Abstract
Genome editing exploiting CRISPR/Cas9 has been adopted widely in academia and in the biotechnology industry to manipulate DNA sequences in diverse organisms. Molecular engineering of Cas9 itself and its guide RNA, and the strategies for using them, have increased efficiency, optimized specificity, reduced inappropriate off-target effects, and introduced modifications for performing other functions (transcriptional regulation, high-resolution imaging, protein recruitment, and high-throughput screening). Moreover, Cas9 has the ability to multiplex, i.e., to act at different genomic targets within the same nucleus. Currently, however, introducing concurrent changes at multiple loci involves: (i) identification of appropriate genomic sites, especially the availability of suitable PAM sequences; (ii) the design, construction, and expression of multiple sgRNA directed against those sites; (iii) potential difficulties in altering essential genes; and (iv) lingering concerns about “off-target” effects. We have devised a new approach that circumvents these drawbacks, as we demonstrate here using the yeast Saccharomyces cerevisiae. First, any gene(s) of interest are flanked upstream and downstream with a single unique target sequence that does not normally exist in the genome. Thereafter, expression of one sgRNA and cotransformation with appropriate PCR fragments permits concomitant Cas9-mediated alteration of multiple genes (both essential and nonessential). The system we developed also allows for maintenance of the integrated, inducible Cas9-expression cassette or its simultaneous scarless excision. Our scheme—dubbed mCAL for “Multiplexing of Cas9 at Artificial Loci”—can be applied to any organism in which the CRISPR/Cas9 methodology is currently being utilized. In principle, it can be applied to install synthetic sequences into the genome, to generate genomic libraries, and to program strains or cell lines so that they can be conveniently (and repeatedly) manipulated at multiple loci with extremely high efficiency.
Keywords: Saccharomyces cerevisiae, CRISPR, essential genes, genome editing, genome engineering
Discovery of CRISPR (Clustered Regularly-Interspaced Short Palindromic Repeats)-based RNA-mediated adaptive immunity in bacteria and archaea (Sorek et al. 2013; Shmakov et al. 2015), and especially the RNA-guided DNA endonuclease Cas9 from the Class II CRISPR system of Streptococcus pyogenes (Jinek et al. 2012; Doudna and Charpentier 2014), has provided a remarkably versatile tool for modifying genomes (Hsu et al. 2014; Wang et al. 2016). Combining the normally separate DNA sequence-binding crRNA with the Cas9-stabilizing tracrRNA into a “single-guide” or “synthetic-guide” (sgRNA) streamlined target site recognition (Jinek et al. 2012; Ran et al. 2013). Changes to the stem-loop architecture of the tracrRNA portion of a sgRNA greatly strengthen its affinity for Cas9 (Chen et al. 2013), and shortening of the crRNA portion of a sgRNA to just 20 nucleotides reduces off-target action while preserving efficiency (Pattanayak et al. 2013). The range of DNA/chromosome-based applications has been further extended by engineering of S. pyogenes Cas9 [or use of Cas9 orthologs from other bacterial species (Jinek et al. 2014)] to relax its requirement for initiating DNA sequence recognition at a so-called PAM (“protospacer adjacent motif”) site (5′-NGG-3′) (Kleinstiver et al. 2015), to inactivate one or both of its two (McrA/HNH-like and RuvC/RNAaseH-like) catalytic sites to create a “nickase” (Fu et al. 2014) or a catalytically “dead” (dCas9) version (Gilbert et al. 2013), or to insert new functionalities (Oakes et al. 2014). Cas9 and associated sgRNAs have been used in diverse organisms for genome editing, both gene knock-outs (Gaj et al. 2013) and gene fusions (Wei et al. 2013), as well as to force biased inheritance of a desired allele within entire populations (“gene drives”) (DiCarlo et al. 2015; Dong et al. 2015; Gantz et al. 2015). Cas9-mediated genome alterations have been achieved in bacterial species (Jiang et al. 2013; Tsarmpopoulos et al. 2016), various fungi (DiCarlo et al. 2013; Wagner and Alper 2015), zebrafish (Hwang et al. 2013), Caenorhabditis elegans (Friedland et al. 2013), Drosophila melanogaster (Gratz et al. 2013), plants (Mao et al. 2013), and human cells (Cho et al. 2013; Jinek et al. 2013; Mali et al. 2013; Ran et al. 2013), including clinical trials to explore Cas9-mediated therapy in infectious and inherited disease (Kaminski et al. 2016; Mendell and Rodino-Klapac 2016; Su et al. 2016). Additional applications include sequence-specific repression or activation of gene expression (Cheng et al. 2013; Gilbert et al. 2013; La Russa and Qi 2015), fluorescent labeling of chromosomal loci (Chen et al. 2013, 2016), and RNA-scaffolded recruitment of proteins to a programmed chromosomal localization (Zalatan et al. 2015).
For genome editing, the Cas9-sgRNA enzyme allows precise placement of a double-strand break (DSB) at any desired location(s) within a genome of interest. The DSB can be sealed in a highly error-prone manner via nonhomologous end-joining (NHEJ) (Richardson et al. 2016; Vriend et al. 2016) or, more usefully, by homologous recombination (HR) (typically with PCR products provided in trans) to substitute a modification (deletion, insertion, allele replacement, fusion to a reporter sequence, etc.) (Shalem et al. 2015; Chandrasegaran and Carroll 2016; Hu et al. 2016). Although accuracy and efficiency are generally high, an sgRNA-guided Cas9 can act at other sites in addition to the intended sequence (Cho et al. 2014; O’Geen et al. 2015; Zhang et al. 2015). To reduce such off-target action, specificity-enhancing alterations of Cas9 (Kleinstiver et al. 2016; Slaymaker et al. 2016) and sgRNA design (Dang et al. 2015; Xu et al. 2015; Doench et al. 2016), and computational methods to search for optimal sgRNA-recognition sites (Bolukbasi et al. 2015; Naito et al. 2015) have been devised. By the same token, when provided with different sgRNAs concomitantly, Cas9 can effect simultaneous alterations at multiple locations within the genome in any given cell (“multiplex” genome engineering) (Cong et al. 2013), and this strategy has been successfully applied in Saccharomyces cerevisiae, but almost exclusively to nonessential genes (Ryan and Cate 2014; Bao et al. 2015; Horwitz et al. 2015; Jakociunas et al. 2015; Laughery et al. 2015; Mans et al. 2015; Ronda et al. 2015; Tsai et al. 2015). Here, we describe a useful alternative strategy—introduction of unique, programmable, artificial target sequences into the genome, thereby permitting multiplex gene manipulation by Cas9 with a single sgRNA.
