Abstract
The lens epithelium-derived growth factor p75 (LEDGF/p75) is a chromatin-bound protein essential for efficient lentiviral integration. Genome-wide studies have located LEDGF/p75 inside actively transcribed genes where it mediates lentiviral integration. Although its role in HIV-1 integration is clearly established, the role of LEDGF/p75-associated proteins in HIV-1 infection remains unexplored. Using protein-protein interaction assays, we demonstrated that LEDGF/p75 complexes with a chromatin remodeling complex, Facilitates Chromatin Transcription (FACT), a heterodimer of the structure specific recognition protein 1 (SSRP1) and the human homolog of suppressor of Ty 16 (hSpt16). Detailed analysis of the interaction of LEDGF/p75 with the FACT complex indicates that LEDGF/p75 interacts with SSRP1 in an hSpt16-independent manner that requires the PWWP domain of LEDGF proteins and the HMG domain of SSRP1. Functional characterizations demonstrate a LEDGF/p75-independent role of SSRP1 in the regulation of HIV-1 replication. shRNA-mediated partial knockdown of SSRP1 reduces HIV-1, but not Murine Leukemia Virus, infection in human CD4+ T cells. Similarly, SSRP1 knockdown affects infection by HIV-1-derived viruses that express genes from the viral LTR but not from an internal immediate-early CMV promoter, suggesting a role of SSRP1 in LTR-driven gene expression but not viral DNA integration. Together, our data demonstrate for the first time the association of LEDGF proteins with the FACT complex and give further support to a role of SSRP1 in HIV-1 infection.
Keywords: HIV-1 cofactor, FACT complex, HIV-1 replication, PWWP domain, HMG domain
Graphical Abstract
INTRODUCTION
LEDGF proteins, p75 and p52, are ubiquitously expressed, chromatin-bound proteins produced by the alternative splicing of the PSIP1 gene transcript 1; 2; 3; 4. These proteins share the N-terminal region that mediates their tight interaction with chromatin at all stages of the cell cycle. The binding of LEDGF to the chromatin occurs mainly through the PWWP domain, which binds to chromatin-bound proteins and DNA, as well as two AT hook motifs that bind the minor groove of AT-rich DNA regions 5; 6.
The ability of LEDGF/p75 to tightly bind chromatin is central for its cellular and virological roles 4; 7; 8. Chromatin-bound LEDGF/p75 tethers, through the C-terminal integrase binding domain (IBD), the menin/MLL histone methyl transferase complex, associated with the co-repressors Bmi1 and Ctbp, to the Hox loci and regulates the transcription of these genes 9; 10. Similarly, LEDGF/p75 recruits C-terminal-binding protein interacting protein (CtIP) to DNA double strand breaks promoting their repair by homologous recombination 11. Lentiviruses, but not other retroviruses, have also exploited the genome-wide location and chromatin-tethering ability of LEDGF/p75 for viral replication 12. Early after infection, the HIV genome is reverse transcribed and the resulting cDNA non-covalently associates with the viral enzyme integrase within the pre-integration complex. This complex is then transported into the nucleus where integrase firmly binds to the IBD of chromatin-bound LEDGF/p75, resulting in the tethering of integrase and the viral cDNA to the host chromatin. LEDGF/p75-mediated chromatin tethering is essential for efficient integration of the HIV-1 cDNA into the host genome, a required step in the retroviral life cycle 4; 7; 8. In addition, the interaction of LEDGF/p75 with integrase favors HIV-1 cDNA integration into actively transcribed genes 13; 14; 15; 16, where LEDGF/p75 is enriched13.
The potential role of LEDGF/p75-associated proteins in HIV-1 replication has not yet been explored. We postulate that HIV-1 could have evolved dependency of LEDGF/p75-associated proteins for efficient viral replication. In support of this hypothesis, we have identified and characterized the interaction of LEDGF/p75 with the chromatin remodeler FACT complex, and defined a specific role of the FACT subunit SSRP1 in HIV-1 replication. Our findings suggest a molecular explanation for the dependency evolved by lentiviruses on LEDGF/p75 for viral replication.
RESULTS
LEDGF/p75 is associated with the FACT complex
Genome-wide LEDGF/p75 is enriched inside actively transcribed genes 17. Therefore, we evaluated the interaction of LEDGF/p75 with several members of the transcriptional apparatus. LEDGF/p75-deficient TL3 cells and TL3 cells engineered to express LEDGF/p75 were lysed in a buffer containing Triton X-100 and 100 mM NaCl, and a chromatin-bound (pellet) and -unbound (supernatant) fractions [S1 fraction 5; 18; 19] were obtained by centrifugation. Then chromatin-bound proteins were solubilized from the pellet by DNase treatment in the presence of 0.27 M (NH4)2SO4 [S2 fraction5; 18; 19]. S2 fractions were then used for immunoprecipitation of LEDGF/p75, and pulled-down proteins were evaluated by immunoblotting with specific antibodies. Using this procedure, we found that LEDGF/p75 co-immunoprecipitated with the chromatin remodeling factor FACT complex, a heterodimer composed of hSpt16 and SSRP1 proteins20. The FACT complex remodels nucleosomes in an ATP-independent fashion allowing RNA polymerase II to access the DNA during transcriptional elongation 20; 21. LEDGF/p75 interacted with the chromatin-bound fraction of FACT complex that comprise approximately fifty percent of the cellular FACT and is likely implicated in chromatin remodeling 20; 21. Although DNase and high salt were used to extract LEDGF/p75 and FACT from chromatin, and DNase was present in the immunoprecipitation reactions, it is possible that nucleosomes where still present in these preparations mediating the interaction of LEDGF/p75 and FACT complex.
To further verify the interaction of LEDGF/p75 with the FACT subunit SSRP1 in intact cells, we evaluated the subcellular co-localization of these proteins by quantitative confocal microscopy analysis, as previously described 22. As a control for specificity, we calculated the co-localization of HIV-1 integrase (IN) with LEDGF/p75 WT or a mutant lacking the integrase binding domain (ΔIBD). In correlation with previously reported data 23; 24, IN fully co-localized with LEDGF/p75 WT but not with the LEDGF/p75 ΔIBD (Figures 1b and 1c). Using this method, we demonstrated that 76% of endogenous LEDGF/p75 and SSRP1 co-localized (Figure 1b and 1c) in intact cells corroborating the results of the immunoprecipitation experiment in Figure 1a. This elevated degree of co-localization is expected considering that FACT is required for transcriptional elongation and nucleosomes in actively transcribed genes are enriched in H3K36me3, a hallmark of elongation, recognized by LEDGF/p75. To exclude the possibility that LEDGF/p75 and SSRP1 co-localize simply because of their enrichment in actively transcribed genes, we also analyzed the co-localization of RNA polymerase II (Pol II) with LEDGF/p75. These two proteins were reported to interact in vitro 1; however, it is known that Pol II is present in the majority of the promoters of protein-encoding genes, although only a sub-set of them are engaged in transcriptional elongation, hence enriched in H3K36me3 marks 25. As expected, only a partial co-localization (53%) was detected between LEDGF/p75 and Pol II, validating the specificity of our procedure.
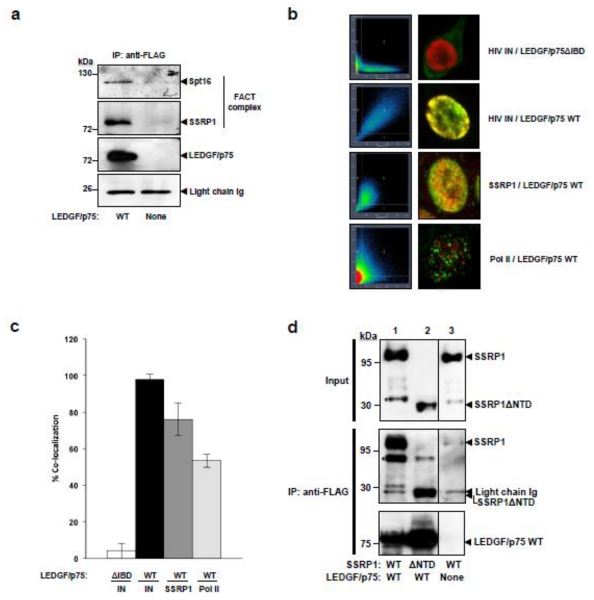
Figure 1. Interaction of LEDGF/p75 with the FACT complex.
(a) Chromatin-bound proteins were isolated from TL3 and TL3 LEDGF/p75 WT cells by DNase treatment and FLAG-tagged LEDGF/p75 was immunoprecipitated from this subcellular fraction with an anti-FLAG mAb antibody. The presence of the FACT complex (hSpt16 and SSRP1) was evaluated in the immunoprecipitated proteins by immunoblotting with specific antibodies. (b) Quantitative confocal microscopy co-localization of LEDGF/p75 with SSRP1. Panels 1 and 2 are controls and represent the co-localization of HIV integrase with LEDGF/p75 mutant lacking the integrase-binding domain (ΔIBD) or LEDGF/p75 wild type (WT). LEDGF/p75-deficient HEK293T cells stably expressing eGFP-tagged HIV-1 integrase were transiently transfected with LEDGF/p75ΔIBD or WT. LEDGF/p75 proteins were detected with an anti-LEDGF antibody. The lower panels represent the co-localization of endogenous LEDGF/p75 with endogenous SSRP1 or Pol II in HeLa cells after immunostaining with specific antibodies. (c) Quantification of co-localization data in (b). Standard deviations indicated co-localization values found in ten different cells randomly selected from a field representative of ten different random areas of the microscope slide. (d) HEK293T cells were co-transfected with plasmids expressing: FLAG-tagged LEDGF/p75 and Myc-tagged SSRP1 WT (lane 1), FLAG-tagged LEDGF/p75 and Myc-tagged SSRP1ΔNTD (lane 2), or Myc-tagged SSRP1 WT and an empty plasmid (lane 3). Samples were analyzed in the same gel, the line dividing lanes 2 and 3 indicate that lanes in between were removed for comparison in this figure. In 1d, (*) indicates degradation products of SSRP1
Collectively, the results presented in Figure 1 clearly indicate that LEDGF/p75 interacts with the FACT complex.
