Abstract
Rationale
Rapid diagnosis of pulmonary tuberculosis (TB) is critical for timely initiation of treatment and interruption of transmission. Yet, despite recent advances, many patients remain undiagnosed. Culture, usually considered the most sensitive diagnostic method, is sub-optimal for paucibacillary disease.
Methods
We evaluated the Totally Optimized PCR (TOP) TB assay, a new molecular test that we hypothesize is more sensitive than culture. After pre-clinical studies, we estimated TOP’s per-patient sensitivity and specificity in a convenience sample of 261 HIV-infected pulmonary TB suspects enrolled into a TB diagnostic study in Mbarara, Uganda against MGIT culture, Xpert MTB/RIF and a composite reference standard. We validated results with a confirmatory PCR used for sequencing M. tuberculosis.
Measurements and Results
Using culture as reference, TOP had 100% sensitivity but 35% specificity. Against a composite reference standard, the sensitivity of culture (27%) and Xpert MTB/RIF (27%) was lower than TOP (99%), with similar specificity (100%, 98% and 87%, respectively). In unadjusted analyses, culture-negative/TOP-positive patients were more likely to be older (P<0·001), female (P<0·001), have salivary sputum (P = 0·05), sputum smear-negative (P<0.001) and less advanced disease on chest radiograph (P = 0.05). M. tuberculosis genotypes identified in sputum by DNA sequencing exhibit differential growth in culture.
Conclusions
These findings suggest that the TOP TB assay is accurately detecting M. tuberculosis DNA in the sputum of culture-negative tuberculosis suspects. Our results require prospective validation with clinical outcomes. If the operating characteristics of the TOP assay are confirmed in future studies, it will be justified as a “TB rule out” test.
Introduction
Despite recent advances, tuberculosis (TB) remains a major global health problem with 9 million new cases and 1.4 million deaths in 2013.[1] Critically, the global incidence is decreasing by less than 2% per year, far from the 20% decline required to reach the World Health Organization (WHO) stated goal of eliminating TB by 2050.[2, 3] Patients with pulmonary TB represent ~75% of the global disease burden and contribute exclusively to transmission. Rapid, accurate and early detection of Mycobacterium tuberculosis (MTB) in the sputum of TB suspects, and active case finding are key components of the WHO strategy.[4, 5]
For decades, the rapid diagnosis of pulmonary TB has relied on sputum acid-fast bacilli (AFB) smear microscopy but its yield is low when compared to mycobacterial culture, which is considered the most sensitive method for diagnosis.[6] Recently developed molecular tests such as Xpert® MTB/RIF and GenoType® MTBDRplus provide a rapid alternative to culture in patients with high bacterial loads (i.e. sputum AFB smear-positive). However, their overall sensitivity (~90% against culture) in programmatic conditions has been lower than initially anticipated,[7] and particularly poor (~50%) in smear-negative/culture-positive individuals.[8–10] Other TB diagnostics under development suffer the common limitation of being less sensitive than cultures.[5, 11, 12]
For definitive diagnosis, reliance on cultures as the reference method is problematic because the process of decontaminating samples prior to culture is inherently detrimental to mycobacterial viability. As a result, the overall sensitivity of cultures is only 80–85% compared to a composite reference standard,[6] but significantly lower in clinical conditions where the bacterial load in sputum is low (i.e. paucibacillary TB disease) such as certain patients with HIV-infection,[13] children,[14] and extra-pulmonary TB.[15] Other individuals with active disease harboring non-culturable organisms in sputum include subjects with unstable latent TB infection and early sub-clinical disease that have “percolating” organisms,[16] and those with old untreated TB.[17, 18] In addition, “persistent” organisms after antituberculous therapy may represent the paucibacillary TB pool for poor treatment outcomes.[17] Without culture confirmation, paucibacillary TB is rarely identified leading to empirical treatment, over- or underdiagnosis, and increased morbidity and mortality.[19]
We have developed the “Totally Optimized PCR (TOP) TB assay”, a new nucleic acid amplification test (NAAT) that utilizes a combination of efficient sample processing, novel gene target selection, modern primer design techniques, and an extended PCR for selective target isolation and amplification. The assay is highly specific for Mycobacteria in the MTB complex and therefore is not affected by background genomic noise, which enables detection with heightened sensitivity. We report here results of in silico and in vitro data. To compare accuracy of the TOP TB assay with culture and the leading molecular test (e.g. Xpert MTB/RIF), we then performed a cross-sectional evaluation using specimens from HIV-infected subjects enrolled into an existing prospective diagnostic study in Uganda.
Materials and Methods
Ethical approvals: The studies were approved by the Institutional Review Boards at Boston University Medical Center, Mbarara University of Science and Technology, and the Uganda National Council for Science and Technology. Samples were shipped to Boston under a Material Transfer Agreement for DNA sequencing.
TOP TB assay
The assay targets a gene (ponA1) involved in the assembly of peptidoglycans in the MTB bacterial wall.[20] The assay’s diagnostic primer set (3-ponA-F/R) targets sequences unique to all species in the MTB complex (Section IA, Fig A and Table A in S1 File). Amplicons generated by 3-ponA were detected using a capture-probe colorimetric assay, and the resultant Optical Densities (OD) provided a semi-quantitative measurement of MTB bacillary load.[21] A more detailed description of the TOP TB assay and its associated laboratory methods, including sample processing and DNA extraction, PCR amplification and amplicon detection are provided in S1 File (Sections IA and IB).
PCR genotyping
To establish the presence of MTB DNA, we tested all specimens with primer set 2-ponA-F/Ra (used for genotyping), which targets a section of ponA1 that is sufficiently distant (~1,100 bp) from the 3-ponA target (used for diagnosis) to remain unaffected by amplicons generated with primer 3-ponA (Section IC, Fig A and Table A in S1 File). 2-ponA PCR products were sequenced to distinguish among five possible genetic variants of MTB (genotypes 0T, 1T, 2, 3 and 4) (Section IC in S1 File). The sequencing nomenclature (Fig B in S1 File) and the genetic correspondence of 2-ponA genotypes to other familiar MTB whole genome genotyping methods are shown in the Appendix (Fig C and Table B in S1 File).
Clinical study
After completing pre-clinical studies, we tested a convenience sample of discarded sputum specimens obtained from participants enrolled into a cross-sectional TB diagnostic study in Uganda. Table C in S1 File summarizes: the study design, a description of the subjects, and methodology (including reference methods).
