Abstract
Balancer chromosomes are multiply inverted chromosomes that suppress meiotic crossing over and prevent the recovery of crossover products. Balancers are commonly used in Drosophila melanogaster to maintain deleterious alleles and in stock construction. They exist for all three major chromosomes, yet the molecular location of the breakpoints and the exact nature of many of the mutations carried by the second and third chromosome balancers has not been available. Here, we precisely locate eight of 10 of the breakpoints on the third chromosome balancer TM3, six of eight on TM6, and nine of 11 breakpoints on TM6B. We find that one of the inversion breakpoints on TM3 bisects the highly conserved tumor suppressor gene p53—a finding that may have important consequences for a wide range of studies in Drosophila. We also identify evidence of single and double crossovers between several TM3 and TM6B balancers and their normal-sequence homologs that have created genetic diversity among these chromosomes. Overall, this work demonstrates the practical importance of precisely identifying the position of inversion breakpoints of balancer chromosomes and characterizing the mutant alleles carried by them.
Keywords: balancer chromosomes, p53, meiosis, whole-genome sequencing, crossing over
Balancer chromosomes are multiply rearranged chromosomes that are used extensively in Drosophila melanogaster for tasks such as stock construction and the maintenance of recessive deleterious alleles in populations (Ashburner et al. 2005). Balancers work by suppressing meiotic crossing over, by creating recombinant chromatids that will not segregate properly during the first meiotic division (Novitski and Braver 1954), or, in the case of pericentric inversions, by creating recombinants that carry duplications or deficiencies large enough to result in zygotic lethality. While all balancer chromosomes carry easily scored dominant marker alleles that allow for visual identification of flies carrying the balancer, most balancers also carry recessive lethal mutations that prevent the balancer from becoming homozygous in stock (Lindsley and Zimm 1992; Ashburner et al. 2005).
A variety of balancers are available for the X, second, and third chromosomes in Drosophila, and they have become increasingly effective as the number of inversions has increased and as visible markers and recessive lethal or sterile alleles have been added. For example, First Multiple one (FM1), an X chromosome balancer, improved upon earlier single-inversion balancers such as In(1)dl-49, In(1)sc, and ClB, by combining the In(1)dl-49 and In(1)sc inversions into one chromosome (Lindsley and Zimm 1992; Ashburner et al. 2005). Further improvements generated a series of FM balancers, and similar series exist for the second (second multiple; SM) and third (third multiple; TM) chromosomes (Lindsley and Zimm 1992). The current study will focus on the third chromosome balancers TM3, TM6, and TM6B.
TM3 was created in the late 1950s by repeated X-raying of a chromosome marked with kniri-1, pp, vvlsep (previously known as sep1), Ubxbx-34e, and e1, and carrying two inversions, In(3LR)sep (65D2-3;85F2-4) and In(3R)C (92D1-E1;100F2-3). The irradiation superimposed three additional inversions on this chromosome, creating a balancer with five total inversions (Lewis 1960) (Figure 1). Tinderholt (1960) introduced the dominant markers Serrate (Ser) and Stubble (Sb) into inverted regions of this chromosome by double crossing over, relying on the increased recombination created by the so-called interchromosomal effect (Schultz and Redfield 1951; Ramel 1966) to obtain these double crossovers (DCOs). Specifically, Tinderholt (1960) performed this synthesis in a female heterozygous for three balancers to increase the likelihood of recombination within the desired inversions. In doing so, he demonstrated that segments could be swapped into inverted segments of TM3—even if such events were uncommon.
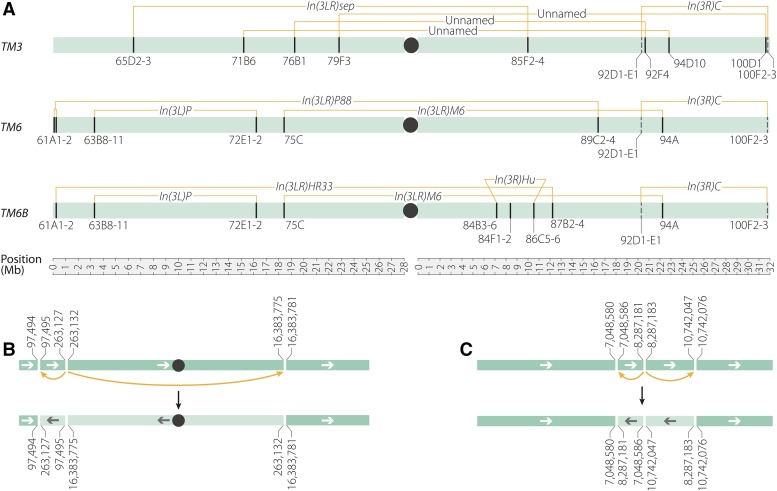
Figure 1.
TM3, TM6, and TM6B inversion breakpoints. Black circles indicate centromeres and left-facing arrows indicate an inverted segment. (A) The inversions carried by the third chromosome balancers TM3, TM6, and TM6B. Breakpoints that have been molecularly identified are shown as solid lines; those that are estimates are shown as dashed lines; numbers are cytological bands of breakpoints given in Lindsley and Zimm (1992). (B) The In(3LR)P88 (61A1-2;89C2-4) rearrangement on TM6 is a previously unreported three-breakpoint rearrangement with a breakpoint at 3L:263,127–263,132 that bisects the gene Tudor-SN, a breakpoint at 3R:16,383,781 that bisects spineless, an allele previously reported to be carried by this chromosome (Duncan et al. 1998), and a breakpoint at 3L:97,494 that is intergenic. (C) In the In(3R)Hu (84B1;84F4;86C7-8) three-breakpoint rearrangement on TM6B, the breakpoint at 3R:8,287,181 bisects the noncoding RNA gene CR44318 while the 3R:10,742,076 breakpoint bisects TkR86C. The breakpoint at 3R:7,048,580 likely causes the AntpHu phenotype.
