Abstract
CircRNAs are a class of endogenous RNA that regulates gene expression at the post-transcriptional or transcriptionallevel through interacting with other molecules or microRNAs. Increasing studies have demonstrated that circRNAs play a crucial role in biology processes. CircRNAs are proved as potentialbiomarkers in many diseases including cancers. However, the role of Cdr1as in Hepatocellular carcinoma (HCC) remains to be elucidated. We demonstrated that Cdr1as expression was upregulated in HCC tissues compared with the adjacent non-tumor tissues. In addtion, miR-7 expression was downregulated in HCC tissues compared with the adjacent non-tumor tissues. Moreover, the expression level of miR-7 was inversely correlated with that in HCC tissues. Knockdown of Cdr1as suppressed the HCC cell proliferation and invasion. Overexpression of miR-7 inhibited the HCC cell proliferation and invasion. Overexpression of miR-7 could suppress the direct target gene CCNE1 and PIK3CD expression. Knockdown of Cdr1as suppressed the expression of miR-7 and also inhibited the CCNE1 and PIK3CD expression. Furthermore, knockdown of Cdr1as suppressed the HCC cell proliferation and invasion through targeting miR-7. These data suggested that Cdr1as acted as an oncogene partly through targeting miR-7 in HCC.
Introduction
Hepatocellular carcinoma (HCC) ranks asthe fifth most common tumor and the third leading cause for cancer related death worldwide [1–4]. It is estimated that 21,000 new patientsare diagnosed as HCC, accounting for about 700,000 deaths every year [5, 6]. Despite several therapeutic advances in recent years, the 5-year survival rate of HCC is still low [7–9]. Increasing studies have showed that various antioncogenes or oncogenes are related with HCC metastasis[10–14]. Therefore, it is useful to understand the exact mechanisms to improve the diagnosis and therapeutic approaches for HCC.
Circular RNAs (circRNAs) are a novel class of noncoding RNA and represent a research hot spot in the RNA field[15–18].CircRNAs demonstrate high tissue special expression and stable structure[19–21]. CircRNAs regulate gene expression at the post-transcriptional or transcription all evel through interacting with other molecules or microRNAs[22–24]. Increasing studies have demonstrated that circRNAs play a crucial role in various biological processes such as cell development, proliferation, metastasis, fate decision,migration and invasion[20, 25–27]. CircRNAs are proved to be potentia lbiomarkers in many diseases especially in cancers [15, 17, 18, 28, 29]. However, the role of circRNAs in HCC remains to be elucidated.
Recent study showed that Cdr1as (also namedas ciRS-7) was theinhibitor and sponge of miR-7 in the embryonic zebrafish [19]. Ectopic expression of Cdr1as induced midbrain brain defects, which was similar with the phenotypes found in the knockdown of miR-7[19]. Cdr1as was demonstrated to act as a powerful miR-7 sponge/inhibitor in developing midbrain of zebrafish, suggesting a novel mechanism for regulating miRNA functions[25]. In this study, we showed that Cdr1as expression was upregulated in HCC tissues compared with adjacent non-tumor tissues. Knockdown of Cdr1as suppressed HCC cell proliferation and invasion. Moreover, knockdown of Cdr1as suppressed the expression of miR-7 and also inhibited the CCNE1 and PIK3CD expression.
Materials and Methods
Ethics Statement
All patients agreed to participate in the study and gave written informed consent. This study was approved by the ethical board of the institute of The Fourth Hospital of Harbin Medical University and complied with Declaration of Helsinki.
Tissue specimens and Cell line culture and transfection
Total of 35 HCC tissues and adjacent non-tumor tissues were collected from patients who underwent surgery in our hospital between 2012 and 2014. None of thesecases received chemotherapy or radiotherapy before surgery. The characteristics of the patients are described in S1 Table.The cell lines were used in our study as following: Hep3B, HepG2, SMMC-7721, Bel7402 and HL-7702. The miR-7mimics, inhibitor and their control oligonucleotide, shCdr1as and its control was obtained from GenePharma (Shanghai, China) and transfected into the SMMC-7721cells using lipofectamine 2000 (Invitrogen, USA).
