Abstract
Brain metastases represent the greatest clinical challenge in treating HER2-positive breast cancer. We report the development of orthotopic patient-derived xenografts (PDXs) of HER2-expressing breast cancer brain metastases (BCBM), and their use for the identification of targeted combination therapies. Combined inhibition of PI3K and mTOR resulted in durable tumor regressions in three of five PDXs, and therapeutic response correlated with reduction of 4EBP1 phosphorylation. The two non-responding PDXs showed hypermutated genomes with enrichment of mutations in DNA repair genes, suggesting an association of genomic instability with therapeutic resistance. These findings suggest that a biomarker-driven clinical trial of PI3K inhibitor plus an mTOR inhibitor should be conducted for patients with HER2-positive BCBM.
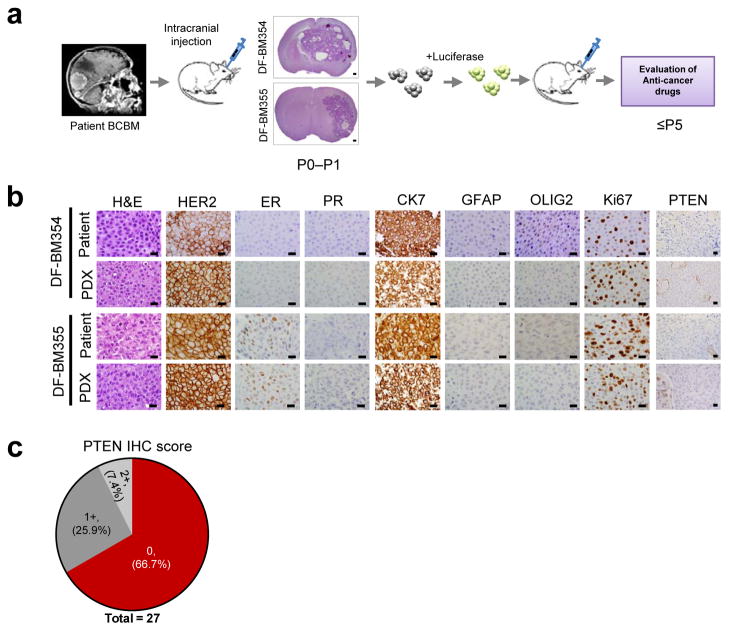
To develop therapeutic strategies for HER2-positive BCBM, we established a panel of orthotopic PDXs (Fig. 1a). Fresh BCBMs from two patients (DF-BM354 and DF-BM355) were grafted directly into the brains of SCID mice with median survival ~2–3 months (Supplementary Fig. 1). The PDXs resembled the parental BCBMs histologically, as well by estrogen receptor (ER), progesterone receptor (PR) and HER2 expression (Fig. 1b). We also verified expression of the epithelial marker cytokeratin 7 (CK7) and the absence of glial markers (GFAP and OLIG2) (Fig. 1b). We subsequently established PDXs using BCBM from three further patients with HER2-positive BCBM. Importantly, none of the five PDXs expressed detectable PTEN protein (Fig 1b, Supplementary Table 1). Consistently, 67% of 27 clinical specimens of HER2-positive BCBMs showed no PTEN staining (Fig. 1c), further confirming that PTEN-loss is widespread in breast cancer brain metastases1,2.
Figure 1. Establishment of orthotopic HER2-positive BCBM PDXs.
(a) Schematic depicting the process of generating orthotopic PDX BCBM models for use in preclinical studies. Fresh brain metastatic tissues from patients with BCBM were grafted directly into the brains of female SCID mice. The xenografts in the brain were explanted, dissociated and transduced with a luciferase gene, and then re-injected into new cohorts of mice. P0, primary graft; P1–P5, passage number in mice. DF-BM: Dana-Farber Brain Metastases samples. (b) Representative histologic and immunophenotypic analyses of two patient surgical biopsies and corresponding PDXs. (Scale bars = 25 μm). (c) Compiled result of PTEN immunohistochemistry performed on 27 human HER2-positive BCBM samples. 0, no staining in > 90% of tumor cells; 1+, weak staining in > 75% of tumor cells; 2+, strong staining in > 75% of tumor cells.
