Abstract
During the life cycle of shellfish, larval development, especially metamorphosis, has a vital influence on the dynamics, distribution, and recruitment of natural populations, as well as seed breeding. Rapana venosa, a carnivorous gastropod, is an important commercial shellfish in China, and is an ecological invader in the United States, Argentina, and France. However, information about the mechanism of its early development is still limited, because research in this area has long suffered from a lack of genomic resources. In this study, 15 digital gene expression (DGE) libraries from five developmental stages of R. venosa were constructed and sequenced on the IIIumina Hi-Sequation 2500 platform. Bioinformaticsanalysis identified numerous differentially and specifically expressed genes, which revealed that genes associated with growth, nervous system, digestive system, immune system, and apoptosis participate in important developmental processes. The functional analysis of differentially expressed genes was further implemented by gene ontology, and Kyoto encyclopedia of genes and genomes enrichment. DGE profiling provided a general picture of the transcriptomic activities during the early development of R. venosa, which may provide interesting hints for further study. Our data represent the first comparative transcriptomic information available for the early development of R. venosa, which is a prerequisite for a better understanding of the physiological traits controlling development.
Keywords: transcriptome, Rapana venosa, gastropod, larva, digital gene expression
Rapana venosa is a species of large predatory sea snail that is naturally distributed in the western Pacific, the Sea of Japan, the Yellow Sea, the Bohai Sea, and the East China Sea (Savini et al. 2004). Since Valenciennes first reported that R. venosa had invaded the northern Adriatic Sea in 1846, it has been established in many different localities around the world in the last few decades. R. venosa is found in countries such as the United States, Argentina, and France (Mann and Harding 2003; Mann et al. 2006; Giberto et al. 2006; Leppäkoski et al. 2013) as a biological invader because of its ecological flexibility, and its tolerance to low salinity, low oxygen and significantly polluted water (Giberto et al. 2006; Çulha et al. 2009). It feeds at night, and is often found in locations that are hard to sample, making early detection of newly established populations difficult (Harding et al. 2007a). R. venosa invasion can have a seriously negative impact on the environment. As R. venosa became more prevalent in the Chesapeake Bay region (VA), Harding et al. (2007a) found significant differences in predation strategies and prey species consumed between R. venosa and the native gastropod Urosalpinx cinerea, which could cause considerable disruption of the trophic structure (feeding relationships among species) in Chesapeake Bay. However, R. venosa is considered as an important commercial gastropod species in China (Liu et al. 2003), and, as a result of intensive research, the industrial aquaculture of R. venosa remains efficient. In recent years, research has been conducted on its reproductive biology (Harding et al. 2007b; Yang et al. 2007), invasion biology (Leppäkoski et al. 2013; Harding et al. 2008; Lanfranconi et al. 2009), and population genetics (Xue et al. 2014; Sun et al. 2014). However, information related to genomic and transcriptome resources, and the mechanism of development of R. venosa, are currently unavailable.
The veliger is a critical life cycle stage of the gastropod, bivalve, and scaphopod taxonomic classes. With the development of veliger larvae, larvae become competent for settlement and metamorphosis, which link free-swimming larva and benthic juveniles. Metamorphosis is a dramatic and irreversible developmental event involving physical, physiological, and behavioral transformations (Leise et al. 2009). Studies on the mechanism of mollusk early development can provide useful information for aquaculture, resource restoration and bio-antifouling. Studies on the mechanism of larval development, especially metamorphosis, have made great progress in recent years. Some genes and proteins involved in these events have been identified and characterized functionally. Hox and paraHox genes with different sequences and gene expression patterns in mollusks are considered to play an important role in shell formation, body plan evolution, and diversification (Biscotti et al. 2014). Nitric oxide synthase (NOS) and Hsp90 are required for metamorphosis of the mollusk Haliotis asinina: their RNA expression levels are upregulated at the initiation of settlement and metamorphosis, and their expression levels show marked differences between competent and precompetent larval stages (Ueda 2013). mRNA differential display of gene expression in the settlement metamorphosis process was performed in Ruditapes philippinarum to elucidate the potential mechanisms underlying larval development and metamorphosis (Lu et al. 2008). However, research in this field has been hampered by a long-term lack of genomic resources, especially useful transcriptome sequences.
Recently, next-generation sequencing (NGS) technologies have become the main method for genome-wide characterization and profiling of mRNA (Ansorge 2009). NGS offers an effective approach to obtaining sequence information for nonmodel species in which genomic sequences are limited. Deep-sequenced transcriptomes of several mollusks, such as oysters Crassostrea virginica (Quilang et al. 2007), Crassostrea gigas (Fleury et al. 2009), the abalone Haliotis midae (Franchini et al. 2011), the scallop Patinopecten yessoensis (Hou et al. 2011), the Antarctic bivalve Laternula elliptica (Clark et al. 2010), and the mussel Mytilus galloprovincialis (Craft et al. 2010) have been published recently. However, in mollusks, very few transcriptome sequences have been reported for their early developmental stages, especially for planktonic larvae. Digital gene expression (DGE) can effectively and efficiently detect the differential and specific gene expression of a specific organization in certain conditions using NGS technology and high performance computing analysis. DGE has been used in such fields as basic scientific research, medical research, and drug development. In mollusks, DGE was applied to detect immune responses to vibrio infection in the oyster C. gigas (De Lorgeril et al. 2011); DGE studies of molluskan development are still limited.
In this study, our objective was to establish a useful database of differentially expressed genes (DEGs) from different developmental stages of R. venosa. These data would provide a better understanding of the mechanism of veliger development. We performed a comparative analysis of the transcriptomes from five different developmental stages of R. venosa by using DGE. Our data should facilitate better understanding of the molecular mechanism of early development of R. venosa larva, and represents a valuable resource for genetic and genomic researches on R. venosa in the future.
Materials and Methods
Larva culture and sample preparation
Rapana venosa larvae were obtained from the Blue Ocean Co. Limited (Laizhou, Shandong, China). Parent culture, spawning, and larva rearing were performed according to the study of Yang et al. (2007). Planktonic larvae were cultured in 3 m × 5 m × 1.5 m cement pools at 24–26°, at a density of 0.5/ml. Platymonas subcordiformis, Isochrysis galbana, and Chlorella vulgaris were mixed as a diet (15 × 104 cell/ml, three or four times) for the Planktonic larvae. Larval samples from five major developmental stages were collected: the one-spiral whorl stage, the two-spiral whorls stage, the three-spiral whorls stage, the four-spiral whorls stage (competent larva), and the postlarval stage. The samples were monitored under a microscope to ensure developmental synchrony. Each sample was then washed with ddH2O, snap-frozen in liquid nitrogen, and stored at −80° until use.
RNA extraction, library preparation, and Hi-sequation 2500 sequencing
Each stage had three replicates for RNA extraction, and each replicate contained approximately 500 mg of larvae. We carried out RNA extraction following the instructions of the TRIzol Kit (Invitrogen, CA). RNA quality was examined by electrophoresis through 1.2% agarose gels to check for possible degradation and contamination. RNA purity was monitored on a NanoPhotometer spectrometer (IMPLEN, CA) using default parameters. Integrity and concentration and of RNA was measured using a Qubit RNA Assay Kit in a Qubit 2.0 Flurometer (Life Technologies, CA), and the RNA 6000 Nano Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA), respectively. Fifteen RNA samples (five stages × three replicates) that satisfied all requirements of the quality test were selected for library preparation.
RNA (3 μg per sample) was used as the input material for the RNA sample preparations. Sequencing libraries were constructed according to the manufacturer’s instructions using a NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB). After clustering of the index-coded samples, we performed RNA sequencing of each library on an Illumina HiSequation 2500 platform to generate 100-bp single-end (SE) reads. This procedure was carried out by Novogene Bioinformatics Technology Co. Ltd (Tianjing, China).