Setting
The study was conducted at the Epicentre/ Médecins sans Frontières Laboratory located at Mbarara University of Science and Technology in Mbarara, Uganda. With an estimated TB incidence of 166 cases per 100,000 inhabitants, Uganda is on the WHO list of high burden TB countries; the prevalence of HIV infection among TB patients is 48%.[1] Mbarara District is situated in the South Western (SW) zone of the Uganda National Tuberculosis and Leprosy Programme (NTLP). According to NTLP laboratory activity reports, 8,423 TB patients were registered in the SW zone (incidence rate 290 per 100,000). Of the 3701 TB suspects from Mbarara District, 668 (18%) were AFB smear-positive, and 68% were HIV infected.[22]
Study population
Participants for this study were enrolled into a prospective cross-sectional study designed to independently evaluate the diagnostic accuracy of a new AFB smear microscopy method [23] and Xpert MTB/RIF, with liquid media culture (manual MGIT 960) as the reference method.[24] From September 4th 2012 to April 11th 2014, the parent study enrolled 1,047 (737 HIV-infected and 310 HIV-uninfected) consecutive TB suspects admitted to the wards or attending any of the outpatient clinics of the Mbarara Regional Referral Hospital or the Municipality Health Centre in Mbarara city. Eligible participants were adult (≥18 years), TB suspects (≥2 weeks of cough + at least one other symptoms of TB) [25] willing to follow the study protocol. Patients were excluded if they had received antituberculous drugs within three days, were too ill to consent, or presented with disseminated or extra-pulmonary TB without cough.
Study design and measurements
Participants had a standardized TB evaluation, HIV testing and provided three spontaneously expectorated (≥2 mL) sputum samples that included one early morning and two spot samples in a 24-hr period. One of the spot samples (selected by randomization) [24] was tested for direct AFB smear, Xpert MTB/RIF and culture.
Sample handling prior to TOP processing and testing
Specimens from the last 261 HIV-infected participants enrolled into the parent study were available for this study. TOP testing was done on the discarded portion of a pellet processed for Xpert MTB/RIF. A ~1mL aliquot was frozen at -80°C for two to six months prior to TOP testing; after thawing, the pellet was washed to remove N-acetyl-L-cysteine / sodium hydroxide solution, [26] processed for TOP and tested in a single batch at the Epicentre laboratory in Mbarara. Study personnel were blind to routine TB results; coded results were later linked via a study identification number.
Standard Laboratory Methods
The Epicentre/ Médecins sans Frontières Laboratory has quality assurance (QA) and quality control (QC) protocols, and well-trained personnel with extensive experience in laboratory based TB research. The appearance of sputa specimens was classified as purulent, mucopurulent, mucosalivary or, salivary by the microbiology technicians according to international laboratory guidelines.[27] We used the light-emitting diode-auramine fluorescence technique (FluorescenS® LED system, Bergman Labora, Danderyd, Sweden) for direct AFB microscopy on each specimen and reported the results according to the WHO grading scale.[28] The specimen was then decontaminated using the N-acetyl-L-cysteine (0.5%) / sodium hydroxide (1.5%) method.[26] For the reference culture method, we inoculated 500 μl into one manual-testing MGIT 960 (Becton, Dickinson, Franklin Lakes, NJ). We reported a negative culture result after 56 days of incubation at 37°C. Contamination in MGIT media was ruled out using Ziehl-Neelsen (ZN) microscopy and culture on blood agar. For all positive MGIT cultures, we differentiated between M. tuberculosis and non-tuberculous mycobacteria (NTM) using the SD TB Ag MPT64 Rapid system (SD Bioline, Kyongi-do, South Korea), following the manufacturer’s instructions. The GenoType Mycobacterium CM/AS identification kit (Hain Lifescience, Nehren, Germany) was used for identification of NTM. The Xpert® MTB/RIF assay (Cepheid, Sunnyvale, CA, U.S.A.) was performed according to the manufacturer’s instructions.
Analytical strategy
We report the results according to the Standards for Reporting of Diagnostic Accuracy (STARD) guidelines.[29, 30] We calculated the diagnostic cut-off for TOP OD using a cut-off value of three standard deviations above the mean of the OD values of negative controls (e.g. laboratory cut-off). [31] As a sensitivity analysis, we used receiver operating curve (ROC) tools to determine the cut-off that simultaneously maximized sensitivity and specificity (e.g. ROC cut-off). We estimated per-patient sensitivity and specificity using culture as the reference standard, using all available results to adjudicate TB status. We also estimated per-patient sensitivity and specificity using a Composite Reference Standard (CRS) that included culture, MTB sequencing (e.g. 2-ponA genotyping), a NAAT other than TOP (e.g. Xpert MTB/RIF) and AFB smear, as described. [15, 32–34] We analyzed patient characteristics according to TOP and culture results using Kruskal-Wallis (for continuous data) and Fisher’s exact test (for categorical data), and compared groups using Wilcoxon and Fisher’s exact tests. Variables with p< 0.1 and those considered to be clinically significant were included in multivariate logistic and ordinal logistic regression models. In the former we consider correlates with culture+/TOP+ compared to culture-/TOP+ individuals. In the latter, we compare all three outcomes ordinally. For these models we group X-ray results into two categories: Normal/Minimal versus Moderate/Far Advanced. The models controlled for age, sex, previous TB treatment, sputum appearance, sputum volume (only for first model), and X-ray (2 category).
Results
Preclinical studies
In the preclinical phase, TOP TB’s primer set 3-ponA (used for diagnosis) demonstrated: i) excellent analytical sensitivity when clinical sputum samples were “spiked” with Mycobacterium bovis Bacille Calmette-Guérin (Fig D, top in S1 File); ii) semi-quantitative detection capability over a range of MTB loads (Fig D, bottom in S1 File); iii) high analytical specificity, testing negative against a panel of 18 common respiratory bacteria and other microorganisms (Fig E in S1 File), and; iv) high specificity against non-tuberculous mycobacteria (Fig F in S1 File). The 2-ponA primer set (used for sequencing) demonstrated a ~8–10% lower analytical sensitivity but similar analytical specificity in the preclinical phase of testing (data not shown).
Clinical study
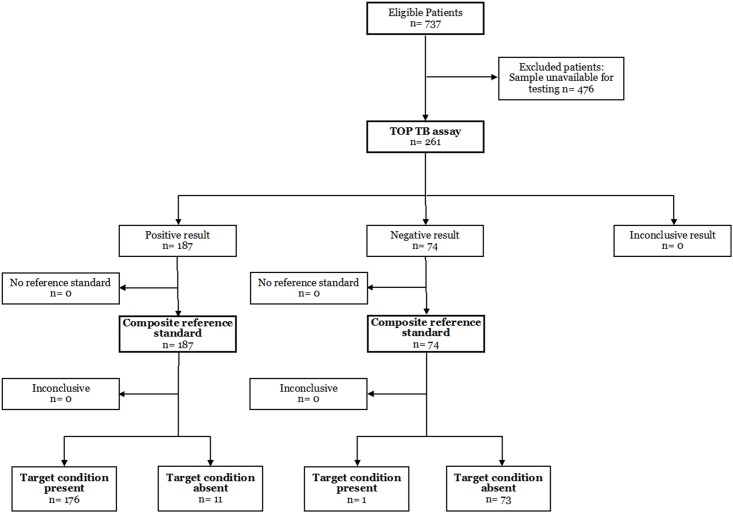
We then evaluated the TOP TB assay in 261 HIV-infected pulmonary TB suspects enrolled into the parent study between October 2, 2013 and April 11, 2014 (Fig 1). Table 1 shows characteristics of the study cohort according to culture and TOP TB assay results.
