Abstract
The plant homeodomain (PHD) finger is found in many chromatin-associated proteins and functions to recruit effector proteins to chromatin through its ability to bind both methylated and unmethylated histone residues. Here, we show that the dual PHD fingers of Rco1, a member of the Rpd3S histone deacetylase complex recruited to transcribing genes, operate in a combinatorial manner in targeting the Rpd3S complex to histone H3 in chromatin. Although mutations in either the first or second PHD finger allow for Rpd3S complex formation, the assembled complexes from these mutants cannot recognize nucleosomes or function to maintain chromatin structure and prevent cryptic transcriptional initiation from within transcribed regions. Taken together, our findings establish a critical role of combinatorial readout in maintaining chromatin organization and in enforcing the transcriptional fidelity of genes.
Keywords: chromatin, gene transcription, histone acetylation, histone deacetylase (HDAC), PHD finger, Rco1, Rpd3S
Introduction
Post-translational modifications on histone proteins play a critical role in many DNA-templated processes, particularly the control of gene transcription. Complexes that modify and remodel chromatin to regulate proper transcription contain proteins with conserved recognition domains that bind either modified or unmodified residues within histone proteins (1–3). Because these post-translational modifications are dynamically regulated and are targeted to specific locations across the open reading frame of genes, effector proteins/complexes that read these post-translational modifications can be recruited in a spatiotemporal manner to control chromatin structure and RNA polymerase II (RNAPII)2 elongation during transcription (4). For example, histones are hyperacetylated in front of elongating RNAPII, allowing for nucleosome disassociation and are hypoacetylated behind RNAPII to maintain chromatin structure and prevent inappropriate cryptic transcription (5). The deacetylation of nucleosomes in transcription is carried out by histone deacetylase complexes, which typically have one or more reader domains that are able to engage chromatin. This multidomain structure allows for recognition of increasingly complex and specific chromatin environments.
Rpd3S, an histone deacetylase that functions in a co-transcriptional manner, has five conserved chromatin-binding domains: a chromodomain in Eaf3, which recognizes Set2-mediated histone H3 lysine 36 methylation (H3K36me) (6–8), and four plant homeodomains (PHDs), two per copy of Rco1, which has recently been shown to form a homodimer in Rpd3S (see Fig. 1A) (9). The chromodomain of Eaf3 and the N-terminal PHD finger of Rco1 (PHD1) have previously been characterized and are necessary for Rpd3S function and nucleosome engagement (6–8, 10). PHD1 is thought to engage H3 on one nucleosome, whereas the chromodomain of Eaf3 recognizes H3K36me on a neighboring nucleosome, allosterically activating the deacetylase activity of Rpd3 (11, 12). This activity is necessary to enforce chromatin integrity and transcriptional fidelity across the transcribed regions of genes, thereby preventing the formation of pervasive cryptic unstable transcripts and stable untranslated transcripts (6, 10, 13, 14). It has been recently shown that the Set2/Rpd3S pathway is particularly important for repressing antisense transcription from divergent promoters (13). Although significant advances have been made in understanding the role of Rpd3S in cells, the precise mechanism by which Rpd3S is targeted to chromatin and mediates its function is still poorly understood.
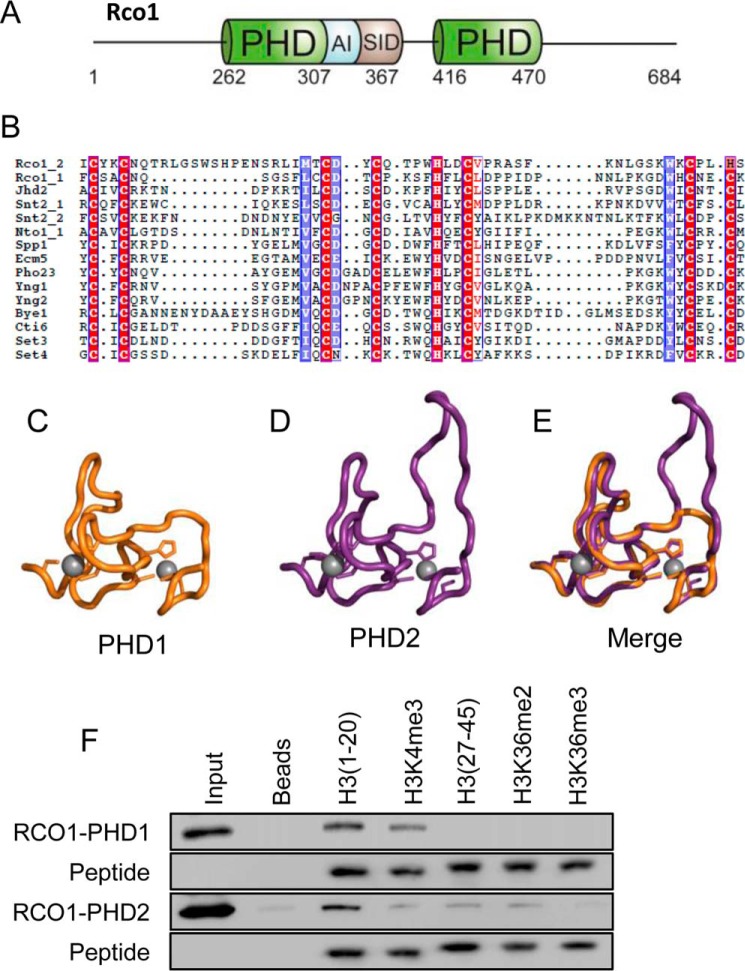
FIGURE 1.