Mapping and analysis of DGE reads
The raw data were deposited in the gene expression omnibus (GEO) database of NCBI with the accession number GSE70548. Raw reads were processed into clean reads by removing adapters, poly-N regions, and low quality reads. Moreover, the Q20, Q30, GC-content, and sequence duplication level of the clean data were calculated. The high quality clean data were aligned to the R. venosa reference transcriptome using RSEM v1.2.15 (Li and Dewey 2011) (the reference transcriptome was de novo sequenced and assembled in our previous work, based on a mixture of RNAs from different developmental larval stages, the raw data of which is available in the SRA with the accession number SRR2086477; the annotation has been deposited at DDBJ/EMBL/GenBank under accession no. GDIA00000000 (Song et al. 2016). To estimate the gene expression level, RSEM analysis was performed to obtain the readcount for each gene of each sample, based on the mapping results. Then readcount was converted into the number of fragments per kilobase per million (FPKM value) (Mortazavi et al. 2008). Differential expression analysis between five developmental stages was implemented using the DESeq R package v1.12.0 (Anders and Huber 2010). The P-value was adjusted using the q-value (or padj) (Anders and Huber 2010). There were three replicates of each samples; therefore, a q-value < 0.05 was set as the threshold to select genes with significant differential expression (Tan et al. 2015).
Functional analysis of differentially expressed genes
Gene ontology (GO) enrichment analysis of DEGs was performed using GOseq v1.10.0, which can eliminate gene length bias in DEGs. The KEGG is a database resource used to aid understanding of the high-level functions and uses of biological systems (http://www.genome.jp/kegg/). Here, we used KOBAS software, version 2.0.12, to analyze the statistical enrichment of DEGs in KEGG pathways.
Based on the normalized, filtered sequences, cluster analysis of gene expression was implemented to identify genes of which the expression level were remarkably different among this five developmental stages of R. venosa. To ensure a more persuasive result, we chose those annotated genes with an average FPKM in all stages of more than 10, a q-value < 0.001 and |log2(foldchange)| > 2 in certain comparisons of two stages as candidates (1794 genes) for analysis using Cluster 3.0 software.
Quantitative real-time reverse transcription PCR
To validate the mRNA expression levels indicated by the DGE analysis, 20 annotated DEGs (including the 10 most upregulated and 10 most downregulated DEGs) from the comparison of competent vs. post larva stages were selected. To make the result more reliable, genes without annotation or annotated with “hypothetical protein” were filtered out, and genes with an FPKM < 1 in these two stages were also filtered out. The DGE input RNA was used as the template for cDNA synthesis, following the instructions of the PrimeScript RT reagent Kit with gDNA Eraser (Takara, Japan). The quantitative analysis of mRNA expression levels by quantitative real-time reverse transcription PCR (qRT-PCR) was implemented using a SYBR PrimeScript RT-PCR Kit II (Takara, Japan), according to the instruction manual, in an Eppendorf Mastercycler ep realplex apparatus (Eppendorf, Hamburg, Germany). GAPDH was chosen as an internal reference gene to normalize the data (Badariotti et al. 2007; Qian et al. 2012), and three replicates were performed. The primers used (Supplemental Material, Table S2) were designed using Primer3 (http://primer3.sour-ceforge.net/r-eleases.php). The PCR reaction volumes comprised 10 × PCR buffer, 2.5 μl; dNTPs, 2 μl (2.5 mmol/l); forward primer, 1 μl (5 μmol/l); reverse primer, 1 μl (5 μmol/l); Ex Taq (TAKARA), 0.2 μl; cDNA template, 0.1 μl (containing 1 ng of cDNA); and ddH2O up to 25 μl. The PCR conditions were set as follows: 95° for 3 min; 95° for 15 sec, 60° for 25 sec, 72° for 15 sec for 40 cycles; 72° for 5 min, with a final 10-min extension step at 72°. The PCR products were detected by electrophoresis through a 1.2% agarose gel. The relative expression level was calculated by the 2–△△Ct method. PASW Statistics 18 (SPSS Inc.) was used for statistical analysis. Significance was tested by one-way ANOVA using Tukey’s test. P < 0.05 was considered significant.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
General characteristics of the 15 DGE libraries
To explore the genes and their networks which control the development of larva in R. venosa, we determined and compared the transcriptome of five larval stages (C1: the one-spiral whorl stage, D2: the two-spiral whorls stage, F3: the three-spiral whorls stage, J4: the four-spiral whorls stage, and Y5: the postlarval stage). As shown in Figure 1, there are visible morphological and body size differences between these five stages (Pan et al. 2013).
Figure 1.
Five developmental stages of R. venosa that were sampled for transcriptome analysis (shown in blue). The one-spiral whorl stage (C1), the two-spiral whorls stage (D2), the three-spiral whorls stage (F3), the four-spiral whorls stage (competent larva, J4), and the postlarval stage (Y5).
For each sample, the high-throughput RNA sequencing obtained 10.51–15.57 million single-ended reads (Table 1). After filtering for low quality reads and adapter sequences, 10.30–15.17 million clean reads (99.26–96.05% from raw data) were generated. Among the clean data of the 15 libraries, about 82.37–87.34% of clean sequences was successfully mapped to the entire reference transcriptome (Table 1).
Table 1. Statistics of DGE sequencing.
| Sample | Raw Reads | Clean Reads | Clean Bases (G) | Error (%) | Q20 (%) | Q30 (%) | GC Content (%) | Total Mapped (% of Clean Data) |
|---|---|---|---|---|---|---|---|---|
| C1_1 | 10,985,377 | 10,893,487 | 0.54 | 0.01 | 98.47 | 97.02 | 47.89 | 9,428,310 (86.55%) |
| C1_2 | 13,169,590 | 12,998,744 | 0.65 | 0.01 | 98.48 | 97.04 | 46.89 | 11,153,576 (85.81%) |
| C1_3 | 12,218,112 | 11,735,699 | 0.59 | 0.01 | 98.48 | 97.04 | 47.58 | 10,123,873 (86.27%) |
| D2_1 | 15,568,814 | 15,171,768 | 0.76 | 0.01 | 98.59 | 97.16 | 48.73 | 13,191,966 (86.95%) |
| D2_2 | 12,006,329 | 11,727,213 | 0.59 | 0.01 | 98.49 | 96.97 | 48.71 | 10,186,346 (86.86%) |
| D2_3 | 14,569,020 | 14,161,517 | 0.71 | 0.01 | 98.59 | 97.17 | 48.91 | 12,368,256 (87.34%) |
| F3_1 | 13,122,713 | 12,689,128 | 0.63 | 0.01 | 98.08 | 95.99 | 48.99 | 11,044,387 (87.04%) |
| F3_2 | 10,972,646 | 10,832,113 | 0.54 | 0.01 | 98.11 | 96.04 | 48.37 | 9,341,753 (86.24%) |
| F3_3 | 12,235,201 | 12,145,123 | 0.61 | 0.01 | 98.12 | 96.06 | 48.32 | 10,514,719 (86.58%) |
| G3_1 | 10,508,213 | 10,302,115 | 0.52 | 0.01 | 97.91 | 95.4 | 47.9 | 8,708,196 (84.53%) |
| G3_2 | 11,352,406 | 11,225,014 | 0.56 | 0.01 | 97.91 | 95.38 | 46.38 | 9,245,843 (82.37%) |
| G3_3 | 12,068,623 | 11,907,135 | 0.6 | 0.01 | 97.89 | 95.34 | 47.03 | 9,846,087 (82.69%) |
| J4_1 | 14,827,351 | 14,665,024 | 0.73 | 0.01 | 98.14 | 96.07 | 46.76 | 12,283,484 (83.76%) |
| J4_2 | 12,026,881 | 11,908,433 | 0.6 | 0.01 | 98.14 | 96.07 | 46.57 | 9,996,051 (83.94%) |
| J4_3 | 12,376,019 | 12,118,735 | 0.61 | 0.01 | 98.14 | 96.08 | 47.26 | 10,146,937 (83.73%) |
| Y5_1 | 13,520,738 | 13,375,042 | 0.67 | 0.01 | 98.27 | 96.53 | 46.72 | 11,220,158 (83.89%) |
| Y5_2 | 12,988,065 | 12,634,083 | 0.63 | 0.01 | 98.22 | 96.45 | 47 | 10,615,299 (84.02%) |
| Y5_3 | 12,780,985 | 12,628,348 | 0.63 | 0.01 | 98.24 | 96.48 | 47.27 | 10,597,512 (83.92%) |
The Pearson correlation coefficients between each DGE library are presented in Figure 2. The Pearson correlation coefficients between replicates remained high, with an average coefficient of r = 0.894 for C1, r = 0.917 for D2, r = 0.897 for F3, r = 0.892 for J4, and r = 0.903 for Y5, which indicates the satisfactory reproducibility of the biological replicates. A volcano plot showing the number and relationship between fold-change and p-value of up/downregulated DEGs in each comparison can be found in Figure S1.