To assess the response of HER2-positive BCBMs to targeted therapy, we treated DF-BM355 PDXs with the EGFR/HER2 kinase inhibitor lapatinib. DF-BM355 PDXs showed no response to lapatinib, consistent with the resistance of the donor patient’s tumor to HER2-directed therapy (Supplementary Fig. 2a, b). Since DF-BM355 lacks PTEN (a key regulator of the PI3K pathway), we tested the combination of lapatinib with BKM120, a pan-PI3K inhibitor that penetrates the blood-brain barrier (BBB)3–5. Again, no response was observed (Supplementary Fig. 2a, b).
To understand the lack of response to combined HER2 / PI3K inhibition, we assessed tumor PI3K pathway signaling in response to lapatinib and/or BKM120. While these treatments reduced AKT and S6RP phosphorlyation, we observed little change in p-4EBP1, an mTORC1 effector that mediates translation (Supplementary Fig. 2c, d). Therefore even combined HER2/PI3K inhibition did not completely suppress mTORC1 activity in the DF-BM355 model.
Notably, persistent mTOR activity in breast cancers can mediate resistance to PI3K inhibition, and this can be overcome by mTORC1 inhibition6. However, the brain microenvironment is unique, and brain metastases are notoriously refractory to systemic therapies that are effective against extracranial metastases. In keeping with this, it is not known whether mTOR inhibition might overcome PI3K inhibitor resistance in BCBMs. To explore this, we combined either lapatinib or BKM120 with RAD001, an mTORC1 inhibitor that penetrates the BBB7,8.
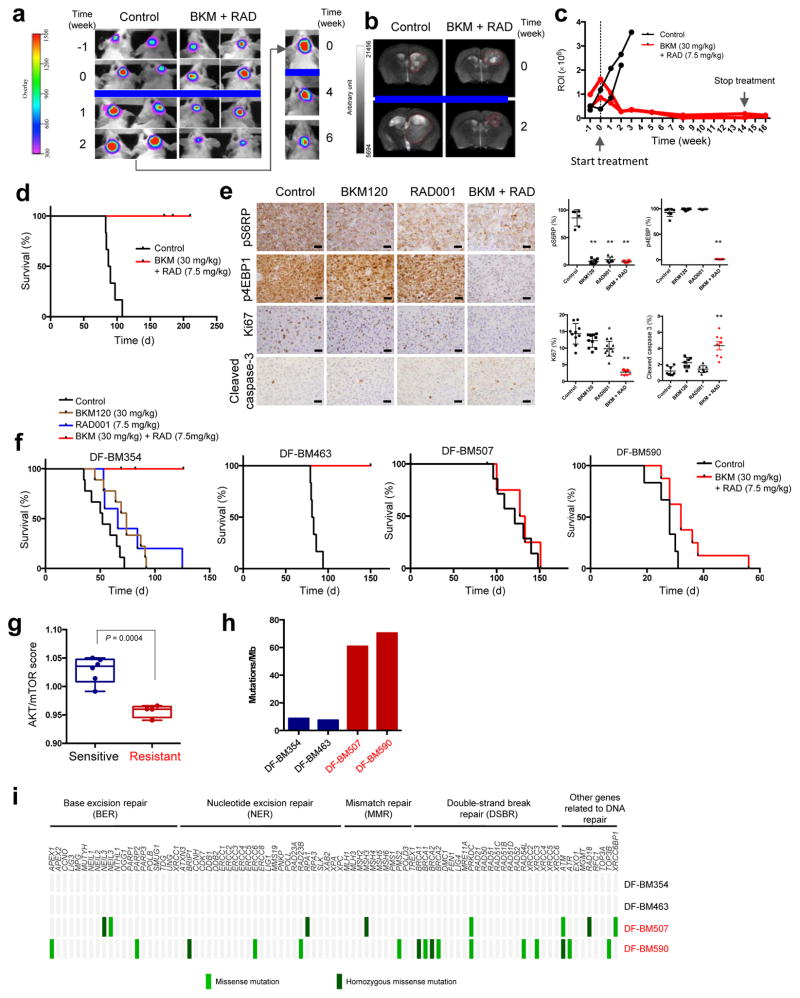
Whereas DF-BM355 PDXs showed limited response to the combination of lapatinib and RAD001 (Supplementary Fig. 2e), BKM120/RAD001 resulted in marked tumor regression as measured by bioluminescence (Fig. 2a). Due to the unprecedented nature of this response, we removed mice from the control group once their tumors were much larger, and introduced BKM120/RAD001. The larger tumors also regressed over time (Fig. 2a). MRI before and after treatment confirmed these results (Fig. 2b). While mice in the control group quickly reached study endpoint with high luciferase signals, reduced mobility and other neurologic signs, the luciferase signal in BKM120/RAD001-treated tumors declined to a nearly undetectable level over the treatment period of 14 weeks, and mice remained healthy and luciferase signal-free for weeks after treatment cessation (Fig. 2c and Supplementary Fig. 2e). All treated mice survived during 210 days of observation whereas all mice in the control group died after approximately 90 days (Fig. 2d).
