Abstract
Artemisia pallens is an important medicinal plant. In-vitro regeneration and multiplication of A. pallens have been established using attached cotyledons. Different growth regulators were considered for regeneration of multiple shoots. An average of 36 shoots per explants were obtained by culturing attached cotyledons on Murashige and Skoog’s medium containing 2 mg/L BAP and 0.1 mg/L NAA, after 45 days. The shoots were rooted best on half Murashige and Skoog’s medium with respect to media containing 1 mg/L IBA or 1 mg/L NAA. Different parameters such as type of bacterial strains, OD600 of bacterial culture, co-cultivation duration, concentration of acetosyringone and explants type were optimized for transient expression of the reporter gene. Agrobacterium tumefaciens harbouring pCambia1301 plasmid carrying β-glucuronidase as a reporter gene and hygromycin phosphotransferase as plant selectable marker genes were used for genetic transformation of A. pallens. Hygromycin lethality test showed concentration of 15 mg/L were sufficient to inhibit the growth of attached cotyledons and multiple shoot buds of nontransgenics in selection media. Up to 83 % transient transformation was found when attached cotyledons were co-cultivated with Agrobacterium strain AGL1 for 2 days at 22 °C on shoot induction medium. The bacterial growth was eliminated by addition of cefotaxime (200 mg/L) in selection media. T0 transgenic plants were confirmed by GUS histochemical assay and further by polymerase chain reaction (PCR) using uidA and hpt gene specific primers. The study is useful in establishing technological improvement in A. pallens by genetic engineering.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-016-0353-3) contains supplementary material, which is available to authorized users.
Keywords: A. pallens, Agrobacterium tumefaciens, Attached cotyledons, Transient transformation, GUS
Introduction
Artemisia pallens Wall (A. pallens) is an aromatic medicinal plant species used as a folk remedy for the treatment of diabetes. There are 32 species belonging to the genus Artemisia L. of family Asteraceae, growing in different zones of India. The anti-pyretic, anti-inflammatory, anthelmintic, anti-malarial, anti-diabetic, and anti-microbial effects of A. pallens make it an medicinally important plant (Subramoniam et al. 1996; Haider et al. 2014). Recently, anti-malarial property has been reported in two other species of Artemisia, A. japonica and A. nilagirica (Shukla et al. 2015).
The aromatic annual medicinal shrub A. pallens has pleasant fragrances, and leaves and flowers are used in decorations. An essential oil “Davana oil” is extracted from this plants, and is mainly used as flavoring agent for eatables, perfumes and some beverages (Mallavarapu et al. 1999; Ruikar et al. 2011). Chemically, Davana oil is a mixture of cis-davanone and seven other sesquiterpene ketone (Mallavarapu et al. 1999). Sesquiterpene lactone is also recovered from the plant, which is a potent drug molecule and is very effective for treatment of inflammation (Mallavarapu et al. 1999).
The aforementioned properties have resulted in this plant being in high demand. The large scale production of the plant will provide substantial amount of biomass for the extraction of the compound of interest. The optimization of genetic transformation of Artemisia species will pave way to engineer a metabolic pathway for in-vivo enhanced production of the compound of our interest. Various genetic transformation methods are available, but most frequently “Biolistic method” (Rech et al. 2008; Bhalothia et al. 2013) and “Agrobacterium-mediated” (Upadhyay et al. 2013; Elfahmi and Chahyadi, 2014) transformation are used. Agrobacterium-mediated genetic transformation is a cost effective and easier method for transformation of plants. The different parameters which affects gene transfer to plants via Agrobacterium tumefaciens (A. tumefaciens) are: type of explants, type of bacterial strains, co-culture duration, temperature of co-cultivation, concentration of acetosyringone, etc. (Tiwari and Tuli 2012).
There are various reports on plant regeneration and transformation of Artemisia L. genus such as A. annua (Han et al. 2005; Elfahmi and Chahyadi, 2014), A. aucheri (Sharafi et al. 2014a), A. sieberi (Sharafi et al. 2014b), and A. vulgaris (Sujatha et al. 2013). However, to the best of our knowledge there is no report on efficient genetic transformation of A. pallens. In this paper, we have examined effects of different growth regulators in Murashige and Skoog’s (MS) medium on direct regeneration of A. pallens using cotyledon and leaf as explants. Further, we have standardized the lethal dose of hygromycin for the selection of transgenic plants. Various factors were optimized for efficient genetic transformation of A. pallens using transient Agrobacterium mediated transformation method. The efficiency of transient transformation was calculated for each parameter by histochemical GUS staining of explants. This report establishes an efficient method for in-vitro regeneration and stable genetic transformation of A. pallens.