The PHD fingers of Rco1 bind to the extreme N terminus of H3. A, a schematic representation of the Rco1 protein. PHD fingers are highlighted in green with the autoinhibitory domain (AI) in blue and the Sin3 interaction domain (SID) in gray. B, an alignment of yeast PHD fingers, highlighting the conserved cysteine and histidine residues. C, a molecular model of PHD1. Zinc atoms are in gray. D, a molecular model of PHD2. Zinc atoms are in gray. E, a merge of the models of PHD1 and PHD2 show a high degree of similarity. F, in-solution peptide pulldown assays with PHD1 and PHD2 were carried out with the indicated histone H3 peptides. Both domains bind the unmodified H3 N terminus and show sensitivity to H3K4me3.
In this report, we interrogated the role of the C-terminal PHD finger in Rco1 (PHD2), which had not been previously investigated. We show that PHD2 is a functional domain and recognizes the unmodified N terminus of H3, as does PHD1. Further, mutational analysis shows that nucleosome binding in vitro and chromatin association of Rpd3S in vivo depend on the function of both PHD fingers. Consistent with this finding, we demonstrate that mutation of either PHD1 or PHD2 leads to chromatin and transcriptional fidelity defects. Together, our data unveil a critical role for two adjacent PHD fingers in coordinating Rpd3S recruitment and function.
Experimental Procedures
Mutagenesis
Conserved residues were identified and specifically mutated using a site-directed mutagenesis kit (Stratagene) and confirmed via Sanger sequencing.
Immunoblot
A single colony was inoculated overnight to saturation and then diluted to an A600 = 0.2 and grown to mid-log phase. Five optical densities of cells were isolated and lysed via bead beating in SUTEB (1% SDS, 8 m urea, 10 mm Tris, pH 6.8, 10 mm EDTA, 0.01% bromphenol blue) for 3 min. Lysates were boiled for 10 min and then isolated. Cell debris were removed via centrifugation, and the supernatant was isolated. Cleared lysates were loaded onto 8% SDS-PAGE gels and then transferred to PVDF membrane. Membranes were probed overnight (4 °C) with anti-HA (UNC Antibody Core) or anti-G6PDH (Sigma). Immunoblots were visualized using HRP-conjugated secondary antibodies and ECL Prime solution (GE Healthcare).
Alignment and Molecular Modeling
Yeast PHD fingers were isolated from the SMART database and aligned using the Espript 3.0 tool (15). PHD1 and PHD2 sequences were then modeled using the HHpred tool (16) and visualized in PyMOL.
Purification of GST-tagged PHD Fingers
The PHD fingers of Rco1 were purified from SOLUBL21 competent Escherichia coli cells. The cells were grown in the presence of 1 mm zinc. Bacteria pellets were lysed in 50 mm Tris-HCl, pH 7.4, 130 mm NaCl, 1 mm DTT, 1 μm ZnCl2, 1 mm PMSF, universal nuclease for cell lysis (Pierce, 1:20,000), 1 Roche protease inhibitor mixture tablet/50 ml, 1 mg/ml lysozyme (Sigma), 0.1% Triton. Cleared lysate was incubated with Pierce glutathione resin for 2 h at 4 °C. Protein was eluted from the resin in 50 mm Tris-HCl, pH 8.0, 130 mm NaCl, 10 mm glutathione, 1 mm DTT, and 1 μm ZnCl2. Protein was then dialyzed overnight into a storage buffer composed of 50 mm Tris-HCl, pH 7.4, 130 mm NaCl, 1 mm DTT, and 1 μm ZnCl2.
