Abstract
Incorporation of the histone variant H2A.Z into nucleosomes by the SWR1 chromatin remodeling complex is a critical step in eukaryotic gene regulation. In Arabidopsis, SWR1c and H2A.Z have been shown to control gene expression underlying development and environmental responses. Although they have been implicated in defense, the specific roles of the complex subunits and H2A.Z in immunity are not well understood. In this study, we analyzed the roles of the SWR1c subunits, PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1), ACTIN-RELATED PROTEIN6 (ARP6), and SWR1 COMPLEX 6 (SWC6), as well as H2A.Z, in defense and gene regulation. We found that SWR1c components play different roles in resistance to different pathogens. Loss of PIE1 and SWC6 function as well as depletion of H2A.Z led to reduced basal resistance, while loss of ARP6 fucntion resulted in enhanced resistance. We found that mutations in PIE1 and SWC6 resulted in impaired effector-triggered immunity. Mutation in SWR1c components and H2A.Z also resulted in compromised jasmonic acid/ethylene-mediated immunity. Genome-wide expression analyses similarly reveal distinct roles for H2A.Z and SWR1c components in gene regulation, and suggest a potential role for PIE1 in the regulation of the cross talk between defense signaling pathways. Our data show that although they are part of the same complex, Arabidopsis SWR1c components could have non-redundant functions in plant immunity and gene regulation.
Key words: arabidopsis, SWR1, chromatin remodeling, H2A.Z, immunity, gene regulation
SWR1 chromatin remodeling complex subunits show functional specialization and have non-overlapping roles. While PIE1, SWC6 and H2A.Z are positive regulator of resistance to pathogens, ARP6 acts as a negative regulator. Our results show that Arabidopsis SWR1c components could have non-redundant functions in plant immunity and gene regulation.
Introduction
Precise regulation of gene expression in response to environmental and endogenous signals is fundamental for reproductive success. The ability to adapt gene expression in response to the changing external conditions is particularly important for plants owing to their sessile nature. In eukaryotes, the organization of genomic DNA into chromatin and the ordered regulation of its accessibility to the transcription machinery are central to gene regulation (Li et al., 2007, Narlikar et al., 2013). Mechanisms for regulation of chromatin structure include ATP-dependent chromatin remodeling as well as post-translational histone modifications. ATP-dependent chromatin remodeling complexes alter nucleosome composition and positioning, and thus can regulate DNA accessibility via chromatin compactness (Li et al., 2007, Narlikar et al., 2013).
The SWR1 complex (SWR1c) is an evolutionarily conserved Swi2/Snf2-related ATPase-containing chromatin remodeling complex that catalyzes the replacement of H2A by the histone variant H2A.Z in nucleosomes (Kobor et al., 2004, Mizuguchi et al., 2004). Through its distinct physicochemical properties, H2A.Z influences nucleosome stability, and therefore chromatin structure, to modulate gene expression (Thambirajah et al., 2006, Zlatanova and Thakar, 2008, Thakar et al., 2009). These properties along with its incorporation into the chromatin out of mitosis have made H2A.Z central to transcriptional regulation underlying development and environmental responses (Talbert and Henikoff, 2014). In budding yeast, SWR1c has been characterized as a large multi-protein structure with ∼13 accessory subunits in complex with the ATPase, SWR1 (Kobor et al., 2004, Mizuguchi et al., 2004, Wu et al., 2005). Several of the non-catalytic subunits including ACTIN-RELATED PROTEIN 6 (ARP6) and SWR1 COMPLEX 6 (SWC6) have been shown to be essential for histone replacement. ARP6, SWC6, and SWC2 act as a sub-complex, where the proteins are mutually essential for each other's association and function within the complex (Wu et al., 2005).
In line with the current understanding, SWR1c is expected to act as a single complex in plants to catalyze H2A.Z incorporation (Figure 1A). The components and function of SWR1c have been shown to be conserved in Arabidopsis (Noh and Amasino, 2003, Deal et al., 2005, Martin-Trillo et al., 2006, Choi et al., 2007, March-Diaz et al., 2007, Lázaro et al., 2008). In contrast to other organisms, SWR1c and H2A.Z are not essential for viability in Arabidopsis (Coleman-Derr and Zilberman, 2012). Forward and reverse genetic analyses have identified the SWR1 homolog PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 (PIE1) as well as ARP6, SWC6, and H2A.Z encoding genes (HTA8, HTA9, HTA11) (Noh and Amasino, 2003, Deal et al., 2005, Martin-Trillo et al., 2006, Lázaro et al., 2008, March-Díaz et al., 2008, Coleman-Derr and Zilberman, 2012). As in other eukaryotes, the non-catalytic subunits of SWR1c in Arabidopsis are considered to have similar functions and to be essential for H2A.Z deposition by the ATPase subunit. Mutants defective in SWR1c are highly pleiotropic (March-Díaz et al., 2008, Coleman-Derr and Zilberman, 2012).
Figure 1.
Developmental and Basal Resistance Phenotypes in SWR1c Component and H2A.Z Mutants.
(A) Current understanding of the SWR1c: the complex subunits function in a single complex to catalyze H2A.Z incorporation. The subunits for which viable mutants exist in Arabidopsis are shown in color.
(B) Phenotype of mutants grown in LD conditions. arp6, swc6-1, and hta9-1 hta11-1 showing early flowering phenotype. The pie1-2 mutant shows a very stunted growth phenotype. In SD conditions, the SWR1c mutants (pie1-2, arp6-1, and swc6-1) and hta9-1 hta11-1 show more similar phenotypes: serrated leaves with elongated petioles. The pie1-2 phenotype is less severe in these conditions.
(C) Trypan blue staining of leaves from SWR1c and H2A.Z mutant plants grown under SD photoperiods showing no cell death, similar to wild-type plants.
(D) Disease symptoms 3 dpi with virulent Pst DC3000 in the indicated genotypes.
(E) Resistance phenotype after spray inoculation with Pst DC3000 bacterial suspension in the indicated genotypes. Bacterial titers were determined 2 h (day 0, white bars) and 3 days (day 3, black bars) post infection. Bars are average of data from four plants (n = 4), and error bars show SD. Asterisks indicate statistically significant differences compared with Col-0, with *P <0.05 and **P < 0.01 analyzed with Student's t-test. Similar results were obtained in at least three independent experiments.
See also Supplemental Figures 1–3.
SWR1c components and H2A.Z have been shown to function in various aspects of plant development, most importantly in the timing of the vegetative to reproductive transition (Noh and Amasino, 2003, Deal et al., 2005, Martin-Trillo et al., 2006, Lázaro et al., 2008). ARP6 has been shown to regulate meiosis during megagametogenesis through the regulation of meiotic gene expression (Qin et al., 2014). In addition, the role of H2A.Z in modulating chromatin structure was recently shown to be also important for crossovers during meiosis. While the chromatin landscape at the crossover hotspots is marked with H2A.Z nucleosomes, the arp6 mutant is compromised in meiotic crossovers (Choi et al., 2013). H2A.Z has recently been proposed to be critical for genome stability and DNA repair in Arabidopsis. The pie1, arp6 and swc6 mutants show constitutive DNA damage and compromised somatic homologous recombination as well as hypersensitivity to genotoxic agents (Rosa et al., 2013). Importantly, SWR1c and H2A.Z have also been implicated in regulating gene expression in response to environmental signals in general (Coleman-Derr and Zilberman, 2012). ARP6 has been shown to be important for repression of genes involved in the phosphate starvation response (Smith et al., 2009). Temperature-dependent H2A.Z nucleosome dynamics have been shown to modulate thermosensory responses in Arabidopsis (Kumar and Wigge, 2010) and in grasses (Boden et al., 2013). Mutations in SWR1c components PIE1 and SWC6 or in H2A.Z have been reported to result in constitutive activation of defense responses, demonstrating their importance in biotic interactions as well (March-Díaz et al., 2008).