Real-time quantitative PCR analysis
Total RNA was isolated from tissues and cells using TRIzol (Invitrogen, USA) following to instructions. The expression of miR-7 and Cdr1as was measure by Real-time quantitative PCR (qRT-PCR). The following primers were shown: GAPDH, forward 5’-GACTCATGACCACAGTCCATGC-3’; reverse 5’-AGAGGCAGGGATGATGTTCTG-3’; CCNE1, forward 5’-GCCAGCCTTGGGACAATAATG-3’; reverse 5’-CTTGCACGTTGAGTTTGGGT-3’; Cdr1as, forward 5’-GTGTCTCCAGTGTATCGGCG-3’; reverse 5’-TACTGGCACCACTGGAAACC-3’; PIK3CD, forward 5’-AAATTTGAACGGTTCCGGGG-3’; reverse 5’-CCTCCTCTGTTTTCCCCAGT-3’;
Cell proliferation assay
CCK-8 analysis kit (Dojindo, USA) was performed to determine the cell proliferation. After cell transfection, cells were cultured in the 96-well plates (Biosciences,USA) for 0h, 24, 48 and 72 hours respectively. CCK-8 kit (10 μL) was put into each well and the absorbance wasrecorded at 450 nm.
Western blotting
Protein was isolated from tissues and cells using RIPA buffer. The total proteins was separated by using 12% SDS-PAGE (SDS polyacrylamide gel electrophoresis) and transferred into the PVDF membrane (Millipore, USA). The PVDF membrane was blocked with 5% milk and then incubated with primary antibody: CCNE1, PIK3CD and GAPDH (Abcam, USA). The membrane was detected using X-ray films by chemiluminescent reagents.
Invasion analysis
The Transwell Matrigel (BD Biosciences, USA) was used to measure the cell invasion. The insert was coated with Matrigel and the cells were culture in the upper chamber while 20% FBS was put to the lower chamberas a chemoattractant. After cultured for 24hours, cells on the lower surface were stained, counted and photographed.
Statistical analysis
Data was shown as mean ± SD and P<0.05 was thought to be statistically significant. Student’s t-test was used to detect the differences between two groups and ANOVA was used to measure the differences between more than two groups. The correlation between Cdr1as and miR-7 was measure by Spearman correlation test.
Results
Cdr1asexpression was upregulated in HCC tissues and was inversely associated with the expression of miR-7
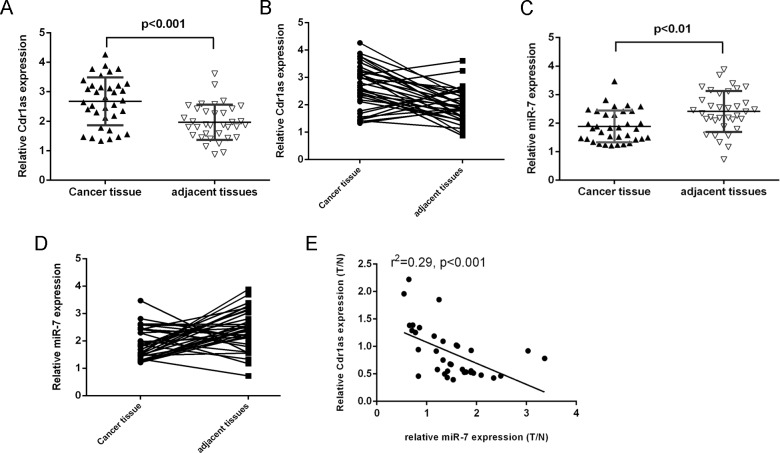
We demonstrated that Cdr1as expression was upregulated in HCC tissues compared with adjacent non-tumor tissues (Fig 1A). Cdr1as expression was upregulated in 74% (26/35) HCC tissues compared with their adjacent non-tumor tissues (Fig 1B). Moreover, miR-7expression was downregulated in HCC tissues compared with the adjacent non-tumor tissues (Fig 1C). Among them, miR-7 expression was downregulated in 66 (23/35) HCC tissues compared with their adjacent non-tumor tissues (Fig 1D). Interestingly, the expression level of miR-7 was inversely correlated with that of Cdr1as (Fig 1E).