SSRP1 and LEDGF proteins interact independently of hSpt16
Gene expression analysis of human cells indicates that SSRP1 has hSpt16-independent and – dependent roles in transcriptional regulation 26. Therefore, we determined the hSpt16-dependency of the interaction of LEDGF/p75 with SSRP1. Myc-tagged SSRP1 wild type or a mutant lacking the N-terminal domain (NTD, Supplementary Figure S1) required for interaction with Spt16 20 were co-expressed with FLAG-tagged LEDGF/p75 in HEK293T cells and immunoprecipitated with an anti-FLAG antibody. SSRP1 interacts with nucleosomes through Spt16 20; 21; 27; 28, and therefore the NTD mutant is fully recuperated (Supplementary Figure S2) in the Triton-soluble chromatin-unbound fraction of cell lysates obtained in the absence of DNase treatment 5. On the contrary, LEDGF/p75 and wild type SSRP1 are chromatin-bound then only present in the S2 fraction obtained after salt and DNase treatment of chromatin-bound proteins. Therefore, in order to evaluate the interaction of LEDGF/p75 with these SSRP1 proteins the immunoprecipitation experiment was performed with S1 and S2 fractions combined (S1/S2). The presence of SSRP1 proteins in the immunoprecipitated fraction was then determined by immunoblot analysis with an anti-Myc antibody. Findings in Figure 1d indicate that LEDGF/p75 strongly binds to the SSRP1ΔNTD mutant confirming the hSpt16-independent interaction of SSRP1 and LEDGF/p75.
SSRP1 interacts with the N-terminal region of LEDGF proteins
Next, we defined what region of LEDGF/p75 (Supplementary Figure S1) was responsible for SSRP1 recognition. The interaction of Myc-tagged SSRP1 with FLAG-tagged LEDGF/p75 or LEDGF/p52 wild type proteins, or a LEDGF/p75 mutant containing only the C-terminal region was evaluated by co-immunoprecipitation experiments. HEK293T-derived LEDGF/p75-deficient cells (si1340/1428 cells) were co-transfected with expression plasmids encoding these proteins, and two days later, S2 fractions were obtained and subjected to immunoprecipitation with an anti-FLAG mAb. The immunoprecipitated proteins were analyzed by immunoblotting with tag-specific antibodies. Results in Figure 2a indicate that LEDGF/p75 and p52 interact with SSRP1 with similar efficiency; however, C-terminal LEDGF/p75 does not bind to SSRP1, demonstrating that the shared N-terminal region of the LEDGF proteins interacts with SSRP1.
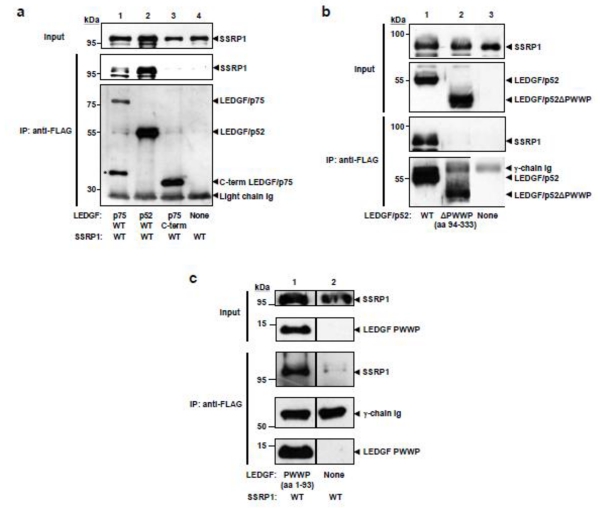
Figure 2. Mapping the LEDGF regions implicated in SSRP1 binding.
(a) si1340/1428 cells were co-transfected with plasmids encoding Myc-SSRP1 and either LEDGF/p75-FLAG (lane 1), LEDGF/p52-FLAG (lane 2), C-terminal LEDGF/p75-FLAG (lane 3), or an empty expression plasmid (lane 4). Immunoprecipitation analyses were performed as previously described in figure legend 1a. (*) Marks degradation products of LEDGF/p52-FLAG. (b) Implication of PWWP domain in the interaction of LEDGF/p52 with SSRP1. Cell lysates were obtained from si1340/1428 cells co-transfected with plasmids expressing Myc-SSRP1 and either LEDGF/p52-FLAG (lane 1), LEDGF/p52ΔPWWP-FLAG (lane 2), or an empty plasmid (lane 3). Immunoprecipitations were performed as previously described. (c) si1340/1428 cells were co-transfected with plasmids expressing Myc-SSRP1 and PWWP-FLAG (lane 1) or an empty plasmid (lane 2). Immunoprecipitations were performed as previously described in figure legend 1a. Results in (c) are representative of two independent experiments. The line separating lanes 1 and 2 indicate that lanes of the gel in between contained irrelevant samples that were removed.
To formally exclude any role of the C-terminal region of LEDGF/p75 in SSRP1 binding, the interaction of SSRP1 with a panel of LEDGF/p75 mutants lacking each of the regions described in the C-terminus of this protein 18 (Supplementary Figure S1) was evaluated by co-immunoprecipitation using S2 fractions, as described above. Co-immunoprecipitated SSRP1 was detected by immunoblotting and quantified by densitometry analysis (Supplementary Figure S3a). In further support of our conclusions that the C-terminal region of LEDGF/p75 is dispensable for SSRP1 binding, FLAG-tagged LEDGF/p75 deletion mutants lacking IBD (aa 340-417), CR5.1 (aa 443-477), or CR6 (aa 478-530) efficiently interacted with SSRP1, whereas the interaction of a LEDGF/p75 mutant lacking CR4 (aa 326-339) was only slightly reduced (Supplementary Figure S3a).
The PWWP domain of LEDGF proteins is necessary and sufficient for SSRP1 interaction
We further mapped the SSRP1-binding domain of LEDGF proteins by co-immunoprecipitation experiments, as described above. FLAG-tagged LEDGF/p75 mutants lacking different regions of the N-terminus 18 were co-transfected with Myc-SSRP1 in si1340/1428 cells and the S2 fractions obtained from these cells subjected to immunoprecipitation with an anti-FLAG mAb. The presence of SSRP1 in the immunoprecipitated proteins was then evaluated by immunoblotting with an anti-SSRP1 antibody. Data in Supplementary Figure S3b indicate that deletion of CR1b (aa 157-177), CR2 (aa 199-266), or CR3 (aa 267-325) of LEDGF/p75 did not impair its binding to SSRP1. LEDGF/p75 mutants lacking CR1 (aa 94-145) and the nuclear localization signal (aa 146-156) were only partially defective in the interaction but the binding to SSRP1 was unambiguously evidenced. However, the LEDGF/p75 mutant lacking the PWWP domain (aa 1-93) was severely impaired in SSRP1 binding (Supplementary Figure S3), indicating that this domain is required for this protein interaction.
To demonstrate further that PWWP domain is the SSRP1-binding domain of LEDGF proteins, we determined by immunoprecipitation experiments the effect of deleting PWWP on the ability of LEDGF/p52 to interact with SSRP1. The same strategy described above was followed. LEDGF/p52ΔPWWP is mostly distributed to the S1 fraction and the positive control LEDGFp52 to the S2 fraction whereas SSRP1 is distributed to both S1 and S2. Therefore, in order to have similar levels of all of these proteins and an identical buffer composition in the immunoprecipitation input, this experiment was performed with S1 and S2 fractions combined (S1/S2). As expected, in these experiments (Figure 2b) we also observed that the deletion of the PWWP domain completely abrogated the interaction of LEDGF/p52 with SSRP1 demonstrating the importance of this domain in the interaction between LEDGF proteins and SSRP1.
The PWWP domain is organized into two independent subdomains. The N-terminal region of the PWWP domain is composed of a five-stranded beta-barrel core (aa 1-60) and the C-terminal is comprised of an alpha-helix bundle (aa 61-93). The beta-barrel is solvent exposed and contains a hydrophobic cavity that harbors a tryptophan (aa 21), which is essential for the interaction of PWWP with H3K36me3 29. To further characterize the surface of interaction of the PWWP domain, S2 fractions from si1340/1428 cells transiently co-expressing Xpress-SSRP1 and either FLAG-tagged LEDGF/p75 WT, LEDGF/p75ΔPWWPβ-barrel (Δaa1-60), LEDGF/p75ΔPWWPα-helix (Δaa61-93), or LEDGF/p75W21A were immunoprecipitated with an anti-FLAG mAb, and the pulled-down proteins evaluated by immunoblot for the presence of SSRP1 or LEDGF/p75 with specific antibodies. Data in Supplementary Figures S4a-c indicate that none of the mutations introduced in the PWWP domain affected the interaction between LEDGF/p75 and SSRP1 proteins. These results suggest that both sub-domains of PWWP importantly contribute to SSRP1 binding and that interaction of LEDGF/p75 with the nucleosomal protein H3K36me3 is not a pre-condition for SSRP1 interaction.
Next, we determined whether the PWWP domain alone was sufficient for SSRP1 binding by co-immunoprecipitation analysis of si1340/1428 cells co-expressing Myc-SSRP1 and FLAG-tagged PWWP domain or only Myc-SSRP1. Chromatin binding of the LEDGF/p75 PWWP domain expressed as a sole protein is weak and consequently this protein is fully recuperated in the Triton-soluble chromatin-unbound fraction (S1) obtained without salt/DNase treatment 5, whereas SSRP1 is distributed to the Triton-soluble chromatin-bound fraction (S2) only after salt/DNase treatment of Triton-insoluble chromatin-bound proteins. Then, immunoprecipitations were performed with S1 and S2 fractions mixed. An anti-FLAG mAb was used for immunoprecipiation and the pulled-down proteins were analyzed for the presence of LEDGF and SSRP1 proteins by immunoblotting with specific antibodies. Findings in Figure 2c clearly demonstrate that the PWWP domain alone firmly interacts with SSRP1, indicating that this domain is sufficient for specific binding to SSRP1.