Although SWR1c and H2A.Z have been implicated in immunity in Arabidopsis, the respective role of the complex subunits is still not well understood in plant defense. Moreover the function of ARP6, a key component of the SWR1c subunit in H2A.Z deposition, has not been previously studied in this respect. In this study, we have carried out a comprehensive analysis to unravel the role of SWR1c components in immunity and gene regulation. Our analyses suggest that SWR1c subunits could have specialized functions in defense. While resistance to biotrophic and necrotrophic pathogens is impaired in pie1, swc6, and hta9 hta11 mutants, arp6 showed wild-type or increased resistance. Loss of PIE1 and SWC6 but not ARP6, leads to impaired effector-triggered immunity (ETI). Genome-wide gene expression analyses have further highlighted the potentially specialized roles of H2A.Z and SWR1c components in gene regulation and thereby in plant defense responses.
Results
Mutants Affected in H2A.Z Incorporation Have Diverse Developmental and Immunity Phenotypes
To study the role of H2A.Z in plant defense processes, we analyzed mutants defective in SWR1 complex components PIE1, ARP6, and SWC6, as well as those depleted of the histone variant H2A.Z. Consistent with previous studies, pie1-2 (Noh and Amasino, 2003), arp6-1 (Deal et al., 2005), and swc6-1 (Lázaro et al., 2008) mutants showed pleiotropic growth and developmental phenotypes including early flowering, elongated petioles, and reduced fertility when grown in long photoperiod (LD) (Figure 1B). The pie1 mutant showed the most severe growth defects and even though it displayed accelerated reproductive transition, bolting was delayed. The arp6 and swc6 mutants phenocopied each other with characteristic early flowering and serrated leaves. This was consistent with the biochemical interaction of ARP6 and SWC6 in yeast, where their existence in the complex is mutually dependent (Mizuguchi et al., 2004). The hta9-1 hta11-1 double mutant (loss of two major H2A.Z encoding genes out of the possible three in Arabidopsis) phenocopied arp6 and swc6 mutants with early flowering and serrated leaves. The severe growth defects in the pie1 mutant have been previously attributed to the de-repression of immune responses characterized by spontaneous cell death and upregulation of defense genes (March-Díaz et al., 2008). Growth in short photoperiods (SD) largely suppressed some of the growth defects of the pie1 mutant, even though the rosette size remained significantly smaller compared with the other SWR1c mutants (Figure 1B). All the studies reported here, unless otherwise specified, were performed on plants grown under SD.
SWR1c components have been proposed to be negative regulators of plant immunity with pie1, swc6, and hta9 hta11 mutants reported to display spontaneous cell death and enhanced resistance to virulent bacterial pathogens (March-Díaz et al., 2008). The role of ARP6 in this process is still not known and has been expected to be similar to PIE1, SWC6, and H2A.Z. In order to investigate this, we characterized the defense responses of these mutants. In our experimental conditions, trypan blue staining revealed no apparent spontaneous cell death in the mutants (Figure 1C). We occasionally observed patches of cell death and leaf torsion in some leaves of the pie1 mutant (Supplemental Figure 1). To investigate their immunity phenotypes further, we challenged 4-week-old mutants with the hemibiotrophic bacterial pathogen P. syringae pv tomato DC3000 (Pst DC3000) by spray inoculation and monitored bacterial growth. We found that pie1, swc6, and hta9 hta11 mutants showed more macroscopic disease symptoms and increased susceptibility toward Pst DC3000 compared with the wild-type Col-0. However, the arp6 mutant showed increased resistance (Figure 1D and 1E). Overall, pie1, swc6, and hta9 hta11 mutants accommodated 10 to 15 times more bacterial titers at 3 days post inoculation (dpi) than the wild type. The pie1, swc6, and hta9 hta11 mutants displayed severely compromised resistance similar to that of the enhanced disease susceptibility1 (eds1-2) mutant, which was used as a susceptible control (Feys et al., 2001). There were no differences in the bacterial titers at day 0 (2 h post inoculation) between the different mutants and the wild-type Col-0 plants. A transgenic SWC6 complemented line showed wild-type resistance to Pst DC3000 (Supplemental Figure 2), suggesting that the resistance phenotype we observe is caused by the mutation. Our observations suggest that H2A.Z, PIE1, and SWC6 are essential for basal resistance in Arabidopsis, whereas ARP6 has an opposite function.
These results were intriguing as a previous study has shown that pie1, swc6, and hta9 hta11 mutants displayed rather constitutive defense activation marked by spontaneous lesions and disease-resistance phenotypes (March-Díaz et al., 2008). As mentioned earlier, the pie1 mutant showed severe growth defects when grown under LD. These phenotypes were partially suppressed by growth under SD (Figure 1B). In order to investigate whether the severe growth defects in pie1 under LD were due to constitutive defense activation and therefore to test if pie1 shows photoperiod-dependent defense phenotypes, we assessed its resistance to Pst DC3000 in these growth conditions. To bypass early flowering induction by constant growth under LD photoperiods, the mutants along with the wild type were pre-grown for two weeks under SD conditions before being shifted to LD for another two weeks prior to infection. Consistent with the results from the SD experiments, pie1 remained increasingly susceptible to Pst DC3000 (Supplemental Figure 3A). Moreover, it showed no spontaneous cell death in the leaves under these conditions, similarly to the other mutants (Supplemental Figure 3B), except for the patches of dead tissue observed as in SD. These results suggest that the growth defects in the pie1 mutant are not necessarily a consequence of enhanced defense activation. The defense phenotypes of arp6, swc6, and hta9 hta11 grown in LD were also similar to the results obtained under SD conditions (Supplemental Figure 3A) further confirming that the immunity phenotypes we observed are not due to specific growth conditions, especially the photoperiod. These results also confirm that the contrasting defense phenotypes observed in this study and by March-Díaz et al. (2008) are unlikely to be caused by growth conditions such as the photoperiod.
Resistance to Avirulent Pathogens Is Compromised in pie1 and swc6
During interaction with avirulent pathogens, recognition of effectors by the receptor proteins in the host plant leads to the activation of ETI. In Arabidopsis, the bacterial effectors AvrRps4 and AvrRpt2 trigger resistance through recognition by the toll interleukin-1 receptor (TIR)-type and the coiled coil (CC)-type nucleotide binding-leucine-rich-repeat (NB-LRR) proteins RPS4/RRS1 and RPS2, respectively (Kunkel et al., 1993, Gassmann et al., 1999, Narusaka et al., 2009). To examine the possible roles of SWR1c and H2A.Z in ETI, we inoculated the corresponding mutants with avirulent Pst DC3000 expressing either AvrRps4 or AvrRpt2 effectors. The pie1 and swc6 mutants are more susceptible compared with wild-type in both interactions (Figure 2A and 2B). They were, however, not as hypersusceptible as eds1 or ndr1 (non-race-specific disease resistance1), used as susceptible controls in response to AvrRps4 and AvRrpt2 (Century et al., 1995), respectively. Consistent with the earlier results, arp6 displayed full resistance like wild-type plants, suggesting that arp6 mutation does not alter RPS4- and RPS2-mediated resistance. The hta9 hta11 double mutant showed mild but not significant susceptibility compared with wild-type. In Arabidopsis, H2A.Z is encoded by three genes HTA9, HTA11, and HTA8. The presence of HTA8, even though generally lowly expressed (Supplemental Figure 4), in the hta9 hta11 double mutant could compensate for the lack of HTA9 and HTA11. These results suggest that PIE1 and SWC6 have a positive function in ETI. Interestingly, ARP6 function appears to be dispensable.
Figure 2.
ETI Responses in the SWR1c Component and H2A.Z Mutants.
(A and B) Resistance phenotype after spray inoculation with avirulent Pst DC3000 AvrRps4 (A) and Pst DC3000 AvrRpt2 (B) bacterial suspension in the indicated genotypes. Bacterial titers were determined a 2 h (day 0, white bars) and 3 days (day 3, black bars). Bars are average from four plants (n = 4), and error bars show SD. Asterisks indicate statistically significant differences compared with Col-0, with **P < 0.01 analyzed with Student's t-test. Similar results were obtained in at least three independent experiments.
