ABSTRACT
Tumor immune escape has been a major problem for developing effective immunotherapy. The human leukocyte antigen G (HLA-G) is a non-classical MHC class I molecule whose primary function is to protect the fetus from the mother's immune system. While HLA-G is hardly found in normal adult tissues, various tumor cells are known to express it, aiding their escape from the immune system. Thus, HLA-G is an attractive immunotherapy target. CD4+ helper T lymphocytes (HTLs) play an important role in the immune reaction against tumors by assisting in the generation and persistence of CD8+ cytotoxic T lymphocytes (CTLs) or by displaying direct antitumor effects. We report here that HLA-G expression in breast cancer significantly correlates with a poor prognosis. Also, we describe that the MHC class II-binding peptide HLA-G26–40 was effective in eliciting tumor-reactive CD4+ T cell responses. Furthermore, treatment with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine increased HLA-G expression in tumors and subsequently enhanced recognition by HLA-G26–40-specific HTLs. These findings predict that a combination immunotherapy targeting HLA-G together with a DNA methyltransferase inhibitor could be useful against some cancers.
KEYWORDS: Human leukocyte antigen G, immune escape, immunotherapy, peptide vaccine, 5-aza-2′-deoxycytidine
Abbreviations
- APCs
antigen-presenting cells
- 5-AZA
5-aza-2′-deoxycytidine
- β2-m
β2-microglobulin
- CTLs
CD8+ cytotoxic T lymphocytes
- DCs
dendritic cells
- ER
estrogen receptor
- E:T
effector-to-target
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- HLA-G
human leukocyte antigen G
- HTLs
CD4+ helper T lymphocytes
- IFNγ
interferon-γ
- KIR2DL4
killer immunoglobulin-like receptor
- LDH
lactate dehydrogenase
- MHC-II
MHC class II
- NK cells
natural killer cells
- PBMCs
peripheral blood mononuclear cells
- PR
progesterone receptor
- TAAs
tumor-associated antigens.
Introduction
While natural killer (NK) cells and CD8+ cytotoxic T lymphocytes (CTLs) are driving effectors in antitumor immune responses, CD4+ helper T lymphocytes (HTLs) and dendritic cells (DCs) are critical in supporting these effectors. However, tumor cells readily escape from immune surveillance through various mechanisms. Immune-checkpoint inhibitors have been developed and approved to target immunosuppressive pathways, and have shown clinical efficacy against several malignancies.1-3 However, relies on existing immune responses and thus, many patients fail to benefit. Thus, it is anticipated that generating immune responses, for example, using therapeutic vaccines targeting tumor-associated antigens (TAAs), will enhance the effectiveness of checkpoint inhibitor therapy.
Human leukocyte antigen G (HLA-G) is a non-classical MHC class I molecule that plays an important tolerogenic function. HLA-G is hardly expressed in normal adult tissues, with the exception of the placenta, where it protects the fetus from the maternal immune system.4,5 Some tumors such as melanoma, head and neck, lung, urogenital, gastrointestinal and breast cancers express HLA-G,6-11 inhibiting effector immune functions via killer immunoglobulin-like receptor (KIR2DL4) and immunoglobulin-like transcripts, ILT2 and ILT4.12-14 Indeed, HLA-G tumor expression is associated with poor prognosis for some malignancies.15-17 Epigenetic factors such as DNA methylation are involved in the regulation of HLA-G expression and demethylating agents such as 5-aza-2′-deoxycytidine (5-AZA) can enhance HLA-G expression in leukemias.18,19 Because of its immune-suppressive activity and its tissue-restricted expression, it is possible that HLA-G could function as a TAA for developing T cell-based immunotherapy. Here, we report that HLA-G specific, MHC class II (MHC-II)-restricted, tumor-reactive CD4+ HTL responses can be generated in both healthy donors and cancer patients. Furthermore, treatment of tumor cells with both 5-AZA and interferon-γ (IFNγ) augmented HLA-G and MHC-II expression in tumor cells, enhancing recognition by HTLs. These findings suggest that T cell-based immunotherapy targeting TAAs that function as immunosuppressive molecules such as HLA-G in combination with demethylation agents could be implemented as a novel strategy for cancer immunotherapy.
Results
HLA-G expression correlates with poor prognosis in breast cancer
We first examined whether HLA-G expression in breast cancer correlates with prognosis. We evaluated HLA-G expression in breast cancer tissue samples from 102 patients by immunohistochemistry, where each section was assessed blindly by three pathologists and classified into four types according to immunostaining intensity (−, no staining; +, weak staining; ++, moderate staining and +++, strong staining). Examples are shown in Fig. 1A–D. The results of the initial evaluation were six patients, −; 58 patients, +; 32 patients, ++ and six patients, +++. For further analysis, the patients were grouped into HLA-Glow (− and +) with 64 patients, and HLA-Ghigh (++ and +++) with 38 patients. A significant correlation was found between HLA-G expression and both estrogen receptor (ER) and progesterone receptor (PR) expression (Table 1). On the other hand, no correlation was observed between HLA-G expression with other factors such as menopause, nuclear grade, tumor vascular invasion, tumor size, lymph node metastasis and pathological stage. Within the follow-up period, there were six cancer-related deaths: one (1.6%) in the HLA-Glow group and five (13.2%) in the HLA-Ghigh group (Fig. 1E). In addition, three patients (4.7%) relapsed in the HLA-Glow group, while six patients (15.8%) relapsed in the HLA-Ghigh group (Fig. 1F). Statistical analysis (Kaplan–Meier) showed that both overall and relapse-free survival rates of HLA-Glow patients were significantly higher than HLA-Ghigh patients. These results suggest that HLA-G could be a potential immune therapy target, if it were to function as a TAA, in patients with progressive cancer. Thus, we evaluated the existence of T cell epitopes in the HLA-G protein capable of stimulating tumor-reactive HTL responses.
Figure 1.
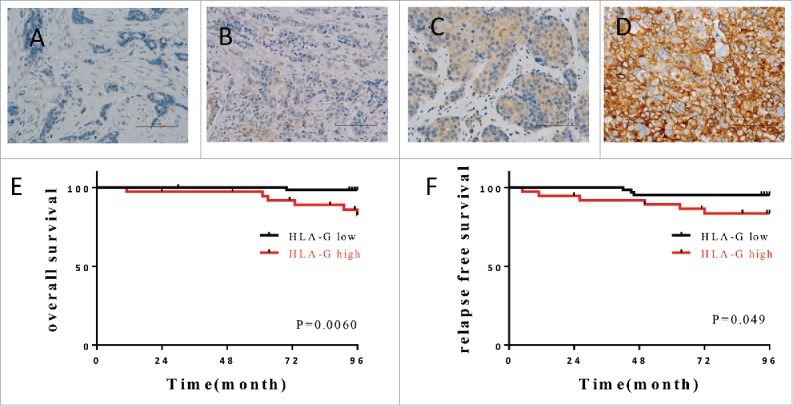
Expression of HLA-G in breast cancer correlates with outcome. Examples of immunostaining intensity: (A) no staining; (B) weak staining; (C) moderate staining and (D) strong staining used for establishing classification of the level of expression. (E, F) The Kaplan–Meier graphs was used to estimate the correlation of HLA-G expression with overall survival and relapse-free survival. Log-rank tests were used to assess significance (p values).