Fig 1. The expression of Cdr1as was upregulated in the HCC tissues and was inverse associated with the expression of miR-7.
(A) The expression of Cdr1as was detected in the HCC tissues and adjacent non-tumor tissues using qRT-PCR. (B) Cdr1as expression was upregulated in the (74%, 26/35) HCC tissues compared with their adjacent non-tumor tissues. (C) The expression of miR-7 was detected in the HCC tissues and adjacent non-tumor tissues using qRT-PCR. (D) miR-7 expression was downregulated in the (66%, 23/35) HCC tissues compared with their adjacent non-tumor tissues. (E) The expression of miR-7 was inversely correlated with the expression of Cdr1as in the HCC tissues.
Knockdown of Cdr1as suppressed the HCC cell proliferation and invasion
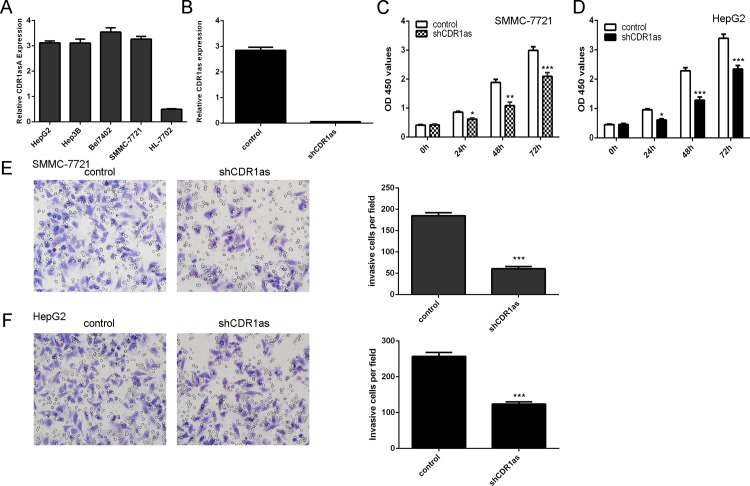
We found that Cdr1as expression was upregulated in HCC cell lines (HepG2, Hep3B, Bel7402 and SMMC-7721) compared with the normal hepatocytes (HL-7792 cells) (Fig 2A). We also confirmed that knockdown of Cdr1as could suppress the Cdr1as expression (Fig 2B). Knockdown of Cdr1as inhibited the HCC cell line (SMMC-7721 and HepG2) proliferation (Fig 2C and 2D). Moreover, inhibition of Cdr1as expression suppressed the SMMC-7721 and HepG2 cell invasion (Fig 2E and 2F)
Fig 2. Knockdown of Cdr1as suppressed the HCC cell proliferation and invasion.
(A) The expression of Cdr1as in the HCC cell lines were measured by using qRT-PCR. (B) Knockdown of Cdr1as expression could suppress the Cdr1as expression. (C) Knockdown of Cdr1as expression inhibited the HCC cell line (SMMC-7721) proliferation. (D) The HepG2 cell proliferation was measured by CCK-8 assay. (E) Knockdown of Cdr1as expression inhibited the SMMC-7721 cell invasion. (F) The HepG2 cell invasion was detected by cell invasion assay. *p<0.05, **p<0.01 and ***p<0.001. The shCDR1as panel of Fig 2F is excluded from this article's CC-BY license. See the accompanying retraction notice for more information.
