Abstract
Wolbachia is an intracellular symbiont of invertebrates responsible for inducing a wide variety of phenotypes in its host. These host-Wolbachia relationships span the continuum from reproductive parasitism to obligate mutualism, and provide a unique system to study genomic changes associated with the evolution of symbiosis. We present the genome sequence from a parthenogenesis-inducing Wolbachia strain (wTpre) infecting the minute parasitoid wasp Trichogramma pretiosum. The wTpre genome is the most complete parthenogenesis-inducing Wolbachia genome available to date. We used comparative genomics across 16 Wolbachia strains, representing five supergroups, to identify a core Wolbachia genome of 496 sets of orthologous genes. Only 14 of these sets are unique to Wolbachia when compared to other bacteria from the Rickettsiales. We show that the B supergroup of Wolbachia, of which wTpre is a member, contains a significantly higher number of ankyrin repeat-containing genes than other supergroups. In the wTpre genome, there is evidence for truncation of the protein coding sequences in 20% of ORFs, mostly as a result of frameshift mutations. The wTpre strain represents a conversion from cytoplasmic incompatibility to a parthenogenesis-inducing lifestyle, and is required for reproduction in the Trichogramma host it infects. We hypothesize that the large number of coding frame truncations has accompanied the change in reproductive mode of the wTpre strain.
Keywords: Trichogramma, gene truncations, symbiosis, genome content, Rickettsiales
Wolbachia is a maternally transmitted, intracellular symbiont of arthropods and nematodes that exhibits a range of complex interactions with its hosts (Werren 1997; Werren et al. 2008; Stouthamer et al. 1999a). It is estimated to infect 40–60% of arthropod species (Zug and Hammerstein 2012; Hilgenboecker et al. 2008). Across the arthropods, Wolbachia is well known for modifying host reproduction, by utilizing various mechanisms that enhance fitness or numbers of infected females. By promoting infected females, Wolbachia ensures its own maternal transmission and has the ability to spread rapidly through a population (Walker et al. 2011; Weeks et al. 2007; Turelli and Hoffmann 1991). These reproductive modifications include: cytoplasmic incompatibility (CI), male killing, feminization, and parthenogenesis-induction (PI) (Werren 1997). In addition to these reproductive phenotypes, some Wolbachia strains protect against pathogens (Chrostek et al. 2013; Moreira et al. 2009; Kambris et al. 2010), supply essential nutrients to their hosts (Nikoh et al. 2014; Hosokawa et al. 2010), are required for successful egg development (Kremer et al. 2009; Timmermans and Ellers 2009; Dedeine et al. 2001), or are essential for the production of female offspring (Russell and Stouthamer 2011; Stouthamer et al. 2010). In filarial nematodes, Wolbachia is an obligate mutualist providing a diversity of benefits to its host, including evasion of the vertebrate immune system (Darby et al. 2012). For these reasons, Wolbachia has captured considerable interest in applied fields as a potential “agent” to modify pest populations, reduce pathogen loads in vectors, and specifically target filarial nematodes by way of their obligate symbionts (Zabalou et al. 2004; Bourtzis et al. 2014; Taylor et al. 2000).
In addition to the practical applications of studying Wolbachia, the complexity of interactions with diverse hosts provides an opportunity to explore genomic changes accompanying the evolution of such unique life histories. Nested within a clade of other symbiotic and pathogenic bacteria, Wolbachia are members of the Rickettsiales, an order of α-proteobacteria (O’Neill et al. 1992; Dumler et al. 2001). The Wolbachia clade is composed of 16 reported supergroups, denoted A–F and H–Q (Ros et al. 2009; Augustinos et al. 2011; Bing et al. 2014; Haegeman et al. 2009; Lo et al. 2002; Glowska et al. 2015), with supergroups A–D being the most well studied. Supergroup G is no longer considered a distinct Wolbachia lineage, as it represents a recombinant between supergroups A and B (Baldo and Werren 2007). Supergroups A and B are a monophyletic assemblage infecting arthropods (Gerth et al. 2014), whereas supergroups C and D are the major nematode-infecting lineages (Bandi et al. 1998). Supergroup F is unique as it contains both nematode and arthropod-infecting strains (Casiraghi et al. 2005), including the bed bug-infecting Wolbachia strain wCle that supplements B vitamins to its obligate blood-feeding hosts (Nikoh et al. 2014; Hosokawa et al. 2010). The less studied supergroups H–Q infect a variety of hosts, including termites, aphids, whiteflies, mites, fleas, and a plant-parasitic nematode (Ros et al. 2009; Augustinos et al. 2011; Bing et al. 2014; Haegeman et al. 2009; Lo et al. 2002; Glowska et al. 2015).
While cocladogenesis of Wolbachia and their hosts does occur (Raychoudhury et al. 2009), it is relatively uncommon, and host-switching is a prominent feature of Wolbachia’s evolutionary history (Vavre et al. 1999; van Meer et al. 1999; Zhou et al. 1998; Baldo et al. 2006). In addition to the incongruence of host and symbiont phylogenies, there is little conservation of the induced phenotypes. For example, independently derived parthenogenesis-inducing (PI) Wolbachia are found in the A and B supergroups (Stouthamer et al. 1993), and likely the F supergroup (Baldo et al. 2007). These PI-Wolbachia strains induce parthenogenesis through different mechanisms including the merging of nuclei (Gottlieb et al. 2002), a failed anaphase during the first embryonic cell division (Stouthamer and Kazmer 1994; Pannebakker et al. 2004), and functional apomixis (Weeks and Breeuwer 2001). Uninfected parasitoid wasps of the genus Trichogramma are arrhenotokous, but infection with PI-Wolbachia strains causes gamete duplication in unfertilized eggs by preventing chromosome segregation during anaphase of the first mitotic division of the egg, resulting in a diploid female (Stouthamer and Kazmer 1994). The PI-Wolbachia strains infecting Trichogramma spp. are unique for at least three reasons: there is a single origin of Wolbachia infection for the genus (Werren et al. 1995; van Meer et al. 1999); the Trichogramma hosts can evolve dependencies upon their Wolbachia infection for the production of females (Russell and Stouthamer 2011; Stouthamer et al. 2010); and, unlike other arthropod-infecting strains, the PI-Wolbachia infecting Trichogramma do not have relationships with phages (Gavotte et al. 2007).
Wolbachia genomes are small in size, ranging from 0.9–1.5 Mbp, and contain a number of unique features. The arthropod infecting genomes have a large number of repetitive and mobile elements, including ankyrin repeat domain-containing (ANK) genes (Iturbe-Ormaetxe et al. 2005; Siozios et al. 2013b; Papafotiou et al. 2011), bacteriophage sequences (Gavotte et al. 2007), transposons, and many copies of short open reading frames (ORFs) of unknown function (Wu et al. 2004). Little is known about the role that these short, unannotated ORFs play in the biology of Wolbachia.
Here, we explore the changes in genome content across Wolbachia, and present a draft genome for the PI-Wolbachia strain, wTpre, infecting the parasitoid wasp Trichogramma pretiosum. The wTpre genome represents the most complete PI-Wolbachia genome assembly to date, and the first B supergroup PI-Wolbachia genome. We show evidence for protein sequence truncation in 20% of the wTpre gene set, and hypothesize that these truncations are a feature of the change in reproductive phenotype.
Materials and Methods
Biological materials
A unisexual colony of naturally Wolbachia-infected T. pretiosum was chosen for genome sequencing. Originally collected in the Puira Valley of Peru, this colony has been maintained in a commercial insectary since 1966 (Beneficial Insectary, Guelph, Ontario, Canada), and herein is referred to as the “Insectary Line.” Species identifications were confirmed by molecular protocols from Stouthamer et al. (1999b), and Wolbachia infection status was confirmed using the protocols from Stouthamer et al. (1990) and Werren and Windsor (2000). Attempts to initiate Wolbachia-free replicates of this colony following antibiotic treatment protocols from Stouthamer et al. (1990) have not been successful due to severe fertility reduction, as seen in Russell and Stouthamer (2011).
