Abstract
Background
Lysosome-associated transmembrane-4 beta (LAPTM4B) is an oncogene that participates tumorgenesis in a variety of human solid tumors, and it has two alleles named as LAPTM4B*1 and *2. The present study aimed to identify the association of LAPTM4B genotype with clinicopathological features and prognosis in colorectal and esophageal cancer patients.
Method
Genotypes of LAPTM4B were determined by PCR in 167 colon cancer cases (72 patients in a discovery cohort and 95 patients in a testing cohort), 160 rectal cancer cases and 164 esophageal cancer cases. Association between the LAPTM4B gene polymorphism and clinicopathological variables was calculated by Chi-square test or Fisher’s exact test. Patient survival differences were calculated by the Kaplan-Meier method. Prognostic factors were determined with Log-rank test and Cox regression model.
Results
LAPTM4B *1/1 was more frequently detected in colon cancer patients with lymph node metastasis and TNM III+IV stages in total colon cancer (discovery + testing cohorts). LAPTM4B *2/2 decreased in recurrent patients in total colon cancer patients (P = 0.045). Kaplan-Meier survival curves and Log-rank test showed that LAPTM4B*1 was correlated with shorter overall survival (OS) in discovery and testing cohorts of colon cancer (P = 0.0254 and 0.0292, respectively), but not in rectal and esophageal cancer cases (P = 0.7669 and 0.9356, respectively). Multivariate analysis showed that LAPTM4B genotype was an independent prognostic factor for OS in total colon cancer [P = 0.004, hazard ratio (HR) = 0.432; 95% confidence interval (CI) = 0.243–0.768], but not in rectal and esophageal cancers (P = 0.791, HR = 1.073, 95% CI = 0.638–1.804 and 0.998, HR = 1.000, 95% CI = 0.663–1.530, respectively).
Conclusion
These findings suggested that LAPTM4B allele *1 was a risk factor associated with poor prognosis in patients with colon cancer, but not in patients with rectal or esophageal cancers. LAPTM4B genotype status might be a useful prognostic indicator for patients that need surgical operation in colon cancer.
Introduction
Colorectal and esophageal cancers are common malignant digestive diseases with high incidence and mortality worldwide [1–3]. Nearly one-half patients are diagnosed at an advanced stage, and there are still lack effective targeted therapies in colorectal and esophageal cancers, so survival rates in these cancers have not been improved markedly compared with hematopoietic and lymphoid malignancies. However, it is beneficial to monitor cancer progression for cancer patients [4].
Lysosomal-associated protein transmembrane-4 beta (LAPTM4B) is an oncogene that is upregulated in various solid cancers [5, 6] and associated with poor prognosis, such as gastric cancer [7–9], hepatocellular cancer [10], lung cancer [11, 12] and [13] and etc. LAPTM4B exists as two allelic genes, which have the same sequence except for one 19 bp segment for LAPTM4B *1 and two tight tandem segments for LAPTM4B *2 in the 5’ untranslated region of exon 1 [14]. Previous studies have demonstrated that LAPTM4B *2 allele was associated with significantly elevated risk of cancers, such as lung [15, 16], breast [17, 18], gastric [19], colon [20], ovarian [21], gallbladder cancer [22] and etc. Recent studies also suggested that LAPTM4B *2 was an independent prognostic biomarker for hepatocellular carcinoma [23], lung [24], breast [25], endometrial cancer patients [26] and etc.
According to our previous report, the LAPTM4B*2 allele frequency was 33.2% in colon cancer group, 25.5% in rectal cancer group, 22.7% in esophageal cancer group and 24.1% in health control group, indicating that LAPTM4B*2 was correlated with increased risk of colon cancer (P = 0.0016), but not with that of rectal and esophageal cancers [20]. However, there was no report about the association between the existence of two variant alleles of LAPTM4B with the prognosis in patients with colorectal and esophageal cancers. The present study aimed to investigate whether there is a correlation of LAPTM4B gene polymorphism with prognosis in colorectal and esophageal cancer patients after surgical resection.
Materials and Methods
Patients and Controls
In this retrospective study, we collected 167 colon cancer cases (a discovery cohort including 72 patients from Department of Gastrointestinal Surgery between 1999 and 2006, and a testing cohort including 95 patients from Department of Clinical Laboratory between 1997 and 2006), 160 rectal cancer cases and 164 esophageal cancer cases who were hospitalized in Beijing Cancer Hospital, Peking University School of Oncology between June 1997 and December 2006. All patients underwent surgical resection and were finally confirmed according to the World Health Organization classification. The blood samples were stored in the biological tissue bank of Peking University Cancer Hospital & Institute. The tumor-node-metastasis (TNM) stage was determined according to the classification of the American Joint Committee on Cancer and International Union against Cancer. All patients who participated in our study underwent tumor resection at Clinical Oncology of Peking University with follow-up time of 1 to 209.2 months (median: 54.0 months) for colon cancer, 1–134.5 months (median: 60.0 months) for rectal cancer and 1–123.5 months (median: 37.5 months) for esophageal cancer. At the end of follow up, 56.9% (95/167), 55.3% (84/152) and 74.6% (97/130) patients died from colon, rectal and esophageal cancers, respectively.
This study had been approved by the Research and Ethical Committee of Peking University School of Oncology. Written informed consent was obtained from each patient participated in this study.
DNA extraction and PCR analysis
Total genomic DNA was isolated from peripheral white cells using Blood Genomic DNA extraction kit following the manufacturer’s instructions (Tiangen Beijing, China). DNA was dissolved in elution buffer, and its concentration was measured with a Nanodrop 2000 spectrophotomer (Thermo Fisher Scientific, Wilmington, Delaware, USA) and stored at -80°C until use.