(C) Quantification of free SA content before (T0, light gray bars) and 24 h after spray inoculation with Pst DC3000 AvrRps4 (black bars) or after mock treatment with 10 mM MgCl2 (dark gray bars). Bars represent the average of three independent biological replicates and error bars show SD. **P < 0.01 analyzed with Student's t-test.
To understand the basis of the failure to mount ETI, we examined the level of salicylic acid (SA) accumulation, an important defense hormone in response to biotrophic pathogens and particularly during ETI (Robert-Seilaniantz et al., 2011). The basal level of SA did not noticeably differ between the the wild type and the mutants except for arp6, where it was higher (Figure 2C). Interestingly, 24 h after infiltration with Pst DC3000 AvrRps4, all the mutants showed increased SA accumulation in comparison with Col-0. Accumulation of SA in pie1, swc6, and hta9 hta11 was 2-fold higher relative to Col-0, and reached 3-fold in arp6. Our results show that in spite of SA accumulation, resistance to virulent and avirulent P. syringae is impaired in pie1 and swc6 mutants, suggesting that SWR1c activity (especially PIE1, SWC6) and H2A.Z are required for SA-dependent downstream signaling processes. These results are further supportive to the resistance phenotypes of the mutants to virulent Pst DC3000 (Figure 1D and 1E).
Role of SWR1c Components in Resistance to Necrotrophic Pathogen
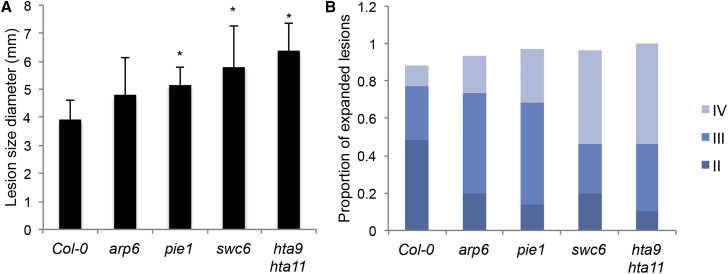
Signaling crosstalks and antagonistic interactions between different defense pathways regulated by hormones have been extensively studied (Robert-Seilaniantz et al., 2011, Thaler et al., 2012). SA-mediated defense is activated during interaction with biotrophic pathogens while jasmonic acid (JA)-mediated defense is active against necrotrophs and herbivores (Glazebrook, 2005, Howe and Jander, 2008). As presented earlier, pie1, swc6, and hta9 hta11 mutant plants showed impaired SA-mediated defense responses even though they accumulated a high level of the SA hormone in response to Pseudomonas infection. In order to investigate whether these mutants are affected in the SA/JA crosstalk, we investigated their resistance to the necrotrophic pathogen Botrytis cineria. Leaves of the wild-type and mutant plants were drop inoculated with spore suspension, and the growth of lesions was monitored. Average lesion size measured 3 dpi was significantly bigger in pie1, swc6, and hta9 hat11. The proportions of outgrowing lesion type IV (lesion over 6 mm in diameter) in pie1, swc6, and hta9 hat11 were higher compared with arp6 mutant plants that showed mild resistance (Figure 3 and Supplemental Figure 5). These results show that perturbation of H2A.Z incorporation results in a compromised JA/ethylene (ET)-mediated immunity. It also suggests that components of SWR1c do not play the same function in this defense pathway, as arp6 showed wild-type resistance to Botrytis, as opposed to the rest of the mutants.
Figure 3.
SWR1c Components Have Different Roles in Resistance to Necrotrophic Pathogen.
(A) Resistance to Botrytis cineria: average lesion diameter for all inoculation sites at 3 dpi. Asterisks indicate statistically significant differences compared with Col-0, with *P < 0.05 analyzed with Student's t-test. Similar results were obtained in at least two independent experiments (n = 30).
(B) Frequency of outgrowing lesions (a size bigger than the inoculation site, 2 mm) in the different mutants background at 3 dpi with Botrytis cinerea. Type II, outgrowing lesion between 2 and 4 mm in diameter; type III, 4 and 6 mm in diameter; and type IV, >6 mm in diameter.
See also Supplemental Figure 5.
SWR1c Components Have Distinct Functions in Gene Regulation
In order to elucidate the molecular basis of the phenotypes described above, and to dissect the role of H2A.Z and SWR1c subunits in gene regulation, we carried out a global transcriptome analysis of the wild-type and mutant lines using RNA-seq. Three replicates of total RNA from 2-week-old seedlings from each mutant were sequenced. Genes showing at least 2-fold change from the wild type with a P value cut-off of 0.05 were used for further analyses. We found that mutations in SWR1c components result in misregulation of a large number of genes (Figure 4A and 4B and Supplemental Dataset 1). The pie1 mutation resulted in the largest number of gene misregulation, with 2295 genes upregulated and 1051 genes downregulated. This was expected, as PIE1 is the catalytic subunit of the SWR1 complex. The genes misregulated in our study showed statistically significant (hypergeometric test) overlap with the previously published data (March-Díaz et al., 2008, Coleman-Derr and Zilberman, 2012).
Figure 4.
SWR1c Subunits Have Distinct Functions in Gene Regulation.
(A and B) Venn diagrams of differentially expressed genes showing overlap in genes whose expression levels are uniquely or concordantly upregulated (≥2-fold) (A) or downregulated (≥2-fold) (B) in the mutants. Total number of ≥2-fold up- or downregulated genes in each mutant is shown in parentheses.
(C) Principal component analysis of the RNA-seq data. The results are depicted three-dimensionally with PC1 (44.4%), PC2 (30.5%), and PC3 (17.1%) as the X, Y, and Z axes, respectively.
(D–G) Scatter plot comparing differential expression of all genes in respective mutant backgrounds. Each point represents the log 2 change in expression of individual genes in the respective mutant. The solid black line shows linear regression representing the correlation coefficient (r2). See also Supplemental Figures 6 and 7.
We found a total of 671 genes to be commonly misregulated among all four mutants (473 genes upregulated and 198 genes downregulated). This accounts for 20% of the genes misregulated in pie1, and 22%, 32%, and 27% in arp6, swc6, and hta9 hta11, respectively. Moreover, the commonly misregulated genes (up and down) showed a statistically significant overlap with H2A.Z enrichment in the gene bodies (Coleman-Derr and Zilberman, 2012). We found that 1262 genes were uniquely upregulated in pie1 accounting for 55% of the total number of genes upregulated in this mutant. In arp6, swc6, and hta9 hta11, however, only 20%, 4%, and 13%, respectively, were uniquely upregulated. Similarly 50% of the genes downregulated in pie1 were unique. The proportions of uniquely downregulated genes were 25% in arp6, 4.5% in swc6, and 18% in hta9 hta11. The large proportion of uniquely misregulated genes in pie1 suggests a potential SWR1c/H2A.Z independent role for PIE1 in gene regulation.
To explore this further and to elucidate the relationship between individual mutants, we performed principal component analysis (PCA) of the RNA-seq data. The first three principal components (PC1, PC2, and PC3) together accounted for 92.1% of the variance in the data (44.44%, 30.55%, and 17.11%, respectively) (Figure 4C). It also revealed that the four mutant genotypes segregate into three distinct groups: swc6 and hta9 hta11 mutants co-segregated while pie1 and arp6 mutants were distinct. The second principal component (PC2) distinguished the mutants from the wild type. PC1 clearly separated pie1 from the rest of the group, whereas PC3 defined the contribution of arp6 (Figure 4C and Supplemental Figure 6). This analysis has further strengthened the possibility of non-overlapping functions for the complex components. In line with the PCA analysis, differential gene expression analyses showed a poor correlation of gene misregulation caused by arp6 and pie1 mutations (r2 = 0.16) (Figure 4D), as well as between hta9 hta11 and pie1 (r2 = 0.11) (Figure 4E), which is surprising as pie1 mutation is expected to phenocopy H2A.Z depletion to a large extent. The arp6 and swc6 mutants in Arabidopsis have similar morphological and developmental phenotypes, especially early flowering and serrated leaves. Moreover, ARP6 and SWC6 have been shown to physically interact in Arabidopsis (March-Diaz et al., 2007, Lázaro et al., 2008). Intriguingly, in our RNA-seq data, the transcriptional changes due to these mutations did not show a strong correlation (r2 = 0.42) (Figure 4F). This observation is consistent with the defense phenotypes described above, suggesting that ARP6 and SWC6 could have distinct functions in Arabidopsis. In agreement with the PCA analysis, swc6 and hta9 hta11 double mutants displayed a strong correlation of transcriptional misregulation (r2 = 0.75), indicating overlapping roles for SWC6 and H2A.Z in gene regulation (Figure 4G). This is in contrast to arp6, which showed only a modest correlation (r2 = 0.38) with hta9 hta11 (Supplemental Figure 7).