The progenitor chromosome that was X-rayed to produce TM3 also carried Dp(1;3)sc260-20, an aberration that replaced the tip of chromosome 3L with the tip of an X chromosome carrying a wild-type allele of the yellow (y) gene (Sutton 1943). However, this y+ marker was frequently lost by a single crossover event between TM3 and normal-sequence chromosomes in the region distal to the 65D inversion breakpoint; consequently, most TM3 chromosomes now carry a normal 3L tip (Shearn 1980). This is one of several observations indicating that the relatively large uninverted region distal to 65D undergoes frequent exchange events—even though recombination is largely suppressed in regions proximal to 65D.
TM6 was created by X-ray mutagenesis of a chromosome marked with Ubxbx-34e and e1 and carrying three preexisting inversions: In(3L)P (63C;72E1-2) lying inside In(3LR)P88 (61A;89CD) with In(3R)C (92D1-E1;100F2-3) to the right (Figure 1). Irradiation resulted in an additional inversion, In(3LR)M6, between bands 75C and 94A (Lindsley and Zimm 1992). TM6B was built from TM6 by replacing the left breakpoint of In(3LR)P88 with the left end of In(3LR)HR33 (61A1-2;87B) (Ashburner 1972) by a single crossover (Figure 1). The three-breakpoint rearrangement In(3R)Hu (84B1;84F4;86C7-8) (Hazelrigg and Kaufman 1983) was carried onto the recombinant chromosome along with the left end of In(3LR)HR33 from a double-aberration progenitor. An internal segment spanning the right breakpoint of In(3LR)P88 was then replaced with a segment spanning the right breakpoint of In(3LR)HR33 by a DCO. Finally, the dominant Tubby (Tb1) marker was added by a DCO event within an inverted segment near the right end of the newly created TM6B (Craymer 1981, 1984; Lindsley and Zimm 1992).
Because balancers are used widely in Drosophila experiments, sometimes as heterozygous controls, it is informative for the community to determine the exact position of their breakpoints and the nature of the alleles carried by them. A recent study reported rare DCO events between the X chromosome balancer FM7 and its normal sequence homologs that were selected for because they conferred an advantage to flies carrying the recombinant chromosome (Miller et al. 2016a). A similar whole-genome analysis of commonly used autosomal balancers has not yet been conducted.
Here, we use whole-genome sequencing to identify all but one shared pair of inversion breakpoints on the TM3, TM6, and TM6B balancer chromosomes (Figure 1). Importantly, we find that the breakpoint at 94D on TM3 splits the highly conserved tumor suppressor gene p53 in half, demonstrating that any stock balanced with TM3 is heterozygous for a p53 loss-of-function allele. We also find evidence of single and double crossover events on more than half of the TM3 chromosomes sampled and on one TM6B chromosome, and we are able to estimate the distance over which inversions suppress exchange by examining single crossover events that occur in an unbalanced region of the TM3 chromosome. These findings demonstrate that, similar to the X chromosome balancer FM7, sequence diversity exists among third chromosome balancers and suggest that this variation may influence experimental outcomes.
Materials and Methods
Stocks used for breakpoint identification and validation
Stocks used in this study, along with their current Bloomington IDs and genotypes, are listed in Supplemental Material, Table S2. Throughout the manuscript we have referred to stocks as <balancer chromosome>-<Bloomington Stock ID> unless they were lab stocks not available at the Bloomington stock center, in which case they are referred to as <balancer chromosome>-lab. For example, TM3-500 is the TM3 chromosome in Bloomington stock 500. The ISO-1 (y1; Gr22biso-1 Gr22diso-1 cn1 CG33964iso-1 bw1 sp1; LysCiso-1 MstProxiso-1 GstD5iso-1 Rh61) stock used to create heterozygous TM3 and TM6B flies for sequencing was obtained from Sue Celniker (Lawrence Berkeley National Laboratory). The single TM6 chromosome used in this study was not sequenced as an ISO-1/TM6 heterozygote, but as a +/TM6 heterozygote. All flies were kept on standard cornmeal-molasses medium and maintained at 25°.
DNA preparation and genome alignment
DNA for sequencing was prepared from either heterozygous males or a combination of heterozygous males and heterozygous females using the Qiagen DNeasy Blood and Tissue Kit. All flies were starved for 1 hr before freezing at –80° for at least 1 hr. Mate pair DNA libraries for stocks CyO-TM3-mp-22239, SM6a-TM3-mp-lab, and TM6B-mp-587 were generated from 1 μg of high-quality genomic DNA. Following the manufacturer’s directions, libraries were generated using the gel-free method of the Illumina Nextera Mate Pair Library Preparation kit, with 10 cycles of PCR amplification. Resulting libraries were checked for quality and quantity using a Bioanalyzer 2100 (Agilent) and Qubit Fluorometer (Life Technologies). All libraries were pooled, requantified, and sequenced as 150-bp paired end on an Illumina NextSeq 500 instrument. Following sequencing, Illumina Real Time Analysis version 2.4.6 was run to demultiplex reads and generate FASTQ files; 100 ng of sample TM6-lab was sheared using the Covaris s220 instrument to 300 bp, and prepared using the KAPA HTP Library Prep Kit for Illumina and Bioo Scientific NEXTflex DNA barcodes. The resulting library was quantified using a LabChip GXII (Perkin Elmer), and a Qubit Fluorometer (Life Technologies). This library was pooled with others, requantified and sequenced as 150-bp paired end on an Illumina HiSeq 2500 in rapid mode. Following sequencing, Illumina Real Time Analysis version 1.17.21.3 and CASAVA version 1.8.2 were run to demultiplex reads and generate FASTQ files. For the remainder of samples used in this study, 500 ng of DNA from each was fragmented to 600-bp fragments using a Covaris S220 sonicator by adjusting the treatment time to 30 sec, except for sample CyO-TM3-504, which was sonicated using 89 ng of DNA and was not size selected. Libraries were prepared using the KAPA HTP Library Prep Kit for Illumina and Bioo Scientific NEXTflex DNA barcodes. The resulting libraries were quantified using a Bioanalyzer (Agilent Technologies) and a Qubit Fluorometer (Life Technologies). All libraries were pooled, requantified, and sequenced as 150-bp paired end on the Illumina NextSeq 500 instrument. Following sequencing, Illumina Real Time Analysis version 2.4.6 was run to demultiplex reads and generate FASTQ files. Alignment to the D. melanogaster reference genome (dm6) was performed using bwa version 0.7.7-r441 (Li and Durbin 2009). SNPs were called using SAMtools and BCFtools (version 0.1.19-44428cd) (Li et al. 2009). Indels were not considered and low quality SNPs (those with quality scores < 200) were filtered out. All TM3 and TM6B chromosomes were sequenced as balancer/ISO-1, allowing us to treat every heterozygous SNP as a SNP present on the balancer chromosome.