Fig 1. Study profile.
Table 1. Characteristics of 261 HIV-infected pulmonary tuberculosis suspects in Mbarara, Uganda by M. tuberculosis culture and TOP TB assay results.
| Characteristic | Overall | Culture positive | Culture negative | P value | ||
|---|---|---|---|---|---|---|
| TOP positive | TOP positive | TOP negative | Overall1 | Two-way2 | ||
| N | 261 | 48 | 139 | 74 | ||
| Age (years) | 39.0 [30.5–47.0] | 33.5 [28.0–40.0] | 42.0 [33.0–49.0] | 38.5 [31.0–48.8] | 0.002 | <0.001 |
| Female sex | 138 (53) | 12 (25) | 78 (56) | 48 (65) | <0.001 | <0.001 |
| Previous TB treatment* | 33 (13) | 3 (6) | 23 (17) | 7 (9) | 0.04+ | 0.05+ |
| Years since previous TB treatment | 9.3 [5.9–10.9] | 5.4 | 9.3 [5.9–10.9] | 11.9 [10.3–12.0] | 0.28# | 0.16# |
| N = 13 | N = 1 | N = 9 | N = 3 | |||
| CD4 (cells/mL)* | 322 [104–495] | 182 [54–338] | 343.5 [93–457] | 355 [158–590] | 0.06 | 0.13 |
| N = 172 | N = 22 | N = 92 | N = 58 | |||
| Sputum volume (mL) | 3 [2–5] | 4 [3–5] | 3 [2–4.5] | 3 [2–5] | 0.33# | 0.16# |
| Sputum appearance^ | <0.001+ | 0.05+ | ||||
| Purulent | 69 (27) | 20 (42) | 36 (26) | 13 (18) | ||
| Mucoid | 36 (13) | 10 (25) | 23 (17) | 3 (4) | ||
| Salivary | 156 (60) | 18 (33) | 80 (58) | 58 (78) | ||
| Chest radiograph* | 0.04+ | 0.05+ | ||||
| Normal | 29/103 (28) | 2/17 (12) | 14/47 (30) | 13/39 (33) | ||
| Minimal | 17/103 (17) | 1/17 (6) | 8/47 (17) | 8/39 (21) | ||
| Moderate | 46/103 (45) | 8/17 (47) | 21/47 (45) | 17/39 (44) | ||
| Far advanced | 11/103 (11) | 6/17 (35) | 4/47 (9) | 1/39 (3) | ||
| Cavitation present | 18/104 (17) | 5/17 (29) | 7/48 (15) | 6/39 (15) | 0.37+ | 0.27+ |
| Sputum AFB smear* | − | <0.001+ | ||||
| Negative | 222 (85) | 11 (23) | 137 (99) | 74 (100) | ||
| Scanty | 9 (3) | 8 (17) | 1 (1) | 0 (0) | ||
| 1+ | 10 (4) | 10 (21) | 0 (0) | 0 (0) | ||
| 2+ | 7 (3) | 6 (13) | 1 (1) | 0 (0) | ||
| 3+ | 13 (5) | 13 (27) | 0 (0) | 0 (0) | ||
| Sputum MGIT culture | ||||||
| Positive | 48 (18) | 48 (100) | 0/139 (0) | 0/74 (0) | - | - |
| Contaminated | 12 (5) | − | 2/139 (1) | 10/74 (14) | ||
| MGIT DTP (days) | 19 [13–33] | 19 [13–33] | NA | NA | - | |
| Xpert Mtb/RIF * | ||||||
| Positive | 50/259 (19) | 45/47 (96) | 3/139 (2) | 2/73 (3) | <0.001+ | <0.001+ |
| Indeterminate | 4/259 (2) | 1/47 (2) | 1/139 (1) | 2/73 (3) | ||
| M. tuberculosis 2-ponA genotype | ||||||
| 0T | 5 (2) | 4 (8) | 1 (1) | 0 | - | 0.005+ |
| 1T | 6 (2) | 3 (6) | 3 (2) | 0 | ||
| 1 | 2 (1) | 0 | 2 (1) | 0 | ||
| 2 | 40 (15) | 16 (33) | 24 (17) | 0 | ||
| 3 | 93 (36) | 18 (38) | 74 (53) | 1 (1) | ||
| 4 | 27 (10) | 3 (6) | 24 (17) | 0 | ||
| Neg | 88 (34) | 4 (8) | 11 (8) | 73 (99) | ||
Values are median [interquartile range] or number (percentage), unless otherwise specified
MGIT = Mycobacterial Growth Indicator Index (BACTEC 960, Becton Dickinson, U.S.A.); DTP = Days-to-positive; AFB = Acid-fast bacilli
1 P overall = Comparison between three groups
2 P two-way = Comparison between culture-positive/ TOP-positive vs. culture-negative/ TOP-positive
* Missing information: Previous TB treatment (1); CD4 cell count (85); Time since previous TB treatment (20); Chest X-ray extent of disease (154), cavitation (153); Sputum AFB smear (1); Xpert MTB/RIF (2)
^ Purulent sputum category includes purulent and muco-purulent; Mucoid category includes mucoid and muco-salivary
+ Fisher’s exact test.
