Abstract
In vivo μCT imaging allows for high-resolution, longitudinal evaluation of bone properties. Based on this technology, several recent studies have developed in vivo dynamic bone histomorphometry techniques that utilize registered μCT images to identify regions of bone formation and resorption, allowing for longitudinal assessment of bone remodeling. However, this analysis requires a direct voxel-by-voxel subtraction between image pairs, necessitating rotation of the images into the same coordinate system, which introduces interpolation errors. We developed a novel image transformation scheme, matched-angle transformation (MAT), whereby the interpolation errors are minimized by equally rotating both the follow-up and baseline images instead of the standard of rotating one image while the other remains fixed. This new method greatly reduced interpolation biases caused by the standard transformation. Additionally, our study evaluated the reproducibility and precision of bone remodeling measurements made via in vivo dynamic bone histomorphometry. Although bone remodeling measurements showed moderate baseline noise, precision was adequate to measure physiologically relevant changes in bone remodeling, and measurements had relatively good reproducibility, with intra-class correlation coefficients of 0.75-0.95. This indicates that, when used in conjunction with MAT, in vivo dynamic histomorphometry provides a reliable assessment of bone remodeling.
Keywords: In vivo μCT, 3D image registration, trabecular microstructure, animal models/rodent
1. Introduction
In vivo μCT imaging allows for high-resolution, longitudinal evaluation of bone properties. Over the past ten years, both preclinical and clinical studies have utilized in vivo μCT to track changes in the trabecular bone microarchitecture as a result of disease states and treatments 5, 7-11, 17, 20, 28-30. In such studies, subjects are scanned at multiple time-points, and the resulting μCT images are compared to detect changes in trabecular microstructure within a defined volume of interest (VOI). Such measurements are subject to various sources of error, including those induced by the partial volume effect, the digitization process, and image interpolation. There is no way to completely avoid these intrinsic error sources, however, steps can be taken to minimize their impact on microstructural measurements.
To ensure accurate identification of changes in trabecular bone quality, registration of the repeated, longitudinal scans is crucial 6, 17, 20. By aligning patterns of trabecular connectivity, image registration allows the same VOI to be identified in each repeated scan, resulting in a highly sensitive assessment of changes in trabecular microarchitecture. In general, a typical image registration procedure involves aligning the baseline (b) and follow-up (f) images, resulting in a transformation matrix (T). This transformation matrix can then be directly applied to f to result in f1 (f1 = Tf), a version of image f which has been rotated and translated so that it is directly aligned with the baseline image, b. By applying a VOI mask to f1 and b, the trabecular parameters of these two time-points can then be directly compared. However, direct application of the transformation matrix T to f results in inherent errors caused by image interpolation 22. To avoid these errors, several studies have used a masking procedure where a VOI mask is defined in one image, and the mask is then rotated and translated, allowing for the identification of an identical VOI in subsequent scans without having to transform the images themselves 17, 20.
In addition to tracking changes in the trabecular microstructure, several recent studies have developed in vivo dynamic bone histomorphometry techniques that utilize registered μCT images to identify regions of bone formation and resorption, allowing for longitudinal assessment of bone remodeling and structural changes in the caudal vertebra 16, 21, 23 and at the proximal tibia 3, 12. In these studies, animals are scanned at multiple time-points, and the resulting μCT images are registered, thresholded, and subtracted to identify, on a voxel-by-voxel basis, locations where bone has formed or resorbed 3, 12, 21. However, this new analysis requires a direct voxel-by-voxel subtraction between image pairs. Because image pairs are directly subtracted from one another, the images must be rotated into the same coordinate system, and it is therefore impossible to utilize a VOI masking technique to avoid image interpolation.
This requirement of directly rotating images to allow for a voxel-by-voxel comparison may introduce interpolation errors and/or affect reproducibility of the in vivo dynamic bone histomorphometry technique, neither of which has been evaluated to date. In the standard transformation (ST) scheme, where the follow-up image, f, is rotated to f1 (f1 = Tf) to align with the baseline image, b, the unequal interpolation effects between b (which remains fixed and is not rotated at all) and f1 (which is transformed to the same coordinate system as b) are of particular concern, as these can introduce biases when directly comparing f1 and b. Such errors can be minimized through selection of an appropriate interpolator 22. However, when comparing effects of several standard interpolators, including nearest neighbor, linear, and B-splines, an error of at least 1.4% remained in measurements of standard trabecular parameters following interpolation of repeated μCT images 22, and the impact of image interpolation on measurements made through in vivo dynamic bone histomorphometry remains unknown.
In this study, we developed a novel image transformation scheme, termed matched-angle transformation (MAT), whereby both the baseline and the follow-up images undergo an equal amount of rotation, resulting in similar interpolation effects between the two images. Although application of MAT will not necessarily reduce the magnitude of the interpolation errors within a single image, by rotating the baseline and follow-up images by the same amount, this technique eliminates the biases that result from directly comparing an image that has been rotated with one that remains fixed. This new transformation method was then applied to two image analysis scenarios: (1) the evaluation of trabecular bone microstructure, and (2) the identification of regions of trabecular bone formation/resorption using in vivo dynamic bone histomorphometry 12 in repeated scans of rat tibiae. We hypothesized that the MAT scheme would result in reduced interpolation bias and improved reproducibility of these measurements as compared to the standard transformation (ST) scheme, where only the follow-up image is rotated. Overall, the purpose of this study was to quantify the effects of image interpolation caused by the two different transformation procedures (ST and MAT), and to assess the reproducibility of in vivo dynamic bone histomorphometry measurements.
2. Methods
2.1 Animals
All experiments were approved by the University of Pennsylvania's Institutional Animal Care and Use Committee. Four-month-old, intact, female, Sprague Dawley rats (n=18) were used in this study. For each rat, a 2.2 mm region of the left proximal tibia was scanned using μCT (VivaCT40, Scanco Medical). Rats were scanned twice at baseline (day 0), then again after one week (day 7). For each scan, rats were placed in a custom holder 17, which minimized motion and allowed for high consistency between repeated scans. Scans were made at 10.5 μm resolution, 145 μA intensity, 55 keV energy, 200 ms integration time, and 1000 projections, resulting in a total scan time of about 10 minutes/scan. A 0.5mm Al filter and a standard, manufacturer-provided beam-hardening correction algorithm were used. For the repeated, same-day scans, rats were removed from the scanner after the first scan, and repositioned prior to the start of the second scan. Three rats were excluded due to motion artifacts in the μCT scans, resulting in a final sample size of n=15.