Identification of a wTpre genome
The genome of the T. pretiosum Insectary Line (GenBank Accession Number: JARR00000000) (A. R. I. Lindsey et al., unpublished results) was sequenced in collaboration with the i5k initiative to sequence 5000 arthropod genomes (www.arthropodgenomes.org/wiki/i5K) and made publicly available prior to publication under the Fort Lauderdale agreement. The T. pretiosum assembly was scanned for evidence of Wolbachia DNA using two methods. First, total DNA was extracted from 10 wasps using a Chelex method (Walsh et al. 1991) as implemented by Stouthamer et al. (1999b). The Wolbachia 16S rRNA gene was amplified and sequenced with W-Specf and W-Specr primers (Werren and Windsor 2000). Sequences were aligned and primer sequences excised in Sequencher 4.9. The 16S rRNA gene was then queried against the T. pretiosum genome assembly using nucleotide BLASTN at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The remaining scaffolds were checked for bacterial DNA sequences by querying them against Bacteria (taxid: 2) in NCBI GenBank with blastn. Second, the T. pretiosum assembly was scanned with the bioinformatics pipeline developed by Wheeler et al. (2013), in order to identify bacterial sequences from a eukaryotic background.
Genome annotation, clusters of orthologous genes, and completeness estimates
The IGS Annotation Engine was used for structural and functional annotation of the wTpre genome (http://ae.igs.umaryland.edu/cgi/index.cgi, Galens et al. 2011). Manatee was used to view annotations (http://manatee.sourceforge.net/). The wTpre genome and 17 other previously published genomes (see Table 1) were used in comparative analyses. Previously published genomes were reannotated with the IGS Annotation Engine, and Clusters of Orthologous Genes (COGs) across all 18 genomes were defined using Sybil (http://sybil.sourceforge.net/index.html, Riley et al. 2012; Crabtree et al. 2007). Genome completeness was assessed with the BUSCO pipeline (Simão et al. 2015) using the 40 core bacterial genes from Mende et al. (2013) compared to the gene set from each Wolbachia genome (-m = OGS).
Table 1. Wolbachia strains used in comparative and phylogenetic analyses.
| Strain | Host | Supergroup | Size (bp) | ORFs | Reference | Accession Number | BUSCO Scorea |
|---|---|---|---|---|---|---|---|
| wGmm | Glossina morsitans morsitans | A | 1,019,687 | 1378 | Brelsfoard et al. (2014) | AWUH00000000 | C: 77.5% [D: 6.4%], F: 5%, M: 17.5%, n: 40 |
| wHa | Drosophila simulans | A | 1,295,804b | 1342 | Ellegaard et al. (2013) | CP003884 | C: 85% [D: 2.9%], F: 5%, M: 10%, n: 40 |
| wMel | Drosophila melanogaster | A | 1,267,782c | 1401 | Wu et al. (2004) | AE017196 | C: 87.5% [D: 2.9%], F: 2.5%, M: 10%, n: 40 |
| wRi | Drosophila simulans | A | 1,445,873c | 1493 | Klasson et al. (2009) | CP001391 | C: 82.5% [D: 3%], F: 5%, M: 12.5%, n: 40 |
| wSuzi | Drosophila suzukii | A | 1,415,350 | 1528 | Siozios et al. (2013a) | CAOU00000000 | C: 87.5% [D: 2.9%], F: 2.5%, M: 10%, n: 40 |
| wAIbB | Aedes albopictus | B | 1,162,431 | 1187 | Mavingui et al. (2012) | CAGB00000000 | C: 82.5% [D: 3%], F: 2.5%, M: 15%, n: 40 |
| wBol1 | Hypolimnas bolina | B | 1,377,933 | 1369 | Duplouy et al. (2013) | CAOH00000000 | C: 80% [D: 3.1%], F: 5%, M: 15%, n: 40 |
| wDi | Diaphorina citri | B | 1,240,904 | 1250 | Saha et al. (2012) | AMZJ00000000 | C: 80% [D: 3.1%], F: 2.5%, M: 17.5%, n: 40 |
| wNo | Drosophila simulans | B | 1,301,823c | 1317 | Ellegaard et al. (2013) | CP003883 | C: 82.5% [D: 3%], F: 2.5%, M: 15%, n: 40 |
| wPip_Pel | Culex quinquefasciatus Pel | B | 1,482,355c | 1461 | Klasson et al. (2008) | AM999887 | C: 80% [D: 3.1%], F: 5%, M: 15%, n: 40 |
| wPip_JBH | Culex quinquefasciatus JBH | B | 1,542,137 | 1556 | Salzberg et al. (2009) | ABZA00000000 | C: 75% [D: 3.3%], F: 2.5%, M: 22.5%, n: 40 |
| wPip_Mol | Culex pipiens molestus | B | 1,340,443c | 1340 | Pinto et al. (2013) | HG428761 | C: 80% [D: 3.1%], F: 2.5%, M: 17.5%, n: 40 |
| wTpre | Trichogramma pretiosum | B | 1,133,709b | 1405 | This study | LKEQ00000000 | C: 77.5% [D: 3.2%], F: 5%, M: 17.5%, n: 40 |
| wVitB | Nasonia vitripennis | B | 1,107,643 | 1245 | Kent et al. (2011) | AERW00000000 | C: 77.5% [D: 3.2%], F: 2.5%, M: 20%, n: 40 |
| wOo | Onchocerca ochengi | C | 957,990c | 1272 | Darby et al. (2012) | HE660029 | C: 75% [D: 3.3%], F: 2.5%, M: 22.5%, n: 40 |
| wBm | Brugia malayi | D | 1,080,084c | 1339 | Foster et al. (2005) | AE017321 | C: 82.5% [D: 3%], F: 5%, M: 12.5%, n: 40 |
| wWb | Wuchereria bancrofti | D | 1,052,327 | 2144 | Desjardins et al. (2013) | ADHD00000000 | C: 45% [D: 0%], F: 20%, M: 35%, n: 40 |
| wCle | Cimex lectularius | F | 1,250,060c | 1357 | Nikoh et al. (2014) | AP013028 | C: 72.5% [D: 3.4%], F: 2.5%, M: 25%, n: 40 |
ORFs, open reading frames; BUSCO, benchmarking universal single-copy orthologs; C, complete; D, duplicated; F, fragmented; M, missing; n, number of genes used.
BUSCO scores in standard BUSCO notation.
Single-scaffold assembly.
Complete assembly.
Phylogenetic analyses
A phylogenetic reconstruction of Wolbachia strains was inferred using the five Multi Locus Sequence Typing (MLST) genes (Baldo et al. 2006), with Anaplasma marginale str. Florida (GenBank Accession Number: PRJNA58577) “Ama” as an outgroup. In addition to the strains in Table 1 (minus wWb, see Results), we included Wolbachia strains from the MLST database (wAjap infecting Asobara japonica, wUni infecting Muscidifurax uniraptor, wDali infecting Diaphorencyrtus aligarhensis, wTdei infecting Trichogramma deion, wEfor infecting Encarsia formosa, wPsiaB infecting Protocalliphora sialia, and wLcla infecting Leptopilina clavipes) and the wTbras strain infecting Trichogramma brassicae (downloaded from GenBank, Accession Numbers: JF920468.1, JF920470.1, JF920472.1, JF920464.1, and JF920466.1). Multiple alignments were created for each gene using the L-INS-i algorithm in MAFFT version 7 (Katoh and Standley 2013), and were concatenated prior to maximum likelihood analyses in RAxML version 8.2.4 (Stamatakis 2014) using the GTRGAMMA substitution model and 1000 bootstrap replicates. A second phylogenetic reconstruction was made using the same methods, but with only the strains used in our comparative analyses. Trees were visualized in FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) and annotated in Inkscape (https://inkscape.org/en/).
Identification of core and unique genomes
Unique and core genome assessments were performed using Sybil results loaded on a Chado relational database (Galens et al. 2011; Mungall et al. 2007). The core genome was determined by identifying all COGs that had at least one gene member from each Wolbachia strain being considered. COGs were considered unique to a monophyletic assemblage when all members of the COG belonged exclusively to the clade, and were found in all members of the clade. To determine the uniqueness of the Wolbachia core, a representative wTpre gene for each of the core COGs was queried against a database of the protein coding sequences of Rickettsia rickettsii, Ehrlichia chaffeensis, and A. marginale, (respective GenBank Accession Numbers CP003318, CP000236, and CP001079) using BLASTP. A cutoff e-value of 1e-10 was used to determine significance. The comparison of the core was done with both the 496-COG core (excluding wWb and wGmm) and the 436-COG core (excluding only wWb, and wGmm included).