Identification and validation of inversion breakpoints
Breakpoints were identified as reported in Miller et al. (2016a). Briefly, split or discordant read pairs were isolated using Samblaster (Faust and Hall 2014), and known regions of repetitive or low-complexity sequence were masked with repeatmasker (Chen 2004). Separately, we used BreakDancer (Chen et al. 2009) to identify candidate inversion breakpoints. Regions where BreakDancer identified large inversion polymorphisms and where rearrangements were previously reported to be present (Lindsley and Zimm 1992) were analyzed in 1-kb windows for regions that contained more than 10 split or discordant read pairs. Breakpoints were visually validated using Integrative Genomics Viewer (Thorvaldsdottir et al. 2013) and the UCSC Genome Browser (Rosenbloom et al. 2015). Original FASTQ reads from each breakpoint were collected and de novo assembled using SOAPdenovo2 with a kmer size of 41 (Luo et al. 2012). Primers for PCR validation were designed using Primer3 (Rozen and Skaletsky 2000). PCR was done with Phusion polymerase, and Sanger sequencing confirmed each breakpoint. PCR primers used to validate inversion breakpoints are listed in Table S1.
Identification of crossover events and generation of heatmaps
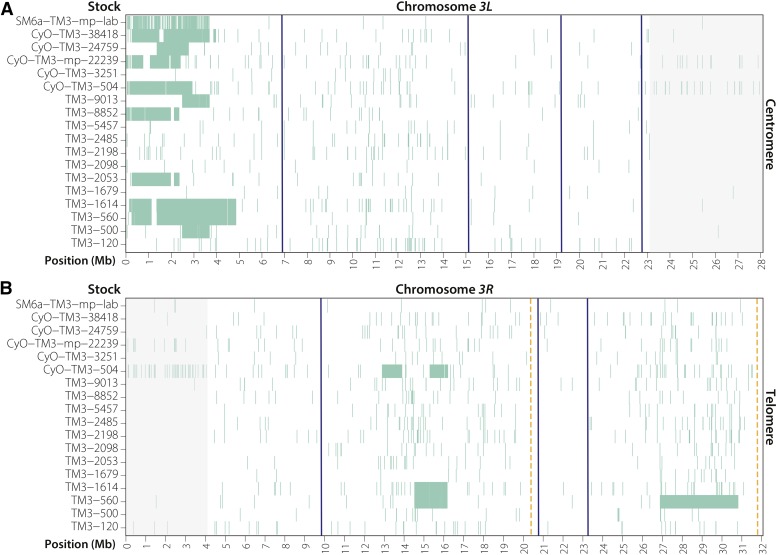
Because no TM3 or TM6B reference sequence exists, we identified crossover tracts by comparing the SNP profile of each individual chromosome to all chromosomes of the same type. Specifically, for each SNP from each stock, we checked all other stocks to see if the same polymorphism was present. If it was, then this was considered a variant carried by all balancers of that type. If a SNP was observed in only one stock, then we considered it a unique polymorphism. To build the heatmap in Figure 2, we plotted SNPs in 10-kb windows if that SNP was present in five or fewer TM3 stocks. The interval on TM3 between the telomere and the 65D breakpoint at 3L:6,925,034 contained ∼43,000 informative SNPs in each of the TM3 stocks sequenced. For the three TM6B stocks represented in Figure 3, we plotted completely unique SNPs, or any SNP not shared by all three chromosomes.
Figure 2.
Visualizing SNPs present in five or fewer TM3 chromosomes reveals numerous single crossover events on 3L and several DCO events on 3R (see Materials and Methods). Blue lines indicate the positions of inversion breakpoints whose precise location is known, orange dashed lines show the approximate positions of the unidentified In(3R)C (92D1-E1;100F2-3) inversion breakpoints. Gray shaded regions are centromere-proximal heterochromatin with low-quality read alignment. (A) Single crossovers are common in the region distal to the 65D inversion breakpoint at position 3L:6,925,034, and occur no closer than 2 Mb from the breakpoint. (B) Several DCOs are apparent on 3R. Stocks TM3-560 and TM3-1614 may be versions of TM3 before Ser1 was added to a TM3, Sb+ Ser+ chromosome (TM3-560), and before Sb1 was added to a TM3, Sb+ Ser1 chromosome (TM3-1614).
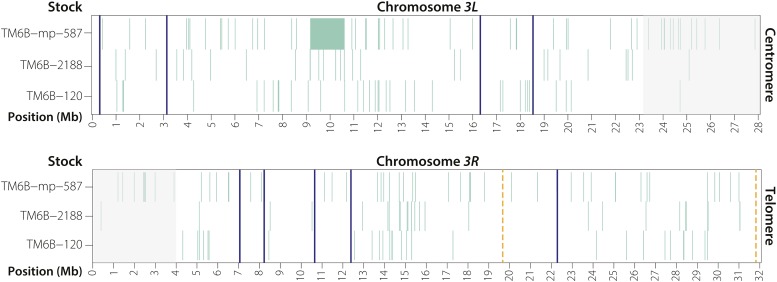
Figure 3.
