Abstract
We analyzed 3,872 common genetic variants across the ESR1 locus (encoding estrogen receptor α) in 118,816 subjects from three international consortia. We found evidence for at least five independent causal variants, each associated with different phenotype sets, including estrogen receptor (ER+ or ER−) and human ERBB2 (HER2+ or HER2−) tumor subtypes, mammographic density and tumor grade. The best candidate causal variants for ER− tumors lie in four separate enhancer elements, and their risk alleles reduce expression of ESR1, RMND1 and CCDC170, whereas the risk alleles of the strongest candidates for the remaining independent causal variant disrupt a silencer element and putatively increase ESR1 and RMND1 expression.
SNPs at 6q25.1 have been reported to be associated with breast cancer susceptibility in genome-wide association studies (GWAS) in women of Chinese1 and European2 ancestry. Subsequent analyses have demonstrated that SNPs in the same region are associated with breast cancer risk for BRCA1 mutation carriers3 and mammographic density4, a strong breast cancer risk factor. Thus far, however, attempts to identify the candidate causal variant(s) underlying the associations have been inconclusive3,5,6. Here we report fine-scale mapping and comprehensive analysis of the genotype-phenotype associations in this region, using dense genotyping and imputed data from the custom-designed iCOGS array, in 118,816 subjects from three consortia: the Breast Cancer Association Consortium (BCAC), the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) and the Markers of Density Consortium (MODE). We additionally demonstrate, through functional analyses, the likely modes of action of the strongest candidate causal variants.
RESULTS
Genetic epidemiological studies
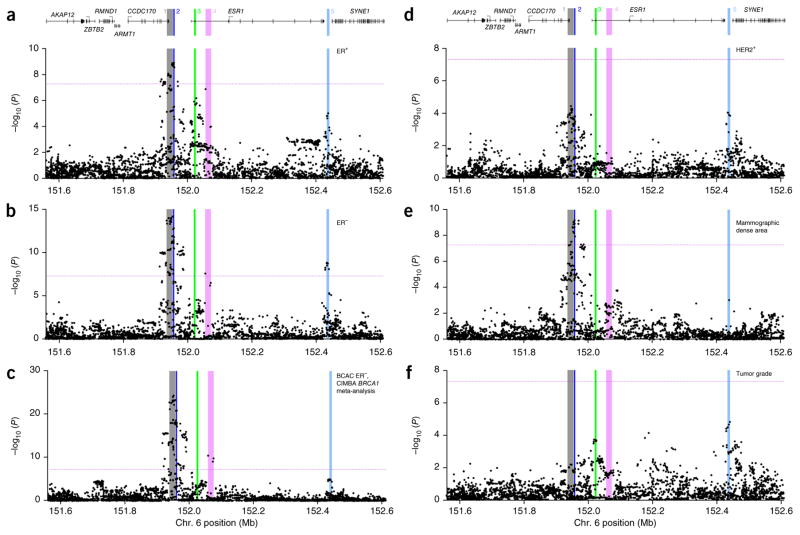
We successfully genotyped 902 SNPs across a 1-Mb region containing ESR1 in 50 case-control studies from populations of European (89,050 participants) and Asian (12,893 participants) ancestry in BCAC, together with 15,252 BRCA1 mutation carriers in CIMBA. Mammographic density measures were available for 6,979 women from the BCAC studies and an additional 1,621 women from the MODE Consortium, who had also been genotyped using the iCOGS array. Subsequently, the genotypes of additional variants with minor allele frequency (MAF) >2% were imputed in all European-ancestry participants, using data from the 1000 Genomes Project as a reference. In total, data from 3,872 genotyped or imputed (imputation info score >0.3) SNPs were analyzed. Results for all SNPs associated with overall breast cancer risk (P < 1 × 10−4) are presented in Supplementary Table 1. Manhattan plots of the associations of these 3,872 SNPs with the main phenotypes are shown in Figure 1.
Figure 1.
Association results for all SNPs with six phenotypes. (a–f) The phenotypes analyzed include risk of ER+ breast cancer in BCAC (a), risk of ER− breast cancer in BCAC (b), risk of triple-negative breast cancer, derived from the CIMBA meta-analysis of BRCA1 mutation carriers with ER− tumors (c), risk of HER2+ breast cancer in BCAC (d), mammographic dense area in MODE (e) and tumor grade after adjustment for ER status in BCAC (f). P values for each SNP (from unconditional logistic regression) are shown plotted as the negative log-transformed P value against relative position across the locus. A schematic of the gene structures is shown above a and d. The physical positions of signals 1–5 are shown as colored, numbered stripes. Dotted horizontal lines indicate the genome-wide significance level.
Conditional analyses
All genotyped and imputed SNPs displaying evidence of association with overall breast cancer risk in women of European ancestry (P < 1 × 10−4) were initially included in forward stepwise logistic regression models for ER− and ER+ breast tumor risk. The most parsimonious models (Online Methods) included four SNPs for ER− breast cancer and four SNPs for ER+ breast cancer, with three SNPs being common to both models. In each model, all selected SNPs fell into a subset of five bins of correlated SNPs (r2 >0.8). Stepwise regression models were independently fitted to breast cancer risk in the CIMBA BRCA1 mutation carriers and to mammographic density (measured as mammographic dense area; see the Online Methods for full details). For the BRCA1 mutation carriers and for mammographic dense areas, the SNPs in the best fitting models also fell within a subset of the five originally defined bins. For further analyses, we selected the directly genotyped SNP that was most significantly associated with the predominant phenotype for that bin. Regression analyses were repeated using just these five SNPs, with each representing an independent signal7. Results are presented in Table 1. Additionally, in the BCAC studies, we were able to examine SNP associations with risks of HER2 (HER2+ and HER2−) and progesterone receptor (PR+ and PR−) tumor subtypes and with tumor grade at diagnosis. There were weak but detectable correlations between the representative SNPs for signals 1–4 (Table 1 and Supplementary Table 2). We therefore modeled the associations with each SNP conditional on the other four; these conditional risk estimates and significance levels are also presented in Table 1. At conditional significance levels of P < 1 × 10−3, four of the lead SNPs (signals 1, 2, 4 and 5) were independently associated with risk of developing ER− breast cancer (Table 1). Another, partially overlapping, set of four SNPs (signals 1–3 and 5) was associated with ER+ tumor risk (Table 2 and Supplementary Table 3), and another subset of SNPs (signals 1–4) was associated with breast cancer risk in BRCA1 mutation carriers (Table 1). The per-allele odds ratios were higher for ER− than for ER+ disease for three lead SNPs (signals 1, 2 and 5), whereas representative SNPs for signal 3 displayed smaller effects of similar magnitude on risk for ER− and ER+ tumors. Mammographic dense area was associated with representative SNPs from signal 2 and less strongly with those from signal 1 (Table 1). We additionally carried out a meta-analysis of the SNP associations with breast cancer risk for CIMBA BRCA1 mutation carriers and risk of ER− tumors in BCAC. We anticipated that this analysis would increase statistical power to detect ER− risk signals, and, indeed, it did strengthen the evidence for association of SNPs representing signals 1–4 but not signal 5, which showed no association with breast cancer risk in BRCA1 mutation carriers (Table 1).
Table 1.
The associations of each signal-representative SNP with tumor risk and mammographic density in the three contributing consortia
| Signal | Representative SNP | Position | Alleles | Freq. | BCAC |
CIMBA |
BCAC ER−
|
MODE |
|
|---|---|---|---|---|---|---|---|---|---|
| ER−
|
ER+
|
BRCA1 mutations |
CIMBA BRCA1
|
Mammographic dense areaa
|
|||||
| OR (95% CI)
Ptrend cOR (95% CI) Pcond |
OR (95% CI)
Ptrend cOR (95% CI) Pcond |
HR (95% CI) P
value cHR (95% CI) Pcond |
Meta-analysis P value Pcond |
β (95%
CI)
Ptrend cβ (95% CI) Pcond |
|||||
| 1 | rs3757322 | 151,942,194 | GT | 0.33 | 1.17 (1.12–1.21) 1.00 × 10−14 | 1.07 (1.04–1.09) 1.10 × 10−7 | 1.15 (1.10–1.20) 3.78 × 10−10 | 2.50 × 10−23 | 0.12 (0.07, 0.17) 1.82 × 10−6 |
| 1.14 (1.10–1.19) 1.51 × 10−9 | 1.06 (1.04–1.09) 1.02 × 10−5 | 1.10 (1.06–1.14) 3.79 × 10−7 | 7.59 × 10−15 | 0.07 (0.01, 0.12) 0.017 | |||||
| 2 | rs9397437 | 151,952,332 | AG | 0.07 | 1.28 (1.19–1.37) 5.29 × 10−12 | 1.15 (1.10–1.20) 1.26 × 10−9 | 1.24 (1.15–1.33) 3.98 × 10−8 | 6.79 × 10−19 | 0.27 (0.18, 0.36) 2.36 × 10−9 |
| 1.18 (1.11–1.26) 1.20 × 10−5 | 1.12 (1.07–1.17) 3.56 × 10−6 | 1.12 (1.05–1.19) 3.60 × 10−4 | 3.29 × 10−8 | 0.22 (0.12, 0.32) 1.66 × 10−5 | |||||
| 3 | rs851984 | 152,023,191 | AG | 0.41 | 1.04 (1.01–1.08) 0.024 | 1.06 (1.03–1.08) 1.97 × 10−6 | 1.05 (1.01–1.10) 0.015 | 9.14 × 10−4 | −0.03 (−0.07, 0.02) 0.29 |
| NA | 1.07 (1.05–1.10) 1.09 × 10−8 | 1.07 (1.03–1.10) 3.60 × 10−4 | 3.12 × 10−5 | 0.01 (−0.04, 0.06) 0.83 | |||||
| 4 | rs9918437 | 152,072,718 | TG | 0.07 | 1.18 (1.11–1.27) 6.20 × 10−7 | 1.08 (1.04–1.13) 1.04 × 10−4 | 1.17 (1.08–1.26) 1.30 × 10−4 | 1.48 × 10−10 | 0.03 (−0.05, 0.12) 0.45 |
| 1.13 (1.06–1.20) 4.46 × 10−4 | NA | 1.10 (1.04–1.17) 0.0015 | 2.61 × 10−6 | 0.03 (−0.06, 0.12) 0.46 | |||||
| 5 | rs2747652 | 152,437,016 | CT | 0.54 | 1.12 (1.08–1.16) 1.83 × 10−9 | 1.05 (1.03–1.08) 9.49 × 10−6 | 1.00 (0.96–1.04) 0.95 | 1.44 × 10−5 | −0.02 (−0.07, 0.03) 0.39 |
| 1.12 (1.08–1.16) 2.32 × 10−9 | 1.05 (1.03–1.08) 6.60 × 10−6 | 1.00 (0.97–1.04) 0.86 | 5.97 × 10−5 | −0.02 (−0.07, 0.03) 0.45 | |||||
For each signal-representative SNP (the best associated genotyped SNP), odds ratios for the minor/major allele and conditional odds ratios (OR and cOR) together with 95% confidence intervals (CIs), hazard ratios (HR and cHR), β coefficients (β and cβ) and P values (Pcond) are from models including the other four signal-representative SNPs. Representative conditional odds ratios and confidence intervals could not be generated from the meta-analysis. NA indicates that the SNP was not included in conditional analysis because its individual effect was not significant at P > 1 × 10−4. Freq., frequency.