Analysis of genome content and ankyrin genes
Role category annotations from the IGS annotation pipeline were used to compare genome content across 17 Wolbachia strains, excluding unannotated genes. The number of genes in each role category for each genome was plotted according to standard deviation, then subjected to a Principle Components Analysis (PCA) based on the standardized proportion of genes in each role category, using prcomp in R version 3.1.2 (R Core Team 2014). Due to the high variance of the hypervariable “mobile and extrachromosomal element functions” category, a second PCA analysis was performed after removing the category and recalculating proportions.
The term “ankyrin” was queried against all gene annotations, and the number of positive matches was tabulated for each genome. The number of ankyrin repeat-containing genes was plotted in R, and a Mann–Whitney U-test was used to test for a significant difference in abundance between supergroups A and B. Supergroups C, D, and F were not included in the statistical analyses due to the small number of sequenced genomes available for those groups.
Identification of truncated ORFs in wTpre
The nucleotide sequence of all wTpre genes determined not to be a member of any orthologous clusters (see Results) were queried against a database of all Wolbachia genes from the remaining 16 genomes using BLASTN. The full nucleotide sequence of the best match was then queried back against the wTpre genome sequence to look for regions of homology beyond the wTpre gene ORF. To be further considered as evidence of protein sequence truncation, the BLASTN best match to the genome was required to meet an 85% identity cutoff, and the best match had to align to wTpre across at least 70% of its length, or at least three times the length of the wTpre gene in question. Alignments that passed these quality measures were scanned for the presence of mutations that would result in premature stop codons, and categorized by mutation type. ORF length comparisons were performed in R and a Mann–Whitney U-test was used to determine significance.
Comparison to inactive genes in Wolbachia strain wAu
The set of wMel genes that were found to be potentially inactive in Wolbachia strain wAu (Sutton et al. 2014) was compared to the wTpre gene set. wAu was not included in previous analyses because it was published after COG assessment was completed. The wMel genes were classified as either: 1) having an ortholog in wTpre (as determined by Sybil COG assessment), 2) being truncated in wTpre (as determined by the homolog of a truncated wTpre gene sharing COG membership with the respective wMel gene), or 3) absent in wTpre.
Data availability
The T. pretiosum colony used for sequencing is available upon request. Supplemental Material, Table S1 contains a detailed breakdown of the counts of genes in each role category and subcategory, for each Wolbachia strain, as annotated by IGS. Table S2 provides complete BUSCO results for all Wolbachia strains. Table S3 is the wTpre “unique genes” considered in truncation analyses. Table S4 contains comparisons of truncated genes in wAu and wTpre. The wTpre Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession LKEQ00000000. The version described in this paper is version LKEQ01000000.1.
Results
The wTpre genome: a parthenogenesis-inducing Wolbachia strain
The genome sequence of wTpre was extracted from a whole genome assembly of its host, T. pretiosum, performed as a part of the i5k genome project (A. R. I. Lindsey et al., unpublished results). The wTpre genome was recovered in a single scaffold, composed of nine contigs. The scaffold was 1,133,709 bp in length, and BLASTN searches against the NCBI GenBank database revealed 97% nucleotide similarity to the Wolbachia symbiont wPip_Pel infecting Culex quinquefasciatus (GenBank accession number: AM999887). No other bacterial sequence was identified in the T. pretiosum assembly. Average scaffold coverage for the Wolbachia scaffold was the lowest of all scaffolds in the i5k genome project assembly, indicating that the recovered genome is not the result of a lateral transfer into the T. pretiosum genome (Wolbachia scaffold = 35.6 × coverage, T. pretiosum assembly = 232.7 × coverage). The wTpre genome was structurally and functionally annotated with the Institute for Genome Sciences (IGS) pipeline at the University of Maryland (http://ae.igs.umaryland.edu/cgi/index.cgi, Galens et al. 2011), revealing 1405 ORFs, 35 tRNA coding genes, and a single set of rRNA genes (one each of 5S, 23S and 16S), giving a coding density of 81.8%. The size and number of coding sequences fell within the range of previously sequenced Wolbachia genomes (Table 1). While the arthropod-infecting Wolbachia genomes are known to carry a large number of mobile elements, the wTpre genome was depauperate in these features. Only nine genes related to prophage function, and 14 transposon function genes were identified in the genome (Table S1).
Genome completeness and phylogenetic relationships
Seventeen previously published Wolbachia genomes, representing supergroups A–D and F, were examined alongside the wTpre genome in phylogenetic and comparative analyses (Table 1). All genomes were reannotated with the same IGS pipeline used to annotate wTpre. BUSCO (Simão et al. 2015) was used to scan for the 40 core bacterial genes defined by Mende et al. (2013) to estimate completeness for each sequenced genome based on the proportion of missing BUSCO genes. Scores from these analyses are reported in Table 1. Notably, none of the Wolbachia strains, including completely sequenced genomes, contained all 40 BUSCO genes. All 18 strains are missing the BUSCO orthologs that encode for ribosomal proteins S7, L11, L4, and L14 (COG0049, COG0080, COG0088, and COG0093, respectively). The wWb strain (from the nematode Wuchereria bancrofti) appeared to be an outlier, as 22 of the 40 orthologs were missing or fragmented (Table S2). Additionally, wWb was missing a duplication of COG0552 (Signal recognition particle GTPase) that is present in all 17 other strains. The draft Wolbachia genomes have BUSCO scores that fall within the range of scores from the complete genomes, with the exception of wWb. The wWb assembly is the expected size for a Wolbachia genome, but has an abnormally large number of ORFs (n = 2144), almost 600 more than the other Wolbachia genomes (Table 1). For these reasons, the wWb strain was excluded from additional analyses.
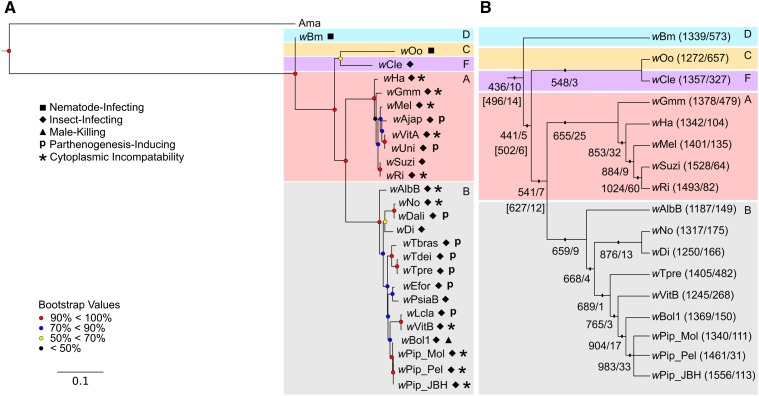
Phylogenetic reconstruction based on maximum likelihood analysis was conducted using Multilocus Sequence Typing (MLST) genes (Baldo et al. 2006) to determine relationships among the PI-Wolbachia. This analysis confirms multiple independent origins of PI-Wolbachia, placement of the wTpre strain in the B supergroup, and the monophyly of the Trichogramma-infecting Wolbachia (Figure 1A). All supergroups with multiple members were recovered as monophyletic. The major arthropod-infecting lineages, supergroups A and B, formed a monophyletic clade, and supergroups C and F also formed a monophyletic clade. The nematode-infecting supergroup D was sister to the rest of the Wolbachia lineage. The wPip strains have identical MLST sequences, and are represented as a polytomy.
Figure 1.