Overnight Peptide Pulldown Method
1 μg of biotinylated histone peptide (supplemental Table S3) was incubated with 1 μg of purified GST fusion protein in 1 ml of peptide binding buffer (50 mm Tris-HCl, pH 7.5, 130 mm NaCl, 0.1% (v/v) Nonidet P-40, 1 mm PMSF, 1 mm DTT, 1 μm ZnCl2, and 1 Roche protease inhibitor mixture tablet/100 ml of peptide binding buffer) overnight at 4 °C. After incubation for 1 h at 4 °C with streptavidin beads (Pierce), the beads were washed three times with 1 ml of peptide binding buffer. Peptide was then eluted from the bead into SDS loading buffer by boiling for 5 min at 98 °C. Samples were then subjected to Western blot analysis, and the membrane was probed with GST antibody (Sigma, 1:4000) for 1 h at room temperature. Peptide loading was assessed by probing the membrane with Streptavidin-HRP (Cell Signaling, 1:5000) for 30 min at room temperature.
Rpd3 Complex Isolation
Recombinant Rpd3S complexes were purified from a Sf21 insect cell-based baculovirus expression system as described previously (9, 11). Briefly, freshly passed Sf21 cells were co-infected with individual virus that encodes each subunit of Rpd3S for 48 h. The cells were collected and lysed in BV lysis buffer (50 mm HEPES, pH 7.9, 300 mm NaCl, 2 mm MgCl2, 0.2% Triton X-100, 10% glycerol, 0.5 mm EDTA, and freshly added protease inhibitors) on ice for 30 min. Cell lysates were clarified by ultracentrifugation and incubated with anti-FLAG M2 resin (Sigma) at 4 °C for 2 h. After extensive washing, each complex was eluted using 500 μg/ml 3×FLAG peptides in BV elution buffer (50 mm HEPES, pH 7.9, 100 mm NaCl, 2 mm MgCl2, 0.02% Nonidet P-40, and 10% glycerol) and concentrated using Amicon concentrators.
EMSA
Mononucleosomes were reconstituted using a 222-bp 601-positioning sequence containing DNA template and purified as described previously (17, 18). EMSA reactions were carried out in a 15-μl system containing 10 mm HEPES, pH 7.8, 50 mm KCl, 4 mm MgCl2, 5 mm DTT, 0.25 mg/ml BSA, 5% glycerol, and 0.1 mm PMSF. The samples were incubated at 30 °C for 45 min and run on a 3.5% acrylamide (37.5:1, acrylamide:bis-acrylamide) gel at 4 °C.
Chromatin Association Assay
Fifty optical densities of mid-log phase cells were isolated and fractionated as previously described (19). Fractions were immunoblotted and probed with anti-HA (UNC Antibody Core), anti-G6PDH (Sigma), or anti-H4 (Millipore).
Rco1 Co-immunoprecipitation
All strains were grown overnight in SC−Leu. Cultures were diluted to an A600 of 0.2 and grown to log phase in 100 ml of SC−Leu. The cells were pelleted and washed with 50 ml of distilled H2O. The pellets were resuspended in 500 μl of lysis buffer (20) and split equally into two tubes. Glass beads were added to bring the total volume to 750 μl, and samples were vortexed for 12 min and rested for 10 min on ice for a total of two times at 4 °C. Lysates were collected into fresh tubes via centrifugation, and the lysates were cleared at maximum speed for 15 min at 4 °C. Protein concentration was quantified via Bradford assay. An aliquot was taken for input, and 1.5 mg/ml of protein was incubated overnight in 1 ml of lysis buffer at 4 °C with 1:1000 dilution of protein A antibody (Sigma). Antibody was conjugated to IgG Sepharose beads (GE Healthcare) for 2 h at 4 °C before being washed with lysis buffer and protein eluted with 100 μl of 5× SDS buffer. Samples were boiled at 95C for 5 min before loaded onto an 8% SDS-PAGE gel.
Spotting Assays
All spotting assays were performed with 5-fold serial dilutions of saturated overnight cultures of the indicated strains. Growth was assayed after 2–5 days. All yeast strains used in this study are described in supplemental Table S1, and all plasmids are described in supplemental Table S2.
Results
Rco1 Contains Two PHD Fingers That Bind to the N Terminus of H3
Rco1 is a unique member of the Rpd3S complex, which is defined by a N-terminal PHD finger followed by an autoinhibitory domain; a Sin3 interaction domain, which associates with the MRG domain of Eaf3 (11); and a second C-terminal PHD finger (Fig. 1A). Although the first PHD finger is required for nucleosome binding and Rpd3S function (10), its second PHD finger (PHD2) remained uncharacterized. To explore whether PHD2 would encode a functional domain, we first performed a sequence alignment of the known PHD fingers from Saccharomyces cerevisiae (Fig. 1B) and created structure prediction models in HHpred of PHD1 and PHD2 using a PHD domain from CHD4 (Protein Data Bank code 1MM2) as a scaffold (Fig. 1, C–E) (21). Our sequence alignments showed that both PHD fingers of Rco1 contain the necessary conserved cysteine and histidine residues needed for the coordination of two zinc ions, a feature that defines functional PHD fingers. In addition, structural modeling predicted PHD2 as being a folded domain with high structural similarities to PHD1. Interestingly, PHD2 contains a small sequence insertion between the first and second grouping of cysteines and has substituted a conserved cysteine residue for an additional histidine at its C terminus, thus suggesting that PHD2 differs slightly from other PHD fingers.
