High UGT2B17 mRNA expression has recently been correlated with poor prognosis in chronic lymphocytic leukemia (CLL).1 In the present study, we investigated the expression of UGT2B17 in a Scandinavian population-based CLL cohort (n=253) and can confirm that high expression of UGT2B17 is associated with advanced clinical stage at diagnosis, unmutated IGHV genes (U-CLL) and poor clinical outcome. That said, we discovered a notable and novel finding based on the expression of UGT2B17, that of identifying patients with a poor prognosis within the IGHV-mutated group (M-CLL) (31/120, 26%), which previously could not be discriminated by any other established molecular marker, including recurrent genomic aberrations, novel mutations (SF3B1, NOTCH1 and TP53) and CD38 expression. Interestingly, high UGT2B17 expression arose as the strongest independent molecular prognostic marker of overall survival (OS) in multivariate analysis within M-CLL. The incorporation of LPL into our expression analysis enabled the further stratification of M-CLL, thus highlighting the potential use of RNA-based markers in the prognostic stratification of CLL, particularly for cases exhibiting an otherwise favorable clinicobiological profile.
During the last decades, a plethora of prognostic factors have been reported which can aid in the identification of CLL patients with a high risk of progression;2–5 however, the usefulness of the majority of these parameters is generally restricted to U-CLL cases. Thus, amongst M-CLL cases, there is a lack of prognostic markers that can identify patients at greater risk of progression at diagnosis. For example, M-CLL patients generally lack poor-prognostic genomic aberrations and recurrent mutations e.g., TP53, NOTCH1 or SF3B1, and CD38 is highly expressed in only a small proportion of M-CLL cases.6–8
It has recently been proposed that RNA-based markers could be used to predict clinical outcome, especially among cases with an otherwise favorable biological profile, namely early-stage, isolated del(13q) or M-CLL. One such example, which we and others have recently identified, is LPL expression, which retained its prognostic significance by multivariate analysis in the presence of other well-established markers, thereby highlighting not only the potential usefulness of RNA markers, but also the need for further investigation using this approach.9,10 A potential candidate marker is UGT2B17, which has previously been associated with cancer and was recently reported to be overexpressed in high-risk CLL patients.1 UGT2B17, a phase II metabolizing enzyme, is a member of the UGT2B super-family which conjugates various endogenous compounds, including steroid hormones as well as several pharmaceutical drugs. By catalyzing the transfer of glucuronic acid from uridine diphosphoglucuronic acid to a variety of substrates, UGT2B17 detoxifies endogenous and exogenous steroid hormones and xenobiotics.1,11,12
