Abstract
Notch-type signaling mediates cell−cell interactions important for animal development. In humans, reduced or inappropriate Notch signaling activity is associated with various developmental defects and disease states, including cancers. Caenorhabditis elegans expresses two Notch-type receptors, GLP-1 and LIN-12. GLP-1 mediates several cell-signaling events in the embryo and promotes germline proliferation in the developing and adult gonad. LIN-12 acts redundantly with GLP-1 in certain inductive events in the embryo and mediates several cell−cell interactions during larval development. Recovery of genetic suppressors and enhancers of glp-1 or lin-12 loss- or gain-of-function mutations has identified numerous regulators of GLP-1 and LIN-12 signaling activity. Here, we report the molecular identification of sog-1, a gene identified in screens for recessive suppressors of conditional glp-1 loss-of-function mutations. The sog-1 gene encodes UBR-5, the sole C. elegans member of the UBR5/Hyd family of HECT-type E3 ubiquitin ligases. Molecular and genetic analyses indicate that the loss of ubr-5 function suppresses defects caused by reduced signaling via GLP-1 or LIN-12. In contrast, ubr-5 mutations do not suppress embryonic or larval lethality associated with mutations in a downstream transcription factor, LAG-1. In the gonad, ubr-5 acts in the receiving cells (germ cells) to limit GLP-1 signaling activity. SEL-10 is the F-box component of SCFSEL-10 E3 ubiquitin–ligase complex that promotes turnover of Notch intracellular domain. UBR-5 acts redundantly with SEL-10 to limit Notch signaling in certain tissues. We hypothesize that UBR-5 activity limits Notch-type signaling by promoting turnover of receptor or limiting its interaction with pathway components.
Keywords: Notch, germ cell, GLP-1, LIN-12, HECT domain
The highly conserved Notch-type signaling process mediates inductive cell interactions during animal development (see reviews by Greenwald and Kovall 2013; Koch et al. 2013; Suresh and Irvine 2015; Yamamoto et al. 2014). Notch signaling is active in many different tissues in any given species, and defective Notch signaling is associated with many human disease conditions, including developmental syndromes and certain cancers (see reviews by Louvi and Artavanis-Tsakonas 2012; Ntziachristos et al. 2014; Penton et al. 2012; Suresh and Irvine 2015). Notch-type signaling is unusual compared with other developmentally important signaling mechanisms in that it is limited to adjacent cells, involves cleavage of the receptor to release a transcription factor, and acts in a relatively diverse set of developmental and physiological contexts. During canonical Notch signaling, summarized in Figure 1A, the DSL (Delta, Serrate, LAG-2) -type ligand on the signaling cell binds membrane-associated Notch-type receptor on the receiving cell, and this interaction triggers proteolytic cleavage of the receptor. Sequential cleavage events, accomplished by ADAM protease (the S2 cleavage) and γ-secretase (the S3 cleavage), release the Notch intracellular domain (NICD) for transport to the nucleus where it interacts with a CSL (CBF1/Su(H)/LAG-1)-type DNA binding protein and a conserved coactivator protein (Mastermind family in mammals and Drosophila, SEL-8/LAG-3 in nematodes), and displaces a corepressor complex. The NICD/CSL activator complex up-regulates transcription of target genes whose identity depends on cell type.
Figure 1.
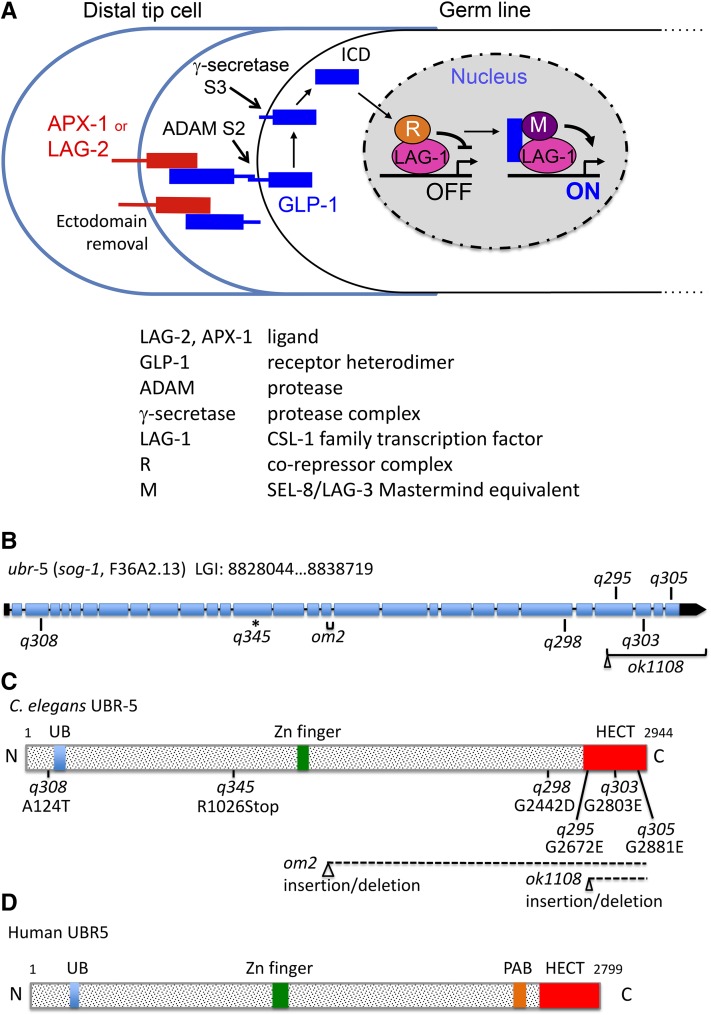
sog-1 encodes UBR-5, a HECT-type E3 ubiquitin ligase. (A) Working model for GLP-1/Notch signaling in the adult C. elegans germline. Interaction of transmembrane LAG-2 and/or APX-1 ligand and GLP-1 heterodimer triggers proteolytic cleavage of GLP-1. The S2 cleavage requires an ADAM family protease and releases the GLP-1 ectodomain (bound to LAG-2); the S3 cleavage requires γ-secretase and releases the GLP-1 intracellular domain (ICD) for transport to the nucleus. Nuclear GLP-1 ICD interacts with the CSL-1-type transcription factor, LAG-1, and the coactivator SEL-8/LAG-3 (M), and displaces the LAG-1-bound corepressor complex (R). Signaling “strength” is modulated by numerous processes, as described in the text. (B) Diagram represents ubr-5 gene structure; mutant lesions associated with suppression of glp-1 are indicated. Nucleotide coordinates refer to genome version WS240. (C) Diagram represents UBR-5 protein structure. Conserved domains are indicated, as are amino acid substitutions and deletions/insertions associated with mutant alleles. The UB domain is variably referred to as the EDD or E3 domain in the literature. See Table S2 for details of sequence insertions and deletions in ok1108 and om2. (D) Domain architecture of human UBR5. Many UBR5 family members contain a poly(A) binding protein (PABP) motif (aka MLLE motif) just upstream of the HECT domain.
Notch-type signaling is modulated by a variety of mechanisms (Louvi and Artavanis-Tsakonas 2012; Yamamoto et al. 2014). In the signaling cell, endocytosis of the ligand−receptor complex causes an essential conformational change that allows the S2 cleavage to occur. This process requires components of the endosomal trafficking machinery and mono-ubiquination of DSL intracellular domain by E3 ubiquitin ligases. In some signaling contexts, endocytosis also plays an earlier role in activation of ligand. In the receiving cell, newly synthesized Notch-type receptor receives a variety of post-translational modifications that modulate its activity prior to reaching the cell surface. As is the case for signaling activity in general, it is important that Notch-type signaling not be continuous, and mechanisms are in place to down-regulate NICD activity (reviewed by Baron 2012; Barth and Kohler 2014; Le Bras et al. 2011; Kandachar and Roegiers 2012; Weinmaster and Fischer 2011). For example, NICD activity is limited by ubiquitin-mediated targeting to the proteasome for degradation (reviewed by Lai 2002; Wang et al. 2011). In this process, ubiquitin is covalently linked to ubiquitin-activating enzyme (E1), transferred to ubiquitin-conjugating enzyme (E2), and finally transferred to the protein substrate (e.g., NICD) via the activity of a ubiquitin–protein ligase (E3) (reviewed by Kipreos 2005; Hershko and Ciechanover 1998). Target specificity is conferred by the E3 ligase, and SEL-10/Fbw7 is a conserved E3 ligase component that limits Notch signaling activity in many species (Hubbard et al. 1997; see reviews by Lai 2002; Wang et al. 2011). In addition to promoting protein turnover, E3-mediated ubiquitination can modulate proteinprotein interactions and impact processes such as nuclear import (Rodriguez 2014). Therefore, E3 ligase activity may also regulate Notch signaling by modulating interactions between signaling components.