Table 1.
The association of HLA-G expression with clinicopathological status was analyzed by Chi-squared tests.
| HLA-G |
||||
|---|---|---|---|---|
| Clinicopathological parameters | Low N = 64 | High N = 38 | p value | |
| Age (32–83) | 56 ± 11 | 55 ± 11 | ||
| Menopausal status | pre | 26 | 14 | 0.7 |
| post | 38 | 24 | ||
| Nuclear grade | 1 | 27 | 10 | 0.25 |
| 2 | 34 | 25 | ||
| 3 | 3 | 3 | ||
| Lymphatic duct invasion (ly) | − | 46 | 24 | 9.36 |
| + | 18 | 14 | ||
| Vascular invasion (v) | − | 42 | 31 | 0.08 |
| + | 22 | 7 | ||
| Pathological T | T1 | 52 | 28 | 0.14 |
| T2 | 9 | 10 | ||
| T3 | 3 | 0 | ||
| Lymph node metastasis | − | 46 | 22 | 0.14 |
| + | 18 | 16 | ||
| Pathological Stage | I | 41 | 18 | 0.2 |
| II | 19 | 18 | ||
| III | 4 | 2 | ||
| ER | − | 6 | 11 | 0.01 |
| + | 58 | 27 | ||
| PR | − | 12 | 16 | 0.01 |
| + | 52 | 22 | ||
| HER2 | − | 56 | 30 | 0.25 |
| + | 8 | 8 | ||
In vitro induction of HLA-G-specific CD4+ T cell responses
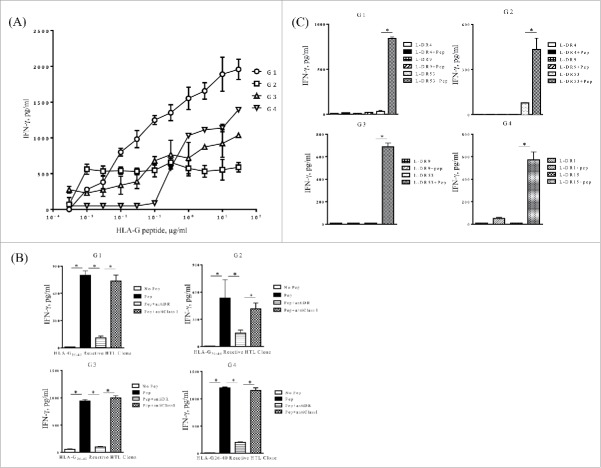
For these experiments, we identified two potential HTL epitopes from HLA-G, HLA-G6–20 (PRTLFLLLSGALTLT) and HLA-G26–40 (SHSMRYFSAAVSRPG), based on high MHC-II binding scores using a comprehensive computer-based algorithms to three common HLA-DR alleles (DRB1*0101, DRB1*0401, DRB1*0701).20 However, we were only able to study the HLA-G26–40 peptide because HLA-G6–20 was not sufficiently soluble to be used in cell culture experiments. To generate HLA-G26–40-specific HTLs, purified CD4+ T cells from four healthy volunteers were repeatedly stimulated with HLA-G26–40 pulsed onto autologous DCs as described in Materials and Methods. T cell clones from each donor were isolated and were first evaluated for their ability to respond to various antigen concentrations. The results shown in Fig. 2A demonstrate that HLA-G26–40 induced IFNγ production in all four HTL clones in a dose-dependent manner and that concentrations as low as 1 µg/mL were sufficient to generate a significant cytokine response.
Figure 2.
Induction of HLA-G26–40-specific CD4+ T cell responses. (A) CD4+ T cell clones (G1; HLA-DR4/DR9/DR53, G2; HLA-DR4/DR9/DR53, G3; HLA-DR9/DR53 and G4; HLA-DR1/DR15) were tested for their capability to respond to various concentrations of HLA-G26–40 peptide using autologous PBMCs as APCs. (B) Inhibition of HLA-G26–40-specific CD4+ T cell responses by anti-HLA-DR mAb L243 but not by anti-HLA class I mAb W6/32 (negative control) (*p < 0.05, Student's t test). (C) CD4+ T cell clones were evaluated using L-cells transfected with individual HLA-DR genes as APCs to determine the restricting HLA class II alleles (*p < 0.05, Student's t test). Supernatants were collected after 48 h of incubation and analyzed by ELISA for IFNγ production (*p < 0.05, one-way ANOVA with the Holm post-hoc test). Bars and error bars indicate the mean and SD, respectively. Experiments were performed in duplicate. (*p < 0.05, Student's t test). Bars and error bars indicate the mean and SD, respectively. Experiments were performed in duplicate.
MHC-II restriction analysis of HLA-G26–40 peptide-specific HTLs
Next, we determined the HLA class II molecules presenting HLA-G26–40 to the four CD4+ T cell clones. We first evaluated the capacity of anti-HLA-DR mAbs (or anti-class I mAbs, as negative control) to inhibit T cell responses to peptide-pulsed autologous PBMCs. The L243 antibody is specific for HLA-DR and does not react with other human MHC-II molecules (HLA-DP or -DQ), facilitating this process. As shown in Fig. 2B, the antigen response of all four CD4+ T cell clones was markedly blocked by the anti-HLA-DR mAb L243, but not by anti-HLA class I mAbs W6/32, indicating that HLA-G26–40 was presented in the context of HLA-DR. Subsequently, a panel of mouse fibroblasts (L-cells) transfected with individual HLA-DR were used as antigen-presenting cells (APCs) to facilitate the identification of the specific DR allele. The results showed that T cell clones G1, G2 and G3 recognized the peptide in the context of HLA-DR53, and that clone G4 used HLA-DR15 (Fig. 2C). The sequence of the HLA-G26–40 is highly homologous to sequences found in the classical HLA class I molecules (Table S1) and because classical HLA class I molecules are ubiquitously expressed in normal tissues, we evaluated the possible cross-reactivity of the four T cell clones. None of the HLA-G26–40-specific CD4+ T cells responded to the homologous peptides derived from classical HLA class I molecules (Fig. S1). These results indicate that T cell responses induced to the HLA-G26–40 epitope would be specific and likely not cause detrimental autoimmunity by reacting with classical HLA class I molecules.