To understand the function of SWR1c and H2A.Z in gene regulation, particularly in immunity, we performed a Gene Ontology (GO) analysis of the mutant transcriptomes. We found a significant over-representation of SA response (P = 4.46 × 10−6) and defense (P = 7.75 × 10−3) related GO terms in the 473 genes commonly misregulated in the mutants (Supplemental Dataset 2). This is in accordance with what was reported earlier (March-Díaz et al., 2008). The commonly downregulated genes where enriched in triterpene (thalianol, P = 1.77 × 10−4 and tricyclic triterpenoid metabolic process, P = 7.04 × 10−4) related GO terms. In addition, pie1 showed a significant over-representation of several biological processes including auxin metabolic processes, membrane organization, and response to heat, indicating the important role of PIE1 in gene expression in general. In swc6 and hta9 hta11, the GO term response to heat was significantly enriched, which is consistent with H2A.Z function in response to temperature (Kumar and Wigge, 2010). We also observed that swc6 and hta9 hta11 mutants display an enrichment of hydrogen peroxide-related GO terms like in arp6 (Supplemental Dataset 2). Taken together, the results of the transcriptome analysis demonstrate that the components of the SWR1c have distinct roles in the regulation of gene expression in Arabidopsis.
PIE1 Functions to Coordinate Distinct Defense Signaling Pathways
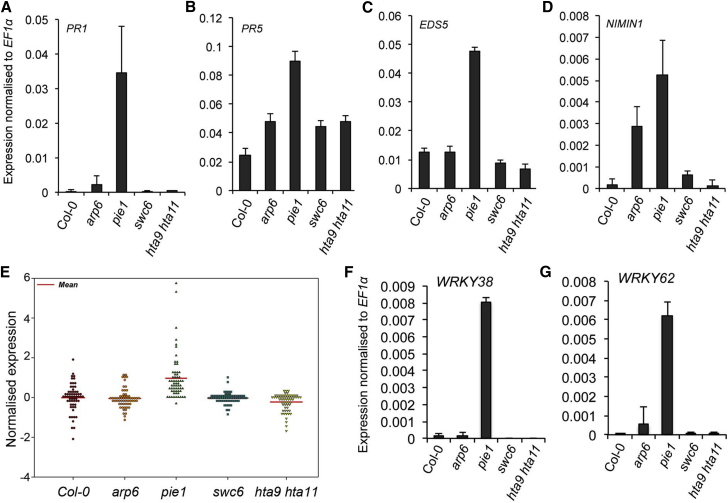
Our RNA-seq analysis revealed that, in the SWR1c mutants, particularly pie1, genes required for disease resistance were misregulated as previously shown (March-Díaz et al., 2008). Genes involved in systemic acquired resistance such as PR1, PR5, EDS5, and NIMIN1 were highly upregulated in pie1 (Figure 5A–5D and Supplemental Dataset 1). The arp6 mutant showed modest upregulation of all the above genes except for EDS5. Only PR5 among these genes was upregulated in swc6 and hta9 hta11, but not to the same extent as in pie1. Interestingly, the pie1 mutant shows severely compromised resistance in spite of the enhanced expression of defense genes (as shown above). To further understand the molecular basis for this paradox, we analyzed the transcriptome further. We observed that genes encoding the WRKY family of transcription factors are upregulated in pie1 (Figure 5E and Supplemental Dataset). WRKY transcription factors are important regulatory components during plant response to pathogen infection and abiotic stresses. Moreover, they have been implicated in plant immunity as both positive and negative regulators (Pandey and Somssich, 2009, Rushton et al., 2010). WRKY38 and WRKY62, in particular, are highly expressed in the pie1 background (Figure 5F and 5G). These two WRKY proteins were shown to be negative regulators of resistance to pathogens. In line with these observations, the uniquely upregulated genes in pie1 are also enriched in GO terms related to negative regulation of the defense response (P = 1.02 × 10−9).
Figure 5.
Defense Gene Expression Is Altered in SWR1c Mutants.
(A–D) Gene expression of SAR genes in SWR1c mutants: PR1(A) is strongly upregulated in the pie1 mutant. PR5(B) shows upregulation in all the mutants, with the strongest effect in pie1. EDS1(C) is specifically upregulated in pie1, while NIMIN1 (D) is upregulated in arp6 and pie1 only.
(E) Dot density plot showing misregulation of the WRKY gene family in the SWR1c component mutants. The pie1 mutant displays a general upregulation of the WRKY genes.
(F)WRKY38 and (G)WRKY62 show strong upregulation in the pie1 mutant. Data represented are expression values from RNA-seq data of three independent biological replicates, normalized to EF1α. Error bars show SD from three biological replicates.
The pie1 mutant displays upregulation of SA-responsive gene expression. SA is a key regulator of signaling networks involved in defense along with other hormones such as JA (Robert-Seilaniantz et al., 2011). SA- and JA-mediated defense responses are triggered in the plant depending on the nature of the pathogen (Van der Does et al., 2013). These two pathways act antagonistically to modulate defense responses. Activation of one pathway usually correlates with attenuation of the other (Thaler et al., 2012). Consistent with this, we found that the SA-inducible glutaredoxin gene GRX480, which has been proposed to antagonize the JA-responsive transcription of PDF1.2 is nearly 10-fold upregulated in pie1 (Figure 6A). Transgenic plants ectopically expressing GRX480 displayed reduced JA-induced gene expression (Ndamukong et al., 2007). Surprisingly, JA-mediated responses were not suppressed in pie1. In fact, uniquely upregulated genes in pie1 are enriched in GO terms related to the JA metabolic process (P = 9.12 × 10−32) and JA-mediated signaling pathway (P = 5.17 × 10−23). For instance, genes involved in JA biosynthesis (Figure 6B–6D), signaling (Figure 6E and 6F), and response (Figure 6G–6I) were upregulated. We further analyzed the expression of JA-induced genes (Nemhauser et al., 2006) in our data and found that these are upregulated in pie1 compared with the other mutants (Figure 6J), confirming that JA-responsive pathways are derepressed in spite of the SAR genes being upregulated. These observations, together with the upregulation of negative regulators of defense (e.g., WRKYs), could explain the unexpected disease susceptibility phenotypes we observe. They also point to a potential role that PIE1 plays in coordinating distinct defense signaling pathways, especially in the SA-JA antagonism.
Figure 6.
PIE1 Is Required to Maintain the Fine Balance between SA and JA Signaling Pathways.
(A–I) Genes involved in SA/JA signaling trade-off are upregulated in the pie1 mutant: (A) SA-inducible GRX480, known to antagonize the JA-responsive transcription of PDF1.2, is strongly upregulated in the pie1 mutant. JA biosynthesis genes AOC2(B), OPR3(C), and LOX2 (D) show strong upregulation in pie1. Genes involved in JA signaling JAZ1(E) and JAZ10(F) are specifically upregulated in pie1, but downregulated in swc6 and hta9 hta11. JA-responsive genes VSP2(G), PDF1.2 (H), and JR2/TAT1(I) are upregulated in pie1.
(J) JA-induced genes show increased expression in pie1 suggesting that PIE1 activity is essential for SA-mediated repression of JA-responsive gene expression.