The HMG domain of SSRP1 is required but not sufficient for the interaction with LEDGF/p75
We next defined the LEDGF-binding domain of SSRP1. Two structural domains, a structure-specific recognition (SSRC) motif (aa 187-437) and an HMG-box domain (aa 548-613), have been previously described in SSRP1, in addition to four evolutionarily conserved regions designated N-terminal domain (NTD, aa 1-186), middle domain (MD, aa 438-514), intrinsically disordered domain (IDD, 515-547), and C-terminal domain (CID, aa 614-709) 21 (Supplementary Figure S1). A panel of Myc-tagged SSRP1 mutants lacking each of these domains and the WT protein were co-expressed with FLAG-tagged LEDGF/p75 in HEK293T cells and S2 fractions obtained from these cells were subjected to immunoprecipitation with an anti-FLAG antibody. The presence of SSRP1 proteins in the immunoprecipitated fraction was then determined by immunoblot analysis with an anti-Myc antibody. Data in Supplementary Figure S5 show that the interaction between LEDGF/p75 and the SSRP1 mutants lacking the SSRC (Fig. S5a), MD (Fig. S5b), IDD (Fig. S5c), and CID (Fig. S5d) domains were not impaired. Similarly, we showed that NTD is also dispensable for LEDGF/p75 binding (Figure 1d). Conversely, the interaction with the mutant lacking the HMG domain was severely defective (Figure 3a), suggesting the implication of this SSRP1 region in the LEDGF/p75 binding. The lack of the HMG domain did not alter the distribution of SSRP1 in the chromatin-bound or -unbound fractions (Supplementary Figure S2), indicating that the defective interaction was not due to a change in the subcellular distribution of the SSRP1 mutant protein.
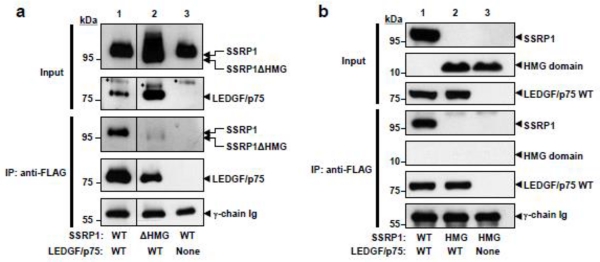
Figure 3. Immunoprecipitation analyses of the interaction of SSRP1 mutants with LEDGF/p75.
(a) HEK293T cells were co-transfected with plasmids expressing: FLAG-tagged LEDGF/p75 and Myc-tagged SSRP1 WT (lane 1), FLAG-tagged LEDGF/p75 and Myc-tagged SSRP1ΔHMG (lane 2), or Myc-tagged SSRP1 WT and an empty plasmid (lane 3). Results in (a) are representative of three independent experiments. The vertical lines separating lanes 1 and 2 indicate that samples were in the same gel but not in adjacent positions. (*) Denotes residual SSRP1 resulting from partial antibody stripping. (b) HEK293T cells were co-transfected with plasmids encoding LEDGF/p75-FLAG and SSRP1 WT-Myc (lane 1) or HMG-Myc (lane 2). Lane 3 was co-transfected with SSRP1 WT-Myc and an empty plasmid. S1 and S2 fractions were obtained from the transfected cells and mixed. Mixed fractions were immunoprecipitated as described in figure legend 1a. Results are representative of two independent experiments.
In order to further clarify the role of HMG SSRP1 in LEDGF/p75 binding, we determined whether this domain is sufficient for the interaction of these proteins. As described above, HEK293T cells were transiently co-transfected with FLAG-tagged LEDGF/p75 WT and Myc-tagged SSRP1 HMG expression plasmids. Their interaction was evaluated by immunoprecipitation from S1/S2 fractions with an anti-FLAG antibody followed by anti-Myc immunoblot analysis. Results in Figure 3b revealed that the HMG domain alone failed to bind to LEDGF/p75, indicating that, although required, this domain is not sufficient for binding. These findings suggest that the surface of interaction of SSRP1 with LEDGF/p75 is complex and other regions of SSRP1 may contribute to the binding.
Partial knockdown of SSRP1 specifically reduces HIV-1 infection
We next investigated the functional significance of the LEDGF/p75-FACT complex association for HIV-1 infection. LEDGF/p75 tethers the HIV-1 pre-integration complex to the host chromatin allowing for efficient integrase-mediated viral cDNA integration into the host genome 4; 7; 8. It is possible that the LEDGF/p75 PWWP-interacting FACT complex could contribute to HIV-1 infection by facilitating the access of integrase to the chromatinized DNA during integration, or at post-integration steps by influencing DNA repair, or transcription of the provirus. Implications of the nucleosome remodeling activity of the FACT complex in facilitating the access of cellular enzymes to the host DNA during DNA repair and transcription have been extensively reported. An important limitation to these studies is the essential role of FACT for cell survival 30, which precludes drastic reduction of the cellular levels of this complex. Similarly, the required role of the LEDGF/p75-binding domain of SSRP1, HMG, in the chromatin remodeling activity of FACT 31 impedes rescuing from apoptosis SSRP1-deficient cells by expressing a LEDGF/p75 non-interacting SSRP1 mutant lacking the HMG domain. To circumvent these problems, we analyzed the effects of transient partial knockdown of SSRP1 on HIV-1 infection. Knockdown was achieved by expressing an SSRP1-specific shRNA in the target cells using a non-integrative HIV-1-derived viral vector that harbors an integrase catalytic site mutant, D64V. Non-integrative vectors transduce cells as efficiently as their integrative counterparts but express lower levels of the transgenes per cell. Therefore, at the same MOI, DNA integration-incompetent lentiviral vectors expressing shRNAs more likely produce a partial instead of the more stringent knockdown observed with integrative lentiviruses.
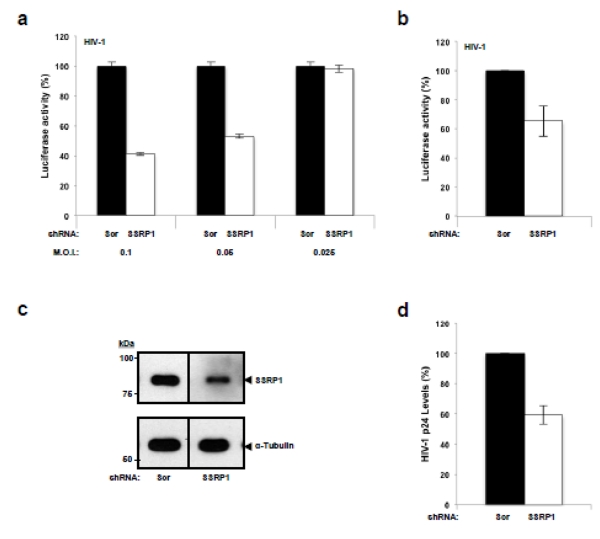
Using the above-mentioned approach, cells of the human CD4+ T cell line SupT1 were transduced at different MOIs with non-integrative HIV-derived viral vectors (TRIP D64V shRNA) expressing an SSRP1-specific or scrambled shRNA sequence, and three days after transduction the cells were infected with a VSV-G pseudotyped, single-round infection, HIV-derived virus expressing LTR-driven luciferase (Hluc). Three days later the luciferase activity and ATP levels were measured. In addition, morphology and growth rate of the infected cells were evaluated by daily microscopy examination during the experiment. Results in Figure 4a indicate a modest but reproducible decrease in the susceptibility of SSRP1-knockdown cells to HIV-1 infection at the highest MOIs evaluated. Importantly, under these conditions, cell viability was unremarkable as indicated by the cellular ATP content (Supplementary Figure S6a), a very sensitive indicator of cell viability, as well as by cellular morphology and growth rate of the infected cells (data not shown). Results in Figure 4a were further verified by multiple experiments performed on different days (Figure 4b) highlighting the reproducibility of this effect. In these experiments, SSRP1 in the knockdown cells was 57% of the levels found in control cells, as indicated by densitometry analysis of immunoblots (Figure 4c). This partial reduction likely correlates with the modest decrease in infectivity observed in the SSRP1-deficient cells. Therefore, findings represented in Figure 4a-c indicate that SSRP1 partial knockdown affects HIV-1 infection without altering cell viability.
Figure 4. Effect of SSRP1 partial knockdown on HIV-1 infection.
(a and b) SupT1 cells were transduced at different MOIs with a lentiviral vector expressing a Scrambled or SSRP1-specific shRNA and three days later were infected with single-round HIV-1 expressing luciferase. Three days post-infection luciferase and ATP levels were measured. Luciferase readings in control cells infected with Hluc in (a) and (b) were around 5×103 relative light untis (RLU)/ml and background readings were approximately 0.02 RLU/ml. Luciferase was normalized to ATP content in the same samples. Standard deviations in (a) represent the variability in luciferase readings of single experiments and in (b) the variability of three independent experiments at MOI 0.1. (c) The levels of SSRP1 in one of the experiments represented in (b) were determined by immunoblot analysis. α-tubulin was measured as a loading control. The level of knockdown achieved is representative of the other two infection experiments. The vertical line separating the lanes indicates that the samples were in the same electrophoresis gel and immunoblot membrane but not in adjacent positions. (d) SupT1 cells were transduced with either the scrambled or SSRP1-specific shRNA expressing lentiviral virus and three days later were infected with HIV-1NL4-3. After twenty-four hours, infected cells were extensively washed to remove input HIV-1NL4-3 virus and 96 hours post-infection the supernatant was harvested and HIV-1 p24 quantified by ELISA. Standard deviations represent data from three different experiments.
To further evaluate the effect of SSRP1 deficiency on HIV-1 infection, we transduced SupT1 cells at MOI 1 with the shRNA expressing retroviral vectors and on the third day post-transduction the cells were infected with HIV-1NL4-3. Twenty-four hours later the infected cells were extensively washed to remove the input HIV-1NL4-3 and the viral supernatant was harvested 72 hours later for HIV-1 p24 quantification by ELISA. In three independent lentiviral transduction/HIV-1 infection experiments performed on different days (Figure 4d) we observed that partial knockdown of SSRP1 caused a modest (~ 40%) but reproducible reduction in HIV-1 p24 levels.