We next interrogated the ability of both PHD1 and PHD2 to directly associate with histones. Each domain was expressed and purified as a GST fusion and assayed in solution peptide pulldowns for their ability to bind differentially modified biotinylated histone peptides from distinct regions of the H3 N terminus. As shown in Fig. 1F, we consistently found that both PHD1 and PHD2 preferentially bound to the N-terminal region (residues 1–20) of the H3 tail. This result is consistent with previous analyses of PHD1 (22, 23). Interestingly, we also found that trimethlyation of K4 (H3K4me3) decreases the ability of both PHD1 and PHD2 to bind N-terminal H31–20 peptides, suggesting that these domains bind to the extreme N terminus of H3 and are affected by N-terminal post-translational modifications. Furthermore, this finding may account, at least in part, for how Rpd3S is restricted from binding to promoter nucleosomes normally marked with H3K4me3.
PHD2 Is Required for Rpd3S Association on Chromatin in Vitro
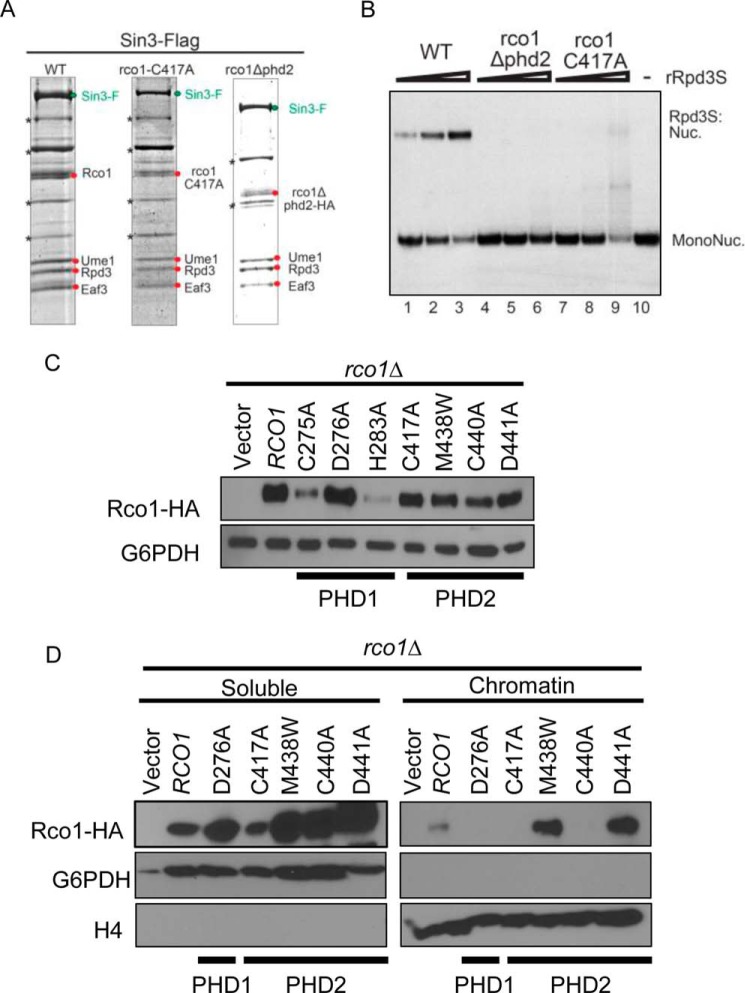
Our previous studies demonstrated that PHD1 was essential in mediating Rpd3S association on nucleosomes (10). Given this, we wondered what the contribution would be, if any, for the second PHD finger of Rco1 in nucleosome binding or the homodimerization of Rco1. To ascertain this, we recombinantly expressed the five Rpd3S members and assembled in vitro complexes competent for nucleosome binding (Fig. 2A). In addition to a complete deletion of PHD2 (rco1Δ phd2), we made a point mutant in PHD2 predicted to disrupt zinc binding and PHD function (rco1-C417A). As shown in Fig. 2A, both the deletion of PHD2 and the C417A point mutation had no effect on Rpd3S complex assembly. Furthermore, mutation of PHD2 showed no defects in Rco1 homodimerization by co-immunoprecipitation analysis (supplemental Fig. S1). Surprisingly, even though the integrity of Rpd3S complexes with PHD2 mutants was fully intact, the ability of these complexes to bind nucleosomes was completely abolished (Fig. 2B), a result that is identical to the loss of PHD1 (10). These results imply that both PHD fingers of Rco1 function in a coordinated fashion to bind nucleosomes.