Phylogenetic relationships of Wolbachia. (A) Phylogeny inferred with RAxML from a nucleotide supermatrix of the five Wolbachia MLST (multi locus sequence typing) genes using 1000 bootstrap replicates. Supergroups are shown in colored boxes, and labeled in the top right corner of each box. Symbols next to taxa denote Wolbachia host and phenotypes. Colors at nodes indicate bootstrap values. Anaplasma marginale str. Florida “Ama” is the outgroup. (B) Cladogram of Wolbachia inferred with RAxML using the same methods as in Figure 1A, but analyzing only the strains with sequenced genomes. Numbers in parentheses next to taxon names represent, on the left, the number of genes in the genome, and on the right, the number of genes unique to that genome. Numbers corresponding to points on internodes represent, on the left, the number of core cluster of orthologous genes (COGs) for that clade, and on the right, the number of COGs unique to that clade. Numbers in square brackets represent alternative core and unique genome sizes for the respective clade, calculated without wGmm. Colored boxes denote supergroups, with labels in the top right corner.
The core Wolbachia genome
The core genome of the 17 Wolbachia strains was made up of 436 COGs (Figure 1B). The core genomes of the A (655 COGs) and B (659 COGs) supergroups were similar in size despite the B supergroup being represented by four more strains than the A supergroup. Together, these two supergroups had a core genome of 541 COGs. As expected, the inclusion of additional supergroups led to a reduction in the size of the core genome. Sampling more heavily among more distantly related groups yielded a decrease in shared similarities. It is important to note that the positions of wGmm and wHa have changed: in the phylogenetic reconstruction including more strains (Figure 1A), wHa is sister to the rest of the A supergroup and wGmm is sister to the rest of the A supergroup when the phylogeny is reconstructed with only the strains for which genomes are available (Figure 1B). That node in both trees is supported by a bootstrap value of 100, so we kept the topologies and calculated core and unique genome sizes with wGmm as sister to the rest of the A supergroup.
The size of the core genome for the eight Wolbachia strains with completely sequenced genomes (wBm, wCle, wMel, wNo, wOo, wPip_Pel, wPip_Mol, and wRi) was 511 COGs. Inclusion of wHa, which has a genome assembly of a single scaffold with two gaps, did not reduce the core size. Addition of wTpre, the remaining single-scaffold assembly, only reduced the core genome by one COG, to 510 COGs, indicating that the wTpre assembly is relatively complete. These 10 complete and single-scaffold genomes were used to determine which genome(s) were having the largest effect on the final core genome size of all 17 strains. One at a time, the core genome was determined for the aforementioned 10 genomes, plus one of the seven remaining assemblies. wDi and wSuzi had a small effect on the core size, each resulting in one less COG in the core. wPip_JBH reduced the core genome by two COGs. wAlbB and wBol1 were each responsible for a loss of three COGs from the core, and wVitB for five COGs. The wGmm strain had the most drastic effect on the size of the Wolbachia core, as the wGmm assembly (infecting the tsetse fly Glossina morsitans morsitans) is missing 63 of the 510 COGs found in the 10 complete and single-scaffold genomes. Its low BUSCO score (Table 1), in combination with the effect on the core genome, indicate that a significant portion of sequence data may be missing or misassembled for wGmm. Elimination of wGmm from the analysis resulted in a core Wolbachia genome of 496 COGs for the remaining 16 strains, which is likely closer to the true size of the Wolbachia core. This 496 COG core was searched against R. rickettsii, E. chaffeensis, and A. marginale. Fourteen Wolbachia core COGs did not have hits to the other Rickettsiales: 11 hypothetical or predicted proteins, a cutA1 divalent ion tolerance family protein, a surface antigen family protein, and a nitroreductase family protein. Four of these 14 Wolbachia-unique COGs, all conserved hypothetical proteins, are missing from the 436-COG core that includes wGmm.
Ordination of Wolbachia strains based on genome content
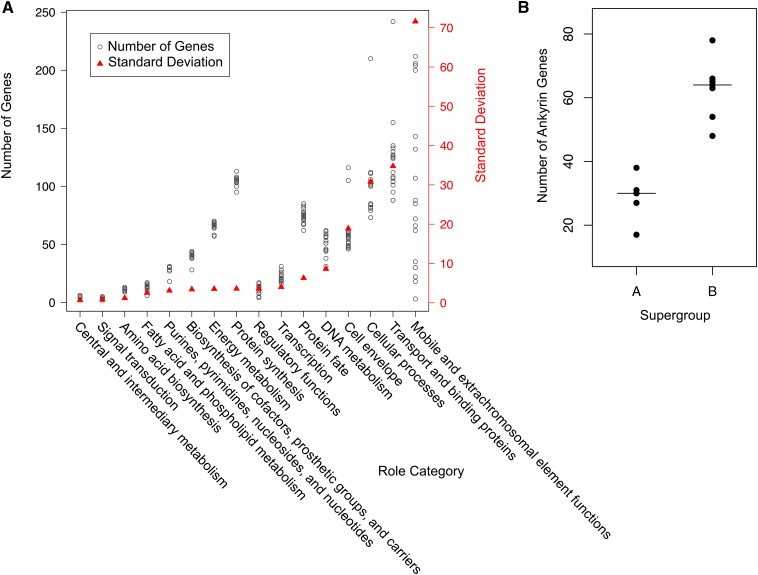
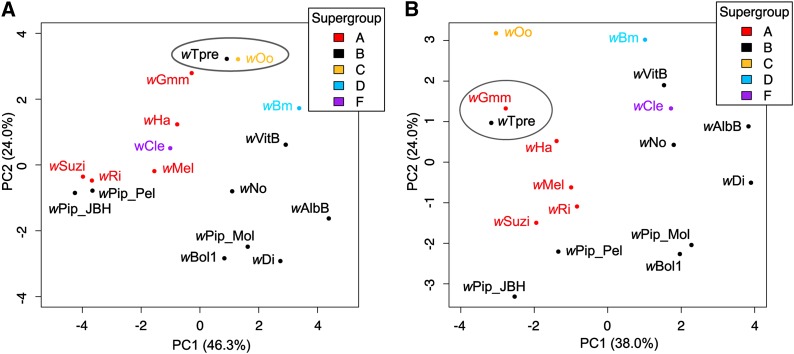
The number of genes in each role category, for each genome, as determined by the IGS annotation pipeline, was used in comparative analyses of genome content. The role categories with the most variation in gene number per genome were: mobile and extrachromosomal element functions, transport and binding proteins, and cell envelope (Figure 2A). Wolbachia genomes showed little variance in the number of genes devoted to central intermediary metabolism, signal transduction, and amino acid biosynthesis. All Wolbachia genomes had a high (median = 106), but relatively conserved number of genes devoted to protein synthesis. Principal Components Analysis (PCA) was used to visualize the similarity of genomes based on the proportion of genes in each of these role categories (Figure 3A). While the A supergroup genomes ordinate to the upper left quadrant, the B supergroup strains showed greater diversity in genome content across strains. Bed bug-infecting wCle clustered with the distantly related, yet also arthropod-infecting, A supergroup strains, although phylogenetically wCle belongs to the F supergroup (Rasgon and Scott 2004). wTpre’s closest neighbor in the genome content-based ordination was the obligate, nematode-infecting wOo strain. We suspect that the highly variable number of genes in the mobile and extrachromosomal element functions role category could strongly influence these patterns. Therefore, proportions were recalculated without this category and again subjected to PCA (Figure 3B). Without the mobile and extrachromosomal element functions role category, the wCle genome neighbored B supergroup strains, and the wTpre genome neighbored the group of A supergroup strains. This category had a dominant effect on the ordination of wTpre and wCle. However, the overall pattern of a loose A supergroup cluster and B supergroup diversity was maintained in the absence of the mobile and extrachromosomal element functions category, indicating support from other role categories for this patterning.
Figure 2.
Gene content of Wolbachia. (A) The numbers of genes in each role category, for each Wolbachia genome are plotted with open circles and correspond to the left axis. Role categories are sorted by standard deviation, represented by the red triangles, and the right axis. (B) Number of ankyrin repeat-containing genes per genome, by supergroup. The B supergroup has a significantly higher number of ankyrin genes than the A supergroup (Mann–Whitney U-test, P = 0.003).
Figure 3.