Two Notch isoforms are present in Caenorhabditis elegans: GLP-1 (germline proliferation defective-1) and LIN-12 (lineage defective-12) (Greenwald and Kovall 2013). GLP-1 and/or LIN-12 mediate numerous cell-signaling events throughout development and in the adult gonad (reviewed by Priess 2005; Sternberg 2005; Greenwald and Kovall 2013). GLP-1 is active in cell−cell interactions in the embryo and in soma-to-germline signaling in the larval and adult gonad. LIN-12 is redundant with GLP-1 for some signaling events in the late embryo and later mediates signaling events in many different somatic cells during larval development. Numerous modulators of Notch signaling – as well as core components of the pathway – have been identified in extensive genetic screens for suppressors or enhancers of loss- or gain-of-function mutations in glp-1, lin-12, or components of the γ-secretase complex (reviewed by Greenwald and Kovall 2013). Most of these modulators also regulate additional developmental processes not known to involve Notch signaling.
GLP-1-mediated inductive signaling from the somatic gonad to the germline is essential for germ cell proliferation in the larva and adult (reviewed by Hansen and Schedl 2013; Kimble and Seidel 2013). All germ cells are proliferative in early larval development; later, proximal germ cells enter meiosis, and germline proliferation becomes restricted to the distal region of the gonad arm. During the early proliferative phase, several somatic gonadal cells signal to the germline via GLP-1; in the later phase, only the distal tip cell signals to the germline via GLP-1. Proliferative germ cells are not maintained if signaling is reduced or abolished by mutation in a GLP-1 pathway component or by ablation of somatic signaling cells; instead, germ cells prematurely exit mitosis, enter meiosis, and form gametes. Additional factors promote a wild-type level of germ cell proliferation, including the distal sheath cells (Killian and Hubbard 2005), gap junctions between the somatic gonad and germline (Starich et al. 2014), and nutritional factors, among others (Hubbard et al. 2013).
We previously recovered suppressor of glp-1 (sog) mutations in genetic screens for suppressors of the glp-1 temperature-sensitive (ts) phenotype (Maine and Kimble 1993). The sog mutations partially suppressed glp-1 maternal effect embryonic lethality and germline proliferation defects. Here, we report the molecular characterization of sog-1. We demonstrate that sog-1 encodes UBR-5, a member of the HECT (homologous to the E6-AP carboxyl terminus)-type E3 protein–ubiquitin ligase family whose closest mammalian relative is UBR5 (Ubiquitin protein ligase E3 component n-recognin 5; also called EDD, E3 identified by differential display). We find that loss of UBR-5 activity causes an increase in both GLP-1 and LIN-12 signaling activity in sensitized genetic backgrounds where the receptor carries a conditional mutation. Genetic analysis suggests that UBR-5 acts in the receiving cell. The SCFSEL-10 E3 ubiquitin−ligase complex is known to limit LIN-12 activity and, to a minor extent, GLP-1 activity by binding to and promoting turnover of the intracellular domain (Sundaram and Greenwald 1993; Hubbard et al. 1997). Our genetic analysis suggests that UBR-5 and SEL-10 function in concert to limit Notch signaling in some tissues.
Materials and Methods
Mutant strains
C. elegans strains were cultured using standard methods (Epstein and Shakes 1995). All strains were derived from the Bristol strain, N2. Mutations used in this study are described in www.wormbase.org unless otherwise noted and include the following:
LGI: sog-1 alleles q295, q298, q303, q305, q308, q345 (all described in Maine and Kimble 1993), om2 (this study), ok1108 (OMRF Knockout Group, see wormbase.org), rrf-1(pk1417), unc-13(e51), nDf25, ccIs4251 [myo-3p::GFP(NLS)::LacZ (pSAK2) + myo-3p::GFP (mitochondrially targeted) (pSAK4) + dpy-20(+)].
LGIII: glp-1(q231ts), glp-1(ar202ts), lin-12(ar170), unc-32(e189).
LGIV: lag-1(om13ts), nT1 [qIs51].
LGV: him-5(e1467), him-5(e1490), sel-10(ok1632).
In addition, syIs50 [cdh-3::GFP + dpy-2(+)] served as an anchor cell marker (Pettitt et al. 1996; Inoue et al. 2002).
The om2 deletion allele was isolated in a noncomplementation screen as follows. L4 unc-13(e51); glp-1(q231) hermaphrodites were treated with 20−30 μg/ml trimethylpsoralen (TMP) in M9 medium, irradiated for 20 sec at a distance of 10 cm with a long-wave UV power source, allowed to recover for several hr, and mated with sog-1(q298);glp-1(q231);him-5(e1490) males at 15°. Mating plates were shifted to 20°, after a substantial number of F1 embryos had been produced, and screened 2−3 d later for the presence of fertile non-Unc cross-progeny, which were presumed to be genotype unc-13(+) sog-1(q298)/unc-13 sog-1(omx); glp-1(q231); him-5(e1490/+). Homozygous unc-13 sog-1(omx); glp-1(q231) animals were recovered and out-crossed to confirm that the new sog allele was linked to unc-13.
Whole genome sequencing
Genomic DNA isolation from strains JK946 [carrying sog-1(q303)] and JK952 [carrying sog-1(q308)], library construction, whole genome sequencing (WGS), and bioinformatics analysis were performed as described (Flibotte et al. 2010; Thompson et al. 2013). For analysis of other sog-1 strains, the F36A2.13 gene region was recovered by DNA amplification and sequenced using standard methods. The sog-1 mutations were originally mapped to the cluster on LGI, and we particularly focused on WGS data from this region.
In the course of this study, we also performed WGS analysis of JK953, a strain previously reported to carry a sog-1 mutation called q309. Unlike other sog-1 alleles, which were recovered in F2 screens for recessive suppressors of glp-1, q309 was recovered in the course of a dominant suppressor screen as described (Maine and Kimble 1993). WGS analysis revealed that JK953 is wild-type for glp-1 and does not contain a mutation in F36A2.13. We conclude that the reduced brood size and temperature sensitivity associated with JK953 result from a combination of other mutations in the genetic background.
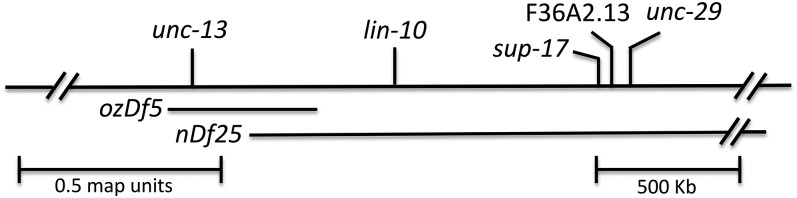
Our molecular data led us to revise how we interpret earlier gene dosage data. Genetic mapping had placed sog-1 mutations in a region uncovered by the deficiencies ozDf5 and nDf25 (Maine and Kimble 1993). Neither sog-1(-)/ozDf5 nor sog-1(-)/nDf25 suppressed glp-1(q231); therefore we speculated that the sog-1 alleles isolated in our glp-1 suppression screen were recessive gain-of-function mutations. Now, based on our molecular identification of sog-1 as located outside the region uncovered by ozDf5, we would not expect sog-1/ozDf5 to suppress glp-1(ts) (Figure 2). In contrast, nDf25 uncovers genes that flank sog-1, suggesting it should uncover sog-1 (Figure 2). To investigate further, we generated an nDf25/unc-13 ccIs4251; glp-1(q231ts) strain and mated hermaphrodites of this genotype with sog-1(om2);glp-1(q231ts) males to generate nDf25/sog-1(om2); glp-1(q231ts) animals. This assay is straightforward compared with our earlier test because use of a GFP-tagged chromosome obviates the need for other marker mutations and allows us to identify unambiguously the very slow growing nDf25/sog-1(om2) cross-progeny. Matings were conducted at 15°, and progeny were shifted to 20° after hatching. nDf25/sog-1(om2); glp-1(q231ts) hermaphrodites were picked to a separate plate and observed to segregate viable embryos, indicating that nDf25/sog-1(om2) suppresses glp-1(q231ts). Hence, nDf25 indeed appears to uncover sog-1. nDf25/sog-1(om2); glp-1(q231ts) hermaphrodites also segregated nonviable embryos, presumably nDf25 homozygotes.
