMD1 and its regulation in mitosis also remain to be defined. Addressing these questions will not only help understand the cellular function of these proteins in mitosis, but also provide insights into their biological significance and underlying mechanisms in cancer development.
Experimental Procedures
Cell Culture and Transfection
HEK293T, HEK293GP, and HeLa cell lines were purchased from American Type Culture Collection (ATCC) and cultured as ATCC instructed. HPNE cells were provided by Dr. Michel Ouellette (University of Nebraska Medical Center, who established and deposited this cell line at ATCC) and were cultured as described (43). The cell lines were authenticated at ATCC and were used at low (<25) passages. The colon cancer cell line RCA was a gift from Dr. Michael Brattain (University of Nebraska Medical Center) (44) and was maintained in minimal essential medium supplemented with 10% FBS and antibiotics. Attractene (Qiagen) was used for transient overexpression of proteins in HEK293T and HEK293GP cells following the manufacturer's instructions. Ectopic expression of Ajuba or its mutants in HPNE cells was achieved by a retrovirus-mediated approach. Retrovirus packaging, infection, and subsequent selection were done as we have described previously (45). Nocodazole (100 ng/ml for 16 h) and Taxol (100 nm for 16 h) (Selleck Chemicals) were used to arrest cells in G2/M phase unless otherwise indicated. VX680 (Aurora-A, -B, and -C inhibitor), ZM447439 (Aurora-B, -C inhibitor), BI2536 (Plk1 inhibitor), Purvalanol A (CDK1/2/5 inhibitor), SB216763 (GSK-3β inhibitor), Rapamycin (mechanistic target of rapamycin inhibitor), and MK2206 (Akt inhibitor) were also from Selleck Chemicals. RO3306 (CDK1 inhibitor) and Roscovitine (CDK1/2/5 inhibitor) were from ENZO Life Sciences. MK5108 (Aurora-A inhibitor) was from Merck. Kinase inhibitors for MEK-ERK (with U0126), p38 (with SB203580), and PI-3K (with LY294002) were from LC Laboratory. All other chemicals were either from Sigma or ThermoFisher.
Expression Constructs
The human Ajuba cDNA clone (ID HSCD00323154) was obtained from Harvard Medical School. To make the retroviral or GFP-tagged Ajuba expression constructs, the above full-length cDNA was cloned into the MaRXTMIV (45) or pEGFP-C1 vector (Clontech), respectively. HA-FRMD6 (HA-EX) was made by cloning FRMD6 cDNA (46) into the pcDNA3.1-HA vector (45). Myc-Lats2 has been described (45). Point mutations were generated by the QuikChange Site-directed PCR mutagenesis kit (Stratagene) and verified by sequencing.
Tet-On-inducible Expression System
Tet-On-inducible shRNA vectors against Ajuba were purchased from GE Healthcare/Dharmacon (V3THS-343741). To make the shRNA-resistant (shR) Ajuba cDNA, the target sequence (5′-ACCGACTACCACAAAAATT-3′) was changed into 5′-ACgGAtTAtCAtAAAAATT-3′ by PCR mutagenesis. The mutated Ajuba cDNA was then cloned into the Tet-All vector (47) to generate a Tet-On-inducible shR-Ajuba construct. Ajuba down-regulation in RCA cells was achieved by lentivirus-mediated Ajuba shRNA expression in a doxycycline-dependent manner. Lentivirus generation and infection were performed as described with slight modifications (48). The transduced cells were selected with puromycin (1 μg/ml) to establish pooled cell lines. The cell line in which the lack of Ajuba expression was confirmed (Tet-inducible knockdown) was then used for transduction/infection with viruses expressing Tet-All-shR Ajuba or mutant constructs. Cells were maintained in medium containing Tet system-approved fetal bovine serum (Clontech Laboratories).
Quantitative Real-time PCR
Total RNA isolation, RNA reverse transcription, and quantitative real time-PCR were done as we have described previously (45).