Figure 2.
Pearson correlation coefficients between samples.
Development dynamics of differential gene expressions
A Venn diagram was drawn to illustrate the number of common and unique DEGs among the five stages (Figure 3). There were 11 common DEGs among the comparison groups, and 7069 specific DEGs in the J4 vs. Y5 comparison.
Figure 3.
Venn diagram of differentially expressed genes. The sum of numbers in each big circle is the total number of differentially expressed genes in this comparison group, and the overlapping part is the number of common differentially expressed genes among the comparison groups.
While comprehensive analysis of the expression level of these sequences still requires much work, a preliminary analysis revealed that some genes showed a rather different expression pattern among different larval stages (Table S3). Some of these data are shown in Table 2. The expression patterns of these genes varied widely. Different genes were abundantly expressed mainly in the precompetent larval stage (e.g., SARP-19), competent larval stage (actin, and sulfotransferase 1A2 and 1C2), or expressed in the postlarval stage (e.g., trypsin and defensin). Their functions were related to growth and development, immunity, signal transduction, etc. This information brings indications revealing their function.
Table 2. Some of the genes that were differentially expressed among the five developmental stages of R. venosa.
| FPKM | ||||||
|---|---|---|---|---|---|---|
| Gene_id | C1 | D2 | C3 | J4 | Y5 | NR/Swissprot Description |
| Growth related genes | ||||||
| c145901_g1 | 28.17 ± 4.11 | 4.89 ± 0.20 | 4.83 ± 0.74 | 1.63 ± 0.17 | 1.13 ± 0.38 | Growth/differentiation factor 8 |
| c137141_g1 | 1.58 ± 0.77 | 0.11 ± 0.19 | 0.10 ± 0.18 | 51.52 ± 6.43 | 145.47 ± 9.43 | Fibropellin |
| c100045_g1 | 1.64 ± 0.16 | 0.65 ± 0.06 | 0.74 ± 0.41 | 0.17 ± 0.02 | 0.34 ± 0.07 | Nodal |
| c145993_g2 | 3.98 ± 1.76 | 0.97 ± 0.60 | 0.32 ± 0.56 | 50.06 ± 2.90 | 1.41 ± 0.04 | Actin |
| c146789_g2 | 9.02 ± 0.17 | 5.68 ± 0.64 | 5.68 ± 0.04 | 13.56 ± 0.67 | 13.01 ± 0.95 | Epidermal growth factor receptor |
| Nervous system related genes | ||||||
| c134668_g1 | 4.14 ± 0.32 | 2.12 ± 0.26 | 1.78 ± 0.24 | 2.32 ± 0.39 | 0.89 ± 0.29 | 5-Hydroxytryptamine receptor 1 |
| c138434_g1 | 2.58 ± 0.42 | 3.09 ± 0.08 | 3.44 ± 0.63 | 0.28 ± 0.20 | 0.00 ± 0.00 | Neuronal acetylcholine receptor |
| c156806_g2 | 4.76 ± 0.11 | 3.67 ± 0.14 | 3.39 ± 0.51 | 2.45 ± 0.04 | 1.22 ± 0.36 | Nitric oxide synthase |
| c90120_g1 | 71.07 ± 2.83 | 50.29 ± 1.91 | 49.18 ± 1.55 | 46.61 ± 1.98 | 101.93 ± 4.53 | GABA(A) receptor-associated protein |
| Digestive system related genes | ||||||
| c124801_g1 | 2083.42 ± 193.38 | 8288.19 ± 320.86 | 9144.35 ± 631.79 | 2226.16 ± 100.74 | 0.13 ± 0.23 | Conotoxin |
| c150903_g1 | 65.50 ± 5.18 | 70.21 ± 3.19 | 69.11 ± 7.10 | 2.64 ± 0.34 | 1.59 ± 0.09 | Exoglucanase XynX |
| c154739_g1 | 35.47 ± 1.81 | 526.05 ± 13.37 | 595.43 ± 28.38 | 22.31 ± 0.91 | 0.05 ± 0.01 | Endoglucanase E-4 |
| c95470_g1 | 3989.92 ± 168.41 | 11536.75 ± 123.56 | 13857.77 ± 358.89 | 2519.76 ± 154.86 | 3.09 ± 1.45 | vdg3 |
| c105120_g1 | 1.95 ± 0.37 | 12.40 ± 0.24 | 17.84 ± 3.84 | 1695.11 ± 50.76 | 6247.54 ± 525.34 | Developmentally-regulated vdg3 |
| c128291_g1 | 0.04 ± 0.03 | 0.02 ± 0.03 | 0.06 ± 0.06 | 29.25 ± 1.51 | 221.63 ± 15.27 | Trypsin |
| c147105_g1 | 0.30 ± 0.26 | 0.10 ± 0.10 | 0.00 ± 0.00 | 91.37 ± 7.91 | 187.18 ± 25.41 | Carboxypeptidase B |
| Immune system related genes | ||||||
| c115222_g1 | 4.27 ± 1.29 | 12.00 ± 0.68 | 12.61 ± 0.66 | 6.71 ± 0.37 | 36.95 ± 0.15 | Tumor Necrosis Factor |
| c137778_g1 | 0.62 ± 0.27 | 0.95 ± 0.17 | 1.35 ± 0.78 | 4.70 ± 0.98 | 163.68 ± 6.87 | Defensin |
| c149483_g1 | 3.29 ± 0.34 | 10.01 ± 2.01 | 10.32 ± 1.35 | 5.20 ± 0.89 | 24.37 ± 4.74 | toll2 |
| Apoptosis related genes | ||||||
| c132048_g1 | 22.78 ± 0.90 | 30.65 ± 1.10 | 26.18 ± 0.92 | 42.81 ± 2.64 | 14.86 ± 0.34 | Apoptosis-inducing factor 1 |
| c135194_g1 | 0.76 ± 0.10 | 0.61 ± 0.06 | 0.78 ± 0.10 | 2.54 ± 0.25 | 1.25 ± 0.19 | Caspase-7 |
| c147256_g2 | 18.34 ± 0.12 | 15.66 ± 0.35 | 15.15 ± 0.55 | 15.41 ± 0.52 | 7.53 ± 0.73 | Caspase-3 |
| c151900_g1 | 2.52 ± 0.69 | 5.06 ± 0.56 | 4.23 ± 0.88 | 4.61 ± 0.65 | 10.52 ± 0.62 | Apoptosis 2 inhibitor |
| Others | ||||||
| c116117_g1 | 7.06 ± 1.78 | 64.45 ± 4.37 | 52.10 ± 2.14 | 15.95 | 99.14 ± 9.69 | Calmodulin |
| c125727_g1 | 0.49 ± 0.12 | 0.37 ± 0.03 | 0.63 ± 0.24 | 62.61 | 3.70 ± 0.06 | Sulfotransferase 1C2 |
| c141012_g1 | 6.10 ± 0.55 | 6.37 ± 0.75 | 6.54 ± 0.97 | 70.58 | 8.93 ± 1.33 | Sulfotransferase 1A2 |
| c112229_g1 | 267.04 ± 8.16 | 223.74 ± 0.946 | 252.36 ± 11.47 | 53.19 ± 1.99 | 5.86 ± 0.62 | SARP-19 |
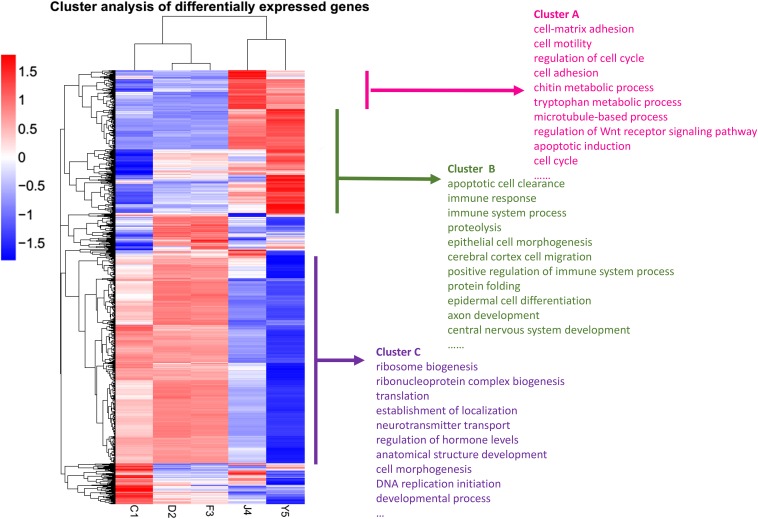
To estimate expression patterns of DEGs under different developmental conditions, 1794 genes selected according to criteria detailed in Materials and Methods were clustered hierarchically. The expression profiles of the 1794 genes are displayed in Figure 4, and were considered reliable by strict control of the p-value and FPKM. The information from the horizontal axis of the gene clustering dendrogram indicated that the developmental stages were aggregated into two distinct clusters. The one-, two- and three-spiral whorls stages formed one cluster; and the four-spiral whorls stage (competent larva), and postlarval stage formed another. The vertical axis of the dendrogram showed that each developmental stage contained genes that were specifically and abundantly expressed. These stage-specific genes predominantly distributed in the two- or three-spiral whorls stages, four-spiral whorls stages, and postlarval stages. For example, expression of genes in cluster A remained quite low during the precompetent larval stage, but increased rapidly in the competent larval stage, and then reduced after metamorphosis. And genes were abundantly expressed in the postlarval stage in cluster B. In cluster C, genes were abundantly expressed during the precompetent larval stage (one-, two- and three-spiral whorl stage), but decreased significantly in the following developmental stages (four-spiral whorl stage, and postlarval stage). More detailed information on these genes of different clusters is available in Table S1. This analysis of gene expression patterns will help discover genes playing a pivotal role in controlling early developmental processes in R. venosa.