FIGURE 2.
Both PHD fingers of Rco1 are necessary for association with chromatin. A, Coomassie Blue-stained gels showing that the Rpd3S complex can be purified intact with Rco1 that is WT, C417A, or lacking PHD2. B, an EMSA assay showing that the PHD2 finger of Rco1 is necessary for Rpd3S association with nucleosomes. C, a Western blot of the indicated strains to examine the stability of the mutated RCO1 constructs. All mutations except C275A and H283A were able to sustain near wild-type levels of Rco1 protein. D, Western blot analysis of soluble and chromatin fractions from the indicated strains. Mutation of conserved residues in either PHD finger renders Rco1 unable to associate with chromatin in vivo.
PHD1 and PHD2 Are Required for Chromatin Association in Vivo
To determine the significance of PHD1 and PHD2 in Rpd3S function in cells, we generated a panel of mutations at conserved residues found in both PHD1 and PHD2 that we predicted to be critical for their function (Cys275, Asp276, and His283 in PHD1 and Cys417, Met438, Cys440, and Asp441 in PHD2). As shown in Fig. 2C, nearly wild-type levels of protein were obtained for all of the mutants made in PHD2, but two mutants in PHD1 proved to be unstable (C275A and H283A) and therefore were not used further in our analyses. We next assessed the ability of these mutants to affect the association of Rpd3S on chromatin in vivo using chromatin association assays. Yeast cells expressing wild-type or mutated versions of Rco1 were fractionated into soluble or chromatin-associated fractions (Fig. 2D). As expected, wild-type Rco1 was predominantly found in the chromatin fraction. In stark contrast, however, the D276A PHD1 mutant and C417A and C440A PHD2 mutants were unable to maintain association with chromatin. Hence, PHD1 and PHD2 are both required for chromatin association of Rpd3S in vitro and in vivo.
Loss of PHD1 or PHD2 Function in Rco1 Leads to Chromatin Structure and Transcriptional Fidelity Defects
One of the key functions of Rpd3S is to restore chromatin to a hypoacetylated state after the passage of RNAPII during gene transcription (5). This locally compacts chromatin structure, thereby preventing bidirectional transcription and RNAPII complexes from binding to cryptic promoter elements along the gene and aberrantly initiating transcription. To monitor cryptic transcription, we employed a yeast strain wherein the HIS3 gene is fused to a naturally occurring cryptic promoter in the FLO8 gene (24). Importantly, HIS3 is out of frame with the normal 5′ promoter and only produces a functional transcript if the 3′ cryptic promoter is used (see schematic in Fig. 3A). Under growth conditions using media lacking histidine, cells will not grow if chromatin structure is normal. As expected, strains deleted for RCO1 resulted in a growth phenotype on plates lacking histidine (Fig. 3B), signifying a significant disruption to the chromatin structure at this locus, resulting in cryptic transcription occurring at the internal promoter. This phenotype is rescued by the addition of wild-type RCO1. In complete contrast, however, mutations of PHD1 and PHD2 that disrupt Rpd3S association to chromatin also result in a cryptic transcription phenotype (Fig. 3B). Comparison of another reporter gene, STE11, whose natural cryptic promoter is also fused to the HIS3 gene, showed identical results, thus verifying that the cryptic transcription defect we were observing is not a gene-specific effect (supplemental Fig. S2). Together, these results show that without PHD1 or PHD2 function, Rpd3S is unable to engage chromatin and properly regulate chromatin structure during gene transcription.
FIGURE 3.