HLA-G tumor expression is enhanced by IFNγ and DNA methyltransferase inhibitors
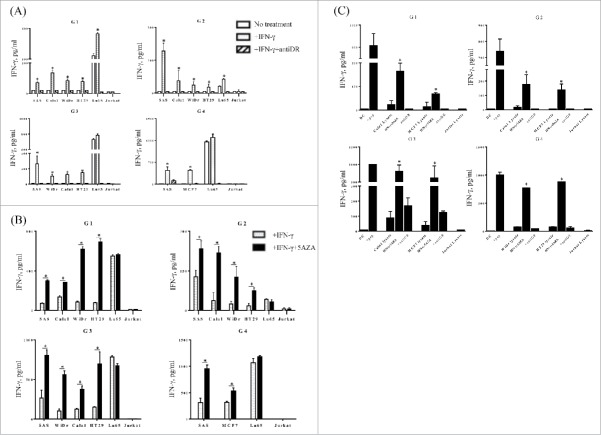
HLA-G can be expressed in a variety of malignancies.6-11 To determine whether the HLA-G26–40 reactive T cell clones that we generated were capable of recognizing tumor cell, we first evaluated HLA-G expression in some available tumor cell lines (SAS, Calu-1, WiDr, HT29, Lu65, MCF-7, HSC-4 and Jurkat). As shown using Western blot analyses, SAS and Lu65 showed constitutive expression of HLA-G (Fig. 3), and Calu-1, WiDr, HT29, MCF7 expressed HLA-G after treatment with IFNγ. Furthermore, treatment with 5-AZA and IFNγ enhanced the expression of HLA-G in SAS, Calu-1, WiDr, HT-29 and MCF-7. It should be noted that we were unable to detect cell surface expression of HLA-G protein in all these tumor cell lines by fluorescence flow cytometry using a commercially available antibody (data not shown). It is possible that levels of HLA-G on the tumor cells were below the limits of detection by this antibody. To examine why HLA-G was not expressed on the cell surface after IFNγ and 5-AZA treatment, we assessed the expression of β2-microglobulin (β2-m) on tumor cells. As shown in Fig. S2, the expression of β2-m was slightly augmented by IFNγ and 5-AZA treatment in some tumor cell lines (MCF7, Calu-1 and SAS) in the same manner as HLA class I but not with HLA-G suggesting that β2-m is not involved in the regulation of HLA-G cell surface expression by this treatment. Also, we observed that the production of soluble HLA-G was not increased by IFNγ and 5-AZA in all tumor cells except HT29 cells (Fig. S3). Notwithstanding these results, cell surface expression or secretion of mature HLA-G protein does not need to be a requirement for CD4+ T cells to recognize a processed peptide such as HLA-G26–40 when presented in the context of HLA-DR.
Figure 3.
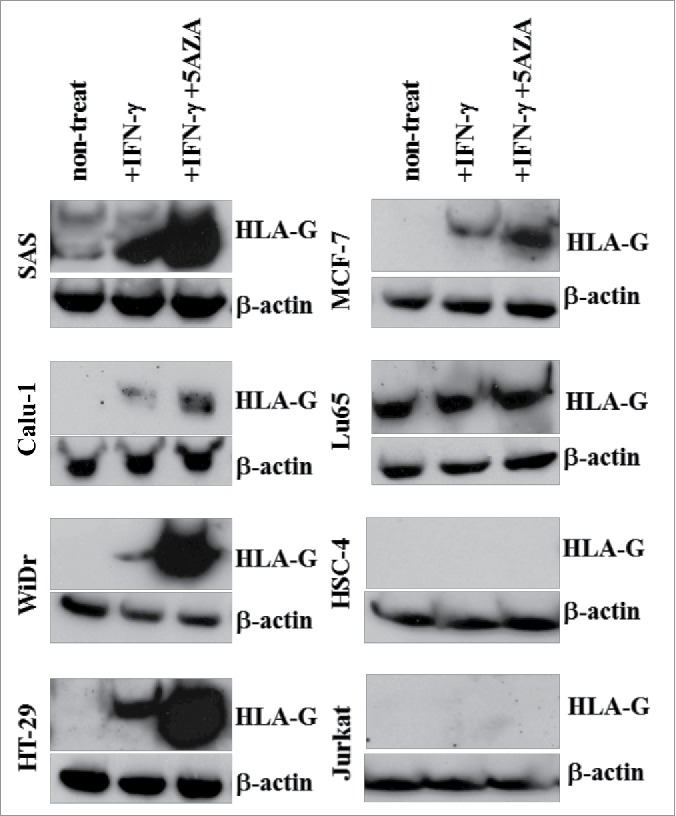
Evaluation of HLA-G expression in tumor cell lines after treatment with IFNγ and 5-AZA. HLA-G expression in the tumor cell lines was evaluated by Western blotting. These tumor cell lines were treated with or without IFNγ alone or in combination with 5-AZA for 72 h before tested.
HLA-G26–40-reactive CD4+ T cells recognize naturally processed antigen on HLA-DR molecules of tumor cells
It is important to determine whether peptide-induced CD4+ T cells can recognize antigens that are naturally processed and expressed as peptides complex to surface MHC-II molecules. To assess the capacity of the HLA-G26–40-specific CD4+ T cell clones to directly react with tumor cells expressing HLA-G and surface MHC-II, the following tumor lines were used as APCs (after IFNγ and 5-AZA treatment): SAS (tongue carcinoma, HLA-DR9/15/53), WiDr (colorectal adenocarcinoma, HLA-DR4/7/53), Calu-1 (non-small cell lung carcinoma, HLA-DR7/14/53), HT29 (colorectal adenocarcinoma, HLA-DR4/7/53), Lu65 (lung giant cell carcinoma, HLA-DR4/15/53), MCF7 (breast cancer, HLA-DR3/15) and Jurkat (T cell leukemia, HLA-DR negative, negative control). The expression of HLA-DR on tumor cells was induced with IFNγ (except for Lu65 cells, which constitutively expressed HLA-DR) and treatment with 5-AZA did not influence the expression level of HLA-DR on the tumor cell surface (data not shown).
With the above information in hand, we proceeded to evaluate the reactivity of the four HLA-G26–40 reactive T cell clones against HLA-DR and HLA-G expressing tumor cells. As shown in Fig. 4A, the HLA-G-specific CD4+ T cell clones directly recognized the HLA-DR subtype-matched tumor cells expressing HLA-G when the cell lines were treated with IFNγ (Lu65, did not have to be treated with IFNγ, to be recognized by the CD4+ T cell clones). The T cell response against the tumor cells was effectively inhibited by anti-HLA-DR mAb, indicating that HLA-G26–40 was presented in the context of HLA-DR. Treatment with 5-AZA increased the T cell responses to the HLA-G-expressing tumors except for Lu65 (Fig. 4B). These results indicate that 5-AZA can act as an adjuvant to increase HLA-G expression and enhance HLA-G-specific CD4+ T cell reactivity.
Figure 4.
HLA-G26–40-specific CD4+ T cell clones directly react with tumor cells. (A) Several tumor cell lines were tested for their ability to be recognized by the HLA-G26–40-specific CD4+ T cell clones and these responses were suppressed by anti-HLA-DR mAb (*p < 0.05, one-way ANOVA with the Holm post-hoc test compared among the same tumor cell lines). (B) The HLA-G26–40-specific CD4+ T cell responses to tumor cells were enhanced by the upregulation of HLA-G molecules after treatment with 5-AZA (*p < 0.05, Student's t test). (C) The HLA-G26–40-specific CD4+ T cells reacted with naturally processed exogenous antigens presented by autologous DCs. Supernatants were collected after 48 h of incubation and analyzed by ELISA for IFNγ production (*p < 0.05, one-way ANOVA with the Holm post-hoc test compared among the same tumor cell lysate used). Bars and error bars indicate the mean and SD, respectively. Experiments were performed in duplicate.