Recombinant Protein Purification and in Vitro Kinase Assay
The GST-tagged Ajuba (amino acids 2–240, cloned in pGEX-5X-1) proteins were bacterially expressed and purified on GSTrap FF affinity columns (GE Healthcare) following the manufacturer's instructions. To make His-tagged Ajuba (amino acids 2–468), the corresponding Ajuba cDNA was subcloned into the pET-28a vector (Novagen/EMD Chemicals). The proteins were expressed and purified on HisPurTM Cobalt spin columns (Pierce) following the manufacturer's instructions.
His- or GST-Ajuba (0.5–1 μg) was incubated with 5–10 units of recombinant CDK1/cyclin B complex (New England Biolabs) or 50–100 ng of CDK1/cyclin B (SignalChem) or HeLa cell total lysates (treated with DMSO or Taxol) in kinase buffer (New England Biolabs) in the presence of 5 μCi of [γ-32P]ATP (3000 Ci/mmol, PerkinElmer Life Sciences). Phosphorylation (32P incorporation) was visualized by autoradiography followed by Western blotting or detected by phospho-specific antibodies.
Antibodies
The polyclonal Ajuba antibodies (4897) from Cell Signaling Technology were used for Western blotting throughout the study. Rabbit polyclonal phospho-specific antibodies against human Ajuba Ser119 and Ser175 were generated and purified by AbMart. The peptides used for immunizing rabbits were TAPAL-pS-PRSSF (Ser119) and DQRHG-pS-PLPAG (Ser175). The corresponding non-phosphorylated peptides were also synthesized and used for antibody purification and blocking assays. HA antibodies were from Sigma (H9658). Anti-β-actin (SC-47778), anti-GFP (SC-9996), and anti-cyclin B (SC-752) antibodies were from Santa Cruz Biotechnology. Aurora-A (A300–070A), glutathione S-transferase (GST) (A190–122A), His (A190–114A), Mst1 (A300–465A), Mst2 (A300–467A), Lats1 (A300–478A), Aurora B (A300–431A), BUB1 (A300–373A), and BubR1 (A300–386A) antibodies were from Bethyl Laboratories. Phospho-Thr288/Thr232/Thr198 Aurora-A/B/C (2914), phospho-Ser10 H3 (3377), phospho-Ser127 YAP (4911), phospho-Ser909 Lats1 (9157), Lats2 (5888), WW45 (3507), TAZ (4883), TEAD1 (12292), NF2 (6995), Vimentin (5741), E-cadherin (3195), PTPN14 (13808), LIMD1 (13245), Zyxin (3553), CDC25C (4688), CDK1 (9116), phospho-Tyr15 CDK1 (9111), cyclin A (4656), cyclin E (4132), p53 (2527), MAD2 (4636), phospho-Ser795 Rb (9301), and phospho-Ser642 Wee1 (4910) antibodies were also from Cell Signaling Technology. The monoclonal antibody against KIBRA has been described (45). Rabbit anti-α-tubulin (Abcam, 15246) and mouse anti-β-tubulin (Sigma, T5293) antibodies were used for immunofluorescence staining.
Phos-tag and Western Blot Analysis
Phos-tagTM was obtained from Wako Pure Chemical Industries, Ltd. (304-93521) and used at 20 μm (with 100 μm MnCl2) in 6 or 8% SDS-acrylamide gels. Prior to transferring, the gels were equilibrated in transfer buffer containing 10 mm EDTA, two times, each for 10 min. The gels were then soaked in transfer buffer (without EDTA) for another 10 min. Western blotting, immunoprecipitation, and λ-phosphatase treatment assays were done as previously described (31).
Immunofluorescence Staining and Confocal Microscopy
Cell fixation, permeabilization, fluorescence staining, and microscopy were done as previously described (35). For peptide blocking, a protocol from the Abcam website was used, as we previously described (12).
Colony Formation and Cell Proliferation Assays
Colony formation assays in soft agar were performed as described (43). Cells (10,000/well) were seeded in a 6-well plate and colonies were counted by ImageJ online. For cell proliferation assays, cells (100,000/well) were seeded in a 6-well plate in triplicate. Cells were counted by a hemacytometer and proliferation curves were made based on the cell number in each well from three independent experiments.