Figure 2.
Genetic and physical map of the ubr-5 region. Genetic mapping previously placed sog-1 between unc-13 and lin-10. Our molecular studies reported here indicate that sog-1 corresponds to F36A2.13/ubr-5, located between lin-10 and unc-29. The positions shown for deficiencies ozDf5 and nDf25 reflect their ability to uncover mutations in genes in the region.
Phenotypic analysis
Brood size assays were carried out using standard methods as follows. All sog-1;unc-32 glp-1(231) strains were maintained at 20°. The unc-32 glp-1(q231ts) control strain was maintained at 15°, and animals to be used for brood size experiments were shifted to 20° as late-stage embryos or newly hatched L1 larvae. Broods were assayed by placing individual L4 larvae onto single plates; once they became gravid adults, they were moved to a fresh plate daily until they no longer produced embryos. Embryos were counted immediately after the mother was moved; viable L4 larvae were counted 2−3 d later.
The anchor cell (AC)-ventral uterine (VU) precursor phenotype was assayed by examining late L2 stage larvae with differential interference contrast and epifluorescence microscopy using a Zeiss Axioscope. Anchor cells were identified based on morphology, position within the gonad primordium, and expression of a cdh-3::gfp transgene included in the strain.
Proliferative zone size was analyzed in adults labeled with DAPI using standard methods (e.g., Qiao et al. 1995). L4 stage hermaphrodites were picked to a fresh culture plate, aged 24−25 hr, fixed for ∼15 min with −20° methanol, stained for ∼15 min with 0.2 μg/ml DAPI, and mounted in Vectashield (Vector Laboratories) for epifluorescence analysis. To assay the proliferative region of each specimen, we counted the number of rows of germ cells distal to the leptotene−zygotene “transition” zone.
RNAi assays
RNA interference (RNAi) was performed by the feeding method (Timmons et al. 2001). L4 larvae were placed onto culture plates seeded with E. coli expressing sog-1 double-stranded RNA (dsRNA), and their F1 progeny were assayed in the first day of adulthood for the presence/absence of embryos. Animals were removed from culture plates as they were counted. Non-RNAi controls were performed by culturing each strain [glp-1(ar202) or rrf-1(pk1417);glp-1(ar202)] on the standard OP50 E. coli food source. Presence of the rrf-1(pk1417) deletion was verified by DNA amplification. In addition, the sensitivity of rrf-1(pk1417);glp-1(ar202) animals to RNAi of various somatic genes was tested to confirm the presence of the RNAi defect as described by Kumsta and Hansen (2012). As a control for an effect of RNAi per se on the glp-1(ar202) phenotype, RNAi was performed with empty L4440 vector, which produces a short dsRNA that does not correspond to C. elegans genomic sequence.
Data availability
Strains are available upon request. Whole genome sequence data are provided in Supplemental Material, Table S1.
Results
sog-1 encodes a HECT-type E3 ubiquitin ligase
Mutations in sog-1 were recovered in a genetic screen for recessive suppressors of glp-1(q224ts) and glp-1(q231ts) (Maine and Kimble 1993). Both of these glp-1(ts) alleles have a Glp-1 null phenotype at 25° and a partial loss-of-function (lf) phenotype at 20° (Maine and Kimble 1989). Consequently, when L1 stage glp-1(ts) larvae are shifted from permissive temperature to 25°, their germ cells exit mitosis, enter meiosis, and undergo spermatogenesis. In contrast, when L1 stage glp-1(ts) larvae are shifted from permissive temperature to 20°, their germ cells proliferate for a period of time before prematurely entering meiosis and undergoing gametogenesis; in most cases, a full complement of sperm and some oocytes form and some embryos are generated. These embryos die due to defects in GLP-1 signaling during embryogenesis. The sog-1 mutations partially suppress the Glp-1(ts) defects at 20°, but not at 25°, and therefore do not bypass the requirement for GLP-1 activity (Maine and Kimble 1993).
We initiated a molecular study of sog-1 by performing whole genome sequence analysis of two sog-1 mutant strains: JK946, containing sog-1(q303); and JK952, containing sog-1(q308) (see Materials and Methods). When we compared the JK946 and JK952 sequence data to the reference C. elegans genome, we identified numerous common mutations that presumably were present in the original unc-32(e189) glp-1(q231ts) strain prior to mutagenesis (Table S1). In addition, we identified a number of mutations unique to either JK946 or JK952. Of note, JK946 and JK952 contain distinct mutations in a common open reading frame (ORF), F36A2.13 (Table S1, Figure 1B). F36A2.13 is predicted to encode a member of the HECT family of E3 ubiquitin ligases (Figure 1). Its closest mammalian relative is UBR5/EDD (Callaghan et al. 1998; Tasaki et al. 2005), and consequently F36A2.13 is listed in Wormbase as ubr-5 (UBR E3 ubiquitin ligase homolog – 5) (www.wormbase.org).
We confirmed that ubr-5 and sog-1 are the same gene by amplifying and sequencing the F36A2.13 genomic region from five additional strains carrying sog-1 alleles recovered following mutagenesis with EMS (q295, q298, q305, q345; Maine and Kimble 1993) or UV (om2; see Materials and Methods). Each strain contained a mutation in F36A2.13 (Figure 1, Table S2). In addition, we obtained an F36A2.13 deletion allele, ok1108, from the Caenorhabditis Genetics Center and tested its ability to suppress glp-1(q231ts). At 20°, ok1108 partially suppresses glp-1(q231ts) (Table 1). We conclude that sog-1 and F36A2.13 are the same gene. Although sog-1 is the original published gene name, “ubr-5” better denotes the gene product. Therefore, we will refer to F36A2.13 as ubr-5 for the rest of this article.
Table 1. Suppression of glp-1(q231ts) by ubr-5 deletion alleles at 20°.
| Genotype | Avg. No. Embryos Produced per Brood | % Viable | n |
|---|---|---|---|
| unc-32 glp-1(q231ts) | 129 ± 8a | 0 | 16 |
| ubr-5(om2);unc-32 glp-1(q231ts) | 174 ± 10 | 40 | 10 |
| ubr-5(ok1108);unc-32 glp-1(q231ts) | 189 ± 6 | 34 | 14 |
Full broods were counted for the indicated (“n”) number of hermaphrodites, including both viable and nonviable embryos.
The baseline No. of embryos produced by unc-32 glp-1(q231ts) controls in these experiments was substantially higher than previously reported (e.g., Maine and Kimble 1989, 1993). Controls were performed with two strains, both of which had been frozen since the early 1990s and were thawed specifically for these assays (see Materials and Methods). As described in Maine and Kimble (1993), ∼98% of unc-32 glp-1(q231ts) controls produced some (nonviable) embryos, and only ∼2% were Glp-1 sterile. >99.9% of ubr-5(-);glp-1(q231ts) animals produced embryos.
Previous analysis of ubr-5 mutants indicated that they were superficially normal (Maine and Kimble 1993). We reevaluated this question with the deletion alleles, ubr-5(om2) and ubr-5(ok1108). These mutants likewise do not have obvious developmental defects in a glp-1(+) background under laboratory conditions. In particular, we considered that ubr-5 mutants might impact the germline stem cell pool. As a measure of proliferative zone size, we counted the number of rows of germ cell nuclei from the distal end of the somatic gonad to the start of the leptotene/zygotene region in animals raised at 20°. When we compared the number of rows of nuclei in the mitotic zone in glp-1(q231ts) and ubr-5(om2);glp-1(q231ts) animals at 24 hr post-L4 stage, we observed an increase in mitotic zone size from an average of four rows in glp-1(q231ts) to an average of 11 rows in ubr-5(om2); glp-1(q231ts) (Table 2). Therefore, the loss of UBR-5 activity leads to increased germ cell proliferation in the sensitized GLP-1(ts) background. In contrast, we did not observe an increase in the number of rows of mitotic germ cells in ubr-5 mutants compared with wild-type at 24 hr post-L4 (Table 2). Hence, the loss of UBR-5 activity did not impact the length of the mitotic region in germlines with wild-type GLP-1 function.