Materials and Methods
Yeast strains and plasmids
All budding yeast strains used in this study can be found in Supplemental Material, Table S1. Standard molecular biology methods were used in this study (Sambrook and Russell 2001). The introduction of the u1 and u2 Cas9 target sites was performed by first cloning vectors using in vivo ligation and homologous recombination harboring a single Cas9 site including the PAM sequence (Finnigan and Thorner 2015). As an example, a vector (pGF-V130) containing the 5′ UTR of CDC11 was digested with a restriction enzyme (NotI) downstream of the promoter sequence, and transformed with a PCR fragment of the CDC11 coding region amplified with oligonucleotides containing overhanging “tails” to insert the Cas9 u1 target sequence in-frame. Two constructs, each with a single flanking u1 site placed upstream or downstream of CDC11, were created separately and then combined by a second round of in vivo ligation to generate the final construct that contained both flanking u1 sites as well as flanking CDC11 5′ and 3′ UTR (330 bp of each). This process was repeated for the shs1∆::HygR cassette harboring flanking u1 sites and two Cas9-expressing cassettes containing either u1 or u2 sites at the HIS3 locus (Table S1). The generated constructs were PCR amplified and integrated into the parent strain in successive yeast transformations. Diagnostic PCRs and Sanger sequencing (Univ. of California, Berkeley Barker Hall Sequencing Facility) of chromosomal DNA were performed to ensure proper integration of all manipulated loci.
Plasmids used in this study can be found in Table S2. Expression of the sgRNA cassettes was modeled after a previous study (DiCarlo et al. 2013), using the snoRNA SNR52 promoter and SUP4 terminator sequences, and they were synthesized as custom genes with flanking XhoI and BamHI restriction sites (GenScript, Piscataway, NJ). The u1 and u2 sequences were chosen from two human genes, SEPT9 and MMP23A, respectively, using the DNA2.0 gRNA Design Tool (DNA2.0, Newark, CA). Putative guide sequences were then examined against the entire yeast genome using a nucleotide BLAST search (National Center for Biotechnology Information) and sequences were considered for having the lowest possible number of matches to the 15 bp sequence (PAM + upstream 12 bp) important for Cas9 “seeding” to minimize off-target effects (Jinek et al. 2012; Jiang et al. 2013). Additionally, the chosen u1 and u2 sequences were checked against the backbone vector sequences of the pRS316 covering vector, the high-copy sgRNA-expressing pRS425/pRS423 vectors, and both the KanR and HygR cassettes (Goldstein and McCusker 1999), to ensure no highly similar matches existed in these exogenous non-yeast sequences.
Culture conditions
Yeast were grown in rich YPD or YPGal medium (2% peptone, 1% yeast extract, and 2% dextrose or 2% galactose), or in synthetic medium containing the necessary amino acids with either 2% dextrose or a 2% raffinose and 0.2% sucrose mixture. For transformation of yeast using the Cas9-mediated system, strains were grown overnight in synthetic medium with a raffinose/sucrose mixture lacking uracil (to select for the CDC11-expressing WT-covering plasmid) to saturation, back-diluted into YPGal (to an OD600 of approximately 0.25–0.35), and grown at 30° for 4.5–5.0 hr. A modified lithium acetate transformation protocol (Eckert-Boulet et al. 2012) was used to transform 10 OD600 of yeast with combinations of purified plasmid DNA and/or PCR products. Yeast were heat shocked for 45–50 min at 42° and recovered in fresh YPGal overnight at 30° prior to plating onto selective media (selection for both plasmids and no selection for integrated knock-in alleles). An identical transformation protocol was used whether Cas9 was integrated at the HIS3 locus or expressed on a CEN-plasmid.
The growth of single yeast colonies on various media (G418, Hygromycin, SD-HIS, etc.) was tested by first selecting isolated colonies, creating a small square “patch” (1 cm2) on an SD-URA plate, incubating overnight at 30°, and then replica-plating to additional plates to be scored after 1 d of additional incubation. For yeast plates containing a significant number of colonies, the total colony count was estimated in several ways. First, several sectors (half, a quarter, or an eighth, etc.) were selected on the agar plate and the total number of colonies in the sample sector was counted and extrapolated to the entire surface area. Second, subsequent repeated experiments plated various dilutions (1/10, 1/20, 1/50, etc.) of the final transformation product and the total colony counts were added, extrapolated, and averaged together. All experiments were performed in at least triplicate.
Fluorescence microscopy
For fluorescence microscopy, yeast were grown to saturation overnight in S+Raff/Suc-LEU, back-diluted into YPGal, grown for 5 hr at 30°, harvested, washed with water, and prepared on a standard microscope slide with a coverslip. Samples were immediately imaged on an Olympus BH-2 upright fluorescence microscope (Olympus, Tokyo, Japan) with a 100 × objective lens. A CoolSNAP MYO CCD camera (Photometrics, Tuscon, AZ), a SOLA light source (Lumencore, Beaverton, OR), Micro-Manager software (Edelstein et al. 2010), and ImageJ software (National Institutes of Health) were used to process fluorescent images. The cell periphery was determined using an overexposed fluorescence image.
Polymerase chain reaction and DNA sequencing
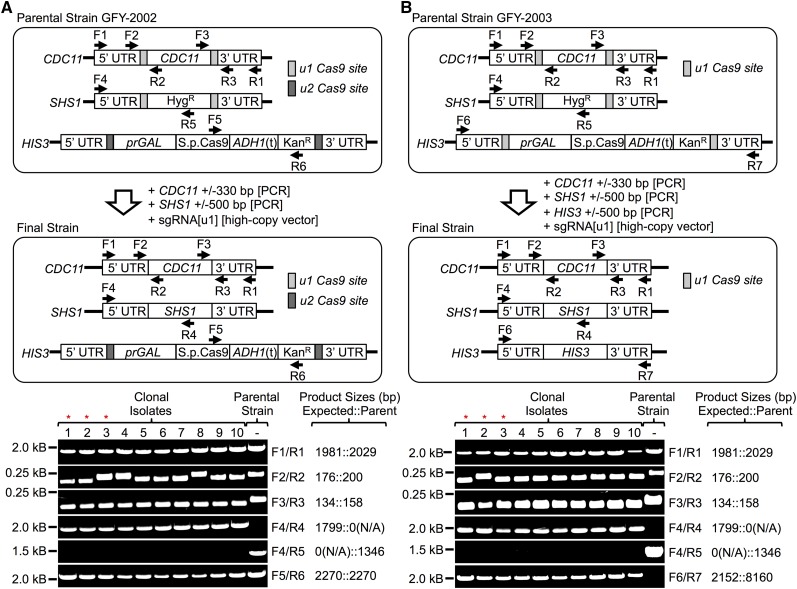
All PCR reactions were performed using either high fidelity KOD Hot Start DNA Polymerase (EMD Millipore, Billerica, MA) or PfuUltra II Fusion Hot Start DNA Polymerase (Agilent Technologies, Santa Clara, CA) according to the recommended manufacturer’s conditions (KOD reactions all contained 3 mM Mg2+) on a PTC-200 Thermal Cycler (MJ Research, Bio-Rad). Oligonucleotides (Integrated DNA Technologies, Coralville, IA) used in this study can be found in Table S5. For PCR reactions used in the Cas9-mediated integrations, the template DNA was from either purified yeast chromosomal DNA or from bacterial-based plasmids (that cannot be propagated in yeast). Products for integration were confirmed to be the correct size on an agarose gel, but were not purified nor gel extracted; amplified DNA was directly added to the yeast transformation reaction. For diagnostic PCRs to confirm various manipulated loci, DNA agarose gels (1% or 2%) containing Ethidium Bromide were used to separate and image (ChemiDoc System, Bio-Rad Laboratories, Hercules, CA) separated products. Sanger DNA sequencing was performed on all constructed vectors and plasmid intermediates. For sequencing of genomic loci, chromosomal DNA was isolated (Amberg et al. 2006) and PCR amplified using a high-fidelity polymerase. The product sizes were confirmed on an agarose gel and the remaining DNA was purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) and sequenced with overlapping coverage at each desired locus. For diagnostic PCRs, chromosomal DNA was first isolated from yeast strains, as follows. Two precautions were taken to avoid isolating DNA that contained the URA3-based covering vector expressing WT CDC11. First, cells were grown to saturation under nonselective conditions in rich (YPD) medium overnight (16 hr) at 30°. Second, DNA was isolated using a procedure that recovers only chromosomal DNA (Amberg et al. 2006). Indeed, control F2/R2 PCR reactions carried out on DNA isolated using these approaches from the plasmid-containing parental strains GFY-2002 and GFY-2003 demonstrated that the preparations obtained generated PCR products diagnostic for the chromosomal CDC11 locus, and did not generate any PCR products diagnostic for the plasmid-borne DNA (see Figure 3, Figure S3, and Figure S5).