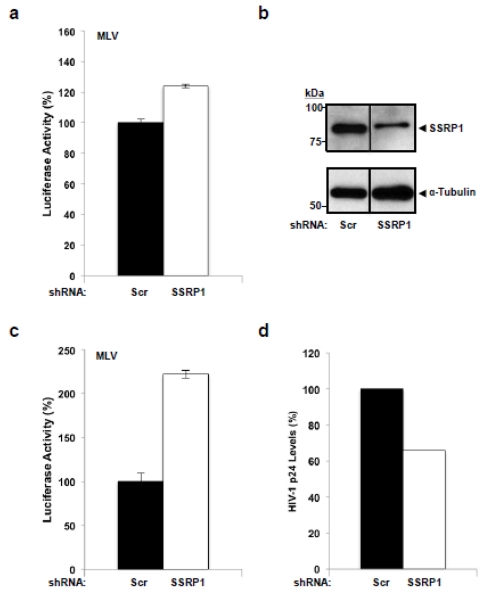
The effect of SSRP1 deficiency on HIV-1 infection could be LEDGF/p75-dependent or - independent. For example, the requirement of the FACT complex for transcription of chromatinized genes in the HIV-1 provirus, a LEDGF/p75-independent process, could explain the effect of SSRP1 knockdown on HIV-1 infection. In order to evaluate these possible mechanisms and define whether this phenomenon is HIV-1 specific, we determined the effect of SSRP1 partial deficiency on the infection by the gamma retrovirus Murine Leukemia Virus (MLV). Similar to HIV-1, the expression of chromatinized MLV provirus depends on FACT activity; however, MLV infection is LEDGF/p75-independent 12. Briefly, SupT1 cells were transduced with the TRIP D64V shRNA expressing either an SSRP1-specific or scrambled shRNA sequence and three days after transduction the cells were infected with a VSV-G pseudotyped, single-round infection, MLV-derived virus expressing luciferase (MLVluc). In these experiments, we observed that MLV infection was not affected (Figure 5a) excluding the possibility that the effect of SSRP1 knockdown on HIV-1 infection is due to the role of the FACT complex in transcription or to unnoticed cellular toxicity. These data also indicate that the effect of SSRP1 on HIV-1 infection is specific.
Figure 5. Influence of partial knockdown of SSRP1 on MLV and HIV-1 infections.
(a) SupT1 cells were transduced with a lentiviral vector encoding a Scrambled or SSRP1-specific shRNA, three days later the cells were infected with an MLV-derived virus expressing luciferase, and seventy-two hours post-infection, luciferase was measured. (b-d) SupT1 cells were transduced at MOI 1 with a lentiviral vector encoding a Scrambled or SSRP1-specific shRNA and three days later one-third of the cells were analyzed for SSRP1 expression by immunoblot (b) and the remaining cells were infected with MLV (c) or HIV-1NL4-3 (d). Three days after infection, luciferase activity was measured in the MLV-infected cells (c) and HIV-1 replication was determined by quantifying HIV-1 p24 in the supernatant of the HIV-1NL4-3 infected cells by ELISA (d). Standard deviation in (a) and (c) represent the variability of luciferase activity readings from single experiments. The vertical line separating the lanes in (b) indicates that the samples, although in the same electrophoresis gel and immunoblot membrane, were not in adjacent positions. Luciferase readings in control cells infected with MLVluc in (a) and (c) were around 8 RLU/ml and background readings were approximately 0.02 RLU/ml.
Although the SupT1 target cells were transduced at the same MOI and with the same lentiviral vector preps in the MLV and HIV-1 infection experiments described in Figures 4b, 4d, and 5a; small variations in SSRP1 knockdown levels in the target cells could account for the differential effect of SSRP1 deficiency on these retroviruses. To control for this potential confounding factor, SupT1 cells were transduced with the shRNA-expressing lentiviruses at MOI 1 and three days later the same cells were used either for infection with HIV-1NL4-3 or with MLVluc, or for immunoblotting analysis of SSRP1 expression. Three days after infection, cellular ATP levels and MLV-encoded luciferase activity were measured in the MLV-infected cells, and HIV-1 p24 quantified in the supernatant of HIV-1NL4-3 infected cells.
Densitometry analysis of the immunoblot in Figure 5b indicates that these knockdown cells expressed 52% of the SSRP1 levels found in control cells. In these cells, HIV-1 replication was impaired (Figure 5d) to levels comparable to those achieved in experiments reported in Figures 4a, 4b, and 4d, which correlate with the partial SSRP1 knockdown obtained in the target cells (Figure 5b). In contrast to HIV-1, MLV infection was not impaired in these cells (Figures 5c), corroborating previous observations (Figures 5a). As expected, cell viability was not affected in the SSRP1-knockdown cells studied (Supplementary Figure S6b). Data in figures 4 and 5 highlight the reproducibility of our results further indicate a specific role for SSRP1 in HIV-1 infection.
Partial knockdown of SSRP1 specifically reduces LTR-driven HIV-1 gene expression in a LEDGF/p75-independent manner
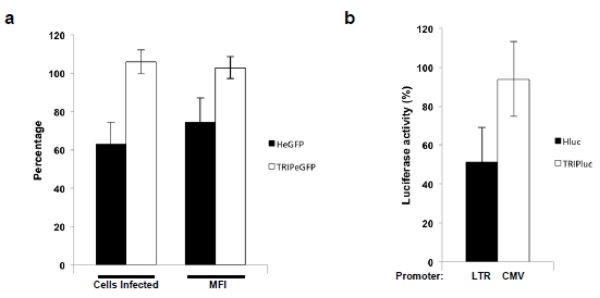
As previously discussed, SSRP1 deficiency could affect HIV-1 infection by reducing LTR-driven transcription or viral DNA integration. In order to distinguish between these potential mechanisms we compared the effect of SSRP1 deficiency on infection by HIV-1-derived viruses that, although replicate through viral DNA integration, use different promoters for transgene expression. TRIP viruses 32 express their transgenes from an internal immediate-early CMV promoter in contrast to HeGFP or Hluc viruses in which the transgenes are expressed from the HIV-1 LTR. As described above, SupT1 cells were transduced with shRNA retroviral vectors expressing a control or an SSRP1 shRNA and three days later were infected with TRIP eGFP, TRIPluc, HeGFP, or Hluc. In these experiments, the shRNAs were delivered using a retroviral vector lacking the eGFP reporter. As shown in Figure 6a and b, SSRP1 knockdown diminished infection of viruses expressing the transgene from the LTR but not from the CMV promoter, suggesting that SSRP1 deficiency affected LTR-driven transgene expression rather than viral DNA integration.
Figure 6. Influence of partial knockdown of SSRP1 on HIV-1 infection carrying different promoters.
(a and b) SupT1 cells were transduced with a lentiviral vector encoding a Scrambled or SSRP1-specific shRNA, three days later the cells were infected with HIV-derived viruses expressing eGFP or luciferase from the HIV-1 LTR (HeGFP/Hluc) or from an internal immediate early CMV promoter (TRIPeGFP/TRIPluc). Seventy-two hours post-infection, luciferase or eGFP were measured. Luciferase activity measured in the cells transduced with control shRNAs was considered 100% values. Standard deviations represent the variability of four independent experiments.
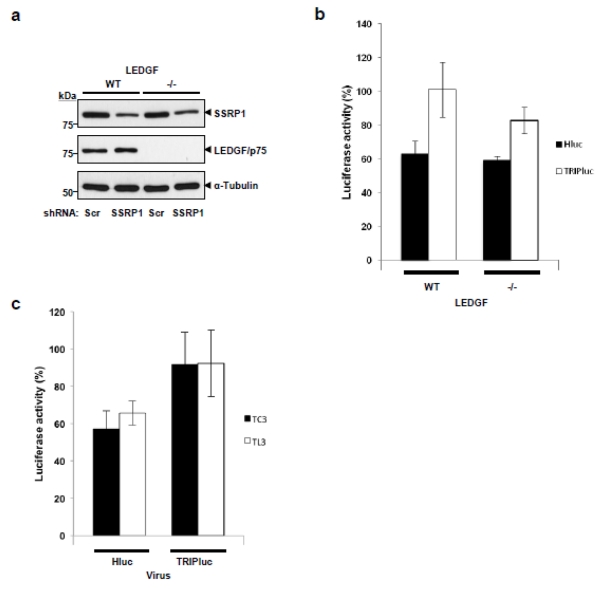
Data in Figure 6a and 6b also suggest that SSRP1 regulates HIV-1 infection independently of LEDGF/p75 since this protein is required for efficient integration of both Hluc and TRIPluc viruses. Therefore, we evaluate this mechanism of action by determining the requirement of LEDGF/p75 in the effect of SSRP1 deficiency on HIV-1 infection. Nalm-6 wild type and LEDGF/p75 knockout cells (Figure 7a) 33 were transduced with shRNA retroviral vectors expressing control or SSRP1 shRNAs. Three days later, these cells were infected with TRIPluc or Hluc, and seventy-two hours afterwards, the luciferase and ATP were measured. Data in Figure 7b indicate that partial SSRP1 knockdown (Figure 7a) similarly reduced Hluc infection independently of the LEDGF/p75 levels of the target cells (Figure 7b), indicating that the effect of SSRP1 on HIV-1 infection is LEDGF/p75-independent. In both, wild type and LEDGF/p75 knockout cells, SSRP1 deficiency affected Hluc infection in a higher magnitude than TRIPluc infection (Figure 7b), as indicated in data shown in Figure 6. These findings (Figure 7b) also indicate that the role of SSRP1 in HIV-1 infection is not exclusive to CD4+ T cells (SupT1 cells) but is also is conserved in a human pre-B lymphoblast (Nalm-6 cells) cell line.
Figure 7. Consequence of SSRP1 partial knockdown in LEDGF/p75-deficient cells on HIV-1 infections.
(a and b) LEDGF/p75 wild type or knockout Nalm-6 cells were transduced with a lentiviral vector expressing a Scrambled or SSRP1-specific shRNA and after three days one-third of these cells were analyzed for SSRP1 and LEDGF/p75 expression by immunoblot in (a). The remaining cells were infected with HIV-1-derived viruses expressing luciferase from the HIV-1 LTR (Hluc) or an internal immediate CMV promoter (TRIPluc). Seventy-two hours post-infection, luciferase and viability were measured in (b). (c) LEDGF/p75-deficent (TL3) and control (TC3) cells were transduced with a lentiviral vector encoding a Scrambled or SSRP1-specific shRNA, three days later the cells were infected with Hluc or TRIPluc and seventy-two hours post-infection, luciferase was measured. Luciferase activity measured in the cells transduced with control shRNAs was considered 100% values. Standard deviations represent the variability of six (b) or four (c) independent transduction/infection experiments.