Figure 2. Differential responses of HER2-positive BCBM PDXs to the combination of BKM120/RAD001.
(a) Representative bioluminescence imaging analysis of mice bearing DF-BM355 tumor before and after treatment with combined BKM120 (30 mg/kg) and RAD001 (7.5 mg/kg), n = 5. (b) Representative MRI of DF-BM355-bearing mice treated with vehicle control or combined BKM120 with RAD001, n = 3. (c) Quantification of the regions of interest (ROI) determined at each imaging time point, n = 2. (d) Kaplan–Meier survival of DF-BM355-bearing mice treated with vehicle control or BKM120 + RAD001, n = 6. (e) IHC analyses of p-4EBP1, p-S6RP, Ki67 and cleaved caspase-3 on DF-BM355 tumors treated for 4 days with indicated treatments (Scale bars = 25 μm). Graphs represent mean ± s.d. (n = 6–10 images per group, *P < 0.05, **P < 0.01, one-way ANOVA followed by Dunnett’s test). (f) Kaplan–Meier survival of mice bearing DF-BM354, DF-BM463, DF-BM507, and DF-BM590 with vehicle control or compound as indicated. n = 5–9. (g) Transcriptome analysis of AKT/mTOR-dependent signature genes from brain xenograft tumor tissues from untreated mice. Boxplots correspond to the first and third quartiles with upper and lower whisker extending to the highest value that is within 1.5 times the interquartile range (n = 4–6 per group, P = 0.0004, Student’s t-test). (h) Number of somatic mutations in HER2-positive BCBM PDXs identified by WES. (i) Mutational profiling of a panel of DNA repair genes.
The unique efficacy of this combination is underscored by results from additional experiments; neither a BKM120/MEK162 (a MEK inhibitor) combination (chosen due to high p-ERK levels in the PDX tumors) nor a BKM120/JQ1 (a BET bromodomain inhibitor that downregulates MYC expression) combination (chosen due to MYC amplifications in the PDX tumors) showed efficacy (Supplementary Figs. 3 and 4).
To understand the mechanism underlying synergy between BKM120 and RAD001, we harvested tumors from mice after 4 days of treatment for pharmacodynamic assessment. While both monotherapies reduced p-S6RP, neither drug alone significantly suppressed p-4EBP1 (Fig. 2e), suggesting that mTORC1 was not completely inhibited. These observations are consistent with reports indicating that whereas rapamycin (an mTORC1 inhibitor) stably inhibits p-S6RP, its effect on p-4EBP1 is short-lived, and that a rapid re-emergence of p-4EBP1 contributes to rapamycin resistance9. Notably, combined BKM120/RAD001 treatment dramatically reduced p-4EBP1 and also significantly decreased proliferation (Ki67 staining), and increased apoptosis (cleaved caspase-3 staining) (Fig. 2e).
To determine if these results could be replicated in other BCBMs, we tested the same therapy in the remaining four PDX models of BCBM (DF-BM354, DF-BM463, DF-BM507, and DF-BM590). Consistent with our findings in DF-BM355, neither BKM120 nor RAD001 monotherapy had meaningful effects in the DF-BM354 model, whereas the combination therapy led to durable tumor regression, significant reductions in p-S6RP and p-4EBP1, and significant decreases in Ki67 staining (Fig. 2f and Supplementary Fig. 5a, b). Notably, DF-BM354 and DF-BM355 show disparate ER status (Fig. 1b), suggesting that the BMK120/RAD001 combination might be effective for HER2-positive BCBMs regardless of hormone receptor expression. The DF-BM463 model also exhibited durable responses and similar changes in p-4EBP1, Ki67, and cleaved caspase-3 levels in response to BKM120/RAD001 therapy (Fig. 2f and Supplementary Fig. 5c, d).