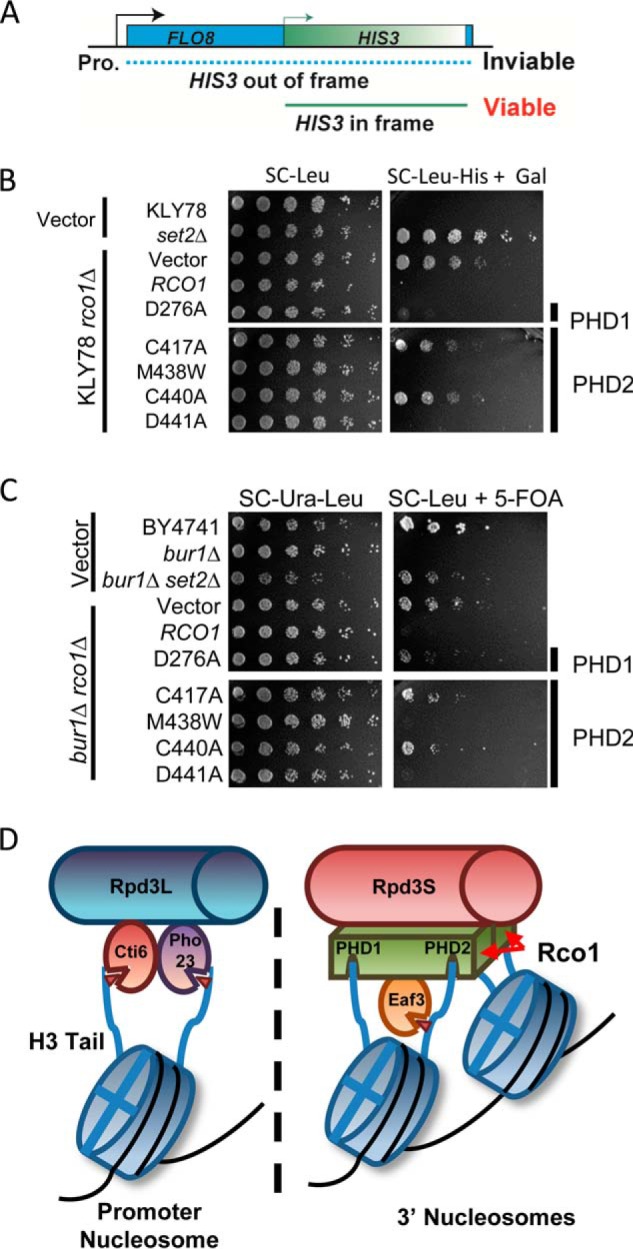
Chromatin structure and transcriptional fidelity requires both PHD fingers in Rco1. A, a schematic of the FLO8-HIS3 fusion gene reporter to detect changes in chromatin structure and cryptic transcription. The cells will only grow in the absence of histidine and if the cryptic promoter in front of HIS3 is utilized. B, cryptic initiation spotting assay using the FLO8-HIS3 fusion gene reporter. The indicated strains are spotted in a 5-fold serial dilution from a starting A600 of 0.5. The plates are imaged after 2–5 days. C, a BUR1 bypass assay reveals transcriptional elongation defects in Rco1 PHD mutations. The indicated strains are spotted in a 5-fold serial dilution from a starting A600 of 2.0. The plates are imaged after 2–3 days. D, a model of how Rpd3S and Rpd3L engage chromatin. Each PHD finger of Rco1 engages the N terminus of H3, whereas the chromodomain of Eaf3 recognizes methylated H3K36, which allosterically activates the histone deacetylase activity of Rpd3. The selectivity of each PHD finger for unmodified H3K4 (H3K4me0) may be an important contributing factor in Rpd3S localization to gene bodies, which are not marked with H3K4me3. In contrast, Rpd3L contains two PHD fingers (one each in Cti6 and Pho23) that do bind to H3K4me3, thus enabling this complex to maintain its localization to promoter regions and not gene bodies. These findings help to further our understanding of combinatorial readout in recruitment of chromatin-associated proteins.
In addition to cryptic transcription, another assay that has been used for the analysis of chromatin and transcription defects is the bur1Δ bypass assay. BUR1 is an essential kinase that acts positively on transcription by phosphorylating several members of the elongating RNAPII including the C-terminal domain of Rpb1 and the C-terminal repeat domain of Spt5 (25–27). However, in the absence of Set2 and other factors in the SET2 genetic pathway (e.g. Rpd3S), cells lacking BUR1 are viable. As expected, loss of RCO1 resulted in a bypass of lethality that was rescued upon restoring wild-type RCO1. Consistent with the role of both PHD1 and PHD2 in Rpd3S function, we observed that mutation of either domain renders cells resistant to the loss of BUR1 (Fig. 3C). Together with the cryptic initiation assay, these data show that combinatorial engagement of histone H3 by the PHD1 and PHD2 is critical for Rpd3S chromatin recruitment and function during gene transcription.
Discussion
Based on the work herein and other recent publications, we propose that Rco1 is a critical scaffolding protein that engages the N termini of histone H3 to stabilize Rpd3S on chromatin, thereby optimally positioning Eaf3 for H3K36me2/me3 binding. With two copies of Rco1 per Rpd3S complex, it is likely that one Rpd3S complex is engaged on two adjacent nucleosomes whereby all four H3 tails are co-occupied to maintain Rpd3S stability on chromatin (see model in Fig. 3D). Future studies will be needed to resolve whether the PHD domains from the same molecule of Rco1 bind both H3 tails in a single nucleosome or whether they bind a separate H3 tail from two neighboring nucleosomes. Regardless, having Rco1 co-occupy two nucleosomes would further highlight the need and role of Isw1b, which utilizes its ATP-dependent chromatin remodeling activities to position adjacent nucleosomes in close proximity for Rpd3S binding (28, 29). Together, these events function to allow Rpd3 to deacetylate histones and maintain chromatin integrity during the transcription process.