Overexpression of miR-7 inhibited HCC cell proliferation and invasion
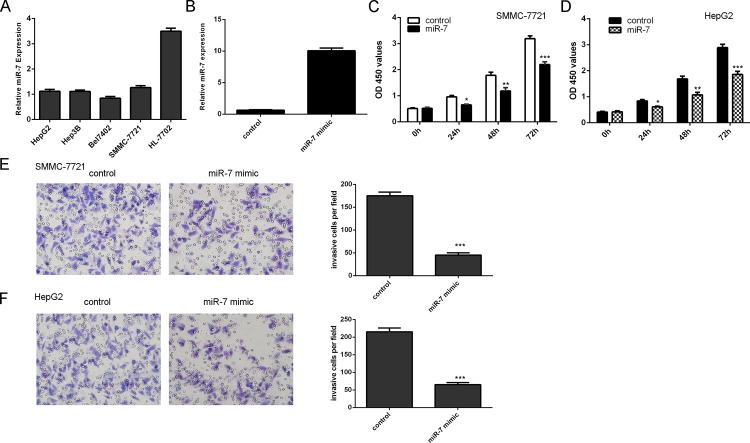
We found that miR-7 expression was downregulated in HCC cell lines (HepG2, Hep3B, Bel7402 and SMMC-7721) compared with the HL-7702 cell (Fig 3A). The expression of miR-7 was increased after treated with miR-7 mimics (Fig 3B). Overexpression of miR-7 suppressed the SMMC-7721 and HepG2 cell proliferation (Fig 3C and 3D). We also found that overexpression of miR-7 inhibited the SMMC-7721 and HepG2 cell invasion (Fig 3E and 3F).
Fig 3. Overexpression of miR-7 inhibited the HCC cell proliferation and invasion.
(A) The expression of miR-7 in the HCC cell lines were measured by using qRT-PCR. (B) miR-7 mimic could increase miR-7 expression in theSMMC-7721 cell. (C) Overexpression of miR-7 inhibited the SMMC-7721 cell proliferation. (D) The HepG2 cell proliferation was measured by CCK-8 assay. (E) Overexpression of miR-7 suppressed the SMMC-7721 cell invasion.(F) The HepG2 cell invasion was detected by cell invasion assay. *p<0.05, **p<0.01 and ***p<0.001. The miR-7 mimic panel of Fig 3E is excluded from this article's CC-BY license. See the accompanying retraction notice for more information.
Knockdown of Cdr1as promoted theexpression of miR-7 as well as its target gene CCNE1 and PIK3CD
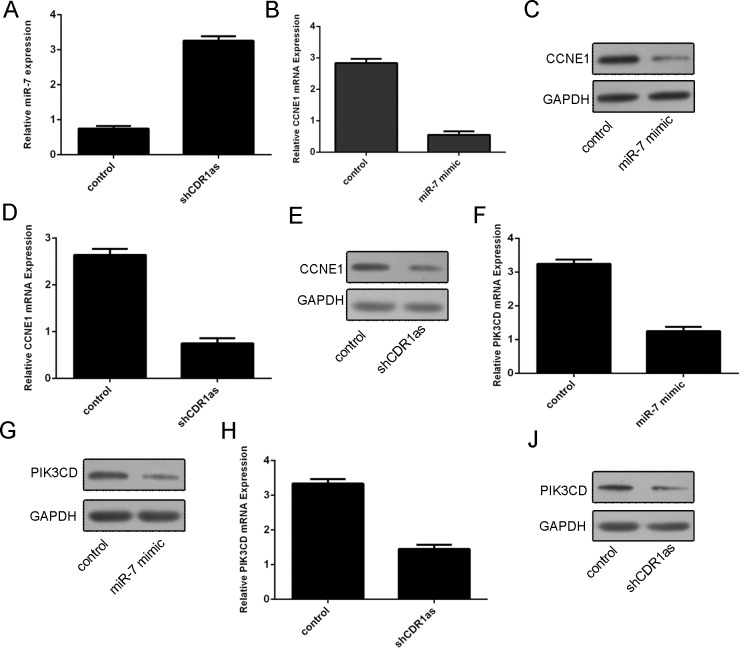
We demonstrated that knockdown of Cdr1as could promote the expression of miR-7 (Fig 4A). Overexpression of miR-7 suppressed the CCNE1 expression (Fig 4B and 4C). In addition, knockdown of Cdr1as suppressed the expression of CCNE1 (Fig 4D and 4E). We also demonstrated the overexpression of miR-7 suppressed thePIK3CD expression (Fig 4F and 4G). Knockdown of Cdr1as inhibited the expression of PIK3CD (Fig 4J and 4H).