Unique SNPs present among the three TM6B chromosomes sequenced in this study. Gray shaded regions are centromere-proximal heterochromatin with low-quality read alignment. Blue lines indicate the positions of inversion breakpoints, orange dashed lines indicate the approximate positions of the unidentified In(3R)C (92D1-E1;100F2-3) inversion breakpoints. A single DCO event was recovered in stock TM6B-mp-587.
Data availability
Raw sequencing data for all samples used in this project have been uploaded to the National Center for Biotechnology Information (NCBI) at ncbi.nlm.nih.gov and can be found under BioProject PRJNA315473. Laboratory strains of wgSp-1/SM6a, DuoxCy; Pr1/TM3, Sb1 Ser1, and +/TM6 are available upon request; all other strains listed in Table S2 are available from the Bloomington Drosophila Stock Center.
Results
Using whole-genome sequencing, we precisely identified eight of the 10 breakpoints on TM3, six of eight breakpoints on TM6, and nine of 11 breakpoints on TM6B (Figure 1A and Table 1). The three balancers share an inversion, In(3R)C, between cytological bands 92D1-E1 and 100F2-3 (Sturtevant 1913; Muller 1918) that we were unable to accurately position because its location near the telomere suggests that it most likely involves highly repetitive sequences. Note that, throughout the manuscript, we refer to breakpoints by the names of the inversions that created them. We also use the observed cytological bands reported in Lindsley and Zimm (1992) rather than the estimated cytological bands that are available on FlyBase or the UCSC genome browser.
Table 1. Molecular details of the TM3, TM6, and TM6B inversion breakpoints.
| Balancer | Inversion | Chr | Reported bandsa | 5′ Break | 3′ Break | Duplication (+) /Deletion (-) | Affected Gene/Region |
|---|---|---|---|---|---|---|---|
| TM3 | In(3LR)sep | 3L | 65D2-3 | 6,925,034 | 6,926,125 | −1090 | Intergenic |
| 3R | 85F2-4 | 9,943,831 | 9,944,040 | −208 | Glut4EF | ||
| TM3 | Unnamed | 3L | 71B6 | 15,150,269 | 15,150,272 | −2 | FucTA |
| 3R | 94D10 | 23,050,763 | 23,050,764 | 0 | p53 | ||
| TM3 | Unnamed | 3L | 76B1 | 19,386,273 | 19,388,151 | −1877 | CG32206, ms(3)76Ba |
| 3R | 92F4 | 20,637,930 | 20,637,930 | +1 | Lrrk | ||
| TM3 | Unnamed | 3L | 79F3 | 22,637,876 | 22,637,952 | −75 | CG14459 |
| 3R | 100D1 | 31,653,695 | 31,653,707 | −11 | kek6 | ||
| TM3, TM6, TM6B | In(3R)C | 3R | 92D1-E1 | Unknown | Unknown | — | Unknown |
| 3R | 100F2-3 | Unknown | Unknown | — | Unknown | ||
| TM6 | In(3LR)P88 | 3L | 61A1-2 | 97,494 | 97,495 | 0 | Intergenic |
| 3L | 61A1-2 | 263,127 | 263,132 | −4 | Tudor-SN | ||
| 3R | 89C2-4 | 16,383,781 | 16,383,775 | +7 | ss | ||
| TM6, TM6B | In(3LR)M6 | 3L | 75C | 18,693,657 | 18,693,663 | −5 | CR43987 |
| 3R | 94A | 22,393,827 | 22,393,828 | 0 | CG13857 | ||
| TM6, TM6B | In(3L)P | 3L | 63B8-11 | 3,173,046 | 3,173,053 | −6 | CG14964 |
| 3L | 72E1-2 | 16,308,841 | 16,308,845 | −3 | Intergenic | ||
| TM6B | In(3LR)HR33 | 3L | 61A1-2 | 233,562 | 233,565 | −2 | Intergenic |
| 3R | 87B2-4 | 12,227,473 | 12,227,471 | +3 | Intergenic | ||
| TM6B | In(3R)Hu | 3R | 86C5-6 | 10,742,047 | 10,742,076 | −28 | TkR86C |
| 3R | 84F1-2 | 8,287,181 | 8,287,183 | −1 | CR44318 | ||
| 3R | 84B3-6 | 7,048,580 | 7,048,586 | −5 | Intergenic |
Reported bands are those found in Lindsley and Zimm (1992), and are not based on estimated genomic position.
Because autosomal balancers carry recessive lethal mutations, the recovery of homozygous progeny for sequencing is not feasible. To circumvent this problem, we crossed males from each TM3 and TM6B balancer stock to females from the ISO-1 stock that was used to construct the Drosophila reference genome and recovered heterozygous individuals for sequencing (see Materials and Methods). We confirmed breakpoints by two methods: first, we whole-genome sequenced large-insert (2–12 kb) library preparations for two TM3 stocks and one TM6B stock (see Materials and Methods); second, we PCR- and Sanger-sequenced all identified breakpoints on TM3 and TM6, and selected breakpoints on TM6B (Table S1).
Third chromosome balancer breakpoints disrupt protein-coding genes
After identifying the exact position of each inversion breakpoint, we found that the breakpoints on TM3 altered six characterized [Glut4EF, FucTA, p53, ms(3)76Ba, Lrrk, and kek6], and two uncharacterized (CG32206 and CG14459) protein-coding genes (Table 1). Perhaps most importantly, we observed that the 94D inversion breakpoint on TM3 at 3R:23,050,763–23,050,764 bisects the fifth intron of the highly conserved tumor suppressor p53 (Jin et al. 2000) and affects all reported p53 isoforms. We also confirmed that the allele Glut4EFTM3 is caused by the inversion at 85F2 on TM3, as reported by Yazdani et al. (2008) (Table 2). Finally, we found that the y+ X chromosome fragment originally present on TM3 (Lewis 1960; Shearn 1980) was the result of a break of the X chromosome at X:416,997 and subsequent attachment to the third chromosome at 3L:149,709, in agreement with its original isolation as a reciprocal translocation affecting the X-linked scute gene (Sutton 1943). This rearrangement deletes or disrupts 10 protein-coding and eight noncoding RNA genes from the third chromosome in the distal 150-kb interval of TM3.