Our studies showed that both PHD fingers of Rco1 have a similar preference for binding to the extreme N terminus of H3 and, further, that this binding is highly sensitive to H3K4 trimethylation. This result may help to provide an explanation for how Rpd3L and Rpd3S binding to discrete regions along the gene are controlled. Rpd3L, which localizes to promoters, does not contain Rco1 but rather two other PHD-containing proteins (Cti6 and Pho23) specific for H3K4me3 (22, 30). This would help to maintain Rpd3L in promoter regions where H3K4me3 is restricted. In contrast, Rpd3S, which lacks these other PHD-containing proteins for Rco1, is repelled by H3K4me3, thereby restricting this complex to gene bodies. Consequently, this would localize the Rpd3S complex in regions with high levels of H3K36me, which is then recognized by the chromodomain of Eaf3 (6–8). Thus, the different PHD fingers found in Rpd3L and Rpd3S likely govern their discrete localizations at genes (see model in Fig. 3D).
Finally, we note that although a previous survey of yeast PHD domains was performed using solution peptide pulldowns (22), our studies differ in regards to the ability of PHD1 and PHD2 to bind H3K36me3, a result that is also true for the characterized PHD1 domain of the human Rco1 counterpart, Pf1 (23). We note that PHD1 and PHD2 expression and maintaining their stability in vitro was found to be extremely difficult, and further, that binding and washing conditions greatly impacted weak interactions and nonspecific binding. These challenges with studying Rco1 may explain how different observations were observed.
Our results showed that both PHD fingers in Rco1 are required for chromatin targeting and Rpd3S function. This finding strongly argues that Rpd3S is targeted to chromatin via combinatorial readout of H3. We propose that in isolation, PHD1 or PHD2 is not robust enough to maintain stable Rpd3S association to nucleosomes but rather is reinforced when both domains operate in unison and further when this occurs as a homodimer capable of binding four H3 tails. These four histone binding events likely enforce a strict reading of the chromatin environment that requires the appropriate spacing of the dinucleosome substrate imposed by the Isw1b chromatin remodeling complex.
Finally, we show that loss of H3 binding by either PHD finger of Rco1 results in a disruption of chromatin structure that leads to cryptic transcription and transcriptional defects. Given the significant role these PHD fingers play, it will be interesting to explore whether other chromatin-associated proteins/complexes with multiple PHD fingers behave similarly. In regards to Rpd3L, we predict that the association of this complex to promoters will require each of the two PHD domains found in the complex that show binding to H3K4me3 (speculated in Fig. 3D). Taken together, our results with Rco1 highlight the significance of combinatorial readout in chromatin function and provide further support for the “histone code” hypothesis (31).
Author Contributions
S. L. M., B. L., and B. D. S. designed the research; S. L. M., J. E. F., C. R., H. C., J. B. B., J. V. D., and A. H. G. performed the research; S. L. M., B. L., and B. D. S. analyzed the data; and S. L. M. and B. D. S. wrote the paper.
Supplementary Material
Acknowledgments
We thank M. Keogh for the original RCO1 yeast expression constructs and J. Martinez and V. Miller for technical assistance. We also thank Brenda Temple for the assistance with protein modeling and Krzysztof Krajewski and the University of North Carolina High Throughput Peptide Synthesis and Array (HTPSA) facility for the peptides used in this study.
This work was supported by NIGMS, National Institutes of Health Grant GM110058 (to B. D. S.), National Institutes of Health Grants T32 GM007092 (to S. L. M. and J. V. D.) and GM090077 (to B. L.), and Welch Foundation Grant I-1713 (to B. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1–S3 and Figs. S1 and S2.
- RNAPII
- RNA polymerase II
- PHD
- plant homeodomain.