# Kruskal-Wallis test or Wilcoxon test.
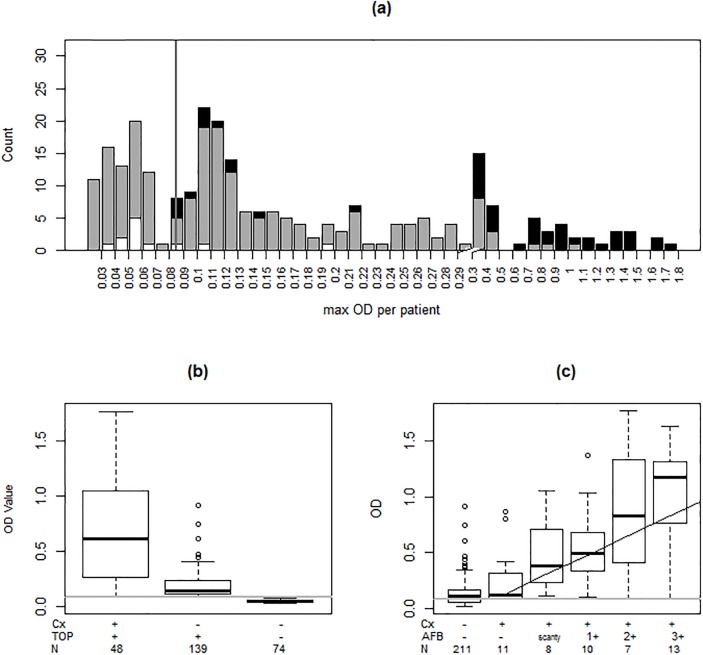
As shown in Fig 2, 48/261 (18%) patients were culture-positive, all of which were also TOP-positive. Seventy-four (28%) were culture-negative (N = 64) or contaminated (N = 10) and were TOP-negative; 139 (53%) were culture-negative (N = 137) or contaminated (N = 2) but TOP-positive (Fig 2a). The distribution of TOP ODs by culture and AFB smear are shown in Fig 2b and 2c, respectively. Of the 139 culture-negative/ TOP-positive samples, 2-ponA sequencing confirmed the presence of MTB DNA in 128 (92%). The sensitivity and specificity of TOP and Xpert MTB/RIF compared to culture or a CRS are shown in Table 2; the breakdown of results included in the CRS is shown in Table D in S1 File. We were unable to sequence MTB from 11/139 (8%) culture-negative/ TOP-positive specimens with low TOP OD values (median 0.13, IQR 0.11–0.34).
Fig 2. TOP TB assay results in 261 HIV-infected TB suspects from Mbarara, Uganda.
(a) Vertical line denotes the laboratory cut-off TOP OD (0.0854) for a positive test. Histograms represent the number of subjects with culture-positive (black), culture-contaminated (white) and culture-negative (grey) results, by TOP OD values. The X-axis is zoomed-in at the lower end of TOP OD values (0.100 to 0.300) to show the large number of subjects in this section of the graph. (b) Group TOP OD values according to culture (Cx) and TOP results (group means are 0.67, 0.19, and 0.05, from left to right). The mean TOP OD of culture-positive/TOP-positive (0.67) samples was higher than in culture-negative/TOP-positive (0.19, P<0.0001), suggesting a low bacterial load content in many HIV-infected TB suspects. (c) Median TOP ODs paralleled sputum AFB grades (P<0.0001), demonstrating the semi-quantitative performance of the TOP TB assay. One subject with a scanty AFB reading and a contaminated culture was excluded (TOP OD 0.103). A smoothing spline fit to the data is shown.
Table 2. Per-patient sensitivity and specificity of the TOP TB assay, Xpert MTB/RIF and culture in 261 HIV-infected tuberculosis suspects according to a reference standard established by M. tuberculosis culture or a Composite Reference Standard (CRS) in Mbarara, Uganda.
| Diagnostic Method | MTB detected (N) | MTB not detected (N) | Sensitivity | Specificity | PPV | NPV | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| n/N | % (95% CI) | n/N | % (95% CI) | n/N | % (95% CI) | n/N | % (95% CI) | |||
| 48 | 2131 | Culture reference standard | ||||||||
| Xpert MTB/RIF 2 | 50 | 209 | 45/47 | 96% (84, 99) | 207/212 | 98% (94, 99) | 45/50 | 90% (77, 96) | 207/209 | 99% (96, 100) |
| TOP TB assay | 187 | 74 | 48/48 | 100% (93, 100) | 74/213 | 35% (28, 41) | 48/137 | 26% (20, 33) | 74/74 | 100% (95, 100) |
| 177 | 84 | Composite reference standard 3 | ||||||||
| Culture | 48 | 213 | 48/177 | 27% (21, 34) | 84/84 | 100% (96, 100) | 48/48 | 100% (93, 100) | 84/211 | 40% (33, 47) |
| Xpert MTB/RIF | 50 | 209 | 48/176 | 27% (21, 35) | 81/83 | 98% (91, 100) | 48/50 | 96% (85, 99) | 81/209 | 39% (32, 46) |
| TOP TB assay | 187 | 74 | 176/177 | 99% (97, 100) | 73/84 | 87% (77, 93) | 176/187 | 94% (89, 97) | 73/74 | 99% (93, 100) |
Definition of abbreviations: CI = Confidence interval; CRS = Composite reference standard; MTB = Mycobacterium tuberculosis; NPV = Negative predictive value; PPV = Positive predictive value
1 Includes 12 patients with contaminated culture results
2 Two Xpert MTB/RIF results were missing and 4 had indeterminate result (N = 259)
3 Composite Reference Standard (CRS) included M. tuberculosis culture, M. tuberculosis sequencing (e.g. 2-ponA genotyping), a NAAT other than TOP (e.g. Xpert MTB/RIF), and AFB smear.[15] The breakdown of CRS results is shown in Table S4 (Appendix)
In univariate analyses that compared culture-positive/TOP-positive vs. culture-negative/ TOP-positive patients (Table 1), the latter were more likely to be older (P<0·001); women (P<0·001); have a salivary sputum (P = 0·05); have a previous history of TB disease (P = 0·05), and have early TB disease as measured by sputum AFB smear grade (P<0·001) and chest radiograph (P = 0·05). In a multivariate analysis comparing culture+/TOP+ to culture-/TOP+ patients, age (p = 0.003) and gender (p = 0.002) remained statistically significant. In a comparison of all three TOP/culture categories from Table 1, multivariate results revealed that age (p = 0.02) and gender (p<0.001) were statistically significant and previous TB treatment was marginally significant (p = 0.10).
The cut-off for TOP OD values was determined to be 0.0854 using the laboratory criterion (e.g. +/- three standard deviations criterion). When we used ROC analysis with 100 random observations, we found the cut-off to be 0.088 leading to reclassification of only 4 individuals (Table E in S1 File). The area under the ROC curve was 0.86 for culture and 0.95 for the CRS using TOP OD as the diagnostic test (Fig G in S1 File).
M. tuberculosis sequencing results
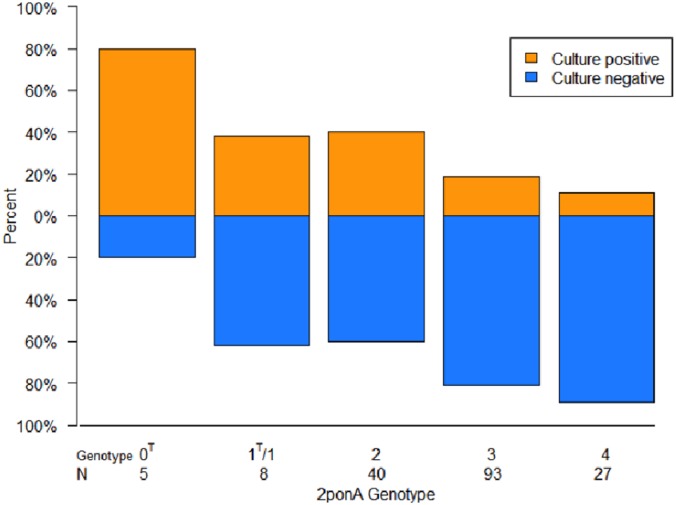
The relative frequency and distribution of 2-ponA genotypes differed significantly according to TOP OD values (Fig 3 and Table 2; P = 0·005); in particular, genotype 4 strains were mostly restricted to culture-negative samples with low ODs (Fig 3). As shown in Fig 4, 2-ponA genotypes had variable growth in culture (P = 0·002).