Genotype of LAPTM4B was identified by PCR analysis using the primers 5’-GCCGACTAGGGGACTGGCGGA-3’ (sense) and 5’-CGAGAGCTCCGAGCTTCTGCC-3’ (antisense). PCR was carried out on a thermo cycler (Gene Cycler TM, Bio-Rad, CA, USA) in 20-μl volumes as follows: denaturation at 95°C for 5 min, followed by 35 cycles of 94°C for 30s, 65°C for 30s, 72°C for 30s. The last cycle was followed by auto-extension 72°C for 7 min with Taq DNA polymerase (Hotstar Taq plus, Qiagen, Valencia, California, USA). The amplified products were analyzed by electrophoresis in 10% polyacrylamide gel (visualized by gel-red).
Statistical analysis
Statistical analysis was carried out by SPSS20.0 software (SPSS Inc., Chicago, IL). Chi-square test or Fisher’s exact test was used to assess the correlation between the genotype and clinical parametric distributions in colorectal and esophageal cancer patients. The association of LAPTM4B gene polymorphism with overall survival (OS) was analyzed using Kaplan-Meier curves and log-rank test. Multivariate analysis determined the potential independent prognostic factors with Cox regression model. All tests of statistical significance were two-sided. P < 0.05 was used as statistically significant level.
Results
Genotypes of the LAPTM4B in colorectal and esophageal cancers
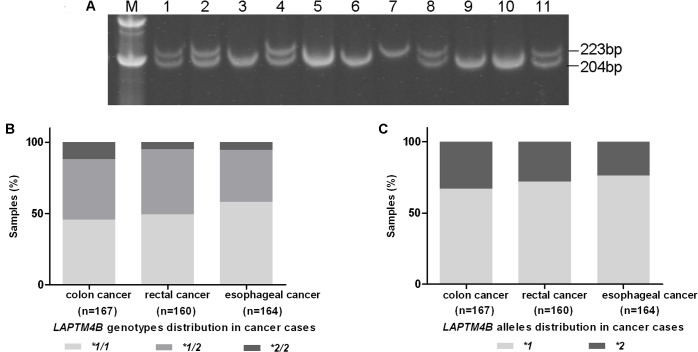
Three different genotypic LAPTM4B polymorphisms: LAPTM4B*1/1, LAPTM4B*2/2 and LAPTM4B*1/2 were shown in Fig 1A. For LAPTM4B*1/1 and LAPTM4B*2/2, we can observe fragments of 204-bp and 223-bp respectively. And both fragments can be observed in LAPTM4B*1/2 heterozygous individual.
Fig 1. Genotypes of LAPTM4B distribution in colorectal and esophageal cancers.
(A) Genotypes of LAPTM4B were separated in 10% polyacrylamide gel electrophoresis. Lanes 1, 2, 4, 8, 11: LAPTM4B *1/2; lanes 3, 5, 6, 9, 10: LAPTM4B *1/1; lanes 7: LAPTM4B *2/2. (B) Distribution of genotypes of LAPTM4B in colorectal and esophageal cancers. LAPTM4B genotypes: *1/1, *1/2 and *2/2 frequencies were 44.6%, 42.3% and 13.1%, respectively, in colon cancer; 49.4%, 45.6% and 5.0%, respectively, in rectal cancer; 57.9%, 36.6% and 5.5%, respectively, in esophageal cancer. (C) Distribution of alleles of LAPTM4B in colorectal and esophageal cancers. LAPTM4B *1 allele frequency was 65.8% in colon cancer; 72.2% in rectal cancer and 76.2% in esophageal cancer. And LAPTM4B *2 allele frequency was 34.2% in colon cancer; 27.8% in rectal cancer and 23.8% in esophageal cancer.
The genotypic and allele frequencies of LAPTM4B in colorectal and esophageal cancers were depicted in Fig 1B. Among 167 colon cancer cases, the LAPTM4B genotypes: *1/1, *1/2 and *2/2 frequencies were 44.6%, 42.3% and 13.1%, respectively. However, the genotype frequencies in 160 rectal cancer cases and 164 esophageal cancer cases were 49.4%, 45.6% and 5.0% vs. 57.9%, 36.6% and 5.5%, respectively (Fig 1B). In colon cancer cases, the LAPTM4B *2 allele frequency was 34.2%, which is different from that in rectal and esophageal cancer cases (27.8% and 23.8%, respectively) (Fig 1C).
Association between LAPTM4B genotypes and clinicopathological parameters in colorectal and esophageal cancers
The distribution of different genotypes of LAPTM4B was analyzed in clinicpathological parameters, including age, gender, lymph node metastasis, depth of invasion, distant metastasis, differentiation degree, gross type, TNM stage, location for colon cancer, CEA (carcinoembryonic antigen) for colorectal cancer and recurrence (Table 1).The association of different genotypes of LAPTM4B with these clinical variables of colorectal and esophageal cancer patients did not reach statistical significance in our study. LAPTM4B *1/1 was more frequently detected in colon cancer patients with lymph node metastasis and TNM III+IV stages compared with non-lymph node metastasis and TNM I+II stages in total colon cancer (discovery + testing cohorts) (P = 0.106, 29.6% vs. 46.6% and P = 0.157 31.4% vs. 46.6%, respectively). LAPTM4B *2/2 decreased in recurrent patients compared with non-recurrent ones in total colon cancer patients (P = 0.045, 7.4% vs. 20.5%) (Table 1) and in discovery and testing cohort (P = 0.203, 7.1% vs. 22.7% and P = 0.368, 7.6% vs. 17.2%, respectively) (S1 Table). However, LAPTM4B*1 was more frequently detected in colon cancer patients with moderate and well differentiation in colon discovery cohort (P = 0.011) (S1 Table). Such association was not found in colon testing cohort and total colon cancer cases.