2.2 Image Registration
To precisely align the trabeculae in each scan, a three-step image registration procedure was performed, as described previously 12, using an open-source image registration toolkit 14. Briefly, the cortical bone in the baseline (b) and follow-up (f) scans was aligned to result in an initial transformation matrix, T1. The trabecular compartment was then aligned using a landmark-initialized registration method, resulting in a second transformation matrix, T2. Lastly, a 150×150×100 voxel VOI in the secondary spongiosa was extracted from the images and aligned precisely, resulting in a final transformation matrix, T3. Combining the resulting three transformation matrices resulted in an overall transformation T = T3T2T1, which describes the rotation and translation required to correctly align the individual trabeculae of the two scans. This registration technique has been previously shown to result in a precise alignment of the trabecular compartment, allowing for accurate measurements of bone formation and resorption 12.
2.3 Effects of Image Interpolation on Measurements of Trabecular Structure
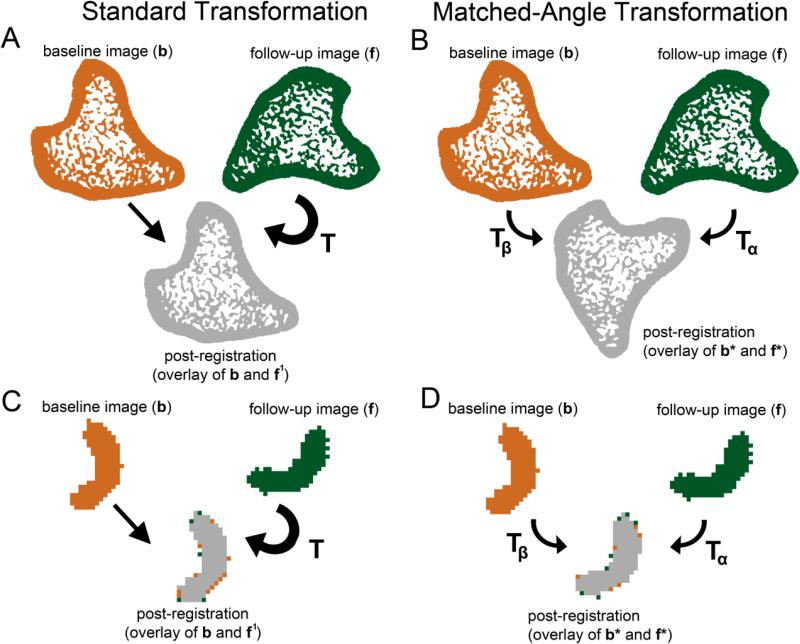
To assess the impact of interpolation artifacts, repeated scans made at baseline (day 0) were used. Each pair of images was registered, as described above, and for each image pair, two image transformation methods were applied. In the Standard Transformation (ST, Fig 1A), the follow-up image, f was rotated to yield f1 (f1 = Tf) so that it was aligned with the baseline image b, which was kept fixed. In the matched-angle transformation (MAT, Fig 1B), on the other hand, the rotation angle described by the transformation matrix T was divided equally and applied to both b and f. Thus, half of the rotation, Tα, was applied to the follow-up image to yield f* (f* = Tαf). Similarly, the inverse of half of the rotation, Tβ (Tβ = Tα−1) was applied to the baseline image, to yield b* (b* = Tβb). Therefore, both the b and f were rotated by the same amount to yield the registered images b* and f*, causing similar interpolation effects to be introduced into both images. Both MAT and ST utilized the same B-splines interpolator, thus it was not expected that application of MAT would reduce the amount of interpolation within a single image. However, as shown by comparing Fig 1C and D, application of the two transformation techniques allowed interpolation artifacts to be introduced only in the follow-up image when using ST, while equal interpolation artifacts were introduced in both images when performing MAT. Computation time for the MAT technique was approximately double that of the ST technique, as MAT required rotation of both the baseline and follow-up images, whereas ST only required rotation of a single image.
Fig. 1.
A schematic illustrating the effect of aligning images using (A) ST vs. (B) MAT. In the ST scheme, f is transformed using transformation T such that f1 (where f1 = Tf) and b are aligned. In the MAT scheme, f and b are both transformed using transformations Tα and Tβ (where Tβ = Tα−1), respectively, such that f* and b* are aligned (where f* = Tαf and b* = Tβb). A close-up of a single trabecula demonstrates the effects of (C) ST and (D) MAT on identification of voxels of bone formation and resorption.
Registered, transformed trabecular subvolumes were Gaussian filtered (sigma=1.2, support=2), and binarized using a global threshold. This threshold was selected by a manufacturer-provided, adaptive thresholding procedure, followed by visual confirmation, resulting in a threshold corresponding to 565 mgHA/cm3. Bone volume fraction (BV/TV), connectivity density (Conn.D), structure model index (SMI), trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular spacing (Tb.Sp) were measured in the repeated scans. For both ST and MAT, interpolation biases were assessed through evaluation of the differences between measurements made in the 1st and 2nd scans, and reproducibility was measured by computing the root mean square average of the standard deviation and coefficient of variance (RMSSD, RMS%CV) and intra-class correlation coefficients (ICCs) as described in 2.7.