Fig 3. Distribution of 2-ponA genotypes (sequencing results) in 261 HIV-infected pulmonary TB suspects in Mbarara, Uganda according to TOP TB assay OD values.
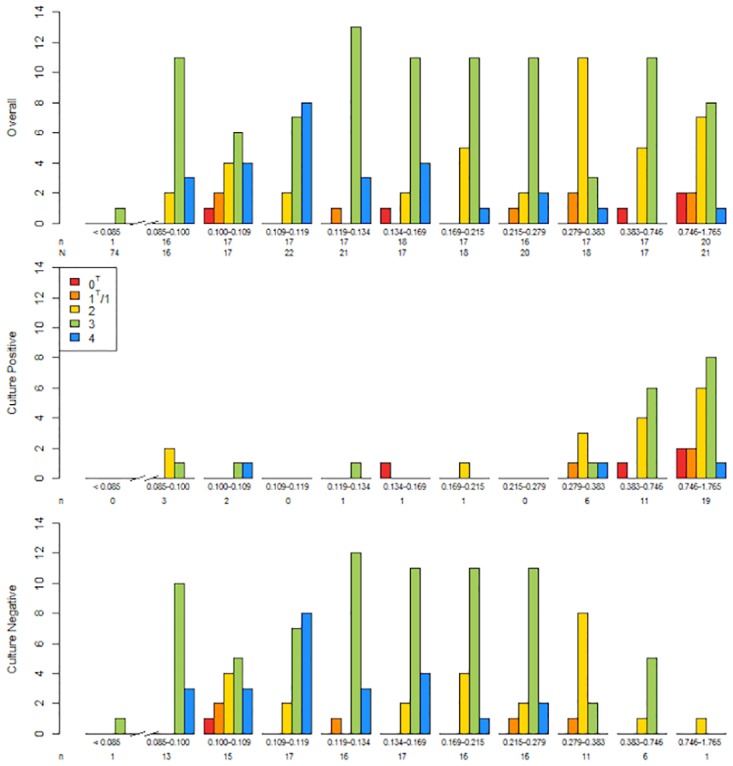
Results are shown for all subjects (top), and then separated by those that were culture-positive (middle) and culture-negative (bottom). The X-axis is divided into groups of subjects with similar bacterial loads as measured by TOP OD values. The first group (far left) includes 74 subjects that were culture-negative and TOP-negative (3-ponA primer). N denotes the number of subjects in each TOP OD group; n denotes the number of subjects with a positive 2-ponA genotype in each group. A 2-ponA genotype could not be identified in 8% (4/48) culture-positive/TOP-positive samples and 8% (11/139) culture-negative/TOP-positive samples. A 2-ponA genotype was identified in 1% (1/74) of culture-negative/TOP-negative samples.
Fig 4. Distribution of 2-ponA genotypes by culture results.
Each histogram represents 100% of strains for each 2-ponA genotype. N denotes the number of strains in each group. The proportion of samples that were culture-positive decreased with the number of Proline codon deletions (e.g. Genotype 4 = 4 Proline deletions) in the poly-Proline track in the ponA1 region targeted by the 2-ponA primer (see Fig B in S1 File) (P = 0.002).
Discussion
Our study provides strong evidence that the TOP TB assay accurately detects trace amounts of MTB DNA in the sputum of HIV-infected TB suspects who otherwise may yield a negative culture. Currently a culture diagnosis is the optimal reference standard for diagnosis. In the absence of culture to determine specificity, we established the validity of positive diagnosis using a composite reference standard and, most importantly, we sequenced MTB from culture-negative specimens. We used a reproducible genotyping method that is supported by the genetic signature of a global collection of MTB clinical isolates representing all major phylogenetic lineages.
The natural history of pulmonary TB in HIV-uninfected adults is traditionally viewed as a sub-acute or chronic illness whose progression is accompanied by increasing bacterial loads in sputum, paralleling worsening disease severity on chest radiography.[4, 6] The paradigm states that patients with early TB are often smear- and culture-negative, and with an increase in severity, most patients become smear-negative/culture-positive first, and eventually, smear- and culture-positive. However, a variety of epidemiologic studies including household contact investigations, molecular epidemiology and TB screening studies have demonstrated that the rate of disease progression in humans can be highly variable, perhaps as a consequence of low or stagnant MTB bacterial loads in sputum.[4, 35, 36] Furthermore, effective diagnostic and treatment programs that seek out cases to identify patients with most advanced disease (i.e. AFB smear- and culture-positive) may produce an epidemiological shift that results in the remaining populations with suspected TB of having a higher prevalence of early TB disease (i.e. smear-negative/culture-negative), that are the most difficult to confirm bacteriologically.[4] For example, in the U.S. during the 1980s, 90% of TB cases were confirmed by culture but this proportion decreased to 77% by 2013; [37] in some settings (e.g. Boston, MA and Alberta, Canada), ~50% of notified TB cases are culture-negative.[38, 39] Therefore, culture-negative TB disease is a global problem resulting from both biological and epidemiological factors, for which there are currently limited solutions beyond the initiation of empirical antituberculous treatment based on clinical algorithms.[40]
TOP TB enables enhanced detection of MTB in the sputum of TB suspects with HIV/AIDS, perhaps one of the largest and most vulnerable (together with children) populations with paucibacillary TB disease. In the early phase of assay development, our in vitro results suggested an analytical sensitivity of 1–4 colony-forming units (CFU) of MTB per mL, a level of detection greater than culture (e.g. 10–100 CFU/ml). With nullification of culture as the reference method, we anticipated challenges in validating the accuracy of our results. Several analytical methods have been recommended when dealing with imperfect reference methods such as mycobacterial cultures.[15, 32, 33, 41] The use of one of these—“discrepant analysis”, or the use of a third test, is limiting because usually it is not applied consistently across all the specimens that are being examined, only the ones where the new test result conflicts with the “gold standard”.[32, 33, 41] Our methods minimize the limitations of using discrepant analysis to evaluate NAATs because we tested all specimens, and because of the complete lack of overlap (lack of ‘dependence’) between the 3-ponA (diagnostic) and 2-ponA (genotyping) primers. Furthermore, the use of both clinical and epidemiologic data to act further as a referee, the latter demonstrating variability of genotypes according to TOP ODs add strength to the interpretation of a positive TOP in the face of a negative culture. Our results may be novel in the TB diagnostic field but the development of molecular tests with sensitivity superior to culture is not new in clinical settings.[42] Based on our results, the inclusion of 2-ponA genotypes into a composite reference standard to evaluate the performance of TOP follows standard practices in the TB diagnostic field [5, 15, 34], and beyond. [43] Importantly, because the sensitivity of the primer used for sequencing is ~8–10% lower that the diagnostic primer, 11/139 (8%) culture-negative/ TOP-positive specimens were adjudicated as false-positive TOP results, lowering the specificity of the assay in this study. In other ongoing studies with non-HIV sputum samples, the specificity of TOP has been 93% to 100% (data not shown).