Figure 3.
Diagnostic PCR confirms efficient multiplex gene replacement. (A) Chromosomal DNA was purified (Amberg et al. 2006) from 10, randomly chosen, clonal isolates from transformations of GFY-2002 in which PCR fragments with 500 bp of flanking genomic homology were provided to restore WT CDC11 and WT SHS1 loci, and which had lost the HygR marker (see Figure 2B), and tested by PCR with the indicated diagnostic primer sets. An identical analysis was performed on 10 isolates in which PCR fragments with only 30 bp of flanking genomic homology were provided and which had lost the HygR marker (see Figure S3). The PCR products were resolved by agarose gel electrophoresis and visualized by staining with ethidium bromide. For CDC11 (top three gels), the entire locus was amplified (primers F1/R1), as well as small fragments flanking the upstream (F2/R2) or downstream (F3/R3) u1 sites to determine whether or not the Cas9 target site was still present. For SHS1 (fourth and fifth gels), PCR was performed using primers unique to either SHS1 itself (F4/R4) or to the HygR cassette (F4/R5). Finally, the HIS3 locus (bottom gel) was testing using a unique primer internal to the Cas9 gene and to the KanR cassette (F5/R6). For optimal separation, 2% agarose was used for the second and third gels, 1% agarose was used for all of the others. Left, nearest DNA size marker (in kb) for each independent gel; right, expected PCR product sizes. (B) The same kind of analysis as in (A) was performed on chromosomal DNA purified from 10, randomly chosen, clonal isolates from transformations of GFY-2003, except that, in addition, PCR diagnostic for the HIS3 locus was performed (F6/R7) to ascertain whether the u1-flanked his3∆::Cas9::KanR cassette had been replaced by the WT HIS3 gene. Left red asterisks, three representative isolates of GFY-2002 that diagnostic PCR indicated carried WT CDC11 and WT SHS1 loci, and retained the u2-flanked his3∆::Cas9::KanR cassette, were confirmed as such by direct DNA sequencing (data not shown). Right red asterisks, three representative isolates of GFY-2003 that diagnostic PCR indicated carried WT CDC11, WT SHS1, and WT HIS3 loci, were also confirmed as such by direct DNA sequencing (data not shown) [for diagnostic PCR and sequencing of surviving colonies from controls (Figure 2B, conditions B–D), see Figure S5]. HygR, hygromycin resistance; KanR, kanamycin resistance; ORF, open reading frame; PCR, polymerase chain reaction; sgRNA, single-guide RNA; UTR, untranslated region; WT, wild-type.
Supplemental Material
All supplemental material (Figures S1–S5, Tables S1–S5, and Supplemental References) can be found in File S1.
Data and reagent availability
We will freely send all plasmids and strains and other research materials and procedures generated from this research to investigators at any and all nonprofit institutions for research purposes upon request.
Results
A new strategy for multiplex Cas9-mediated gene editing
When bound to an appropriate sgRNA, Cas9 is able to recognize repeated sequences within a genome, such as telomeres (Chen et al. 2013) or the long terminal repeats (LTRs) (δ elements) of the yeast Ty1 retrotransposon (Shi et al. 2016). Given that fact, and that current limitations on genome editing by Cas9 include the necessity for an adjacent PAM sequence, the individuality of the desired target sequence itself (to avoid off-target effects), and unknown influences of local chromatin structure, we considered useful ways to circumvent these limitations.
In brief, we first integrate, both upstream and downstream of any locus of interest, a unique 23-nucleotide sequence (a 20 bp target sequence plus a PAM) that has no detectable counterpart in the genome of interest. Second, we integrate, at a safe harbor locus, a cassette that expresses from an inducible Pol II promoter S. pyogenes Cas9 bearing a potent universal nuclear localization signal (NLS), which is also flanked by the same or a different unique 23-nucleotide sequence. Third, introduction by DNA-mediated transformation of a plasmid that expresses, from a Pol III promoter, a single sgRNA that matches the unique 23 bp target, along with PCR fragments to replace the excised loci by HR, completes the system.
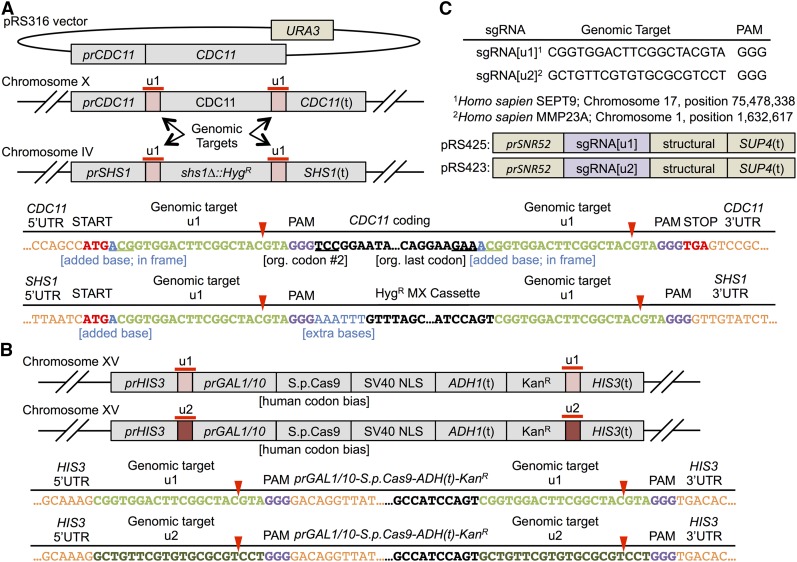
As proof of principle, we chose genes encoding two members of the family of mitotically-expressed septins, CDC11 and SHS1, to illustrate the utility of our method for exploiting the features of Cas9-mediated gene manipulation. CDC11 is an essential gene, whereas cells lacking SHS1, although not normal, are viable (Hartwell 1971; Iwase et al. 2007; Garcia et al. 2011; McMurray et al. 2011; Finnigan et al. 2015). At the genomic loci for both CDC11 (Chromosome X) and an shs1∆::HygR allele (Chromosome IV), we used standard techniques to insert (see Materials and Methods), both upstream and downstream of these two ORFs, a human, 23 bp (or 24 bps if necessary to maintain the reading frame) PAM-containing sequence (designated “u1”), which does not match any other site in the S. cerevisiae genome by more than a few nucleotides (Figure 1A). To flank the genes of interest with the u1 (or u2) sequence, two successive rounds of in vivo homologous recombination-mediated plasmid assembly in yeast (Finnigan and Thorner 2015) were used to separately introduce these target sites at each end of the desired genes. The resulting constructs were then PCR-amplified and used to transplace the endogenous chromosomal locus of interest by integrative recombination, as described in Materials and Methods. Of course, alternative methods could be used to insert the same (or other) Cas9 target sequences upstream and downstream of a gene of interest, including in vitro Gibson cloning (Gibson 2011), inverse PCR with extended oligonucleotide tails (Hartl and Ochman 1996), or artificial gene synthesis (Stemmer et al. 1995). Moreover, “traditional” Cas9-introduced double-strand breaks (Jinek et al. 2012) and appropriate PCR products for their repair could be used to introduce unique sites at desired locations in the genome. The directed placement of the u1 and/or u2 sites can be at any position flanking or within a gene (its UTR sequences, coding sequence, or introns). In our test cases, we inserted the u1 motifs as part of the coding sequence of one gene of interest (CDC11) and flanking a HygR-marked deletion of another (SHS1). The former resulted in an eight-residue insertion at the N-terminal end and an eight-residue insertion at the C-terminal end of the Cdc11 polypeptide. Complementation tests revealed that, at least for Cdc11, such small N- and C-terminal extensions are tolerated in vivo (data not shown).