2.4 Effects of Image Interpolation on In Vivo Dynamic Histomorphometry Measurements
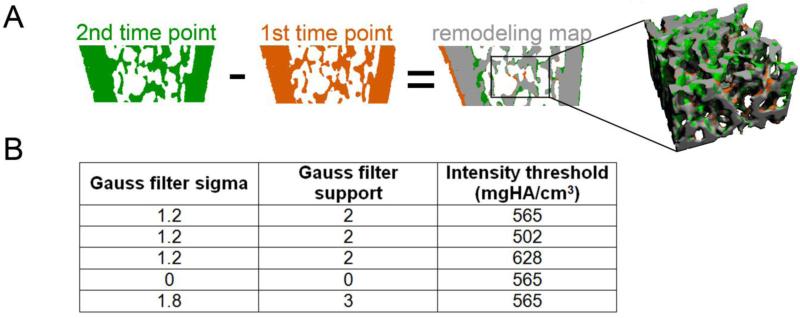
The repeated, same-day scans were registered, transformed by ST and MAT, and thresholded, as described in 2.3. The images were then subtracted in order to identify voxels of bone resorption (i.e., bone voxels that were present in the baseline image but absent in the follow-up scan), and voxels of new bone formation (i.e., bone voxels that were present in the follow-up image but absent at baseline) (Fig. 2A) 12. As was validated in our previous study 12, the surface layer of all bone formation and resorption sites was then removed, in order to eliminate incorrectly identified regions of bone remodeling caused by the partial volume effect. Finally, the resulting images of bone formation and resorption were subjected to a cluster analysis, whereby all clusters of bone formation/resorption that were smaller in size than 10 voxels (equal to 11,576 μm3) were excluded from future analyses. This was performed, because we anticipated that clusters below this size would be unlikely to correspond to actual bone remodeling 26 and would be more likely to correspond to noise. Based on the resulting map of bone formation and resorption, the bone formation volume (volume of bone formed normalized by bone surface) and bone resorption volume (volume of bone resorbed normalized by bone surface) were quantified. Additionally, mineral apposition thickness (average thickness of newly formed bone), mineral erosion thickness (average thickness of resorbed bone), mineralizing surface (MS/BS = surface of newly formed bone / total bone surface), and eroding surface (ES/BS = surface area of resorbed bone / total bone surface) were also determined. The deviation of these measures (based on the same-day, repeated scans) from zero was used to quantify the magnitude of noise and interpolation artifacts using both the ST and MAT methods.
Fig. 2.
(A) The in vivo dynamic histomorphometry technique involved subtracting two registered, binarized images to obtain a map of bone formation (green), resorption (orange), and constant bone (gray). A trabecular subvolume was then extracted from this map. (B) Several combinations of Gauss filter sigma and support, and global intensity thresholds were investigated to determine the effects of the thresholding and filter parameters on noise and reproducibility of the in vivo dynamic bone histomorphometry measurements. Please note, the presence of bone formation on the endosteal surface and resorption on the periosteal surface of the cortical bone in the remodeling map cross-section is due to longitudinal growth of the bone 2, 12, 17, and was not investigated: evaluations were focused on a trabecular bone subvolume.
2.5 Reproducibility of In Vivo Dynamic Bone Histomorphometry Measurements
Reproducibility was determined by individually registering each of the repeated, same-day scans to the scan made one week later, resulting in two sets of registered images. The images were then transformed (using either ST or MAT), Gaussian filtered, thresholded, subtracted, and processed as described in 2.4 to yield a map of bone formation and resorption sites. Standard in vivo dynamic histomorphometry parameters 12, 21 were calculated based on the resulting map of bone formation and resorption over a one-week period. Bone formation rate (BFR/BS = bone formation volume / # of days between scans), mineral apposition rate (MAR = mineral apposition thickness / # of days between scans), and MS/BS were measured. Similarly, measures of bone resorption, including bone resorption rate (BRR/BS = bone resorption volume / # of days), mineral erosion rate (MER = mineral erosion thickness / # of days), and ES/BS were calculated. The reproducibility of the in vivo dynamic bone histomorphometry technique was then determined by computing the root mean square average of the standard deviation and coefficient of variance (RMSSD, RMS%CV) and intra-class correlation coefficients (ICC) based on the repeated measurements (as described in 2.7).
2.6 Effects of Filter and Threshold on Noise in In Vivo Dynamic Histomorphometry
The registered, repeated day 0 scans were transformed using MAT, and were then Gaussian filtered and thresholded. Adaptive thresholding followed by visual comparison of the grayscale and binarized images indicated that filtration using a Gauss filter with sigma=1.2 and support=2, followed by application of a global threshold corresponding to 565 mgHA/cm3 resulted in the most accurate segmentation of the trabecular subvolume. Therefore, the filter and threshold analysis was centered on these parameters. To test the effects of filter and threshold on noise introduced in the dynamic measurements, several filter kernels and thresholds (listed in Fig. 2B) were tested. Following each set of image filtration and thresholding, bone formation and resorption volumes, mineral apposition and erosion thicknesses, MS/BS, and ES/BS were measured as described in 2.4. Noise was quantified by determining the deviation of each measurement from zero.
2.7 Statistical Analysis
Reproducibility was evaluated by computing the root mean square average of the standard deviation and coefficient of variance (RMSSD, RMS%CV) and intra-class correlation coefficients (ICC), as described in 17 and 15. Briefly, the individual coefficient of variance (%CV) was evaluated, and the root mean square average of the %CV (RMS%CV) was derived for each parameter as follows:
Paired Student's t-tests were performed to compare the %CV of repeated scans based on ST and MAT. As described in 24 and 15, ICC is computed as a ratio of the variability due only to differences among samples, over the total variability (including variability caused by differences in the measurement technique used during repeated measurements in addition to that caused by inherent differences among samples). Thus an ICC close to 1 indicates good test-retest repeatability. ICCs were calculated based on the mean squares determined through a two-way ANOVA, as follows 15, 24:
where:
To determine the effects of the image transformation scheme on interpolation biases in measurements of trabecular microstructure made in the repeated, same-day scans, a two-way, repeated-measures analysis of variance (ANOVA), correlated for baseline measures was used. This analysis used transformation method (ST vs. MAT) as the between-factor, and measurement number (first scan vs. second scan) as the repeated, within-factor.
To assess the effects of the image transformation scheme on noise introduced in in vivo dynamic bone histomorphometry measurements, a repeated-measures, 2-way ANOVA was performed, with transformation method (ST vs. MAT) as the between-factor, and the comparison between bone formation and resorption measures as the repeated, within-group factor. Noise present in bone formation and resorption measures after applying ST and MAT was then compared by examining the interaction effects.