Our results suggest that the TOP assay is sufficiently sensitive to overcome well-recognized difficulties with sputum procurement, such as inadequate specimen volumes and/or poor quality of specimens (e.g. excess saliva)–a problem that is thought to diminish diagnosis of TB in women disproportionately.[44–47] This also raises the possibility of using sputum to diagnose paucibacillary TB when non-pulmonary clinical specimens (blood, gastric aspirates, urine, stool) have otherwise been thought necessary to establish the diagnosis.[5, 12, 48] Interestingly, our sequencing data establish a potential link between diagnosis, epidemiology and pathogenic behavior of MTB in humans. In particular, a high frequency of genotype 4 MTB isolates was noted in HIV-infected patients with low TOP ODs in Uganda. We had rarely observed this variant in the global collection of clinical isolates before these studies were started, raising the possibility that genotype 4 strains are uniquely adapted to HIV-infected hosts and cause predominantly culture-negative TB disease.
Our study has limitations. Our results were obtained by testing a convenience sample of low-volume, discarded, stored sputum specimens from an existing diagnostic clinical study; therefore, performance of the TOP assay may have been underestimated. The selection of the study population was solely based on when the TOP TB assay was ready for clinical testing (October 2013) rather than selection bias, as shown by the results of the parent study that included the entire population of HIV-infected patients. [24] The lack of clinical follow-up of subjects limits the clinical interpretation of certain results. For example, TOP was positive in numerous patients with a current or past history of treatment for TB, which complicated the clinical interpretation of negative cultures; interestingly, a similar phenomenon has recently been described with Xpert MTB/RIF, although a discrepant analysis was not performed.[49, 50] A positive TOP result likely represented either residual (dead) MTB DNA, viable but non-culturable organisms or bacterial persistence after treatment, the latter a potential harbinger of TB recurrence and/or risk of drug resistance.[17, 51, 52] Admittedly, detection of trace amounts of MTB DNA may be due to bacterial “spillage” from a dormant lung foci or low level bacterial replication that may not require treatment. The colorimetric readout uses a study-specific cut-off value to establish the “Limit of Blank”, a key assay parameter.[31] Finally, in its current embodiment, the TOP TB assay does not include provisions for detecting drug-resistant TB. However, the primary global need is for a rapid and reliable triage test.[5, 12]
Conclusions
Culture-negative TB is widespread, resulting from several biologic and epidemiologic factors. By shifting diagnostic emphasis to early detection, the TOP TB assay broadens sensitive and accurate detection of MTB across the entire clinical spectrum of TB disease. Our findings will require validation with clinical outcomes obtained prospectively. If the operating characteristics of the TOP assay are confirmed in future studies, it would be justified as a triage or “TB rule out” test.
Supporting Information
- Experimental Laboratory methods
- Technical aspects of TOP TB assay
- TOP TB assay laboratory methods
- M. tuberculosis sequencing and genotype nomenclature
- Supporting Figures
- Fig A: DNA sequence alignment of M. tuberculosis ponA1
- Fig B: Sequence representation of five possible 2-PonA genotypes of M. tuberculosis clinical isolates
- Fig C: Genetic correspondence of M. tuberculosis 2-ponA genotypes with other common whole-genome genotyping methods
- Fig D: Analytical sensitivity of 3-ponA primer
- Fig E: Analytical specificity of 3-ponA primer
- Fig F: Specificity of TOP TB assay against non-tuberculous mycobacteria
- Fig G: Receiver operating curve (ROC) analysis of TOP TB assay results according to culture or composite reference standard
- Supporting Tables
- Table A: Description of 3-ponA and 2-ponA primers
- Table B: Genetic correspondence of M. tuberculosis 2-ponA genotypes with other common whole-genome genotyping methods
- Table C: Summary of Uganda clinical study
- Table D: Breakdown of results for the Composite Reference Standard
- Table E: Sensitivity analysis using TOP TB assay cut-off determined by ROC analysis
- References
(DOC)
Acknowledgments
The authors would like to acknowledge Dr. Joel Bazira, MBChB, MMED, PhD, Head Department of Microbiology Faculty of Medicine Mbarara University of Science and Technology for his support. We thank personnel at Boston Medical Center’s offices of Development (Kirsten Hinsdale and Hugh Keeping) and Boston University’ Office of Technology Development (Michael Pratt) for their continued support. We also wish to thank Kathy Eisenach, PhD and Kevin Fennelly, MD, MPH for their critical input and reviewing the manuscript, and David Hamer, MD, Tamar Barlam, MD, Peter Rice, MD, Jerrold Ellner, MD and Robert Wilkinson, MD for reviewing the manuscript.