Figure 1.
Installation of programmed non-yeast Cas9 target sites at multiple loci. (A) Haploid yeast strains were constructed in which the endogenous CDC11 gene and a shs1∆::HygR allele were flanked by an identical 23 bp sequence containing a Cas9 target site (including a 5′-NGG-3′ PAM sequence) from the human SEPT9 gene, designated “unique Cas9 site 1,” u1. At CDC11, the upstream u1 site was placed in-frame with the initiator Met of the ORF, and the downstream u1 was kept in-frame with the stop codon (via addition of an A to the 5′-end of each u1). Because CDC11 is an essential gene, a URA3-marked CEN plasmid expressing WT CDC11 (but with no 3′-UTR) was also present. Red triangles, site of Cas9-directed DSB (+ 3 upstream of the PAM). (B) A cassette for inducible GAL1/10 promoter-driven expression of S.p.Cas9 bearing a C-terminal SV40 NLS and a ADH1 transcriptional terminator was used to replace the ORF at the endogenous HIS3 locus. In one variant (strain GFY-2002), this cassette was flanked by u2, a different 23 bp human sequence containing a Cas9 target from the human MMP23A locus. In another variant (strain GFY-2003), the cassette was flanked by u1. (C) The corresponding sgRNA[u1] and sgRNA[u2] sequences were expressed using the constitutive yeast pol III snoRNA SNR52 promoter and yeast pol III tRNA SUP4 terminator on high-copy (2 μm DNA) plasmids. DSB, double-strand break; HygR, hygromycin resistance; KanR, kanamycin resistance; NLS, nuclear localization signal; ORF, open reading frame; PAM, protospacer adjacent motif; sgRNA, single-guide RNA; tRNA, transfer RNA; UTR, untranslated region; WT, wild-type.
Such short “foreign” sequences are far below the length necessary for spontaneous loop-out from a yeast chromosome by HR, as observed, for example, with introduced Salmonella hisG repeats (1100 bp) (Alani et al. 1987) or the LTRs of retrotransposons (323–424 bp) (Neuveglise et al. 2002). In the same strain (Table S1), a cassette expressing S.p.Cas9 bearing an SV40 NLS (DiCarlo et al. 2013) under control of the inducible GAL1/10 promoter was integrated at the HIS3 locus marked by a KanR gene (Chromosome XV), flanked by u1 or by a different (u2) unique 23 bp PAM-containing sequence (Figure 1B). To demonstrate how this method can be used to replace essential genes with a desired construct, the strain also contained a “covering” plasmid carrying WT CDC11 and URA3 (a marker that can be counterselected on 5-FOA medium) (Boeke et al. 1984) (Figure 1A). To initiate genome editing, a 2 μm DNA plasmid expressing sgRNA[u1] (Figure 1C) and PCR fragments to integrate at each locus are introduced by transformation into cells in which Cas9 expression has been induced.
The rationale for flanking the target genes with identical sites for Cas9-catalyzed DSB formation is to demand repair of the resulting chromosomal lesions by HR with the PCR fragments provided, permit concurrent replacement of multiple loci using just a single sgRNA, allow for concomitant self-excision of the Cas9-expressing cassette, when desired, and avoid the spurious events that can occur upon standard multiplex Cas9 genome editing (see Figure S1).
Multiplexing Cas9 to a programmed genomic target sequence using a single sgRNA
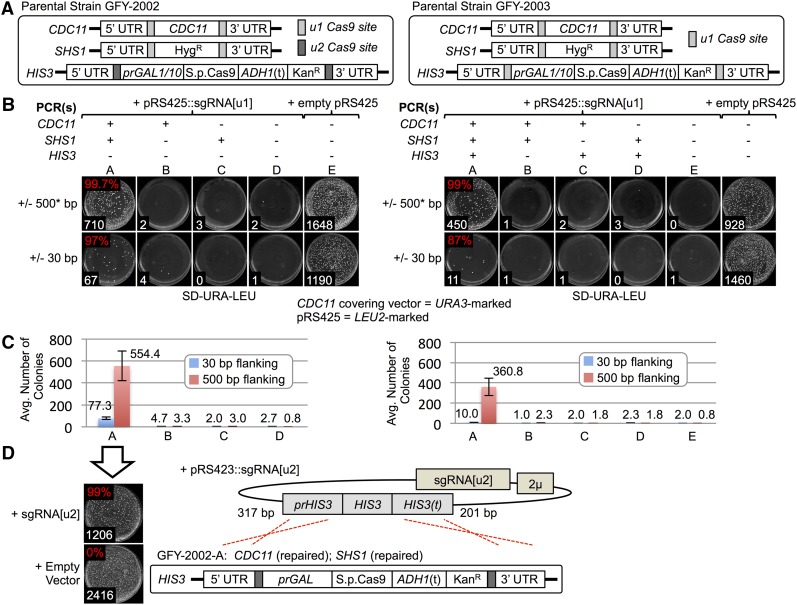
We confirmed, first, that Cas9 is expressed in a galactose-inducible manner and properly localized to the nucleus (Figure S2A) and, second, that expression of neither Cas9 alone, nor sgRNA[u1] or sgRNA[u2] alone (Table S2), nor coexpression of Cas9 with either guide RNA, in otherwise WT cells (i.e., lacking u1 or u2 sequences), caused any detectable loss of viability or transformation efficiency (Figure S2B). We constructed two tester strains, one (GFY-2002) for simultaneous manipulation of two loci (CDC11 and shs1∆::HygR loci) (Figure 2A, left) and one (GFY-2003) for simultaneous manipulation of three loci (CDC11, shs1∆::HygR, and his3∆::Cas9::KanR) (Figure 2A, right). After induction of Cas9, these strains were transformed with either empty vector or the same plasmid expressing sgRNA[u1] in the absence or presence of PCR fragments bearing homology to the genomic sequence upstream and downstream of each locus (Figure 2B, upper). Additional control reactions were conducted in the absence of Cas9 expression or in the absence of sgRNA[u1] (Table S3). The PCR fragments used contained either 500 or 30 bp of flanking genomic sequence homology (Figure 2B, lower). Control strains, in which Cas9 cleavage at the u1 sites produces DSBs that have no corresponding PCR fragment(s) for their repair, yielded very few viable colonies [Figure 2, B and C (conditions B–D in figure part B)], even though the intrinsic transformability of the cells was robust (Figure 2B, condition E), because Cas9-mediated DSB formation (with no subsequent repair of the locus) is lethal in yeast. By contrast, we observed a ≥ 300-fold increase in the recovery of viable colonies when the PCR fragments present to mediate repair by HR had 500 bp of homology to the genomic sequence flanking each locus, and a 20–40-fold increase in recovery of viable colonies even when the homology was only 30 bp [Figure 2, B and C (condition A in figure part B)]. For the GFY-2002 strain (Figure 2, left), the viable colonies recovered correspond to successful HR-mediated repair of two loci (CDC11 and SHS1), and for strain GFY-2003 (Figure 2, right), the viable colonies recovered correspond to simultaneous successful repair at all three loci (CDC11, SHS1, and HIS3). Phenotypic characterization showed that, as expected, the vast majority (≥ 97%) of ∼200 randomly-chosen GFY-2002 survivors had become hygromycin-sensitive, and the vast majority (≥ 87%) of ∼200 randomly-chosen GFY-2003 survivors had become both hygromycin-sensitive and G418-sensitive (the status of CDC11 had to be scored by other means; see below).