We further evaluated the effect of SSRP1 knockdown on infectivity of Hluc and TRIPluc in SupT1-derived TC3 and TL3 cell lines that were generated by stable expression of shRNAs, control (scrambled sequence, TC3) or LEDGF/p75-specific (TL3). These cells were transduced with control or SSRP1 shRNA expressing retroviral vectors, infected with Hluc or TRIPluc three days later, and analyzed for luciferase expression seventy-two hours after infection (Figure 7c). As expected, SSRP1 knockdown reduced infectivity of Hluc but not of TRIPluc viruses in both LEDGF/p75-deficient and control cells to a similar extent, further supporting the LEDGF/p75-independent role of SSRP1 in HIV-1 infection.
Chromatin binding of LEDGF/p75 and SSRP1 occur independently of each other
To characterize further the LEDGF/p75-SSRP1 interaction, we determined whether the binding of these proteins to chromatin is influence by their interaction. With this purpose, SupT1 cells were transduced with the SSRP1 and control shRNA-expressing viral vector at an MOI 2.5-folds higher than the MOI used in the HIV-1 and MLV infection experiments described in Figures 4 and 5, and three days later these cells were used to determine the chromatin binding strength of LEDGF/p75 by the salt extraction method 18. This assay measures the effect of salts on the chromatin binding strength of LEDGF/p75. Briefly, the transduced cells were lysed in a buffer containing Triton X-100 (CSKI buffer) supplemented with NaCl to a final concentration of 175 mM, under these conditions LEDGF/p75 is only partially extracted from chromatin 18. Then, the cell lysates were fractionated by centrifugation into a Triton-soluble, salt-extracted fraction, which contains chromatin non-bound proteins (S1), and in a Triton-insoluble salt-resistant fraction (P1), which includes chromatin-bound proteins and other Triton X-100 insoluble proteins (i.e. cytoskeleton proteins). Subsequently, these cellular fractions were evaluated by immunoblotting for the presence of LEDGF/p75 and SSRP1. As expected, at the higher MOI used in these experiments, a more stringent SSRP1 knockdown than in the retroviral infection experiments reported above, was reached (compare total fraction in supplementary Figure S7a with Figures 4c and 5b). Densitometry analysis of immunoblot in supplementary Figure S7a indicated that knockdown cells express only 13% of the SSRP1 levels found in the control cells. Nevertheless, the salt extraction pattern of LEDGF/p75 in these more stringent SSRP1 knockdown cells was identical to the pattern observed in control cells (Supplementary Figure S7a) which indicates that cellular levels of SSRP1 lower than those that affect HIV-1 infection do not affect the chromatin binding strength of LEDGF/p75. These results suggest that LEDGF/p75 binds to chromatin independently of SSRP1.
Similarly, the chromatin binding strength of SSRP1 in LEDGF/p75-deficient cells was evaluated using the salt extraction method described above. SupT1-derived LEDGF/p75-knockdown (TL3) and control cells (TC3) were subjected to salt extraction with CSKI buffer supplemented with NaCl to a final concentration of 150 mM. Then, the distribution of SSRP1 and LEDGF/p75 was determined in the salt-extracted and -resistant fractions by immunoblotting. As represented in (Supplementary Figure S7b), the SSRP1 salt extraction pattern was similar in control and LEDGF/p75-deficient cells indicating that LEDGF/p75 does not influence the binding strength of SSRP1 to chromatin.
DISCUSSION
A role of LEDGF proteins in transcriptional regulation at promoter regions has been described 1; 4, yet the participation of LEDGF/p75 in other steps of transcriptional regulation has not been evaluated. A genome-wide study indicates that LEDGF/p75 is enriched downstream of the transcription start sites of actively transcribed genes 17 suggesting a potential role for this regulator in transcriptional elongation. However, the mechanism associated with this phenomenon remains unknown. The data presented in this study demonstrate for the first time that LEDGF/p75 associates with the FACT complex, suggesting a role for LEDGF/p75 in transcriptional elongation. We demonstrated that LEDGF/p75 specifically co-immunoprecipitates with the chromatin-bound molecular form of the FACT complex that is likely engaged in transcriptional activity. These findings correlate with the global effects of LEDGF/p75 on transcription 15 and with the high degree of co-localization of LEDGF/p75 with SSRP1 under basal conditions. Our findings also suggest that the location of LEDGF/p75 in transcriptional elongation complexes could determine its role in targeting HIV-1 into actively transcribed genes13.
In this study, we also characterized in detail the interaction of LEDGF/p75 with the FACT complex. The FACT complex is a histone chaperone comprised of a heterodimer of Spt16 and SSRP1 proteins. It has been found to be essential in DNA replication, transcriptional elongation, and DNA damage repair 20; 21; 27; 28; 34; 35; 36; 37; 38; 39; 40; 41 and the targeted disruption of one of its components, SSRP1, is embryonic lethal 30. In any function, FACT works through nucleosome reorganization by disrupting core histone-histone and histone-DNA interactions 20; 21; 27; 28; 34; 35; 36; 37; 38; 39; 40; 41. Specifically, the FACT complex attaches to the nucleosome through the binding of the Spt16 subunit to the H2A-H2B dimer, facilitating its displacement and allowing SSRP1 to interact with an H3-H4 tetramer and nucleosomal DNA. After destabilization, the FACT complex has been shown to reassemble the nucleosome 27.
We demonstrated that LEDGF proteins complex with SSRP1 independently of hSpt16 requiring their PWWP and HMG domains, respetively. Intriguingly, the fragment of TOX4 that interacts with PWWP LEDGF/p75 also contains an HMG domain, although its relevance in this interaction is unknown 42. PWWP and HMG domains are present in multiple proteins involved in transcriptional regulation, DNA repair, and epigenetics 43; 44; 45; 46; 47; 48. PWWP is a nucleosomal-binding domain that interacts specifically with H3K36me3 and H4K20me histone codes and non-specifically with DNA, whereas HMG domain binds to DNA and several proteins implicated in transcriptional regulation. HMG binds to the minor groove of DNA inducing local changes in the DNA structure that triggers different DNA-dependent functions 47; 49. In addition, the HMG domain of Sox proteins have been demonstrated to bind to different protein domains present in transcription factors, including homeodomain, paired domain, POU domains, zinc finger, basic helix-loop-helix and leucine zipper. These HMG-interacting domains mediate the binding of transcription factors to DNA, modulating their transcriptional activity 50; 51; 52; 53. Therefore, our findings expand to PWWP the diversity of protein domains that interact with HMG domains.
Similar to the interaction of HMG Sox with other protein domains 52, the interaction of HMG SSRP1 with LEDGF/p75 seems to be strengthened by other SSRP1 protein regions since HMG is required but not sufficient for LEDGF/p75 binding. In contrast, the PWWP domain is required and sufficient for LEDGF association with SSRP1. A potential mechanism for this phenomenon is the binding of other regions of SSRP1 to PWWP LEDGF/p75. Alternatively, other protein regions in SSRP1 could stabilize a particular conformation of HMG required for PWWP association. In support of the latter possibility, it has been described that the binding of HMG in SSRP1 of Drosophila melanogaster to nucleosomal DNA is modulated by intramolecular interactions between the Middle (aa 438-514), the Intrinsically Disordered (aa 515-547), and the HMG (aa 548-613) domains of SSRP1 54. Therefore, it is possible that a similar intramolecular crosstalk between HMG and other regions of SSRP1 could influence the interaction with PWWP in LEDGF. The potential contribution of other SSRP1 regions to the interaction of HMG with PWWP domain also suggests that the binding of these two protein domains could be protein-specific rather than universal.
The implications of nucleosome or other chromatin-bound proteins to the association of LEDGF proteins with the FACT complex is not clear yet, and deserves further characterization. Data showing that LEDGF/p75 can interact with SSRP1ΔNTD suggest that the histone core of the nucleosome is not necessary for this interaction. Binding of SSRP1 to the nucleosome is dependent on its binding to Spt16 whose in turn binds to the H2A-H2B dimer 27. Similarly, LEDGF/p75 PWWP domains mutants W21A or those lacking the N- or C-terminal subdomains that are defective in their interaction with methylated histones, in particular H3K36me3 and H4K20me3 in the nucleosome 55; 56; 57; 58; 59; 60; 61 still interact with SSRP1, suggesting that direct binding of LEDGF proteins to the nucleosome core proteins is not a pre-requisite for association to SSRP1. However, the implication of DNA and/or other chromatin-bound proteins in the formation of the complex containing LEDGF proteins and the FACT complex is still possible. For example, the interaction of LEDGF/p75 with SSRP1ΔNTD could still be mediated by binding of the HMG domain to DNA that survived the DNase treatment through its tightly association with proteins in the chromatin.
The association of LEDGF proteins with the FACT complex is predicted to have important functional implications. LEDGF/p75 have a role in the repair of DNA double-strand breaks by the homologous recombination pathway 11 whereas LEDGF proteins are implicated in transcriptional regulation 4, and these processes require the chromatin-remodeling function of the FACT complex. Therefore, the interaction of LEDGF proteins with the FACT complex could facilitate transcription and DNA repair of chromatin regions engaged in active transcription. Similarly, the presence of SSRP1 in a complex with LEDGF proteins, in particular with LEDGF/p75 a known HIV-1 cellular cofactor 4, could facilitate HIV-1 infection. In support of the latter hypothesis, we have observed a modest, reproducible, and specific defect in HIV-1 infection in cells partially knocked down for SSRP1. The modest effect of SSRP1 deficiency on HIV-1 replication observed could be due to the high levels of residual SSRP1 in the knockdown cells as detected by immunoblot analyses. Unfortunately, the essential role of SSRP1 and, more specifically its HMG domain that is required for association to LEDGF/p75, preclude analyses of the susceptibility to HIV-1 infection of cells significantly depleted of SSRP1 or only expressing SSRP1 mutants unable to interact with LEDGF/p75 30; 31.
Most likely, the effect of SSRP1 deficiency on HIV-1 is specific and not related to a global effect on cellular transcription since infection by the gamma retrovirus MLV was not affected. In addition, SSRP1 deficiency affected the expression of proviral transgenes transcribed from the HIV-1 LTR but not from an internal immediate-early CMV promoter, suggesting that SSRP1 influences LTR activity rather than viral integration. This conclusion is also supported by the LEDGF/p75-independence of the mechanism implicated in the effect of SSRP1 on HIV-1 infection.