Table 1. Correlation of Distribution of various genotypes of LAPTM4B with clinicopathological parameters in colorectal and esophageal cancer patients.
| Variables | Colon cancer | Rectal cancer | Esophagus cancer | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| *1/1 | *1/2 | *2/2 | P value | *1/1 | *1/2 | *2/2 | P value | *1/1 | *1/2 | *2/2 | P value | ||
| Age | |||||||||||||
| ≤60 | 37 | 31 | 8 | 0.528 | 39 | 44 | 3 | 0.256 | 46 | 27 | 6 | 0.473 | |
| >60 | 38 | 40 | 14 | 40 | 29 | 5 | 49 | 33 | 3 | ||||
| Gender | |||||||||||||
| Male | 44 | 41 | 14 | 0.886 | 45 | 45 | 4 | 0.738 | 80 | 42 | 7 | 0.114 | |
| Female | 30 | 30 | 8 | 34 | 28 | 4 | 15 | 18 | 2 | ||||
| Lymph node metastasis | |||||||||||||
| N0 | 16 | 27 | 11 | 0.106 | 29 | 25 | 4 | 0.820 | 33 | 20 | 1 | 0.371 | |
| N1+2 | 34 | 31 | 8 | 35 | 31 | 3 | 61 | 39 | 7 | ||||
| Undetermined | 24 | 13 | 2 | 15 | 17 | 1 | 1 | 1 | 1 | ||||
| Depth of invasion | |||||||||||||
| T1+2 | 5 | 5 | 3 | 0.691 | 15 | 11 | 1 | 0.815 | 26 | 17 | 1 | 0.579 | |
| T3+4 | 46 | 53 | 16 | 52 | 47 | 6 | 70 | 42 | 7 | ||||
| Undetermined | 23 | 13 | 2 | 12 | 15 | 1 | 0 | 1 | 1 | ||||
| Distant metastasis | |||||||||||||
| M0 | 44 | 43 | 17 | 0.298 | 54 | 45 | 6 | 0.954 | 80 | 47 | 8 | 0.149 | |
| M1 | 14 | 20 | 3 | 15 | 14 | 2 | 16 | 13 | 0 | ||||
| Undetermined | 16 | 8 | 2 | 10 | 14 | 0 | 0 | 0 | 1 | ||||
| Differentiation | |||||||||||||
| Poor | 12 | 21 | 4 | 0.267 | 10 | 12 | 3 | 0.196 | 21 | 18 | 2 | 0.498 | |
| Moderate+Well | 52 | 49 | 17 | 65 | 60 | 4 | 62 | 34 | 8 | ||||
| Undetermined | 10 | 1 | 1 | 4 | 1 | 1 | 13 | 8 | 1 | ||||
| Gross type | |||||||||||||
| Ulcerative type | 31 | 34 | 11 | 0.673 | 40 | 40 | 4 | 0.152 | |||||
| Protrude type | 10 | 13 | 3 | 8 | 2 | 1 | |||||||
| Others | 3 | 8 | 3 | 31 | 31 | 3 | |||||||
| Undetermined | 30 | 16 | 4 | ||||||||||
| TNM stage | |||||||||||||
| I+II | 16 | 25 | 10 | 0.157 | 26 | 23 | 4 | 0.797 | 34 | 20 | 1 | 0.363 | |
| III+IV | 41 | 37 | 10 | 43 | 35 | 4 | 62 | 40 | 7 | ||||
| Undetermined | 17 | 9 | 2 | 10 | 15 | 0 | 0 | 0 | 1 | ||||
| Location | |||||||||||||
| Proximal | 24 | 18 | 9 | 0.627 | |||||||||
| Distal | 33 | 34 | 11 | ||||||||||
| Undetermined | 17 | 19 | 2 | ||||||||||
| Recurrence | |||||||||||||
| No | 30 | 28 | 15 | 0.045 | 44 | 35 | 5 | 0.618 | 80 | 47 | 5 | 0.140 | |
| Yes | 44 | 43 | 7 | 36 | 37 | 3 | 16 | 13 | 4 | ||||
| CEA | |||||||||||||
| Negative | 33 | 31 | 7 | 0.984 | 40 | 26 | 3 | 0.365 | |||||
| Positive | 35 | 32 | 8 | 32 | 32 | 5 | |||||||
| Undetermined | 6 | 8 | 7 | 8 | 14 | 0 | |||||||
Data was calculated by Chi-square test or Fisher’s exact test.
CEA, carcinoembryonic antigen; LAPTM4B, lysosome-associated protein transmembrane 4 beta.
*: Genotype.