Mammographic dense area was square-root transformed and adjusted for age, BMI, menopausal status, study and relevant principal components.
Table 2.
The association of each signal-representative SNP with the main tumor subtype combinations and tumor grade
| Signal Representative SNP | n cases | 1 | 2 | 3 | 4 | 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3757322 |
rs9397437 |
rs851984 |
rs9918437 |
rs2747652 |
|||||||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||
| ER+ | |||||||||||
| IHC classification | |||||||||||
| ER+PR+/PR−HER2− | 10,834 | 1.07 (1.03–1.11) | 3.93 × 10−4 | 1.14 (1.07–1.21) | 9.54 × 10−5 | 1.03 (0.92–1.06) | 1.40 × 10−1 | 1.10 (1.03–1.16) | 4.16 × 10−3 | 1.04 (1.00–1.07) | 3.67 × 10−2 |
| ER+PR+/PR−HER2+ | 1,616 | 1.10 (1.02–1.19) | 1.68 × 10−2 | 1.25 (1.09–1.43) | 1.05 × 10−3 | 1.05 (0.98–1.13) | 1.88 × 10−1 | 1.05 (0.92–1.21) | 4.75 × 10−1 | 1.07 (0.99–1.15) | 7.22 × 10−2 |
| Case-only P | 6.60 × 10−1 | 1.80 × 10−1 | 3.90 × 10−1 | 8.40 × 10−1 | 4.00 × 10−1 | ||||||
| Grade classification | |||||||||||
| Grade 1 | 5,331 | 1.05 (1.00–1.10) | 4.04 × 10−2 | 1.04 (0.96–1.14) | 3.17 × 10−1 | 1.00 (0.96–1.05) | 8.79 × 10−1 | 1.07 (0.99–1.16) | 7.54 × 10−2 | 0.98 (0.95–1.03) | 5.30 × 10−1 |
| Grade 2 | 11,498 | 1.08 (1.04–1.11) | 8.77 × 10−6 | 1.16 (1.09–1.23) | 1.48 × 10−6 | 1.05 (1.02–1.08) | 3.51 × 10−3 | 1.08 (1.02–1.14) | 6.57 × 10−3 | 1.06 (1.03–1.10) | 7.91 × 10−5 |
| Grade 3 | 4,702 | 1.06 (1.01–1.11) | 1.37 × 10−2 | 1.17 (1.08–1.28) | 2.22 × 10−4 | 1.11 (1.06–1.16) | 6.30 × 10−6 | 1.16 (1.07–1.26) | 2.17 × 10−4 | 1.21 (1.08–1.17) | 6.11 × 10−7 |
| Case-only P | 9.20 × 10−1 | 2.00 × 10−2 | 2.60 × 10−4 | 6.00 × 10−2 | 7.97 × 10−6 | ||||||
| ER− | |||||||||||
| IHC classification | |||||||||||
| ER−PR−HER2− (TN) | 2,840 | 1.20 (1.12–1.28) | 7.17 × 10−8 | 1.25 (1.11–1.40) | 1.50 × 10−4 | 1.05 (0.98–1.12) | 1.40 × 10−1 | 1.17 (1.04–1.32) | 7.00 × 10−3 | 1.08 (1.01–1.15) | 1.65 × 10−2 |
| ER−PR−HER2+ | 858 | 1.19 (1.07–1.32) | 8.80 × 10−4 | 1.25 (1.04–1.5) | 1.55 × 10−2 | 1.00 (0.91–1.11) | 9.40 × 10−1 | 1.18 (0.99–1.40) | 6.80 × 10−2 | 1.24 (1.12–1.37) | 2.41 × 10−5 |
| Case-only P | 7.80 × 10−1 | 4.20 × 10−1 | 1.40 × 10−1 | 9.20 × 10−1 | 2.08 × 10−2 | ||||||
| ER−PR+HER2− | 268 | 1.17 (0.97–1.40) | 9.00 × 10−2 | 1.14 (0.83–1.58) | 4.10 × 10−1 | 1.30 (1.10–1.55) | 2.50 × 10−3 | 1.14 (0.82–1.56) | 4.40 × 10−1 | 1.10 (0.92–1.31) | 2.90 × 10−1 |
| Case only vs. TN | 8.00 × 10−1 | 7.80 × 10−1 | 3.00 × 10−2 | 6.50 × 10−1 | 8.30 × 10−1 | ||||||
| Case only vs. ER−PR−HER2+ | 6.40 × 10−1 | 6.10 × 10−1 | 3.00 × 10−2 | 1.60 × 10−1 | 3.70 × 10−1 | ||||||
| Case only vs. ER+PR+/PR−HER2+ | 7.90 × 10−1 | 7.60 × 10−1 | 1.20 × 10−1 | 9.90 × 10−1 | 3.80 × 10−1 | ||||||
| Grade classification | |||||||||||
| Grade 1 | 218 | 1.23 (1.00–1.5) | 4.40 × 10−2 | 1.35 (0.96–1.91) | 8.60 × 10−2 | 0.87 (0.71–1.07) | 1.60 × 10−1 | 0.87 (0.6–1.26) | 4.70 × 10−1 | 1.01(0.84–1.23) | 9.00 × 10−1 |
| Grade 2 | 1,204 | 1.14 (1.05–1.24) | 2.88 × 10−3 | 1.19 (1.02–1.39) | 2.63 × 10−2 | 1.09 (0.99–1.18) | 5.30 × 10−2 | 1.26 (1.09–1.45) | 1.79 × 10−3 | 1.12 (1.03–1.22) | 5.93 × 10−3 |
| Grade 3 | 3,463 | 1.20 (1.13–1.26) | 6.10 × 10−11 | 1.30 (1.19–1.43) | 1.88 × 10−8 | 1.05 (0.995–1.10) | 7.49 × 10−2 | 1.19 (1.09–1.31) | 1.24 × 10−4 | 1.12 (1.05–1.17) | 4.36 × 10−5 |
| Grade polytomous adjusted for ER, constrained | 9.18 × 10−1 | 6.42 × 10−3 | 5.43 × 10−4 | 2.96 × 10−1 | 1.82 × 10−5 | ||||||
| Subtypes with strongest association | ER− | High grade | High grade | ER− | ER−HER2+ and high grade | ||||||
Bold text highllights the most significant phenotype associations within each signal. IHC, immunohistochemistry; TN, triple negative.
Tumor subtype and grade analyses
We next explored the associations of each signal with specific tumor subtype combinations and with tumor grade (Fig. 1f, Table 2 and Supplementary Tables 3–5). The representative SNPs at two signals (3 and 5) were strongly associated with high-grade disease, after adjusting for ER status (P < 1 × 10−3; Table 2 (bottom line) and Supplementary Table 5). Among ER− tumors, three signals (1, 2 and 4) were associated with triple-negative (ER−PR−HER2−) and high-grade tumors, as well as the rarer ER−PR−HER2+ subtype, with similar odds ratios (Table 2 and Supplementary Tables 3 and 5). However, signal 5 was more strongly associated with ER−PR−HER2+ disease (odds ratio (OR) = 1.24, 95% confidence interval (CI) = 1.12–1.37; P = 2.4 × 10−5; Table 2) than with the triple-negative subtype (OR = 1.08, 95% CI = 1.01–1.15; P = 0.016; Table 2, case-only P = 0.021; Supplementary Table 5), consistent with the lack of association for breast cancer in BRCA1 mutation carriers, in whom tumors are predominantly triple negative8.
Haplotype analysis
We next explored the combined effects of the same five signal-representative genotyped SNPs (Supplementary Table 6). Haplotype-specific effects were consistent with additive effects of the individual signal-representative SNPs. In particular, haplotype 22221 (all minor alleles except for signal 5; frequency = 0.005) was associated with the largest increased risks of both ER+ (OR = 1.38, 95% CI = 1.11–1.71; P = 3.3 × 10−3) and ER− (OR = 2.34, 95% CI = 1.76–3.10; P = 3.5 × 10−9) tumors; this group includes the triple-negative (ER−PR−HER2−) tumor subtype (detected via the meta-analysis of BCAC subjects with ER− tumors and CIMBA BRCA1 mutation carriers; P = 8 × 10−10). Haplotype 22111 (frequency = 0.02) was associated with the highest risk of HER2+ tumors (OR = 1.5, 95% CI = 1.21–1.87; P = 3 × 10−4) and with mammographic dense area (β coefficient = 0.45, 95% CI = 0.20 to 0.69; P = 3 × 10−4).