Lastly, one-way ANOVA was performed to evaluate the effects of different filter and threshold parameters on noise introduced in bone remodeling measurements. Planned comparisons were made to the standard filter and threshold (Gaussian filter with sigma=1.2, support=2, followed by application of a global threshold corresponding to 565 mgHA/cm3). All analyses were conducted using NCSS 7.1.14 (NCSS, LLC, Kaysville, UT) and Microsoft Excel 2007 (Microsoft Office, Redmond, WA), Bonferroni post hoc corrections were applied for all individual comparisons, and two-sided p values <0.05 were considered to indicate statistical significance. All error bars in graphs represent the standard error of mean (SEM).
3. Results
3.1 Reproducibility of Standard Measurements of Trabecular Microstructure
Image registration followed by interpolation, either by ST or MAT, resulted in highly reproducible measurements of trabecular microarchitecture (Table 1). Specifically, precision error (as measured through RMS%CV) ranged from 0.7-3.1% when applying the ST interpolation scheme, and was significantly reduced for each parameter except SMI when using MAT (resulting in RMS%CV of 0.3-2.4%). ICCs were greater than 0.97 for both interpolation schemes, indicating good test-retest reliability.
Table 1.
Reproducibility of trabecular structure measurements when using ST and MAT
| ST | MAT | |||||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SD | RMSSD | RMS%CV | ICC | Mean ± SD | RMSSD | RMS%CV | ICC | |
| BV/TV | 0.23 ± 0.06 | 0.001 | 0.707% | 0.9996 | 0.23 ± 0.06 | 0.001 | 0.483%* | 0.9993 |
| Conn.D (1/mm3) | 100.5 ± 30.8 | 3.395 | 3.165% | 0.9937 | 106.7 ± 30.8 | 2.367 | 2.438%* | 0.9855 |
| SMI | 1.61 ± 0.46 | 0.023 | 1.594% | 0.9985 | 1.61 ± 0.48 | 0.019 | 1.321% | 0.9975 |
| Tb.N (1/mm) | 4.68 ± 0.54 | 0.060 | 1.275% | 0.9960 | 4.68 ± 0.54 | 0.022 | 0.509%* | 0.9768 |
| Tb.Th (mm) | 0.069 ± 0.007 | 0.001 | 0.688% | 0.9983 | 0.069 ± 0.007 | 0.000 | 0.411%* | 0.9944 |
| Tb.Sp (mm) | 0.20 ± 0.03 | 0.001 | 0.709% | 0.9993 | 0.19 ± 0.03 | 0.001 | 0.263%* | 0.9963 |
%CV significantly different between ST and MAT, p<0.05
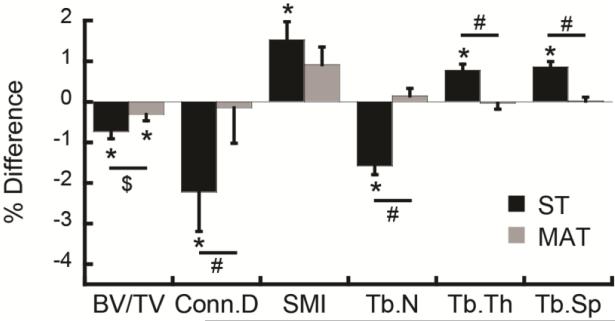
When comparing trabecular bone parameters from the 2nd scan (transformed) to the 1st scan (fixed) following ST, the 2nd scan yielded significantly lower BV/TV, Conn.D, and Tb.N (−0.74 to −2.23%), and significantly higher SMI, Tb.Th, and Tb.Sp (0.77 to 1.53%) than the 1st scan (Fig. 3), suggesting that ST induces critical interpolation biases. In contrast, transformation through MAT showed no significant differences in these measures except for BV/TV which was 0.31% lower in the 2nd than the 1st scan. Moreover, ANOVA indicated that the % difference measured through ST in Conn.D, Tb.N, Tb.Th, and Tb.Sp was significantly greater than the percent difference in these measures determined following MAT (p<0.05), and the % difference in BV/TV measured through ST tended to be greater than that measured following MAT (p<0.1).
Fig. 3.
Percent difference in trabecular microstructure measurements over two repeated, same-day scans as measured after applying ST or MAT. *:significant difference between the two repeated measurements (p<0.05), #: significant difference between % difference measured through ST vs. MAT (p<0.05), $: trend towards difference between % difference measured through ST vs. MAT (p<0.1).
3.2 Effects of Image Interpolation on Noise in In Vivo Dynamic Histomorphometry Measurements
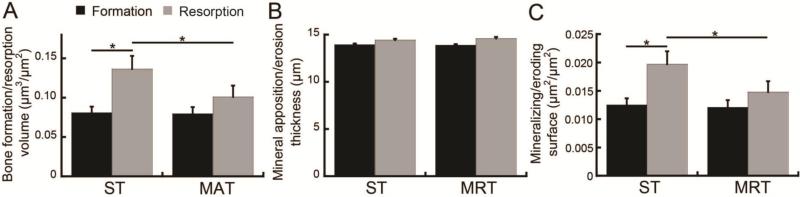
In vivo dynamic bone histomorphometry measurements made based on repeated, same-day scans resulted in an average bone formation/resorption volume, thickness, and surface area of about 0.09 μm3/μm2, 14 μm, and 0.013 μm2/μm2, respectively (Fig. 4). These results indicate that the noise levels of bone formation/resorption volume and surface area were equivalent to 51% and 56% of measures made over a one-week period in a situation of low bone remodeling (as occurred in this study). Percentage of the formation/resorption thickness due to noise is not given, as this parameter corresponds to the average thickness of bone formation/resorption sites, and thus the percentage due to noise is not a relevant measure.
Fig. 4.
Effect of image interpolation scheme on noise in the in vivo dynamic bone histomorphometry measurements (as measured based on repeated, day 0 scans) of (A) bone formation/resorption volume, (B) average thickness of apposition/erosion sites, and (C) formation/resorption surface area. *: p<0.05.