Association of LAPTM4B genotypes with overall survival in colorectal and esophageal cancer patients
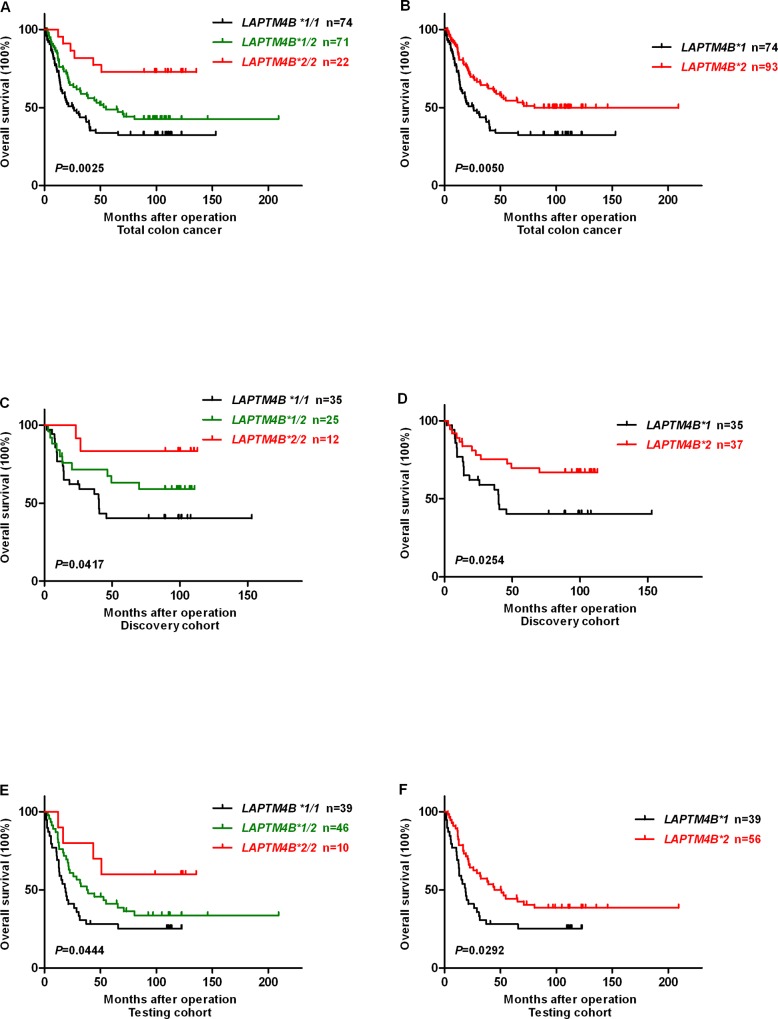
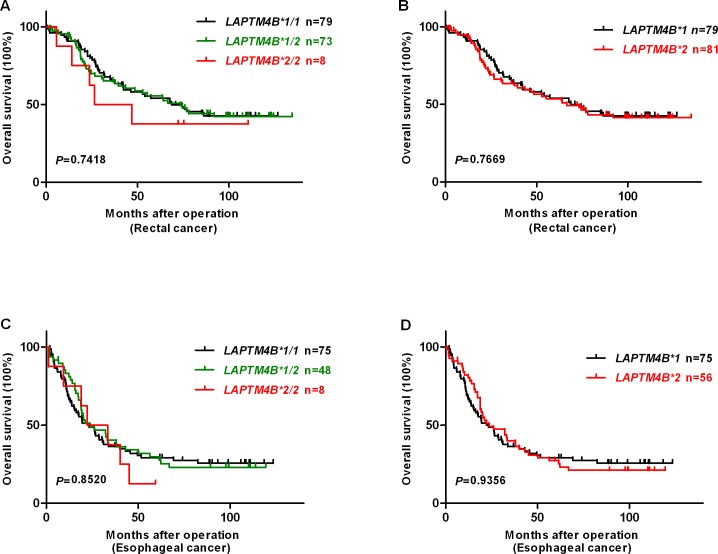
Kaplan-Meier survival analysis and log-rank test indicated that colon cancer patients with LAPTM4B *1/1 genotype showed a shorter overall survival (OS) when compared with those with LAPTM4B *1/2 and LAPTM4B *2/2 genotypes (OS rate 33.8% vs. 43.7% and 72.7%, P = 0.0025) (Fig 2A). Moreover, patients with LAPTM4B *1 allele had a poorer prognosis than LAPTM4B *2 allele in total colon cancer cases (OS rate 33.8% vs. 50.5%, P = 0.0050) (Fig 2B). The same tendency of LAPTM4B *1/1 and LAPTM4B *1 was observed in the colon discovery and testing cohorts of colon cancer (P = 0.0417 and 0.0444, P = 0.0254 and 0.0292, respectively) (Fig 2C–2F). However, we did not find a marked relation between LAPTM4B genotypes or alleles and OS for rectal and esophageal cancer cases (P = 0.7418 and 0.8520 for various genotypes vs. P = 0.7669 and 0.9356 for different alleles, respectively) (Fig 3A–3D).
Fig 2. Kaplan-Meier survival curves for survival comparison of colon cancer patients after surgery resection based on LAPTM4B genotypes and alleles.
Overall survival (OS) after surgery resection based on: (A) LAPTM4B genotypes in total colon cancer (P = 0.025). (B) LAPTM4B alleles in total colon cancer (P = 0.0050). (C and D) LAPTM4B genotypes and alleles in colon discovery cohort (P = 0.0417 and 0.0254), respectively. (E and F) LAPTM4B genotypes and alleles in colon testing cohort (P = 0.0444 and 0.0292), respectively.
Fig 3. Kaplan-Meier survival curves for survival comparison of rectal and esophageal cancer patients undergone surgery resection based on LAPTM4B genotypes and alleles.
Overall survival (OS) after surgery resection based on: (A and B) LAPTM4B genotypes and alleles in rectal cancer (P = 0.7418 and 0.7669), respectively. (C and D) LAPTM4B genotypes and alleles in esophageal cancer (P = 0.8520 and 0.9356), respectively.