Table 2.
Genomic aberrations of previously characterized mutations and recessive lethal alleles carried by TM3, TM6, and TM6B
| Gene | Allele | Balancer(s) | Observed aberration | Previous reports |
|---|---|---|---|---|
| ebony | e1 | TM3, TM6, TM6B | TE (family: 412) at 3R:21,231,832–21,231,838, 6 nt into the 2nd exon | — |
| Ultrabithorax | Ubxbx-34e | TM3, TM6 | TE (family: DMIS176) insertion in the first intron of Ubx at approximately 3R:16,731,980 | Gypsy insertion (Bender et al. 1983) |
| knirps | kniri-1 | TM3 | 252-bp deletion at 3L:20,707,101-20,707,352. | (Lunde 2003) |
| pink | pp | TM3 | 1-bp deletion at 3R:6,661,619 resulting in a frameshift | 1-bp deletion at 3R:6,661,624 (Syrzycka et al. 2007) |
| lethal (3) 89Aa | l(3)89Aa1 | TM3 | Unknown | Mapped to 89A2-89A5 |
| ventral veins lacking | vvlsep | TM3 | Unknown | — |
| Stubble | Sb1 | TM3 | TE (family: 412) insertion in 4th exon of Sb at 3R:16,141,939-16,141,942. | TE insertion (Hammonds and Fristrom 2006) |
| Serrate | Ser1 | TM3 | TE (family: TIRANT) insertion at 3R:27,172,910-27,172,913 in the 3’ UTR of Ser | TE insertion (Fleming et al. 1990) |
| Ultrabithorax | UbxP15 | TM6 | Unknown | — |
| Henna | HnP | TM6 | Multiple deletions within the first intron and a G->A mutation at splice acceptor site (AG becomes AA) in the third intron of the gene. | — |
| spineless | ssaP88 | TM6 | Gene is split by the In(3LR)P88 (61A1-2;89C2-4) rearrangement. | Break in the transcription unit (Duncan et al. 1998) |
| Antennapedia | AntpHu | TM6B | Unknown. Phenotype may be a result of the In(3R)Hu triple rearrangement (Figure 1). | — |
| Tubby | Tb1 | TM6B | An in-frame 15-nt deletion in the 2nd exon from 3R:26,656,728-26,656,742; a 69-nt in-frame deletion of 23 amino acids from 3R:26,657,089-26,657,157; and a T->G mutation (Ser->Ala) at 3R:26,657,334. | — |
The breakpoints on TM6 affected four protein-coding genes (Tudor-SN, ss, CG13857, and CG14964) and one noncoding RNA gene (CR43987) (Table 1). Using whole-genome data, we confirmed that the previously reported spineless allele (ssaP88) on TM6, reported as a break in the transcription unit (Duncan et al. 1998), is indeed caused by the inversion at 89C4 (Table 2). We also observed that the In(3LR)P88 (61A;89CD) inversion on TM6, which had been reported to be a simple inversion of 61A to 89C, is instead a three-breakpoint rearrangement that creates a previously unknown 165-kb inversion (Figure 1B and Table 1).
Finally, the TM6B breakpoints affect three protein-coding genes (CG13857, CG14964, and TkR86c) and two noncoding RNA genes (CR43987 and CR44318) (Table 1). We also characterized the three-breakpoint In(3R)Hu (84B1;84F4;86C7-8) rearrangement on TM6B, and found that it consists of 1.2-Mb and 2.5-Mb inverted segments (Figure 1C and Table 1). Based on the positions of these breakpoints, it appears that the gain-of-function mutation AntennapediaHu (AntpHu) is a regulatory mutation caused by the 84B1 inversion breakpoint that lies ∼50 kb away from Antp, as suggested by previous breakpoint mapping (Hazelrigg and Kaufman 1983; Scott et al. 1983).
In addition to the mutations caused by inversion breakpoints, balancer chromosomes carry a number of mutations that provide visible markers for easy identification, as well as recessive lethal alleles that prevent balancers from becoming homozygous in stock. Some of these markers are shared by more than one balancer—such as ebony (e1), present on TM3, TM6, and TM6B—while others are present on only one balancer—such as Tubby (Tb1), present only on TM6B (Table 2). The general nature of many of these alleles has been described previously [such as that a transposable element (TE) insertion in Ultrabithorax gives rise to the Ubxbx-34e allele carried by TM3 and TM6 (Bender et al. 1983), or that a TE insertion is responsible for Ser1 on TM3 (Fleming et al. 1990)], but the specific lesions that convey their respective phenotypes are unknown for most alleles. Using our whole-genome sequencing data, we were able to identify the precise nature of nine of 13 visible or lethal alleles carried by the three balancers analyzed in this study. These data are summarized in Table 2.
The TM3 balancer allows single crossover events distal to 65D
Inversion breakpoints are known to suppress exchange in nearby regions, but the mechanism by which they do this, and over what distance they act, is unknown (Sturtevant and Beadle 1936; Novitski and Braver 1954). Previous work has shown that balancer chromosomes pair along their lengths with their normal-sequence homologs (Gong et al. 2005) and that both crossover-associated and noncrossover gene conversion events occur between balancers and their normal-sequence homologs (Blumenstiel et al. 2009; Miller et al. 2016a). Because the distal-most inversion breakpoint on the left arm of TM3 is 6.9 Mb from the telomere (estimated cytological band 65D3), we hypothesized that single crossover events would be common in this region (Figure 1A). Evidence of recombination within this interval would manifest as tracts of unique SNPs within a pool of TM3 chromosomes; thus, we sequenced a panel of 17 stocks from the Bloomington Drosophila Stock Center and one laboratory stock carrying the TM3 chromosome (Table S2) to identify how close to the inversion breakpoint these crossovers occurred.