Our data in correspondence with previous observations 62 indicate a role for SSRP1 in HIV-1 LTR-driven expression. It has been demonstrated that TNF-α-induced activation of HIV-1 LTR-mediated transcription increased FACT occupancy at nucleosome 1 of the LTR and nucleosomes in the Env region, and removal of H2A/H2B dimers from these locations 62. This role of SSRP1 is expected considering the reported function of this protein in chromatin remodeling 20; 21; 27 and the fact that HIV-1 proviral gene expression is subjected to the same mechanisms governing transcriptional regulation of host genes 63; 64; 65. In addition, the FACT subunit hSpt16 has been reported to interact with Tat 66; 67; 68. In apparent contradiction with our findings, down regulation of SSRP1 and/or hSpt16 has been reported to modestly increase HIV-1 expression 67; 68. The inhibitory role of hSpt16 on HIV-1 expression could illustrate FACT-independent hSpt16 functions previously described 26. Alternatively, down regulation of FACT levels in the cell could globally alter the chromatin structure, leading to transcriptional initiation from cryptic initiation sites within the coding region of genes, including HIV-1, as previously reported in yeast 39; 69.
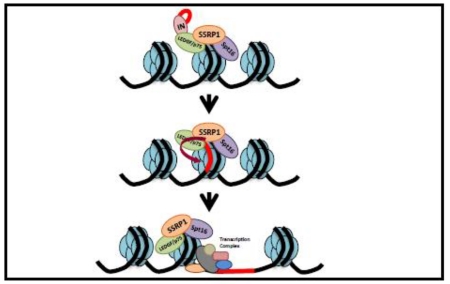
Finally, our data suggest a model that indicates that HIV-1 hijacks a cellular complex in which LEDGF/p75 is associated with FACT. This complex promotes efficient proviral formation and gene expression through LEDGF/p75 and SSRP1 functions, respectively. We predict that this SSRP1 function could be relevant during the Tat-independent stage of the viral life cycle.
MATERIALS AND METHODS
Expression and shRNA constructs
LEDGF/p75 expression plasmids
pFLAG LEDGF/p75 and pFLAG LEDGF/p52 previously described 18; 22 were used in transient expression experiments and to generate the LEDGF mutants described in this work.
SSRP1 expression plasmids
pSSRP1-Myc or pXpress-SSRP1 were generated by cloning the SSRP1 cDNA into pCMV, which contains the promoter and intron A, the immediate early gene promoter of the human Cytomegalovirus (CMV promoter) 70, with a C-terminal Myc or Xpress tag. SSRP1 cDNA was PCR amplified from a human lymph node cDNA library. These SSRP1 expression plasmids were further used to generate a panel of SSRP1 mutants.
Retroviral vector expression plasmids
The plasmids used to generate retroviral vectors were described previously 12. HIV-1-derived vectors were produced using pHIV luc, pMD.G, pTRIP eGFP 32, pCMVΔR8.91, and pCMVΔR8.91 IN D64V (a gift of Che Serguera, INSERM MIRCen LMB). pHIVLuc was derived from pNL4-3.Luc.R–E–71 by introducing a deletion in the Env open reading frame. pMD.G encodes the Vesicular Stomatitis Virus glycoprotein G (VSV-G). pTRIP eGFP was derived from pTRIP by substituting the LacZ open reading by the eGFP cDNA 12. eGFP is transcribed from an internal immediate-early CMV promoter in pTRIP-derived vectors 32. pCMVΔR8.91 and pCMVΔR8.91 IN D64V express the HIV-1 gag-pol polyprotein harboring a wild type or a D64V integrase mutant, respectively. Murine Leukemia Virus (MLV)-derived vectors were produced using pMLV luc 12 that was obtained by cloning firefly luciferase cDNA into pLPCX (Clontech) and the packaging plasmid pCS+mGP (a gift of M. Emerman. Fred Hutchinson Cancer Research Center).
shRNA constructs
pTRIP eGFP shRNA SSRP1 and pTRIP eGFP shRNA Scr express SSRP1-specific and scrambled sequences, respectively. They were constructed by cloning the U6 promoter and the respective shRNA sequences at a unique PpuMI site in pTRIP eGFP. The U6-shRNA expression cassettes were PCR amplified from pSilencer 2.1-U6 hygro Negative Control (Ambion) and pSilencer 2.1-U6 hygro SSRP1. The latter plasmid was generated by annealing oligonucleotides 5′-GATCCGCACCACAGTACTGCGTCTGTTTTCAAGAGAAACAGACGCAGTACTGTGGTGTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAACACCACAGTACTGCGTCTGTTTCTCTTGAAAACAGACGCAGTACTGTGGTGCG-3′ 72 and cloning them into the pSilencer 2.1-U6 hygro (Ambion). This shRNA targets nucleotides 708-728 in SSRP1. In experiments in which the effect of SSRP1 knockdown on HIV-derived viruses expressing eGFP was evaluated, the shRNAs were delivered using a modified TRIP vector that lacks the eGFP cDNA.
All the mutants characterized in this work were generated using the Phusion™ Site-Directed Mutagenesis Kit (F-541, Life Technologies). The sequences of the primers used are available upon request. The identity of these mutants was verified by overlapping DNA sequencing of the entire cDNAs.
Cell lines, culture, and transfection conditions
The LEDGF/p75-deficient human CD4+ T cell lines studied were previously described 12; 18. TL3 and TC3 cell lines were derived from SupT1 cells by stable expression of a shRNA sequence targeting LEDGF/p75 or a control scrambled sequence, respectively. TL3 cells express 97% less LEDGF/p75 mRNA than TC3 cells as determined by qPCR. LEDGF/p75-deficient TL3 cells were later engineered to re-express a C-terminally FLAG-tagged LEDGF/p75 wild type (TL3 LEDGF/p75 WT). LEDGF/p75 knockout Nalm-6 cells 33 were obtained from Z. Debyser (Katholieke Universiteit Leuven, Leuven, Flanders, Belgium). HeLa cells were used for confocal co-localization studies. HEK293T and si1340/1428 cells were used for transient transfection experiments. si1340/1428 cells (a gift of E. Poeschla. University of Colorado Denver) are derived from HEK293T cells by stable expression of two LEDGF/p75-specific siRNAs, targeting sequences at nt 1340 and 1428 70. The 2LKD-IN-eGFP cells are derived from HEK293T cells by stable expression of a LEDGF/p75-specific shRNA and the HIV-1 integrase C-terminally tagged with eGFP 18. SupT1-derived cell lines were grown in RPMI 1640, and HeLa and HEK293T-derived cell lines in DMEM. All culture mediums were supplemented with 10% of heat-inactivated fetal calf serum, 2 mM L-glutamine and 1% penicillin/streptomycin. LEDGF and SSRP1 expression plasmids were transfected in HEK293T or si1340/1428 cells using the calcium-phosphate co-transfection method. Briefly, cells were plated at 0.45 × 106 cells / well in a six-well plate and transfected with 2 μg of DNA of the corresponding plasmids. Transfection medium was replaced 18 hours later with fresh culture medium and cells were cultured for an additional 18 hours until they were harvested for analysis.
Generation of retroviruses
Procedures previously described 18 were followed. Briefly, 3×106 HEK293T cells were plated in a T75 cm2 tissue culture flask and co-transfected the next day with the corresponding plasmids by the calcium-phosphate precipitation method. Eighteen hours later the transfection medium was replaced with fresh medium and the cells cultured for forty-eight hours until the viral supernatant was harvested and filtered. Single-round infection viral vectors were further concentrated by ultracentrifugation at 124,750 g for two hours on a 20% sucrose cushion. Viral preparations were stored at −80°C until use.
VSV-G pseudotyped HIV-derived reporter virus expressing firefly luciferase (HIVluc) was prepared by co-transfection of 15 μg of pHIV luc and 5 μg of pMD.G. VSV-G pseudotyped HIV-1-derived viral vectors expressing shRNAs were produced by transfection of 15 μg of the corresponding pTRIP-derived plasmids, 15 μg of pCMVΔR8.91 D64V, and 5 μg of pMD.G. HIV-1NL4-3 was produced by transfection of 15 μg of pNL4-3 HIV-1 expression plasmid. VSV-G pseudotyped MLV-derived vector was produced in Phoenix A packaging cells as described above. Cells were cotransfected with of 75 μg of pCS+mGP (packaging plasmid), 75 μg of pMLVLuc, and 15 μg of pMD.G. Expression of pCS+mGP into Phoenix A was intended to boost MLV vector production.
Single-round infectivity assay
SupT1 cells were plated at 1×105 cells in 500μl of RPMI 1640 culture medium in 24-well plates and infected with HIVluc or MLVluc viral vectors. Three to four days post-infection, cells were collected by centrifugation at 1000 g for six minutes and the pellet resuspended in 200 μl of PBS. Half of the sample was mixed with 100 μl of luciferase substrate (Bright-Glow™ Luciferase Assay System, Promega) and the other half with 100 μl of cell viability substrate (CellTiter-Glo® Assay, Promega). Cell lysates were incubated for 10 minutes at room temperature in the dark and then luminescence measured in triplicate in 50 μl-samples using a microplate luminometer reader (Thermo Scientific, Luminoskan Ascent).
Retroviral infection of SSRP1 partially knockdown cells
SupT1 cells 1×105 were transduced with Scrambled and SSRP1 shRNA-expressing HIV-derived viral vectors at different MOIs and three days later were infected with HIVluc, MLVluc, or HIV-1NL4-3 (2.1 ng of HIV-1 p24). Twenty-four hours after infection, the cells infected with HIV-1NL4-3 were washed three times by centrifugation in 10 mls of culture medium each time to remove the input virus. Cell supernatant was then collected three to four days after HIV-1NL4-3 infection, and used for HIV-1 p24 quantification by ELISA. Cells infected with HIVluc or MLVluc were harvested and analyzed as described for single-round infectivity assays three to four days after infection.
Enzyme-linked immunosorbent assay (ELISA)
HIV-1 p24 levels were determined by a sandwich ELISA (ZeptoMetrix Corporation, Cat. # 0801008) following manufacturer instructions. Briefly, 200 μl of the viral samples were diluted appropriately and incubated on the ELISA wells overnight at 37°C. Unbound proteins were removed by washing the wells six times with 200 μl of washing buffer, and bound HIV-1 p24 was detected by incubating each well with 100 μl of the anti-HIV-1 p24 secondary antibody for one hour at 37°C. Unbound antibodies were removed by washing as described above and bound antibodies were detected by incubating each well with 100 μl of substrate buffer for 30 minutes at room temperature until the reaction was stopped by adding 100 μl of stop solution into each well. The absorbance of each well was determined at 450 nm using a microplate reader (Molecular Devices, Versa max microplate reader).