Associations in Asian-ancestry studies
We examined the associations of the five signal-representative SNPs in the nine Asian-ancestry studies in BCAC (Supplementary Table 7). All five displayed allelic associations in the same direction as in Europeans, with overlapping confidence intervals, consistent with the hypothesis that the same candidate causal variants determine risk in both populations.
Determining the candidate SNPs within each signal
To identify the potential causal variants to be taken forward for functional analysis, we determined the most significant SNP association within each signal and then calculated the likelihood ratio of every other SNP relative to that SNP. We assumed that SNPs with a likelihood of <1:100 (ref. 9) in comparison with the most significant SNP for each signal could be excluded from consideration as potentially causative variants. On the basis of the assumption that, within a given signal, the same variant(s) would be driving all observed phenotype associations, we derived the list of most likely causal SNPs for each signal. We used the results from one of two analyses to define the list of potentially causal SNPs for each signal: the meta-analysis of BCAC subjects with ER− disease and CIMBA BRCA1 mutation carriers for signals 1, 2 and 4, which were most strongly associated in this analysis, and overall breast cancer risk in BCAC for signals 3 and 5. These lists of unexcluded variants are presented in Table 3 and are highlighted in Supplementary Table 1.
Table 3.
Remaining candidate causal variants within each independent signal after likelihood-ratio testing based on exclusion phenotype
| Signal Representative SNP | 1 |
2 |
3 |
4 |
5 |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3757322 | rs9397437 | rs851984 | rs9918437 | rs2747652 | ||||||||||||||||
|
|
|
|
|
|
||||||||||||||||
| Exclusion phenotype | Meta-analysis (BCAC ER− CIMBA) | Meta-analysis (BCAC ER− CIMBA) | Overall breast cancer (BCAC) | Meta-analysis( BCAC ER− CIMBA) | Overall breast cancer (BCAC) | |||||||||||||||
| Lead SNP | rs2046210 | rs12173570 | rs851985 | rs9918437 | rs2747652 | |||||||||||||||
| Position | 151,948,366 | 151,957,714 | 152,020,390 | 152,072,718 | 152,437,016 | |||||||||||||||
| A1/A2 | A/G | T/C | C/A | G/T | T/C | |||||||||||||||
| Freq. | 0.35 | 0.10 | 0.41 | 0.07 | 0.54 | |||||||||||||||
| Conditional OR (95% CI) | 1.07 (1.05–1.09) 3.09 × 10−9 | 1.12 (1.08–1.15) 1.64 × 10−10 | 1.08 (1.05–1.10) 9.65 × 10−12 | 1.05 (1.01–1.09) 1.27 × 10−2 | 1.07 (1.05–1.09) 1.23 × 10−12 | |||||||||||||||
| Ptrend overall breast cancer risk in BCAC | ||||||||||||||||||||
|
| ||||||||||||||||||||
| Unexcluded candidates | rs75859313 | 1.52 × 108 | 1.83 × 10−15 | 0.96 | rs60954078 | 1.52 × 108 | 1.32 × 10−8 | 0.75 | rs851985 | 1.52 × 108 | 9.65 × 10−12 | 1 | rs6904031 | 1.52 × 108 | 1.66 × 10−5 | 0.82 | rs910416 | 1.52 × 108 | 2.70 × 10−12 | 0.99 |
| Chromosome position | rs11155803 | 1.52 × 108 | 3.67 × 10−15 | 0.89 | rs12173570 | 1.52 × 108 | 2.92 × 10−10 | 1 | rs851984 | 1.52 × 108 | 1.11 × 10−11 | 1 | rs1361024 | 1.52 × 108 | 8.74 × 10−6 | 0.99 | 6–152434275 | 1.52 × 108 | 2.24 × 10−12 | 0.99 |
| Pcond, r2 with lead SNP | rs11155804 | 1.52 × 108 | 2.13 × 10−16 | 0.89 | rs17081533 | 1.52 × 108 | 3.85 × 10−10 | 1 | rs851983 | 1.52 × 108 | 1.43 × 10−11 | 1 | rs9918437 | 1.52 × 108 | 2.61 × 10−6 | 1 | rs34133739 | 1.52 × 108 | 2.28 × 10−12 | 0.99 |
| rs11155805 | 1.52 × 108 | 2.83 × 10−15 | 0.99 | rs851982 | 1.52 × 108 | 1.45 × 10−11 | 1 | rs66485058 | 1.52 × 108 | 5.81 × 10−12 | 0.99 | |||||||||
| rs7740686 | 1.52 × 108 | 2.28 × 10−15 | 0.9 | rs2747652 | 1.52 × 108 | 1.23 × 10−12 | 1 | |||||||||||||
| rs2046210 | 1.52 × 108 | 4.38 × 10−17 | 1 | rs11345553 | 1.52 × 108 | 4.69 × 10−11 | 0.97 | |||||||||||||
| rs7763637 | 1.52 × 108 | 2.60 × 10−15 | 0.9 | |||||||||||||||||
| rs6557160 | 1.52 × 108 | 2.58 × 10−15 | 0.9 | |||||||||||||||||
| rs6557161 | 1.52 × 108 | 6.51 × 10−16 | 0.96 | |||||||||||||||||
| rs6900157 | 1.52 × 108 | 4.72 × 10−16 | 0.96 | |||||||||||||||||
Bold text highlights the top candidate SNPs within each signal. Freq., frequency.
In signal 1, the most strongly associated variant was rs2046210 (the original Asian GWAS hit1,10), with nine other variants (likelihood ratios <100:1, r2 ≥0.89 with rs2046210; spanning 151,935,539–151,954,127) remaining as strong causal candidates. In signal 2, the best causal candidate was SNP rs12173570, with two other candidates remaining (likelihood ratios <100:1, r2 ≥0.75 with rs12173570; spanning 151,955,914–151,958,815). The European GWAS SNP, rs3757318l (ref. 2), is most strongly correlated with rs12173570 (r2 >0.45). In signal 3, the best causal candidate was rs851984, with three other candidates remaining (likelihood ratios <100:1, r2 = 0.99; in two ESR1 introns spanning 152,020,390–152,024,985). In signal 4, the top candidate was rs9918437, and two other candidates spanned another segment of an ESR1 intron at 152,055,978–152,072,718 (approximately 30 kb telomeric of signal 3; likelihood ratios <100:1, r2 > 0.81 with rs9918437). In signal 5, the strongest candidate causal SNP was rs2747652 (also the representative SNP for signal 5 in Table 1), and there were five other candidates (likelihood ratios <100:1, r2 >0.97 with rs2747652; spanning 152,432,902–152,440,522) in the intergenic region between ESR1 and SYNE1. Across the five signals, we were able to exclude all but 26 of the original 3,872 variants from being potentially causal.
Local gene expression analyses
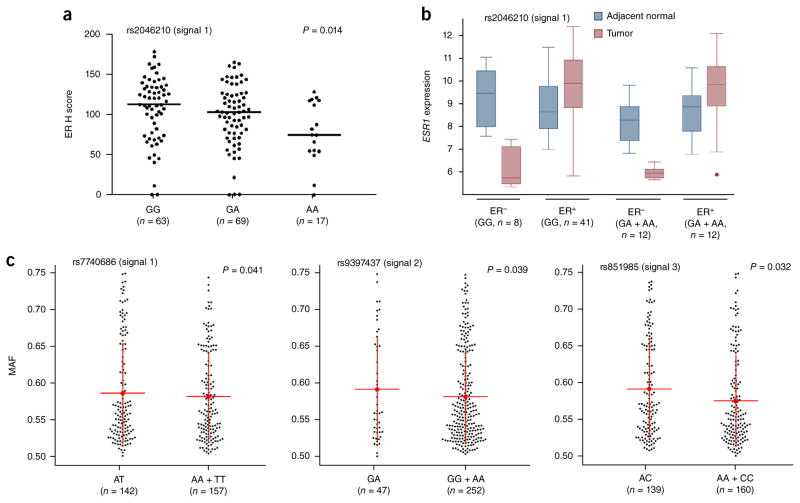
We used four techniques to assess associations between candidate causal variants (or available proxy SNPs) in the five signals and local gene expression. (i) ER protein expression, measured by immunohis-tochemistry in normal breast tissue samples from 150 postmenopausal donors, identified a significant correlation of the risk alleles of signal 1 SNPs with reduced ER levels (Fig. 2a and Supplementary Figs. 1 and 2). (ii) ESR1 expression in breast tumors and adjacent normal breast tissue from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) study was compared relative to signal-representative SNP allele (Fig. 2b and Supplementary Table 8). In patients with ER− tumors, risk allele carriers had lower median ESR1 expression in normal, tumor-adjacent tissue than homozygotes for the protective allele at signals 1, 4 and 5, although none of the differences were statistically significant. By contrast, in patients with ER+ tumors, risk allele carriers had higher median ESR1 expression in normal, tumor-adjacent tissue than homozygotes for the protective allele at signals 1, 3 and 5. (iii) Allele-specific expression (ASE) analysis, using RNA sequencing (RNA-seq) data from breast tumor samples and SNP array genotype data from The Cancer Genome Atlas (TCGA)11, showed allelic imbalances in ESR1 expression among heterozygotes for proxy SNPs in signals 1–3 (Fig. 2c and Supplementary Table 9). Similar imbalances in CCDC170 expression were detected among heterozygotes for signal 2 SNP rs9397437 and in RMND1 expression with signal 3 SNP rs851983 (Supplementary Table 9). Such allelic imbalances indicate that risk alleles at these signals are associated with expression differences in local genes, but they do not indicate the directions of association. (iv) Expression quantitative trait locus (eQTL) analysis using the Gene-Tissue Expression (GTEx) database identified a significant association for SNPs in signal 3 with CCDC170 expression in normal breast tissues (Supplementary Table 10). We also performed cis-eQTL analyses on the 12 flanking genes in 135 normal breast tissue samples from the METABRIC study; however, no additional associations were detected (Supplementary Table 11).