Table 2. Suppression of the glp-1(q231ts) germline proliferation defect by ubr-5(om2).
| Genotype | No. Rows of Nuclei in Proliferative Zonea (Range) | n |
|---|---|---|
| Wild type (N2) | 21 ± 0.6 (16–24) | 18 |
| ubr-5(om2) | 21 ± 0.8 (16–25) | 13 |
| glp-1(q231ts)b | 4 ± 0.7 (0–11) | 36 |
| ubr-5(om2);glp-1(q231ts) | 11 ± 0.4 (5–16) | 32 |
| sel-10(ok1632)c | 15 ± 0.6 (11–20) | 14 |
| glp-1(q231ts); sel-10(ok1632) | 6 ± 0.9 (0–14) | 24 |
| ubr-5(om2);glp-1(q231ts);sel-10(ok1632) | 12 ± 0.6 (8–19) | 21 |
| ubr-5(om2);sel-10(ok1632) | 19 ± 1.2 (13–28) | 16 |
Assays were conducted at 20°. L4 stage larvae were picked to a fresh plate and DAPI-stained 24 hr later. n, number of gonad arms evaluated.
± represents standard error of the mean. The number of rows of proliferative nuclei was rounded to the nearest whole number.
The unc-32(e189) glp-1(q231ts) strain was maintained at 15°; late-stage embryos were shifted to 20° for growth at restrictive temperature. All glp-1(q231ts) strains listed here carry the unc-32(q231) marker mutation.
We note that the sel-10(ok1632) strain, RB1432, contains additional mutations that may reduce mitotic zone size. See text.
UBR-5 negatively regulates GLP-1 activity
The predicted ubr-5 product is a 2944 amino acid protein with domain architecture characteristic of the UBR5/EDD HECT protein subfamily. It contains three conserved motifs: a UB (also called E3) domain located near the N terminus, a zinc-finger domain located in the middle of the protein (the UBR motif), and a C-terminal HECTc domain (Figure 1C). Many members of this protein family also contain a PABP (poly A binding protein; also called MLLE) domain within ∼100 amino acids of the HECT domain (Callaghan et al. 1998; Tasaki et al. 2005; Scheffner and Kumar 2014) (Figure 1D). C. elegans UBR-5 appears to lack this domain. Among the eight ubr-5 alleles we characterized, three contain a premature stop codon. The ubr-5(om2) allele is likely to be null; it contains a deletion of 105 nucleotides in the center of the gene just downstream of the zinc-finger domain that is predicted to shift the ORF, inserting 45 amino acids and deleting the C-terminal half of the protein, including the entire HECT domain (Figure 1, Table S2). Similarly, ubr-5(q345) may be null as it contains a single nucleotide change that converts residue 1026 to a stop codon (Figure 1, Table S2). It is predicted to encode a truncated protein lacking the zinc-finger and HECT domains. ubr-5(ok1108) is a complex mutation with a 1360 nucleotide deletion and 73 nucleotide insertion; most of the HECT domain is deleted as well as the 3′ UTR and some downstream sequence (Figure 1, Table S2). The net result of the mutation is to insert nine amino acids downstream of residue 2631. This allele is expected to lack E3 ligase activity, as well. The nature of these alleles suggests that a reduction in ubr-5 function suppresses the loss of glp-1 activity, and therefore UBR-5 is a negative regulator of GLP-1. The other ubr-5 alleles we characterized contain missense mutations predicted to cause single amino acid substitutions as follows: q308 just upstream of the E3 domain; q298 just upstream of the HECT domain; and q295, q303, and q305 within the HECT domain (Figure 1, Table S2).
We characterized glp-1 suppression by the ubr-5 deletion alleles, om2 and ok1108 (Table 1), and compared the results with data obtained previously for other alleles (Maine and Kimble 1993). We evaluated the total number of progeny (viable and nonviable) produced and the number of progeny that hatched and developed to adulthood. As observed previously (Maine and Kimble 1993), we do not see a simple relationship between suppression of the brood size defect and embryonic lethality, and the likely null alleles do not show identical suppression. We suspect that suppression is influenced by other, unique mutations present in different ubr-5 strains. For example, WGS data revealed a suite of shared mutations and a number of unique mutations in ubr-5(q303); unc-32(e189) glp-1(q231) and ubr-5(q308); unc-32(e189) glp-1(q231) (Table S1). We hypothesize that some of these mutations may influence the degree of suppression by ubr-5 in one or more tissues. Indeed, it was noted during previous three-factor mapping experiments that suppression by ubr-5 is abrogated to a large extent in the presence of certain marker mutations (Maine and Kimble 1993). As detailed in the Materials and Methods, data obtained in the course of our studies allow us to make corrections to the literature with respect to (i) gene dosage requirements and (ii) the identity of a previously reported allele, q309.
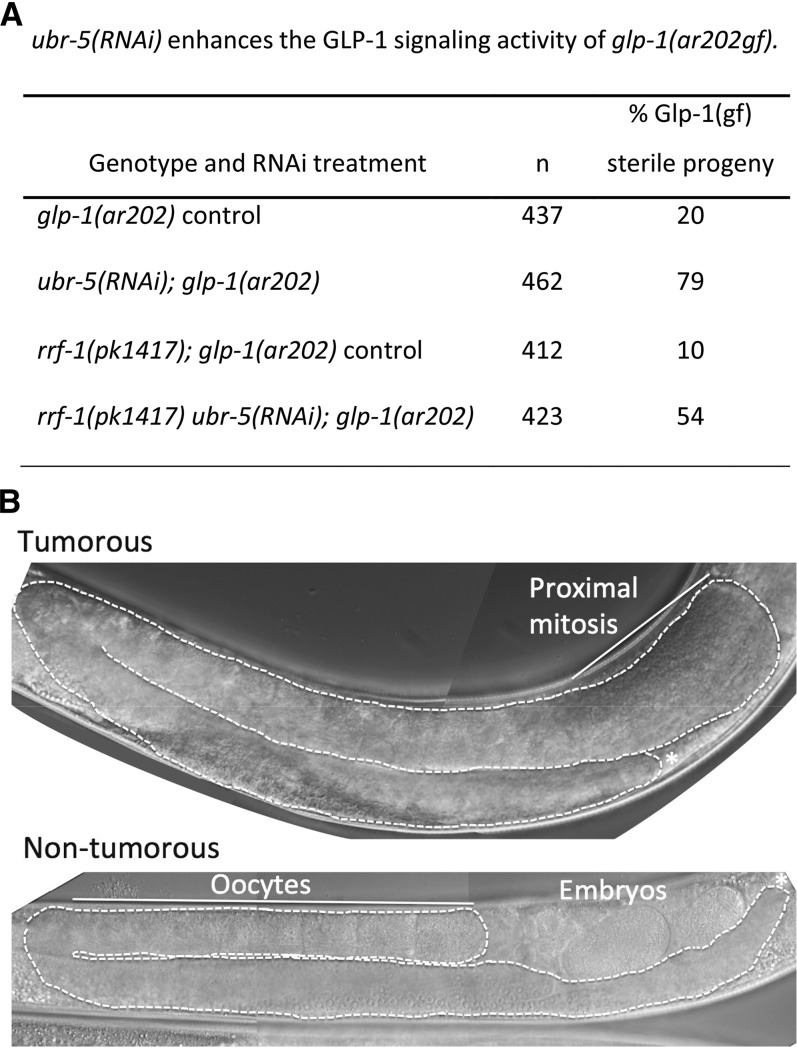
To confirm further that the ubr-5 mutations are loss-of-function, we knocked down UBR-5 protein in a glp-1 gain-of-function background using RNAi and examined the consequences for GLP-1 signaling. Gain-of-function glp-1 mutations have elevated GLP-1 signaling, resulting in germline over-proliferation and eventual formation of a germline tumor (Berry et al. 1997; Pepper et al. 2003). The glp-1(ar202gf) allele is temperature sensitive, producing a more elevated level of GLP-1 signaling at higher temperatures (Pepper et al. 2003). At 25°, germ cells form a tumor relatively early in development, and the animal does not produce oocytes. At lower (semipermissive) temperatures, overproliferation occurs more slowly, and a sizable proportion of glp-1(ar202) mutants are fertile (Pepper et al. 2003).