LAPTM4B genotype was an independent prognostic marker in patients with colon cancer, but not for rectal and esophageal cancer patients
The univariate Cox’s model for OS of colorectal and esophageal cancer patients displayed that LAPTM4B genotype was one of the prognostic factors in total colon cancer patients (P = 0.006; HR: 0.565, 95% CI: 0.377–0.846), not in rectal and esophageal cancer patients (P = 0.692 and 0.958, respectively) (Table 2).
Table 2. Univariate analysis of the prognostic factors in colorectal and esophageal cancer patients by Log-rank test.
| Variables | Colon cancer | Rectal cancer | Esophageal cancer | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Univariate analysis | Univariate analysis | |||||||
| HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | |
| Age | |||||||||
| ≤60 | 1.468 | 0.969–2.222 | 0.070 | 1.069 | 0.702–1.628 | 0.757 | 1.415 | 0.948–2.112 | 0.090 |
| >60 | |||||||||
| Gender | |||||||||
| Male | 0.782 | 0.515–1.186 | 0.247 | 1.196 | 0.783–1.827 | 0.409 | 0.584 | 0.336–1.015 | 0.057 |
| Female | |||||||||
| Lymph node metastasis | |||||||||
| N0 | 4.385 | 2.375–8.096 | <0.001 | 1.838 | 1.115–3.031 | 0.017 | 1.628 | 1.041–2.548 | 0.033 |
| N1+2 | |||||||||
| Depth of invasion | |||||||||
| T1+2 | 1.841 | 0.669–5.076 | 0.237 | 1.903 | 0.997–3.630 | 0.051 | 1.900 | 1.110–3.255 | 0.019 |
| T3+4 | |||||||||
| Distant metastasis | |||||||||
| M0 | 6.687 | 4.134–10.815 | <0.001 | 4.990 | 3.012–8.266 | <0.001 | 2.306 | 1.390–3.825 | 0.001 |
| M1 | |||||||||
| Differentiation | |||||||||
| Poor | 1.116 | 0.679–1.836 | 0.665 | 0.779 | 0.438–1.384 | 0.394 | 1.348 | 0.825–2.201 | 0.233 |
| Moderate+Well | |||||||||
| Gross type | |||||||||
| Ulcerative type | 1.035 | 0.717–1.496 | 0.853 | 0.427 | 0.133–1.374 | 0.154 | |||
| Protrude type | |||||||||
| Others | |||||||||
| TNM stage | |||||||||
| I+II | 6.434 | 3.290–12.583 | <0.001 | 2.415 | 1.428–4.084 | 0.001 | 1.627 | 1.048–2.526 | 0.030 |
| III+IV | |||||||||
| Location | |||||||||
| Proximal | 1.194 | 0.726–1.692 | 0.485 | ||||||
| Distal | |||||||||
| Recurrence | |||||||||
| No | 5.943 | 3.533–9.999 | <0.001 | 4.073 | 2.557–6.488 | <0.001 | 1.882 | 1.219–2.904 | 0.004 |
| Yes | |||||||||
| CEA | |||||||||
| Negative | 1.651 | 1.072–2.545 | 0.023 | 1.865 | 1.184–2.936 | 0.007 | |||
| Positive | |||||||||
| LAPTM4B genotype | |||||||||
| *1/1 | 0.565 | 0.377–0.846 | 0.006 | 1.089 | 0.715–1.658 | 0.692 | 0.989 | 0.665–1.472 | 0.958 |
| *1/2+ *2/2 | |||||||||
Data was calculated by Log-rank test. HR, hazard ratio; CI, confidence interval.
CEA, carcinoembryonic antigen; LAPTM4B, lysosome-associated protein transmembrane 4 beta.
*: Genotype.
Furthermore, LAPTM4B genotype was a novel independent prognostic factor of OS for colon cancer (P = 0.004, HR = 0.432, 95% CI: 0.243–0.768), even in the colon discovery cohorts (P = 0.007, S2 Table), but not for rectal and esophageal cancer patients (P = 0.791, HR = 1.073, 95% CI: 0.638–1.804 vs. P = 0.998, HR = 1.000, 95% CI: 0.663–1.530, respectively). Depth of invasion, distant metastasis and recurrence were also independent prognostic factors for colon cancer (P = 0.014, P<0.001 and P<0.001, respectively). For rectal cancer patients, distant metastasis and recurrence were also independent prognostic factors (P = 0.021 and P<0.001, respectively). In addition, distant metastasis (P = 0.025) was also an independent prognosis factor in esophageal cancer patients (Table 3).
Table 3. Multivariate analysis of the prognostic factors in colorectal and esophageal cancer patients by Cox proportional hazard regression model.