Figure 2.
ER expression and allelic imbalance correlate with signal 1 SNPs. (a) Negative correlation between the signal 1 SNP rs2046210 and ER protein expression. Black dots represent ER expression from individual samples measured by immunohistochemistry (H score). Horizontal lines represent the mean H score for each genotype. The P value was calculated using a Spearman rank correlation test. (b) Box plots of ESR1 gene expression (log2 transformed) in breast tumor and adjacent normal samples. Boxes extend from the 25th to the 75th percentile, horizontal bars represent the median, whiskers indicate the full range of ESR1 expression and outliers are represented as circles. (c) Allelic imbalance in ESR1 expression by genotypic status at breast cancer risk variants. Data are classified according to the genotypes at risk SNPs (heterozygous versus homozygous). Black dots represent the average major allele fraction of the marker SNPs across ESR1 for an individual from TCGA with breast cancer. Red lines and whiskers correspond to means ± 1 s.d. For rs7740686 (signal 1) and rs9397437 (signal 2), Levene’s test (equality of variances) was used to calculate the P values; for rs851985 (signal 3), a two-tailed t test (equality of means) was used to calculate the P value.
Bioinformatic and chromatin analyses
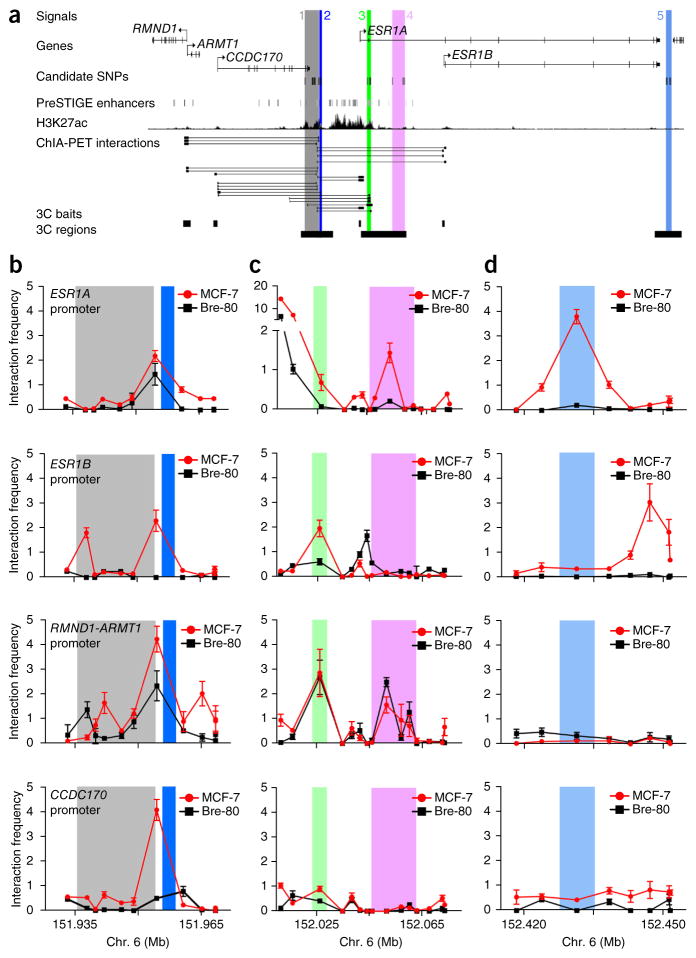
Analysis of cis enhancer–gene interactions using PreSTIGE12 showed evidence of multiple regulatory elements coinciding with signals 1–3 in ER+ MCF-7 breast cancer cells (Fig. 3a and Supplementary Fig. 3). A ‘super-enhancer’, associated with high levels of acetylation of his-tone H3 at lysine 27 (H3K27ac), was also identified in MCF-7 cells and encompasses the top risk-associated SNPs in these three signals (Fig. 3a and Supplementary Fig. 3)13. This super-enhancer was most readily detectable in MCF-7 cells and was not observed in other breast cancer cell lines, normal mammary epithelial cells or other tissues analyzed (Supplementary Fig. 4). Chromatin conformation capture (3C) experiments demonstrated that elements within signals 1 and 2 physically interacted with the promoters of ESR1, RMND1-ARMT1 and CCDC170 in MCF-7 and T-47D cells (Fig. 3b and Supplementary Fig. 5a,b). Furthermore, we detected interactions between signals 3–5 and ESR1 and/or RMND1-ARMT1 promoters (Fig. 3c,d and Supplementary Fig. 5c,d). The majority of these interactions were restricted to MCF-7 and T-47D cells (ER+ breast cancer cell lines), but the RMND1-ARMT1 interactions were also detected in either Bre-80 or MCF10A cells (ER− ‘normal’ breast cell lines; Fig. 3b–d and Supplementary Fig. 5b–d). The 3C-identified interactions for each signal are summarized in Supplementary Table 12.
Figure 3.
Chromatin interactions across the 6q25.1 risk region. (a) Signals 1–5 are numbered and shown as colored stripes. RMND1, ARMT1, CCDC170 and ESR1 gene structures are depicted with exons (vertical bars) joined by introns (lines). Gene-enhancer predictions from PreSTIGE12, ChIP-seq binding profiles for H3K27ac13 and Encyclopedia of DNA Elements (ENCODE) RNA polymerase II ChIA-PET interactions in MCF-7 cells are shown. (b–d) 3C anchor points (3C baits) and interrogated sequences (3C regions) are depicted as black boxes and gray shading, respectively. 3C interaction profiles in ER+ MCF-7 and ER− Bre-80 breast cell lines are shown for signals 1 and 2 (b), signals 3 and 4 (c) and signal 5 (d). 3C libraries were generated with EcoRI, with the anchor point set at the ESR1, RMND1-ARMT1 or CCDC170 promoter region. Graphs present the results from three biological replicates; error bars, s.d.
Prioritizing candidate SNPs for functional assays
We applied a combination of in silico and in vitro analyses to prioritize candidate causal SNPs for functional follow-up, using previous observations that common cancer susceptibility alleles are enriched in cis-regulatory elements14–16. First, Table 3 showed that 19 of the 26 top candidates overlapped DNase I–sensitive sites and were associated with enhancer-enriched histone marks such as dimethylation of histone H3 at lysine 4 (H3K4me2) and H3K27ac in MCF-7 and HMEC breast cells, indicative of putative regulatory elements (PREs) (Supplementary Fig. 6). In electromobility shift assays (EMSAs), 11 of these 19 SNPs altered the binding affinity of transcription factors in vitro (Supplementary Fig. 7). Of these, seven fell within promoter-specific long-range interactions identified by 3C (Fig. 3 and Supplementary Fig. 5). The 7 SNPs prioritized for further detailed analyses included 2 of 10 remaining candidates in signal 1 (rs7763637 and rs6557160), 1 of 3 candidates in signal 2 (rs17081533), 2 of 4 candidates in signal 3 (rs851982 and rs851983), 1 of 3 candidates in signal 4 (rs1361024) and 1 of 6 candidates in signal 5 (rs910416) (Supplementary Table 12).
Luciferase reporter assays
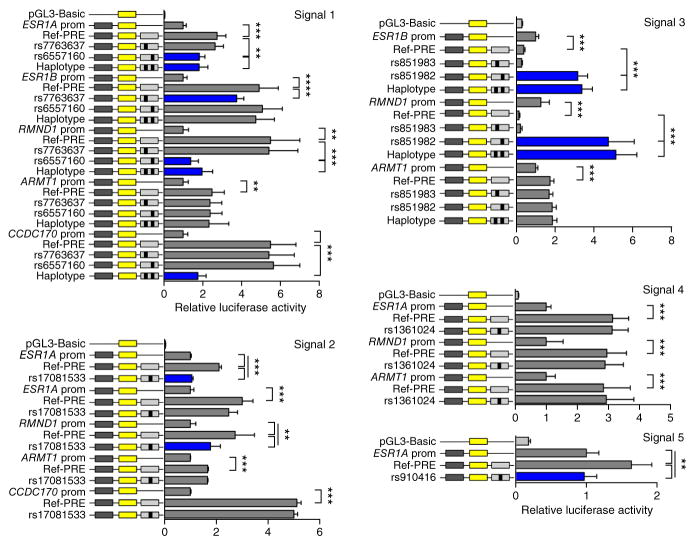
The regulatory capabilities of the PREs overlapping each signal and the effects of the seven prioritized candidate SNPs were examined in luciferase reporter assays in the ER+ MCF-7 and BT-474 and the ER− Bre-80 breast cell lines. PRE constructs containing the reference alleles of the prioritized SNPs for signals 1, 2, 4 and 5 significantly increased associated target gene promoter activity when cloned in either direction, indicating that they act as orientation-independent transcriptional enhancers. In contrast, a PRE containing the reference alleles of the signal 3 candidates ablated target gene promoter activity, but only when cloned in the forward direction, suggesting that this region acts as an orientation-dependent silencer (Fig. 4 and Supplementary Figs. 8–10). Notably, inclusion of the minor (risk) alleles of individual candidate SNPs in signals 1, 2 and 5 (rs6557160, rs17081533 and rs910416) significantly reduced ESR1 and RMND1 promoter activity but had no effect on the ARMT1 or CCDC170 promoters. However, inclusion of the signal 1 haplotype significantly decreased ESR1, RMND1 and CCDC170 promoter activity (Fig. 4 and Supplementary Figs. 8 and 9). Inclusion of the individual minor (risk) allele of signal 4 SNP rs1361024 or signal 3 SNP rs851983 in the respective constructs had no additional effects. In contrast, inclusion of the signal 3 minor (risk) allele of rs851982 or the haplotype construct increased ESR1 promoter activity in ER+ MCF-7 and BT-474 cells and RMND1 promoter activity in all three cell lines (Fig. 4, Supplementary Figs. 8 and 9, and Supplementary Table 12).