Recognition of naturally processed exogenous HLA-G as presented by DCs
Professional APCs such as DCs are able to capture and process antigens from dead tumor cells and present them as peptide/MHC-II complexes to CD4+ T cells. Thus, we evaluated whether DCs could present the HLA-G26–40 peptide epitope to the CD4+ T cell clones, after pulsing the DCs with IFNγ/5-AZA-treated HLA-G-expressing tumor cell lysates. (Jurkat cell lysate was used as a negative control). As shown in Fig. 4C, all T cell clones responded to DCs pulsed with the HLA-G-expressing tumor cell lysates indicating that the HLA-G was processed by the exogenous MHC-II pathway. In addition, the T cell reactivity was significantly increased when IFNγ/5-AZA-treated tumor cell lysates were used to pulse the DCs and this reactivity was blocked by anti-HLA-DR mAb.
HLA-G26–40-specific CD4+ T cells exhibit cytotoxic activity
Having observed that the CD4+ T cell clones could directly recognize HLA-G-positive tumor cells, we evaluated whether these T cells displayed any cytotoxic activity against the tumor cell lines. As shown in Fig. 5, all the T cells were able to efficiently kill the tumor cell lines that were treated with IFNγ alone, and more effectively when the tumor cells were treated with both IFNγ and 5-AZA. The clone G3 did not show any cytotoxic activity against tumor cell lines (data not shown).
Figure 5.
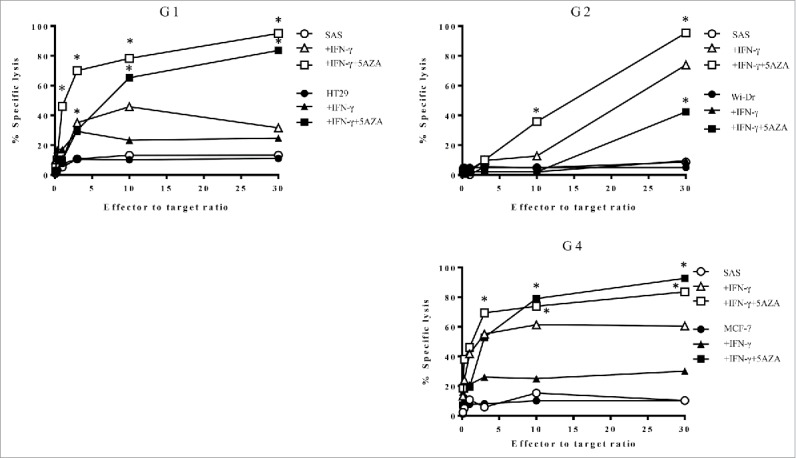
HLA-G26–40-specific CD4+ T cell clones were evaluated for their cytotoxicity against HLA-G-expressing tumor cells. The cytotoxic activity of the T cell clones (G1, G2 and G4) against HLA-DR-matched HLA-G-positive tumor cells was assessed. The HLA-G26–40-specific CD4+ T cell clones effectively lysed the tumor cells treated with IFNγ and 5-AZA. Supernatants were collected after 6 h of incubation and analyzed using a colorimetric CytoTox 96 assay. (*p < 0.05, one-way ANOVA with the Holm post-hoc test compared at the same E:T ratio among the same target).
T cell responses to HLA-G26–40 in cancer patients
It would be important to determine whether HLA-G26–40-reactive lymphocytes are present in cancer patients to assess the potential use of this peptide as a cancer vaccine. To measure the responses of CD4+ T cells from cancer patients to HLA-G26–40, we stimulated PBMCs from 10 cancer patients (seven breast cancer patients and three lung cancer patients) with peptide in short-term cultures. We were unable to produce T cell lines or clones for subsequent more detailed studies or HLA restriction analyses because the limitations of obtaining sufficient blood. A tetanus toxoid peptide (TT830–843) was used as a positive control since this peptide generates robust CD4+ T cell responses in the majority of people regardless of their HLA-DR alleles. Seven days after the second peptide stimulation, IFNγ production was measured in the culture supernatants. Substantial T cell responses to the HLA-G26–40 peptide were observed in the breast and lung cancer patients (Fig. 6) and these responses were inhibited by anti-HLA-DR mAb, indicating that peptide recognition by the T cells is mediated by HLA-DR molecules.
Figure 6.
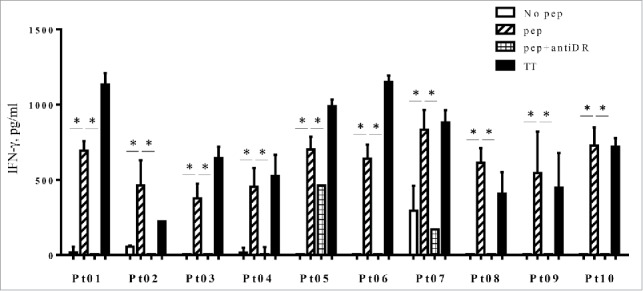
PBMCs from three lung cancer patients (Pt01, Pt02 and Pt03) and seven breast cancer patients (Pt04 to Pt10) were stimulated with HLA-G26–40 peptide and re-stimulated with the peptide on day 7. The supernatant of each culture was collected on day 14 and analyzed by ELISA for IFNγ production. Bars and error bars indicate the mean and SD, respectively. Experiments were performed in duplicate. (*p < 0.05, Student's t test). Bars and error bars indicate the mean and SD, respectively. Experiments were performed in duplicate.
Discussion
Here, we report that the non-classical MHC class I molecule, HLA-G can function as a TAA for MHC-II-restricted T cells. Specifically, we have identified the novel T cell epitope HLA-G26–40, which elicits effective antitumor CD4+ T cell responses against HLA-G-positive tumors. Although emphasis has been put on CD8+ CTLs for tumor immunotherapy because of their direct cytotoxicity against tumor cells, CD4+ T cells also play an important role in antitumor immunity. For example, not only has it been reported that CD4+ T cells have cytotoxic activity in addition to their helper functions for NK cells and CTLs, but in some instances, they can be more efficient in eliminating tumors than CD8+ T cells regardless of whether the tumors express MHC-II.21 In the current study, HLA-G26–40-specific CD4+ T cells showed cytotoxic activity against HLA-G-positive tumor cells, suggesting that CD4+ T cell-based immunotherapy has a strong therapeutic potential. Furthermore, HLA-G-specific CD8+ T cells against renal cell carcinoma have been described, 22 reinforcing the notion that HLA-G is a valid immunological target for cancer immunotherapy.