In contrast, BKM120/RAD001 combination therapy had little effect on the survival of mice bearing DF-BM507 and DF-BM590 PDXs, and p-S6RP and p-4EBP1 levels were also not suppressed in these models (Fig. 2f and Supplementary Fig. 5e, f). To understand the molecular basis for the differential therapeutic responses between models, we performed transcriptome analyses on tumors from untreated mice. The three responding models showed significantly higher expression of AKT-mTOR-dependent signature genes10 (Fig. 2g), suggesting that some but not all HER2-positive BCBMs depend on the AKT-mTOR pathway.
We also performed whole-exome-sequencing (WES) of all five PDX tumors and matched samples from the donor patients’ blood (patient blood was unavailable for DF-BM355). Copy number variations (CNVs) were frequent in all five tumor models (Supplementary Fig. 6) and each PDX and its matched patient tumor shared almost identical CNVs (Supplementary Fig. 7a), suggesting conservation of genetic alterations in these PDXs. The rate of non-synonymous somatic mutations in responding tumors was ~7–8 per Mb (Fig. 2h), in line with recently reported data that the mutation rate in HER2-positive BCBMs is ~10 mutations per Mb11. The mutation rate in non-responsive BCBMs, in contrast, was ~60–70 mutations per Mb (Fig. 2h). Perhaps relevant to this hypermutation phenotype is the knowledge that the two non-responding PDXs were derived from patients who had more cycles of chemotherapy and/or radiation therapy than the patients from whom the responding PDXs were derived (Supplementary Table 2). Hyper-mutation has been linked to mutations in DNA repair genes12, and indeed therapy-resistant PDXs and their matched patient specimens harbored mutations in several DNA repair genes (Fig. 2i, Supplementary Fig. 7b, and Supplementary Tables 3 and 4). In agreement with this observation, a recent analysis of metastatic breast cancer samples from the BOLERO-2 trial revealed that higher genomic instability correlates with resistance to everolimus (RAD001)13.
Since the combination of BKM120 plus RAD001 is already under clinical evaluation in advanced solid malignancies, the translation of our preclinical findings could be fast-tracked into the clinic for patients with HER2-positive BCBM. More broadly, our study demonstrates that the use of brain metastasis-specific PDX models facilitates the integration of phenotypic and genotypic analyses and advances precision medicine in cancer.
Online Methods
Patient-derived xenografts
The informed consent was obtained from patients and fresh brain metastases were acquired from patients with breast cancer brain metastases undergoing neurosurgery at the Brigham and Women’s Hospital as part of Dana-Farber Institutional Review Board (IRB) approved protocol (DFCI IRB 93-085 and 10-417) within the Dana-Farber/Harvard Cancer Center (DF/HCC) Living Tissue Bank program. To establish patient-derived metastatic breast models, fresh tumor tissue was dissociated in gentleMACS C Tubes using mechanical and enzymatic methods (Miltenyi Biotech). A suspension of metastatic breast cells was prepared at a concentration of 100,000 viable cells per microliter and temporarily incubated on ice prior to intracranial injections. 8–10 weeks old female severe combined immunodeficiency (SCID) mice acquired from Taconic (IcrTac:ICR-Prkdcscid) were anesthetized with oxygen-diluted isoflurane or ketamine/xylazine and positioned into a stereotactic frame, whereby the head was secured by gentle pressure from ear bars while maintaining deep anesthesia. A one centimeter scalp incision was made to identify the bregma which served as the zero coordinates (x = 0 mm, y = 0 mm, z = 0 mm). A burr hole was created in the skull at right hemisphere at coordinates x=0 mm, y=2 mm, z= 0 mm and each animal was injected with 100,000 viable tumor cells into the right striatum (z=2 mm). The scalp was closed with 9 mm Autoclips (BD Diagnostic Systems). Mice bearing xenografts were housed under standard conditions and monitored closely for the development of systemic symptoms of morbidity or neurologic phenotypes, including rapid body weight loss, hunched posture, inability to obtain food or water, or other humane endpoints necessitating euthanasia. Following euthanasia, brain tumors from symptomatic animals were collected by dissection, dissociated and re-injected intracranially into additional animals (serial passaging in vivo). Luciferase expression was introduced in tumor cells from primary grafts (P0–1) and low passage PDXs (at passage 5 or lower) were used for all the therapeutic experiments. Alternatively for neuropathologic evaluation of brain tumors, euthanized animals bearing xenografts were perfused by intra-cardiac injection of 4% paraformaldehyde (PFA) in Phosphate-Buffered Saline (PBS, pH 7.0) and the brain tissue with tumor lesions were processed by standard methods for paraffin embedding. Hematoxylin and Eosin (H&E) stained sections were generated and the tumors were then evaluated. All the animal experiments were performed according to protocols approved by the Dana-Farber Cancer Institute Animal Care and Use Committee in compliance with NIH animal guidelines.