Principal components (PC) analysis of Wolbachia genomes based on proportion of annotated genes devoted to each role category, with wTpre and closest neighbor circled. (A) All annotated role categories analyzed. The strongest factor loadings along PC1 (46.3% of total variance) and PC2 (34.0% of total variance), respectively, are energy metabolism and regulatory functions. (B) Mobile and extrachromosomal elements functions category excluded. The strongest factor loadings along PC1 (38.0% of total variance) and PC2 (24.0% of total variance), respectively, are cellular processes and DNA metabolism.
Supergroup B has significantly more ankyrin repeat-containing genes
We specifically looked at the number of ankyrin repeat-containing (ANK) genes in each of the Wolbachia genomes. ANK genes are involved in protein-protein interactions and are rare in bacteria, but are found in Wolbachia, where they may modulate host phenotypes (Iturbe-Ormaetxe et al. 2005; Papafotiou et al. 2011). The wTpre strain has 54 ANK genes. With 48 ANK genes, the wAlbB strain has the fewest number of ANK genes in the B supergroup. We demonstrate a significant difference in the number of ANK genes between supergroups A and B (Mann Whitney-U, P = 0.003) (Figure 2B). The B supergroup has, on average, more than double the number of ANK genes than any other supergroup. The median number of ANK genes in supergroup A is 30, and in supergroup B is 64. While supergroups C, D, and F were not subjected to statistical analysis due to the low number of representative genomes available, the numbers of ANK genes present in those genomes was low when compared to supergroup B. The wOo (C), wBm (D), and wCle (F) genomes have 3, 20, and 39 ANK genes, respectively.
“Unique” wTpre genes are derived from truncated versions of Wolbachia genes
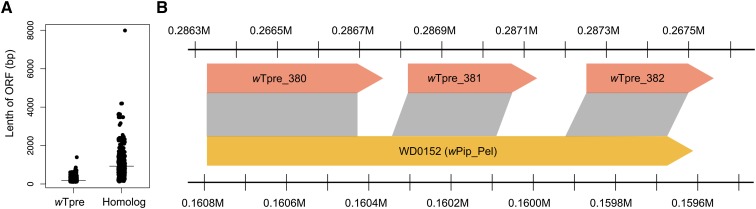
The newly sequenced wTpre strain has one of the largest sets of “unique genes,” and the largest of all the arthropod-infecting Wolbachia strains, with 482 genes not assigned any orthologs (Figure 1B). This represents 34% of the total genes in the wTpre genome. Nucleotide BLAST searches of the wTpre “unique genes” against a database of all the other coding sequences from the other Wolbachia genomes in Table 1 reveal that 367 of wTpre “unique genes” show similarity with other Wolbachia genes (Table 2). However, the predicted coding regions of wTpre “unique genes” were on average 77.5% shorter than their corresponding homologs in other Wolbachia genomes (Mann–Whitney U-test, P < 0.0001) (Figure 4A). The significant difference in size could indicate that these genes are truncated versions of the coding sequence, either due to deletions, or premature stop codons. To explore this, the nucleotide sequences of the best matches were aligned to the wTpre genome sequence to look for homology of the wTpre “unique gene” up- and downstream of the ORF. Of the 367 wTpre “unique genes” with sequence similarity to other Wolbachia genes, 86 genes were excluded from analyses based on low identity values and/or lack of evidence for up/downstream homology, and 281 genes showed evidence of truncation of the predicted protein sequence and potential pseudogenization due to nonsense and frameshift mutations (Table 2 and Table S3). Many of the wTpre “unique genes” occur in tandem, where an early frameshift or nonsense mutation resulted in a premature stop codon, and subsequent annotation of additional short, downstream ORFs with sequence homology to the downstream portions of the same ORF in the other Wolbachia genome. Figure 4B shows a schematic representation of this phenomenon, where the wTpre “unique genes” wTpre_380, wTpre_381, and wTpre_382 all align to sequential portions of the wPip_Pel gene, WD0152. A single base pair deletion at position 421 in wTpre_380, relative to wPip_167, resulted in a premature stop codon. The intergenic spaces between these wTpre “unique genes” also showed sequence similarity to corresponding locations in the wPip_Pel gene. The short ORFs downstream of the nonsense or frameshift mutation are hereafter referred to as “postnonsense” or “postframeshift” ORFs, respectively. In the wTpre genome, 52% (n = 146) of these “unique genes” with evidence of truncation were postframeshift ORFs (Table 2 and Table S3). The coding frame truncated wTpre genes were more likely to have a hypothetical annotation than their counterparts from other Wolbachia genomes (Chi-Square, P < 0.0001). Of the 281 truncated wTpre genes, 149 (53%) had a hypothetical annotation. This contrasts to the 188 genes that the truncated wTpre genes match to, where only 62 (33%) had a hypothetical annotation. Of the truncated wTpre genes, 57 are of phage or transposon origin, and 45 are homologs of ANK genes. Therefore, we conclude that the majority of these “unique genes” are artifacts of ORF prediction, and are actually degenerated protein coding sequences of genes found in other Wolbachia.
Table 2. Classification of wTpre “unique genes”.
| “Unique Genes” with Evidence of Truncation | “Unique Genes” Without Evidence of Truncation | ||
|---|---|---|---|
| Nonsense mutation | 26 | No match to other Wolbachia genes | 115 |
| Postnonsense | 76 | Low identity score of alignment | 7 |
| Frameshift mutation | 30 | Homolog is shorter than wTpre gene | 11 |
| Postframeshift | 139 | No up/downstream homology | 68 |
| Poststart codon mutation | 10 | ||
| Total truncations | 281 | Total excluded | 201 |
Figure 4.
Evidence for truncation in wTpre genes. (A) Length of wTpre “unique genes” and their homologous genes from other Wolbachia genomes. There is a significant difference in the size of the wTpre unique gene set as compared to their homologous counterparts (Mann–Whitney U-test, P < 0.0001). (B) Schematic representation of wTpre coding frame truncation and fragmentation. The wTpre “unique genes,” wTpre_380, wTpre_381, and wTpre_382, are homologous to sequential locations in the WD0152 gene from wPip_Pel. A frameshift mutation at base pair 421 in wTpre_380 resulted in a premature stop codon and the subsequent annotation of downstream ORFs (open reading frames), or “postframeshift” ORFs.
Comparison to inactive genes in Wolbachia strain wAu
The genome for the wAu strain infecting Drosophila simulans was recently sequenced, and also found to be missing or have potentially inactive versions of homologous genes present in the closely related wMel strain (Sutton et al. 2014). While wMel induces strong CI, wAu has lost this function (Hoffmann et al. 1996). All of the 46 wMel genes found to be inactive in wAu were members of COGs, and were not unique to wMel. Of these 46 wMel genes, 36 were either absent (n = 24), truncated (n = 9), or “unique genes” that did not meet criteria to be considered truncations (n = 3) in the wTpre genome (Table S4). Ten of the wMel genes shared the same fate in both the wTpre and wAu genomes. Five hypothetical proteins, an ANK protein, and DNA repair protein RadC, are absent in both wTpre and wAu. Multidrug resistance protein D and a hypothetical protein both have frameshift mutations in wTpre and wAu. Lastly, a prophage gene has a nonsense mutation in both strains.
Discussion
The wTpre assembly represents the most complete genome sequence of a parthenogenesis-inducing Wolbachia to date. This particular PI-Wolbachia strain is required for reproduction in its host; attempts to initiate Wolbachia-free replicates of this Trichogramma colony, following protocols from Stouthamer et al. (1990), have not been successful (e.g., Russell and Stouthamer 2011). The only other available PI-Wolbachia genome is strain wUni from the parasitic wasp Muscidifurax uniraptor, an A supergroup Wolbachia (Klasson et al. 2009). wUni was not included in analyses as the record contains only partial genome data that was generated by amplification with primers based on the wMel genome.