As demonstrated in Fig. 4, ST resulted in a significant overestimation of bone resorption over bone formation activities in the repeated, same-day scans. Bone resorption volume was 68% greater than the bone formation volume, and ES/BS was 57% greater than the MS/BS in the ST group (p<0.05), indicating a significant interpolation bias. Conversely, there were no significant differences between measures of bone formation and resorption made in the repeated, same-day scans when processed using MAT. In addition to differences between bone resorption and formation measurements within each transformation method, ST-based measures of bone resorption volume and ES/BS were also 35% and 33% greater, respectively, than equivalent MAT-based measures (Fig. 4, p<0.05), indicating that MAT resulted in reduced noise levels.
3.3 Reproducibility of In Vivo Dynamic Bone Histomorphometry Measurements
Over one week, an average BFR/BS and BRR/BS of 0.027 and 0.022 μm3/μm2/day, respectively, an average MAR and MER of 2.22 and 2.05 μm/day, respectively, and an average MS/BS and ES/BS of 0.026 and 0.021 μm2/μm2 were measured (Table 2). Reproducibility analysis indicated that repeated measures of bone remodeling over a one-week period were consistent when using both MAT and ST (Table 2), with precision error (as measured through RMS%CV) ranging from 2.1-23.5%, and ICCs ranging from 0.75-0.95. Precision errors for MAR and MER were the lowest (~3%), while measurements of remodeling surface and volume resulted in precision errors of around 16-23%. ICCs were fairly consistent over the different bone remodeling measurements.
Table 2.
Reproducibility of in vivo dynamic bone histomorphometry measurements
| ST | MAT | |||||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SD | RMSSD | RMS%CV | ICC | Mean ± SD | RMSSD | RMS%CV | ICC | |
| BFR/BS (μm3/μm2) | 0.032 ± 0.018 | 0.0064 | 17.43% | 0.875 | 0.027 ± 0.014 | 0.0062 | 23.51% | 0.822 |
| BRR/BS (μm3/μm2) | 0.025 ± 0.011 | 0.0052 | 17.49% | 0.819 | 0.022 ± 0.011 | 0.0051 | 21.62% | 0.807 |
| MAR (μm) | 2.18 ± 0.20 | 0.0454 | 2.12% | 0.948 | 2.22 ± 0.24 | 0.0701 | 3.07%* | 0.917 |
| MER (μm) | 2.05 ± 0.14 | 0.0755 | 3.60% | 0.751 | 2.05 ± 0.15 | 0.0631 | 3.07% | 0.836 |
| MS/BS (μm2/μm2) | 0.031 ± 0.016 | 0.0056 | 16.01% | 0.890 | 0.026 ± 0.012 | 0.0055 | 23.15% | 0.817 |
| ES/BS (μm2/μm2) | 0.025 ± 0.010 | 0.0046 | 16.32% | 0.819 | 0.021 ± 0.009 | 0.0045 | 20.53% | 0.792 |
%CV significantly different between ST and MAT, p<0.05
3.4 Effect of Thresholding Technique on Noise in In Vivo Dynamic Bone Histomorphometry Measurements
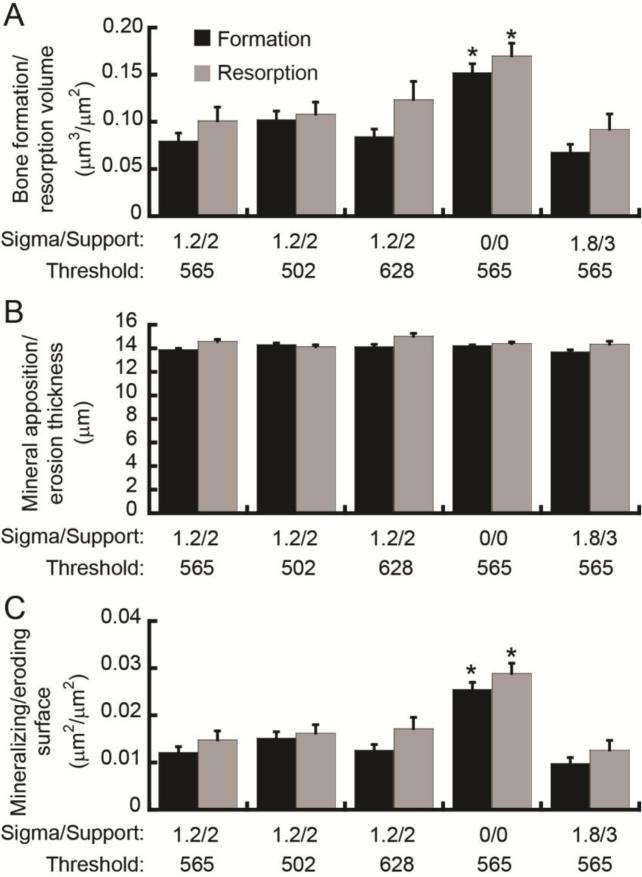
Because in vivo dynamic bone histomorphometry measurements showed reduced noise and bias when obtained following interpolation through MAT than when obtained following ST, MAT was used for the remainder of the study. After MAT, application of a filter kernel with sigma=1.2 and support=2, and threshold of 565 mgHA/cm3 resulted in a noise measurement of approximately 0.09 μm3/μm2, 14 μm , and 0.013 μm2/μm2 for measures of bone formation/resorption volume, mineral apposition/erosion thickness, and mineralizing/eroding surfaces, respectively (Fig. 5). Moderate changes in the applied threshold (to 502 and 628 mgHA/cm3) did not affect the noise present in the in vivo dynamic bone histomorphometry measurements.
Fig. 5.
Effect of thresholding technique on noise in the in vivo dynamic bone histomorphometry measurements (as measured based on repeated, day 0 scans) of (A) bone formation/resorption volume, (B) average thickness of apposition/erosion site, and (C) formation/resorption surface area. *: significantly different from original filter and threshold (sigma=1.2, support=2, threshold=565 mgHA/cm3).
However, Gaussian filtration had a significant impact on the noise present in measures of bone formation/resorption volume and surface area. Elimination of the filtration step (Gauss filter with sigma=0, support=0) resulted in a 91% and 68% increase in the noise present in measures of bone formation volume and bone resorption volume, respectively, and a 111% and 95% increase in the noise present in measurements of MS/BS and ES/BS (p<0.05). On the other hand, excessive smoothing of the image by applying a Gauss filter with sigma=1.8, support=3 did not significantly affect the noise present in bone remodeling measurements. Measures of mineral apposition thickness and mineral erosion thickness were not affected by the Gaussian filter.