Figure 2.
Multiplex Cas9-mediated scarless gene replacement (including an essential gene) and optional concurrent elimination of Cas9. (A) Otherwise isogenic yeast strains containing six programmed Cas9 target sites. In strain GFY-2002, the CDC11 and shs1∆::HygR loci are flanked by u1, whereas the Cas9 expression cassette at the HIS3 locus is flanked by u2. In strain GFY-2003, all three loci are flanked by u1. Both strains also carried a URA3-marked CEN plasmid harboring WT CDC11. (B) Cas9 expression was induced in strains GFY-2002 (left) or GFY-2003 (right), and then the cells were transformed with an empty LEU2-marked vector (pRS425) or with the same plasmid expressing sgRNA[u1] in the absence or presence of various combinations of PCR fragments that span each of the genomic loci of interest, as indicated. The PCR fragments contained either 500 bp (upper plates) or just 30 bp (lower plates) of homology to the genomic sequence flanking each locus. Asterisk, for the CDC11 PCR fragment, the flanking homology was 330 bp. After recovery in rich medium containing galactose (to support continued Cas9 expression), the cells were plated on SD-Ura-Leu medium. The plates were imaged and the number of colonies recovered were counted after incubation at 30° for 3 d. Each independent trial was performed in triplicate. Representative plates are shown; white numbers, total colony count. The empty vector control confirmed that these conditions allowed for efficient transformation and selection for the LEU2- and URA3-marked plasmids. Individual colonies from Condition A, where all of the PCR fragments necessary to heal the Cas9-sgRNA[u1]-generated DSBs were provided, were tested for growth on various diagnostic media to ascertain whether successful gene replacement occurred (see Table S3). Red values, percentage of colonies scored that exhibited successful gene replacement at all loci tested. (C) The average colony count over all experimental trials for each condition (A–D), as indicated. Error bars, SEM. (D) An isolate of GFY-2002 from Condition A (B and C) in which both the u1-flanked CDC11 locus and u1-flanked shs1∆::HygR allele were successfully replaced with WT CDC11 (see Figure 3) and WT SHS1, respectively, was grown in galactose to induce Cas9 expression, and then transformed with either empty vector (pRS423) or the same plasmid expressing sgRNA[u2], plated on SD-Ura-His medium, and grown at 30° for 3 d. The selectable marker in the sgRNA[u2]-expressing plasmid is the S. cerevisiae HIS3 gene with 317 bp of 5′- and 201 bp of 3′-flanking genomic sequence. Therefore, this plasmid not only provides sgRNA[u2] to target Cas9 cleavage at the u2 sites flanking the his3∆::Cas9::KanR cassette, but it also serves as a source of WT HIS3 DNA to repair the cleaved locus. Representative plates are shown; white numbers, total colony count. To assess conversion of the u2-flanked his3∆::Cas9::KanR cassette to WT HIS3, the His+ Ura+ colonies obtained were scored for loss of G418 resistance and complete elimination of the entire cassette (Table S4). Red values, percentage of colonies scored that exhibited successful elimination of the his3∆::Cas9::KanR cassette. HygR, hygromycin resistance; KanR, kanamycin resistance; ORF, open reading frame; PCR, polymerase chain reaction; sgRNA, single-guide RNA; UTR, untranslated region; WT, wild-type.
Unlike strain GFY-2003, where removal and replacement of the u1-flanked Cas9-expressing cassette occurs concomitantly with multiplex substitution at the other u1-flanked loci, strain GFY-2002 contains a Cas9 expression cassette flanked by u2, a different unique target site. This arrangement allows for additional Cas9-dependent integration (or deletion) events at other loci, if desired, but also allows for excision of the his3∆::Cas9::KanR cassette upon introduction of a plasmid expressing sgRNA[u2]. To test the efficacy of this sequential scheme for removal of Cas9, an isolate of GFY-2002, in which direct DNA sequence analysis showed that sgRNA[u1]-driven genome editing had resulted in restoration of WT CDC11 and SHS1 at both loci (Figure 2, B and C, condition A), was transformed with an empty HIS3-marked vector (pRS423) or a derivative-expressing sgRNA[u2] cassette (Table S2). In this plasmid, the HIS3 gene is flanked with significant lengths of genomic sequence (Figure 2D, right); therefore, in theory, it serves both as the source of sgRNA[u2] to catalyze Cas9-mediated excision of the Cas9-expressing cassette and the source of the homologous DNA needed to repair the cleaved locus, without the necessity of cotransforming any PCR fragment or oligonucleotide. Indeed, reassuringly, nearly all (99%) of ∼200 His+ colonies obtained from cells exposed to sgRNA[u2] were G418-sensitive, indicating loss of the Cas9-expressing cassette, whereas all of ∼200 His+ colonies exposed to the empty vector were KanR, as expected for retention of the Cas9-expressing cassette (Figure 2D, left). The 2 μm DNA-based plasmids used to express sgRNA[u1] or sgRNA[u2] are themselves rapidly lost when not subjected to selection for the appropriate marker (Table S4).
Confirmation of successful multiplex gene replacement
Genomic DNA from 10 randomly-chosen colonies from transformations with PCR fragments containing 500 bp of homology (Figure 2, condition A) was analyzed by diagnostic PCR (Figure 3) and direct nucleotide sequencing (data not shown) to examine each manipulated locus. Diagnostic PCR was also performed on colonies from transformations with PCR fragments containing only 30 bp of homology (Figure 2, condition A) with very similar results (Figure S3). For GFY-2002, PCR analysis showed that all 10 isolates replaced the shs1∆::HygR allele with the WT SHS1 gene (and, as expected, still harbored the Cas9-expression cassette), and seven of 10 also properly replaced the u1-flanked CDC11 locus with the WT CDC11 gene, which was further confirmed by sequencing. Multiple PCRs tested for the presence or absence of the upstream and downstream u1 sites present at the CDC11 locus; DNA of the covering plasmid expressing WT CDC11 was not present, since amplification of the parental strains (control lanes) only displayed single PCR bands corresponding to the chromosomal locus (Figure 3 and Figure S3).