Animal Studies
For in vivo xenograft studies, RCA cells (with Tet-shRNA-Ajuba) expressing Tet-All-shR-Ajuba or Tet-All-shR-Ajuba-2A (non-phosphorylatable mutant) (2.0 × 106 cells each line) were subcutaneously injected into the left or right flank of 6-week-old male athymic nude mice (Ncr-nu/nu, Harlan). Ten animals were used per group. Tumor sizes were measured once a week using an electronic caliper starting at 3 weeks after injection (when tumors in the Ajuba-2A group are palpable). Tumor volume (V) was calculated by the formula: V = 0.5 × length × width2 (43). Mice were euthanized at 6 weeks post-injection and the tumors were excised for subsequent analysis. The animals were housed in pathogen-free facilities. All animal experiments were approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee.
Statistical Analysis
Statistical significance was analyzed using a two-tailed, unpaired Student's t test.
Author Contributions
J. D. and X. C. designed and wrote the paper. X. C., S. S., and Y. C. performed the experiments, analyzed the data, and interpreted the results. Y. C. also provided technical support. All authors reviewed and approved the manuscript prior to submission.
Acknowledgments
All fluorescence images were acquired by Zeiss LSM 710 confocal microscope at the Advanced Microscopy Core at the University of Nebraska Medical Center. The core is supported in part by Grant P30 GM106397 from the National Institutes of Health. We also thank Dr. Joyce Solheim for critical reading and comments on the manuscript.
Footnotes
This work was supported by Grants P30 GM106397 and R01 GM109066 from the National Institutes of Health, and Grant W81XWH-14-1-0150 from the Department of Defense Health Program (to J. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Pan D. (2007) Hippo signaling in organ size control. Genes Dev. 21, 886–897 [DOI] [PubMed] [Google Scholar]
- 2.Pan D. (2010) The Hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey K. F., Zhang X., and Thomas D. M. (2013) The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 [DOI] [PubMed] [Google Scholar]
- 4.Zhao B., Tumaneng K., and Guan K. L. (2011) The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 13, 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu F. X., Zhao B., and Guan K. L. (2015) Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu F. X., and Guan K. L. (2013) The Hippo pathway: regulators and regulations. Genes Dev. 27, 355–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moroishi T., Hansen C. G., and Guan K. L. (2015) The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piccolo S., Dupont S., and Cordenonsi M. (2014) The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 94, 1287–1312 [DOI] [PubMed] [Google Scholar]
- 9.Ma B., Chen Y., Chen L., Cheng H., Mu C., Li J., Gao R., Zhou C., Cao L., Liu J., Zhu Y., Chen Q., and Wu S. (2015) Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat. Cell Biol. 17, 95–103 [DOI] [PubMed] [Google Scholar]
- 10.Wang W., Xiao Z. D., Li X., Aziz K. E., Gan B., Johnson R. L., and Chen J. (2015) AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat. Cell Biol. 17, 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mo J. S., Meng Z., Kim Y. C., Park H. W., Hansen C. G., Kim S., Lim D. S., and Guan K. L. (2015) Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat. Cell Biol. 17, 500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S., Zhang L., Liu M., Chong R., Ding S. J., Chen Y., and Dong J. (2013) CDK1 phosphorylation of YAP promotes mitotic defects and cell motility and is essential for neoplastic transformation. Cancer Res. 73, 6722–6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S., Zhang L., Chen X., Chen Y., and Dong J. (2015) Oncoprotein YAP regulates the spindle checkpoint activation in a mitotic phosphorylation-dependent manner through up-regulation of BubR1. J. Biol. Chem. 290, 6191–6202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganem N. J., Cornils H., Chiu S. Y., O'Rourke K. P., Arnaud J., Yimlamai D., Théry M., Camargo F. D., and Pellman