Fig 4. Knockdown of Cdr1as promoted the miR-7 and its target gene CCNE1 and PIK3CD expression.
(A) The mRNA expression of miR-7 in the SMMC-7721 cell was measured by qRT-PCR. (B) The mRNA expression of CCNE1 was detected by qRT-PCR in the SMMC-7721 cell. (C) The protein expression of CCNE1 was detected by western blot. (D) The mRNA expression of CCNE1 in the SMMC-7721 cell was detected by qRT-PCR. (E) The protein expression of CCNE1 was detected by western blot. (F) The mRNA expression of PIK3CD was detected by qRT-PCR in the SMMC-7721 cell. (G) The protein expression of PIK3CD was measured using western blot. (H) The mRNA expression of PIK3CD was detected by qRT-PCR in the SMMC-7721 cell. (J) The protein expression of PIK3CD was determined by western blot.
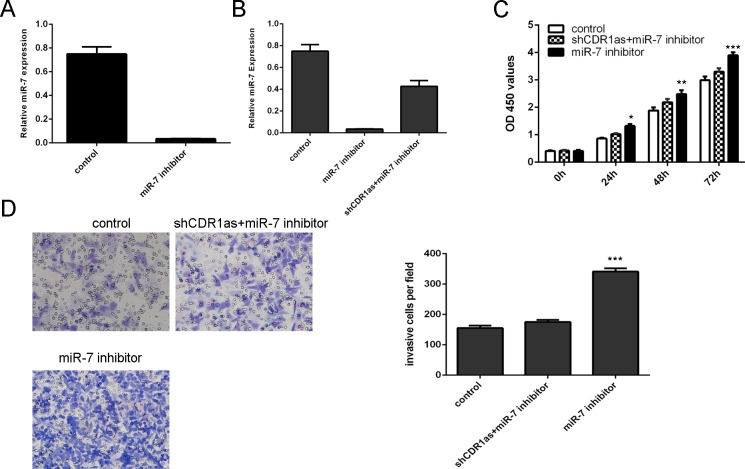
Knockdown of Cdr1as suppressed HCC cell proliferation and invasion through targeting miR-7
We confirmed that miR-7expression was decreased after treated with miR-7 inhibitor (Fig 5A). Knockdown of Cdr1as could rescue this effect (Fig 5B). Inhibition of miR-7 could promote the SMMC-7721 cell proliferation, however, this effect can be abolished by shCdr1as (Fig 5C). Inhibition of miR-7 could promote the SMMC-7721 cell invasion, however, this effect could be abolished by shCdr1as (Fig 5D).
Fig 5. Knockdown of Cdr1as suppressed the HCC cell proliferation and invasion through targeting miR-7 expression.
(A) The expression of miR-7 in the SMMC-7721 cell was determined by qRT-PCR. (B) The expression of miR-7 was measured by qRT-PCRinthe SMMC-7721 cell. (C) Cell proliferation was detected by CCK-8 analysis in the SMMC-7721 cell. (D) The cell invasive ability was measured by invasion assay in the SMMC-7721 cell. *p<0.05, **p<0.01 and ***p<0.001. Fig 5D is excluded from this article's CC-BY license. See the accompanying retraction notice for more information.
Discussion
In this study, we demonstrated that Cdr1asexpression was upregulated in HCC tissues compared with the adjacent non-tumor tissues. Among them,Cdr1as expression was upregulated in 74% (26/35) HCC tissues compared with their adjacent non-tumor tissues. MiR-7 expression was downregulated in HCC tissues compared with adjacent non-tumor tissues. Interestingly, the expression level of miR-7 was inversely correlated with the expressionlevel of Cdr1as. Knockdown of Cdr1as suppressed the HCC cell proliferation and invasion. Overexpression of miR-7 inhibited HCC cell proliferation and invasion. We confirmed that overexpression of miR-7 could suppress the expression of CCNE1 and PIK3CD. Knockdown of Cdr1as suppressed the expression of miR-7 and also inhibited the CCNE1 and PIK3CD expression. Furthermore, knockdown of Cdr1as suppressed HCC cell proliferation and invasion through targeting miR-7. These data suggested that Cdr1as acted as an oncogene partly through targeting miR-7.