| Variables | Colon cancer | Rectal cancer | Esophageal cancer | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Multivariate analysis | Multivariate analysis | Multivariate analysis | |||||||
| HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | |
| Age | |||||||||
| ≤60 vs >60 | 1.019 | 0.565–1.838 | 0.949 | 1.211 | 0.712–2.062 | 0.480 | 1.313 | 0.852–2.022 | 1.313 |
| Gender | |||||||||
| Male vs Female | 0.864 | 0.498–1.500 | 0.603 | 1.225 | 0.705–2.129 | 0.471 | 0.765 | 0.408–1.434 | 0.765 |
| Depth of invasion | |||||||||
| T1+2 vs T3+4 | 4.240 | 1.275–14.102 | 0.018 | 1.494 | 0.689–3.242 | 0.309 | 1.528 | 0.848–2.751 | 0.158 |
| Lymph node metastasis | |||||||||
| N0 vs N1+3 | 0.950 | 0.614–1.470 | 0.819 | 1.186 | 0.654–2.148 | 0.575 | 1.200 | 0.723–1.992 | 0.480 |
| Distant metastasis | |||||||||
| M0 vs. M1 | 4.517 | 2.281–8.945 | <0.001 | 2.173 | 1.122–4.208 | 0.021 | 1.988 | 1.089–3.629 | 0.025 |
| Recurrence | |||||||||
| No vs. Yes | 6.898 | 3.061–15.543 | <0.001 | 3.748 | 1.896–7.411 | <0.001 | 1.577 | 0.980–2.537 | 0.061 |
| CEA | |||||||||
| Negative vs. Positive | 0.828 | 0.466–1.471 | 0.519 | 1.539 | 0.895–2.648 | 0.120 | |||
| LAPTM4B genotype | |||||||||
| *1/1 vs *1/2+*2/2 | 0.432 | 0.243–0.768 | 0.004 | 1.073 | 0.638–1.804 | 0.791 | 1.000 | 0.663–1.530 | 0.998 |
Data was calculated by Cox regression test. HR, hazard ratio; CI, confidence interval.
CEA, carcinoembryonic antigen; LAPTM4B, lysosome-associated protein transmembrane 4 beta.
*: Genotype.
Discussion
Previous studies have demonstrated that LAPTM4B can play critical roles in various solid tumors, including proliferation, migration, invasion, apoptosis and angiogenesis [12, 27–29]. It also motivated multidrug resistance through promoting drug efflux by interacting with P-gp and activating PI3K/AKT signaling pathway [30]. In addition, new evidence has also revealed that LAPTM4B can participate in the autophagy initiation through binding with inactive epidermal growth factor receptor (EGFR) [31, 32].
In the present study, we revealed an independent prognostic role of LAPTM4B gene polymorphism in colon cancer patients who received surgical resection, but not in rectal and esophageal cancers. In our study, the LAPTM4B *2 allele frequency rate are 33.3%, 27.8% and 23.8%, nearly the same as previous report in colon, rectal and esophageal cancers [20], respectively. For the clinicopathological parameters, LAPTM4B genotype was correlated with recurrence in total colon cancer, especially for LAPTM4B *2/2 which decreased in recurrent colon cancer patients. There was not a relationship between LAPTM4B genotype and other clinical factors in colorectal and esophageal cancer patients. However, LAPTM4B*1 was more frequently detected in colon cancer patients with moderate and well differentiation in colon discovery cohort. This phenomenon might be caused by incomplete clinicopathological parameters in the present work. Furthermore, LAPTM4B *1/1 tended to be frequently detected in patients with lymph node metastasis and TNM III+IV stages in total colon cancer cases.
Patients with LAPTM4B *1 (genotypes *1/1) had a significantly poorer overall survival when compared with LAPTM4B *2 (genotypes *1/2 or *2/2) patients in colon cancer (discovery and testing cohorts), but not in rectal and esophageal cancers. This is the first time demonstrating the LAPTM4B *1 allele as a poor prognostic indicator. The association of LAPTM4B *2 allele with colon cancer prognosis is not consistent with recent reports including in hepatocellular [23], ovary [27], lung [23], breast cancer [25] and etc. In hepatocellular carcinoma, Yang et al indicated that LAPTM4B*2 was correlated with tumor recurrence, poor histopathological differentiation and also an independent prognostic factor. Previous studies indicated that the 19-bp difference in 5’ untranslated region of the first exon of the LAPTM4B gene can alter the ORF, resulting in two different protein isoforms: LAPTM4B-35 and -40 [14]. It might suggest that the 19-bp sequence plays an important role in transcriptional regulation or new isoform produced by LAPTM4B*2 may influence physiological activity and function of cancer cells.
Whereas in our study, LAPTM4B *1 allele shows a significant correlation with overall survival of colon cancer patients, but not in rectal and esophageal cancer patients. One explanation might be: the diverse expression patterns or isoforms of LAPTM4B in epithelial cells might demonstrate the difference of LAPTM4B genotype in prognosis in colon cancer vs rectal and esophageal cancers; the other explanation might be: the 19-bp sequence may play an important role in transcriptional regulation such as binding with the transcription factors or non-coding linker RNA in different cancers. Furthermore, in patients with gastric cancer, we have found that even though LAPTM4B genotype was correlated with susceptibility of gastric cancer, this polymorphism did not correlate with prognosis (data unpublished). The phenomenon illustrated the tumor heterogenicity between LAPTM4B genotype and its function, which discriminates with that in hepatocellular carcinoma, breast cancer and etc.
LAPTM4B was obviously up-regulated in various types of cancers [6]. Its overexpression might be caused by gene amplification and transcriptional up-regulation. However, the specific reason remains unknown. LAPTM4B has two different protein isoforms: LAPTM4B-24 (226 aa) and LAPTM4B-35 (317 aa). Li et al. has indicated that LAPTM4B-35 isoform can activate PI3K/Akt to participate multidrug resistance of cancer cells and anti-apoptosis [30]. Previous studies have proved that LAPTM4B-35 and -24 have different expression status and different roles in tissues and various cell lines of hepatocellular carcinoma [29, 33, 34]. Their balance may affect malignant transformation. However, a recent report has shown that LAPTM4B-24 isoform can stimulate mTORC1 via V-ATPase by influx of leu through binding with LAT1-4F2hc to lysosomes [35]. LAPTM4B-24 can also promote cell growth and proliferation [35].