Immunohistochemistry
Diaminobenzidine (DAB), brightfield staining was performed according to standard protocols using DAB EnVision+ System (Dako) on paraffin sections. Briefly five micron thick sections were deparaffinized with xylene, followed by gradation washes in 100%, 95%, 80% ethyl alcohol before performing heat antigen retrieval in 10 mM Sodium Citrate buffer (pH 6.0) for 20 minutes. Subsequently sections were treated with peroxidase block (Dako) for ten minutes followed by overnight incubation at 4°C with the following primary antibodies: PTEN (Cell Signaling #9559), p-S6RP-Ser235/236 (Cell Signaling #2211), p-4EBP1-Thr37/46 (Cell Signaling #2855), cleaved caspase-3 (Cell Signaling #9664), Estrogen Receptor (ER) (ThermoScientific SP1), Progesterone Receptor (PR) (Dako PgR 636), HER2 (ThermoScientific SP3), CK7 (Dako), GFAP (Dako #z0334), OLIG2 (Dako) and monoclonal mouse anti-Ki67 (Dako MIB-1 or Vector lab #VP K-451). After multiple washes with 1X Tris-buffered saline and Tween 20 (TBST) solution, slides were incubated at room temperature for two hours with corresponding species- specific horseradish peroxidase (HRP) conjugated secondary antibody from EnVision+ System (Dako). Signal was visualized by the HRP-DAB reaction. Counterstaining for nuclei was performed using Mayer’s hematoxylin stain followed by graded dehydration and xylene washes. Coverslips were mounted with Permount (Fisher Scientific). Antibody validation is provided on the manufacturers’ website.
Ki67 and cleaved caspase-3 indexes were calculated as a percentage of positive cells in 4–5 random areas of each sample. Images were captured at 40x or 60x magnification and quantifications of Ki67 and cleaved caspase-3 immunoreactivity were performed using the Image J software with ImmunoRatio plugin. P-S6RP and p-4EBP indexes were calculated as a percentage of positive areas in 3–5 random areas of each sample. Images were captured at 40x or 60x magnification and quantification of p-S6RP and p-4EBP immunoreactivity was performed using the Cellvigene software (VigeneTech). The IHC experiments and data analyses were done by two investigators blinded to the group allocation.
Lentiviral production and transduction
HEK293T cell line was obtained from ATCC (Manassa, VA). The cells were maintained in DMEM supplemented with 10% FBS and 100 μg/ml penicillin-streptomycin. The cells were frozen after receipt and were used at early passages. No authentication or mycoplasma contamination test was done. The pLenti-blasticidin-Luciferase vector was co-transfected with pCMV-delta8.9 and pMD.G at the ratio of 4:3:1 into HEK293T cells by PEI (1 μg/μl) (4:1 to DNA). The culture medium was replaced 1 day after transfection and the viral supernatants were collected 1 day and 2 day later. The viral supernatants were filtered through 0.45 μm filter and were then concentrated by ultracentrifugation (SW28, 16,600 rpm, 2h). Viral pellets were resuspended into PBS and aliquoted and stored at −80 °C for future use. Viral titers were determined by qPCR Lentivirus titration Kit (Applied Biological Materials Inc.)