SWC6 Is Epistatic to ARP6
Our analyses revealed that mutants of the SWR1c components show considerable differences in physiological and molecular phenotypes, leading to the assumption that the complex components may be functionally specialized. It is also likely that the non-enzymatic subunits might not necessarily be essential for complex function, and their depletion or inclusion in the complex could modulate its function. The components could also have functions independent of H2A.Z incorporation. To further elucidate the possible specialization and to understand their role in relation to each other, we analyzed double mutant plants. Since arp6 and swc6 mutants show similar morphological and developmental phenotypes but contrasting immunity traits, we analyzed arp6 swc6 double mutants to study disease resistance without the confounding effects of contrasting developmental phenotypes. The double mutants did not show any noticeable additional morphological phenotypes than the single mutants suggesting overlapping functions in development. As shown above, arp6 showed resistance to DC3000 while the swc6 mutant showed increased susceptibility compared with wild-type Col-0. Interestingly, the arp6 swc6 double mutants displayed increased susceptibility to Pst DC3000 phenocopying swc6 (Figure 7). These results indicate that SWC6 is epistatic to ARP6 in disease-resistance traits. Despite the overlapping functions in development, it seems that loss of SWC6 significantly compromises the regulation of immunity genes by SWR1c more than the loss of ARP6.
Figure 7.
SWC6 Is Epistatic to ARP6 in Disease Resistance to P. syringae.
Analysis of the arp6 swc6 double mutant for resistance to Pst DC3000. Bacterial titers were determined 3 days post spray inoculation with Pst DC3000 bacterial suspension in the indicated genotypes. Bars are average of six plants (n = 6), and error bars show SD. Asterisks indicate statistically significant differences compared with Col-0, with **P < 0.01 analyzed with Student's t-test. Similar results were obtained in at least two independent experiments.
SWR1c Mutants Show Compromised Defense Gene Induction in Response to Pseudomonas
Our gene expression analyses have shown that genes involved in systemic acquired resistance such as PR1 and PR5 were derepressed in the SWR1c mutants and therefore showed increased basal expression (Figure 5). Interestingly, however, pie1, swc6, and hta9 hta11 mutants were severely compromised in resistance to both virulent and avirulent P. syringae strains in spite of the enhanced basal expression of defense genes. To understand the molecular basis for these paradoxical phenotypes, we analyzed defense gene expression in the mutants in response to Pst DC3000. Gene expression was analyzed in 4-week-old wild-type Col-0 and the mutants by quantitative RT–PCR 6 h post inoculation with Pst DC3000 or mock treatment (10 mM MgCl2) (Figure 8). As expected, PR1 showed very low basal expression in Col-0 under control conditions, whereas bacterial infection resulted in a strong induction of gene expression (Figure 8A). The arp6 mutant showed derepression of PR1 and a stronger induction in response to Pst DC3000. On the other hand, the pie1 mutant showed a strong constitutive expression of PR1; however, inoculation with Pst DC3000 inoculation resulted in strong downregulation (Figure 8A). In swc6 and hta9 hta11, where basal expression of PR1 was modestly altered, DC3000 infection resulted in its repression (Figure 8A). Similarly, analysis of PR5 expression showed that while it was strongly induced in arp6 in response to Pst DC3000, there was little or no induction in pie1, swc6, and hta9 hta11 mutants (Figure 8B). These results show that induction of PR1 and PR5 was compromised upon pathogen inoculation in pie1, swc6, and hta9 hta11 mutants despite the higher basal expression. Together, these results show that while it is required to maintain defense genes in repressed state in the absence of pathogen signals, SWR1c function, and therefore H2A.Z are essential for strong pathogen-induced defense gene activation that is essential for disease resistance.
Figure 8.
SWR1c Function Is Required for Pathogen-Responsive Gene Expression.
Analysis of PR1(A) and PR5(B) gene expression in SWR1c mutants in response to bacterial infection by Pst DC3000. Four-week-old Col-0, arp6, pie1, swc6, and hta9 hta11 were inoculated with Pst DC3000 or mock treated with MgCl2. RNA was isolated 6 h post treatment, and gene expression was analyzed by qRT–PCR (three biological replicates and three technical replicates). Insets show log fold induction upon Pst DC3000 treatment. Asterisks indicate significant difference from respective Col-0 treatment as analyzed by Student's t-test (*P <0.05, **P < 0.01, and ***P < 0.001).
Discussion
SWR1c is an evolutionarily conserved ATP-dependent chromatin remodeling complex involved in the post mitotic incorporation of the histone variant H2A.Z into nucleosomes. SWR1c and H2A.Z have been implicated in gene regulation in eukaryotes. In Arabidopsis, putative SWR1c subunits have been identified and shown to be involved in plant growth and development as well as environmental adaptation. Although they have been implicated in immunity, their exact role is yet to be understood. Above all, the respective roles that each subunit has in gene regulation and their influence in major physiological outcomes remain elusive. We undertook a comprehensive analysis of SWR1c and H2A.Z mutants to systematically analyze their function in plant immunity and gene regulation. Our results indicate that perturbing SWR1c function or depleting H2A.Z levels results in severely impaired immune responses: PIE1, SWC6, and H2A.Z are positive regulators of resistance in Arabidopsis against both biotrophic and necrotrophic pathogens. ARP6 is a negative regulator of defense against biotrophs. Transcriptome analysis revealed further complexities as the SWR1c subunits showed overlapping but, more importantly, distinct functions in the regulation of gene expression.
SWR1c Components and H2A.Z Have Non-overlapping Functions in Immunity in Arabidopsis
In this study, we found that mutations of the SWR1c subunits, PIE1 and SWC6, and depletion of H2A.Z severely impaired immunity suggesting that they are essential for defense responses in Arabidopsis. PIE1, SWC6, and H2A.Z have been implicated in immunity in an earlier study, where mutations in the respective genes were reported to result in constitutive activation of defense responses including spontaneous cell death and increased resistance to virulent P. syringae (March-Díaz et al., 2008). While the developmental phenotypes we observed were in agreement with the previous reports (Deal et al., 2005, March-Diaz et al., 2007, Lázaro et al., 2008), defense phenotypes were contrasting in our experiments. While P. syringae infection assays were performed under the same growth conditions in our study and in that of March-Díaz et al. (2008), the contradictory resistance phenotypes are intriguing. We used appropriate genotypes as controls (eds1, ndr1, etc.) in all our experiments and have shown the expected phenotypes, suggesting that the phenotypic differences are not due to the experimental conditions. We have also extended our analyses to test if the SWR1c mutants show photoperiod-dependent defense phenotypes and performed disease resistance assays using Pst DC3000 under long photoperiod. Strikingly pie1, swc6, and hta9 hta11 were all increasingly susceptible independent of the photoperiod, suggesting that the difference in the resistance phenotype is likely not due to growth conditions, particularly day length. It is still likely that other environmental factors such as nutrient availability associated with plant growth conditions could underlie the discrepancy. However, our reproducible results with both virulent and avirulent P. syringae strains, as well as the expected phenotypes for both positive and negative control genotypes, support our conclusions that the SWR1c mutants pie1, swc6, and hta9 hta11 are compromised in disease resistance.
As reported previously (Noh and Amasino, 2003, March-Díaz et al., 2008), we found that the pie1 mutant is highly pleiotropic, displaying stunted growth and low fertility. Moreover, as described above, it showed increased susceptibility even under long photoperiod conditions, where its growth defects were more severe. These observations suggest that the pleiotropic phenotypes of pie1 are not necessarily a result of constitutive defense activation. It is, however, still likely that defective regulation of certain defense-associated pathways underlies these phenotypes despite the lack of apparent disease resistance. Similar examples where defense pathways are turned on without apparent enhanced resistance have been described previously. Mutation in CPK28 genes for instance, resulted in growth defects and constitutively enhanced JA accumulation and activation of JA-responsive genes, but not enhanced resistance to necrotrophs (Matschi et al., 2015).