We tested the extent to which reducing ubr-5 activity would increase the level of GLP-1 signaling in the glp-1(ar202) background at the semipermissive temperature, 22°. We assayed the increase in GLP-1 signaling by quantifying the increase in % tumorous sterility in the population at ∼24 hr post-L4 stage. Under conditions used in our assay, on average ∼20% of glp-1(ar202) control animals were tumorous and lacked embryos, and this phenotype increased nearly fourfold to ∼79% in ubr-5(RNAi); glp-1(ar202) animals (Figure 3). The tumorous glp-1(ar202) controls and ubr-5(RNAi); glp-1(ar202) double mutants contained mitotic nuclei in the proximal germline (Figure 3B). We conclude that ubr-5(RNAi) significantly increases the level of GLP-1 signaling in glp-1(ar202) mutants. This result supports the conclusion that reduced UBR-5 activity leads to elevated GLP-1 signaling activity.
Figure 3.
ubr-5(RNAi) enhances glp-1(ar202gf). (A) Assays were performed at 22°, a semipermissive temperature for glp-1(ar202gf). L4 larvae were placed onto ubr-5(RNAi) or control plates, and their adult progeny were evaluated for fertility or sterility. Three replicate sets of experiments were performed, where all four treatments were run in parallel. In the rrf-1(+) background, ubr-5 RNAi is active in all tissues; in the rrf-1(0) background, ubr-5 RNAi is active in the germline, but not in the somatic gonad (Kumsta and Hansen 2012). The Glp-1 gf phenotype is significantly enhanced in both rrf-1(+) and rrf-1(0) backgrounds relative to the appropriate non-RNAi control assay (animals cultured on OP50 bacteria). A paired t-test indicates no difference between enhancement of glp-1(ar202gf) in rrf-1(+) and rrf-1(0) samples. In contrast to the 4- to 5.5-fold increase in % tumorous animals upon ubr-5(RNAi), negative controls performed with “empty vector” (L4440 plasmid without an insert) increased the % tumorous animals by 0.4- to 0.5-fold relative to controls grown in parallel [n = 449 glp-1(ar202), n = 78 rrf-1;glp-1(ar202)]. n, total number of animals scored in replicate treatments. (B) Examples of the glp-1(ar202gf) Tumorous and non-Tumorous phenotypes observed under our assay conditions.
UBR-5 acts in the germ cells to limit GLP-1 pathway activity in the gonad
To evaluate whether UBR-5 activity is important in the signaling and/or receiving cell, we assayed the ability of ubr-5(RNAi) to enhance GLP-1 signaling activity in an rrf-1(0) mutant background. In rrf-1(pk1417) mutants, RNAi is severely impaired in many somatic tissues, including the somatic gonad (Kumsta and Hansen 2012; Sijen et al. 2001). We performed ubr-5 RNAi in parallel in rrf-1(pk1417);glp-1(ar202) and rrf-1(+); glp-1(ar202) hermaphrodites and assayed enhancement of the glp-1(ar202gf) sterile phenotype (Figure 3). glp-1(ar202gf) sterility was enhanced significantly in rrf-1(pk1417) animals (Figure 3). Statistical analysis indicates no significant difference in the degree of enhancement in the rrf-1(+) vs. rrf-1(pk1417) backgrounds; therefore we conclude that UBR-5 acts in the germline to limit GLP-1 signaling and germ cell proliferation.
UBR-5 negatively regulates LIN-12 activity
As a result of LIN-12-mediated lateral signaling between cells in the L2 stage gonad primordium, Z1.ppp and Z4.aaa, one cell becomes the AC and the other cell becomes a VU precursor cell. In the absence of LIN-12 activity, both Z1.ppp and Z4.aaa take on an AC fate (Greenwald et al. 1983; Seydoux and Greenwald 1989). To determine if UBR-5 activity limits signaling via LIN-12/Notch, we evaluated the ability of ubr-5(om2) and ubr-5(q345) to suppress the 2-AC phenotype in lin-12(ar170ts) mutants at 25°. We included a cdh-3::gfp transgene in our strains to aid in identification of ACs (Karp and Greenwald 2003). CDH-3 is a cadherin protein expressed by the AC but not by other nearby cells during late L2/early L3 stage (Pettitt et al. 1996). At 25°, we observed two ACs in ∼64–68% of lin-12(ar170) controls scored at late L2 stage (Table 3). In contrast, we observed two ACs in ∼33% of ubr-5(om2); lin-12(ar170) and ubr-5(q345); lin-12(ar170) larvae grown in parallel with controls (Table 3). This statistically significant reduction in the 2-AC phenotype indicates that ubr-5(om2) and ubr-5(q345) partially suppress the loss of lin-12 function. We conclude that wild-type UBR-5 activity limits signaling via the LIN-12 receptor in the Z1.ppp–Z4.aaa interaction.
Table 3. Loss of ubr-5 function suppresses the lin-12 2-AC defect.
| Strain | % 2 AC | n |
|---|---|---|
| unc-32(e189) lin-12(ar170); cdh-3::gfp | 67.6 | 108 |
| ubr-5(om2); unc-32(e189) lin-12(ar170); cdh-3::gfp | 33.3 | 96 |
| unc-32(e189) lin-12(ar170); cdh-3::gfp | 63.6 | 110 |
| ubr-5(q345); unc-32(e189) lin-12(ar170); cdh-3::gfp | 33.0 | 100 |
Assays were conducted at 25°. Control unc-32(e189) lin-12(ar170); cdh-3::gfp animals were assayed in parallel with each ubr-5(-);unc-32(e189) lin-12(ar170);cdh-3::gfp strain. In each case, the value for “% 2 AC” was significantly different in control vs. experimental strains, P < 0.03 (Z-test). n, number of larvae evaluated; AC, anchor cell.
ubr-5 mutations do not suppress lag-1(ts) lethality
We hypothesize that UBR-5, as an E3 ligase, may limit GLP-1 signaling by promoting turnover of a pathway component, e.g., full-length GLP-1 or processed GLP-1 intracellular domain, or a positive modulator of pathway activity. By this scenario, an elevated level of the partially active GLP-1(q231) protein would accumulate in ubr-5 mutants and GLP-1 signaling activity would increase. To investigate potential UBR-5 targets, we tested whether ubr-5 mutations can suppress a ts mutation in the Notch pathway transcription factor, LAG-1. The lag-1(om13ts) mutation strongly enhances the glp-1(bn18ts) germline proliferation defect at semipermissive temperatures and, in a glp-1(+) background, causes embryonic and early larval lethality (Qiao et al. 1995). Larval lethality occurs at the L1 stage shortly after hatching and is characteristic of the Lag phenotype (Lambie and Kimble 1991). CSL-type transcription factors also function independently of Notch (reviewed by Johnson and MacDonald 2011; Ghai and Gaudet 2008), and we hypothesize that at least some aspects of the lag-1(om13ts) phenotype may be independent of GLP-1 and LIN-12 signaling.
We evaluated the ability of ubr-5(ok1108) to suppress the lag-1(om13ts) embryonic and larval defects at 20°. The lag-1(om13ts) single mutant and ubr-5(ok1108);lag-1(om13ts) double mutant strains had similar viability at 20° (4–5%; Table 4). In addition, brood sizes were similar for the two strains, and ubr-5(ok1108) did not appreciably change the proportion of progeny that died as embryos or as L1 stage larvae (Table 4). We interpret these data to indicate that ubr-5 does not suppress the lag-1(om13ts) embryonic or larval lethality.
Table 4. Loss of ubr-5 function does not suppress lag-1(ts) phenotypic defects.
| Genotype | Avg No. Embryos Produced (± SE) | % Dead Embryos | % Dead Larvae | % Viable Progeny | n |
|---|---|---|---|---|---|
| lag-1(om13ts) | 188 ± 18 | 39.6 | 57.2 | 3.2 | 6 |
| ubr-5(ok1108);lag-1(om13ts) | 225 ± 11 | 48.6 | 47.7 | 3.7 | 10 |
Tests were performed at 20°. Most nonviable larvae died at early L1 stage, as is characteristic of the Lag phenotype (Lambie and Kimble 1991). n, number of full broods counted.
ubr-5 and sel-10 interact synergistically to suppress glp-1 embryonic lethality
SEL-10 is the F-box component of an SCF (Skp1-Cullin-F-box) E3 ubiquitin–protein ligase complex containing SKR-1 as the Skp1 ortholog. SEL-10 and SKR-1 have been shown independently to limit Notch signaling (Sundaram and Greenwald 1993; Hubbard et al. 1997; Killian et al. 2008). Members of the SEL-10 family also negatively regulate Notch signaling in other organisms (reviewed by Lai 2002). SEL-10 directly binds the LIN-12 and GLP-1 intracellular domains and is hypothesized to promote their turnover (Hubbard et al. 1997). Sundaram and Greenwald (1993) reported that sel-10(ar41) very weakly suppressed the glp-1(q231ts) maternal effect lethality at 20°, producing an average of 1.5 viable progeny per hermaphrodite. At 25°, sel-10(ar41) did not suppress the glp-1(q231ts) germline proliferation defect to an appreciable extent. It should be noted that their assay was performed with a glp-1(q231ts);sel(arX) sel-10(ar41) strain, which contained a linked, uncharacterized suppressor, sel(arX), that may have contributed to the suppression phenotype.