References
- 1.Lalonde M. E., Cheng X., and Côté J. (2014) Histone target selection within chromatin: an exemplary case of teamwork. Genes Dev. 28, 1029–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musselman C. A., Lalonde M. E., Côté J., and Kutateladze T. G. (2012) Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yun M., Wu J., Workman J. L., and Li B. (2011) Readers of histone modifications. Cell Res. 21, 564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister A. J., and Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner E. J., and Carpenter P. B. (2012) Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., and Workman J. L. (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 [DOI] [PubMed] [Google Scholar]
- 7.Joshi A. A., and Struhl K. (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20, 971–978 [DOI] [PubMed] [Google Scholar]
- 8.Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., Collins S. R., Schuldiner M., Chin K., Punna T., Thompson N. J., Boone C., Emili A., Weissman J. S., Hughes T. R., Strahl B. D., Grunstein M., Greenblatt J. F., Buratowski S., and Krogan N. J. (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123, 593–605 [DOI] [PubMed] [Google Scholar]
- 9.Ruan C., Cui H., Lee C. H., Li S., and Li B. (2016) Homodimeric PHD-domain containing Rco1 constitutes a critical interaction hub within the Rpd3S histone deacetylase complex. J. Biol. Chem. 291, 5428–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B., Gogol M., Carey M., Lee D., Seidel C., and Workman J. L. (2007) Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316, 1050–1054 [DOI] [PubMed] [Google Scholar]
- 11.Ruan C., Lee C. H., Cui H., Li S., and Li B. (2015) Nucleosome contact triggers conformational changes of Rpd3S driving high-affinity H3K36me nucleosome engagement. Cell Reports 10, 204–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C. H., Wu J., and Li B. (2013) Chromatin remodelers fine-tune H3K36me-directed deacetylation of neighbor nucleosomes by Rpd3S. Mol. Cell 52, 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Churchman L. S., and Weissman J. S. (2011) Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lickwar C. R., Rao B., Shabalin A. A., Nobel A. B., Strahl B. D., and Lieb J. D. (2009) The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One 4, e4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gouet P., Courcelle E., Stuart D. I., and Metoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 16.Söding J., Biegert A., and Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh J. W., Wu J., Lee C. H., Yun M., Gilada D., Brautigam C. A., and Li B. (2012) Multivalent di-nucleosome recognition enables the Rpd3S histone deacetylase complex to tolerate decreased H3K36 methylation levels. EMBO J. 31, 3564–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun M., Ruan C., Huh J. W., and Li B. (2012) Reconstitution of modified chromatin templates for in vitro functional assays. Methods Mol. Biol. 833, 237–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert T. M., McDaniel S. L., Byrum S. D., Cades J. A., Dancy B. C., Wade H., Tackett A. J., Strahl B. D., and Taverna S. D. (2014) A PWWP domain-containing protein targets the NuA3 acetyltransferase complex via histone H3 lysine 36 trimethylation to coordinate transcriptional elongation at coding regions. Mol. Cell. Proteomics 13, 2883–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keogh M. C., Mennella T. A., Sawa C., Berthelet S., Krogan N. J., Wolek A., Podolny V., Carpenter L. R., Greenblatt J. F., Baetz K., and Buratowski S. (2006) The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20, 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwan A. H., Gell D. A., Verger A., Crossley M., Matthews J. M., and Mackay J. P. (2003) Engineering a protein scaffold from a PHD finger. Structure 11, 803–813 [DOI] [PubMed] [Google Scholar]
- 22.Shi X., Kachirskaia I., Walter K. L., Kuo J. H., Lake A., Davrazou F., Chan S. M., Martin D. G., Fingerman I. M., Briggs S. D., Howe L., Utz P. J., Kutateladze T. G., Lugovskoy A. A., Bedford M. T., and Gozani O. (2007) Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282, 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar G. S., Chang W., Xie T., Patel A., Zhang Y., Wang G. G., David G., and Radhakrishnan I. (2012) Sequence requirements for combinatorial recognition of histone H3 by the MRG15 and Pf1 subunits of the Rpd3S/Sin3S corepressor complex. J. Mol. Biol. 422, 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva A. C., Xu X., Kim H. S., Fillingham J., Kislinger T., Mennella T. A., and Keogh M. C. (2012) The replication-independent histone H3-H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J. Biol. Chem. 287, 1709–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu Y., Sutton A., Sternglanz R., and Prelich G. (2006) The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 26, 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keogh M. C., Podolny V., and Buratowski S. (2003) Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 23, 7005–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y., Warfield L., Zhang C., Luo J., Allen J., Lang W. H., Ranish J., Shokat K. M., and Hahn S. (2009) Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29, 4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smolle M., Venkatesh S., Gogol M. M., Li H., Zhang Y., Florens L., Washburn M. P., and Workman J. L. (2012) Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 19, 884–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesh S., Smolle M., Li H., Gogol M. M., Saint M., Kumar S., Natarajan K., and Workman J. L. (2012) Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 [DOI] [PubMed] [Google Scholar]
- 30.Wang S. S., Zhou B. O., and Zhou J. Q. (2011) Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol. Cell. Biol. 31, 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strahl B. D., and Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.