4. Discussion
This study investigated the effects of image interpolation on longitudinal, μCT-based measures of trabecular bone structure and dynamics, and presented a novel image transformation scheme to minimize the impact of interpolation artifacts on trabecular measurements. The newly developed matched-angle transformation (MAT) scheme was applied to repeated, in vivo μCT scans of rat tibiae, and its effects on the repeatability of trabecular microstructural measures as well as the repeatability and noise associated with bone remodeling parameters measured using in vivo dynamic bone histomorphometry were determined. Comparison of the MAT scheme with the standard transformation (ST) method indicated that using MAT in place of ST resulted in a significant reduction in the precision error and interpolation bias associated with repeated measurements of trabecular structure. Most importantly, MAT resulted in reduced noise and biases in bone remodeling measurements.
For evaluation of trabecular bone microstructure, both ST and MAT resulted in precision errors (as measured through RMS%CV) and ICCs similar to those measured through the gold standard of rotating a mask to identify a consistent VOI in longitudinal scans 17, 20, 22. Overall, MAT resulted in a small but significant decrease in precision error as compared to ST. Similar to what others have found, the derived measures of Conn.D and SMI showed slightly lower repeatability than other structural measures, suggesting that these may be more sensitive to interpolation effects and noise 17, 20. However, although both ST and MAT allowed good reproducibility, the application of ST caused significant interpolation-induced biases in measurements of trabecular microstructure. As shown in Fig. 3, although the differences between the two scans were small, the consistent rotation of only the follow-up image, while the baseline image remained fixed, resulted in a biased result after the two images were digitized, as significant, directional changes between the repeated, same-day scans were detected in every parameter. Taken together, these changes suggested compromised trabecular bone structure and elevated trabecular thickness in the follow-up scan when compared with the earlier scan conducted in the same day. If observed over a non-zero timespan, such measurements would lead to the conclusion that the trabecular microarchitecture was deteriorated in the second scan compared to the first. Thus, the interpolation biases introduced during ST could lead to false conclusions in longitudinal studies of trabecular bone changes. In contrast, these interpolation biases were nearly abolished when applying MAT.
Similar to measurements of trabecular microstructure, in vivo dynamic bone histomorphometry measurements were also significantly affected by the transformation scheme used. Due to noise introduced during scanning and image processing, both ST and MAT resulted in non-zero measurements of bone remodeling determined based on the same-day, repeated scans. Of greater concern, images transformed using ST had up to a 68% difference between measures of bone “formation” and “resorption” based on the repeated, same-day scans, whereas no difference was found between measures bone “formation” and “resorption” based on repeated scans when using MAT. This difference between bone resorption and formation parameters when using ST is most likely due to the consistent rotation of the follow-up image, while the baseline image remained static. Rotation of the follow-up image causes voxels on the edges of trabeculae to take on an intensity intermediate between bone and background. As a result of their reduced intensity, a fraction of these voxels were likely identified as background when the image was thresholded. Because they were labeled as bone in the initial (non-transformed) scan, these voxels were then identified as bone resorption, resulting in an overestimation of bone resorption over bone formation when using ST. On the other hand, MAT results in approximately equal levels of interpolation of both images, thus reducing any biases caused by unequal interpolation.
The presence of non-zero levels of noise in measures of both bone formation and resorption needs to be considered when interpreting in vivo dynamic bone histomorphometry measurements. Comparisons of repeated, same-day scans with scans made one week apart indicate that basal levels of bone formation/resorption volumes in healthy, 4-month-old rats are only two-fold greater than the noise levels that occur when scanning rats twice within the same day, highlighting a limitation of the in vivo dynamic bone histomorphometry method. Namely, the sensitivity of the bone remodeling measurements is limited by the resolution of the in vivo μCT scans. Because the μCT scanner has a maximum resolution of 10.5 μm, variations in bone remodeling thickness below this length scale cannot be detected. However, the scans made one week apart in this study demonstrate the lowest possible rates of bone remodeling, and when using the in vivo dynamic bone histomorphometry method to identify the effects of interventions such as ovariectomy surgery, pharmacological treatment, and mechanical stimulation on bone remodeling, changes in bone formation and resorption rates are well above the noise levels measured here. For example, rats undergoing high levels of bone remodeling induced by anabolic treatment show bone formation rates more than 1200% greater than noise levels quantified in this study 12. Even in rats with lower rates of bone remodeling, the level of noise quantified in the current study remains significantly lower than previously measured differences in remodeling due to osteoporosis or drug treatment 1, 12. Combined with previous studies by other groups 3, 16, 18, 21, 23, this indicates that the in vivo dynamic bone histomorphometry technique has adequate precision to detect physiologically relevant changes in bone remodeling. In vivo dynamic bone histomorphometry has many strengths, including its ability to provide noninvasive, longitudinal measures. However, for applications where differences in bone remodeling rate are too low to be accurately distinguished from noise, alternate techniques should be used, such as the recently developed serial milling-based 3D histomorphometry technique 13, 19, 25-27, which results in an in-plane resolution of 0.7 μm, allowing greater sensitivity.
In vivo dynamic bone histomorphometry measurements were found to have fairly good reproducibility, with precision errors ranging from 2-23% and ICCs ranging from 0.75-0.95 (Table 2). MAR and MER were found to have higher reproducibility (as indicated by lower precision errors and higher ICCs) than all other in vivo dynamic bone histomorphometry measurements. This is likely due to the nature of these measurements. Specifically, MAR and MER are measures of the average thickness of regions of bone formation/resorption. Because the young, healthy rats in this study underwent low rates of bone remodeling during the one-week inter-scan period, the average thickness of regions of bone formation and resorption was generally quite low, and on the order of a single voxel. Because of the finite resolution of the μCT scanner, this resulted in minimal variation of the average thickness of bone formation/resorption regions, resulting in a lower sensitivity but higher reproducibility of these measurements.