As a result, the different LAPTM4B isoforms may play diverse functions in LAPTM4B *1 patients when compared with LAPTM4B *2 patients with colon cancer. Our findings on LAPTM4B alleles in colon cancer provide additional evidence that different LAPTM4B isoforms might play various roles, that is, LAPTM4B-35 can activate PI3K/Akt pathway and LAPTM4B -24 can activate mTORC1 pathway. Different isoform pattern might induce the function of 19 bp sequence in various cancers. Further studies should be carried out to elucidate this phenomenon. However, LAPTM4B genotype will be a useful biomarker for colon cancer patients when considering curative surgical resection.
Supporting Information
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The National Nature Science Foundation of China (No. 81402308), the National High Technology Research and Development Program of China (863 Program, No. 2014AA020603), the capital health research and development of special (No. 2016-2-2151), Beijing municipal natural science foundation, (No. 7153161), International science and technology cooperation program of China (No. 2013DFG32720).
References
- 1.Chen W, Zheng R, Zeng H, Zhang S. The updated incidences and mortalities of major cancers in China, 2011. Chinese journal of cancer. 2015;34(3):53. Epub 2015/09/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65(1):5–29. Epub 2015/01/07. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87–108. Epub 2015/02/06. [DOI] [PubMed] [Google Scholar]
- 4.Welch HG, Schwartz LM, Woloshin S. Are increasing 5-year survival rates evidence of success against cancer? Jama. 2000;283(22):2975–8. Epub 2000/06/24. [DOI] [PubMed] [Google Scholar]
- 5.Shao GZ, Zhou RL, Zhang QY, Zhang Y, Liu JJ, Rui JA, et al. Molecular cloning and characterization of LAPTM4B, a novel gene upregulated in hepatocellular carcinoma. Oncogene. 2003;22(32):5060–9. Epub 2003/08/07. [DOI] [PubMed] [Google Scholar]
- 6.Kasper G, Vogel A, Klaman I, Grone J, Petersen I, Weber B, et al. The human LAPTM4b transcript is upregulated in various types of solid tumours and seems to play a dual functional role during tumour progression. Cancer letters. 2005;224(1):93–103. Epub 2005/05/25. [DOI] [PubMed] [Google Scholar]
- 7.Cheng X, Zheng Z, Bu Z, Wu X, Zhang L, Xing X, et al. LAPTM4B-35, a cancer-related gene, is associated with poor prognosis in TNM stages I-III gastric cancer patients. PloS one. 2015;10(4):e0121559 Epub 2015/04/08. 10.1371/journal.pone.0121559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Tian B, Yu H, Yao H, Gao Z. LAPTM4B-35 protein as a potential therapeutic target in gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(12):12737–42. Epub 2014/11/25. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Xu X, Jing L, Zhou G, Cao Z, Han Y, et al. Lysosomal-associated protein transmembrane 4 Beta-35 overexpression is a novel independent prognostic marker for gastric carcinoma. PloS one. 2015;10(2):e0118026 Epub 2015/02/18. 10.1371/journal.pone.0118026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H, Xiong FX, Lin M, Yang Y, Nie X, Zhou RL. LAPTM4B-35 overexpression is a risk factor for tumor recurrence and poor prognosis in hepatocellular carcinoma. Journal of cancer research and clinical oncology. 2010;136(2):275–81. Epub 2009/08/20. 10.1007/s00432-009-0659-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maki Y, Fujimoto J, Lang W, Xu L, Behrens C, Wistuba II, et al. LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells. Scientific reports. 2015;5:13846 Epub 2015/09/08. 10.1038/srep13846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang H, Tian H, Yue W, Li L, Li S, Gao C, et al. Overexpression of LAPTM4B is correlated with tumor angiogenesis and poor prognosis in non-small cell lung cancer. Med Oncol. 2014;31(6):974 Epub 2014/05/13. 10.1007/s12032-014-0974-8 [DOI] [PubMed] [Google Scholar]
- 13.Yin M, Li C, Li X, Lou G, Miao B, Liu X, et al. Over-expression of LAPTM4B is associated with poor prognosis and chemotherapy resistance in stages III and IV epithelial ovarian cancer. Journal of surgical oncology. 2011;104(1):29–36. Epub 2011/03/19. 10.1002/jso.21912 [DOI] [PubMed] [Google Scholar]
- 14.Shao GZ. Molecular cloning and characterization of LAPTM4B, a novel gene unregualted in hepatocellular carcinoma. Philosophy Degree Thesis, Beijing: Peking University Health Science Center. 2003.