The results shown here were presented in part at the 1st Massachusetts-South Africa Conference for Technology Transfer & Global Innovation, Stellenbosch, South Africa 8–11 June 2015.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by internal funds, The Uganda Research Student Support Fund (URSSF), and in part by Thisis Diagnostics, Inc. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. Global Tuberculosis Report 2014. Geneva, Switzerland: World Health Organization, 2014. Contract No.: WHO/HTM/TB/2014.08. [Google Scholar]
- 2.WHO. The End TB Strategy Global strategy and targets for tuberculosis prevention, care and control after 2015. Geneva: World Health Organization, 2014. [Google Scholar]
- 3.Lonnroth K, Castro KG, Chakaya JM, Chauhan LS, Floyd K, Glaziou P, et al. Tuberculosis control and elimination 2010–50: cure, care, and social development. Lancet. 2010;375(9728):1814–29. Epub 2010/05/22. 10.1016/S0140-6736(10)60483-7 . [DOI] [PubMed] [Google Scholar]
- 4.WHO. Systematic screening for active tuberculosis: principles and recommendations. Geneva, Switzerland: World Health Organization, 2013. Contract No.: WHO/HTM/TB/2013.04. [PubMed] [Google Scholar]
- 5.Pai M, Schito M. Tuberculosis Diagnostics in 2015: Landscape, Priorities, Needs, and Prospects. J Infect Dis. 2015;211(suppl 2):S21–S8. Epub 2015/03/15. 10.1093/infdis/jiu803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ATS/CDC/IDSA. Diagnostic standards and classification of tuberculosis in adults and children. Am J Respir Crit Care Med. 2000;161:1376–95. [DOI] [PubMed] [Google Scholar]
- 7.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363(11):1005–15. Epub 2010/09/10. 10.1056/NEJMoa0907847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, Dendukuri N. Xpert(R) MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2014;1:CD009593 Epub 2014/01/23. 10.1002/14651858.CD009593.pub3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang K, Lu W, Wang J, Zhang K, Jia S, Li F, et al. Rapid and effective diagnosis of tuberculosis and rifampicin resistance with Xpert MTB/RIF assay: a meta-analysis. J Infect. 2012;64(6):580–8. Epub 2012/03/03. 10.1016/j.jinf.2012.02.012 . [DOI] [PubMed] [Google Scholar]
- 10.Barnard M, Gey van Pittius NC, van Helden PD, Bosman M, Coetzee G, Warren RM. The Diagnostic Performance of the GenoType MTBDRplus Version 2 Line Probe Assay Is Equivalent to That of the Xpert MTB/RIF Assay. J Clin Microbiol. 2012;50(11):3712–6. Epub 2012/09/14. 10.1128/JCM.01958-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.UNITAID. Tuberculosis Diagnostics Technology and Market Landscape. 4th Edition 2015. Switzerland: World Health Organization, 2015. [Google Scholar]
- 12.Dorman S. Advances in the diagnosis of tuberculosis: current status and future prospects. Int J Tuberc Lung Dis. 2015;19(5):504–16. Epub 2015/04/14. 10.5588/ijtld.15.0048 . [DOI] [PubMed] [Google Scholar]
- 13.Getahun H, Harrington M, O'Brien R, Nunn P. Diagnosis of smear-negative pulmonary tuberculosis in people with HIV infection or AIDS in resource-constrained settings: informing urgent policy changes. Lancet. 2007;369(9578):2042–9. Epub 2007/06/19. 10.1016/S0140-6736(07)60284-0 . [DOI] [PubMed] [Google Scholar]
- 14.Perez-Velez CM, Marais BJ. Tuberculosis in children. N Engl J Med. 2012;367(4):348–61. Epub 2012/07/27. 10.1056/NEJMra1008049 . [DOI] [PubMed] [Google Scholar]
- 15.Denkinger CM, Schumacher SG, Boehme CC, Dendukuri N, Pai M, Steingart KR. Xpert MTB/RIF assay for the diagnosis of extrapulmonary tuberculosis: a systematic review and meta-analysis. Eur Respir J. 2014;44(2):435–46. Epub 2014/04/04. 10.1183/09031936.00007814 . [DOI] [PubMed] [Google Scholar]
- 16.Esmail H, Barry CE 3rd, Young DB, Wilkinson RJ. The ongoing challenge of latent tuberculosis. Philos Trans R Soc Lond B Biol Sci. 2014;369(1645):20130437 Epub 2014/05/14. 10.1098/rstb.2013.0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salgame P, Geadas C, Collins L, Jones-Lopez E, Ellner JJ. Latent tuberculosis infection—Revisiting and revising concepts. Tuberculosis (Edinb). 2015;95(4):373–84. 10.1016/j.tube.2015.04.003 . [DOI] [PubMed] [Google Scholar]
- 18.Stead WW. Pathogenesis of the sporadic case of tuberculosis. N Engl J Med. 1967;277(19):1008–12. Epub 1967/11/09. 10.1056/NEJM196711092771906 . [DOI] [PubMed] [Google Scholar]
- 19.Sterling T JC, Jayathilake K, Gotuzzo E, Veloso V, Cortes C, Padgett D, Crabtree-Ramirez B, Shepherd B, McGowan C., editor Culture-negative TB is associated with increased mortality in HIV-infected persons. Conference on Retroviruses and Opportunistic Infections; 2015. February 23–26, 2015; Seattle, Washington. [Google Scholar]
- 20.Kieser KJ, Boutte CC, Kester JC, Baer CE, Barczak AK, Meniche X, et al. Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria. PLoS Pathog. 2015;11(6):e1005010 10.1371/journal.ppat.1005010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denis M, Soumet C, Legeay O, Arnauld C, Bounaix S, Thiery R, et al. Development of a semiquantitative PCR assay using internal standard and colorimetric detection on microwell plate for pseudorabies virus. Mol Cell Probes. 1997;11(6):439–48. Epub 1998/03/17. 10.1006/mcpr.1997.0139 . [DOI] [PubMed] [Google Scholar]
- 22.Ministry of Health RoU. National tuberculosis and leprosy programme, South Western Uganda annual report. 2008.
- 23.Fennelly KP, Morais CG, Hadad DJ, Vinhas S, Dietze R, Palaci M. The small membrane filter method of microscopy to diagnose pulmonary tuberculosis. J Clin Microbiol. 2012;50(6):2096–9. Epub 2012/03/17. 10.1128/JCM.00572-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boum Y 2nd, Kim S, Orikiriza P, Acuna-Villaorduna C, Vinhas S, Bonnet M, et al. Diagnostic Accuracy of the Small Membrane Filtration Method for Diagnosis of Pulmonary Tuberculosis in a High HIV Prevalence Setting. J Clin Microbiol. 2016. 10.1128/JCM.00017-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.WHO. Treatment of tuberculosis guidelines. 2010. [PubMed]
- 26.Kent PT, Kubica GP. Public Health Mycobacteriology—A Guide for the Level III Laboratory. Atlanta, GA: U. S. Department of Health and Human Services, Public Health Service, Centers for Disease Control, 1985. [Google Scholar]
- 27.Rieder HL C T, Myking H, Urbanczik R, Laszlo A, Kim SJ, Van Deun A T A. The public health service national tuberculosis reference laboratory and the national laboratory network: minimum requirements, role and operation in a low-income country. Paris, France: International Union Against Tuberculosis and Lung Disease, 1998. [Google Scholar]
- 28.WHO. Strategic and Technical Advisory Group for Tuberculosis: report on conclusions and recommendations. Geneva, Switzerland: World Health Organization, 2009. [Google Scholar]
- 29.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: The STARD Initiative. Ann Intern Med. 2003;138(1):40–4. Epub 2003/01/07. . [DOI] [PubMed] [Google Scholar]
- 30.Fontela PS, Pant Pai N, Schiller I, Dendukuri N, Ramsay A, Pai M. Quality and reporting of diagnostic accuracy studies in TB, HIV and malaria: evaluation using QUADAS and STARD standards. PLoS One. 2009;4(11):e7753 10.1371/journal.pone.0007753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29 Suppl 1:S49–52. Epub 2008/10/15. [PMC free article] [PubMed] [Google Scholar]
- 32.Dendukuri N. Evaluating Diagnostic Tests in the Absence of a Gold Standard. Advanced TB diagnostics course, Montreal, July 2011. [Google Scholar]
- 33.Reitsma JB, Rutjes AW, Khan KS, Coomarasamy A, Bossuyt PM. A review of solutions for diagnostic accuracy studies with an imperfect or missing reference standard. J Clin Epidemiol. 2009;62(8):797–806. Epub 2009/05/19. 10.1016/j.jclinepi.2009.02.005 . [DOI] [PubMed] [Google Scholar]
- 34.Naaktgeboren CA, Bertens LC, van Smeden M, de Groot JA, Moons KG, Reitsma JB. Value of composite reference standards in diagnostic research. BMJ. 2013;347:f5605 10.1136/bmj.f5605 . [DOI] [PubMed] [Google Scholar]
- 35.Guwatudde D, Nakakeeto M, Jones-Lopez EC, Maganda A, Chiunda A, Mugerwa RD, et al. Tuberculosis in household contacts of infectious cases in Kampala, Uganda. Am J Epidemiol. 2003;158(9):887–98. Epub 2003/10/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sloot R, Schim van der Loeff MF, Kouw PM, Borgdorff MW. Risk of tuberculosis after recent exposure. A 10-year follow-up study of contacts in Amsterdam. Am J Respir Crit Care Med. 2014;190(9):1044–52. Epub 2014/09/30. 10.1164/rccm.201406-1159OC . [DOI] [PubMed] [Google Scholar]
- 37.CDC. Reported Tuberculosis in the United Sates, 2013. Atlanta, GA, U.S: In: Department of Health and Human Services C, editor. Atlanta: 2014. [Google Scholar]
- 38.BPHC. Tuberculosis Impact in Boston Residents: 2013. In: Commission. BPH, editor. Boston: 2013. [Google Scholar]
- 39.Health A. Tuberculosis in Alberta Surveillance Report (2010 to 2012). In: Health OotCMOo, editor. 2014.