Tumor cells were isolated from passage 0 or 1 PDXs and transduced with a lentivirus encoding Luciferase (pLenti-blasticidin-Luciferase) at ~MOI 5 in suspension overnight with polybrene 8 μg/ml, and then subjected to 3-day antibiotic selection with blasticidin 2 μg/ml to enrich the population of blasticidin-Luciferase expressing cells in NeuroCult NS-A media (Stemcell Technologies) supplemented with heparin sulfate (2 mg/mL), Epidermal Growth Factor (EGF, 20 ng/ml), basic Fibroblast Growth Factor (bFGF, 20 ng/ml), and Hydrocortisone (0.5 μg/ml). These tumor cells were then propagated in mice.
Bioluminescence imaging
For imaging, mice were injected i.p. with D-luciferin (Promega) (~60 mg/kg) together with anesthetized reagents ketamine (100 mg/kg) and xylazine (7 mg/kg). Ten minutes later, luciferase expression was recorded and images were obtained with Kodak Image Station 4000MM for 20 minutes (DF-BM355) or 5 minutes (DF-BM354). The signals were analyzed with CareStream MI Software.
In vivo treatment
BKM120 was dissolved in 10% NMP with 90% PEG300 and given orally once/day at 30 mg/kg. RAD001 was freshly prepared from a microemulsion pre-concentrate (Novartis) with 5% glucose dilution or dissolved in 10% NMP with 90% PEG300 and daily oral delivery to mice at 7.5 mg/kg. Lapatinib was dissolved in 0.5% hydroxypropyl methylcellulose (HPMC) with 0.1% Tween80 and administered at 100 mg/kg body weight once a day by oral gavage. All compounds were purchased from Haoyuan ChemExpress Co.
Treatment started at 3–6 weeks following injections of tumor cells when mice developed luciferase signals with ROI 0.3–1.0 × 106 as baseline levels of tumor lesion. Tumor-bearing mice were randomized into control and treatment groups and 5–6 mice were used for each experimental cohort. Mice were followed to their end points (30–100 days) or the treated mice when they survived 50%–100% longer than mice in the control groups. The sample size for the experiments was determined by the pilot studies in the laboratories and the StatsToDo program based on probability of type I error (α) at 0.05, power at 0.8, expected difference between two means and standard deviation within the group. The drug treatments with the outcome measurements and data analysis were done by two investigators blinded to the group allocations.
MRI imaging
MRI experiments were performed on a Bruker BioSpec 7T/30 cm USR horizontal bore Superconducting Magnet System, equipped with the B-GA12S2 gradient and integrated with up to 2nd order room temperature shim system, which provides a maximum gradient amplitude of 440 mT/m and slew rate of 3440 T/m/s. The Bruker made 23 mm ID birdcage volume radiofrenquancy (RF) coil was used for both RF excitation and receiving. AutoPac with laser was used for precise animal positioning. Animals were anesthetized throughout the imaging procedure through inhalation of a mixture of 1.5% isoflurane into medical supplied air. Animal respiration and temperature were monitored and regulated by the SAII (Stony Brook, NY) monitoring and gating system model 1025T.
Bruker Paravision 5.1 was used for MRI data acquisition. Once animals were positioned in the magnet, a three orthogonal scout imaging protocol was loaded and run with the traffic light meaning it would run the automatic center frequency, automatic shim, reference RF gain, receive gain and then acquire the reference images. T2 weighted images were obtained from fast spin echo (RARE) with fat suppression sequence and the following parameters: TE = 33 ms, TR = 2500 ms, Rare Factor = 8, Number of Averages = 2, total acquisition time 2m40s, FOV = 20×20 mm2, matrix size = 256×256, spatial resolution = 78 × 78 μm2, slice thickness = 1.0 mm, number of slices = 12. 3D volume reconstructions were obtained using OsiriX software.