Breast cancer patients with high HLA-G expression levels had a poorer prognosis and higher recurrence rates as compared to those with low HLA-G expression (Figs. 1E–F); these results agree with those previously reported.23 However, de Kruijf et al. published that HLA-G expression had no significant prognosis correlation in breast cancer.17 This discrepancy could be due to differences in the classification method of HLA-G expression. In our study, we classified HLA-G expression into two groups according to expression intensity, while de Kruijf et al. classified the patients as either being positive or negative for HLA-G expression. Owing to our classification method, we also found that the HLA-G expression intensity had an inverse association with the expression of ER and PR (Table 1). Because ER/PR-negative breast cancer patients have a poorer prognosis when compared to ER- and/or PR-positive breast cancer patients,24 HLA-G may be a suitable target for the subgroup of breast cancer patients with poor prognosis. It should be noted that, because HLA-G is one of the fetal immune defense mechanisms during pregnancy, HLA-G-targeted immunotherapy would not be used in pregnant patients. Also, HLA-G peptide vaccine would not be suitable for autoimmune patients because it could aggravate their disease in instances where HLA-G is expressed.25
Homologous sequences very similar to HLA-G26–40 were found on the classical MHC-I HLA-A, -B and -C molecules (Table S1). Nevertheless, the HLA-G26–40-specific CD4+ T cells did not cross-react with these analogous peptides. This suggests that the unique residues in HLA-G26–40 (two alanines at position 34 and 35) may play an important role in binding to MHC-II or as T cell receptor contact residues. Whichever the cause is for the lack of cross-reactivity, our results and the restricted expression of HLA-G indicate that T cell responses to HLA-G26–40 would not results in widespread autoimmunity when used in the clinical setting. Moreover, we found a substantial amount of IFNγ production from patient PBMCs by HLA-G25–33 peptide stimulation suggesting that this peptide can potentially induce Th1 rather than regulatory T cells. However, recent studies show that the polarization of CD4+ T cell is independent of the peptide sequence26 and thus the combinatorial use of Th1-skewed adjuvants will be important for a peptide vaccine to polarize CD4+ T cells into Th1 cells instead of T regulatory cells.
In recent years, immune-checkpoint inhibitors have gained attention in the field of oncology and immunology because of their remarkable clinical effects, which are achieved by blocking tumor immune escape function. Several studies indicate that the immune system can eliminate tumors when immune suppression is removed.1-3,27,28 HLA-G is another inhibitory molecule utilized by tumor cells to protect themselves against host's immune system. Although the exact mechanism of HLA-G-associated immune tolerance has not yet been elucidated, some studies point to various possibilities. For example, HLA-G downregulates the effector function of NK cells, CTLs and CD4+ T cells through its interaction with the KIR2DL4 and ILT2 receptors.29-35 Interestingly, in some circumstances, DCs express HLA-G after they acquire a regulatory function by interacting with tumor-derived MUC1.36,37 From this perspective, it is possible that an HLA-G26–40 vaccine could target regulatory DCs in addition to HLA-G expressing tumors.
Previous studies have revealed that the regulation of HLA-G gene activity involves epigenetic mechanisms such as histone acetylation or DNA demethylation38,39 and that 5-AZA treatment enhances HLA-G expression at both the mRNA and protein levels.19 In accordance with these reports, we showed here that 5-AZA treatment of tumor cell lines increased HLA-G protein expression and subsequently enhanced HLA-G26–40-specific T cell responses. Interestingly, 5-AZA treatment did not upregulate the expression levels of HLA-G on the tumor cell surface, indicating that the production of the HLA-G26–40 peptide epitope via antigen processing does not require protein cell surface expression. Because intracellular antigens can be processed as CD4+ epitopes in endosomal compartments40 or by cytoplasmic proteases,41 it is not that all surprising that tumor cells solely expressed intracellular HLA-G protein were capable of processing and presenting the HLA-G-derived CD4+ epitope on their surface MHC class II molecules (Figs. 4, 5). In addition, our data indicates that 5-AZA treatment does not enhance the immune inhibitory activity of tumor cells. Because the surface expression of HLA-G is required to express the suppressive activity by binding to its receptor such as LT2 and KIR2DL4 on effector cells, our HLA-G-reactive CD4+ responses appeared not to be affected by the inhibitory properties of HLA-G molecules, whose expression was not induced on the cell surface by 5-AZA treatment. The exact mechanism inhibiting the transport of HLA-G to cell membrane remains unclear. The expression of β2-m, which is a necessary component to form mature MHC complex on the cell surface, was slightly augmented by IFNγ and 5AZA treatment in some tumor cells in accordance with HLA-class I expression but not with HLA-G (Fig. S2), suggesting that lack of β2-m was not the reason for the decrease of cell surface expression of HLA-G. Because tapasin is a prerequisite for cell surface expression of HLA-G42 and we previously showed that tapasin is downregulated in many tumors,43 it is probable that lack of tapasin expression is preventing the migration of HLA-G to the cell membrane. In support of our findings, Odunsi et al. demonstrated in the clinical setting that treatment with 5-AZA and the NY-ESO-1 vaccine efficiently increased immune responses against this TAA in the majority of their patients.44 In view of this, we believe that 5-AZA could be used in vivo to enhance HLA-G-based immunotherapy.
Materials and methods
Cell lines
L-cells (mouse fibroblasts) expressing transfected HLA class II molecules were obtained from Dr. R. Karr (Karr Pharma, St. Louis, MO) and Dr. T. Sasazuki (Kyushu University, Fukuoka, Japan). The tumor cell lines WiDr and HT29 (human colon adenocarcinoma), Calu-1 (human lung squamous cell carcinoma), MCF7 (human breast cancer), SAS (human tongue squamous cell carcinoma) and Jurkat (HLA-DR-negative T cell lymphoma) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The human lung large cell carcinoma cell line Lu65 and the human tongue SCC cell line HSC-4 were supplied by the RIKEN Bio-Resource Center (Tsukuba, Japan). All cells were cultured in RPMI-1640 medium (Nacalai Tesque, #30264–85) supplemented with 10% fetal bovine serum (FBS) (Biowest, #S1650), penicillin (100 U/mL) and streptomycin (100 μg/mL) at 37°C in a 5% CO2 incubator. All cell lines were meticulously cultured and used up within 6 mo. No authentication assay was performed since we obtained all cell lines from reputable sources.
Synthetic peptides
Potential HLA-DR-restricted CD4+ T cell epitopes were selected from the amino acid sequence of HLA-G using computer-based algorithms developed by Southwood et al.20 for peptide/MHC binding of three common HLA-DR alleles (DRB1*0101, DRB1*0401 and DRB1*0701). The predicted peptide epitopes were synthesized by solid-phase organic chemistry and purified by high-performance liquid chromatography (HPLC). The purity (>80%) and identity of the peptides were assessed by HPLC and mass spectrometry, respectively. The synthetic peptides HLA-G6–20 (PRTLFLLLSGALTLT) and HLA-G26–40 (SHSMRYFSAAVSRPG) were used throughout this study. In addition, peptides analogous to HLA-G26–40, i.e., HLA-A26–40 (SHSMRYFYTSVSRPG), HLA-B26–40 (SHSMRYFHTSVSRPG) and HLA-C26–40 (SHSMRYFSTSVSRPG), were also synthesized (Table S1). Tetanus toxoid (TT830–843; QYIKANSKFIGITE) peptide was used as a universal epitope-peptide control that can bind to multiple HLA class II molecules.45
In vitro stimulation of antigen-specific CD4+ helper T cells with synthetic peptides
The method utilized for the generation of HLA-G-specific CD4+ T cell lines using peptide-induced lymphocytes from fresh peripheral blood derived from healthy individuals was described previously.46 In short, CD4+ T cells were firstly activated with peptide-pulsed autologous DCs and stimulated repeatedly with γ-irradiated autologous peripheral blood mononuclear cells (PBMCs) and peptides. Furthermore, CD4+ T cell clones were obtained by limiting dilution to examine the responses against antigens. All blood materials were acquired after informed consent was obtained. After 2–3 rounds of stimulation, the microcultures were tested for IFNγ production upon stimulation with HLA-G26–40 peptides. Positive microcultures exhibiting at least a threefold increase in IFNγ production after peptide stimulation when compared to the non-stimulated ones were subsequently expanded, and the T cell clones were isolated. As such, we were able to induce HLA-G26–40-specific CD4+ T cell clones using the PBMCs of four healthy donors.