Recent study showed that Cdr1as (also namedas ciRS-7) was theinhibitor and sponge of miR-7 in the embryonic zebrafish [19]. Ectopic expression of Cdr1as induced midbrain brain defects, which was similar with the phenotypes found in the knockdown of miR-7[19]. Xu et al[30]. demonstrated that Cdr1as expression was upregulated in islet cells after treated with PMA stimulation or long-term forskolin through targeting PKC and cAMP pathways. Overexpression of Cdr1as promoted insulin secretion and content in islet cells. However, the expression and role of Cdr1as in the development of HCC are still unknown. In this study, we found that the expression of Cdr1as was upregulated in HCC tissues compared with the adjacent non-tumor tissues. Among them, Cdr1as expression was upregulated in 74% (26/35) HCC tissues compared with their adjacent non-tumor tissues. We also confirmed that the expression level of miR-7 was downregulated in HCC tissues compared with the adjacent non-tumor tissues. Furthermore, the expression levelof miR-7 was inversely correlated with the expressionlevel of Cdr1as.
We further demonstrated that knockdown of Cdr1as suppressed the expression of miR-7. Previous studies demonstrated that miR-7 acted as a tumor suppressor gene in HCC [31–34]. For example, Fang et al[31]. demonstrated that overexpression of miR-7 suppressed HCC cell proliferation and metastasis both in vivo and in vitro through targeting phosphoinositide 3-kinase catalytic subunit delta (PIK3CD). They also found there was a negative correlation between PIK3CD and miR-7 expression in HCC tissues. Ma et al[33]. found that the expression of miR-7 was decreased in HCC tissues compared withthe non-tumor tissues. Ectopic expression of miR-7 suppressed HCC cell colony formation and induced cell arrest in the G1/S phase through targeting Cullin 5 (CUL5). Furthermore, Zhang et al[34]. showed that overexpression of miR-7 arrested HCC cell cycle at G1 to S phase through inhibiting CCNE1 expression. In line with this, we found that miR-7 expression was downregulated in HCC tissues and overexpression of miR-7 suppressed HCC invasion and proliferation. MiR-7 overexpression suppressed the expression of CCNE1 and PIK3CD. Knockdown of Cdr1as suppressed the expression of miR-7, as well as the expression of CCNE1 and PIK3CD. In addition, knockdown of Cdr1as suppressed the HCC cell proliferation and invasion through targeting miR-7.
In conclusion, we demonstrated that Cdr1asexpression was upregulated in HCC tissues compared with the adjacent non-tumor tissues. The expression level of Cdr1as was inversely correlated with that of miR-7 in HCC tissues. Knockdown of Cdr1as suppressed HCC cell proliferation and invasion. These data suggested Cdr1as acted as an oncogene partly through targeting miR-7 in HCC.