Figure 4.
Clusters of expression levels of the candidate genes. Dendrograms showing the gene expression patterns for 1794 genes. The clustering indicates similar expression patterns among the developmental stages (x-axis) and among the genes (y-axis). The bar color reflects the gene expression level from blue (low) to white (medium) to red (high). For each cluster, some of the enriched terms of biological process in GO enrichment are listed.
GO enrichment and KEGG enrichment of upregulated DEGs after metamorphosis
To further understand the function of DEGs, GO term enrichment analysis, and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment were performed. We focused mainly on the DEGs of the J4 and Y5 comparison, because metamorphosis, which involves velum degeneration, diet transformation, growth of the secondary shell, and other morphological and ecological habit changes, is the critical point of the R. venosa life cycle. For genes that were upregulated in Y5, “RNA-dependent DNA replication” (72 DEGs), “immune system process” (51 DEGs), and “immune response” were most enriched in the biological process (GOBP) group (red in Figure 5). GO enrichment analysis of the other developmental comparison groups is shown in Figure S2.
Figure 5.
Bar graph of GO enrichment of genes that were upregulated after metamorphosis. The horizontal axis shows the GO term in the next level of the three main GO classifications; the vertical axis shows the number of the differentially expressed genes annotated in the GO term, and the total number of annotated differentially expressed genes. The three main GO classifications are (from left to right): biological process, cell composition, and molecular function.
Gene products usually interact with each other in vivo to play roles in certain biological functions. KEGG pathway enrichment was carried out to identify DEGs involved in the main biochemical pathways and signal transduction pathways. Scatterplots of DEGs showed the analysis results of KEGG pathway enrichment. We display the top 20 significantly enriched pathways of the J4 vs. Y5 comparison in Figure 6. KEGG enrichment analyses of other developmental comparison groups are shown in Figure S3. Some immune-related pathways, including “Toll-like receptor (TLR) signaling,” “bacterial invasion of epithelial cells,” and “Leukocyte transendothelial migration,” were highly enriched as upregulated pathways after metamorphosis.
Figure 6.
KEGG enrichment scatterplots of the upregulated genes after metamorphosis. The vertical axis represents the pathway name; the horizontal axis represents the corresponding enrichment factor. The q-value is corresponds to a points’ color: the redder the color, the smaller the q-value. Finally, a points’ size indicates the number of DEG in each pathway.
qPCR validation
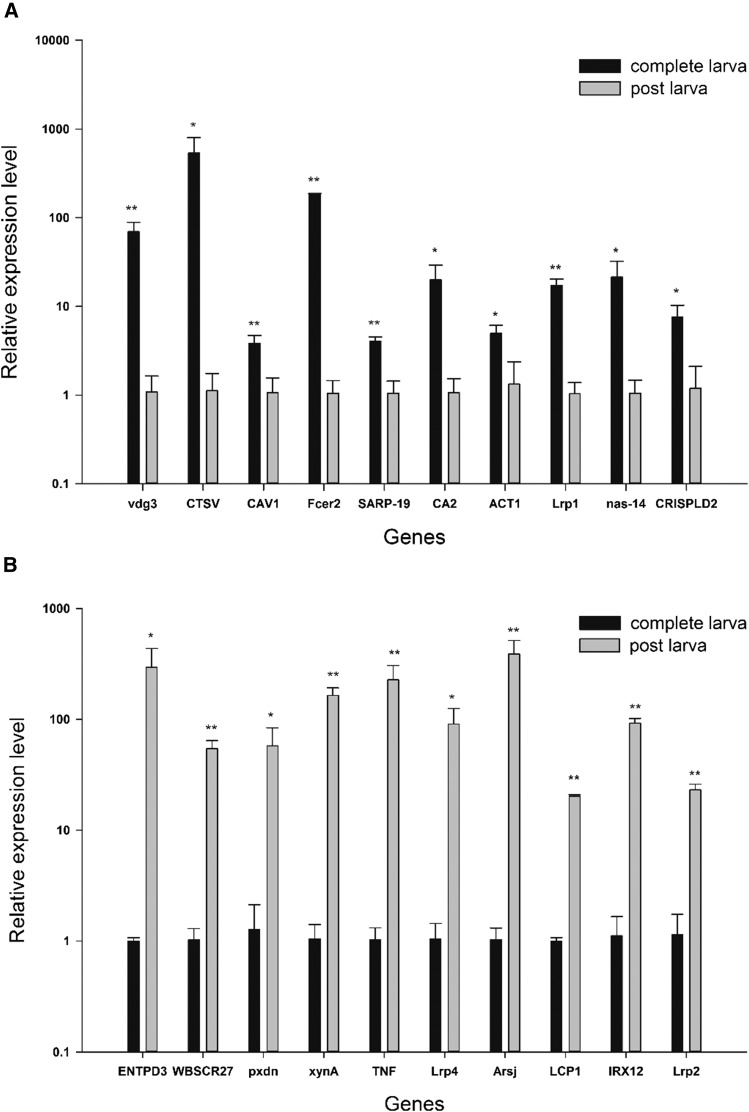
To validate the RNA-seq results, 20 DEGs between J4 and Y5 were selected and analyzed by qRT-PCR, including the 10 most upregulated DEGs, and the 10 most downregulated DEGs. Results in Figure 7 show that the DGE tag profiling had a good consistency with the qRT-PCR results, although the fold changes in expression differed substantially in some cases, likely caused by differences in the sensitivity of estimating gene expression (Zhao et al. 2014).
Figure 7.
Relative expression levels for 20 differentially expressed genes in competent larva and postlarval stages, as determined by qRT-PCR. (A) Ten genes that were downregulated after metamorphosis. vdg3, vdg3; CTSV, cathepsin L2; CAV, caveolin-1; Fcer2, low affinity immunoglobulin epsilon Fc receptor; SARP-19, SARP-19; CA2, carbonic anhydrase 2; ACT1, actin; Lrp1, prolow-density lipoprotein receptor-related protein 1; nas-14, zinc metalloproteinase nas-14; and CRISPLD2, cysteine-rich secretory protein LCCL domain-containing 2. (B) Ten genes that were upregulated after metamorphosis. ENTPD3, ectonucleoside triphosphate diphosphohydrolase 3; WBSCR27, Williams-Beuren syndrome chromosomal region 27 protein; pxdn, peroxidasin; xynA, beta-1,4-xylanase; TNF, tumor necrosis factor; Lrp4, low-density lipoprotein receptor-related protein 4; Arsj, arylsulfatase J; LCP1, digestive cysteine proteinase 1; IRX12, Laccase-4; Lrp2, low-density lipoprotein receptor-related protein 2. Values are the mean ± SD (n = 3). * P < 0.05, ** P < 0.01.
Discussion
A knowledge of the developmental biology of R. venosa is essential for scientific research, invasion control, and aquaculture. To promote work in this field, we constructed a digital gene expression profile on the different larval stages of R. venosa, and thus could globally identify genes participating in important early developmental bioprocesses.