Whole-exome sequencing
The exome was sequenced on the Ion Torrent Proton platform (Life Technologies, Thermo Fisher) according to the manufacturer’s instructions. Briefly, genomic DNA was extracted from the patient peripheral blood or PDX tumors using DNeasy blood & tissue kit (Qiagen). DNA libraries were constructed from 100 ng gDNA using the Ion AmpliSeq Exome kit (Life Technologies, Thermo Fisher) that provides targeted regions of greater than 97% the coding exons of the human genome. The final exome libraries were quantitated by Ion Library Quantitation Kit (Life Technologies, Thermo Fisher). 2–3 libraries were multiplexed and clonally amplified to obtain template-positive ion sphere particles by using the Ion OneTouch 2 System (Life Technologies, Thermo Fisher), and were then sequenced on an Ion Torrent Proton using one PI chip kit V2 (Life Technologies, Thermo Fisher). Alignment of sequencing reads was performed using Torrent Suite Software and Torrent Server. Further data analysis, variant calling and annotation of variants were carried out by AmpliSeq Exome single sample (Somatic) workflow and Tumor-Normal pair workflow using Ion reporter software (Life Technologies, Thermo Fisher). Variant calls with less than 10 reads for normal samples and 20 reads for tumor samples were removed. For the analyses of somatic mutations of DNA repair genes in the tumors, mutation calls with less than 20 reads were excluded. R and Bioconductor packages14 were used to prioritize and visualize the sequencing data. The segment plotting tool from the readDepth package for R was further modified to visualize CNV alterations15. The sequencing data have been deposited in The National Center for Biotechnology Information (NCBI) database of Genotypes and Phenotypes (dbGaP) with an accession number phs001063.v1.p1.
Transcriptome analysis
AmpliSeq human transcriptome libraries were constructed and sequenced in technical duplicate using the Ion Proton platform according to manufacturer instructions and as previously described16. Briefly, 10 ng of total RNA samples from brain xenograft tumor tissue was used for cDNA library preparation. 8 libraries were multiplexed and clonally amplified by using the Ion OneTouch 2 System (Life Technologies, Thermo Fisher), and were then sequenced on an Ion Torrent Proton machine. Data was first analyzed by Torrent Suite and ampliSeqRNA analysis plugin (Life Technologies, Thermo Fisher) to generate count data. Count data were transformed using the R-bioconductor packages DESeq217, then log2 transformed, then mean-normalized by gene to allow comparison. Given that the two agents in this study target the PI3K-Akt-mTOR pathway, we searched MSigDB (http://software.broadinstitute.org/gsea/msigdb) for gene expression signatures representing this pathway using the terms ‘Akt AND mTOR.’ Of the results, only one signature was experimentally derived10 (PMID: 17213801). We reviewed the primary manuscript and used the 35 ‘up’ genes in the original manuscript as the ‘Akt-mTOR’ signature. The AKT-mTOR signature score represents the mean of the 35 upregulated genes induced by Akt in a transgenic mouse model and sensitive to mammalian target of rapamycin (mTOR) inhibitor RAD001 in a previously published study (AKT1, BIK, BSG, DDR1, CDC34, CLDN3, CYB561, GPX4, HNRPAB, LASP1, MMP15, MVK, NEDD8, NEU1, PCTK1, POR, PRKCD, PVRL2, SPINT1, UBE2M, TMED10, DUSP10, CLSTN1, PMPCA, BRMS1, TJP3, ARHGEF16, ADIPOR1, SLC37A1, KCTD5, TOLLIP, SYNJ2BP, RNF126, CORO1B)10. Boxplots correspond to the first and third quartiles (the 25th and 75th percentiles) with upper/lower whisker extending to the highest value that is within 1.5 times the interquartile range. Code is available upon request. The transcriptome data have been deposited in The National Center for Biotechnology Information (NCBI) database of Gene Expression Omnibus (GEO) with the accession number GSE80722. The sequencing data have been deposited in The National Center for Biotechnology Information (NCBI) database of Genotypes and Phenotypes (dbGaP) with an accession number phs001063.v1.p1.
Statistical analysis
Statistical significance was determined using unpaired Student’s t-test or ANOVA by GraphPad Prism 6 (GraphPad Software). Data are considered significant when P values are < 0.05. Sample sizes and animal numbers were chosen on the basis of power calculations of 0.8 and pilot studies performed in the laboratory. No animals were excluded from the analysis.