For GFY-2003, all 10 isolates replaced the shs1∆::HygR allele with the WT SHS1 gene and also replaced the Cas9-expression cassette with the WT HIS3. For both, the PCR fragments used for gene replacement shared homology only with the genomic sequences flanking these two loci. In the same 10 isolates, nine also properly replaced the u1-flanked CDC11 locus with the WT CDC11 gene and, in the remaining one, only the upstream u1 site was retained. In 36 total isolates tested from all experimental trials, 32 replaced both the upstream and downstream u1 sites with WT CDC11 and only four retained just the upstream u1 site. The most likely explanation for these few exceptions arises from the fact that the CDC11-containing PCR fragment we used for replacement shares homology across its entire coding region with the u1-flanked chromosomal CDC11 locus, and that the upstream u1 site lies just downstream and in-frame with the Met codon need to initiate Cdc11 translation. Thus, crossovers between the PCR fragment and the chromosome that occur within the CDC11 ORF and in the 3′-UTR will heal a DSB at the downstream u1 site and yield a viable cell that can produce Cdc11, yet retain the upstream u1 (Figure S4). These rare exceptions can be readily avoided by eliminating the internal homology by (i) starting with a genomic cdc11∆ null allele (covered by WT CDC11 on a plasmid) or (ii) installing in the chromosome a synthetic ORF with codon alterations that minimize its nucleotide sequence identity to the authentic CDC11 ORF on the PCR fragment.
Prior work has shown that repair of a DSB via HR in yeast is orders of magnitude more frequent than by NHEJ (Storici et al. 2003), which is extremely inefficient (Rattray et al. 2001; Daley et al. 2005; Storici et al. 2006). Indeed, in 16 of the very rare survivors obtained from the controls where the transformations lacked one or more PCR fragments to repair the DSBs (Figure 2, conditions B–D), phenotypic analysis (Table S4) and diagnostic PCR (Figure S5) showed that the majority did not have any replacements and likely escaped any Cas9-induced DSBs and a few isolates replaced one locus, but failed to cut and remove the u1 sites elsewhere. In only three (from Figure 2, condition B), the shs1∆::HygR allele was excised, but left a single intact u1 site, most consistent with repair of the Cas9-induced DSBs via HR between the u1 repeats rather than by NHEJ. We conclude from our data that correct replacement at all loci examined is nearly three orders of magnitude more frequent than any other event.
Discussion
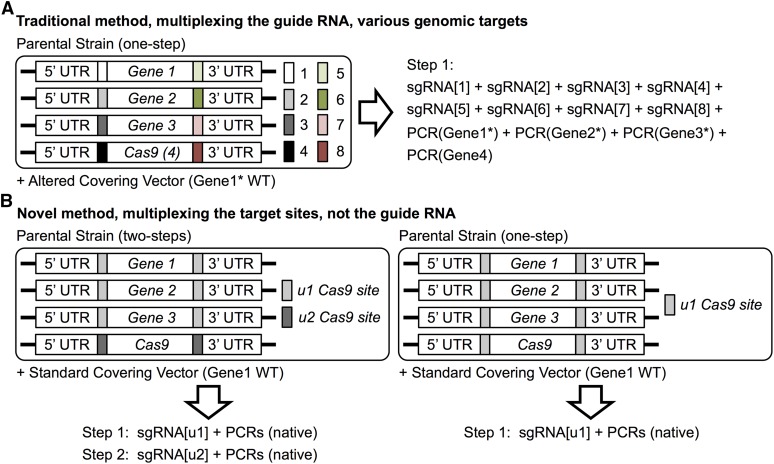
The crux of our method is first programming the desired Cas9 cleavage sites at will by installing a unique sequence of the investigator’s own choosing, rather than relying on naturally-occurring genomic sequences (Figure 4). By flanking any number of selected genes with the same “alien” sequence, and providing PCR fragments homologous to the loci of interest, expression of just a single sgRNA initiates multiplex Cas9-mediated removal and scarless replacement of these targeted genes. Here, we achieved concurrent replacement of three ORFs on three different chromosomes, including one essential gene; however, the same approach can be used to excise or alter exons, introns, splice junctions, transcription factor-binding sites, locus control regions, etc. Our method also eliminates the need for codon alterations to the integrated allele(s) [or any WT covering plasmid(s)] to prevent recutting of the newly substituted DNA by Cas9 (Figure S1).
Figure 4.
Comparison of Cas9-mediated genome editing by multiplexing sgRNAs vs. multiplexing loci with a unique target site. (A) Traditional targeting of Cas9 to multiple genomic loci (including one locus where Cas9 is integrated). Each of four loci is illustrated as requiring Cas9 action at two distinct sites. Hence, concurrent action of Cas9 at these four genes would require the selection of eight individual PAM-containing genomic sequences and the production of eight corresponding sgRNAs. In addition, it should be noted that, in this scenario, at least one target site lies within the coding sequence of each gene; therefore, PCR fragments used to replace Genes(1–3) would also require alterations of the coding sequence to avoid recutting by Cas9 (also see Figure S1). Finally, for manipulation of any essential genes (e.g., Gene1), a counterselectable plasmid expressing a WT copy will also need to be altered to not include the genomic target site(s), again to avoid its Cas9-mediated cleavage (Figure S1). (B) The approach of multiplexing the target site(s) has a number of useful advantages. First, there is no need to restrict the target for Cas9 cleavage to sequences that exist within the genome of interest, which may be suboptimal (with regard to off-target effects) or may have a limited number or inopportune placements of available PAM sites. Second, the artificial target site chosen for insertion may be any stretch of 23 nucleotides (20 plus a 5′-NGG-3′ PAM) taken from any known species (or designed de novo), as long as it has no counterpart in the genome of interest. In fact, such a programmed target site sequence should greatly reduce or eliminate off-target effects, and also has the virtue that it can be inserted at a precise location (down to the base pair) to optimally facilitate recombination and precisely control the placement of the Cas9-mediated DSBs. The limiting step in this approach is, of course, introduction of these unique target site insertions into the parental genome at the desired locations. Once created, however, such an engineered parental strain can be used repeatedly to install various different alterations at one or many loci using only a single sgRNA, allowing for rapid construction of multiple strain variants. Moreover, in this approach, the Cas9 expression cassette can be retained, targeted for simultaneous excision in parallel with the manipulations of other loci (right), or eliminated at a later time, if the Cas9 expression cassette is flanked with a separate unique target site (left). Finally, because the sequence of the target sites flanking each locus are distinct from any of the elements of the targeted genes themselves, no modifications to the sequence of the PCR fragments used for gene editing (or of a covering plasmid carrying the corresponding WT gene) are required to make them immune to the further action of Cas9. DSB, double-strand break; PAM, protospacer adjacent motif; PCR, polymerase chain reaction; sgRNA, single-guide RNA; UTR, untranslated region; WT, wild-type.