- 15.Li C, Zhou Q, Wang Y, Chen X, Yang X, Zhu D. [Relationship between LAPTM4B gene polymorphism and susceptibility of lung cancer]. Zhongguo fei ai za zhi = Chinese journal of lung cancer. 2006;9(2):109–12. Epub 2006/04/20. 10.3779/j.issn.1009-3419.2006.02.02 [DOI] [PubMed] [Google Scholar]
- 16.Deng LJ, Zhang QY, Liu B, Zhou RL. [Relationship between LAPTM4B gene polymorphism and susceptibility of lung cancer]. Beijing da xue xue bao Yi xue ban = Journal of Peking University Health sciences. 2005;37(3):302–5. Epub 2005/06/22. [PubMed] [Google Scholar]
- 17.Fan M, Liu Y, Zhou R, Zhang Q. Association of LAPTM4B gene polymorphism with breast cancer susceptibility. Cancer epidemiology. 2012;36(4):364–8. Epub 2012/01/25. 10.1016/j.canep.2011.12.004 [DOI] [PubMed] [Google Scholar]
- 18.Hashemi M, Amininia S, Ebrahimi M, Hashemi SM, Yousefi J, Eskandari-Nasab E, et al. Association between LAPTM4B gene polymorphism and breast cancer susceptibility in an Iranian population. Med Oncol. 2014;31(8):111 Epub 2014/07/09. 10.1007/s12032-014-0111-8 [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Zhang QY, Qian N, Zhou RL. Relationship between LAPTM4B gene polymorphism and susceptibility of gastric cancer. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2007;18(2):311–6. Epub 2006/11/01. [DOI] [PubMed] [Google Scholar]
- 20.Cheng XJ, Xu W, Zhang QY, Zhou RL. Relationship between LAPTM4B gene polymorphism and susceptibility of colorectal and esophageal cancers. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2008;19(3):527–32. Epub 2007/10/30. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Liu Y, Zhou R, Meng F, Gao Y, Yang S, et al. LAPTM4B polymorphisms is associated with ovarian cancer susceptibility and its prognosis. Japanese journal of clinical oncology. 2012;42(5):413–9. Epub 2012/03/14. 10.1093/jjco/hys026 [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Zhai G, Ji X, Xiong F, Su J, McNutt MA. Correlation of LAPTM4B polymorphisms with gallbladder carcinoma susceptibility in Chinese patients. Med Oncol. 2012;29(4):2809–13. Epub 2012/02/04. [DOI] [PubMed] [Google Scholar]
- 23.Yang H, Zhai G, Ji X, Xiong F, Su J, McNutt MA. LAPTM4B allele *2 is a marker of poor prognosis following hepatic tumor resection for hepatocellular carcinoma. PloS one. 2012;7(4):e34984 Epub 2012/04/18. 10.1371/journal.pone.0034984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang H, Tian H, Yue W, Li L, Li S, Gao C, et al. LAPTM4B polymorphism is associated with nonsmall cell lung cancer susceptibility and prognosis. Oncology reports. 2014;31(5):2454–60. Epub 2014/03/29. 10.3892/or.2014.3116 [DOI] [PubMed] [Google Scholar]
- 25.Li X, Kong X, Chen X, Zhang N, Jiang L, Ma T, et al. LAPTM4B allele *2 is associated with breast cancer susceptibility and prognosis. PloS one. 2012;7(9):e44916 Epub 2012/09/18. 10.1371/journal.pone.0044916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng F, Li H, Zhou R, Luo C, Hu Y, Lou G. LAPTM4B gene polymorphism and endometrial carcinoma risk and prognosis. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 2013;18(2):136–43. Epub 2013/01/15. [DOI] [PubMed] [Google Scholar]
- 27.Meng F, Chen X, Song H, Lou G. LAPTM4B down regulation inhibits the proliferation, invasion and angiogenesis of HeLa cells in vitro. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2015;37(3):890–900. Epub 2015/09/19. [DOI] [PubMed] [Google Scholar]
- 28.He J, Shao G, Zhou R. [Effects of the novel gene, LAPTM4B, highly expression in hepatocellular carcinoma on cell proliferation and tumorigenesis of NIH3T3 cells]. Beijing da xue xue bao Yi xue ban = Journal of Peking University Health sciences. 2003;35(4):348–52. Epub 2003/09/02. [PubMed] [Google Scholar]
- 29.Li L, Shan Y, Yang H, Zhang S, Lin M, Zhu P, et al. Upregulation of LAPTM4B-35 promotes malignant transformation and tumorigenesis in L02 human liver cell line. Anat Rec (Hoboken). 2011;294(7):1135–42. Epub 2011/05/28. [DOI] [PubMed] [Google Scholar]
- 30.Li L, Wei XH, Pan YP, Li HC, Yang H, He QH, et al. LAPTM4B: a novel cancer-associated gene motivates multidrug resistance through efflux and activating PI3K/AKT signaling. Oncogene. 2010;29(43):5785–95. Epub 2010/08/17. 10.1038/onc.2010.303 [DOI] [PubMed] [Google Scholar]
- 31.Tan X, Thapa N, Sun Y, Anderson RA. A kinase-independent role for EGF receptor in autophagy initiation. Cell. 2015;160(1–2):145–60. Epub 2015/01/17. 10.1016/j.cell.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan X, Sun Y, Thapa N, Liao Y, Hedman AC, Anderson RA. LAPTM4B is a PtdIns(4,5)P2 effector that regulates EGFR signaling, lysosomal sorting, and degradation. The EMBO journal. 2015;34(4):475–90. Epub 2015/01/16. 10.15252/embj.201489425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng C, Zhou RL, Shao GZ, Rui JA, Wang SB, Lin M, et al. Expression of lysosome-associated protein transmembrane 4B-35 in cancer and its correlation with the differentiation status of hepatocellular carcinoma. World journal of gastroenterology: WJG. 2005;11(18):2704–8. Epub 2005/05/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu XR, Zhou RL, Zhang QY, Zhang Y, Jin YY, Lin M, et al. Structure analysis and expressions of a novel tetratransmembrane protein, lysosoma-associated protein transmembrane 4 beta associated with hepatocellular carcinoma. World journal of gastroenterology: WJG. 2004;10(11):1555–9. Epub 2004/05/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milkereit R, Persaud A, Vanoaica L, Guetg A, Verrey F, Rotin D. LAPTM4b recruits the LAT1-4F2hc Leu transporter to lysosomes and promotes mTORC1 activation. Nature communications. 2015;6:7250 Epub 2015/05/23. 10.1038/ncomms8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.