We asked whether SEL-10 might be partially redundant with UBR-5 with respect to limiting Notch-type signaling activity. To do so, we evaluated suppression of glp-1(q231ts) by ubr-5 and sel-10 alone and in combination. sel-10(ar41) contains a premature stop codon at residue 323 and is predicted to encode a truncated protein (Hubbard et al. 1997). In our experiments, we used sel-10(ok1632), which contains a deletion/insertion close to the 5′ end of the ORF that is predicted to remove all but the first 18 amino acids of SEL-10 (Killian et al. 2008). As previously reported for sel-10(ar41), we find that sel-10(ok1632) very weakly suppresses the glp-1(q231ts) embryonic lethality at 20°, resulting in <1% viability (Table 5). Interestingly, when we examine the ubr-5(om2);glp-1(q231ts);sel-10(ok1632) triple mutant, we observe substantially higher offspring viability compared to ubr-5(om2);glp-1(q231ts) despite the poor suppression by sel-10 alone (Table 5). Embryonic viability was ∼75% for ubr-5(om2);glp-1(q231ts);sel-10(ok1632) triple mutants as opposed to ∼40% for ubr-5(om2);glp-1(q231ts) and <1% for glp-1(q231ts);sel-10(ok1632) double mutants.
Table 5. Tests for redundancy between UBR-5 and SEL-10.
| Genotype | Avg No. Embryos Produced (± SE) | N | % Viable Progeny | n |
|---|---|---|---|---|
| ubr-5(om2); unc-32(e189) glp-1(q231ts); sel-10(ok1632) | 124 ± 5 | 10 | 74.8 | 1243 |
| ubr-5(om2); unc-32(e189) glp-1(q231ts) | 174 ± 10 | 10 | 40.0 | 1740 |
| unc-32(e189) glp-1(q231ts); sel-10(ok1632)a | 119 ± 15 | 12 | 0.6 | 1428 |
| unc-32(e189) glp-1(q231ts)b | 129 ± 8 | 16 | 0.0 | 2065 |
| ubr-5(om2); sel-10(ok1632)a | 248 ± 6 | 12 | 97.2 | 2972 |
| ubr-5(om2) | 276 ± 17 | 7 | 98.4 | 1901 |
| sel-10(ok1632) original RB1432a,c | 157 ± 17 | 8 | 32.0 | 1441 |
| sel-10(ok1632) reisolated from ubr-5(om2); sel-10(ok1632)c | 225 ± 7 | 5 | 98.6 | 1125 |
Assays were conducted at 20°. N, number of full broods counted; n, number of individuals counted.
The reported broods were produced by animals with a functional vulva. Some animals of these genotypes have a defective vulva and consequently fail to lay eggs and/or die prematurely, in each case producing a limited number of offspring that does not reflect the degree of germline proliferation. Hence, the effective brood size of this strain is smaller than the value listed here.
These data also are listed in Table 1.
The embryonic lethality and reduced brood size of strain RB1432 do not appear to be caused by sel-10(ok1632). See text.
We also observe a very weak suppression of the germline proliferation defect by sel-10(ok1632). When we compare the average mitotic zone size at 24 hr post-L4 stage, we observe an average of six rows of proliferative nuclei in glp-1(q231ts);sel-10(ok1632) compared with four rows in glp-1(q231ts) alone (Table 2). Unlike the case for embryonic viability, analysis of mitotic zone size and brood size suggested that sel-10 and ubr-5 do not interact synergistically to suppress the glp-1(q231ts) germline proliferation defect. Mitotic zone size in the ubr-5(om2);glp-1(q231ts);sel-10(ok1632) triple mutant and the ubr-5(om2);glp-1(q231ts) double mutant are essentially the same (12 ± 0.6 rows vs. 11 ± 0.4 rows of mitotic germ cells) (Table 2). Moreover, ubr-5(om2);glp-1(q231ts);sel-10(ok1632) triple mutants produce approximately the same number of embryos as do glp-1(q231ts);sel-10(ok1632) double mutants and glp-1(q231ts) controls, and substantially fewer embryos than ubr-5(om2);glp-1(q231ts) double mutants (Table 5).
In the course of our work, we observed that the sel-10(ok1632) strain we obtained from the Caenorhabditis Genetics Center, RB1432, has highly penetrant defects not reported for sel-10 in the literature, including substantial embryonic lethality and a reduced brood size of ∼160 (Table 5). When we examined the ubr-5(om2);sel-10(ok1632) double mutant phenotype, we noted that embryonic viability and brood size approximated that of ubr-5(om2) single mutants (Table 5). To determine whether ubr-5(om2) suppressed these defects or, alternatively, they might be caused by mutations in the RB1432 strain background and were eliminated when we constructed the ubr-5(om2);sel-10(ok1632) double, we reisolated the sel-10(ok1632) allele from the ubr-5(om2);sel-10(ok1632) strain and examined the phenotype. We found that the brood size and % viability of the reisolated sel-10(ok1632) single mutant were essentially the same as the ubr-5(om2);sel-10(ok1632) double (Table 5). We conclude that strain RB1432 indeed carries one or more additional mutation(s) distinct from sel-10(ok1632) causing embryonic lethality and reduced brood size.
Discussion
Here, we demonstrate that the sog-1 gene, previously shown to interact genetically with glp-1, encodes UBR-5, the sole C. elegans member of a conserved E3 ubiquitin ligase family. Mammalian UBR5 and the Drosophila ortholog, hyperplastic discs (hyd), function in diverse aspects of development including cell proliferation (reviewed by Shearer et al. 2015; Mansfield et al. 1994). Our data indicate that UBR-5 activity limits GLP-1/Notch signaling in the embryo and larval/adult gonad and LIN-12/Notch signaling in the AC/VU decision. Moreover, UBR-5 acts autonomously to repress germline proliferation, suggesting its primary impact on Notch signaling is within the receiving cell. In contrast, UBR-5 activity does not suppress the brood size or viability defects associated with reduced function of LAG-1, the CSL-type transcription factor component of the GLP-1 signaling pathway in the germline. Given these findings, we hypothesize that UBR-5 functions in the turnover of GLP-1/LIN-12 receptor and/or other proteins responsible for receptor production. Alternatively, UBR-5-mediated ubiquitination may modulate signaling by reducing the ability of GLP-1/LIN-12 to interact with other pathway components, e.g., LAG-1 or LAG-3. Additional experiments will be required to determine if UBR-5 restricts Notch-type signaling in all tissues.
Extensive genetic and molecular analysis has revealed that the outcome of Notch-type signaling is extremely sensitive to the level (“strength”) of signaling activity in a wide variety of contexts (Louvi and Artavanis-Tsakonas 2012; Yamamoto et al. 2014). Our data are consistent with these previous observations. Eliminating just one negative regulator of GLP-1 and LIN-12 activity, UBR-5, increased signaling activity sufficiently to rescue moderately severe loss-of-function phenotypes and strongly enhance a weak gain-of-function phenotype.