D. (2014) Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell 158, 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Chen X., Stauffer S., Yang S., Chen Y., and Dong J. (2015) CDK1 phosphorylation of TAZ in mitosis inhibits its oncogenic activity. Oncotarget 6, 31399–31412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen C. G., Moroishi T., and Guan K. L. (2015) YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 25, 499–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das Thakur M., Feng Y., Jagannathan R., Seppa M. J., Skeath J. B., and Longmore G. D. (2010) Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 20, 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy B. V., and Irvine K. D. (2013) Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev. Cell 24, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun G., and Irvine K. D. (2013) Ajuba family proteins link JNK to Hippo signaling. Sci. Signal. 6, ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rauskolb C., Sun S., Sun G., Pan Y., and Irvine K. D. (2014) Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell 158, 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schimizzi G. V., and Longmore G. D. (2015) Ajuba proteins. Curr. Biol. 25, R445–446 [DOI] [PubMed] [Google Scholar]
- 22.Liang X. H., Zhang G. X., Zeng Y. B., Yang H. F., Li W. H., Liu Q. L., Tang Y. L., He W. G., Huang Y. N., Zhang L., Yu L. N., and Zeng X. C. (2014) LIM protein JUB promotes epithelial-mesenchymal transition in colorectal cancer. Cancer Sci. 105, 660–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka I., Osada H., Fujii M., Fukatsu A., Hida T., Horio Y., Kondo Y., Sato A., Hasegawa Y., Tsujimura T., and Sekido Y. (2015) LIM-domain protein AJUBA suppresses malignant mesothelioma cell proliferation via Hippo signaling cascade. Oncogene 34, 73–83 [DOI] [PubMed] [Google Scholar]
- 24.Gao Y. B., Chen Z. L., Li J. G., Hu X. D., Shi X. J., Sun Z. M., Zhang F., Zhao Z. R., Li Z. T., Liu Z. Y., Zhao Y. D., Sun J., Zhou C. C., Yao R., Wang S. Y., et al. (2014) Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 46, 1097–1102 [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Network (2015) Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharp T. V., Al-Attar A., Foxler D. E., Ding L., de A Vallim T. Q., Zhang Y., Nijmeh H. S., Webb T. M., Nicholson A. G., Zhang Q., Kraja A., Spendlove I., Osborne J., Mardis E., and Longmore G. D. (2008) The chromosome 3p21.3-encoded gene, LIMD1, is a critical tumor suppressor involved in human lung cancer development. Proc. Natl. Acad. Sci. U.S.A. 105, 19932–19937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., and Saya H. (2003) Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114, 585–598 [DOI] [PubMed] [Google Scholar]
- 28.Abe Y., Ohsugi M., Haraguchi K., Fujimoto J., and Yamamoto T. (2006) LATS2-Ajuba complex regulates γ-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 580, 782–788 [DOI] [PubMed] [Google Scholar]
- 29.Ferrand A., Chevrier V., Chauvin J. P., and Birnbaum D. (2009) Ajuba: a new microtubule-associated protein that interacts with BUBR1 and Aurora B at kinetochores in metaphase. Biol. Cell 101, 221–235 [DOI] [PubMed] [Google Scholar]
- 30.Ji M., Yang S., Chen Y., Xiao L., Zhang L., and Dong J. (2012) Phospho-regulation of KIBRA by CDK1 and CDC14 phosphatase controls cell-cycle progression. Biochem. J. 447, 93–102 [DOI] [PubMed] [Google Scholar]
- 31.Xiao L., Chen Y., Ji M., Volle D. J., Lewis R. E., Tsai M. Y., and Dong J. (2011) KIBRA protein phosphorylation is regulated by mitotic kinase aurora and protein phosphatase 1. J. Biol. Chem. 286, 36304–36315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aylon Y., Michael D., Shmueli A., Yabuta N., Nojima H., and Oren M. (2006) A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 20, 2687–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nigg E. A. (1993) Cellular substrates of p34(cdc2) and its companion cyclin-dependent kinases. Trends Cell Biol. 3, 296–301 [DOI] [PubMed] [Google Scholar]