Figure 4.
Risk alleles reduce ESR1 and RMND1 promoter activity. Luciferase reporter assays were performed following transient transfection of ER+ MCF-7 breast cancer cells. PREs containing the major SNP alleles were cloned downstream of target gene promoter-driven luciferase constructs (prom) for the creation of reference (Ref-PRE) constructs. Minor SNP alleles were engineered into the constructs and are designated by the rsID of the corresponding SNP. “Haplotype” denotes a construct that contains the minor alleles of both candidate SNPs within signal 1 or 3. Error bars, 95% confidence intervals from three independent experiments. P values were determined by two-way ANOVA followed by Dunnett’s multiple-comparisons test: **P < 0.01, ***P < 0.001.
Transcription factor binding analyses
We used both bioinformatic analyses and functional studies to examine DNA-protein interactions for the seven prioritized SNPs. In silico prediction tools including intragenomic replicates (IGR)17, HaploReg18 and Alibaba2 (ref. 19) predicted that all seven SNPs alter transcription factor binding (Supplementary Fig. 11 and Supplementary Table 13).
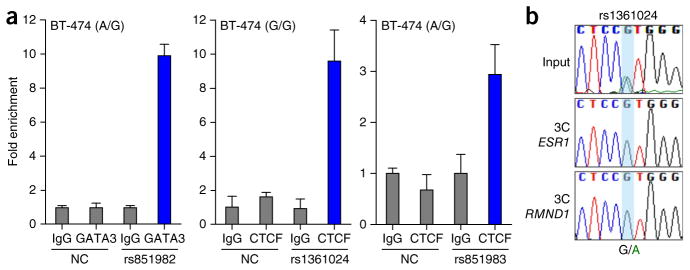
Competition with known transcription factor binding sites suggested the identity of bound proteins for four of the prioritized SNPs, including GATA3 binding to the minor (risk) allele of signal 3 SNP rs851982 and CTCF binding to the minor allele of a second signal 3 candidate, rs851983, as well as the common (protective) allele of signal 4 candidate rs1361024 and MYC binding to the common allele of signal 5 candidate rs910416 (Supplementary Fig. 12 and Supplementary Table 12). Additional well-established breast cell transcription factors, such as ER itself and FOXA1, were also assessed but did not display competitive binding to any prioritized SNP sites (Supplementary Fig. 13). Chromatin immunoprecipitation (ChIP) confirmed enrichment of GATA3 binding to DNA overlapping signal 3 candidate rs851982, but no difference between the alleles, and confirmed CTCF binding to the region overlapping signal 4 candidate rs1361024 in BT-474 cells (Fig. 5a and Supplementary Fig. 14). CTCF also bound to the region encompassing signal 3 candidate rs851983 (Fig. 5a, Supplementary Fig. 14 and Supplementary Table 12). CTCF mediates long-range chromatin looping; therefore, to assess the potential impact of signal 4 candidate rs1361024 and signal 3 candidate rs851983 on chromatin interactions, we performed allele-specific 3C in heterozygous cell lines. Sequence profiles indicated that the protective G allele of signal 4 candidate rs1361024 increases looping between this enhancer and the ESR1 and RMND1 promoters (Fig. 5b and Supplementary Fig. 15a). We found no evidence for allele-specific looping between the silencer overlapping signal 3 and local gene promoters (Supplementary Fig. 15b).
Figure 5.
GATA3 and CTCF binding in vivo. (a) ChIP and quantitative PCR (qPCR) assays using antibody against GATA3 or CTCF in ER+ BT-474 breast cancer cells. A region within the second intron of ESR1 served as a negative control (NC). Normal rabbit IgG was used as a non-specific antibody control. Graphs present the results of two biological replicates; error bars, s.d. (b) 3C followed by sequencing for the signal 4 PRE containing rs1361024 in heterozygous ER+ MCF-7 breast cancer cells shows allele-specific chromatin looping. The chromatograms represent one of three independent 3C libraries generated and sequenced.
DISCUSSION
The fine-scale mapping, bioinformatic and functional analyses presented here provide evidence for the existence of at least five different genetic variants, each with a direct effect on breast cancer risk in Europeans, findings also supported by the limited available data in Asian populations. These variants are distributed upstream, within introns and downstream of ESR1, each in a region, which we have demonstrated via reporter assays, is regulatory for ESR1. Some may additionally regulate other local genes, such as RMND1, ARMT1 and CCDC170, previously reported to be co-regulated with ESR1 (ref. 20). Of note, the four sites more strongly associated with risks of ER− than ER+ tumors (signals 1, 2, 4 and 5) all overlap enhancer regions, and our evidence indicates that the minor (risk) alleles of candidate causal variants, within each of these enhancers, act to reduce expression of ESR1, RMND1 and CCDC170. In contrast, signal 3—which is associated with smaller but equal risks of developing both ER− and ER+ tumors—overlaps a putative gene silencer, and the risk alleles of the candidate causal variants here increase ESR1 and RMND1 expression. Furthermore, we have demonstrated altered binding of looping factor CTCF to candidate causal SNPs in signals 3 and 4, with evidence that the risk allele of signal 4 candidate rs1361024 abrogates binding and reduces chromatin looping between this enhancer element and the promoters of ESR1 and RMND1. We also provided evidence that signal 5 candidate rs910416 may display allele-specific binding of MYC.
Notably, the previously unrecognized signal 5 candidates, downstream of ESR1, significantly increase the risk of developing ER−PR−HER2+ tumors (a specific subtype shown to be more responsive to the drug trastuzumab) in contrast to the triple- negative (ER−PR−HER2−) tumor subtype, which has already been reported to be associated with other signals at 6q25 as well as 19p13 (ref. 21) and 5p15 (TERT)22. We also found evidence that the candidate causal variants at signals 3 and 5 predispose to aggressive, high-grade breast cancer, independently of ER status.
Mammographic density adjusted for age and body mass index (BMI), which describes the variation in epithelial and stromal tissue on a mammogram, is one of the strongest known risk factors for breast cancer23 and has been shown to have a shared genetic basis with breast cancer, mediated through a large number of common variants24. Associations between ESR1 SNPs and mammographic density have previously been reported25–27, but, in this detailed analysis, only signal 2 was significantly associated with mammographic dense area (P = 1.7 × 10−5), although signal 1 also showed some evidence of an effect in the conditional analysis (P = 0.017). Although adjusting the breast cancer analysis of signal 2 for mammographic dense area produced some attenuation of the associated effect, the lead SNP remained significantly associated with breast cancer risk (unconditional OR = 1.30, 95% CI = 1.13–1.49; P = 0.00024; OR conditional on dense area = 1.24, 95% CI = 1.08–1.43; P = 0.0025), suggesting either that the mechanism by which the signal 2 candidate causal variant affects breast cancer risk is not mediated through mammographic density or, alternatively, that dense area, as measured here, is unable to capture the association with breast composition that is most relevant to risk. This phenomenon, whereby the association with risk appears to be partially independent of mammographic density, has also been observed for the 10q21.2 breast cancer locus4.
SNPs in the ESR1 region have previously been reported to be associated with bone mineral density28,29. These include SNPs within signal 1 (rs6930633, r2 = 0.73 with rs3757322) and signal 3 (rs2982575, r2 = 0.57 with rs851984), although the SNP with the most significant reported association with bone density measures, rs4870044, was not associated with breast cancer risk (P > 1 × 10−4) in our analysis nor correlated with any signal-representative SNPs (r2 <0.06). Similarly, SNP rs6933669, recently reported as associated with age at menarche30, is uncorrelated with these five signals (r2 <0.02) and was not associated with breast cancer (P = 0.1). Thus, although there is a known relationship between age at menarche and breast cancer risk, these phenotypes do not appear to share candidate causal variants in this region.
Our findings help address the question of the role of ERα in establishing breast cancer. Notably, the candidate causal SNPs identified here all increase risks of both ER+ and ER− tumor subtypes by varying degrees. ERα is a ligand-activated transcription factor that mediates the effect of estrogen through altering gene expression, and the links between estrogen, ERα and ER+ breast cancer are well documented, with adjuvant endocrine therapy considered standard treatment for ER+, early-stage breast cancer. Other studies have also reported 6q25 associations with ER− subtypes1,2,5, but the mechanisms by which ER− tumors develop are still debated. There is speculation that ER− tumors may arise from ER+ precursors by potentially reversible mechanisms, and our findings may lend support to this hypothesis. However, several recent studies have indicated that most tumors in BRCA1 mutation carriers arise from ER− luminal progenitor cells; thus, estrogen may be working indirectly through paracrine regulation in the mammary epithelium, possibly stimulating the Notch or epidermal growth factor receptor (EGFR) signaling pathways of adjacent ER+ cells31,32. Our analyses unexpectedly suggested that, whereas signals 1–4 increased risks of all ER− tumor subtypes, the signal 5 candidate causal variant increased risks of ER−HER2+ breast cancer subtypes but not of triple-negative tumor development or of tumors in BRCA1 mutation carriers (Table 1). This further complicates present understanding and underlines the need for further studies to address this issue.