Supporting Information
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This work was supported by the Postdoctoral scientific research developmental fund of Heilongjiang Province (No. LBH-Q14115), and sponsored by National Science and Technology Major Project (2014ZX10002002) and Natural Science Foundation of Heilongjiang Province (ZD2015019), URL: http://jj.hljkj.cn/zr/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yu L, Ding GF, He C, Sun L, Jiang Y, Zhu L. MicroRNA-424 is down-regulated in hepatocellular carcinoma and suppresses cell migration and invasion through c-Myb. PloS one. 2014;9(3):e91661. Epub 2014/03/29. doi: 10.1371/journal.pone.0091661 ; PubMed Central PMCID: PMC3968007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Yu L, Gong X, Sun L, Yao H, Lu B, Zhu L. miR-454 functions as an oncogene by inhibiting CHD5 in hepatocellular carcinoma. Oncotarget. 2015;6(36):39225–34. Epub 2015/08/20. doi: 10.18632/oncotarget.4407 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu L, Zhou L, Cheng Y, Sun L, Fan J, Liang J, et al. MicroRNA-543 acts as an oncogene by targeting PAQR3 in hepatocellular carcinoma. American journal of cancer research. 2014;4(6):897–906. Epub 2014/12/19. ; PubMed Central PMCID: PMC4266721. [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Yan J, Zhou C, Ma Q, Jin Q, Yang Z. miR-1285-3p acts as a potential tumor suppressor miRNA via downregulating JUN expression in hepatocellular carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014. Epub 2014/09/19. doi: 10.1007/s13277-014-2622-5 . [DOI] [PubMed] [Google Scholar]
- 5.Zheng Q, Sheng Q, Jiang C, Shu J, Chen J, Nie Z, et al. MicroRNA-452 promotes tumorigenesis in hepatocellular carcinoma by targeting cyclin-dependent kinase inhibitor 1B. Molecular and cellular biochemistry. 2014;389(1–2):187–95. Epub 2014/01/02. doi: 10.1007/s11010-013-1940-z . [DOI] [PubMed] [Google Scholar]
- 6.Li QJ, Zhou L, Yang F, Wang GX, Zheng H, Wang DS, et al. MicroRNA-10b promotes migration and invasion through CADM1 in human hepatocellular carcinoma cells. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33(5):1455–65. Epub 2012/04/25. doi: 10.1007/s13277-012-0396-1 . [DOI] [PubMed] [Google Scholar]
- 7.Furuta M, Kozaki K, Tanimoto K, Tanaka S, Arii S, Shimamura T, et al. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PloS one. 2013;8(3):e60155. Epub 2013/04/02. doi: 10.1371/journal.pone.0060155 ; PubMed Central PMCID: PMC3609788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Yu J, Yin J, Xiang Q, Tang H, Lei X. MiR-195 regulates cell apoptosis of human hepatocellular carcinoma cells by targeting LATS2. Die Pharmazie. 2012;67(7):645–51. Epub 2012/08/15. . [PubMed] [Google Scholar]
- 9.Zhou X, Zhang CZ, Lu SX, Chen GG, Li LZ, Liu LL, et al. miR-625 suppresses tumour migration and invasion by targeting IGF2BP1 in hepatocellular carcinoma. Oncogene. 2014. Epub 2014/03/19. doi: 10.1038/onc.2014.35 . [DOI] [PubMed] [Google Scholar]
- 10.Bae HJ, Noh JH, Kim JK, Eun JW, Jung KH, Kim MG, et al. MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene. 2014;33(20):2557–67. Epub 2013/06/04. doi: 10.1038/onc.2013.216 . [DOI] [PubMed] [Google Scholar]
- 11.Giray BG, Emekdas G, Tezcan S, Ulger M, Serin MS, Sezgin O, et al. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Molecular biology reports. 2014;41(7):4513–9. Epub 2014/03/07. doi: 10.1007/s11033-014-3322-3 . [DOI] [PubMed] [Google Scholar]
- 12.Liao CG, Kong LM, Zhou P, Yang XL, Huang JG, Zhang HL, et al. miR-10b is overexpressed in hepatocellular carcinoma and promotes cell proliferation, migration and invasion through RhoC, uPAR and MMPs. Journal of translational medicine. 2014;12(1):234. Epub 2014/09/23. doi: 10.1186/s12967-014-0234-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. Journal of hepatology. 2014;60(3):590–8. Epub 2013/11/12. doi: 10.1016/j.jhep.2013.10.028 . [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Feng Z, Huang R, Xia Z, Xiang G, Zhang J. MicroRNA-449 suppresses proliferation of hepatoma cell lines through blockade lipid metabolic pathway related to SIRT1. International journal of oncology. 2014;45(5):2143–52. Epub 2014/08/15. doi: 10.3892/ijo.2014.2596 . [DOI] [PubMed] [Google Scholar]