The relationship between UBR-5 and SEL-10
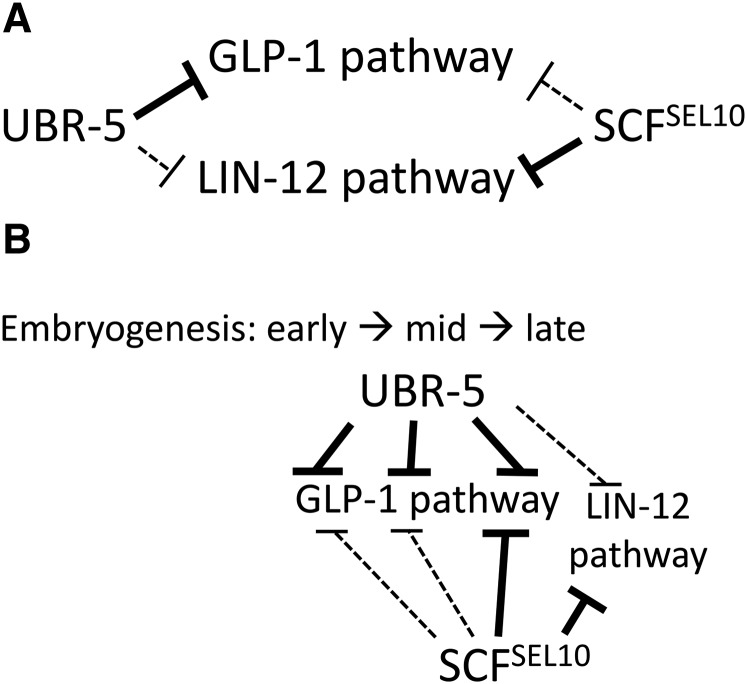
Our data suggest a complex relationship between UBR-5 and SEL-10 with respect to Notch signaling. These findings may reflect, in part, the equivalent receptor activity of GLP-1 and LIN-12 (Fitzgerald et al. 1993) and the fact that both GLP-1 and LIN-12 are active in some tissues, e.g., in the embryo, whereas only GLP-1 or LIN-12 is active in other tissues, e.g., GLP-1 in the gonad. Whereas UBR-5 has a major role in limiting Notch signaling in the embryo and gonad, SEL-10 appears to have a larger role in limiting Notch signaling in the embryo than in the gonad. Our observation of a synergistic genetic interaction between ubr-5 and sel-10 with respect to suppression of the glp-1(q231ts) embryonic lethality indicates that UBR-5 and SEL-10 do not function in a simple linear pathway, at least in embryonic tissues. One model for the relationship between these factors in the embryo is that UBR-5 restricts GLP-1 activity broadly during embryogenesis, whereas SEL-10 restricts GLP-1 primarily in the late embryo – where it also restricts LIN-12 (Figure 4B). Only rarely would development of the glp-1(q231ts) embryo be rescued by sel-10(ok1632) alone. An alternative hypothesis is that SEL-10 primarily restricts LIN-12 signaling activity and has little impact on GLP-1 signaling activity. This would explain the very minor role of SEL-10 in the gonad, where LIN-12 signaling is not active. One way to think about these data is that UBR-5 may primarily limit GLP-1 signaling, SEL-10 may primarily limit LIN-12 signaling, and reduced UBR-5 or SEL-10 activity may better suppress reduced glp-1 or lin-12 activity in those cells or tissues where increased activity of one receptor can compensate for reduced activity of the other, e.g., in the embryo (Figure 4A).
Figure 4.
Hypothetical relationships between UBR-5 and SEL-10 activity in the embryo. (A) UBR-5 and SEL-10 may each primarily limit signaling activity via one Notch-type receptor and only play a minor role in limiting signaling via the other receptor. (B) UBR-5 and SEL-10 may limit signaling via both GLP-1 and LIN-12, with UBR-5 having a primary role in the early embryo and both factors acting in the late embryo.
UBR5 activity in development and disease
UBR5 proteins in mammals and Drosophila have been implicated in numerous developmental processes (reviewed by Shearer et al. 2015). Vertebrate UBR5 regulates cell cycle progression, and misregulation of UBR5 activity is linked to cancer in many tissues (Scheffner and Kumar 2014; Shearer et al. 2015). UBR5 activity appears to promote cell proliferation in some tissues and limit it in others, as the loss of UBR5 activity promotes cancer in some tissues whereas UBR5 overexpression promotes cancer in other tissues. Presumably these differences reflect the multitude of UBR5 targets that may contribute to oncogenesis. The Drosophila ortholog, Hyperplastic discs (Hyd), regulates cell proliferation in developing imaginal discs and promotes development of other tissues, as well (Mansfield et al. 1994).
UBR5 appears to contribute to cell proliferation control in a number of ways. One role for UBR5 is in modulating activity of the mitotic spindle assembly checkpoint (SAC) mechanism, thereby impacting the ability of cells to enter anaphase. SAC activity ensures cells remain in metaphase until all chromosomes have attached to the mitotic spindle; once chromosomes have done so, then SAC activity must be reduced in order to allow anaphase entry. Evidence suggests that UBR5 functions both to promote SAC activity when needed, e.g., if microtubules are disrupted (Scialpi et al. 2015), and to reduce SAC activity once chromosomes have attached to the mitotic spindle (Jiang et al. 2015). UBR5 targets different SAC components in these different situations, and its subcellular localization changes during the cell cycle (Scialpi et al. 2015; Jiang et al. 2015).
Another function of UBR5 is in modulating stability of nuclear myosin 1 (NM1), a factor required for RNA polymerase I transcriptional activity. NM1 is stabilized by GSK3β-mediated phosphorylation, which prevents UBR5-mediated ubiquitination and subsequent degradation (Sarshad et al. 2014). Progression beyond G1 stage requires NM1 activity, and hence modulation of NM1 is one means by which UBR5 limits cell cycle progression. Antagonistic activity of GSK3β vs. Hyd/UBR5 appears to be a general regulatory mechanism as it also modulates Hedgehog signaling in Drosophila (Lee et al. 2002; Moncrieff et al. 2015). As we observed for Notch signaling, Hedgehog signaling is limited by Hyd/UBR5 activity. Interestingly, mammalian FBW7/SEL10 is recruited to certain targets upon GSK3β−mediated phosphorylation (Flugel et al. 2012).
Other studies have implicated UBR5 in modulating the DNA damage response (DDR), where it participates at several steps, and in regulation of certain transcription factors (Shearer et al. 2015). One aspect of UBR5 activity is that it limits activity of the DDR machinery to sites of double-strand breaks and prevents inappropriate/unnecessary activity elsewhere on the chromosome. In regulating transcription factor activity, UBR5 functions in many cell types, positively regulating certain transcription factors and negatively regulating others. We present the first evidence, to our knowledge, of a UBR5 family E3 ligase modulating Notch signaling activity. In the future, it will be informative to identify the targets of UBR-5 activity.