Supplementary Material
Acknowledgments
We thank D. Livingston for reading the manuscript. We thank R. Modiste and G. Dai at Dana-Farber Lurie Family Image Center for MRI imaging. We thank R. Bronson and the Dana-Farber/Harvard Cancer Center Rodent Histoplathology Core for histopathological analyses. We thank F. Pan, D. Light, and R. Qi (Life Technologies, Thermo Fisher) for assistance of WES and transcriptome analyses with the Ion Torrent sequencing system. We thank J. Ruan and M. Ruan (VigeneTech) for quantification of pS6RP and p4EBP IHC data by Cellvigene data analysis program. This work was supported by Breast Cancer Research Foundation (N.U.L., E.P.W., Z.W. and J.J.Z.), Aid for Cancer Research (E.P.W. and J.J.Z.), Breast Cancer Alliance (J.J.Z.), NIH grants R01 CA187918 (T.M.R. and J.J.Z.), CA172461 (J.J.Z.), 1K08NS087118 (S.H.R.), P50 CA165962 (T.M.R., K.L.L. and J.J.Z.), P01 CA142536 (J.J.Z.), 1P50CA168504 (T.M.R., I.E.K., E.P.W., N.U.L., and J.J.Z.).
Footnotes
ACCESSION CODES
Whole-exome sequencing data have been deposited in The National Center for Biotechnology Information (NCBI) database of Genotypes and Phenotypes (dbGaP) with an accession number phs001063.v1.p1. The transcriptome data have been deposited in The National Center for Biotechnology Information (NCBI) database of Gene Expression Omnibus (GEO) with the accession number GSE80722.
AUTHOR CONTRIBUTIONS
J.N., S.H.R., S.X., E.P.W, N.U.L., K.L.L., and J.J.Z. conceived and designed experiments. J.N., S.H.R., S.X., K.L.L., and J.J.Z. developed methodology. J.N., S.H.R., S.X., H.G., V.L., Y.J.K., and M.H. performed the surgeries and in vitro and in vivo experiments. J.N., S.H.R., S.X., S.G., D.G.S., E.M., K.L.L., and J.J.Z. analyzed and interpreted data. J.N., S.H.R., S.X., S.G., J.D.I., I.E.K., G.-C.Y., T.M.R., E.P.W., N.U.L., K.L.L., and J.J.Z. wrote/revised the manuscript. J.N., S.H.R., S.X., L.A.R., Q.-D.N., A.H.L., R.D., E.B.C., B.M.A., and Z.C.W. provided administrative, technical, or material support. H.G., V.L., Y.J.K., and M.H. provided technical assistance. E.P.W., N.U.L., K.L.L., and J.J.Z. supervised and coordinated all the aspects of the work.
Competing Financial Interests
T.M.R. is a consultant of Novartis and has received a research grant from Novartis. E.P.W. has received research grants from Genentech and Roche. I.E.K. is a consultant of Amgen and has received research funding from Genentech. N.U.L. has received research grants from Genentech, Array Biopharma, GlaxoSmithKline, Kadmon, and Novartis. The remaining authors declare no competing financial interests.
References
- 1.Wikman H, et al. Breast cancer research : BCR. 2012;14:R49. doi: 10.1186/bcr3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, et al. Nature. 2015;527:100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filbin MG, et al. Nature medicine. 2013;19:1518–1523. doi: 10.1038/nm.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maire CL, et al. Stem cells. 2014;32:313–326. doi: 10.1002/stem.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorpe LM, Yuzugullu H, Zhao JJ. Nature reviews Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elkabets M, et al. Science translational medicine. 2013;5:196ra199. doi: 10.1126/scitranslmed.3005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krueger DA, et al. The New England journal of medicine. 2010;363:1801–1811. doi: 10.1056/NEJMoa1001671. [DOI] [PubMed] [Google Scholar]
- 8.O’Reilly T, et al. Cancer chemotherapy and pharmacology. 2010;65:625–639. doi: 10.1007/s00280-009-1068-8. [DOI] [PubMed] [Google Scholar]
- 9.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creighton CJ. Oncogene. 2007;26:4648–4655. doi: 10.1038/sj.onc.1210245. [DOI] [PubMed] [Google Scholar]
- 11.Brastianos PK, et al. Cancer discovery. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shlien A, et al. Nature genetics. 2015;47:257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 13.Hortobagyi GN, et al. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015 doi: 10.1200/JCO.2007.10.9397. [DOI] [PubMed] [Google Scholar]
- 14.Gentleman RC, et al. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller CA, Hampton O, Coarfa C, Milosavljevic A. PloS one. 2011;6:e16327. doi: 10.1371/journal.pone.0016327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, et al. Cell. 2015;163:174–186. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love MI, Huber W, Anders S. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.