Collectively, our evidence supports a hypothesis that ESR1 is the major target gene of the enhancer and silencer elements in which we have identified candidate causal variants. In addition to ESR1, we provide evidence that the regions overlapping signals 1–4 cooperatively regulate RMND1, raising the possibility that candidate causal SNPs act by altering both ESR1 and RMND1 expression. RMND1 (required for meiotic nuclear division 1; C6orf96) has not been well characterized but is reported to localize to mitochondria and be involved in mitochondrial translation33. We additionally identified enhancer activity and chromatin interactions with two other genes, ARMT1 and CCDC170, but the actions of the candidate causal SNPs on these genes remain unclear. ARMT1 encodes Armt1, a protein carboxyl methyltransferase that targets PCNA and differentially regulates cancer cell survival in response to DNA damage34. Nothing is known about the function of CCDC170 (coiled-coil domain–containing protein 170), but recurrent ESR1-CCDC170 rearrangements have been characterized in an aggressive subset of ER+ breast cancers35. A recent study also showed that higher CCDC170 expression correlated with ER negativity, highly proliferative features and worse clinical outcomes36. There are some data to suggest that these genes may cooperatively contribute to the increased proliferative capacity of ER+ tumors20, and it is tempting to speculate that these may be additional target genes for the candidate causal variants at a subset of the five signals identified here and perhaps responsible for their differential phenotype associations. A greater understanding of these genes may also provide novel targets for breast cancer prevention or therapies.
URLs
1000 Genomes Project, http://www.1000genomes.org/; Breast Cancer Association Consortium (BCAC), http://ccge.medschl.cam.ac.uk/consortia/bcac/index.html; Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA), http://ccge.medschl.cam.ac.uk/consortia/cimba/index.html; Collaborative Oncological Gene-environment Study (COGS), http://www.cogseu.org/; iCOGS, http://ccge.medschl.cam.ac.uk/research/consortia/icogs/; SNAP, https://www.broadinstitute.org/mpg/snap/; The Cancer Genome Atlas (TCGA), https://tcga-data.nci.nih.gov/; Cancer Genomics Hub (CGHub), https://cghub.ucsc.edu/; eMAP, http://www.bios.unc.edu/~weisun/software.htm.
ONLINE METHODS
Study populations and genotyping
Epidemiological data were obtained from three separate consortia that had all conducted genotyping using the iCOGS array, a custom array comprising approximately 200,000 SNPs. (i) Data on overall breast cancer risk, tumor subtypes and grade came from 50 breast cancer case-control studies participating in BCAC; these comprised 41 studies from populations of European ancestry and 9 studies from populations of East Asian ancestry3. Details of the participating studies, genotype calling and quality control are given elsewhere3. After quality control exclusions, we analyzed data from 46,451 cases and 42,599 controls of European ancestry and 6,269 cases and 6,624 controls of Asian ancestry. A further 23 SNPs were directly genotyped in two case-control studies (CCHS and SEARCH). The ER status of the primary tumor was available for 34,539 European and 4,972 Asian cases; of these, the tumor was ER− for 7,465 (22%) European and 1,610 (32%) Asian cases3. (ii) Data on BRCA1 mutation carriers were obtained through CIMBA. Eligibility is restricted to females 18 years or older with pathogenic mutations in BRCA1 or BRCA2. The majority of the participants were sampled through cancer genetics clinics37, including some related participants. Fifty-one studies from 25 countries contributed data on BRCA1 mutation carriers who were genotyped using the iCOGS array38. After quality control of the phenotypes and genotypes, data were available on 15,252 BRCA1 mutation carriers, of whom 7,797 had been diagnosed with breast cancer, all of European ancestry. Analyses in BRCA1 mutation carriers assessed associations with breast cancer risk. (iii) Mammographic density information was available for 7,025 women from ten studies in BCAC and, in addition, 1,621 women from the Mayo Mammographic Health Study (MMHS). All were additionally participants in the MODE Consortium. Forty-six women were excluded because of missing BMI information, leaving 8,600 women with mammographic density information, relevant covariates and iCOGS genotyping (2,955 breast cancer cases and 5,645 controls). Study details are given in Supplementary Table 14 and in Lindstrom et al.26. Mammographic density measurements were performed on digitized analog mammographic films using ‘Cumulus’ software39. This applies a thresholding technique to measure the total area of the breast and the absolute dense area, from which the absolute non-dense area and percent dense area are derived. Dense areas and non-dense areas were converted to cm2 according to the pixel size used in the digitization. Readers blinded to genotype, case status and risk factor data conducted all measures. For cases, mammograms before the diagnosis of breast cancer were used or, where this was not possible, measures from the contralateral breast were used.
SNP selection, genotyping and imputation
We first defined a mapping interval of ~1 Mb (chr. 6: 151,600,000–152,650,000; NCBI Build 37 assembly). We catalogued 2,821 variants with a MAF >2% using the 1000 Genomes Project (March 2010 Pilot version 60 CEU project data); of these variants, we selected 277 SNPs correlated with the 3 previously reported associated SNPs (rs2046210 (ref. 1), rs3757318 (ref. 2) and rs3020314 (ref. 40)) at r2 >0.1, plus a set of 698 SNPs designed to tag all remaining SNPs with r2 >0.9. Of the SNPs, 902 that passed quality control were included in this analysis. After completion of iCOGS genotyping, this initial set was supplemented with a further 23 SNPs selected from the October 2010 (Build 37) release of the 1000 Genomes Project, to improve coverage. These SNPs were genotyped in two large BCAC studies (CCHS and SEARCH) comprising 12,273 cases and controls, using a Fluidigm array according to the manufacturer’s instructions. Using the above data, results for all the additional known common variants (MAF >0.02 in Europeans) on the January 2012 release of the 1000 Genomes Project were imputed using IMPUTE version 2.0. Quality control and imputation steps were carried out separately in the different consortia, leading to slight differences in the numbers of SNPs with available data. In addition to the 902 successfully genotyped SNPs, genotypes at 2,972 SNPs were imputed in BCAC and 2,907 SNPs were imputed in CIMBA (imputation r2 score >0.3 in each case). In total, 3,872 genotyped or imputed SNPs were available for the combined BCAC ER− and CIMBA BRCA1 mutation carrier meta-analysis
Statistical analysis
Case-control analysis, logistic regression and retrospective cohort analyses
For the case-control analysis in BCAC, per-allele odds ratios and standard errors were estimated for each SNP using logistic regression, separately for subjects of European and Asian ancestry and for each tested phenotype. Principal components were included as covariates as previously described21. The statistical significance of each SNP was derived using a Wald test. To evaluate evidence for multiple association signals, we performed conditional analyses in which the association for each SNP was reevaluated after including other associated SNPs in the model. SNPs with a P value <1 × 10−4 and MAF >2% in the single-SNP analysis were included in this analysis21. Haplotype-specific odds ratios and confidence limits were estimated using haplo.stats22.
Associations between genotypes and breast cancer risk in BRCA1 mutation carriers in CIMBA were evaluated using a per-allele trend test with 1 degree of freedom (Ptrend), based on modeling the retrospective likelihood of the observed genotypes conditional on breast cancer phenotypes41. To allow for non-independence among related individuals, an adjusted test statistic was used that took into account the correlation in genotypes21. Per-allele hazard ratio estimates were obtained by maximizing the retrospective likelihood. All analyses were stratified by country of residence.
Conditional analyses were performed to identify SNPs independently associated with each phenotype. To identify the most parsimonious model, all SNPs with a marginal P value <1 × 10−4 were included in forward selection regression analyses with a threshold for inclusion of P < 1 × 10−4 and including terms for principal components and study. Similarly, forward selection Cox regression analysis was performed for BRCA1 mutation carriers, stratified by country of residence, using the same P-value thresholds. This approach provides valid significance tests of the associations, although the estimates quantifying the association can be biased41,42. Parameter estimates for the most parsimonious model were obtained using the retrospective likelihood approach.
Within MODE, mammographic dense area, non-dense area and percent dense area were each square-root transformed to fit a normal distribution. For the ten MODE and BCAC studies, a linear regression assuming a multiplicative per-allele model adjusting for study, age at mammogram, BMI, menopausal status (pre or post) and the first six principal components was carried out for each trait and for each SNP. The MMHS participants were analyzed separately in the same way but without the principal-components covariates, and the results were combined with those from BCAC using a standard inverse variance–weighted fixed-effects meta-analysis.
Expression analysis
eQTL analyses were conducted in 57 normal breast samples from the GTEx Project43 and 135 adjacent normal breast samples from women of European origin in the METABRIC study44. For the METABRIC analyses, matched gene expression (Illumina HT-12 v3 microarray) and germline SNP data from either genotyping (Affymetrix SNP 6.0) or imputation (1000 Genomes Project, March 2012 data using IMPUTE version 2.0) were used. Correlations between the five signal-representative SNPs and expression levels of nearby genes (500 kb upstream and downstream of the SNPs) were assessed using a linear regression model in which an additive effect on expression level was assumed for each copy of the rare allele. Calculations were carried out using the eMAP library in R.
Allele-specific expression analysis
ASE analysis has been described previously11. Three SNPs for signal 1, two SNPs for signal 3 and a proxy SNP for signal 2 (r2 = 0.85) were on Affymetrix SNP Array 6.0. TCGA genotype calls and corresponding confidence scores were retrieved using level 2 TCGA SNP array Birdseed data downloaded from the TCGA portal. Genotyping data with a confidence score of 0.1 were excluded. We selected 742 breast cancer samples with European ancestry. The corresponding RNA-seq BAM files and metadata are available from the Cancer Genomics Hub (CGHub). Marker SNPs, the exonic SNPs of the target genes, were extracted from dbSNP human Build 142 (collectively ~800 SNPs for ESR1, RMND1, ARMT1 and CCDC170), and RNA-seq read counts on SNP sites for reference and alternative alleles were computed. Homozygote marker SNPs and those with low coverage (less than 15×) were excluded. Major allele fraction (μ) representing allelic imbalance for each marker SNP was computed, and an average of allelic imbalances for each gene was calculated for individual tumor samples. Marker SNPs with extreme μ values (μ >0.75) were not included in the analysis. Level 3 SNP array data were downloaded from the TCGA portal, and GISTIC version 2.0.16 was used to identify copy number variations (CNVs) for each sample. Samples with low or high CNV levels, as presented in the gene-based GISTIC module report, were excluded from the analysis of the corresponding gene. For each risk SNP, allelic imbalance for the target transcripts was compared between heterozygote (AB) and homozygote (AA and BB) samples. For a given risk SNP and target gene, we used Levene’s test, a more robust test than the F test, for equality of variances when the risk SNP was not in linkage disequilibrium with any of the marker SNPs on that gene (r2 <0.5). Otherwise, a two-tailed t test was used for equality of means45.