There are visible morphological and body size differences between these five stages. We observed that planktonic larvae (C1, D2, F3, and J4) could swim freely and feed on microalgae via their ciliary structure velum. The competent larvae (J4) of R. venosa quickly undergo metamorphosis when they find suitable habitats, with specific cues being most mollusk-like. Thus, we speculated that the nervous system participates in reacting with chemical cues, and activating metamorphosis. This pelagic-to-benthic transition took only 3 d, but involved complicated morphological and physiological changes. First, the velum for swimming and ingestion was degenerated and reabsorpbed during metamorphic transition. Second, secondary shell, foot, and extensile tentacles with eyes develop further in postlarvae. Third, phytophagous pelagic larvae develop into carnivorous juveniles. Based on these morphological and physiological changes, DEGs related to growth, nervous system, digestive system, immune system, and apoptosis were considered to play important roles in R. venosa development, especially in metamorphosis. Thus, a preliminary differentiated analysis of these interesting DEGs was performed, revealing that several genes had quite different expression patterns at different stages, as displayed in Table 3. Some useful information can be obtained as follows.
Table 3. Genes selected for real-time PCR validation and the fold change in the postlarval stage (Y5) compared with the four-spiral whorls stage (competent larvae J4) for both RNA-seq and RT-PCR analysis.
| log2.fold_change (Y5 vs. J4) | |||||
|---|---|---|---|---|---|
| Gene ID | Gene Name | Exp | RNA-Seq | RT-PCR | Same Trend |
| c95470_g1 | vdg3 | ↓ | −9.55** | −6.00** | Y |
| c131243_g1 | CTSV | ↓ | −8.40** | −8.90* | Y |
| c139260_g1 | CAV1 | ↓ | −7.39** | −1.84** | Y |
| c129336_g1 | Fcer2 | ↓ | −6.30** | −7.46** | Y |
| c125484_g1 | SARP-19 | ↓ | −5.75** | −2.01** | Y |
| c128565_g1 | CA2 | ↓ | −5.63** | −4.23* | Y |
| c145993_g2 | ACT1 | ↓ | −5.02** | −1.91* | Y |
| c127784_g1 | Lrp1 | ↓ | −4.71** | −4.06** | Y |
| c145412_g1 | nas-14 | ↓ | −4.59** | −4.35* | Y |
| c119967_g1 | CRISPLD2 | ↓ | −4.29** | −2.67* | Y |
| c150152_g1 | ENTPD3 | ↑ | 4.56** | 8.20* | Y |
| c113545_g2 | WBSCR27 | ↑ | 4.50** | 5.73** | Y |
| c94349_g1 | pxdn | ↑ | 4.49** | 5.50* | Y |
| c156029_g2 | xynA | ↑ | 4.43** | 7.30** | Y |
| c117375_g2 | TNF | ↑ | 4.38** | 7.79** | Y |
| c147483_g1 | Lrp4 | ↑ | 4.34** | 6.44* | Y |
| c149073_g1 | Arsj | ↑ | 4.31** | 8.56** | Y |
| c143535_g1 | LCP1 | ↑ | 3.89** | 4.33** | Y |
| c135964_g1 | IRX12 | ↑ | 3.85** | 6.37** | Y |
| c118662_g1 | Lrp2 | ↑ | 3.81** | 4.34** | Y |
Genes with the same trend (up or downregulation) as shown by both RNA-seq and RT-PCR analysis are indicated by “Y” (yes). * P < 0.05, ** P < 0.01 (significant difference between J4 and Y5 stages). Exp, expression trend; “↑”upregulated expression; “↓” downregulated expression.
Growth
The abundance levels of Actin (c145993_g2) increased by nearly 150-fold when the larvae transformed from precompetent larvae into competent larva. Expression dropped by 36-fold from the competent larval stage to the postlarval stage. The actin protein forms microfilaments, and functions in retraction and resorption of the sand dollar larval arms during metamorphosis (Burke 1985). The competent stage-specific expression of these mRNAs might be important for the initiation of metamorphosis. The expression of fibropellin-1 was detected at a low level in precompetent larvae, but increased significantly in competent larvae, and was further upregulated after metamorphosis. Fibropellin-1 was first identified in the sea urchin, and is also called epidermal growth factor-related protein 1. It is thought to regulate species-specific signal transduction pathways in the early development of sea urchin, and loss of fibropellin-1 results in development defects (Kamei et al. 2000; Yang et al. 1989).
Nervous system
The expression level of 5-hydroxytryptamine receptor, neuronal acetylcholine receptor, and nitric oxide synthase were found declined after metamorphosis. Serotonin and its receptors are involved in neuronal functions in mollusks, including the circadian rhythm of Aplysia californica (Levenson et al. 1999), locomotion of Lymnaea stagnalis (Filla et al. 2004), feeding of L. stagnalis (Kawai et al. 2011), and development of Haliotis rubra (Panasophonkul et al. 2009). Pharmacological and ecological experiments suggested that the 5-hydroxytryptamine receptor mediates settlement and metamorphosis of many mollusks, such as Ilyanassa obsolete (Leise et al. 2001) and Helisoma trivolvis (Glebov et al. 2014). The expression level of the 5-HT receptor was downregulated in H. trivolvis during the transition from the premetamorphic stage to the metamorphic stage (Glebov et al. 2014). Nitric oxide synthase, and its synthetic product nitric oxide (NO), were purported to play a part in metamorphosis of mollusks (Laudet et al. 2013). It was also observed in the mud snail I. obsolete that expression of NOS was downregulated after the initiation of metamorphosis (Froggett and Leise 1999). The suppression of NOS activity by NOS-inhibiting pharmacological agents has been reported to induce the initiation of metamorphosis, leading to the hypothesis that NOS plays a negative regulatory role (Froggett and Leise 1999). Against that, NOS and NO were considered to be positive regulators for the initiation of metamorphosis in some gastropods, such as H. asinina (Ueda and Degnan 2014).
Digestive system
Consistently, we also identified genes that are involved in the histogenesis and digestion processes of the digestive system. The expression level of developmentally regulated vdg3 (c105120_g1) increased rapidly during the development of R. venosa, and was significantly upregulated after metamorphosis. This expression pattern was similar to its counterparts in H. diversicolor and H. asinina, which are specifically expressed in the digestive tract and glands, and increased by over 10-fold when larvae undergo the transition from pelagic to benthic lifestyle, suggesting that the gene is involved in gut morphogenesis and digestion (He et al. 2014; Jackson et al. 2005). Interestingly, expression pattern of vdg3 (c95470_g1) is the complete opposite of the developmentally regulated vdg3. We speculated that these two genes have antagonistic effects on the regulation of development and metamorphosis in R. venosa. The expression pattern of vdg3 was also contrary to its counterpart in H. diversicolor (He et al. 2014); therefore, this DGE profile is an interesting phenomenon that requires further study.
The carnivorous digestive enzymes trypsin (c128291_g1) and carboxypeptidase B (c147105_g1) had an extremely low expression level in the precompetent larval stage, a slight increase in the competent larva stage, and reached a very high level in the postlarva stage. Trypsin is a type of serine protease. In the digestive system of many vertebrates, trypsin participates in the hydrolysis of protein. Carboxypeptidase B is known as a protease enzyme that can cleave a peptide bond at the C-terminal end of proteins or peptides. A high level of these enzymes in the post larval stage indicates that the digestion process is active. Instead, the gene expression level of some phytophagous digestive enzymes such as exoglucanase (c150903_g1) and endoglucanase (c154739_g1) remained at a high level in precompetent larvae, but obviously dropped when larvae went through metamorphic transition. The pelagic larvae of R. venosa (precompetent and competent) feed on microalgae, while the juveniles prey on bivalve mollusks. We hypothesized that the upregulation of these proteases is responsible for the change in diet after metamorphosis, and that competent larvae is an important interim period during which the expression of different kinds of digestive enzymes is regulated.
Conotoxin (c124801_g1) was found highly expressed in the pelagic larval stage, and quickly declined to almost zero. Conotoxin is one of a group of neurotoxic peptides isolated from the venom of mollusks of the genus Conus, which enables the marine cone snail to carry out its unique predatory lifestyle (Olivera et al. 1991). Conotoxins have promising applications as drugs (Terlau and Olivera 2004). The abundant expression of conotoxin in this study indicated that the pelagic larvae of R. venosa and Conus were homologous, but conotoxin in R. venosa might degenerate after metamorphosis. The conotoxin of R. venosa warrants further research, because there is no report of conotoxins in this species.