Precision errors found in this study were higher than those reported by Schulte et al., who found precision errors of 0.9-6.5% for μCT-based bone remodeling measurements 21. Additionally, Schulte et al. found higher ICCs than those found in this study 21. This discrepancy is likely due to differences in the way reproducibility was measured in the two studies. All μCT scans in this study were performed in vivo, while Schulte et al. 21 performed the repeated μCT scans after sacrifice, eliminating any animal motion, which is an important cause of error in μCT imaging studies. In addition, the repeated measurements of bone remodeling in our study were made over a fairly short, one-week time interval, in a healthy animal model that was undergoing minimal bone remodeling, leading to low quantities of bone formation and resorption. Precision errors (measured as RMS%CV) are directly related to the amount of scanner and user error, and inversely related to the magnitude of the parameter being measured. The amount of scanner and user error should be fairly constant; thus we anticipate that an animal model or experimental condition with greater volumes, surface areas, and thicknesses of bone formation and resorption (e.g., a longer time interval, or an experimental condition that accelerates bone remodeling activities above basal levels, as shown in Schulte et al.), would have resulted in significantly lower precision error. Thus the RMS%CV reported here represents the maximum precision error that can be expected for the in vivo dynamic bone histomorphometry technique. Another limitation of the current study is that it provides no comparison between the μCT-based in vivo dynamic bone histomorphometry technique and the gold standard of fluorescent labeled histomorphometry, and thus interpretation is limited to the precision and reproducibility of the technique and cannot provide information on its accuracy. However, the accuracy of the in vivo dynamic bone histomorphometry technique has been previously established through a direct, specimen-specific comparison of the μCT-based technique with standard dynamic histomorphometry 12.
The effects of image filtration and thresholding on noise levels in the MAT-transformed images were also investigated in this study. Our results suggested that in vivo dynamic bone histomorphometry measurements are not sensitive to variations in the global threshold that was used to separate voxels of bone from background. In contrast, the Gaussian filtration of the images prior to thresholding did significantly impact the amount of noise present in in vivo dynamic histomorphometry measurements, as noise was increased when no filtration was performed (Fig. 5). However, although a larger kernel size (sigma=1.8, support=3) tended to result in lower noise levels, differences between the two kernel sizes tested (sigma=1.2, support=2 vs. sigma=1.8, support=3) were not significant. Because a large Gaussian filter can result in reduced sensitivity of the measurement due to excess smoothing of the image, these results indicate that the use of a Gaussian filter with a kernel size of sigma=1.2, support=2 results in an adequate balance between the sensitivity and noise present in the measurement.
In summary, this study developed an image transformation scheme based on matched rotations of both the follow-up and baseline images, and performed a robust characterization to determine the precision and reproducibility of μCT-derived, in vivo dynamic bone histomorphometry measurements. Bone dynamic measurements were found to have good reproducibility, and evaluation of noise present in these measurements provided valuable insight into the effects of transformation scheme, threshold, and filter on measurement precision. Furthermore, the new MAT scheme effectively eliminated the interpolation bias associated with the standard transformation scheme for both bone microstructure and dynamic measurements.
Acknowledgements
This study was supported by NIH/NIAMS R03-AR065145 (to XSL), NIH/NIAMS T32-AR007132 (to CMJdB), National Science Foundation Graduate Research Fellowship (to CMJdB), Penn Institute on Aging Pilot Grant (to XSL), and Penn Center for Musculoskeletal Disorders (PCMD; NIH/NIAMS P30-AR050950
Footnotes
Conflict of Interest:
The authors declare that they have no conflict of interest.
References
- 1.Altman AR, Lott C, De Bakker CM, Tseng WJ, Qin L, Liu XS. Intermittent PTH after Prolonged Bisphosphonate Treatment Improves Bone Structure by Inducing Substantial New Bone Formation with Decoupled, Inhibited Bone Resorption in Ovariectomized Rats.. Orthopaedic Research Society Annual Meeting; Las Vegas, NV. 2015. [Google Scholar]
- 2.Altman AR, Tseng WJ, de Bakker CM, Chandra A, Lan S, Huh BK, Luo S, Leonard MB, Qin L, Liu XS. Quantification of skeletal growth, modeling, and remodeling by in vivo micro computed tomography. Bone. 2015;81:370–9. doi: 10.1016/j.bone.2015.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birkhold AI, Razi H, Duda GN, Weinkamer R, Checa S, Willie BM. The influence of age on adaptive bone formation and bone resorption. Biomaterials. 2014;35:9290–301. doi: 10.1016/j.biomaterials.2014.07.051. [DOI] [PubMed] [Google Scholar]
- 4.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010;25:1468–86. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 5.Boyd SK, Davison P, Muller R, Gasser JA. Monitoring individual morphological changes over time in ovariectomized rats by in vivo micro-computed tomography. Bone. 2006;39:854–62. doi: 10.1016/j.bone.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 6.Boyd SK, Moser S, Kuhn M, Klinck RJ, Krauze PL, Muller R, Gasser JA. Evaluation of three-dimensional image registration methodologies for in vivo micro-computed tomography. Ann Biomed Eng. 2006;34:1587–99. doi: 10.1007/s10439-006-9168-7. [DOI] [PubMed] [Google Scholar]
- 7.Brouwers JE, Lambers FM, Gasser JA, van Rietbergen B, Huiskes R. Bone degeneration and recovery after early and late bisphosphonate treatment of ovariectomized wistar rats assessed by in vivo micro-computed tomography. Calcif Tissue Int. 2008;82:202–11. doi: 10.1007/s00223-007-9084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouwers JE, Lambers FM, van Rietbergen B, Ito K, Huiskes R. Comparison of bone loss induced by ovariectomy and neurectomy in rats analyzed by in vivo micro-CT. J Orthop Res. 2009;27:1521–7. doi: 10.1002/jor.20913. [DOI] [PubMed] [Google Scholar]