Cell-mediated cytotoxicity assay
The cytotoxic activity of CD4+ T cell clones was measured by a colorimetric CytoTox 96 assay (Promega, #G1780). This system quantifies the lactate dehydrogenase (LDH) that is released from the target cells. T cells were mixed with 2 × 104 target cells at different effector-to-target (E:T) ratios in 96-well round-bottomed plates. After 6 h of incubation at 37°C, 50 μL of supernatant was collected from each well to measure the LDH content.
Assessment of HLA-G expression in tumor cells
The tumor cell lines were treated with or without 10 μM 5-AZA (Sigma, #A3656) and 500 IU/mL IFNγ (Peprotech, #300–02) or with IFNγ alone for 72 h before assessment of HLA-G expression by Western blotting. The tumor cell lines were lysed in LDS sample buffer (Invitrogen, #NP0008) and the lysates were submitted to electrophoresis in NuPAGE bi-Tris SDS-PAGE gels (Invitrogen, #NP0322PK2). The antibodies used to detect the expression of specific proteins were mouse anti-human HLA-G mAb (4H84) (1:1,000 in blocker; Abcam, #ab025455) and anti-β-actin (C4) (1:2,000; Santa Cruz Biotechnology, #sc-47778). HLA-G, HLA-A,B,C and β2m expression on the tumor cell surface was evaluated by flow cytometry using anti-HLA-G mAb (MEM-G/9) conjugated with fluorescein isothiocyanate (FITC) (Enzo Life Sciences, # ALX-805-700F-C10), anti-HLA-A, B, C mAb (W6/32) conjugated with allophycocyanin (BioLegend, #311409) and anti- β2m mAb (2M2) conjugated with phycoerythrin (BioLegend, #316305), respectively. Soluble HLA-G was measured in the tumor supernatants, which were collected after 72 h of incubation with or without IFNγ (500IY/μλ) and 5-AZA (10 µM), using sHLA-G ELISA kit following the manufacture's instruction (Enzo Life Sciences, # ALX-850-309-KI01).
Measurement of antigen-specific responses with CD4+ T cell clones
Assessment of CD4+ T cell responses to various antigens was performed as described previously.47 To augment HLA-G expression in tumor cells, the tumor cells were treated with 5-AZA and IFNγ as described above. The expression of HLA-DR molecules on the tumor cell surface was evaluated by flow cytometry using anti-HLA-DR mAb conjugated with FITC (BD PharMingen, #55581). Tumor cell lysates were prepared by freeze-thawing 1 × 108 tumor cells three times, and were resuspended in 1 mL of serum-free AIM-V medium. The tumor cell lysates were used as antigens at 5 × 105 cell equivalents per mL. Culture supernatants were collected after 48 h of incubation for measuring IFNγ production by the CD4+ T cells using ELISA kits (BD PharMingen, #555142). To demonstrate antigen-specificity and HLA-DR restriction, CD4+ T cell responses were blocked by adding anti-HLA-DR mAb L243 (IgG2a prepared from the supernatants of the hybridoma HB-55 obtained from the ATCC) or anti-HLA-A/B/C mAb W6/32 (IgG2a; ATCC) at 10 μg/mL throughout the 48 h incubation period.
Immunohistochemistry
Immunohistochemistry was performed using the Envision™ HRP System (Dako, #K5361) as described previously.48 Formalin-fixed sections were obtained from breast cancer patients. Samples were boiled in pH 9.0 EDTA buffer to retrieve antigens and endogenous peroxidase activity was inhibited according to the manufacturer's instructions. The sections were then incubated with mouse anti-human HLA-G mAb (clone 4H84; Abcam, #ab025455)) at a dilution of 1:50 overnight at 4°C, followed by incubation with an HRP-conjugated secondary antibody and substrate. The institutional ethics committee approved this study (approval numbers 14029 and 15131), and written informed consent was obtained from all patients who provided tissue samples.
Measurement of peptide-specific responses in cancer patients
Peripheral blood lymphocytes were separated from fresh heparinized blood by gradient centrifugation and cultured with peptides in 96-well plates as described previously.47 The production level of IFNγ by PBMCs derived from patients with breast or lung cancer was evaluated by ELISA (BD PharMingen, #555142). The institutional ethics committee approved this study protocol (approval number 14029).
Statistical analysis
Kaplan–Meier analysis for survival curves was performed with GraphPad Prism 5 and p values were examined with a long-rank Mantel-Cox test. The other data were analyzed by the Student's t test or one-way ANOVA with the Holm post-hoc test and p < 0.05 was used to indicate statistical significance.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from JSPS KAKENHI Grant Number 24791735, the National Cancer Institute of the National Institutes of Health, R01CA136828 and R01CA157303, and by start-up funds from Augusta University Cancer Center and the Georgia Research Alliance.