Immune system
Many immune-related genes, such as toll 2 (c149483_g1), defensin (c137778_g1), and tumor necrosis factor (c115222_g1), were also abundantly expressed during post larval stage. We speculated that the benthic larvae are more exposed to pathogens when acquiring food in their environment, therefore a stronger immune response is stimulated (Huang et al. 2012). Furthermore, “immune system process” and “immune response” items were significantly upregulated in the GOBP group of GO enrichment, and “alpha-l-fucosidase activity” and “fucosidase activity” were significantly upregulated in the GOMF group. Fucosidase and peptidase were reported to function in the digestive system, as well as playing an important role in host/parasite interactions in Biomphalaria glabrata (Perrella et al. 2015). This may explain the changes in the immune response after the metamorphic conversion from microalgae diet to the bivalve diet of R. venosa. We estimated that the immune response changes before and after metamorphosis were significantly different because the benthic post larva feeds on bivalves, which are more susceptible to parasites than planktonic competent larva feeding on microalgae. Besides, the KEGG enrichment also showed that the TLR signaling pathway was the most significantly enriched (q-value = 7.39e–07). The TLR is a member of pattern-recognition receptors (PRRs), and TLR, together with its signaling pathway, play a critical role in recognizing various pathogen-associated molecular patterns, and represent the front line of defense against invading pathogens (Wang et al. 2011). Interestingly, other immune-related pathways, including “bacterial invasion of epithelial cells,” and “leukocyte transendothelial migration,” were highly enriched in the postlarval stage, indicating that the changes in the immune system were an important first-line defense in the response against the transformation of habitat and predatory feeding habit after metamorphosis. Metamorphosis of M. galloprovincialis was suggested as a pivotal stage when postlarvae respond to environmental signals and upregulated the expression level of immune system-related genes (Balseiro et al. 2013). The expression of most genes involved in immune response in the postlarval stage was significantly upregulated compared to oocytes, trochophores and veligers, suggesting active immune activity in the mussel postlarvae stage. This pattern of gene expression is always followed by the expression of some immune or nonimmune related proteins, based on previous ontogeny studies in mollusks (Ge et al. 2011; Tirapé et al. 2007). In the ascidian Boltenia villosa, immune-related genes were also upregulated during metamorphosis, highlighting the specific role of innate immunity during metamorphosis (Davidson and Swalla 2002). This observed upregulation during metamorphosis might simply reflect immune system maturation, but might also involve the resorption and reorganization of larval tissues, or involve the larvae’s ability to detect and respond to bacterial cues for settlement and metamorphosis (Balseiro et al. 2013).
Apoptosis
As a distinguishing feature of metamorphosis in R. venosa, the velum of competent larvae will be lost by programmed cell death. Therefore, we investigated the expression pattern of some apoptosis genes, as shown in Table 2. The apoptosis inducer (c132048_g1) and executors (c135194_g1, c147256_g2) were found to be downregulated when switched from competent to postlarvae. And the apoptosis 2 inhibitor (c151900_g1) was upregulated after metamorphosis. This conformed well to the morphological transformation during metamorphosis in R. venosa. Many studies have shown that caspases are significantly expressed in larvae such as those of C. angulate (Yang et al. 2012), M. galloprovincialis (Romero et al. 2011), and Ilyanassa obsolete (Gifondorwa and Leise 2006), prior to metamorphosis, suggesting caspases play an important role in larval settlement and metamorphosis of mollusks.
Others
The expression of calmodulin (c116117_g1) was at a low level in competent larval stages, but increased significantly after metamorphosis. It is considered a multifunctional metabolic regulator that can interact with diverse signaling proteins, including phosphatases, kinases, and membrane receptors. Calmodulin and its associated binding proteins play important roles in many biological processes, such as cell growth, neuronal development, and bacterial pathogenesis (O’Day 2003). This protein showed high expression in 12 hr postmetamorphic juveniles of Hydroides elegans, and remained at a high level in adults. In situ hybridization revealed that, in putative growth zones, the calmodulin gene was continuously expressed, indicating its function in tissue differentiation and development (Chen et al. 2012). It also was reported to participate in the biomineralization process in Pinctada fucata (Fang et al. 2008).
c112229_g1 is highly similar to SARP 19, which was discovered in the marine snail Littorina littorea as a protein function in the anoxia response of the snail (Larade and Storey 2004). In contrast to this study, SARP 19 increased during the development of H. diversicolor, and its expression showed a sharp increase after settlement (He et al. 2014).
In addition, c125727_g1 and c141012_g1, which were highly similar to Sulfotransferase 1C2 and Sulfotransferase 1A2, respectively, had a stage-specific high expression at competent larval stages. Using 3′-phosphoadenosine-5′-phosphosulfate as the most common sulfo group donor, the sulfotransferase 1 family functions in detoxification of xenobiotics, and modulation of endogenous hormones, bile acids, and neurotransmitter activities, such as estrogens, iodothyronines, and catecholamines (Blanchard et al. 2004; Gamage et al. 2006). These proteins are suggested to participate in development and metamorphosis in the bullfrog, Rana catesbeiana (Rahman and Yamauchi 2010); however, there is little information on their function in gastropods.
To estimate expression patterns of DEGs under different developmental conditions, 1794 genes selected according to the criteria detailed above were clustered hierarchically. Through clustering genes with similar expression patterns, groups of genes with similar patterns but unknown functions could be recognized, because transcripts with similar expression patterns might have similar roles in the same cellular pathways or metabolic processes. The information from the horizontal axis of the gene clustering dendrogram indicated that the developmental stages were aggregated into two distinct clusters. The one-, two- and three-spiral whorls stages formed one cluster, and the four-spiral whorls stage (competent larva) and postlarval stage formed another. This clustering pattern is in accordance with Haliotis diversicolor (Huang et al. 2012) and H. asinina (Williams et al. 2009). This also explained the anticipatory development phenomenon that some postlarval structures and organs were actually formed before metamorphosis (Degnan and Morse 1995), and also indicated that the later competent larvae require more specific transcripts preparing for settlement and metamorphosis than precompetent larvae.
Conclusions
Research on the larval development and metamorphosis mechanisms of R. venosa remains scarce due to limited genomic resources. Thus, in a previous work, we constructed a comprehensive transcriptome database by de novo sequencing of a mixed RNA pool from pelagic larvae and juveniles (Song et al. 2016). This work identified 212,049 unigenes, of which 70,877 were functionally annotated. However, it did not reveal the dynamic changes and expression patterns of different genes.
Using the previous transcriptome database as reference, in this study we generated a transcriptomic DGE sequencing strategy with a careful design and strict quality controls, and constructed a global gene expression profile of R. venosa during early development, from planktonic larval stages to the post larval stage. Our global DGE profiles provided the useful temporal dynamics of the expressions of thousands of genes. It is the first digital gene profile to cover the early developmental stages of R. venosa. By cluster analysis, we also identified a series of candidate genes that might play an important role in early development of R. venosa, especially in metamorphosis regulation. Interestingly, we observed that some DEGs associated with growth, nervous system, digestive system, immune system, and apoptosis participate in important developmental processes. These data provide significant clues for a better understanding of the regulatory mechanisms of development in R. venosa, and identified some key genes worthy of further research.
Supplementary Material
Acknowledgments
The authors thank Mu-Yan Chen of the Fisheries College of Ocean University of China and Bao-Zhong Liu of the Institute of Oceanology, Chinese Academy of Sciences for suggesting experiments. This research was supported through a project supported by the National Natural Science Foundation of China (grant no. 31572636), the National Natural Science Foundation of China-Shandong Joint Fund for Marine Science Research Centers (Grant No.U1406403), the National Key Technology R&D Program of the Ministry of Science and Technology (grant no. 2011BAD13B01), and the Agricultural Major Application Technology Innovation Project of Shandong Province. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no conflict of interest. The founding sponsors had no role in the design of the study.