- 40.Lawn SD, Ayles H, Egwaga S, Williams B, Mukadi YD, Santos Filho ED, et al. Potential utility of empirical tuberculosis treatment for HIV-infected patients with advanced immunodeficiency in high TB-HIV burden settings. Int J Tuberc Lung Dis. 2011;15(3):287–95. . [PubMed] [Google Scholar]
- 41.Hadgu A, Dendukuri N, Hilden J. Evaluation of nucleic acid amplification tests in the absence of a perfect gold-standard test: a review of the statistical and epidemiologic issues. Epidemiology. 2005;16(5):604–12. Epub 2005/09/02. . [DOI] [PubMed] [Google Scholar]
- 42.Caliendo AM, Gilbert DN, Ginocchio CC, Hanson KE, May L, Quinn TC, et al. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis. 2013;57 Suppl 3:S139–70. 10.1093/cid/cit578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salimnia H, Fairfax MR, Lephart PR, Schreckenberger P, DesJarlais SM, Johnson JK, et al. Evaluation of the FilmArray Blood Culture Identification Panel: Results of a Multicenter Controlled Trial. J Clin Microbiol. 2016;54(3):687–98. 10.1128/JCM.01679-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan MS, Dar O, Sismanidis C, Shah K, Godfrey-Faussett P. Improvement of tuberculosis case detection and reduction of discrepancies between men and women by simple sputum-submission instructions: a pragmatic randomised controlled trial. Lancet. 2007;369(9577):1955–60. Epub 2007/06/15. 10.1016/S0140-6736(07)60916-7 . [DOI] [PubMed] [Google Scholar]
- 45.Holmes CB, Hausler H, Nunn P. A review of sex differences in the epidemiology of tuberculosis. Int J Tuberc Lung Dis. 1998;2(2):96–104. . [PubMed] [Google Scholar]
- 46.Boum Y 2nd, Atwine D, Orikiriza P, Assimwe J, Page AL, Mwanga-Amumpaire J, et al. Male Gender is independently associated with pulmonary tuberculosis among sputum and non-sputum producers people with presumptive tuberculosis in Southwestern Uganda. BMC Infect Dis. 2014;14:638 10.1186/s12879-014-0638-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nhamoyebonde S, Leslie A. Biological differences between the sexes and susceptibility to tuberculosis. J Infect Dis. 2014;209 Suppl 3:S100–6. Epub 2014/06/27. 10.1093/infdis/jiu147 . [DOI] [PubMed] [Google Scholar]
- 48.Nicol MP, Spiers K, Workman L, Isaacs W, Munro J, Black F, et al. Xpert MTB/RIF testing of stool samples for the diagnosis of pulmonary tuberculosis in children. Clin Infect Dis. 2013;57(3):e18–21. 10.1093/cid/cit230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyles TH, Hughes J, Cox V, Burton R, Meintjes G, Mendelson M. False-positive Xpert(R) MTB/RIF assays in previously treated patients: need for caution in interpreting results. Int J Tuberc Lung Dis. 2014;18(7):876–8. 10.5588/ijtld.13.0853 . [DOI] [PubMed] [Google Scholar]
- 50.Theron G, Venter R, Calligaro G, Smith L, Limberis J, Meldau R, et al. Xpert MTB/RIF Results in Patients With Previous Tuberculosis: Can We Distinguish True From False Positive Results? Clin Infect Dis. 2016;62(8):995–1001. 10.1093/cid/civ1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, et al. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol. 2008;67(3):672–84. 10.1111/j.1365-2958.2007.06078.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nathan C, Barry CE 3rd. TB drug development: immunology at the table. Immunol Rev. 2015;264(1):308–18. 10.1111/imr.12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
- Experimental Laboratory methods
- Technical aspects of TOP TB assay
- TOP TB assay laboratory methods
- M. tuberculosis sequencing and genotype nomenclature
- Supporting Figures
- Fig A: DNA sequence alignment of M. tuberculosis ponA1
- Fig B: Sequence representation of five possible 2-PonA genotypes of M. tuberculosis clinical isolates
- Fig C: Genetic correspondence of M. tuberculosis 2-ponA genotypes with other common whole-genome genotyping methods
- Fig D: Analytical sensitivity of 3-ponA primer
- Fig E: Analytical specificity of 3-ponA primer
- Fig F: Specificity of TOP TB assay against non-tuberculous mycobacteria
- Fig G: Receiver operating curve (ROC) analysis of TOP TB assay results according to culture or composite reference standard
- Supporting Tables
- Table A: Description of 3-ponA and 2-ponA primers
- Table B: Genetic correspondence of M. tuberculosis 2-ponA genotypes with other common whole-genome genotyping methods
- Table C: Summary of Uganda clinical study
- Table D: Breakdown of results for the Composite Reference Standard
- Table E: Sensitivity analysis using TOP TB assay cut-off determined by ROC analysis
- References
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.