Estrogen receptor protein expression
Normal breast samples derived from 150 postmenopausal donors (non-Hispanic, mean age of 62 years) and identified through the Susan G. Komen for the Cure Tissue Bank at the Indiana University Simon Cancer Center were used in this study46. DNA was extracted from blood cells at the Indiana CTSI Specimen Storage Facility using an AutogenFlex Star instrument (Autogen) and the FlexiGene AGF3000 blood kit for DNA extractions (Qiagen). SNP analysis was performed with 1 ng of DNA using TaqMan genotyping assays for rs2046210 (C_12034236_10), rs3757322 (C_27475059_10), rs9397437 (C_11556300_10), rs851984 (C_2496819_10), rs9918437 (C_29496189_10) and rs2747652 (C_2823750_10) from Life Technologies, following the manufacturer’s protocol. ER protein abundance was measured by immunohistochemical semiquantitation using an antibody to ERα (clone 6F11; 1:40 dilution; Leica Microsystems) and quantified with (i) an H score consisting of the sum of the percent of tumor cells staining, multiplied by an ordinal value corresponding to the intensity level (0, none; 1, weak; 2, moderate; 3, strong; Supplementary Fig. 2), and (ii) the percentage of positive cells. Correlations between the H scores and ER immunohistochemistry values were calculated using Spearman’s rank correlation analysis. All P values reported are two-sided, and values <0.05 were considered statistically significant.
Cell lines
Breast cancer cell lines MCF-7 (ER+; American Type Culture Collection (ATCC) HTB22), T-47D (ER+; ATCC HTB133) and BT-474 (ER+; ATCC HTB20) were grown in RPMI medium with 10% FCS and antibiotics. Normal breast epithelial cell lines MCF10A (ATCC CRL 10317) and Bre-80 (provided as a gift from R. Reddel, Children’s Medical Research Institute, Sydney) were grown in DMEM/F12 medium with 5% horse serum (HS), 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 20 ng/ml epidermal growth factor, 100 ng/ml cholera toxin and antibiotics. Cell lines were maintained under standard conditions, routinely tested for mycoplasma and short tandem repeat (STR) profiled.
Chromatin conformation capture
3C libraries were generated using EcoRI, HindIII or BglII as described previously15. 3C interactions were quantified by RT-PCR (qPCR) using primers designed within restriction fragments (Supplementary Table 15). qPCR was performed on a RotorGene 6000 instrument using MyTaq HS DNA polymerase (Bioline) with the addition of 5 mM Syto9, an annealing temperature of 66 °C and an extension time of 30 s. 3C analyses were performed in three independent 3C libraries from each cell line, with each experiment quantified in duplicate. BAC clones (RP11-108N8, RP11-713G5, RP11-450E24 and RP11-55K19) covering the 6q25 region were used to create artificial libraries of ligation products to normalize for PCR efficiency. Data were normalized to the signal from the BAC clone library and, between cell lines, by reference to a region within GAPDH. All qPCR products were electrophoresed on 2% agarose gels, gel purified and sequenced to verify the 3C product.
Electromobility shift assays
Gel shift assays were performed with ER+ MCF-7 or ER− Bre-80 nuclear lysates and biotinylated oligonucleotide duplexes (Supplementary Table 16). Nuclear lysates were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific) according to the manufacturer’s instructions. Total protein concentrations in nuclear lysates were determined by Bradford’s method. Duplexes were prepared by combining sense and antisense oligonucleotides in NEBuffer2 (New England BioLabs) and heat annealing at 80 °C for 10 min followed by slow cooling to 25 °C for 1 h. Binding reactions were performed in binding buffer (10% glycerol, 20 mM HEPES (pH 7.4), 1 mM DTT, protease inhibitor cocktail (Roche), 0.75 μg poly(dI:dC) (Sigma-Aldrich)) with 7.5 μg of nuclear lysate. For competition assays, binding reactions were preincubated with 1 pmol of competitor duplex (Supplementary Table 17) at 25 °C for 10 min before the addition of 10 fmol of biotinylated oligonucleotide duplex and a further incubation at 25 °C for 15 min. Reactions were separated on 10% Tris-borate-EDTA (TBE) polyacr-ylamide gels (Bio-Rad) in TBE buffer at 160 V for 40 min. Duplex-bound complexes were transferred onto Zeta-Probe positively charged nylon membranes (Bio-Rad) by semidry transfer at 25 V for 20 min and then cross-linked onto the membranes under 254-nm ultraviolet light for 10 min. Membranes were processed with the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Chemiluminescent signals were visualized with the C-DiGit blot scanner (LI-COR).
Plasmid construction and reporter assays
Promoter-driven luciferase reporter constructs were generated by the insertion of PCR-amplified fragments containing ESR1, ARMT1, RMND1 and CCDC170 promoters into the KpnI and MluI sites of pGL3-Basic. To assist in cloning, AgeI and SbfI sites were inserted into the BamHI and SalI sites downstream of the luciferase gene. A 1,496-bp signal 1 PRE fragment, a 997-bp signal 2 PRE fragment, a 1,566-bp signal 3 PRE fragment, a 1,463-bp signal 4 PRE fragment and a 1,349-bp signal 5 PRE fragment were generated by PCR or gBlocks (Integrated DNA Technologies) and cloned into the AgeI and SbfI sites of the modified pGL3-Promoter constructs. The minor alleles of individual SNPs were introduced into the PRE sequences by overlap extension PCR or gBlocks. Sequencing of all constructs confirmed variant incorporation (AGRF). ER+ MCF-7 and BT-474 or ER− Bre-80 cells were transfected with equimolar amounts of luci-ferase reporter plasmids and 50 ng of pRL-TK transfection control plasmid with Lipofectamine 3000. The total amount of transfected DNA was kept constant at 600 ng for each construct by the addition of pUC19 as a carrier plasmid. Luciferase activity was measured 24 h after transfection by the Dual-Glo Luciferase Assay System. To correct for any differences in transfection efficiency or cell lysate preparation, firefly luciferase activity was normalized to Renilla luciferase activity, and the activity of each construct was measured relative to the promoter-only construct, which had a defined activity of 1. Statistical significance was tested by log transforming the data and performing two-way ANOVA followed by Dunnett’s multiple-comparisons test in GraphPad Prism.
Chromatin immunoprecipitation
ER+ MCF-7 and BT-474 breast cancer cells were cross-linked with 1% formaldehyde at 37 °C for 10 min, rinsed once with ice-cold PBS containing 5% BSA and once with PBS, and collected in PBS containing 1× protease inhibitor cocktail (Roche). The cells were centrifuged for 2 min at 900g. Cell pellets were resuspended in 0.35 ml of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1, 1× pro-tease inhibitor cocktail) and sonicated three times for 15 s each with a 70% duty cycle (Branson SLPt) followed by centrifugation at 15,000g for 15 min. Supernatants were collected and diluted in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1). Two micrograms of antibody was prebound for 6 h to Protein G Dynabeads (Life Technologies) and then added to the diluted chromatin for overnight immunoprecipitation. The magnetic bead–chromatin complexes were collected and washed six times in RIPA buffer (50 mM HEPES, pH 7.6, 1 mM EDTA, 0.7% sodium deoxy-cholate, 1% NP-40, 0.5 M LiCl) and then twice with TE buffer. To reverse the cross-linking, the magnetic bead complexes were incubated overnight at 65 °C in elution buffer (1% SDS, 0.1 M NaHCO3). DNA fragments were purified using a QIAquick Spin kit (Qiagen). For qPCR, 2.0 μl from a 100-μl immu-noprecipitated chromatin extraction was subjected to 40 cycles of amplification. All PCR products were sequenced by Sanger sequencing (AGRF). The antibodies used were to CTCF (C-20; sc-15914) and GATA3 (HG3-31; sc268) or control IgG (sc-2027) (all from Santa Cruz Biotechnology). ChIP primers are listed in Supplementary Table 18.
Supplementary Material
Acknowledgments
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. This study would not have been possible without the contributions of the following: A. Berchuck (OCAC), R.A. Eeles, A.A. Al Olama, Z. Kote-Jarai and S. Benlloch (PRACTICAL), C. Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, the staff of the CNIO genotyping unit, D.C. Tessier, F. Bacot, D. Vincent, S. LaBoissière, F. Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, S.F. Nielsen, B.G. Nordestgaard and the staff of the Copenhagen DNA laboratory, and J.M. Cunningham, S.A. Windebank, C.A. Hilker, J. Meyer and the staff of the Mayo Clinic Genotyping Core Facility. Normal human tissues from the Susan G. Komen for the Cure Tissue Bank at the Indiana University Simon Cancer Center (Indianapolis) were used in this study. We thank the contributors, including Indiana University who collected samples used in this study, as well as the donors and their families, whose help and participation made this work possible. We also acknowledge National Institute for Health Research (NIHR) support to the Royal Marsden Biomedical Research Centre. Funding for the iCOGS infrastructure came from the European Community’s Seventh Framework Programme under grant agreement 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692 and C8197/A16565), the US National Institutes of Health (NIH; CA128978, CA192393, CA116167, CA176785 and an NIH Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201)) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112, the GAME-ON initiative), the US Department of Defense (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, the Komen Foundation for the Cure, the Breast Cancer Research Foundation and the Ovarian Cancer Research Fund. Full acknowledgments are given in the Supplementary Note.
Footnotes
Accession codes. The relevant SNP genotype data underpinning these analyses can be accessed by applying to the BCAC and CIMBA consortia (see URLs).