Supplementary Material
Acknowledgments
We thank Jim Lissemore and Mark Edgley for invaluable assistance in the WGS analysis, Yiqing Guo, Bing Yang, and Yini Li for technical advice, and two reviewers for insightful comments on the manuscript. This study was supported by funds from Syracuse University, including the Coronat Scholars Program and the Korczynski-Lundgren Fund. Work in the laboratory of D.G.M. was supported by a grant from the Canadian Institute for Health Research. D.G.M. is a Senior Fellow of the Canadian Institute for Advanced Research. Some strains used in this study were obtained from the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.027805/-/DC1
Communicating editor: S. Lee
Literature Cited
- Baron M., 2012. Endocytic routes to Notch activation. Semin. Cell Dev. Biol. 23: 437–442. [DOI] [PubMed] [Google Scholar]
- Barth J. M., Kohler K., 2014. How to take autophagy and endocytosis up a notch. BioMed Res. Int. 10.1155/2014/960803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry L. W., Westlund B., Schedl T., 1997. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124: 925–936. [DOI] [PubMed] [Google Scholar]
- Callaghan M. J., Russell A. J., Woollatt E., Sutherland G. R., Sutherland R. L., et al. , 1998. Identification of a human HECT family protein with homology to the Drosophila tumour suppressor gene hyperplastic discs. Oncogene 17: 3479–3491. [DOI] [PubMed] [Google Scholar]
- Epstein, H. F., and D. C. Shakes, 1995 Caenorhabditis elegans: Biological Analysis of an Organism, in Methods in Cell Biology, Vol. 48. Academic Press, San Diego. [Google Scholar]
- Fitzgerald K., Wilkinson H. A., Greenwald I., 1993. glp-1 can substitute for lin-12 in specifying cell fate decisions in Caenorhabditis elegans. Development 119: 1019–1027. [DOI] [PubMed] [Google Scholar]
- Flibotte S., Edgley M. L., Chaudhry I., Taylor J., Neil S. E., et al. , 2010. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics 185: 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flugel D., Gorlach A., Kietzmann T., 2012. GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIG-1α. Blood 119: 1292–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai V., Gaudet J., 2008. The CSL transcription factor LAG-1 directly represses hlh-1 expression in C. elegans. Dev. Biol. 322: 334–344. [DOI] [PubMed] [Google Scholar]
- Greenwald, I., and R. Kovall, 2013 Notch signaling: genetics and structure (January 17, 2013), WormBook, ed. The C. elegans Research Community Wormbook, /10.1895/wormbook.1.10.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Greenwald I. S., Sternberg P. W., Horvitz H. R., 1983. The lin-12 locus specific cell fates in Caenorhabditis elegans. Cell 34: 435–444. [DOI] [PubMed] [Google Scholar]
- Hansen D., Schedl T., 2013. Stem cell proliferation versus meiotic fate decision in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., 1998. The ubiquitin system. Annu. Rev. Biochem. 67: 425–479. [DOI] [PubMed] [Google Scholar]
- Hubbard E. J., Wu G., Kitajewski J., Greenwald I., 1997. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11: 3182–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E. J., Korta D. Z., Dalfo D., 2013. Physiological control of germline development. Adv. Exp. Med. Biol. 757: 101–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Sherwood D. R., Aspöck G., Butler J. A., Gupta B. P., et al. , 2002. Gene expression marks for Caenorhabditis elegans vulval cells. Mech. Dev. Suppl. 1: S203–S209. [DOI] [PubMed] [Google Scholar]
- Jiang H., He X., Feng D., Zhu X., Zheng Y., 2015. RanGTP aids anaphase entry through UBR5-mediated protein turnover. J. Cell Biol. 211: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. E., MacDonald R. J., 2011. Notch-independent functions of CSL. Curr. Top. Dev. Biol. 97: 55–74. [DOI] [PubMed] [Google Scholar]
- Kandachar V., Roegiers F., 2012. Endocytosis and control of Notch signaling. Curr. Opin. Cell Biol. 24: 534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., Greenwald I., 2003. Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 17: 3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian D. J., Hubbard E. J., 2005. Caenorhabditis elegans germline patterning requires coordinated development of the somatic gonadal sheath and the germ line. Dev. Biol. 279: 322–335. [DOI] [PubMed] [Google Scholar]
- Killian D. J., Harvey E., Johnson P., Otori M., Mitani S., et al. , 2008. SKR-1, a homolog of Skp1 and a member of the SCFSEL-10 complex, regulates sex-determination and LIN-12/Notch signaling in C. elegans. Dev. Biol. 322: 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble, J., and H. Seidel, 2013 C. elegans germline stem cells and their niche (November 15, 2013), StemBook, ed. The Stem Cell Research Community StemBook, /10.3824/stembook.1.95.1, http://www.stembook.org.
- Kipreos, E. T., 2005 Ubiquitin-mediated pathways in C. elegans (December 1, 2005), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.36.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Koch U., Lehal R., Radtke F., 2013. Stem cells living with a Notch. Development 140: 689–704. [DOI] [PubMed] [Google Scholar]
- Kumsta C., Hansen M., 2012. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One 7: e35428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E. C., 2002. Protein degradation: four E3s for the Notch pathway. Curr. Biol. 12: R74–R78. [DOI] [PubMed] [Google Scholar]
- Lambie E., Kimble J., 1991. Two homologous regulator genes, lin-12 and glp-1, have overlapping functions. Development 112: 231–240. [DOI] [PubMed] [Google Scholar]
- Le Bras S., Loyer N., Le Borgne R., 2011. The multiple facets of ubiquitination in the regulation of notch signaling pathway. Traffic 12: 149–161. [DOI] [PubMed] [Google Scholar]
- Lee J. D., Amanai K., Shearn A., Treisman J. E., 2002. The ubiquitin ligase Hyperplastic discs negatively regulates hedgehog and decapentaplegic expression by independent mechanisms. Development 129: 5697–5706. [DOI] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S., 2012. Notch and disease: a growing field. Semin. Cell Dev. Biol. 23: 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine E. M., Kimble J., 1989. Identification of genes that interact with glp-1, a gene required for inductive cell interactions in Caenorhabditis elegans. Development 105: 133–143. [DOI] [PubMed] [Google Scholar]
- Maine E. M., Kimble J., 1993. Suppressors of glp-1, a gene required for cell communication during development in Caenorhabditis elegans, define a set of interacting genes. Genetics 135: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield E., Hersperger E., Biggs J., Shearn A., 1994. Genetic and molecular analysis of hyperplastic discs, a gene whose product is required for regulation of cell proliferation in Drosophila melanogaster imaginal discs and germ cells. Dev. Biol. 165: 507–526. [DOI] [PubMed] [Google Scholar]
- Moncrieff S., Moncan M., Scialpi F., Ditzel M., 2015. Regulation of hedgehog ligand expression by the N-end rule ubiquitin protein ligase Hyperplastic Discs and the Drosophila GSK3β homologue, Shaggy. PLoS One 10: e0136760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P., Lim J. S., Sage J., Aifantis I., 2014. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 25: 318–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penton A. L., Leonard L. D., Spinner N. B., 2012. Notch signaling in human development and disease. Semin. Cell Dev. Biol. 23: 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A. S., Killian D. J., Hubbard E. J., 2003. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163: 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt J., Wood W. B., Plasterk R. H., 1996. cdh-3, a gene encoding a member of the cadherin superfamily, functions in epithelial cell morphogenesis in Caenorhabditis elegans. Development 122: 4149–4157. [DOI] [PubMed] [Google Scholar]
- Priess, J. R., 2005 Notch signaling in the C. elegans embryo (June 25, 2005), WormBook, ed. The C. elegans Research Community Wormbook, /10.1895/wormbook.1.4.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Qiao L., Lissemore J. L., Shu P., Smardon A., Gelber M. B., et al. , 1995. Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics 141: 551–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. A., 2014. Interplay between nuclear transport and ubiquitin/SUMO modifications in the regulation of cancer-related proteins. Semin. Cancer Biol. 27: 11–19. [DOI] [PubMed] [Google Scholar]
- Sarshad A. A., Corcoran M., Al-Muzzaini B., Borgonovo-Brandter L., Von Euler A., et al. , 2014. Glycogen synthase kinase (GSK) 3β phosphorylates and protects nuclear myosin 1c from proteasome-mediated degradation to activate rDNA transcription in early G1 cells. PLoS Genet. 10: e1004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M., Kumar S., 2014. Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim. Biophys. Acta. 843: 61–74. [DOI] [PubMed] [Google Scholar]
- Scialpi F., Mellis D., Ditzel M., 2015. EDD, a ubiquitin-protein ligase of the N-end rule pathway, associates with spindle assembly checkpoint components and regulates the mitotic response to nocodazole. J. Biol. Chem. 290: 12585–12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G., Greenwald I., 1989. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell 57: 1237–1245. [DOI] [PubMed] [Google Scholar]
- Shearer R. F., Iconomou M., Watts C. K., Saunders D. N., 2015. Functional roles of the E3 ubiquitin ligase UBR5 in cancer. Mol. Cancer Res. 13: 1523−1532. [DOI] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., et al. , 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476. [DOI] [PubMed] [Google Scholar]
- Starich T. A., Hall D. H., Greenstein D., 2014. Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis elegans. Genetics 198: 1127–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, P., 2005 Vulval development (June 25, 2005), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.6.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Sundaram M., Greenwald I., 1993. Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics 135: 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh S., Irvine A. E., 2015. The NOTCH signaling pathway in normal and malignant blood cell production. J. Cell Commun. Signal. 9: 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T., Mulder L. C., Iwamatsu A., Lee M. J., Davydov I. V., et al. , 2005. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 25: 7120–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson O., Edgley M., Strasbourger P., Flibotte S., Ewing B., et al. , 2013. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23: 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A., 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- Wang Z., Inuzuka H., Fukushima H., Wan L., Gao D., et al. , 2011. Emerging roles of the FBW7 tumour suppressor in stem cell differentiation. EMBO Rep. 13: 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmaster G., Fischer J. A., 2011. Notch ligand ubiquitylation: what is it good for? Dev. Cell 21: 124–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S., K. L. Schulze, and H. J. Bellen, 2014 Introduction to Notch signaling, pp. 1–14 in Notch Signaling: Methods and Protocols, in Methods in Molecular Biology, Vol. 1187, edited by H. Belen and S. Yamamoto. Springer Science + Business Media, New York. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. Whole genome sequence data are provided in Supplemental Material, Table S1.