Other methods for modifying essential genes in yeast (Toh-e and Oguchi 2000; Cross and Pecani 2011; Horwitz et al., 2015) have been described . In our view, our approach provides an alternative that is, in the long run, substantially less cumbersome and markedly more efficient. For example, our strategy does not depend on the fortuitous presence of a unique restriction endonuclease site, as required by the “integration replacement/disruption” method of Toh-e and Oguchi (2000) to “loop-in” a mutagenized plasmid copy of the gene of interest. Although the HO endonuclease-based method of Cross and Pecani (2011) does not require selection, it does require the construction of strain backgrounds with inducible HO expression and demands the exclusive use of the pRS400 series of ARS-less and CEN-less integration vectors. In the use of Cas9 for editing of essential genes described by Horwitz et al. (2015), the endogenous target sequence used for DSB formation needs to lie as close as possible to the desired nucleotide change to prevent inappropriate HR downstream of the mutation resulting in repair of the DSB without incorporation of the desired allele, as we already pointed out (Figure S1). Our approach circumvents all of the above issues, as well as increases the ease and efficiency by which essential genes may be manipulated.
Our methodology is complementary to “traditional” Cas9-mediated multiplex gene editing that requires the design and expression of multiple sgRNAs. Our approach expands how Cas9-based genome editing technology can be deployed and, hence, enhances its utility. Although our strategy first requires the initial installation of a unique target site(s) within the genome to be manipulated, there are several long-term benefits of constructing strains with programmed Cas9 target sites that we feel outweigh the traditional Cas9 approach (Figure 4 and Figure S1). Our method is especially useful (i) when repeated targeting of a locus, or groups of genes (e.g., paralogs or entire genetic pathways) is needed, (ii) for manipulation of essential genes, (iii) where minimizing off-target effects is critical, and (iv) in cases were an “alien” target sequence is required/desired, such as in the design of gene drives. Given the rapid movement toward programmable toolkits in synthetic biology, we envision that it would be worth investing the effort to flank every gene in a genome of interest with such synthetic Cas9 target sites. In fact, traditional Cas9 editing could be used to do so.
Moreover, as we have demonstrated, Cas9 action at the artificially introduced sites can eliminate its own expression cassette without compromising its ability to mediate efficient gene editing elsewhere in the genome of the same cell. In addition, our method can be used to interrogate the effects of chromosome position and local chromatin structure on Cas9 action, because the same 23 bp sequence can be installed at any location in a genome. In this way, apparent differences between species with regard to the efficiency with which Cas9 can access and cleave at sites within heterochromatin (Yu et al. 2013; Wu et al. 2014; Knight et al. 2015; Feng et al. 2016) could be systematically explored. Finally, application of this approach should be extremely useful in generating strain libraries, constructing synthetic genomes, and introducing in a multiplex manner genomic changes to study multiple genes in a signaling pathway, the subunits of a multi-protein complex, paralogous gene sets, or any combination or collection of genes of interest.
Supplementary Material
Acknowledgments
We thank the staff of the University of California (UC) Berkeley DNA Sequencing Facility, especially its supervisor Dr. Hitomi Asahara, for assistance with our nucleotide sequence analysis, and Dana Carroll (University of Utah) and UC Berkeley Innovative Genomics Initiative (IGI) for helpful advice and constructive suggestions. This work was supported by a Miller Research Fellowship from the UC Berkeley Miller Institute for Basic Research in Science Berkeley (to G.C.F.) and by National Institutes of Health research grants GM21841 (to J.T.) and GM101314 (to J.T. and colleague Eva Nogales).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.029801/-/DC1
Communicating editor: B. J. Andrews
Literature Cited
- Alani E., Cao L., Kleckner N., 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116: 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D. C., D. J. Burke, and J. N. Strathern, 2006 2006 Yeast DNA isolation: miniprep, CSH Protoc. DOI: 10.1101/pdb.prot4148. [DOI] [PubMed] [Google Scholar]
- Bao Z., Xiao H., Liang J., Zhang L., Xiong X., et al. , 2015. Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth. Biol. 4: 585–594. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R., 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197: 345–346. [DOI] [PubMed] [Google Scholar]
- Bolukbasi M. F., Gupta A., Wolfe S. A., 2015. Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat. Methods 13: 41–50. [DOI] [PubMed] [Google Scholar]
- Chandrasegaran S., Carroll D., 2016. Origins of programmable nucleases for genome engineering. J. Mol. Biol. 428: 963–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Gilbert L. A., Cimini B. A., Schnitzbauer J., Zhang W., et al. , 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Hu J., Almeida R., Liu H., Balakrishnan S., et al. , 2016. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. DOI: 10.1093/nar/gkv1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. W., Wang H., Yang H., Shi L., Katz Y., et al. , 2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim J. M., Kim J. S., 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim Y., Kweon J., Kim H. S., et al. , 2014. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R., Pecani K., 2011. Efficient and rapid exact gene replacement without selection. Yeast 28: 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley J. M., Palmbos P. L., Wu D., Wilson T. E., 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451. [DOI] [PubMed] [Google Scholar]
- Dang Y., Jia G., Choi J., Ma H., Anaya E., et al. , 2015. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 16: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., et al. , 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41: 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E., Chavez A., Dietz S. L., Esvelt K. M., Church G. M., 2015. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33: 1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., et al. , 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34: 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Lin J., Held N. L., Clem R. J., Passarelli A. L., et al. , 2015. Heritable CRISPR/Cas9-mediated genome editing in the yellow fever mosquito, Aedes aegypti. PLoS One 10: e0122353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Charpentier E., 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Eckert-Boulet N., Pedersen M. L., Krogh B. O., Lisby M., 2012. Optimization of ordered plasmid assembly by gap repair in Saccharomyces cerevisiae. Yeast 29: 323–334. [DOI] [PubMed] [Google Scholar]
- Edelstein, A., N. Amodaj, K. Hoover, R. Vale, and N. Stuurman, 2010 Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. 92: 14.20.1–14.20.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C., Yuan J., Wang R., Liu Y., Birchler J. A., et al. , 2016. Efficient Targeted Genome Modification in Maize Using CRISPR/Cas9 System. J. Genet. Genomics 43: 37–43. [DOI] [PubMed] [Google Scholar]
- Finnigan G. C., Takagi J., Cho C., Thorner J., 2015. Comprehensive Genetic Analysis of Paralogous Terminal Septin Subunits Shs1 and Cdc11 in Saccharomyces cerevisiae. Genetics 200: 841–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan G. C., Thorner J., 2015. Complex in vivo Ligation Using Homologous Recombination and High-efficiency Plasmid Rescue from Saccharomyces cerevisiae. Bio Protoc. 5: e1521 http://www.bio-protocol.org/e1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J. D., Reyon D., Cascio V. M., Joung J. K., 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F. I., 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz V. M., Jasinskiene N., Tatarenkova O., Fazekas A., Macias V. M., et al. , 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 112: E6736–E6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G., 3rd, Bertin A., Li Z., Song Y., McMurray M. A., et al. , 2011. Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J. Cell Biol. 195: 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., et al. , 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., Ochman H., 1996. Inverse polymerase chain reaction. Methods Mol. Biol. 58: 293–301. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., 1971. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69: 265–276. [DOI] [PubMed] [Google Scholar]