Immunoprecipitation
The chromatin binding assay 5; 18 was used to obtain different subcellular fractions that were then used in immunoprecipitation reactions. Briefly, cells (~ 3×106) were harvested and lysed for 15 minutes on ice in 300 μl of CSK I buffer (10 mM Pipes pH 6.8, 100 mM NaCl, 1 mM EDTA, 300 mM sucrose, 1 mM MgCl2, 1 mM DTT, 0.5% Triton X-100) containing protease inhibitors (final concentration: leupeptine 2 μg/ml, aprotinin 5 μg/μl, PMSF 1 mM, pepstatin A 1 μg/ml). Cellular lysates were centrifuged at 1000 g for 6 minutes at 4°C and separated into the supernatant fraction (S1), that contains the Triton X-100-soluble chromatin non-bound proteins, and a pellet (P1), that contains Triton X-100-insoluble proteins and chromatin-bound proteins. P1 was further treated with DNAse to solubilize the chromatin-bound proteins. To this end, P1 was re-suspended in 20 μl of CSK II buffer (10 mM Pipes pH 6.8, 100 mM NaCl, 300 mM sucrose, 6 mM MgCl2, 1 mM DTT) supplemented with protease inhibitors, 16 units of turbo DNase (Ambion™), 3.4 μl of 2.5 M (NH4)2SO4, and 3.1 μl of 10× turbo DNase reaction buffer. DNase treatment was conducted at 37°C for 30 minutes. After incubation, 300 μl of CSK I buffer was added to the DNase/(NH4)2SO4 treated sample to dilute the (NH4)2SO4 and centrifuged at 22,000 g for 3 minutes. Then the supernatant (S2 fraction) was pre-clear twice to remove any unspecific binding by incubation under rotation for 30 minutes at 4°C with 150 μl of goat anti-mouse IgG-coated magnetic beads, referred hereafter as beads (Thermo Scientific, Cat. # 21354). From the pre-cleared S2 fraction 50 μl of sample were obtained and mixed with 10 μl of 6× Laemmli sample buffer, and saved as input. Beads (150 μl), that were previously loaded for 3 hours at 4°C with 30 μg of anti-FLAG mAb (Sigma, F3165) diluted in TBS-5% skim milk-0.05% Tween 20, were magnetically separated from the unbound antibodies, mixed with the remaining pre-cleared S2 fraction (note that S2 contains DNAse), and rotated for 2 hours at 4°C. After this incubation, beads were washed five times for 5 minutes each in CSK I buffer and bound proteins were the eluted by boiling in 60 μl of 2× Laemmli sample buffer. Immunoprecipitated proteins were then analyzed by immunoblotting.
In some experiments cells were sub-fractionated in S1 and S2 fractions but then these fractions were mixed (S1/S2), pre-cleared, and used in immunoprecipitation experiments as described above (note that DNAse was also present during these experiments).
Immunoblotting
Protein samples were resolved by SDS-PAGE and transferred overnight to PDVF membranes (Bio-Rad, Cat. # 163-0177) at 100 mAmp at 4°C. Membranes were blocked in TBS containing 10% skim milk for one hour and then incubated in the corresponding primary antibody diluted in TBS-5% skim milk-0.05% Tween 20 (antibody dilution buffer). FLAG-tagged LEDGF/p75 was detected with anti-FLAG mAb (1/500, M2, Sigma), non-tagged LEDGF/p75 was detected with an anti-LEDGF/p75 (1/500, clone 26/LEDGF, BD Biosciences, 611714), Myc-tagged SSRP1 mutants were detected with anti-Myc mAb (1/500, clone 9E10, Covance, MMS-150P), SSRP1 was detected with anti-SSRP1 (1/500 Santa Cruz, sc-56782), hSpt16 was detected with anti-Spt16 (1/400, Santa Cruz, sc-28734), alpha tubulin was detected with anti-alpha tubulin mAb (1/4000, Clone B-5-1-2 Sigma), and beta actin was detected with anti-beta actin mAb (1/5000, Thermo Scientific, MA5-15739). Membranes were incubated overnight at 4°C with anti-FLAG, -LEDGF, -Myc, -SSRP1, or -Spt16 antibodies, whereas anti-alpha tubulin mAb and anti-beta actin were incubated for 2 hours at 25°C. Primary antibody-bound membranes were washed in TBS-0.1% Tween 20 and all bound antibodies detected with goat anti-mouse IgG-HRP (1/2000, KPL, 074-1806) or anti-mouse Ig light chain-HRP (1/500, Millipore, AP200P), except for anti-Spt16 which was detected with goat anti-rabbit IgG-HRP (1/2000, Calbiochem, 401315), followed by chemo-luminescence detection.
Salt extraction assay
Previously described procedures 18 were followed with minor modifications. Briefly, 3×106 cells were lysed in CSK I buffer containing different concentrations of NaCl and fractionated by centrifugation at 1000 g for 6 minutes to obtain a supernatant containing non-chromatin-bound proteins (S1) and a pellet (P1) that represents the chromatin-bound fraction and triton-insoluble proteins. P1 was then re-suspended in 100 ul of 2× Laemmli sample buffer and S1 was mixed with 15 ul of 6× Laemmli sample buffer. A total fraction (T) was obtained by lysing 3×106 cells in 100 μl of 2× Laemmli sample buffer. Samples were heated at 100°C for 10 minutes and then S1 (17.7 μl), P1 (15 μl of a 1/5 dilution), and T (15 μl of a 1/5 dilution) were separated by SDS-PAGE and analyzed by immunoblotting.
Quantitative confocal co-localization assay
Methods previously described 22 were followed. In order to set up this method we evaluated co-localization of LEDGF/p75 with HIV-1 integrase. These proteins interact during all the phases of the cell cycle 70; 73. As a negative control a LEDGF/p75 mutant lacking the integrase binding domain (IBD) was used 23; 24. LEDGF/p75-deficient HEK293T cells stably expressing eGFP-tagged HIV-1 integrase (2LKD-IN-eGFP cells) 18 were plated at 2×105 cells in LabTek II chambered coverglasses and transfected the next day with 2 ug of pFLAG-LEDGF/p75 WT or ΔIBD. Eighteen hours after transfection fresh culture medium was added and forty-eight hours later cells were washed three times in PBS and fixed with 4% formaldehyde-PBS for 10 minutes at 37°C. Then, cells were washed twice in PBS and immunostained for 2 hours at 37°C. LEDGF/p75 was detected with an anti-LEDGF mouse monoclonal antibody diluted 1/100 (clone 26, BD Transduction Laboratories) followed by incubation with anti-mouse Ig coupled to Alexa™ Fluor 594 (10 μg/ml, Invitrogen A21203). Immunolabeled cells were washed and stained with DAPI. Co-localization of LEDGF/p75 with HIV-1 eGFP-integrase was analyzed with a confocal microscope and the Zeiss Zen software. In order to measure co-localization of LEDGF/p75 with SSRP1 2×105 HeLa cells were plated in LabTek II chambered coverglasses and immunostained as described above with an anti-LEDGF rabbit polyclonal antibody diluted 1/100 (Bethyl Laboratories, A300-848A) and a mouse monoclonal antibody against SSRP1 (Santa Cruz, sc-56782). Secondary antibodies anti-mouse Ig coupled to Alexa™ Fluor 594 (10 μg/ml, Invitrogen A21203) and anti-rabbit Ig coupled to Alexa Flour 488 (10 μg/ml, Invitrogen A21206) were used as described above. DAPI staining and co-localization analysis was performed as described above.
Bioinformatics analysis
Densitometry analysis of immunoblots was performed with the gel analysis software UN-SCAN-IT gel 6.1 (Silkscientific).
Supplementary Material
Highlights.
The potential role of LEDGF/p75-interacting proteins in HIV-1 infection has not been explored yet.
LEDGF/p75 specifically associates with the heterodimer SSRP1/Spt16 (FACT complex) through interaction with SSRP1 independently of Spt16
PWWP and HMG domains of LEDGF/p75 and SSRP1, respectively, are required for their interaction.
SSRP1-partial deficiency specifically impairs LTR-driven gene expression of HIV-1 provirus in a LEDGF/p75-independent manner reducing HIV-1 replication in human CD4 T cells.
ACKNOWLEDGEMENTS
We thank Armando Varela and George Steven Martinez (UTEP) for assistance with the confocal microscope and (UTEP) and the densitometry analyses, respectively. This work was supported by Grant Number 5 SC1 AI098238-02 from the National Institute of General Medical Sciences (NIGMS), National Institutes of Health (NIH), (to ML). M.E.F.-Z. was supported by the Mayo Clinic Cancer Center, Division of Oncology Research, NCI CA136526, and, Mayo Clinic Center for Cell Signaling in Gastroenterology (P30 DK084567), J.R.K. was supported by HHMI grant 52005908. A.P.L. and S.A.S. were supported by grant 5R25GM069621-11 from NIGMS. University of Texas at El Paso (UTEP) core facilities are funded by the BBRC grant 5G12RR008124.