Similar to the impaired basal immunity in response to the virulent pathogen P. syringae, pie1 and swc6 mutants showed compromised resistance to Pst DC3000 carrying AvrRps4 and AvrRpt2 effectors, suggesting that PIE1 and SWC6 are required for RPS4- and RPS2-mediated immunity. The hta9 hta11 showed only a mild susceptibility, likely due to the compensatory activity of HTA8. ARP6, however, was dispensable for ETI as the mutant showed wild-type resistance. These discrepancies in NB-LRR-mediated resistance between the different mutants were not due to an altered expression of either RPS4 or RPS2 (Supplemental Dataset) and further highlight the functional specialization of the complex subunits. Activation of R-mediated defense results in SA accumulation and signaling (Vlot et al., 2009). SA acts as an important intracellular signal for defense gene activation, and mutants impaired in SA accumulation usually display compromised basal defense and ETI. Although pie1 and swc6 mutants were clearly defective in mounting resistance to Pst DC3000 AvrRps4, we notice a robust increase in SA accumulation in response to this effector, suggesting that RPS4-mediated recognition and downstream signaling are unlikely to be altered. In response to Pst DC3000 AvrRps4, these mutants accumulated SA at higher levels compared with the wild type. This accumulation was similarly higher in arp6 and hta9 hta11. Compromised ETI in pie1 and swc6 in spite of SA accumulation suggests that SA-mediated downstream responses might be affected in these mutant backgrounds. SA-induced gene expression and defense activation are dependent on NPR1 (Cao et al., 1994, Delaney et al., 1995). Similar to the results observed for pie1 and swc6, the npr1 mutant accumulates a high level of SA but fails to mount SAR (Delaney et al., 1995, Clarke et al., 1998, Zhang et al., 2003). NPR1 is also required for the negative feedback regulation of SA production (Delaney et al., 1995). The hyperaccumulation of SA following infection with Pst DC3000 AvrRps4 in spite of enhanced susceptibility suggests a compromised NPR1 activity in pie1 and swc6. It is therefore tempting to speculate that SWR1c activity and H2A.Z might be required for proper functioning of NPR1-mediated regulation of immunity. This possibility, however, remains to be tested experimentally. Our results showing that PR gene induction following Pst DC3000 infection was perturbed in pie1, swc6, and hta9 hta11 mutants also support this hypothesis. In addition, PIE1 and SWC6 could also be important for NPR1- and SA-independent processes required for resistance.
We found that SWR1c and H2A.Z are also important for resistance to the necrotrophic pathogen Botrytis cineria. Depletion of PIE1, SWC6, and H2A.Z, not ARP6, in mutants led to increased susceptibility. This was rather intriguing given the antagonistic interaction between SA- and JA-mediated defenses (Robert-Seilaniantz et al., 2011, Thaler et al., 2012). The contrasting phenotypes of the mutants show that PIE1, SWC6, and H2A.Z, not ARP6 have a positive function in JA/ET-mediated defense. This result was not expected especially in the case of pie1, which showed an increase in the JA-responsive genes, while SA-responsive genes are also upregulated. Moreover, pie1 mutation also resulted in a higher level of expression of GRX480, which is a negative regulator of JA-mediated immunity. These results indicate that mutations of the SWR1c components alter plant resistance and gene expression in general. Taken together, our data point to the importance of SWR1c function, and therefore H2A.Z, in coordinating transcriptional responses appropriately in response to the biotic environment. The pleiotropic effect of compromised SWR1c function leads to misregulation of a large subset of defense genes, including both positive and negative regulators, giving rise to complex physiological outcomes as a consequence of epistatic interactions. Further molecular dissection would unravel the precise regulatory role of H2A.Z in transcriptional reprogramming in immunity. The discrepant phenotypes observed when ARP6 is depleted from SWR1c suggest that the complex could regulate different defense pathways depending on the components recruited to it. Our double mutant analysis for resistance to P. syringae supports an epistatic interaction of ARP6 and SWC6, which have contrasting roles in immunity, as we observed in the mutants. Extending the genetic dissection would further elucidate the complex genetic interactions between the complex components in relation to resistance to necrotrophs and the coordination of potentially antagonistic signaling pathways.
SWR1c Subunits Have Distinct Roles in Gene Regulation
Our study has revealed that the SWR1c subunits are functionally specialized and have non-overlapping functions in immunity. The transcriptomic analysis that we performed further established this. In our analyses, we found that there were more differences than similarities between the SWR1c mutants. Expectedly, pie1 mutation resulted in the largest transcriptional reprograming. PIE1 is the Arabidopsis homolog of Swr1 in yeast, the enzymatic subunit of the complex (Noh and Amasino, 2003). Surprisingly, transcriptional misregulation in pie1 and hta9 hta11 double mutant showed a poor correlation (Figure 4E), while hta9 hta11 still has a functional HTA8, likely explaining the poor agreement. A recent study has also shown similar non-overlapping changes while comparing the transcriptome of pie1 with that of h2a.z triple mutant (Coleman-Derr and Zilberman, 2012). It is likely that PIE1 and H2A.Z have also independent roles of one another, and that PIE1 could functions independently of H2A.Z incorporation. Consistent with their contrasting effects in plant immunity, arp6 and swc6 mutants clearly maintained distinct molecular phenotypes as supported by the gene expression analysis. In budding yeast, ARP6 and SWC6 have been shown to be essential subunits in SWR1c for H2A.Z deposition (Mizuguchi et al., 2004, Wu et al., 2005). Along with SWC2, another component of the SWR1c in yeast, they act as a sub-complex that requires all three proteins for their association with the complex and for histone exchange (Mizuguchi et al., 2004, Wu et al., 2005). The contrasting immunity phenotypes and the divergent transcriptome we observed in our analyses suggest that ARP6 and SWC6 do not necessarily always act as part of the same complex and that they could have independent functions in Arabidopsis, either related or not to H2A.Z. It has been previously proposed that ARP6 could have H2A.Z-independent roles in gene expression (Yoshida et al., 2010). Of all the mutant pairs studied, swc6 and hta9 hta11 showed the strongest correlation in gene expression regulation (Figure 4G) suggesting that SWC6 functions solely as a subunit of SWR1c. It remains to be seen how much of the gene expression changes that we observe in the mutants are due to changes in H2A.Z incorporation. Further investigations are required to fully elucidate the role that each subunit could have in histone exchange and therefore gene regulation.
PIE1 Functions to Maintain the Fine Balance between Defense Signaling Pathways
Detailed analysis of the RNA-seq data revealed that the pie1 mutant showed significant upregulation of both positive and negative regulators of defense. For example, enrichment in WRKY transcription factors was observed. Pathogen-induced WRKY transcription factors function as positive or negative regulators of immunity (Eulgem and Somssich, 2007, Pandey and Somssich, 2009), and epistatic interaction between the two types of regulators is required for the proper control and fine tuning of defense. Of particular interest was the strong upregulation of WRKY38 and WRKY62, which have been shown to be negative regulators of immunity, and their overexpression has led to reduced PR1 gene induction in response to bacterial infection (Kim et al., 2008). This is consistent with the compromised pathogen-induced PR1 expression in pie1 mutant (Figure 8). Our data have shown that the pie1 mutant showed severely compromised immunity in spite of upregulation of a remarkable number of genes involved in SAR such as PR1. While constitutive activation of SAR genes is indicative of PIE1 playing a repressive role in SA-mediated gene expression in the absence of pathogens, compromised SA-dependent resistance against biotrophic pathogens was intriguing. Data shown in Figure 2C show that lack of resistance is not due to the inability of pie1 to accumulate SA. In addition, compromised defense gene induction in response to P. syringae (Figure 8) indicates that while PIE1 has a clear role in maintaining SAR genes repressed in the absence of pathogens, it is essential for proper gene induction and resistance upon infection.