- 15.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A new star of noncoding RNAs. Cancer letters. 2015;365(2):141–8. Epub 2015/06/09. doi: 10.1016/j.canlet.2015.06.003 . [DOI] [PubMed] [Google Scholar]
- 16.Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochimica et biophysica acta. 2015. Epub 2015/07/15. doi: 10.1016/j.bbagrm.2015.07.007 . [DOI] [PubMed] [Google Scholar]
- 17.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. American journal of cancer research. 2015;5(2):472–80. Epub 2015/05/15. ; PubMed Central PMCID: PMC4396047. [PMC free article] [PubMed] [Google Scholar]
- 18.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clinica chimica acta; international journal of clinical chemistry. 2015;444:132–6. Epub 2015/02/18. doi: 10.1016/j.cca.2015.02.018 . [DOI] [PubMed] [Google Scholar]
- 19.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. Epub 2013/03/01. doi: 10.1038/nature11928 . [DOI] [PubMed] [Google Scholar]
- 20.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–42. Epub 2014/11/19. doi: 10.1261/rna.047126.114 ; PubMed Central PMCID: PMC4238349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21(12):2076–87. Epub 2015/10/16. doi: 10.1261/rna.052282.115 ; PubMed Central PMCID: PMC4647462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao ZJ, Shen J. Circular RNA Participates in the Carcinogenesis and the Malignant Behavior of Cancer. RNA biology. 2015:0. Epub 2015/12/10. doi: 10.1080/15476286.2015.1122162 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley interdisciplinary reviews RNA. 2015;6(5):563–79. Epub 2015/08/01. doi: 10.1002/wrna.1294 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C, Shan G. What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription. 2015;6(4):61–4. Epub 2015/07/17. doi: 10.1080/21541264.2015.1071301 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. Epub 2013/03/01. doi: 10.1038/nature11993 . [DOI] [PubMed] [Google Scholar]
- 26.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome biology. 2014;15(7):409. Epub 2014/07/30. doi: 10.1186/s13059-014-0409-z ; PubMed Central PMCID: PMC4165365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boeckel JN, Jae N, Heumuller AW, Chen W, Boon RA, Stellos K, et al. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circulation research. 2015;117(10):884–90. Epub 2015/09/18. doi: 10.1161/CIRCRESAHA.115.306319 . [DOI] [PubMed] [Google Scholar]
- 28.Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome biology. 2015;16(1):245. Epub 2015/11/07. doi: 10.1186/s13059-015-0801-3 ; PubMed Central PMCID: PMC4635978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Frontiers in genetics. 2013;4:307. Epub 2014/01/16. doi: 10.3389/fgene.2013.00307 ; PubMed Central PMCID: PMC3875874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Scientific reports. 2015;5:12453. Epub 2015/07/28. doi: 10.1038/srep12453 ; PubMed Central PMCID: PMC4515639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55(6):1852–62. Epub 2012/01/12. doi: 10.1002/hep.25576 . [DOI] [PubMed] [Google Scholar]
- 32.Chen YJ, Chien PH, Chen WS, Chien YF, Hsu YY, Wang LY, et al. Hepatitis B Virus-Encoded X Protein Downregulates EGFR Expression via Inducing MicroRNA-7 in Hepatocellular Carcinoma Cells. Evidence-based complementary and alternative medicine: eCAM. 2013;2013:682380. Epub 2013/07/11. doi: 10.1155/2013/682380 ; PubMed Central PMCID: PMC3693120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma C, Qi Y, Shao L, Liu M, Li X, Tang H. Downregulation of miR-7 upregulates Cullin 5 (CUL5) to facilitate G1/S transition in human hepatocellular carcinoma cells. IUBMB life. 2013;65(12):1026–34. Epub 2013/12/18. doi: 10.1002/iub.1231 . [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Hu S, Wang L, Yan B, Zhao J, Yang A, et al. MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochemical and biophysical research communications. 2014;443(3):1078–84. Epub 2013/12/29. doi: 10.1016/j.bbrc.2013.12.095 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.