Author contributions: T.Z. and H.W. conceived and designed the experiments. H.S. and Z.Y. performed the experiments. H.S. analyzed the data. D.X., T.Z., and H.W. contributed reagents, materials, and analysis tools. H.S. and L.S. wrote the paper.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.029314/-/DC1
Communicating editor: W. S. Davidson
Literature Cited
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11(10): R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge W. J., 2009. Next-generation DNA sequencing techniques. Nat. Biotechnol. 25(4): 195–203. [DOI] [PubMed] [Google Scholar]
- Badariotti F., Thuau R., Lelong C., Dubos M.-P., Favrel P., 2007. Characterization of an atypical family 18 chitinase from the oyster Crassostrea gigas: evidence for a role in early development and immunity. Dev. Comp. Immunol. 31(6): 559–570. [DOI] [PubMed] [Google Scholar]
- Balseiro P., Moreira R., Chamorro R., Figueras A., Novoa B., 2013. Immune responses during the larval stages of Mytilus galloprovincialis: metamorphosis alters immunocompetence, body shape and behavior. Fish Shellfish Immunol. 35(2): 438–447. [DOI] [PubMed] [Google Scholar]
- Biscotti M.A., Canapa A., Forconi M., Barucca M., 2014. Hox and ParaHox genes: a review on molluscs. Genesis 52(12): 935–945. [DOI] [PubMed] [Google Scholar]
- Blanchard R. L., Freimuth R. R., Buck J., Weinshilboum R. M., Coughtrie M. W., 2004. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenet. Genomics 14(3): 199–211. [DOI] [PubMed] [Google Scholar]
- Burke R. D., 1985. Actin-mediated retraction of the larval epidermis during metamorphosis of the sand dollar, Dendraster excentricus. Cell Tissue Res. 239(3): 589–597. [DOI] [PubMed] [Google Scholar]
- Chen Z.-F., Wang H., Qian P.-Y., 2012. Characterization and expression of calmodulin gene during larval settlement and metamorphosis of the polychaete Hydroides elegans. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 162(4): 113–119. [DOI] [PubMed] [Google Scholar]
- Clark M. S., Thorne M. A., Vieira F. A., Cardoso J. C., Power D. M., et al. , 2010. Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genomics 11(1): 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft J. A., Gilbert J. A., Temperton B., Dempsey K. E., Ashelford K., et al. , 2010. Pyrosequencing of Mytilus galloprovincialis cDNAs: tissue-specific expression patterns. PLoS One 5(1): e8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çulha M., Bat L., Doğan A., Dağlı E., 2009. Ecology and distribution of the veined rapa whelk Rapana venosa (Valenciennes, 1846) in Sinop peninsula (Southern Central Black Sea), Turkey. J. Anim. Vet. Adv. 8(1): 51–58. [Google Scholar]
- Davidson B., Swalla B. J., 2002. A molecular analysis of ascidian metamorphosis reveals activation of an innate immune response. Development 129(20): 4739–4751. [DOI] [PubMed] [Google Scholar]
- Degnan B. M., Morse D. E., 1995. Developmental and morphogenetic gene regulation in Haliotis rufescens larvae at metamorphosis. Integr. Comp. Biol. 35(4): 391–398. [Google Scholar]
- De Lorgeril J., Zenagui R., Rosa R. D., Piquemal D., Bachère E., 2011. Whole transcriptome profiling of successful immune response to Vibrio infections in the oyster Crassostrea gigas by digital gene expression analysis. PLoS One 6(8): e23142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Yan Z., Li S., Wang Q., Cao W., et al. , 2008. Localization of calmodulin and calmodulin-like protein and their functions in biomineralization in P. fucata. Prog. Nat. Sci. 18(4): 405–412. [Google Scholar]
- Filla A., Hiripi L., Elekes K., 2004. Serotonergic and dopaminergic influence of the duration of embryogenesis and intracapsular locomotion of Lymnaea stagnalis L. Acta Biol. Hung. 55(1–4): 315–321. [DOI] [PubMed] [Google Scholar]
- Fleury E., Huvet A., Lelong C., De Lorgeril J., Boulo V., et al. , 2009. Generation and analysis of a 29,745 unique expressed sequence tags from the Pacific oyster (Crassostrea gigas) assembled into a publicly accessible database: the GigasDatabase. BMC Genomics 10(1): 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini P., van der Merwe M., Roodt-Wilding R., 2011. Transcriptome characterization of the South African abalone Haliotis midae using sequencing-by-synthesis. BMC Res. Notes 4: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froggett S. J., Leise E. M., 1999. Metamorphosis in the marine snail Ilyanassa obsoleta, yes or NO? Biol. Bull. 196(1): 57–62. [DOI] [PubMed] [Google Scholar]
- Gamage N., Barnett A., Hempel N., Duggleby R. G., Windmill K. F., et al. , 2006. Human sulfotransferases and their role in chemical metabolism. Toxicol. Sci. 90(1): 5–22. [DOI] [PubMed] [Google Scholar]
- Ge H., Wang G., Zhang L., Zhang Z., Wang S., et al. , 2011. Molecular cloning and expression of interleukin-1 receptor-associated kinase 4, an important mediator of Toll-like receptor signal pathway, from small abalone Haliotis diversicolor. Fish Shellfish Immunol. 30(4): 1138–1146. [DOI] [PubMed] [Google Scholar]
- Giberto D. A., Bremec C. S., Schejter L., Schiariti A., Mianzan H., et al. , 2006. The invasive Rapa Whelk Rapana venosa (Valenciennes 1846): status and potential ecological impacts in the Río de la Plata estuary, Argentina-Uruguay. J. Shellfish Res. 25(3): 919–924. [Google Scholar]
- Gifondorwa D. J., Leise E. M., 2006. Programmed cell death in the apical ganglion during larval metamorphosis of the marine mollusc Ilyanassa obsoleta. Biol. Bull. 210(2): 109–120. [DOI] [PubMed] [Google Scholar]
- Glebov K., Voronezhskaya E. E., Khabarova M. Y., Ivashkin E., Nezlin L. P., et al. , 2014. Mechanisms underlying dual effects of serotonin during development of Helisoma trivolvis (Mollusca). BMC Dev. Biol. 14: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding J. M., Kingsley-Smith P., Savini D., Mann R., 2007a Comparison of predation signatures left by Atlantic oyster drills (Urosalpinx cinerea Say, Muricidae) and veined rapa whelks (Rapana venosa Valenciennes, Muricidae) in bivalve prey. J. Exp. Mar. Biol. Ecol. 352(1): 1–11. [Google Scholar]
- Harding J. M., Mann R., Kilduff C. W., 2007b The effects of female size on fecundity in a large marine gastropod Rapana venosa (Muricidae). J. Shellfish Res. 26(1): 33–42. [Google Scholar]
- Harding J. M., Mann R., Kilduff C. W., 2008. Influence of environmental factors and female size on reproductive output in an invasive temperate marine gastropod Rapana venosa (Muricidae). Mar. Biol. 155(6): 571–581. [Google Scholar]
- He T.-F., Chen J., Zhang J., Ke C.-H., You W.-W., 2014. SARP19 and vdg3 gene families are functionally related during abalone metamorphosis. Dev. Genes Evol. 224(4–6): 197–207. [DOI] [PubMed] [Google Scholar]
- Hou R., Bao Z., Wang S., Su H., Li Y., et al. , 2011. Transcriptome sequencing and de novo analysis for Yesso scallop (Patinopecten yessoensis) using 454 GS FLX. PLoS One 6(6): e21560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. X., Chen Z. S., Ke C. H., Zhao J., You W. W., et al. , 2012. Pyrosequencing of Haliotis diversicolor transcriptomes: insights into early developmental molluscan gene expression. PLoS One 7(12): e51279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. J., Ellemor N., Degnan B. M., 2005. Correlating gene expression with larval competence, and the effect of age and parentage on metamorphosis in the tropical abalone Haliotis asinina. Mar. Biol. 147(3): 681–697. [Google Scholar]
- Kamei N., Swanson W. J., Glabe C. G., 2000. A rapidly diverging EGF protein regulates species-specific signal transduction in early sea urchin development. Dev. Biol. 225(2): 267–276. [DOI] [PubMed] [Google Scholar]
- Kawai R., Kobayashi S., Fujito Y., Ito E., 2011. Multiple subtypes of serotonin receptors in the feeding circuit of a pond snail. Zoolog. Sci. 28(7): 517–525. [DOI] [PubMed] [Google Scholar]