References
- 1.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med 2015; 372:320-30; PMID:25399552; http://dx.doi.org/ 10.1056/NEJMoa1412082 [DOI] [PubMed] [Google Scholar]
- 2.Brahmer J, Reckamp KL, Baas P, Crin∫ L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E. Nivolumab versus docetaxel in advanced squamous-cell non–small-cell lung cancer. N Engl J Med 2015; 373:123-35; PMID:26028407; http://dx.doi.org/ 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363:711-23; PMID:20525992; http://dx.doi.org/ 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onno M, Guillaudeux T, Amiot L, Renard I, Drenou B, Hirel B, Girr M, Semana G, Le Bouteiller P, Fauchet R. The HLA-G gene is expressed at a low mRNA level in different human cells and tissues. Hum Immunol 1994; 41:79-86; PMID:7836069; http://dx.doi.org/ 10.1016/0198-8859(94)90089-2 [DOI] [PubMed] [Google Scholar]
- 5.McMaster MT, Librach CL, Zhou Y, Lim KH, Janatpour MJ, DeMars R, Kovats S, Damsky C, Fisher SJ. Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. J Immunol 1995; 154:3771-8; PMID:7706718 [PubMed] [Google Scholar]
- 6.Zhou J-h, Ye F, Chen H-z, Zhou C-y, Lu W-g, Xie X. Altered expression of cellular membrane molecules of HLA-DR, HLA-G and CD99 in cervical intraepithelial neoplasias and invasive squamous cell carcinoma. Life Sci 2006; 78:2643-9; PMID:16434060; http://dx.doi.org/ 10.1016/j.lfs.2005.10.039 [DOI] [PubMed] [Google Scholar]
- 7.Urosevic M, Kurrer MO, Kamarashev J, Mueller B, Weder W, Burg G, Stahel RA, Dummer R, Trojan A. Human leukocyte antigen G up-regulation in lung cancer associates with high-grade histology, human leukocyte antigen class I loss and interleukin-10 production. Am J Pathol 2001; 159:817-24; PMID:11549573; http://dx.doi.org/ 10.1016/S0002-9440(10)61756-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinberg L, Flørenes VA, Skrede M, Dong HP, Nielsen S, McMaster MT, Nesland JM, Shih I-M, Davidson B. Expression of HLA-G in malignant mesothelioma and clinically aggressive breast carcinoma. Virchows Archiv 2006; 449:31-9; PMID:16541284; http://dx.doi.org/ 10.1007/s00428-005-0144-7 [DOI] [PubMed] [Google Scholar]
- 9.Guerra N, Lacombe M-JT, Angevin E, Chouaib S, Carosella ED, Caignard A, Paul P. Tumor-specific up-regulation of the nonclassical class I HLA-G antigen expression in renal carcinoma. Cancer Res 2001; 61:6838-45; PMID:11559559 [PubMed] [Google Scholar]
- 10.El-Chennawi F, Auf F, El-Diasty A, El-Daim M, El-Sherbiny S, Ali A, El-Baz M, El-Hameed M, Paul P, Ibrahim E. Expression of HLA-G in cancer bladder. Egyp J Immunol/Egyp Associat Immunol 2004; 12:57-64; PMID:1673414012874014 [PubMed] [Google Scholar]
- 11.Bukur J, Rebmann V, Grosse-Wilde H, Luboldt H, Ruebben H, Drexler I, Sutter G, Huber C, Seliger B. Functional role of human leukocyte antigen-G up-regulation in renal cell carcinoma. Cancer Res 2003; 63:4107-11; PMID:12874014 [PubMed] [Google Scholar]
- 12.Rajagopalan S, Long EO. A Human Histocompatibility Leukocyte Antigen (HLA)-G–specific receptor expressed on all natural killer cells. J Exp Med 1999; 189:1093-100; PMID:10190900; http://dx.doi.org/ 10.1084/jem.189.7.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colonna M. Specificity and function of immunoglobulin superfamily NK cell inhibitory and stimulatory receptors. Immunol Rev 1997; 155:127-33; PMID:9059888; http://dx.doi.org/ 10.1111/j.1600-065X.1997.tb00945.x [DOI] [PubMed] [Google Scholar]
- 14.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu ML. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 1997; 7:273-82; PMID:9285411; http://dx.doi.org/ 10.1016/S1074-7613(00)80529-4 [DOI] [PubMed] [Google Scholar]
- 15.Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM. Expression of human leucocyte antigen G (HLA-G) is associated with prognosis in non-small cell lung cancer. Lung Cancer 2007; 58:267-74; PMID:17673327; http://dx.doi.org/ 10.1016/j.lungcan.2007.06.011 [DOI] [PubMed] [Google Scholar]
- 16.Yie S-m, Yang H, Ye S-r, Li K, Dong D-d, Lin X-m. Expression of human leukocyte antigen G (HLA-G) correlates with poor prognosis in gastric carcinoma. Ann Surg Oncol 2007; 14:2721-9; PMID:17564748; http://dx.doi.org/ 10.1245/s10434-007-9464-y [DOI] [PubMed] [Google Scholar]
- 17.de Kruijf EM, Sajet A, van Nes JGH, Natanov R, Putter H, Smit VTHBM, Liefers GJ, van den Elsen PJ, van de Velde CJH, Kuppen PJK. HLA-E and HLA-G expression in classical HLA class I-Negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol 2010; 185:7452-9; PMID:21057081; http://dx.doi.org/ 10.4049/jimmunol.1002629 [DOI] [PubMed] [Google Scholar]
- 18.Wastowski IJ, Simoes RT, Yaghi L, Donadi EA, Pancoto JT, Poras I, Lechapt-Zalcman E, Bernaudin M, Valable S, Carlotti CG Jr. et al.. Human leukocyte antigen-G is frequently expressed in glioblastoma and may be induced in vitro by combined 5-aza-2′-deoxycytidine and interferon-gamma treatments: results from a multicentric study. Am J Pathol 2013; 182:540-52; PMID:23219427; http://dx.doi.org/ 10.1016/j.ajpath.2012.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polakova K, Bandzuchova E, Kuba D, Russ G. Demethylating agent 5-aza-2′-deoxycytidine activates HLA-G expression in human leukemia cell lines. Leuk Res 2009; 33:518-24; PMID:18823661; http://dx.doi.org/ 10.1016/j.leukres.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 20.Southwood S, Sidney J, Kondo A, del Guercio M-F, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several Common HLA-DR Types Share Largely Overlapping Peptide Binding Repertoires. J Immunol 1998; 160:3363-73; PMID:9531296 [PubMed] [Google Scholar]
- 21.Perez-Diez A, Joncker NT, Choi K, Chan WF, Anderson CC, Lantz O, Matzinger P. CD4 cells can be more efficient at tumor rejection than CD8 cells. Blood 2007; 109:5346-54; PMID:17327412; http://dx.doi.org/ 10.1182/blood-2006-10-051318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komohara Y, Harada M, Ishihara Y, Suekane S, Noguchi M, Yamada A, Matsuoka K, Itoh K. HLA-G as a target molecule in specific immunotherapy against renal cell carcinoma. Oncol Rep 2007; 18:1463-8; PMID:17982631; http://dx.doi.org/ 10.3892/or.18.6.1463 [DOI] [PubMed] [Google Scholar]
- 23.He X, Dong DD, Yie SM, Yang H, Cao M, Ye SR, Li K, Liu J, Chen J. HLA-G expression in human breast cancer: implications for diagnosis and prognosis, and effect on allocytotoxic lymphocyte response after hormone treatment in vitro. Ann Surg Oncol 2010; 17:1459-69; PMID:20052552; http://dx.doi.org/ 10.1245/s10434-009-0891-9 [DOI] [PubMed] [Google Scholar]
- 24.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 2007; 109:1721-8; PMID:17387718; http://dx.doi.org/ 10.1002/cncr.22618 [DOI] [PubMed] [Google Scholar]