In some ways, the wTpre genome is similar to the other arthropod-infecting strains. wTpre contains a large number of ANK genes, as is common in the Wolbachia clade. With regards to the number of phage genes, the wTpre genome is more similar to the obligate, nematode-infecting Wolbachia: wTpre contains nine annotated phage genes and 14 transposon function genes. As a comparison, the same annotation pipeline identified 55 prophage function genes and 132 transposon function genes in the wPip_Pel genome, and 30 prophage and 81 transposon genes in the wMel strain (infecting Drosophila melanogaster). This corroborates previous analyses that discovered a diversity of phages in many other arthropod-infecting Wolbachia, but no evidence of functional bacteriophages in the Trichogramma-infecting Wolbachia (Gavotte et al. 2007). Phylogenetic analyses confirmed the multiple origins of PI-Wolbachia, and monophyly of the Trichogramma-infecting strains (van Meer et al. 1999). The relationship of the supergroups using the five MLST genes (Baldo et al. 2006) replicated results from phylogenomic analyses using 90 informative loci (Gerth et al. 2014).
We attempted to assess the completeness of the Wolbachia genomes using the BUSCO pipeline and 40 core bacterial genes. Completely sequenced genomes varied widely in the number of genes recovered, indicating that this gene set may not be ideal for assessing completeness in Wolbachia. Four ribosomal proteins were absent from all Wolbachia genomes. Genome sequencing of the primary-symbionts of insects has revealed that not all ribosomal proteins are retained in these highly reduced genomes (McCutcheon 2010). While Wolbachia is not considered a primary-symbiont, and is not strictly maternally transmitted (Raychoudhury et al. 2009), some degree of genome reduction has taken place. There was a trend toward lower BUSCO scores in the obligate Wolbachia strains, indicating more extensive reductions in genomic content.
Due to the draft status of some of the Wolbachia genomes, we relied on the proportions of genes in role categories to assess similarity of genome content. The wTpre strain clusters with the nematode infecting strains when mobile and extrachromosomal elements are included, likely driven by the similarity in the number of phage genes. Without this category of genes, wTpre neighbors A supergroup Wolbachia. The ordination of wCle also changes drastically when the mobile and extrachromosomal element genes are removed from the analysis, going from neighboring A supergroup strains to neighboring B supergroup strains. While the mobile and extrachromosomal elements role category appears to have a dominant effect on ordination for certain strains, the overall pattern of the A and B supergroups was more strongly supported.
The size of the core genome here (496 COGs) was lower than estimates from previous studies. Duplouy et al. (2013) estimated a core of 654 genes based on five strains (from three supergroups): wBol1, wPip_Pel, wMel, wRi, and wBm. Similarly, Ishmael et al. (2009) used exponential regression to estimate a core genome size of 621 genes, but their study examined only Drosophila-infecting Wolbachia strains. It is likely that our inclusion of additional Wolbachia strains, from more diverse hosts and supergroups, is responsible for the smaller core genome size. Comparison of the core Wolbachia genome to other members of the Rickettsiales revealed that only 2.8% of the core is unique to Wolbachia. This finding parallels the discovery of high conservation of two-component systems across 12 Wolbachia strains, A. phagocytophilum, and E. chaffeensis (Christensen and Serbus 2015). These similarities with other closely related rickettsial pathogens may indicate that the core genome comprises genes required for life within an arthropod host, and that the accessory genomes are responsible for the phenotypes that various strains induce.
In wTpre, 482 (34%) of the ORFs were apparently unique: the largest number of any of the arthropod-infecting strains. Only the two nematode-infecting strains, wBm and wOo, had more “unique genes” than wTpre. This may be a feature of the obligate nature of the symbiotic relationships that these strains share with their hosts. However, wBm and wOo are the only representatives from their respective supergroups, and it is likely that inclusion of additional C and D supergroup members would result in a reduction in the number of “unique genes” found in these strains. The wGmm strain also contained a high number of “unique genes”. This may be a result of a problematic assembly, as wGmm had one of the lower BUSCO scores and was responsible for a drastic effect on the size of the core Wolbachia genome.
Examination of the wTpre “unique genes” showed evidence for coding frame truncation in 281 genes, representing 20% of the ORFs in the genome. This is likely an underestimate of the amount of truncation in wTpre. Stringent filtering of sequence similarity, and of up- and downstream homology, did not allow for identifying truncation in rapidly evolving genes, or genes that may have been truncated or fragmented through genomic rearrangements or deletions. Mutations resulting in downstream postnonsense and postframeshift ORFs were not exclusively located in genes identified as unique to wTpre. If the mutation occurred too early in the coding sequence, the ORF was too short to be considered a gene by the IGS pipeline. Conversely, mutations that occurred more 3′ in the coding sequence left an ORF long enough to be considered orthologous with other Wolbachia genes, but could still result in the annotation of short downstream wTpre “unique” ORFs. In wTpre, truncated genes were more likely to carry a hypothetical annotation, despite the fact that homologs from other Wolbachia genomes were often assigned a function. One explanation for this may be the frameshift mutations that result in a change of amino acid sequence, and the loss of recognized functional domains or motifs that would assist in assigning function to the gene. Additionally, the fragmentation of a gene into several ORFs would lead to a functional domain or motif only being associated with one of the resulting ORFs, thus making functional assignments difficult for the other ORFs. Therefore, we conclude that the majority of “unique genes” in wTpre are actually truncated orthologs of known Wolbachia genes from other strains, and likely are not active protein coding genes, but artifacts of ORF prediction machinery.
A relatively small number of inactive or truncated genes were identified in wAu, a Wolbachia strain infecting D. simulans that does not induce strong CI, but does provide viral protection to its host. While the wTpre genome contains a larger number of truncated genes, 78% of the inactive wAu genes were also missing or truncated in wTpre, providing an overlapping set of 36 genes. Both wAu and presumably wTpre have lost the capacity for CI induction. This overlap may indicate an important feature of the transition away from a strong CI phenotype. However, many of these genes have hypothetical gene annotations, and therefore we cannot comment on their potential functions.
We identified a significantly higher number of ANK genes in the B supergroup Wolbachia strains. ANK genes are unusual in bacteria, and it has been hypothesized that phages, transposons, and recombination may have played a role in proliferation of the ANK gene repertoire in Wolbachia (Siozios et al. 2013b; Iturbe-Ormaetxe et al. 2005). The wTpre strain has 54 ANK genes, despite not having associated bacteriophages and having a reduced number of mobile elements. wTpre may have lost its mobile elements and bacteriophages more recently. Indeed, 57 of the 281 truncated wTpre genes (20.2%) are versions of Wolbachia genes with phage or transposon function.
We hypothesize that the extensive protein coding frame truncations present in wTpre reflect the change in reproductive phenotype from CI to PI. In Trichogramma, fixation of asexual reproduction can occur through changes in the host genome, which makes Wolbachia essential to the production of female offspring; so called virginity mutations (Russell and Stouthamer 2011; Stouthamer et al. 2010). While this wTpre strain does infect a host that is dependent upon wTpre’s parthenogenesis-induction, not all Trichogramma, or even all T. pretiosum, have this dependent relationship with their resident Wolbachia strains. Sequencing of additional Trichogramma-infecting Wolbachia strains is necessary to determine whether or not these coding frame truncations are pervasive across all PI-Wolbachia, just the Trichogramma-infecting Wolbachia, or are unique to strains such as wTpre that infect irreversibly asexual hosts.