- Lanfranconi A., Hutton M., Brugnoli E., Muniz P., 2009. New record of the alien mollusc Rapana venosa (Valenciennes 1846) in the Uruguayan coastal zone of Río de la Plata. Pan-Am. J. Aquat. Sci. 4(2): 216–221. [Google Scholar]
- Larade K., Storey K. B., 2004. Anoxia-induced transcriptional upregulation of sarp-19: cloning and characterization of a novel EF-hand containing gene expressed in hepatopancreas of Littorina littorea. Biochem. Cell Biol. 82(2): 285–293. [DOI] [PubMed] [Google Scholar]
- Laudet V., Ueda N., Degnan S. M., 2013. Nitric oxide acts as a positive regulator to induce metamorphosis of the ascidian Herdmania momus. PLoS One 8(9): e72797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leise E. M., Froggett S. J., Nearhoof J. E., Cahoon L. B., 2009. Diatom cultures exhibit differential effects on larval metamorphosis in the marine gastropod Ilyanassa obsoleta (Say). J. Exp. Mar. Biol. Ecol. 379(1): 51–59. [Google Scholar]
- Leise E. M., Thavaradhara K., Durham N. R., Turner B. E., 2001. Serotonin and nitric oxide regulate metamorphosis in the marine snail Ilyanassa obsoleta. Am. Zool. 41(2): 258–267. [Google Scholar]
- Leppäkoski E., Gollasch S., Olenin S., 2013. Invasive Aquatic Species of Europe. Distribution, Impacts and Management, Springer Science & Business Media, New York. [Google Scholar]
- Levenson J., Byrne J. H., Eskin A., 1999. Levels of serotonin in the hemolymph of aplysia are modulated by light/dark cycles and sensitization training. J. Neurosci. 19(18): 8094–8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N., 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12(1): 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ren F., Yang H., 2003. Preliminary study on the ecological characteristic of Rapana venosa (Valenciennes). Fish. Sci. 22: 17–18. [Google Scholar]
- Lu S., Bao Z., Hu J., Hu X., Mu C., et al. , 2008. mRNA differential display on gene expression in settlement metamorphosis process of Ruditapes philippinarum larvae. High Technol. Lett. 14(3): 332–336. [Google Scholar]
- Mann R., Harding J. M., 2003. Salinity tolerance of larval Rapana venosa: implications for dispersal and establishment of an invading predatory gastropod on the North American Atlantic coast. Biol. Bull. 204(1): 96–103. [DOI] [PubMed] [Google Scholar]
- Mann R., Harding J. M., Westcott E., 2006. Occurrence of imposex and seasonal patterns of gametogenesis in the invading veined rapa whelk Rapana venosa from Chesapeake Bay, USA. Mar. Ecol. Prog. Ser. 310: 129–138. [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5(7): 621–628. [DOI] [PubMed] [Google Scholar]
- O’Day D. H., 2003. CaMBOT: profiling and characterizing calmodulin-binding proteins. Cell. Signal. 15(4): 347–354. [DOI] [PubMed] [Google Scholar]
- Olivera B. M., Cruz L. J., Gray W. R., Rivier J. E. F., 1991. Conotoxins. J. Biol. Chem. 266(33): 22067–22137. [PubMed] [Google Scholar]
- Pan Y., Qiu T., Zhang T., Wang P., Ban S., 2013. Morphological studies on the early development of Rapana venosa. Journal of Fisheries of China 37(10): 1503–1510. [Google Scholar]
- Panasophonkul S., Apisawetakan S., Cummins S. F., York P. S., Degnan B. M., et al. , 2009. Molecular characterization and analysis of a truncated serotonin receptor gene expressed in neural and reproductive tissues of abalone. Histochem. Cell Biol. 131(5): 629–642. [DOI] [PubMed] [Google Scholar]
- Perrella N. N., Cantinha R. S., Nakano E., Lopes A. R., 2015. Characterization of α-l-fucosidase and other digestive hydrolases from Biomphalaria glabrata. Acta Trop. 141: 118–127. [DOI] [PubMed] [Google Scholar]
- Qian Z.-J., Kim S.-A., Lee J. S., Kim H.-J., Choi I.-W., et al. , 2012. The antioxidant and anti-inflammatory effects of abalone intestine digest, Haliotis discus hannai in RAW 264.7 macrophages. Biotechnol. Bioprocess Eng.; BBE 17(3): 475–484. [Google Scholar]
- Quilang J., Wang S., Li P., Abernathy J., Peatman E., et al. , 2007. Generation and analysis of ESTs from the eastern oyster, Crassostrea virginica Gmelin and identification of microsatellite and SNP markers. BMC Genomics 8(1): 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman F. B., Yamauchi K., 2010. Characterization of iodothyronine sulfotransferase activity in the cytosol of Rana catesbeiana tadpole tissues. Gen. Comp. Endocrinol. 166(2): 396–403. [DOI] [PubMed] [Google Scholar]
- Romero A., Estevez-Calvar N., Dios S., Figueras A., Novoa B., 2011. New insights into the apoptotic process in mollusks: characterization of caspase genes in Mytilus galloprovincialis. PLoS One 6(2): e17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savini D., Castellazzi M., Favruzzo M., Occhipinti-Ambrogi A., 2004. The alien mollusc Rapana venosa (Valenciennes, 1846; Gastropoda, Muricidae) in the northern Adriatic Sea: population structure and shell morphology. Chem. Ecol. 20(20): 411–424. [Google Scholar]
- Song H., Yu Z.-L., Zhang T., Wang H.-Y., 2016. De novo transcriptome sequencing and analysis of Rapana venosa from six different developmental stages using Hi-seq 2500. Comp. Biochem. Physiol. Part D Genomics Proteomics 17: 48–57. [DOI] [PubMed] [Google Scholar]
- Sun X., Yu H., Yu R., Li Q., 2014. Characterization of 57 microsatellite loci for Rapana venosa using genomic next generation sequencing. Conserv. Genet. Resour. 6(4): 941–945. [Google Scholar]
- Tan F.-Q., Tu H., Liang W.-J., Long J.-M., Wu X.-M., et al. , 2015. Comparative metabolic and transcriptional analysis of a doubled diploid and its diploid citrus rootstock (C. junos cv. Ziyang xiangcheng) suggests its potential value for stress resistance improvement. BMC Plant Biol. 15(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau H., Olivera B. M., 2004. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol. Rev. 84(1): 41–68. [DOI] [PubMed] [Google Scholar]
- Tirapé A., Bacque C., Brizard R., Vandenbulcke F., Boulo V., 2007. Expression of immune-related genes in the oyster Crassostrea gigas during ontogenesis. Dev. Comp. Immunol. 31(9): 859–873. [DOI] [PubMed] [Google Scholar]
- Ueda, N., 2013 A transcriptional perspective of NOS and HSP90 gene activity during phylogenetically divergent marine invertebrate metamorphosis. Ph.D. Thesis, The University of Queensland, Australia.
- Ueda N., Degnan S.M., 2014. Nitric oxide is not a negative regulator of metamorphic induction in the abalone Haliotis asinina. Front. Mar. Sci. 1: 21. [Google Scholar]
- Wang M., Yang J., Zhou Z., Qiu L., Wang L., et al. , 2011. A primitive Toll-like receptor signaling pathway in mollusk Zhikong scallop Chlamys farreri. Dev. Comp. Immunol. 35(4): 511–520. [DOI] [PubMed] [Google Scholar]
- Williams E. A., Degnan B. M., Gunter H., Jackson D. J., Woodcroft B. J., et al. , 2009. Widespread transcriptional changes pre‐empt the critical pelagic–benthic transition in the vetigastropod Haliotis asinina. Mol. Ecol. 18(5): 1006–1025. [DOI] [PubMed] [Google Scholar]
- Xue D., Zhang T., Liu J.-X., 2014. Microsatellite evidence for high frequency of multiple paternity in the marine gastropod Rapana venosa. PloS One 9: e86508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Qin J., Shi B., Han G., Chen J., et al. , 2012. Molecular characterization and functional analysis of adrenergic like receptor during larval metamorphosis in Crassostrea angulata. Aquaculture 366–367: 54–61. [Google Scholar]
- Yang D., Zhou Y., Guan Z., Zhao F., 2007. Technique for industrial breeding in Rapana venosa Valenciennes. Fish. Sci. 4: 012 (in Chinese). [Google Scholar]
- Yang Q., Angerer L. M., Angerer R. C., 1989. Unusual pattern of accumulation of mRNA encoding EGF-related protein in sea urchin embryos. Science 246(4931): 806–808. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Yang H., Storey K. B., Chen M., 2014. Differential gene expression in the respiratory tree of the sea cucumber Apostichopus japonicus during aestivation. Mar. Genomics 18: 173–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.