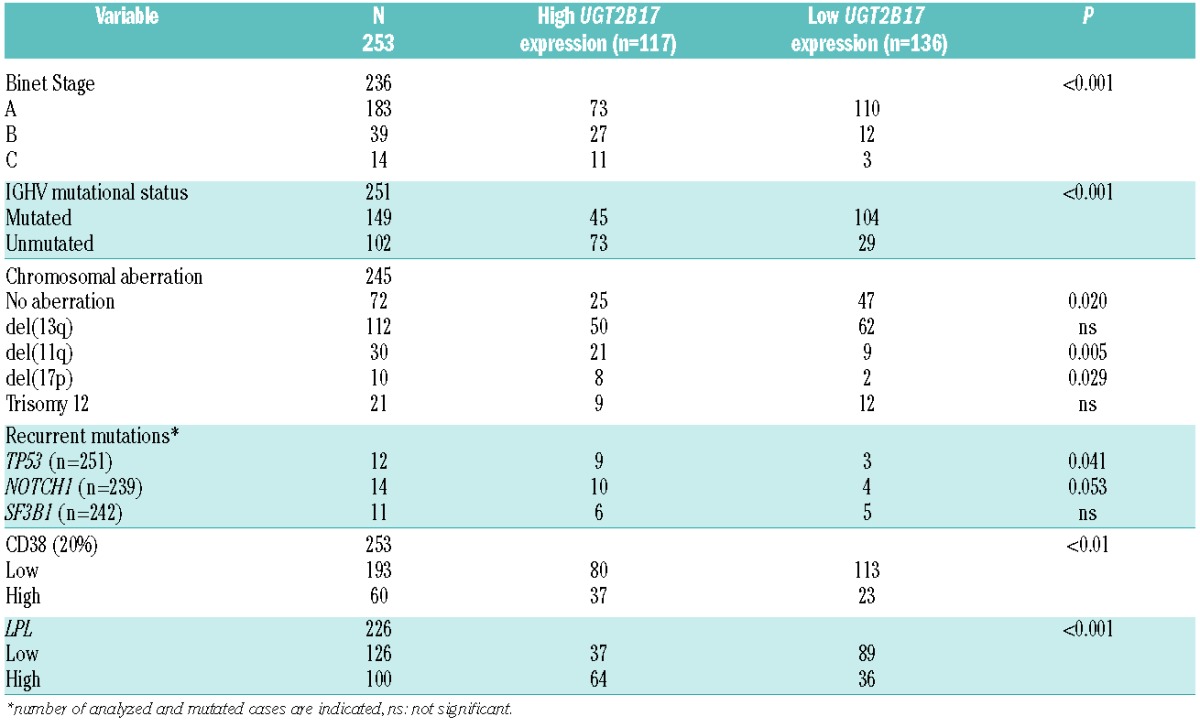
In this study, we analyzed the expression of UGT2B17 in 253 diagnosed CLL patients from a Scandinavian population-based CLL cohort (Table 1A, Online Supplementary Methods). One hundred and seventeen patients (46%) had high UGT2B17 expression, as defined by ROC curve analysis and median survival (Online Supplementary Methods), while the remaining cases (n=136, 54%) exhibited either low (88/136, 65%) or no detectable expression (48/136, 35%) (Table 1A). A significant association was observed between high UGT2B17 expression and U-CLL along with advanced clinical stage (Binet B/C) compared to cases with low expression (P<0.001 each, Table 1A). Of note, a considerable proportion of patients carrying mutated IGHV genes (45/149, 30%) or belonging to Binet stage A (73/183, 40%) displayed high UGT2B17 expression (Table 1A). Thus, while confirming the results reported by Gruber et al., i.e. that high expression of UGT2B17 correlated to poor-risk CLL,1 several differences between these two studies were also noted. More specifically, in our study, high expression of UGT2B17 correlated significantly with del(11q) and del(17p) (P=0.005 and P=0.029, respectively) (Table 1A), whereas Gruber et al. reported a negative correlation with del(17p) and no association between UGT2B17 expression and del(11q). In addition, we detected no correlation between UGT2B17 expression and recurrently mutated genes such as SF3B1 and NOTCH1 (Table 1A).
Table 1A.
Main clinicobiological characteristics of the population-based CLL cohort (n=253).
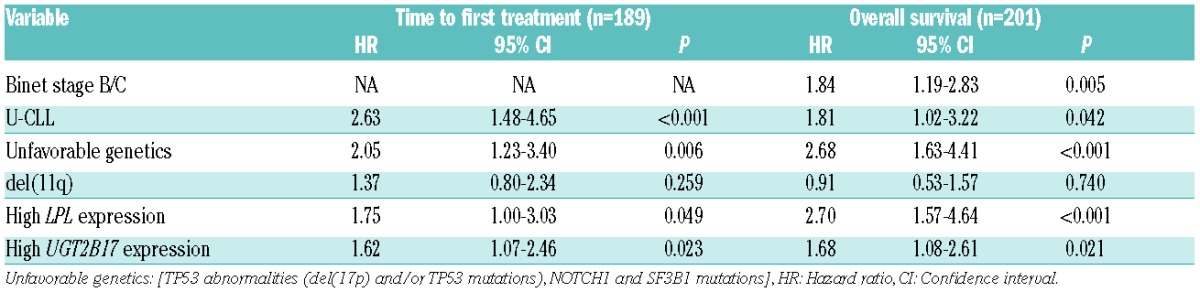
Next, we assessed the prognostic significance of UGT2B17 expression by studying time to first treatment (TTFT) and OS both within the entire cohort and also in specific subgroups of patients, most notably the group of M-CLL patients. Median follow-up for assessment of TTFT and OS was 4.3 and 10.1 years, respectively. Analysis of the entire cohort revealed that high expression of UGT2B17 correlated with shorter TTFT and OS (P<0.001 each, Figure 1A,B). In multivariate analysis, UGT2B17 expression, IGHV mutational status, LPL expression, and “unfavorable genetics” [TP53 abnormalities (del(17p) and/or TP53 mutations), NOTCH1 and SF3B1 mutations] were significant for TTFT (n=189), while UGT2B17 expression, clinical stage, IGHV mutational status, LPL expression, and “unfavorable genetics” remained as independent prognostic factors for OS (n=201) (Table 1B, Online Supplementary Table S1). Genomic lesions, which have previously been associated with unfavorable clinical outcome (TP53 abnormalities, NOTCH1 and SF3B1 mutations), were grouped together due to their low frequency in the present series, which predominantly comprised early stage cases.