- 25.Torres MI, Le Discorde M, Lorite P, Rios A, Gassull MA, Gil A, Maldonado J, Dausset J, Carosella ED. Expression of HLA-G in inflammatory bowel disease provides a potential way to distinguish between ulcerative colitis and Crohn's disease. Int Immunol 2004; 16:579-83; PMID:15039388; http://dx.doi.org/ 10.1093/intimm/dxh061 [DOI] [PubMed] [Google Scholar]
- 26.Clemente-Casares X, Blanco J, Ambalavanan P, Yamanouchi J, Singha S, Fandos C, Tsai S, Wang J, Garabatos N, Izquierdo C et al.. Expanding antigen-specific regulatory networks to treat autoimmunity. Nature 2016; 530:434-40; PMID:26886799; http://dx.doi.org/ 10.1038/nature16962 [DOI] [PubMed] [Google Scholar]
- 27.Munir S, Andersen GH, Svane IM, Andersen MH. The immune checkpoint regulator PD-L1 is a specific target for naturally occurring CD4+ T cells. Oncoimmunology 2013; 2:e23991; PMID:23734334; http://dx.doi.org/ 10.4161/onci.23991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen MH, Svane IM. Indoleamine 2, 3-dioxygenase vaccination. OncoImmunology 2015; 4:e983770; PMID:25949864; http://dx.doi.org/ 10.4161/2162402X.2014.983770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ketroussi F, Giuliani M, Bahri R, Azzarone B, Charpentier B, Durrbach A. Lymphocyte cell-cycle inhibition by HLA-G is mediated by phosphatase SHP-2 and acts on the mTOR pathway. PLoS One 2011; 6:e22776; PMID:21887223; http://dx.doi.org/ 10.1371/journal.pone.0022776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faure M, Long EO. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J Immunol 2002; 168:6208-14; PMID:12055234; http://dx.doi.org/ 10.4049/jimmunol.168.12.6208 [DOI] [PubMed] [Google Scholar]
- 31.Lila N, Rouas-Freiss N, Dausset J, Carpentier A, Carosella ED. Soluble HLA-G protein secreted by allo-specific CD4+ T cells suppresses the allo-proliferative response: a CD4+ T cell regulatory mechanism. Proc Natl Acad Sci USA 2001; 98:12150-5; PMID:11572934; http://dx.doi.org/ 10.1073/pnas.201407398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahri R, Hirsch F, Josse A, Rouas-Freiss N, Bidere N, Vasquez A, Carosella ED, Charpentier B, Durrbach A. Soluble HLA-G inhibits cell cycle progression in human alloreactive T lymphocytes. J Immunol 2006; 176:1331-9; PMID:16424159; http://dx.doi.org/ 10.4049/jimmunol.176.3.1331 [DOI] [PubMed] [Google Scholar]
- 33.Riteau B, Menier C, Khalil-Daher I, Martinozzi S, Pla M, Dausset J, Carosella ED, Rouas-Freiss N. HLA-G1 co-expression boosts the HLA class I-mediated NK lysis inhibition. Int Immunol 2001; 13:193-201; PMID:11157852; http://dx.doi.org/ 10.1093/intimm/13.2.193 [DOI] [PubMed] [Google Scholar]
- 34.Kapasi K, Albert SE, Yie S, Zavazava N, Librach CL. HLA-G has a concentration-dependent effect on the generation of an allo-CTL response. Immunology 2000; 101:191-200; PMID:11012772; http://dx.doi.org/ 10.1046/j.1365-2567.2000.00109.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen BG, Xu DP, Lin A, Yan WH. NK cytolysis is dependent on the proportion of HLA-G expression. Hum Immunol 2013; 74:286-9; PMID:23238216; http://dx.doi.org/ 10.1016/j.humimm.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 36.Monti P, Leone BE, Zerbi A, Balzano G, Cainarca S, Sordi V, Pontillo M, Mercalli A, Di Carlo V, Allavena P et al.. Tumor-derived MUC1 mucins interact with differentiating monocytes and induce IL-10highIL-12low regulatory dendritic cell. J Immunol 2004; 172:7341-9; PMID:15187110; http://dx.doi.org/ 10.4049/jimmunol.172.12.7341 [DOI] [PubMed] [Google Scholar]
- 37.Gregori S, Tomasoni D, Pacciani V, Scirpoli M, Battaglia M, Magnani CF, Hauben E, Roncarolo MG. Differentiation of type 1 T regulatory cells (Tr1) by tolerogenic DC-10 requires the IL-10-dependent ILT4/HLA-G pathway. Blood 2010; 116:935-44; PMID:20448110; http://dx.doi.org/ 10.1182/blood-2009-07-234872 [DOI] [PubMed] [Google Scholar]
- 38.Mouillot G, Marcou C, Rousseau P, Rouas-Freiss N, Carosella ED, Moreau P. HLA-G gene activation in tumor cells involves cis-acting epigenetic changes. Int J Cancer 2005; 113:928-36; PMID:15514928; http://dx.doi.org/ 10.1002/ijc.20682 [DOI] [PubMed] [Google Scholar]
- 39.Moreau P, Mouillot G, Rousseau P, Marcou C, Dausset J, Carosella ED. HLA-G gene repression is reversed by demethylation. Proc Natl Acad Sci 2003; 100:1191-6; PMID:12552087; http://dx.doi.org/12731050 10.1073/pnas.0337539100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, Mautner J. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol 2003; 33:1250-9; PMID:12731050; http://dx.doi.org/ 10.1002/eji.200323730 [DOI] [PubMed] [Google Scholar]
- 41.Lich JD, Elliott JF, Blum JS. Cytoplasmic processing is a prerequisite for presentation of an endogenous antigen by major histocompatibility complex class II proteins. J Exp Med 2000; 191:1513-24; PMID:10790426; http://dx.doi.org/ 10.1084/jem.191.9.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park B, Ahn K. An essential function of tapasin in quality control of HLA-G molecules. J Biol Chem 2003; 278:14337-45; PMID:12582157; http://dx.doi.org/ 10.1074/jbc.M212882200 [DOI] [PubMed] [Google Scholar]
- 43.Ogino T, Bandoh N, Hayashi T, Miyokawa N, Harabuchi Y, Ferrone S. Association of tapasin and HLA class I antigen down-regulation in primary maxillary sinus squamous cell carcinoma lesions with reduced survival of patients. Clin Cancer Res 2003; 9:4043-51; PMID:14519625 [PubMed] [Google Scholar]
- 44.Odunsi K, Matsuzaki J, James SR, Mhawech-Fauceglia P, Tsuji T, Miller A, Zhang W, Akers SN, Griffiths EA, Miliotto A et al.. Epigenetic potentiation of NY-ESO-1 vaccine therapy in human ovarian cancer. Cancer Immunol Res 2014; 2:37-49; PMID:24535937; http://dx.doi.org/ 10.1158/2326-6066.CIR-13-0126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol 1989; 19:2237-42; PMID:2481588; http://dx.doi.org/ 10.1002/eji.1830191209 [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi H, Wood M, Song Y, Appella E, Celis E. Defining promiscuous MHC class II helper T-cell epitopes for the HER2/neu tumor antigen. Cancer Res 2000; 60:5228-36; PMID:11016652 [PubMed] [Google Scholar]
- 47.Kumai T, Matsuda Y, Oikawa K, Aoki N, Kimura S, Harabuchi Y, Celis E, Kobayashi H. EGFR inhibitors augment antitumour helper T-cell responses of HER family-specific immunotherapy. Br J Cancer 2013; 109:2155-66; PMID:24045666; http://dx.doi.org/ 10.1038/bjc.2013.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumai T, Matsuda Y, Ohkuri T, Oikawa K, Ishibashi K, Aoki N, Kimura S, Harabuchi Y, Celis E, Kobayashi H. c-Met is a novel tumor associated antigen for T-cell based immunotherapy against NK/T cell lymphoma. OncoImmunology 2015; 4:e976077; PMID:25949874; http://dx.doi.org/ 10.4161/2162402X.2014.976077 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.