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
Manuscript writing group: A.M.D., K. Michailidou, K.B.K., D. Thompson, J.D.F., K.A.P., J. Beesley, C.S.H., G.C.-T., A.C.A., D.F.E. and S.L.E. Locus SNP selection: A.M.D., C.S.H. and E.D. iCOGS genotyping, genotype calling and quality control: A.M.D., J. Beesley, C.S.H., M.G., G.C.-T., K.A.P. and D.F.E. Imputation: K. Michailidou, K.B.K., A.C.A. and D.F.E. Statistical analyses and programming: K. Michailidou, K.B.K., A.C.A. and D.F.E. Functional analysis and bioinformatics: S.L.E., J.D.F., K.M.H., S. Kaufmann, H.S., M.M.M., J.S.L., E.L.-K., M. Hills, M.J., S.D., J. Beesley, S. Kar, N.A.S.-A., R.C.S., S.C. and S.N. COGS coordination: P.H., D.F.E., J. Beesley and A.M.D. BCAC coordination: D.F.E., G.C.-T., P.D.P.P. and J. Stone. BCAC data management: M.K.B. and Q.W. CIMBA coordination: A.C.A., G.C.-T., J. Stone and F.J.C. CIMBA data management: L.M. and D.B. MODE coordination: D. Thompson, C.V. and F.J.C. Provided participant samples and phenotype information and read and approved the manuscript: A.M.D., K. Michailidou, K.B.K., D. Thompson, J.D.F., J. Beesley, C.S.H., S. Kar, K.A.P., E.L.-K., E.D., D.B., N.A.S.-A., R.C.S., K.M.H., S. Kaufmann, H.S., M.M.M., J.S.L., M. Hills, M.J., S.D., S.C., M.K.B., J.D., Q.W., J.L.H., M.C.S., A. Broeks, M.K.S., A. Lophatananon, K. Muir, M.W.B., P.A.F., I.d.-S.-S., J.P., E.J.S., I.T., B. Burwinkel, F.M., P.G., T.T., S.E.B., H.F., A.G.-N., J.I.A.P., H.A.-C., L.E., V.A., H. Brenner, A. Meindl, R.K.S., H. Brauch, U.H., K.A., C.B., H.I., K. Matsuo, N.B., T.D., A. Lindblom, S. Margolin, V.-M.K., A. Mannermaa, C.T., A.H.W., D.L., H.W., J.C.-C., A.R., P.P., P.R., J.E.O., G.G.G., R.L.M., C.A.H., B.E.H., M.S.G., S.H.T., C.H.Y., S.N., A.-L.B.-D., V.K., J. Long, W.Z., K.P., R.W., I.L.A., J.A.K., P.D., C. Seynaeve, J.F., M.E.S., K.C., H.D., A.Hollestelle, A.M.W.v.d.O., K.H., Y.-T.G., X.-O.S., A.C., S.S.C., W.B., Q.C., B.J.P., M.S., J.-Y.C., D.K., S.C.L., M. Hartman, M. Kabisch, D. Torres, A.J., J. Lubinski, P.B., S.S., C.B.A., A.E.T., C.-Y.S., P.-E.W., N.O., A.S., L.M., S.H., A. Lee, M. Kapuscinski, E.M.J., M.B.T., M.B.D., D.E.G., S.S.B., R.J., L.T., N.T., C.M.D., E.J.v.R., S.L.N., B.E., T.V.O.H., A.O., J. Benitez, R.R., J.N.W., B. Bonanni, B.P., S. Manoukian, L.P., L.O., I.K., P.A., J. Garber, M.U.R., D.F., L.I., S.E., A.K.G., N.A., D.N., K.R., N.B.-M., C. Sagne, D.S.-L., F.D., O.M.S., S. Mazoyer, C.I., K.B.M.C., K.D.L., M.d.l.H., T.C., H.N., S. Khan, A.R.M., M.J.H., M.A.R., A.K., E.O., O.D., J. Brunet, M.A.P., J. Gronwald, T.H., R.B.B., R. Laframboise, P.S., M.M., S.A., M.R.T., S.K.P., N.L., F.J.C., M. Tischkowitz, L.F., J.V., K.O., C.F.S., C.R., C.M.P., M.H.G., P.L.M., G.R., E.N.I., P.J.H., K.-A.P., M.P., A.M.M., G.G., A. Bojesen, M. Thomassen, M.A.C., S.-Y.Y., E.F., Y.L., A. Borg, A.v.W., H.E., J.R., O.I.O., P.A.G., R.L.N., S.A.G., K.L.N., S.M.D., B.K.A., G. Mitchell, B.Y.K., J. Lester, G. Maskarinec, C.W., C. Scott, J. Stone, C.A., R.T., R. Luben, K.-T.K., Å.Helland, V.H., M.D., P.D.P.P., J. Simard, P.H., M.G.-C., C.V., G.C.-T., A.C.A., D.F.E. and, S.L.E.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Zheng W, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoniou AC, et al. Common alleles at 6q25.1 and 1p11.2 are associated with breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum Mol Genet. 2011;20:3304–3321. doi: 10.1093/hmg/ddr226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindström S, et al. Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nat Genet. 2011;43:185–187. doi: 10.1038/ng.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stacey SN, et al. Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet. 2010;6:e1001029. doi: 10.1371/journal.pgen.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hein R, et al. Comparison of 6q25 breast cancer hits from Asian and European genome wide association studies in the Breast Cancer Association Consortium (BCAC) PLoS One. 2012;7:e42380. doi: 10.1371/journal.pone.0042380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavaddat N, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Biomarkers Prev. 2012;21:134–147. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spencer AV, Cox A, Walters K. Comparing the efficacy of SNP filtering methods for identifying a single causal SNP in a known association region. Ann Hum Genet. 2014;78:50–61. doi: 10.1111/ahg.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Q, et al. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of Chinese, Japanese, and European ancestry. Cancer Res. 2011;71:1344–1355. doi: 10.1158/0008-5472.CAN-10-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corradin O, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French JD, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghoussaini M, et al. Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat Commun. 2014;4:4999. doi: 10.1038/ncomms5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glubb DM, et al. Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am J Hum Genet. 2015;96:5–20. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowper-Sal·lari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 2002;2:S1–S15. [PubMed] [Google Scholar]
- 20.Dunbier AK, et al. ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25.1. PLoS Genet. 2011;7:e1001382. doi: 10.1371/journal.pgen.1001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antoniou AC, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor–negative breast cancer in the general population. Nat Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haiman CA, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCormack VA, dos Santos Silva I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:1159–1169. doi: 10.1158/1055-9965.EPI-06-0034. [DOI] [PubMed] [Google Scholar]
- 24.Varghese JS, et al. Mammographic breast density and breast cancer: evidence of a shared genetic basis. Cancer Res. 2012;72:1478–1484. doi: 10.1158/0008-5472.CAN-11-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crandall CJ, et al. Sex steroid metabolism polymorphisms and mammographic density in pre- and early perimenopausal women. Breast Cancer Res. 2009;11:R51. doi: 10.1186/bcr2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindström S, et al. Genome-wide association study identifies multiple loci associated with both mammographic density and breast cancer risk. Nat Commun. 2014;5:5303. doi: 10.1038/ncomms6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stone J, et al. Novel associations between common breast cancer susceptibility variants and risk-predicting mammographic density measures. Cancer Res. 2015;75:2457–2467. doi: 10.1158/0008-5472.CAN-14-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estrada K, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koller DL, et al. Meta-analysis of genome-wide studies identifies WNT16 and ESR1 SNPs associated with bone mineral density in premenopausal women. J Bone Miner Res. 2013;28:547–558. doi: 10.1002/jbmr.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry JR, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 32.Molyneux G, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Janer A, et al. An RMND1 mutation causes encephalopathy associated with multiple oxidative phosphorylation complex deficiencies and a mitochondrial translation defect. Am J Hum Genet. 2012;91:737–743. doi: 10.1016/j.ajhg.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry JJ, et al. Human C6orf211 encodes Armt1, a protein carboxyl methyltransferase that targets PCNA and is linked to the DNA damage response. Cell Rep. 2015;10:1288–1296. doi: 10.1016/j.celrep.2015.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veeraraghavan J, et al. Recurrent ESR1-CCDC170 rearrangements in an aggressive subset of oestrogen receptor–positive breast cancers. Nat Commun. 2014;5:4577. doi: 10.1038/ncomms5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto-Ibusuki M, et al. C6ORF97-ESR1 breast cancer susceptibility locus: influence on progression and survival in breast cancer patients. Eur J Hum Genet. 2015;23:949–956. doi: 10.1038/ejhg.2014.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chenevix-Trench G, et al. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9:104. doi: 10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Couch FJ, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyd NF, et al. Mammographic density and the risk and detection of breast cancer. N Engl J Med. 2007;356:227–236. doi: 10.1056/NEJMoa062790. [DOI] [PubMed] [Google Scholar]
- 40.Dunning AM, et al. Association of ESR1 gene tagging SNPs with breast cancer risk. Hum Mol Genet. 2009;18:1131–1139. doi: 10.1093/hmg/ddn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnes DR, Lee A, Easton DF, Antoniou AC. Evaluation of association methods for analysing modifiers of disease risk in carriers of high-risk mutations. Genet Epidemiol. 2012;36:274–291. doi: 10.1002/gepi.21620. [DOI] [PubMed] [Google Scholar]
- 42.Antoniou AC, et al. A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet Epidemiol. 2005;29:1–11. doi: 10.1002/gepi.20074. [DOI] [PubMed] [Google Scholar]
- 43.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao R, Scott LJ. Detection of cis-acting regulatory SNPs using allelic expression data. Genet Epidemiol. 2011;35:515–525. doi: 10.1002/gepi.20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman ME, et al. The Susan G. Komen for the Cure Tissue Bank at the IU Simon Cancer Center: a unique resource for defining the “molecular histology” of the breast. Cancer Prev Res (Phila) 2012;5:528–535. doi: 10.1158/1940-6207.CAPR-11-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.