We saw evidence of crossing over between the telomere and the most distal 3L inversion breakpoint in 11 of 18 TM3 stocks (Figure 2). Because the ancestral SNP profile of the TM3 chromosome is unknown, we can infer recombination events by identifying SNPs that are unique to only one of the TM3 chromosomes. However, because of the potential relatedness of some chromosomes, we plotted SNPs that were shared by five or fewer TM3 chromosomes. Several stocks in Figure 2A appear to have large gaps lacking SNPs in the intervals of recombination. This is likely evidence of multiple single exchange events in which a TM3 and a normal-sequence third chromosome exchanged distal regions, suggesting that exchange distal to 65D is an ongoing process in stocks.
Several of the crossover tracts recovered are shared among multiple stocks, highlighting the relatedness of these chromosomes. For example, stocks TM3-560 and TM3-1614 share identical SNPs in the 1- to 5-Mb interval of 3L, and stocks TM3-500 and TM3-9013 are nearly identical over 2.5–3.5 Mb in this same region (Figure 2A). Finally, crossovers in stocks TM3-560 and TM3-1614 are observed as close as ∼2 Mb from the inversion breakpoint, the first evidence of the distance over which an inversion breakpoint may suppress exchange.
Double crossover events can occur on TM3 and TM6B
We were also able to identify DCOs that had occurred within inverted segments on both the TM3 and TM6B balancers. We found three DCO events that replaced a mutant copy of Stubble (Sb1) with a wild-type copy in stocks that are phenotypically Sb+ (Figure 2B). Based on shared SNPs, the 1.7-Mb segment between 14.5 Mb and 16.2 Mb in TM3-1614 and TM3-560 appears to have originated from a single DCO in a common progenitor, while the 900-kb segment in CyO-TM3-500 likely arose by an independent DCO event. In addition, we also found a 3.9-Mb DCO that replaced a mutant copy of Serrate (Ser1) with a wild-type copy (Figure 2B). While difficult to confirm, TM3-560 may be directly related to the original isolate of TM3 before Ser1 and Sb1 were added by Tinderholt (1960), and TM3-1614 may be the Sb+ Ser1 version of the chromosome after Ser1 was added and before Sb1 was added (Tinderholt 1960). Ser1 and Sb1 were introduced intentionally by screening for DCOs in the presence of additional balancers, which strengthen the interchromosomal effect on recombination.
The TM3 chromosome carried by the CyO-TM3-504 stock carries a second 1-Mb DCO event near the DCO that replaced Sb1 with Sb+ (Figure 2B). Analysis of this region using SnpEFF (Cingolani et al. 2012) finds no obvious deleterious mutations in this interval on any other TM3 chromosome. We do, however, find a 10-kb tandem duplication within this DCO that fully duplicates CG31157 and CG7966, two uncharacterized genes highly expressed in a variety of tissues, which may confer a competitive advantage to flies carrying the duplication. Interestingly, CG7966, which encodes a selenium-binding protein, is conserved from Drosophila to humans (SELENBP1), which makes this duplication a provocative candidate for further study.
The two presumed DCO events on CyO-TM3-504 are also interesting because of their sizes. At 900 kb and 1 Mb, these are likely the smallest DCO events yet reported in Drosophila—even smaller than the 1.5-Mb DCO observed in a recent study (Miller et al. 2016b). It is unlikely these two DCOs are the result of a single larger DCO at coordinates 12.9–16.2 Mb followed by a second DCO at coordinates 13.9–15.3 Mb, because the second DCO would have had to occur with a homologous TM3 or TM3 progenitor chromosome. A simpler explanation is that these were two independent DCOs.
Finally, we identified a single 1.4-Mb DCO at 3L:9,216,999–10,625,261 on TM6B-587 (Figure 3). It replaces three separate frameshifting deletions in the uncharacterized genes CG46121, CG16711, and CG32055 with wild-type copies—a potential advantage for flies carrying this chromosome. Overall, our findings provide molecular evidence that, while rare, DCO events do occur between TM3 or TM6B balancers and their normal-sequence homologs.
Discussion
We have identified the precise locations of all inversion breakpoints from the Drosophila third chromosome balancers TM3, TM6, and TM6B except for the In(3R)C (92D1-E1;100F2-3) inversion shared by all three chromosomes. We find that one of the TM3 inversion breakpoints bisects all transcripts of the tumor suppressor gene p53, with implications for a wide range of studies in Drosophila. As hypothesized, we identified evidence of single crossover events in the 6.9-Mb interval between the telomere and the most distal inversion breakpoint on TM3 in nearly two-thirds of the stocks we sequenced. These single crossover events provide the first evidence for the distance over which inversion breakpoints can suppress meiotic exchange in Drosophila.
Eleven of 18 TM3 stocks carried evidence of a recombination event between the 65D breakpoint and the telomere, with the closest exchange event occurring ∼2 Mb from the 65D breakpoint. Do all inversion breakpoints suppress exchange in a similar way and over a similar distance? Perhaps the most instructive case is that of the X chromosome inversion In(1)dl-49. The distal-most breakpoint of the inversion lies ∼4.9 Mb from the telomere (2 Mb closer to the telomere than the 65D breakpoint on TM3). Recombination in a single generation was previously measured between the distal-most breakpoint of In(1)dl-49 and the telomere using yellow, a marker near the telomere, and echinus, a marker ∼1 Mb from the most distal In(1)dl-49 breakpoint, and was found to be ∼10% of what it would be in the absence of the inversion (Stone and Thomas 1935; Sturtevant and Beadle 1936). Although we did recover a substantial number of TM3 chromosomes that had undergone distal exchanges, it must be remembered that these could have occurred at any point in the history of each TM3 balancer. While not examined here, it would be interesting to see if recombination is reduced between 65D and the telomere on TM3 within a single generation; we would indeed predict such a reduction. Alternatively, future studies using methods similar to ours could determine exactly how close to other inversions, such as In(1)dl-49, recombination can occur. Either way, the consequence for balanced chromosomes remains the same—crossing over is possible within this region. One feasible explanation for the high diversity in the region distal to 65D observed among the panel of TM3 chromosomes we sampled is that exchange events may confer a competitive advantage in this region and can propagate throughout a stock, although the exact advantage of a recombinant TM3 chromosome remains unclear.