- Horwitz A. A., Walter J. M., Schubert M. G., Kung S. H., K. Hawkins et al, 2015. Efficient Multiplexed Integration of Synergistic Alleles and Metabolic Pathways in Yeasts via CRISPR-Cas. Cell Syst. 1: 88–96. [DOI] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S., Zhang F., 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. H., Davis K. M., Liu D. R., 2016. Chemical biology approaches to genome editing: understanding, controlling, and delivering programmable nucleases. Cell Chem. Biol. 23: 57–73. [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M., Luo J., Bi E., Toh-e A., 2007. Shs1 plays separable roles in septin organization and cytokinesis in Saccharomyces cerevisiae. Genetics 177: 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakociunas T., Bonde I., Herrgard M., Harrison S. J., Kristensen M., et al. , 2015. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab. Eng. 28: 213–222. [DOI] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A., 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., et al. , 2013. RNA-programmed genome editing in human cells. eLife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Jiang F., Taylor D. W., Sternberg S. H., Kaya E., et al. , 2014. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343: 1215–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski R., Chen Y., Fischer T., Tedaldi E., Napoli A., et al. , 2016. Elimination of HIV-1 Genomes from Human T-lymphoid Cells by CRISPR/Cas9 Gene Editing. Sci. Rep. 6: 22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P., M. S. Prew, S. Q. Tsai, V. V. Topkar, N. T. Nguyen et al, 2015. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P., Pattanayak V., Prew M. S., Tsai S. Q., Nguyen N. T., et al. , 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S. C., Xie L., Deng W., Guglielmi B., Witkowsky L. B., et al. , 2015. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science 350: 823–826. [DOI] [PubMed] [Google Scholar]
- La Russa M. F., Qi L. S., 2015. The new state of the art: Cas9 for gene activation and repression. Mol. Cell. Biol. 35: 3800–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughery M. F., Hunter T., Brown A., Hoopes J., Ostbye T., et al. , 2015. New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 32: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R., van Rossum H. M., Wijsman M., Backx A., Kuijpers N. G., et al. , 2015. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. DOI: . 10.1093/femsyr/fov004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Zhang H., Xu N., Zhang B., Gou F., et al. , 2013. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 6: 2008–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray M. A., Bertin A., Garcia G., 3rd, Lam L., Nogales E., et al. , 2011. Septin filament formation is essential in budding yeast. Dev. Cell 20: 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. R., Rodino-Klapac L. R., 2016. Duchenne muscular dystrophy: CRISPR/Cas9 treatment. Cell Res. 26: 513–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Hino K., Bono H., Ui-Tei K., 2015. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31: 1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuveglise C., Feldmann H., Bon E., Gaillardin C., Casaregola S., 2002. Genomic evolution of the long terminal repeat retrotransposons in hemiascomycetous yeasts. Genome Res. 12: 930–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes B. L., Nadler D. C., Savage D. F., 2014. Protein engineering of Cas9 for enhanced function. Methods Enzymol. 546: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H., Yu A. S., Segal D. J., 2015. How specific is CRISPR/Cas9 really? Curr. Opin. Chem. Biol. 29: 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V., Lin S., Guilinger J. P., Ma E., Doudna J. A., et al. , 2013. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., et al. , 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A. J., McGill C. B., Shafer B. K., Strathern J. N., 2001. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics 158: 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. D., Ray G. J., DeWitt M. A., Curie G. L., Corn J. E., 2016. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34: 339–344. [DOI] [PubMed] [Google Scholar]
- Ronda C., Maury J., Jakociunas T., Jacobsen S. A., Germann S. M., et al. , 2015. CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microb. Cell Fact. 14: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan O. W., Cate J. H., 2014. Multiplex engineering of industrial yeast genomes using CRISPRm. Methods Enzymol. 546: 473–489. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W., 2001. Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shalem O., Sanjana N. E., Zhang F., 2015. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Liang Y., Zhang M. M., Ang E. L., Zhao H., 2016. A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab. Eng. 33: 19–27. [DOI] [PubMed] [Google Scholar]
- Shmakov S., Abudayyeh O. O., Makarova K. S., Wolf Y. I., Gootenberg J. S., et al. , 2015. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 60: 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I. M., Gao L., Zetsche B., Scott D. A., Yan W. X., et al. , 2016. Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Lawrence C. M., Wiedenheft B., 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82: 237–266. [DOI] [PubMed] [Google Scholar]
- Stemmer W. P., Crameri A., Ha K. D., Brennan T. M., Heyneker H. L., 1995. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164: 49–53. [DOI] [PubMed] [Google Scholar]
- Storici F., Durham C. L., Gordenin D. A., Resnick M. A., 2003. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl. Acad. Sci. USA 100: 14994–14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F., Snipe J. R., Chan G. K., Gordenin D. A., Resnick M. A., 2006. Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol. Cell. Biol. 26: 7645–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Hu B., Shao J., Shen B., Du J., et al. , 2016. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 6: 20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohe A., Oguchi T., 2000. An improved integration replacement/disruption method for mutagenesis of yeast essential genes. Genes. Genet. Syst. 75: 33–39. [DOI] [PubMed] [Google Scholar]
- Tsai C. S., Kong I. I., Lesmana A., Million G., Zhang G. C., et al. , 2015. Rapid and marker-free refactoring of xylose-fermenting yeast strains with Cas9/CRISPR. Biotechnol. Bioeng. 112: 2406–2411. [DOI] [PubMed] [Google Scholar]
- Tsarmpopoulos I., Gourgues G., Blanchard A., Vashee S., Jores J., et al. , 2016. In-Yeast Engineering of a Bacterial Genome Using CRISPR/Cas9. ACS Synth. Biol. 5: 104–109. [DOI] [PubMed] [Google Scholar]
- Vriend L. E., Prakash R., Chen C. C., Vanoli F., Cavallo F., et al. , 2016. Distinct genetic control of homologous recombination repair of Cas9-induced double-strand breaks, nicks and paired nicks. Nucleic Acids Res. DOI: 10.1093/nar/gkw179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. M., Alper H. S., 2015. Synthetic biology and molecular genetics in non-conventional yeasts: Current tools and future advances. Fungal Genet. Biol. 89: 126–136. [DOI] [PubMed] [Google Scholar]
- Wang H., La Russa M., Qi L. S., 2016. CRISPR-Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85: 22.1–22.38. [DOI] [PubMed] [Google Scholar]
- Wei C., Liu J., Yu Z., Zhang B., Gao G., et al. , 2013. TALEN or Cas9— rapid, efficient and specific choices for genome modifications. J. Genet. Genomics 40: 281–289. [DOI] [PubMed] [Google Scholar]
- Wu X., Scott D. A., Kriz A. J., Chiu A. C., Hsu P. D., et al. , 2014. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Xiao T., Chen C. H., Li W., Meyer C. A., et al. , 2015. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Ren M., Wang Z., Zhang B., Rong Y. S., et al. , 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 195: 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan J. G., Lee M. E., Almeida R., Gilbert L. A., Whitehead E. H., et al. , 2015. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. H., Tee L. Y., Wang X. G., Huang Q. S., Yang S. H., 2015. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol. Ther. Nucleic Acids 4: e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.