Supplementary Material
Acknowledgments
We thank Amanda Redding for running the Wheeler et al. (2013) pipeline, Evette Skinner for GenBank submission assistance, and also Paul Rugman-Jones, Eric A. Smith, and two anonymous reviewers for suggestions on the manuscript. We thank the staff at the Baylor College of Medicine Human Genome Sequencing Center for their contributions. We thank the Institute for Genome Sciences Annotation Engine service at the University of Maryland School of Medicine for providing structural and functional annotation of the sequences. This work was supported by the National Science Foundation (DEB 1501227 to A.R.I.L. and DEB 1257053 to J.H.W.); the United States Department of Agriculture (NIFA 194617 to R.S. and NIFA 2016-67011-24778 to A.R.I.L.); the National Human Genome Research Institute (U54 HG003273 to Richard A. Gibbs); and a Robert and Peggy van den Bosch Memorial Scholarship to A.R.I.L.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.028449/-/DC1
Communicating editor: R. Kulathinal
Literature Cited
- Augustinos A. A., Santos-Garcia D., Dionyssopoulou E., Moreira M., Papapanagiotou A., et al. , 2011. Detection and characterization of Wolbachia infections in natural populations of aphids: Is the hidden diversity fully unraveled? PLoS One 6(12): e28695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L., Werren J. H., 2007. Revisiting Wolbachia supergroup typing based on WSP: spurious lineages and discordance with MLST. Curr. Microbiol. 55(1): 81–87. [DOI] [PubMed] [Google Scholar]
- Baldo L., Hotopp J. C. D., Jolley K. A., Bordenstein S. R., Biber S. A., et al. , 2006. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72(11): 7098–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L., Prendini L., Corthals A., Werren J. H., 2007. Wolbachia are present in southern african scorpions and cluster with supergroup F. Curr. Microbiol. 55(5): 367–373. [DOI] [PubMed] [Google Scholar]
- Bandi C., Anderson T. J. C., Genchi C., Blaxter M. L., 1998. Phylogeny of Wolbachia in filarial nematodes. Proc. Biol. Sci. 265(1413): 2407–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing X. L., Xia W. Q., Gui J. D., Yan G. H., Wang X. W., et al. , 2014. Diversity and evolution of the Wolbachia endosymbionts of Bemisia (Hemiptera: Aleyrodidae) whiteflies. Ecol. Evol. 4(13): 2714–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis K., Dobson S. L., Xi Z., Rasgon J. L., Calvitti M., et al. , 2014. Harnessing mosquito–Wolbachia symbiosis for vector and disease control. Acta Trop. 132(Suppl.): S150–S163. [DOI] [PubMed] [Google Scholar]
- Brelsfoard C., Tsiamis G., Falchetto M., Gomulski L. M., Telleria E., et al. , 2014. Presence of extensive Wolbachia symbiont insertions discovered in the genome of its host Glossina morsitans morsitans. PLoS Negl. Trop. Dis. 8(4): e2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiraghi M., Bordenstein S. R., Baldo L., Lo N., Beninati T., et al. , 2005. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology 151: 4015–4022. [DOI] [PubMed] [Google Scholar]
- Christensen S., Serbus L. R., 2015. Comparative analysis of Wolbachia genomes reveals streamlining and divergence of minimalist two-component systems . G3: (Bethesda) 5(5): 983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrostek E., Marialva M. S. P., Esteves S. S., Weinert L. A., Martinez J., et al. , 2013. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet. 9(12): e1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree J., Angiuoli S. V., Wortman J. R., White O. R., 2007. Sybil: methods and software for multiple genome comparison and visualization. Methods Mol. Biol. 408: 93–108. [DOI] [PubMed] [Google Scholar]
- Darby A. C., Armstrong S. D., Bah G. S., Kaur G., Hughes M. A., et al. , 2012. Analysis of gene expression from the Wolbachia genome of a filarial nematode supports both metabolic and defensive roles within the symbiosis. Genome Res. 22(12): 2467–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeine F., Vavre F., Fleury F., Loppin B., Hochberg M. E., et al. , 2001. Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc. Natl. Acad. Sci. USA 98(11): 6247–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins C. A., Cerqueira G. C., Goldberg J. M., Hotopp J. C. D., Haas B. J., et al. , 2013. Genomics of Loa loa, a Wolbachia-free filarial parasite of humans. Nat. Genet. 45(5): 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumler J. S., Barbet A. F., Bekker C., Dasch G. A., Palmer G. H., et al. , 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51(6): 2145–2165. [DOI] [PubMed] [Google Scholar]
- Duplouy A., Iturbe-Ormaetxe I., Beatson S., Szubert J., Brownlie J., et al. , 2013. Draft genome sequence of the male-killing Wolbachia strain wBol1 reveals recent horizontal gene transfers from diverse sources. BMC Genomics 14(1): 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegaard K. M., Klasson L., Naslund K., Bourtzis K., Andersson S. G. E., 2013. Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet. 9(4): e1003381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J., Ganatra M., Kamal I., Ware J., Makarova K., et al. , 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3(4): 599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galens K., Orvis J., Daugherty S., Creasy H. H., Angiuoli S., et al. , 2011. The IGS standard operating procedure for automated prokaryotic annotation. Stand. Genomic Sci. 4(2): 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavotte L., Henri H., Stouthamer R., Charif D., Charlat S., et al. , 2007. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol. Biol. Evol. 24(2): 427–435. [DOI] [PubMed] [Google Scholar]
- Gerth M., Gansauge M. T., Weigert A., Bleidorn C., 2014. Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nat. Commun. 5: 5117. [DOI] [PubMed] [Google Scholar]
- Glowska E., Dragun-Damian A., Dabert M., Gerth M., 2015. New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect. Genet. Evol. 30: 140–146. [DOI] [PubMed] [Google Scholar]
- Gottlieb Y., Zchori-Fein E., Werren J. H., Karr T. L., 2002. Diploidy restoration in Wolbachia-infected Muscidifurax uniraptor (Hymenoptera: Pteromalidae). J. Invertebr. Pathol. 81(3): 166–174. [DOI] [PubMed] [Google Scholar]
- Haegeman A., Vanholme B., Jacob J., Vandekerckhove T. T. M., Claeys M., et al. , 2009. An endosymbiotic bacterium in a plant-parasitic nematode: member of a new Wolbachia supergroup. Int. J. Parasitol. 39(9): 1045–1054. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K., Hammerstein P., Schlattmann P., Telschow A., Werren J. H., 2008. How many species are infected with Wolbachia? - A statistical analysis of current data. FEMS Microbiol. Lett. 281(2): 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Clancy D., Duncan J., 1996. Naturally-occurring Wolbachia infection in Drosophila simulans that does not cause cytoplasmic incompatibility. Heredity 76(1): 1–8. [DOI] [PubMed] [Google Scholar]
- Hosokawa T., Koga R., Kikuchi Y., Meng X. Y., Fukatsu T., 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 107(2): 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishmael N., Hotopp J. C. D., Ioannidis P., Biber S., Sakamoto J., et al. , 2009. Extensive genomic diversity of closely related Wolbachia strains. Microbiology 155: 2211–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I., Burke G. R., Riegler M., O’Neill S. L., 2005. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J. Bacteriol. 187(15): 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris Z., Blagborough A. M., Pinto S. B., Blagrove M. S. C., Godfray H. C. J., et al. , 2010. Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS Pathog. 6(10): e1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30(4): 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent B. N., Salichos L., Gibbons J. G., Rokas A., Newton I. L. G., et al. , 2011. Complete bacteriophage transfer in a bacterial endosymbiont (Wolbachia) determined by targeted genome capture. Genome Biol. Evol. 3: 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L., Walker T., Sebaihia M., Sanders M. J., Quail M. A., et al. , 2008. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol. Biol. Evol. 25(9): 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L., Westberg J., Sapountzis P., Nasiund K., Lutnaes Y., et al. , 2009. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc. Natl. Acad. Sci. USA 106(14): 5725–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer N., Charif D., Henri H., Bataille M., Prevost G., et al. , 2009. A new case of Wolbachia dependence in the genus Asobara: evidence for parthenogenesis induction in Asobara japonica. Heredity 103(3): 248–256. [DOI] [PubMed] [Google Scholar]
- Lo N., Casiraghi M., Salati E., Bazzocchi C., Bandi C., 2002. How many Wolbachia supergroups exist? Mol. Biol. Evol. 19(3): 341–346. [DOI] [PubMed] [Google Scholar]
- Mavingui P., Moro C. V., Tran-Van V., Wisniewski-Dyé F., Raquin V., et al. , 2012. Whole-genome sequence of Wolbachia strain wAlbB, an endosymbiont of tiger mosquito vector Aedes albopictus. J. Bacteriol. 194(7): 1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon J. P., 2010. The bacterial essence of tiny symbiont genomes. Curr. Opin. Microbiol. 13(1): 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende D. R., Sunagawa S., Zeller G., Bork P., 2013. Accurate and universal delineation of prokaryotic species. Nat. Methods 10(9): 881–884. [DOI] [PubMed] [Google Scholar]
- Moreira L. A., Iturbe-Ormaetxe I., Jeffery J. A., Lu G., Pyke A. T., et al. , 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and Plasmodium. Cell 139(7): 1268–1278. [DOI] [PubMed] [Google Scholar]
- Mungall C. J., Emmert D. B., FlyBase Consortium , 2007. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics 23(13): i337–i346. [DOI] [PubMed] [Google Scholar]
- Nikoh N., Hosokawa T., Moriyama M., Oshima K., Hattori M., et al. , 2014. Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. USA 111(28): 10257–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill S. L., Giordano R., Colbert A. M. E., Karr T. L., Robertson H. M., 1992. 16S ribosomal-RNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. USA 89(7): 2699–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannebakker B. A., Pijnacker L. P., Zwaan B. J., Beukeboom L. W., 2004. Cytology of Wolbachia-induced parthenogenesis in Leptopilina clavipes (Hymenoptera: Figitidae). Genome 47(2): 299–303. [DOI] [PubMed] [Google Scholar]
- Papafotiou G., Oehler S., Savakis C., Bourtzis K., 2011. Regulation of Wolbachia ankyrin domain encoding genes in Drosophila gonads. Res. Microbiol. 162(8): 764–772. [DOI] [PubMed] [Google Scholar]
- Pinto S. B., Stainton K., Harris S., Kambris Z., Sutton E. R., et al. , 2013. Transcriptional regulation of Culex pipiens mosquitoes by Wolbachia influences cytoplasmic incompatibility. PLoS Pathog. 9(10): e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2014 R: A language and environment for statistical computing. Available at: http://www.R-project.org/, R Foundation for Statistical Computing, Vienna, Austria.