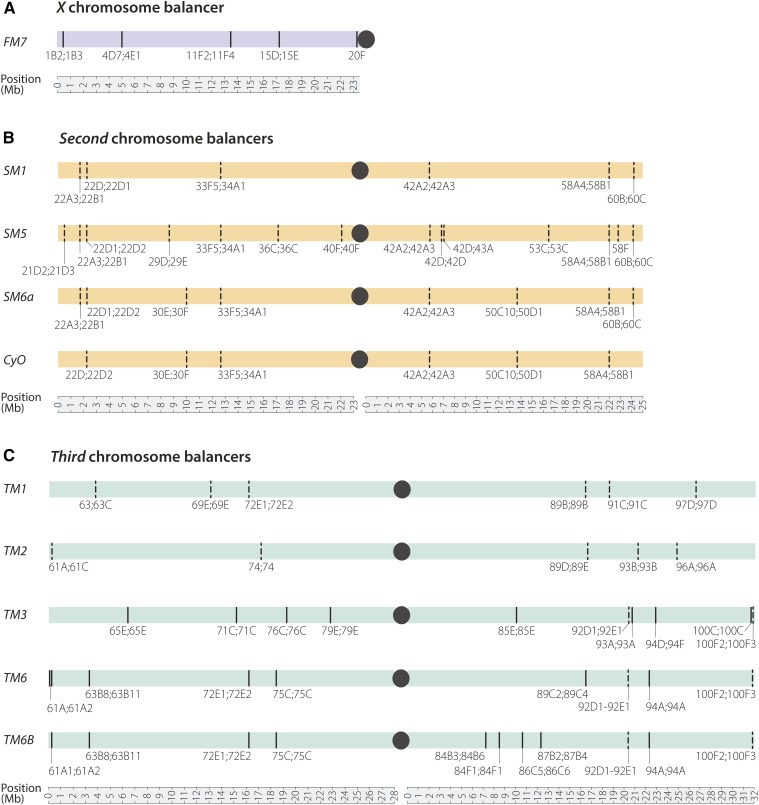
An appreciation that single crossovers can occur distal to the 65D inversion on TM3 also has practical purposes for long-term maintenance of deleterious alleles in stocks. At least 550 stocks at the Bloomington Stock Center have a mutation, transgene insertion, or chromosomal deletion distal to 65D that could be lost by recombination with the TM3 present. Although this number assumes that recombination can occur anywhere from the tip of 3L to the 65D breakpoint, our data suggests an ∼2-Mb buffer over which recombination may be suppressed, potentially reducing the number of vulnerable alleles. Yet the practical implication remains that genetic components thought to be present on all nonbalancer chromosomes in a population may be present in only a subset of individuals in the population, or may have been moved to the balancer chromosome itself. Therefore, it would be prudent for researchers to check for the presence of the desired genetic element distal to 65D in any TM3 stock before undertaking experiments. Furthermore, poorly balanced regions exist at the ends of other popular balancers—including CyO, In(2LR)Gla, and TM1—and these balancers should generally be avoided in constructing stocks with distally located genetic components (Figure 4).
Figure 4.
Inversion breakpoints for commonly used X, second, and third chromosome balancers. Breakpoints that have been molecularly identified are shown as solid lines; those that are estimates are shown as dashed lines; centromeres are represented by black dots; coordinates are based on release 6 of the D. melanogaster genome. (A) Inversion breakpoints of the X chromosome balancer FM7 (Miller et al. 2016a). (B) Inversion breakpoints of four commonly used second chromosome balancers. (C) Inversion breakpoints of five commonly used third chromosome balancers, including the three balancers sequenced in this study.
We also recovered evidence of double crossing over between TM3 and TM6B and their normal sequence homologs. Two of the stocks with DCO events, TM3-560 and TM3-1614, are unique in that they appear to be examples of the TM3 balancer before Sb1 and Ser1 (TM3-1614) or before Sb1 (TM3-560) were added to TM3 through double crossing over in a triple-balanced female (Tinderholt 1960). Recovery of DCO events on these balancer chromosomes was not surprising, as similar exchanges were recently shown to occur within the inverted In(1)dl-49 segment of the X chromosome balancer FM7c. DCO events on FM7c always replaced the female sterile singedX2 (snX2) allele with a wild-type copy of the gene, resulting in sn+ progeny with reproductive advantages (Miller et al. 2016a). Similarly, the DCO events recovered in the TM3-504 and TM6B-587 stocks created a small duplication and the elimination of three frameshifting deletions, respectively, each of which may confer selective advantages.
The precise identification of inversion breakpoints, and the knowledge that rare DCO events are possible within inverted segments, should encourage researchers to carefully consider the proper balancer to use when keeping any allele over a balancer for a long period of time. We suggest using a balancer with an inversion breakpoint as close to the allele of interest as possible to prevent loss through double crossing over (Figure 4). In cases when this is not feasible, then keeping multiple copies of a stock along with periodic validation of the allele is likely in order.
Drosophila has a rich history. It has been over 100 yr since Morgan began to demonstrate the power of this tiny fly as a potent tool for scientific inquiry (Muller 1946; Sturtevant 2001). The success and rapid progress of experimentation in Drosophila today relies on genetic tools that have been built over the past century. Balancers have been especially important to the development of Drosophila as a genetic model organism. Molecular characterization of balancers helps explain how they work, how they vary, and what their inherent limitations are. This study endeavors to help Drosophila geneticists make better use of these invaluable tools.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.029330/-/DC1
Acknowledgments
We thank Angela Miller for assistance with figure preparation and manuscript editing; Anoja Perera, Kate Hall, Rhonda Egidy, and Michael Peterson for expert assistance with DNA sequencing; Sue Celniker for the ISO-1 stock; and Thom Kaufman and members of the Hawley lab for helpful discussions. R.S.H. is supported by the Stowers Institute for Medical Research, and is an American Cancer Society Research Professor supported by the award 118857-RP-05-086-06-COUN. K.R.C. is supported by National Institutes of Health (NIH) grant P40 OD018537.