A complex network of signaling pathways underlies defense responses in plants. Defense signaling elicited by the plant hormones SA and JA are mutually antagonistic and determine defense against biotrophic and necrotrophic pathogens, respectively (Thaler et al., 2012). JA biosynthesis and JA-responsive genes are highly upregulated in pie1, suggesting that PIE1 is also a negative regulator of JA-mediated responses. Remarkably, these results also implicate PIE1 activity in regulating SA/JA signaling trade-off. This possibility, however, remains to be tested experimentally. Our data suggest that SWR1c and H2A.Z might be playing an important role in maintaining the fine balance between signaling pathways in plant immunity. Therefore, in the context of pie1, where activating and repressive factors are similarly de-repressed, the physiological outcome will be the consequence of epistatic interactions. In support of this, we found that pie1 is compromised in resistance to both biotrophic and necrotrophic pathogens. Compromised defense gene induction in the susceptible mutant backgrounds such as pie1, swc6, and hta9 hta11 in response to Pst DC3000 further supports this hypothesis. Further molecular dissection is needed to elucidate the regulatory dynamics of defense gene expression in the context of chromatin remodeling.
We have shown that, although they are as part of the same complex, SWR1c subunits have divergent functions in gene regulation and therefore in important biological processes such as immunity. In this comprehensive study, we investigated defense responses and gene expression and have clearly shown the involvement of SWR1c and H2A.Z in immunity as positive regulators. Although the SWR1c component proteins are not involved in defense signaling events directly, they exert their roles through gene regulation. The genome-wide transcriptomic analysis of the corresponding mutants is consistent with the early reports showing that SWR1c and H2A.Z are involved in regulating defense-related genes (March-Díaz et al., 2008, Coleman-Derr and Zilberman, 2012). This is particularly in line with the observation that responsive genes are enriched in H2A.Z on gene bodies (Coleman-Derr and Zilberman, 2012). Chromatin state and architecture have been implicated in the repression of SAR and stress-related gene expression in general (Bartsch, 2006, Hamon and Cossart, 2008, van den Burg and Takken, 2009, Ma et al., 2011, Talbert and Henikoff, 2014). Our extensive analyses highlight that, despite the evolutionary conservation of fundamental molecular processes such as chromatin remodeling, in Arabidopsis SWR1c components carry out non-overlapping roles. This can be either due to some of the non-enzymatic subunits having roles outside the SWR1c or the potential variation of the degree to which depletion or inclusion of each of the subunits affects the complex function. It is likely that some of the non-enzymatic components such as SWC6 and ARP6 are non-essential for complex activity and that their depletion will affect the complex differently. Our double mutant data provide further support for this. However, further molecular dissection is required to comprehensively define the role that each component protein plays in the assembly, recruitment to specific target, and histone exchange activity.
Importantly, our results that H2A.Z and SWR1c are positive regulators of plant resistance to different pathogens are particularly significant in the context of environmental modulation of defense responses. Elevated temperatures have been shown to suppress plant immunity (Wang et al., 2009, Alcázar and Parker, 2011). H2A.Z nucleosome dynamics have been shown to underlie temperature responses in Arabidopsis (Kumar and Wigge, 2010). Therefore, compromised immunity in the SWR1c subunit mutants where H2A.Z deposition is perturbed could be reminiscent of the influence of temperature response. Further studies will unravel the role of H2A.Z-mediated chromatin dynamics in the environmentally modulated defense gene expression.
Methods
Plant Material and Growth Conditions
All the experiments were performed in Arabidopsis thaliana accession Col-0. We used the previously described mutants arp6-1 (Deal et al., 2005), pie1-2 (Noh and Amasino, 2003), swc6-1 (Lázaro et al., 2008), hta9-1, hta11-1 (March-Díaz et al., 2008), ndr1-1 (Century et al., 1995), and eds1-2 (Bartsch, 2006), and the transgenic complementation line swc6 35S-myc:AtSWC6 (Choi et al., 2007) in our experiments. For bacterial infection assays, the ROS burst assay, and trypan blue leaf staining, Arabidopsis plants were grown on soil for 5 weeks in SD conditions (8 h light/16 h dark) in Sanyo growth cabinets at 22°C and 70% relative humidity. Otherwise 2-week-old seedlings grown in half-strength Murashige and Skoog (MS) plates after seed-surface sterilization were used.
Pathogen Assays
Bacterial infections were performed as described before in Berriri et al. (2012). Briefly, spray inoculations were performed with bacterial suspension at 2 × 107 and 2 × 108 cfu/ml of P. syringae pv tomato DC3000 and Pst DC3000 carrying AvrRps4 or AvrRpt2 genes, respectively, in 10 mM MgCl2 with 0.04% Silwet L-77. Infiltration experiments were performed with bacterial suspensions at 105 cfu/ml in 10 mM MgCl2 using a needless syringe. Bacterial titers where determined at day 0 (2 h) and/or at day 3 post inoculation. Significant differences to Col-0 are expressed using Student's t-test at P < 0.05.
The Botrytis cinerea infection assay was performed as previously described in Schoonbeek et al. (2007). A Botrytis spore suspension (2.5 × 105 spore/ml) prepared in quarter-strength potato dextrose broth (PDB, 6 g/l) was used for drop inoculation on the leaves of 5-week-old Arabidopsis plants. Lesion diameters were measured at 3 dpi. On each plant, five leaves were inoculated with one droplet of spores of Botrytis cinerea isolate B05.10.
SA Measurements
SA contents were determined 24 h after Pst DC3000 AvrRps4 infiltration. Free SA was extracted twice from 10 mg of freeze-dried leaves with 10% methanol and 1% acetic acid in the presence of 125 nM 2-hydroxybenzoic-3,4,5,6-d4 acid as internal standard (CDN Isotopes). The samples were run on a Xevo TQS tandem mass spectrometer attached to an Acquity UPLC system. Separation was on a 50 × 2.1 mm 2.7μ Kinetex XB C18 column (Phenomenex) running in a gradient of acetonitrile versus 0.1% formic acid in water.
Gene Expression Analysis
For RNA-seq analysis, total RNA was extracted from 2-week-old seedlings grown on ½ MS solid medium using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer instructions. Sequencing was performed at The Genome Analysis Center with Illumina HiSeq 2500. The 50-bp-long single-end reads were mapped to the Arabidopsis genome (TAIR10) and quantified to generate RPKM (reads per kilobase of exon per million fragments mapped) values. The Tophat-cufflinks pipeline was used to align RNA-seq reads to Arabidopsis thaliana reference genome assembly. Differential gene expression and PCA analysis was performed with the RPKM values generated using Strand NGS 2.0 (Agilent). The expression values for the three biological replicates were analyzed for statistical significance using a one-way analysis of variance test applying the Benjamini-Hochberg false discovery rate (FDR) and the multiple testing correction (Benjamini and Hochberg, 1995) and asymptotic P value computation with an adjusted P value cut-off of P < 0.05. Genes with absolute fold change values of at least two were included in our analysis as differentially expressed genes. For the comparison of our transcriptome data with the published results, we considered the 15 199 genes that passed the FDR cut-off of <0.05 as the global number. The hypergeometric probability was calculated to reveal significant overlap between the different genes sets.
Pathogen-Responsive Gene Expression Analysis
Four-week-old wild-type and mutant plants were used for analyzing defense gene expression in response to bacterial inoculation. Plants were either spray inoculated with Pst DC3000 or were mock treated with 10 mM MgCl2. Leaf samples were collected 6 h post treatments. Total RNA was isolated as above, and 1 μg of RNA was used as a template for reverse transcription with Superscript III reverse transcriptase (Invitrogen) and oligo dT according to the manufacturer's instructions. Ef1α (AT5G60390) transcript levels were used as internal reference (oligos: 5′ TACGCCCCAGTTCTCGATTG 3′ and 5′ GGCTTGGTTGGGGTCATCTT 3′) for analyzing PR1 (oligos: 5′ ACCAGGCACGAGGAGCGGTA 3′ and 5′ TCCCCGTAAGGCCCACCAGA 3′) and PR5 (oligos: 5′ ACCCACAGCACAGAGACACACA 3′ and 5′ TGGCCATAACAGCAATGCCGC 3′) expression. Quantitative RT–PCR experiments were performed in a Light Cycler LC480 using Light Cycler 480 SYBR Green I Master (Roche).
Funding
This work was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/I019022/1 and Institute Strategic Program grants BB/J004588/1 and BB/J004553/1.