- Rasgon J. L., Scott T. W., 2004. Phylogenetic characterization of Wolbachia symbionts infecting Cimex lectularius L. and Oeciacus vicarius Horvath (Hemiptera: Cimicidae). J. Med. Entomol. 41(6): 1175–1178. [DOI] [PubMed] [Google Scholar]
- Raychoudhury R., Baldo L., Oliveira D. C. S. G., Werren J. H., 2009. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution 63(1): 165–183. [DOI] [PubMed] [Google Scholar]
- Riley D. R., Angiuoli S. V., Crabtree J., Dunning Hotopp J. C., Tettelin H., 2012. Using Sybil for interactive comparative genomics of microbes on the web. Bioinformatics 28(2): 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros V. I. D., Fleming V. M., Feil E. J., Breeuwer J. A. J., 2009. How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae). Appl. Environ. Microbiol. 75(4): 1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. E., Stouthamer R., 2011. The genetics and evolution of obligate reproductive parasitism in Trichogramma pretiosum infected with parthenogenesis-inducing Wolbachia. Heredity 106(1): 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Hunter W. B., Reese J., Morgan J. K., Marutani-Hert M., et al. , 2012. Survey of endosymbionts in the Diaphorina citri metagenome and assembly of a Wolbachia wDi draft genome. PLoS One 7(11): e50067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg S. L., Puiu D., Sommer D. D., Nene V., Lee N. H., 2009. Genome sequence of the Wolbachia endosymbiont of Culex quinquefasciatus JHB. J. Bacteriol. 191(5): 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Siozios S., Cestaro A., Kaur R., Pertot I., Rota-Stabelli O., et al. , 2013a Draft genome sequence of the Wolbachia endosymbiont of Drosophila suzukii. Genome Announc. 1(1): e00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siozios S., Ioannidis P., Klasson L., Andersson S. G. E., Braig H. R., et al. , 2013b The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS One 8(2): e55390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer R., Kazmer D. J., 1994. Cytogenetics of microbe-associated parthenogenesis and its consequences for gene flow in Trichogramma wasps. Heredity 73: 317–327. [Google Scholar]
- Stouthamer R., Luck R. F., Hamilton W. D., 1990. Antibiotics cause parthenogenetic Trichogramma (Hymenoptera, Trichogrammatidae) to revert to sex. Proc. Natl. Acad. Sci. USA 87(7): 2424–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer R., Breeuwer J. A. J., Luck R. F., Werren J. H., 1993. Molecular-identification of microorganisms associated with parthenogenesis. Nature 361(6407): 66–68. [DOI] [PubMed] [Google Scholar]
- Stouthamer R., Breeuwer J. A. J., Hurst G. D. D., 1999a Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53: 71–102. [DOI] [PubMed] [Google Scholar]
- Stouthamer R., Hu J. G., van Kan F., Platner G. R., Pinto J. D., 1999b The utility of internally transcribed spacer 2 DNA sequences of the nuclear ribosomal gene for distinguishing sibling species of Trichogramma. BioControl 43(4): 421–440. [Google Scholar]
- Stouthamer R., Russell J. E., Vavre F., Nunney L., 2010. Intragenomic conflict in populations infected by Parthenogenesis Inducing Wolbachia ends with irreversible loss of sexual reproduction. BMC Evol. Biol. 10: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton E., Harris S., Parkhill J., Sinkins S., 2014. Comparative genome analysis of Wolbachia strain wAu. BMC Genomics 15(1): 928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. J., Bandi C., Hoerauf A. M., Lazdins J., 2000. Wolbachia bacteria of filarial nematodes: a target for control? Parasitol. Today 16(5): 179–180. [DOI] [PubMed] [Google Scholar]
- Timmermans M., Ellers J., 2009. Wolbachia endosymbiont is essential for egg hatching in a parthenogenetic arthropod. Evol. Ecol. 23(6): 931–942. [Google Scholar]
- Turelli M., Hoffmann A. A., 1991. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353(6343): 440–442. [DOI] [PubMed] [Google Scholar]
- van Meer M. M. M., Witteveldt J., Stouthamer R., 1999. Phylogeny of the arthropod endosymbiont Wolbachia based on the wsp gene. Insect Mol. Biol. 8(3): 399–408. [DOI] [PubMed] [Google Scholar]
- Vavre F., Fleury F., Lepetit D., Fouillet P., Bouletreau M., 1999. Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol. Biol. Evol. 16(12): 1711–1723. [DOI] [PubMed] [Google Scholar]
- Walker T., Johnson P. H., Moreira L. A., Iturbe-Ormaetxe I., Frentiu F. D., et al. , 2011. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476(7361): 450–453. [DOI] [PubMed] [Google Scholar]
- Walsh P. S., Metzger D. A., Higuchi R., 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10(4): 506–513. [PubMed] [Google Scholar]
- Weeks A., Breeuwer J., 2001. Wolbachia–induced parthenogenesis in a genus of phytophagous mites. Proc. Biol. Sci. 268(1482): 2245–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks A. R., Turelli M., Harcombe W. R., Reynolds K. T., Hoffmann A. A., 2007. From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 5(5): e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., 1997. Biology of Wolbachia. Annu. Rev. Entomol. 42: 587–609. [DOI] [PubMed] [Google Scholar]
- Werren J. H., Windsor D. M., 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. Biol. Sci. 267(1450): 1277–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Zhang W., Guo L. R., 1995. Evolution and phylogeny of Wolbachia - reproductive parasites of arthropods. Proc. Biol. Sci. 261(1360): 55–63. [DOI] [PubMed] [Google Scholar]
- Werren J. H., Baldo L., Clark M. E., 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6(10): 741–751. [DOI] [PubMed] [Google Scholar]
- Wheeler D., Redding A. J., Werren J. H., 2013. Characterization of an ancient Lepidopteran lateral gene transfer. PLoS One 8(3): e59262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Sun L. V., Vamathevan J., Riegler M., Deboy R., et al. , 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2(3): 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalou S., Riegler M., Theodorakopoulou M., Stauffer C., Savakis C., et al. , 2004. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc. Natl. Acad. Sci. USA 101(42): 15042–15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. G., Rousset F., O’Neill S., 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265(1395): 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zug R., Hammerstein P., 2012. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7(6): e38544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The T. pretiosum colony used for sequencing is available upon request. Supplemental Material, Table S1 contains a detailed breakdown of the counts of genes in each role category and subcategory, for each Wolbachia strain, as annotated by IGS. Table S2 provides complete BUSCO results for all Wolbachia strains. Table S3 is the wTpre “unique genes” considered in truncation analyses. Table S4 contains comparisons of truncated genes in wAu and wTpre. The wTpre Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession LKEQ00000000. The version described in this paper is version LKEQ01000000.1.