- 34.Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Iyer J., Chowdhury A., Ji M., Xiao L., Yang S., Chen Y., Tsai M. Y., and Dong J. (2012) KIBRA regulates aurora kinase activity and is required for precise chromosome alignment during mitosis. J. Biol. Chem. 287, 34069–34077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamouille S., Xu J., and Derynck R. (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kisseleva M., Feng Y., Ward M., Song C., Anderson R. A., and Longmore G. D. (2005) The LIM protein Ajuba regulates phosphatidylinositol 4,5-bisphosphate levels in migrating cells through an interaction with and activation of PIPKIα. Mol. Cell. Biol. 25, 3956–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holland A. J., and Cleveland D. W. (2009) Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mardin B. R., Lange C., Baxter J. E., Hardy T., Scholz S. R., Fry A. M., and Schiebel E. (2010) Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 12, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishio M., Hamada K., Kawahara K., Sasaki M., Noguchi F., Chiba S., Mizuno K., Suzuki S. O., Dong Y., Tokuda M., Morikawa T., Hikasa H., Eggenschwiler J., Yabuta N., Nojima H., et al. (2012) Cancer susceptibility and embryonic lethality in Mob1a/1b double-mutant mice. J. Clin. Invest. 122, 4505–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yabuta N., Okada N., Ito A., Hosomi T., Nishihara S., Sasayama Y., Fujimori A., Okuzaki D., Zhao H., Ikawa M., Okabe M., and Nojima H. (2007) Lats2 is an essential mitotic regulator required for the coordination of cell division. J. Biol. Chem. 282, 19259–19271 [DOI] [PubMed] [Google Scholar]
- 42.Hirota T., Morisaki T., Nishiyama Y., Marumoto T., Tada K., Hara T., Masuko N., Inagaki M., Hatakeyama K., and Saya H. (2000) Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J. Cell Biol. 149, 1073–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., and Pan D. (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 130, 1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brattain M. G., Levine A. E., Chakrabarty S., Yeoman L. C., Willson J. K., and Long B. (1984) Heterogeneity of human colon carcinoma. Cancer Metastasis Rev. 3, 177–191 [DOI] [PubMed] [Google Scholar]
- 45.Xiao L., Chen Y., Ji M., and Dong J. (2011) KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J. Biol. Chem. 286, 7788–7796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunn-Moore F. J., Welsh G. I., Herron L. R., Brannigan F., Venkateswarlu K., Gillespie S., Brandwein-Gensler M., Madan R., Tavaré J. M., Brophy P. J., Prystowsky M. B., and Guild S. (2005) A novel 4.1 ezrin radixin moesin (FERM)-containing protein, “Willin.” FEBS Lett. 579, 5089–5094 [DOI] [PubMed] [Google Scholar]
- 47.Yang S., Ji M., Zhang L., Chen Y., Wennmann D. O., Kremerskothen J., and Dong J. (2014) Phosphorylation of KIBRA by the extracellular signal-regulated kinase (ERK)-ribosomal S6 kinase (RSK) cascade modulates cell proliferation and migration. Cell. Signal. 26, 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., and Guan K. L. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaedcke J., Grade M., Jung K., Camps J., Jo P., Emons G., Gehoff A., Sax U., Schirmer M., Becker H., Beissbarth T., Ried T., and Ghadimi B. M. (2010) Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer 49, 1024–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiser S., Park Y. K., Franklin J. L., Halberg R. B., Yu M., Jessen W. J., Freudenberg J., Chen X., Haigis K., Jegga A. G., Kong S., Sakthivel B., Xu H., Reichling T., Azhar M., et al. (2007) Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 8, R131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong Y., Downey T., Eu K. W., Koh P. K., and Cheah P. Y. (2010) A “metastasis-prone” signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin. Exp. Metastasis 27, 83–90 [DOI] [PubMed] [Google Scholar]
- 52.Skrzypczak M., Goryca K., Rubel T., Paziewska A., Mikula M., Jarosz D., Pachlewski J., Oledzki J., and Ostrowski J. (2010) Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS ONE 5, e13091. [DOI] [PMC free article] [PubMed] [Google Scholar]