Figure 1.
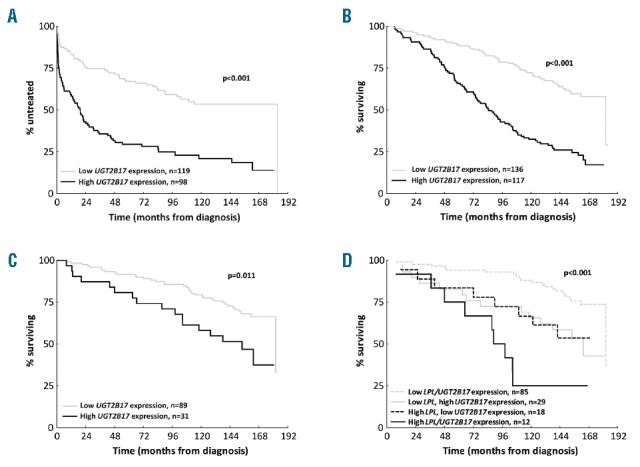
UGT2B17 expression in CLL. Impact of high UGT2B17 expression on (A) Time to first treatment (TTFT); (B) Overall survival (OS). (C) OS for M-CLL cases negative for poor prognostic recurrent genomic aberrations (del(17p), del(11q) and trisomy 12), novel mutations (SF3B1, NOTCH1 and TP53) and CD38 expression. (D) UGT2B17 and LPL expression subdivides OS in M-CLL for OS.
Table 1B.
Multivariate Cox regression analysis of UGT2B17 expression and established prognostic markers in CLL.
A novel finding with potential prognostic impact relates to the observation that 45/149 (30%) of M-CLL cases exhibited high expression of UGT2B17 and displayed poor clinical outcome (P<0.001, Online Supplementary Figure S1). Since the majority of these cases were negative for CD38 expression (134/149, 90%), carried only favorable genomic lesions (del(13q) or no recurrent aberrations (133/145, 92%) and did not display mutations in TP53 (145/149, 97%), NOTCH1 (139/142, 98%) or SF3B1 (140/143, 98%), quantification of UGT2B17 mRNA expression identified a subgroup of progressive M-CLL cases (31/120, 26%) for which, to date, no established prognostic marker has been successful in identifying (Figure 1C). Notably, within M-CLL, high UGT2B17 expression remained as the strongest independent molecular prognostic marker for OS in multivariate analysis (Online Supplementary Table S2). Further evaluation of UGT2B17 expression on clinical outcome in subgroups of CLL with favorable prognosis revealed high expression to be associated with both shorter TTFT and OS in Binet stage A cases (P<0.001 and P<0.001, respectively; Online Supplementary Figure S2A,B) as well as in cases with isolated del(13q) (P=0.012 and P<0.001, respectively; Online Supplementary Figure S3 A,B).
Taking a step further, we explored the co-expression of UGT2B17 and LPL in 226 cases and found that 64/226 (28%) had high expression of both genes, 52/64 (81% of cases concerned U-CLL), while 36 and 37 cases displayed either high LPL or UGT2B17 expression (15.9% and 16.4%, respectively, P<0.001, Table 1A). Interestingly, in contrast to cases with isolated LPL expression, the isolated UGT2B17 high expressing subgroup was predominantly comprised of M-CLL (50% vs. 83%, P=0.003). Cases with high expression for both genes exhibited a median survival of 7.4 years, M-CLL cases with low expression for both genes displayed a median survival of 15.2 years, while cases displaying isolated expression of either gene had an intermediate survival rate (Figure 1D). Taking into consideration the indolent nature of the disease experienced by the majority of M-CLL patients, analysis of UGT2B17 and LPL expression, which could distinguish those cases with the highest risk for progression and therefore shorter OS, is particularly appealing; especially if such markers can be assessed using standardized methodology such as RQ-PCR analysis.