Author Contributions
S.B., S.N.G., and S.V.K. designed experiments; S.B. performed most of the experiments and data analyses; S.N.G. performed experiments and data analysis for Figures 7 and 8; S.B. and S.V.K. analyzed the data and wrote the paper with inputs from S.N.G.
Acknowledgments
We thank L. Hill for help with SA analysis and H.J. Schoonbeek for help with the Botrytis infection assays. We are grateful to J. Jones, S. Boden, J. Murray, J. Colcombet, A.V. Garcia, and R. Sablowski for critical reading of the manuscript and for discussions. We thank the members of the Kumar lab for helpful discussions. No conflict of interest declared.
Published: April 27, 2016
Footnotes
Published by the Molecular Plant Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, SIBS, CAS.
Supplemental Information is available at Molecular Plant Online.
Supplemental Information
References
- Alcázar R., Parker J.E. The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 2011;16:666–675. doi: 10.1016/j.tplants.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Bartsch M. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- Berriri S., Garcia A.V., Frei dit Frey N., Rozhon W., Pateyron S., Leonhardt N., Montillet J.-L., Leung J., Hirt H., Colcombet J. Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell. 2012;24:4281–4293. doi: 10.1105/tpc.112.101253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden S.A., Kavanová M., Finnegan E.J., Wigge P.A. Thermal stress effects on grain yield in Brachypodium distachyon occur via H2A.Z-nucleosomes. Genome Biol. 2013;14:R65. doi: 10.1186/gb-2013-14-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon A.S., Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century K.S., Holub E.B., Staskawicz B.J. NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA. 1995;92:6597–6601. doi: 10.1073/pnas.92.14.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Park C., Lee J., Oh M., Noh B., Lee I. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development. 2007;134:1931–1941. doi: 10.1242/dev.001891. [DOI] [PubMed] [Google Scholar]
- Choi K., Zhao X., Kelly K.A., Venn O., Higgins J.D., Yelina N.E., Hardcastle T.J., Ziolkowski P.A., Copenhaver G.P., Franklin F.C.H. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 2013;45:1327–1336. doi: 10.1038/ng.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J.D., Liu Y., Klessig D.F., Dong X. Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell. 1998;10:557–569. doi: 10.1105/tpc.10.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D., Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Kandasamy M.K., McKinney E.C., Meagher R.B. The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell. 2005;17:2633–2646. doi: 10.1105/tpc.105.035196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney T.P., Friedrich L., Ryals J.A. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Feys B.J., Moisan L.J., Newman M.A., Parker J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W., Hinsch M.E., Staskawicz B.J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Hamon M.A., Cossart P. Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe. 2008;4:100–109. doi: 10.1016/j.chom.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Howe G.A., Jander G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- Kim K.-C., Lai Z., Fan B., Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008;20:2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J. A protein complex containing the conserved Swi2/Snf2-Related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:e131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Kunkel B.N., Bent A.F., Dahlbeck D., Innes R.W., Staskawicz B.J. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro A., Gómez-Zambrano A., López-González L., Piñeiro M., Jarillo J.A. Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 2008;59:653–666. doi: 10.1093/jxb/erm332. [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Ma K.W., Flores C., Ma W. Chromatin configuration as a battlefield in plant-bacteria interactions. Plant Physiol. 2011;157:535–543. doi: 10.1104/pp.111.182295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Diaz R., Garcia-Dominguez M., Florencio F.J., Reyes J.C. SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 2007;143:893–901. doi: 10.1104/pp.106.092270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Díaz R., García-Domínguez M., Lozano-Juste J., León J., Florencio F.J., Reyes J.C. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 2008;53:475–487. doi: 10.1111/j.1365-313X.2007.03361.x. [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M., Lázaro A., Poethig R.S., Gómez-Mena C., Piñeiro M.A., Martinez-Zapater J.M., Jarillo J.A. EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development. 2006;133:1241–1252. doi: 10.1242/dev.02301. [DOI] [PubMed] [Google Scholar]
- Matschi S., Hake K., Herde M., Hause B., Romeis T. The calcium-dependent protein kinase CPK28 regulates development by inducing growth phase-specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in Arabidopsis. Plant Cell. 2015;27:591–606. doi: 10.1105/tpc.15.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W.-H., Sen S., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J., Sundaramoorthy R., Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M., Shirasu K., Noutoshi Y., Kubo Y., Shiraishi T., Iwabuchi M., Narusaka Y. RRS1and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- Ndamukong I., Abdallat A.A., Thurow C., Fode B., Zander M., Weigel R., Gatz C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50:128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F., Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Noh Y.-S., Amasino R.M. PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell. 2003;15:1671–1682. doi: 10.1105/tpc.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S.P., Somssich I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Zhao L., Skaggs M.I., Andreuzza S., Tsukamoto T., Panoli A., Wallace K.N., Smith S., Siddiqi I., Yang Z. ACTIN-RELATED PROTEIN6 regulates female meiosis by modulating meiotic gene expression in Arabidopsis. Plant Cell. 2014;26:1612–1628. doi: 10.1105/tpc.113.120576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D.G. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Rosa M., Harder, Von M., Aiese Cigliano R., Schlogelhofer P., Mittelsten Scheid O. The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell. 2013;25:1990–2001. doi: 10.1105/tpc.112.104067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Schoonbeek H.J., Jacquat-Bovet A.C., Mascher F., Métraux J.P. Oxalate-degrading bacteria can protect Arabidopsis thaliana and crop plants against Botrytis cinerea. Mol. Plant Microbe Interact. 2007;20:1535–1544. doi: 10.1094/MPMI-20-12-1535. [DOI] [PubMed] [Google Scholar]
- Smith A.P., Jain A., Deal R.B., Nagarajan V.K., Poling M.D., Raghothama K.G., Meagher R.B. Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 2009;152:217–225. doi: 10.1104/pp.109.145532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Henikoff S. Environmental responses mediated by histone variants. Trends Cell Biol. 2014;24:642–650. doi: 10.1016/j.tcb.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Thakar A., Gupta P., Ishibashi T., Finn R., Silva-Moreno B., Uchiyama S., Fukui K., Tomschik M., Ausió J., Zlatanova J. H2A.Z and H3.3 histone variants affect nucleosome structure: biochemical and biophysical studies. Biochemistry. 2009;48:10852–10857. doi: 10.1021/bi901129e. [DOI] [PubMed] [Google Scholar]
- Thaler J.S., Humphrey P.T., Whiteman N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012;17:260–270. doi: 10.1016/j.tplants.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Thambirajah A.A., Dryhurst D., Ishibashi T., Li A., Maffey A.H., Ausió J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 2006;281:20036–20044. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Takken F.L.W. Does chromatin remodeling mark systemic acquired resistance? Trends Plant Sci. 2009;14:286–294. doi: 10.1016/j.tplants.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Van der Does D., Leon-Reyes A., Koornneef A., Van Verk M.C., Rodenburg N., Pauwels L., Goossens A., Korbes A.P., Memelink J., Ritsema T. Salicylic Acid Suppresses Jasmonic Acid Signaling Downstream of SCFCOI1-JAZ by Targeting GCC Promoter Motifs via Transcription Factor ORA59. Plant Cell. 2013;25:744–761. doi: 10.1105/tpc.112.108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Wang Y., Bao Z., Zhu Y., Hua J. Analysis of temperature modulation of plant defense against biotrophic microbes. Mol. Plant Microbe Interact. 2009;22:498–506. doi: 10.1094/MPMI-22-5-0498. [DOI] [PubMed] [Google Scholar]
- Wu W.-H., Alami S., Luk E., Wu C.-H., Sen S., Mizuguchi G., Wei D., Wu C. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 2005;12:1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Shimada K., Oma Y., Kalck V., Akimura K., Taddei A., Iwahashi H., Kugou K., Ohta K., Gasser S.M. Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet. 2010;6:e1000910. doi: 10.1371/journal.pgen.1000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Goritschnig S., Dong X., Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell. 2003;15:2636–2646. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J., Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.