Abbreviations
- LEDGF/p75
Lens epithelium-derived growth factor p75
- FACT
Facilitates Chromatin Transcription complex
- SSRP1
Structure Specific Recognition Protein 1
- hSpt16
human homolog of the Suppressor of Ty 16
- HMG
High Mobility Group
- IN
integrase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ge H, Si Y, Roeder RG. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. Embo J. 1998;17:6723–9. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ge H, Si Y, Wolffe AP. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol Cell. 1998;2:751–9. doi: 10.1016/s1097-2765(00)80290-7. [DOI] [PubMed] [Google Scholar]
- 3.Kubo E, Singh DP, Fatma N, Shinohara T, Zelenka P, Reddy VN, Chylack LT. Cellular distribution of lens epithelium-derived growth factor (LEDGF) in the rat eye: loss of LEDGF from nuclei of differentiating cells. Histochem Cell Biol. 2003;119:289–99. doi: 10.1007/s00418-003-0518-3. [DOI] [PubMed] [Google Scholar]
- 4.Llano M, Morrison J, Poeschla EM. Virological and cellular roles of the transcriptional coactivator LEDGF/p75. Curr Top Microbiol Immunol. 2009;339:125–46. doi: 10.1007/978-3-642-02175-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol. 2006;360:760–73. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 6.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1653–75. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 2008;4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poeschla EM. Integrase, LEDGF/p75 and HIV replication. Cell Mol Life Sci. 2008;65:1403–24. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pradeepa MM, Grimes GR, Taylor GC, Sutherland HG, Bickmore WA. Psip1/Ledgf p75 restrains Hox gene expression by recruiting both trithorax and polycomb group proteins. Nucleic Acids Res. 2014;42:9021–32. doi: 10.1093/nar/gku647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daugaard M, Baude A, Fugger K, Povlsen LK, Beck H, Sorensen CS, Petersen NH, Sorensen PH, Lukas C, Bartek J, Lukas J, Rohde M, Jaattela M. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat Struct Mol Biol. 2012;19:803–10. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
- 12.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–4. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 13.Ciuffi A. Mechanisms governing lentivirus integration site selection. Curr Gene Ther. 2008;8:419–29. doi: 10.2174/156652308786848021. [DOI] [PubMed] [Google Scholar]
- 14.Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–95. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–9. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 16.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS ONE. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Rijck J, Bartholomeeusen K, Ceulemans H, Debyser Z, Gijsbers R. High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res. 2010;38:6135–47. doi: 10.1093/nar/gkq410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Rivera JA, Bueno MT, Morales E, Kugelman JR, Rodriguez DF, Llano M. Implication of serine residues 271, 273, and 275 in the human immunodeficiency virus type 1 cofactor activity of lens epithelium-derived growth factor/p75. J Virol. 2010;84:740–52. doi: 10.1128/JVI.01043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meehan AM, Saenz DT, Morrison JH, Garcia-Rivera JA, Peretz M, Llano M, Poeschla EM. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog. 2009;5:e1000522. doi: 10.1371/journal.ppat.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler DD, Luger K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem. 2011;286:18369–74. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler DD, Muthurajan UM, Hieb AR, Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011;286:41883–92. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bueno MT, Garcia-Rivera JA, Kugelman JR, Morales E, Rosas-Acosta G, Llano M. SUMOylation of the lens epithelium-derived growth factor/p75 attenuates its transcriptional activity on the heat shock protein 27 promoter. J Mol Biol. 2010;399:221–39. doi: 10.1016/j.jmb.2010.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem. 2004;279:48883–92. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 24.Vanegas M, Llano M, Delgado S, Thompson D, Peretz M, Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J Cell Sci. 2005;118:1733–43. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 25.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Zeng SX, Landais I, Lu H. Human SSRP1 has Spt16-dependent and -independent roles in gene transcription. J Biol Chem. 2007;282:6936–45. doi: 10.1074/jbc.M603822200. [DOI] [PubMed] [Google Scholar]
- 27.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–3. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 28.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–8. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 29.Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, Poirier MG, Foster MP, Kvaratskhelia M. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013;41:3924–36. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao S, Bendall H, Hicks GG, Nashabi A, Sakano H, Shinkai Y, Gariglio M, Oltz EM, Ruley HE. The high-mobility-group box protein SSRP1/T160 is essential for cell viability in day 3.5 mouse embryos. Mol Cell Biol. 2003;23:5301–7. doi: 10.1128/MCB.23.15.5301-5307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarnell AT, Oh S, Reinberg D, Lippard SJ. Interaction of FACT, SSRP1, and the high mobility group (HMG) domain of SSRP1 with DNA damaged by the anticancer drug cisplatin. J Biol Chem. 2001;276:25736–41. doi: 10.1074/jbc.M101208200. [DOI] [PubMed] [Google Scholar]
- 32.Zennou V, Serguera C, Sarkis C, Colin P, Perret E, Mallet J, Charneau P. The HIV-1 DNA flap stimulates HIV vector-mediated cell transduction in the brain. Nat Biotechnol. 2001;19:446–50. doi: 10.1038/88115. [DOI] [PubMed] [Google Scholar]
- 33.Schrijvers R, De Rijck J, Demeulemeester J, Adachi N, Vets S, Ronen K, Christ F, Bushman FD, Debyser Z, Gijsbers R. LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog. 2012;8:e1002558. doi: 10.1371/journal.ppat.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abe T, Sugimura K, Hosono Y, Takami Y, Akita M, Yoshimura A, Tada S, Nakayama T, Murofushi H, Okumura K, Takeda S, Horikoshi M, Seki M, Enomoto T. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem. 2011;286:30504–12. doi: 10.1074/jbc.M111.264721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 36.Keller DM, Lu H. p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J Biol Chem. 2002;277:50206–13. doi: 10.1074/jbc.M209820200. [DOI] [PubMed] [Google Scholar]
- 37.Keller DM, Zeng X, Wang Y, Zhang QH, Kapoor M, Shu H, Goodman R, Lozano G, Zhao Y, Lu H. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell. 2001;7:283–92. doi: 10.1016/s1097-2765(01)00176-9. [DOI] [PubMed] [Google Scholar]
- 38.LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–4. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 39.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–33. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–16. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 41.Wittmeyer J, Formosa T. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol. 1997;17:4178–90. doi: 10.1128/mcb.17.7.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morchikh M, Naughtin M, Di Nunzio F, Xavier J, Charneau P, Jacob Y, Lavigne M. TOX4 and NOVA1 proteins are partners of the LEDGF PWWP domain and affect HIV-1 replication. PLoS One. 2013;8:e81217. doi: 10.1371/journal.pone.0081217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huen MS, Huang J, Leung JW, Sy SM, Leung KM, Ching YP, Tsao SW, Chen J. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol Cell. 2010;37:854–64. doi: 10.1016/j.molcel.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Izumoto Y, Kuroda T, Harada H, Kishimoto T, Nakamura H. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem Biophys Res Commun. 1997;238:26–32. doi: 10.1006/bbrc.1997.7233. [DOI] [PubMed] [Google Scholar]
- 45.Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000;473:1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- 46.Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF, Altherr MR, den Dunnen JT. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–82. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- 47.Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007;64:2590–606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Reddy B, Thompson J, Wang H, Noma K, Yates JR, 3rd, Jia S. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell. 2009;33:428–37. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malarkey CS, Churchill ME. The high mobility group box: the ultimate utility player of a cell. Trends Biochem Sci. 2012;37:553–62. doi: 10.1016/j.tibs.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–86. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lang D, Epstein JA. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum Mol Genet. 2003;12:937–45. doi: 10.1093/hmg/ddg107. [DOI] [PubMed] [Google Scholar]
- 52.Wissmuller S, Kosian T, Wolf M, Finzsch M, Wegner M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006;34:1735–44. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–45. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 54.Tsunaka Y, Toga J, Yamaguchi H, Tate S, Hirose S, Morikawa K. Phosphorylated intrinsically disordered region of FACT masks its nucleosomal DNA binding elements. J Biol Chem. 2009;284:24610–21. doi: 10.1074/jbc.M109.001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–20. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin S, Min J. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem Sci. 2014;39:536–47. doi: 10.1016/j.tibs.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Qiu Y, Zhang W, Zhao C, Wang Y, Wang W, Zhang J, Zhang Z, Li G, Shi Y, Tu X, Wu J. Solution structure of the Pdp1 PWWP domain reveals its unique binding sites for methylated H4K20 and DNA. Biochem J. 2012;442:527–38. doi: 10.1042/BJ20111885. [DOI] [PubMed] [Google Scholar]
- 59.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–80. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 60.Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17:617–9. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 61.Wu H, Zeng H, Lam R, Tempel W, Amaya MF, Xu C, Dombrovski L, Qiu W, Wang Y, Min J. Structural and histone binding ability characterizations of human PWWP domains. PLoS One. 2011;6:e18919. doi: 10.1371/journal.pone.0018919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Easley R, Carpio L, Dannenberg L, Choi S, Alani D, Van Duyne R, Guendel I, Klase Z, Agbottah E, Kehn-Hall K, Kashanchi F. Transcription through the HIV-1 nucleosomes: effects of the PBAF complex in Tat activated transcription. Virology. 2010;405:322–33. doi: 10.1016/j.virol.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spina CA, Anderson J, Archin NM, Bosque A, Chan J, Famiglietti M, Greene WC, Kashuba A, Lewin SR, Margolis DM, Mau M, Ruelas D, Saleh S, Shirakawa K, Siliciano RF, Singhania A, Soto PC, Terry VH, Verdin E, Woelk C, Wooden S, Xing S, Planelles V. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 2013;9:e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Lint C, Bouchat S, Marcello A. HIV-1 transcription and latency: an update. Retrovirology. 2013;10:67. doi: 10.1186/1742-4690-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xing S, Siliciano RF. Targeting HIV latency: pharmacologic strategies toward eradication. Drug Discov Today. 2013;18:541–51. doi: 10.1016/j.drudis.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gautier VW, Gu L, O’Donoghue N, Pennington S, Sheehy N, Hall WW. In vitro nuclear interactome of the HIV-1 Tat protein. Retrovirology. 2009;6:47. doi: 10.1186/1742-4690-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu J, Davoli T, Perriera JM, Chin CR, Gaiha GD, John SP, Sigiollot FD, Gao G, Xu Q, Qu H, Pertel T, Sims JS, Smith JA, Baker RE, Maranda L, Ng A, Elledge SJ, Brass AL. Comprehensive identification of host modulators of HIV-1 replication using multiple orthologous RNAi reagents. Cell Rep. 2014;9:752–66. doi: 10.1016/j.celrep.2014.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang H, Santoso N, Power D, Simpson S, Dieringer M, Miao H, Gurova K, Giam CZ, Elledge SJ, Zhu J. FACT Proteins, SUPT16H and SSRP1, Are Transcriptional Suppressors of HIV-1 and HTLV-1 That Facilitate Viral Latency. J Biol Chem. 2015;290:27297–310. doi: 10.1074/jbc.M115.652339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–9. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 70.Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78:9524–37. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–11. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dejmek J, Iglehart JD, Lazaro JB. DNA-dependent protein kinase (DNA-PK)-dependent cisplatin-induced loss of nucleolar facilitator of chromatin transcription (FACT) and regulation of cisplatin sensitivity by DNA-PK and FACT. Mol Cancer Res. 2009;7:581–91. doi: 10.1158/1541-7786.MCR-08-0049. [DOI] [PubMed] [Google Scholar]
- 73.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278:33528–39. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.