Finally, in order to evaluate UGT2B17 expression stability and reproducibility, we investigated follow-up samples from 91 cases (range: 5.0–8.1 years, median 6.7 years). UGT2B17 expression remained stable over time (range at diagnosis: 0–3.9, average 0.29 normalized units vs. range at follow-up: 0–4.17, average 0.25 normalized units, P=0.54), a finding which also held true when including only those patients who had received treatment prior to second sampling (n=33, P=0.53). In the latter group, 31 out of 33 patients retained the same expression classification after treatment, while 2 of the patients changed from a low expression of UGT2B17 to a high expression following treatment, thereby highlighting the potential role of UGT2B17 as a suitable follow-up marker in CLL within clinical practice.
Data on the exact mechanism of the action of UGT2B17 in CLL which could provide a rational explanation for the association with progressive disease, especially in M-CLL, is currently lacking. Gruber et al. have reported high expression of UGT2B17 in cases with stable or progressive disease after fludarabine-based therapy administration, implying that UGT2B17 expression could be linked to drug metabolism, either within the cancer cell population or in other sites where degradation of the drug occurs. Suboptimal treatment responses could possibly explain the impact of UGT2B17 expression on OS.
In conclusion, we not only confirm that UGT2B17 expression is associated with advanced clinical stage at diagnosis, U-CLL and a poor clinical outcome, but also note for the first time its prognostic role among M-CLL patients, whereby UGT2B17 expression was able to identify poor risk cases which previously could not be discriminated using established markers, and within which it represented the most significant molecular factor affecting OS. Incorporation of LPL expression into our analysis enabled the further stratification of M-CLL, thus providing a rationale for the use of RNA-based markers in the prognostic stratification of CLL. In order to adopt such an approach into everyday clinical practice, the investigation of RNA markers in a large series of patients is imperative, while elucidation of the biological background and the exact mechanisms of action may provide novel therapeutic targets.
Acknowledgments
The authors thank Dr. Richard Rosenquist for helpful discussions and guidance as well as valuable comments when writing this manuscript.
Footnotes
Funding: this research project was supported by the Swedish Cancer Society, the Swedish Research Council, Uppsala University, Uppsala University Hospital, the Lion’s Cancer Research Foundation (Uppsala), and Selander’s Foundation, Uppsala.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Gruber M, Bellemare J, Hoermann G, et al. Overexpression of uridine diphospho glucuronosyltransferase 2B17 in high-risk chronic lymphocytic leukemia. Blood. 2013;121(7):1175–1183. [DOI] [PubMed] [Google Scholar]
- 2.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–1847. [PubMed] [Google Scholar]
- 3.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–1854. [PubMed] [Google Scholar]
- 4.Mansouri L, Cahill N, Gunnarsson R, et al. NOTCH1 and SF3B1 mutations can be added to the hierarchical prognostic classification in chronic lymphocytic leukemia. Leukemia. 2013;27(2):512–514. [DOI] [PubMed] [Google Scholar]
- 5.Rossi D, Rasi S, Spina V, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121(8):1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thunberg U, Johnson A, Roos G, et al. CD38 expression is a poor predictor for VH gene mutational status and prognosis in chronic lymphocytic leukemia. Blood. 2001;97(6):1892–1894. [DOI] [PubMed] [Google Scholar]
- 7.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. [DOI] [PubMed] [Google Scholar]
- 8.Baliakas P, Hadzidimitriou A, Sutton LA, et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29(2):329–336. [DOI] [PubMed] [Google Scholar]
- 9.Kaderi MA, Kanduri M, Buhl AM, et al. LPL is the strongest prognostic factor in a comparative analysis of RNA-based markers in early chronic lymphocytic leukemia. Haematologica. 2011;96(8):1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sevov M, Rosenquist R, Mansouri L. RNA-based markers as prognostic factors in chronic lymphocytic leukemia. Expert Rev Hematol. 2012;5(1):69–79. [DOI] [PubMed] [Google Scholar]
- 11.Hirata H, Hinoda Y, Zaman MS, et al. Function of UDP-glucuronosyltransferase 2B17 (UGT2B17) is involved in endometrial cancer. Carcinogenesis. 2010;31(9):1620–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angstadt AY, Berg A, Zhu J, et al. The effect of copy number variation in the phase II detoxification genes UGT2B17 and UGT2B28 on colorectal cancer risk. Cancer. 2013;119(13):2477–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]