Footnotes
Communicating editor: H. K. Salz
Literature Cited
- Ashburner M., 1972. New Mutants Report. D. I. S. 49: 34. [Google Scholar]
- Ashburner M., Golic K., Hawley R. S., 2005. Drosophila—a laboratory handbook, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., et al. , 1983. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science 221: 23–29. [DOI] [PubMed] [Google Scholar]
- Blumenstiel J. P., Noll A. C., Griffiths J. A., Perera A. G., Walton K. N., et al. , 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wallis J. W., McLellan M. D., Larson D. E., Kalicki J. M., et al. , 2009. Breakdancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N., 2004 Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics Chapter 4: Unit 4.10. [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craymer L., 1981. Techniques for manipulating chromosomal rearrangements and their application to Drosophila melanogaster. I. Pericentric inversions. Genetics 99: 75–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craymer L., 1984. New Mutants Report. D.I.S. 60: 234–236. [Google Scholar]
- Duncan D. M., Burgess E. A., Duncan I., 1998. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 12: 1290–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust G. G., Hall I. M., 2014. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30: 2503–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming R. J., Scottgale T. N., Diederich R. J., Artavanis-Tsakonas S., 1990. The gene Serrate encodes a putative EGF-like transmembrane protein essential for proper ectodermal development in Drosophila melanogaster. Genes Dev. 4: 2188–2201. [DOI] [PubMed] [Google Scholar]
- Gong W. J., McKim K. S., Hawley R. S., 2005. All paired up with no place to go: pairing, synapsis, and DSB formation in a balancer heterozygote. PLoS Genet. 1: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammonds A. S., Fristrom J. W., 2006. Mutational analysis of Stubble-stubbloid gene structure and function in Drosophila leg and bristle morphogenesis. Genetics 172: 1577–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelrigg T., Kaufman T. C., 1983. Revertants of dominant mutations associated with the antennapedia gene complex of Drosophila melanogaster: cytology and genetics. Genetics 105: 581–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Martinek S., Joo W. S., Wortman J. R., Mirkovic N., et al. , 2000. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97: 7301–7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1960. New Mutants Report. DIS 34: 51. [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al., 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., Zimm G. G., 1992. The Genome of Drosophila melanogaster, Academic Press, San Diego, CA. [Google Scholar]
- Lunde K., 2003. Activation of the knirps locus links patterning to morphogenesis of the second wing vein in Drosophila. Development 130: 235–248. [DOI] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., et al. , 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Cook K. R., Yeganeh Kazemi N., Smith C. B., Cockrell A. J., et al. , 2016a Rare recombination events generate sequence diversity among balancer chromosomes in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 113: E1352–E1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Smith C. B., Yeganeh Kazemi N., Cockrell A. J., Arvanitakis A. V., et al. , 2016b Whole-genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere effect. Genetics 203: 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1918. Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics 3: 422–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1946. Thomas Hunt Morgan 1866–1945. Science 103: 550–551. [DOI] [PubMed] [Google Scholar]
- Novitski E., Braver G., 1954. An analysis of crossing over within a heterozygous inversion in Drosophila melanogaster. Genetics 39: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel C., 1966. The interchromosomal effect of inversions on crossing-over in relation to non homologous pairing in Drosophila melanogaster. Hereditas 54: 293–306. [DOI] [PubMed] [Google Scholar]
- Rosenbloom K. R., Armstrong J., Barber G. P., Casper J., Clawson H., et al. , 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43: D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Schultz J., Redfield H., 1951. Interchromosomal effects on crossing over in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 16: 175–197. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J., Hazelrigg T. I., Polisky B. A., Pirrotta V., et al. , 1983. The molecular organization of the Antennapedia locus of Drosophila. Cell 35: 763–776. [DOI] [PubMed] [Google Scholar]
- Shearn A., 1980. Reintroduction of y+ onto a TM3 chromosome. DIS 55: 167. [Google Scholar]
- Stone W., Thomas I., 1935. Crossover and disjunctional properties of X chromosome inversions in Drosophila melanogaster. Genetica 17: 170–184. [Google Scholar]
- Sturtevant A. H., 1913. A third group of linked genes in Drosophila ampelophila. Science 37: 990–992. [DOI] [PubMed] [Google Scholar]
- Sturtevant, A. H., 2001 Reminiscences of T. H. Morgan. Genetics 159: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., Beadle G. W., 1936. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics 21: 554–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton E., 1943. A Cytogenetic Study of the yellow-scute Region of the X Chromosome in Drosophila melanogaster. Genetics 28: 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrzycka M., McEachern L. A., Kinneard J., Prabhu K., Fitzpatrick K., et al. , 2007. The pink gene encodes the Drosophila orthologue of the human Hermansky-Pudlak syndrome 5 (HPS5) gene. Genome 50: 548–556. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T., Mesirov J. P., 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinderholt V., 1960. New Mutants Report. D.I.S. 34: 53–54. [Google Scholar]
- Yazdani U., Huang Z., Terman J. R., 2008. The glucose transporter (GLUT4) enhancer factor is required for normal wing positioning in Drosophila. Genetics 178: 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data for all samples used in this project have been uploaded to the National Center for Biotechnology Information (NCBI) at ncbi.nlm.nih.gov and can be found under BioProject PRJNA315473. Laboratory strains of wgSp-1/SM6a, DuoxCy; Pr1/TM3, Sb1 Ser1, and +/TM6 are available upon request; all other strains listed in Table S2 are available from the Bloomington Drosophila Stock Center.