- 9.Brouwers JE, van Rietbergen B, Huiskes R. No effects of in vivo micro-CT radiation on structural parameters and bone marrow cells in proximal tibia of wistar rats detected after eight weekly scans. J Orthop Res. 2007;25:1325–32. doi: 10.1002/jor.20439. [DOI] [PubMed] [Google Scholar]
- 10.Buie HR, Moore CP, Boyd SK. Postpubertal architectural developmental patterns differ between the L3 vertebra and proximal tibia in three inbred strains of mice. J Bone Miner Res. 2008;23:2048–59. doi: 10.1359/jbmr.080808. [DOI] [PubMed] [Google Scholar]
- 11.Campbell GM, Buie HR, Boyd SK. Signs of irreversible architectural changes occur early in the development of experimental osteoporosis as assessed by in vivo micro-CT. Osteoporos Int. 2008;19:1409–19. doi: 10.1007/s00198-008-0581-7. [DOI] [PubMed] [Google Scholar]
- 12.de Bakker CM, Altman AR, Tseng WJ, Tribble MB, Li C, Chandra A, Qin L, Liu XS. muCT-based, in vivo dynamic bone histomorphometry allows 3D evaluation of the early responses of bone resorption and formation to PTH and alendronate combination therapy. Bone. 2015;73C:198–207. doi: 10.1016/j.bone.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goff MG, Slyfield CR, Kummari SR, Tkachenko EV, Fischer SE, Yi YH, Jekir MG, Keaveny TM, Hernandez CJ. Three-dimensional characterization of resorption cavity size and location in human vertebral trabecular bone. Bone. 2012;51:28–37. doi: 10.1016/j.bone.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson HJ, McCormick M, Ibanez L, Consortium IS. The ITK Software Guide. 2013 [Google Scholar]
- 15.Kohler T, Beyeler M, Webster D, Muller R. Compartmental bone morphometry in the mouse femur: reproducibility and resolution dependence of microtomographic measurements. Calcif Tissue Int. 2005;77:281–90. doi: 10.1007/s00223-005-0039-2. [DOI] [PubMed] [Google Scholar]
- 16.Lambers FM, Kuhn G, Schulte FA, Koch K, Muller R. Longitudinal assessment of in vivo bone dynamics in a mouse tail model of postmenopausal osteoporosis. Calcif Tissue Int. 2012;90:108–19. doi: 10.1007/s00223-011-9553-6. [DOI] [PubMed] [Google Scholar]
- 17.Lan S, Luo S, Huh BK, Chandra A, Altman AR, Qin L, Liu XS. 3D image registration is critical to ensure accurate detection of longitudinal changes in trabecular bone density, microstructure, and stiffness measurements in rat tibiae by in vivo microcomputed tomography (muCT). Bone. 2013;56:83–90. doi: 10.1016/j.bone.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukas C, Ruffoni D, Lambers FM, Schulte FA, Kuhn G, Kollmannsberger P, Weinkamer R, Muller R. Mineralization kinetics in murine trabecular bone quantified by time-lapsed in vivo micro-computed tomography. Bone. 2013;56:55–60. doi: 10.1016/j.bone.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Matheny JB, Slyfield CR, Tkachenko EV, Lin I, Ehlert KM, Tomlinson RE, Wilson DL, Hernandez CJ. Anti-resorptive agents reduce the size of resorption cavities: a three-dimensional dynamic bone histomorphometry study. Bone. 2013;57:277–83. doi: 10.1016/j.bone.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishiyama KK, Campbell GM, Klinck RJ, Boyd SK. Reproducibility of bone micro-architecture measurements in rodents by in vivo micro-computed tomography is maximized with three-dimensional image registration. Bone. 2010;46:155–61. doi: 10.1016/j.bone.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Schulte FA, Lambers FM, Kuhn G, Muller R. In vivo micro-computed tomography allows direct three-dimensional quantification of both bone formation and bone resorption parameters using time-lapsed imaging. Bone. 2011;48:433–42. doi: 10.1016/j.bone.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Schulte FA, Lambers FM, Mueller TL, Stauber M, Muller R. Image interpolation allows accurate quantitative bone morphometry in registered micro-computed tomography scans. Comput Methods Biomech Biomed Engin. 2014;17:539–48. doi: 10.1080/10255842.2012.699526. [DOI] [PubMed] [Google Scholar]
- 23.Schulte FA, Ruffoni D, Lambers FM, Christen D, Webster DJ, Kuhn G, Muller R. Local mechanical stimuli regulate bone formation and resorption in mice at the tissue level. PLoS One. 2013;8:e62172. doi: 10.1371/journal.pone.0062172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86:420–8. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 25.Slyfield CR, Jr., Niemeyer KE, Tkachenko EV, Tomlinson RE, Steyer GG, Patthanacharoenphon CG, Kazakia GJ, Wilson DL, Hernandez CJ. Three-dimensional surface texture visualization of bone tissue through epifluorescence-based serial block face imaging. J Microsc. 2009;236:52–9. doi: 10.1111/j.1365-2818.2009.03204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slyfield CR, Tkachenko EV, Wilson DL, Hernandez CJ. Three-dimensional dynamic bone histomorphometry. J Bone Miner Res. 2012;27:486–95. doi: 10.1002/jbmr.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tkachenko EV, Slyfield CR, Tomlinson RE, Daggett JR, Wilson DL, Hernandez CJ. Voxel size and measures of individual resorption cavities in three-dimensional images of cancellous bone. Bone. 2009;45:487–92. doi: 10.1016/j.bone.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voor MJ, Yang S, Burden RL, Waddell SW. In vivo micro-CT scanning of a rabbit distal femur: repeatability and reproducibility. J Biomech. 2008;41:186–93. doi: 10.1016/j.jbiomech.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waarsing JH, Day JS, van der Linden JC, Ederveen AG, Spanjers C, De Clerck N, Sasov A, Verhaar JA, Weinans H. Detecting and tracking local changes in the tibiae of individual rats: a novel method to analyse longitudinal in vivo micro-CT data. Bone. 2004;34:163–9. doi: 10.1016/j.bone.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Waarsing JH, Day JS, Verhaar JA, Ederveen AG, Weinans H. Bone loss dynamics result in trabecular alignment in aging and ovariectomized rats. J Orthop Res. 2006;24:926–35. doi: 10.1002/jor.20063. [DOI] [PubMed] [Google Scholar]