Materials and methods
Plant materials, explants preparation and culture conditions
Dry seeds of A. pallens were initially washed with autoclaved water mixed with 4–5 drop of Tween-20. The seeds were surface-sterilized in 0.1 % aqueous mercuric chloride for 5 min and rinsed thrice with sterile water. The surface sterilized seeds were kept on semisolid MS basal medium and moist filter paper in petriplates and incubated at two different temperatures (22 °C and 26 °C) for germination. Attached cotyledons and leaves were dissected from in-vitro grown plants and used as explants for direct shoot regeneration. The cultures were incubated at 26 ± 2 °C in 200 μmol photon/m2/s light intensity with a 16/8-h light/dark period.
Optimization of shoot and root induction media
The cotyledons and leaves were placed with abaxial surface towards media. M.S. basal medium supplemented with different combination of benzylaminopurine (BAP), Thidiazuron (TDZ), Gibberellic acid (GA), Indole-3-acetic acid (IAA), and Zeatin (Z) as mentioned in Table 1 were used. The explants were subcultured in the respective medium at an interval of 15 days. For root initiation, individual shoots were separated and transferred to half MS, MS, MS with 1 mg/l IBA and MS with1 mg/l IAA. Finally, the plantlets were transferred to soil for hardening. The media used in this study and their composition are mentioned in Table 1.
Table 1.
Different media and composition
| Name of Media | Composition of media |
|---|---|
| 2B0.5I or SIM | 4.33 g/L Murashige and Skoog, 3 % (w/v) sucrose, 2 mg/L BAP and 0.1 mg/L NAA, 0.8 % (w/v) agar, pH 5.8 (KOH) |
| 2B2G | 4.33 g/L Murashige and Skoog, 3 % (w/v) sucrose, 2 mg/L BAP and 2 mg/L GA3, 0.8 % (w/v) agar, pH 5.8 (KOH) |
| 5B0.5G | 4.33 g/L Murashige and Skoog, 3 % (w/v) sucrose, 2 mg/L BAP and 0.5 mg/L GA3, 0.8 % (w/v) agar, pH 5.8 (KOH) |
| 2B0.5 T | 4.33 g/L Murashige and Skoog, 3 % (w/v) sucrose, 2 mg/L BAP and 0.5 mg/L TDZ, 0.8 % (w/v) agar, pH 5.8 (KOH) |
| 1Z0.1I | 4.33 g/L Murashige and Skoog, 3 % (w/v) sucrose, 2 mg/L Zeatin and 0.1 mg/L IAA 0.8 % (w/v) agar, pH 5.8 (KOH) |
| Half MS | 2.2 g/L Murashige and Skoog, 3 % (w/v) sucrose, 0.8 % (w/v) agar, pH 5.8 (KOH) |
| MS | 4.33 g/L Murashige and Skoog, 3 % (w/v) sucrose, 0.8 % (w/v) agar, pH 5.8 (KOH) |
| SIM(H) | 4.33 g/L Murashige and Skoog, 3 % (w/v) sucrose, 2 mg/L BAP and 0.1 mg/L NAA, 15 mg/L hygromycin, 200 mg/L cefotaxime, 0.8 % (w/v) agar, pH 5.8 (KOH) |
| RIM(H) | 2.2 g/L Murashige and Skoog, 3 % (w/v) sucrose, 20 mg/L hygromycin, 200 mg/L cefotaxime, 0.8 % (w/v) agar, pH 5.8 (KOH) |
| MSL | 4.33 g/L Murashige and Skoog, 20 mg/L glucose, 195.2 mg/L MES, pH 5.6 (KOH) |
Effect of cefotaxime on A. tumefaciens
To inhibit bacterial overgrowth during co-cultivation of explants with Agrobacterium, antibiotic cefotaxime (Lupin, India) was used. Disc-diffusion assay was performed to determine optimal concentration of cefotaxime. Whatman filter paper was used to prepare 5 mm discs and theses disc were sterilized by autoclaving. Different strains of Agrobacterium (Blank AGL1, EHA101, GV3101, and LBA4404) were grown in Luria broth (LB) with their respective antibiotics (Supplementary Table 1) at 28 °C till OD600 reached 1.4. The discs were imbibed in different concentration of cefotaxime (100, 200, 300, 400 and 500 mg/L). A. tumefaciens culture (75 μl) was taken aseptically and spread on the surface of shoot induction media (SIM) plate. Five discs in duplicate were fully saturated with different concentration of cefotaxime and placed on SIM plates and incubated at room temperature for 72 h.
Hygromycin leathality test
The attached cotyledons and multiple shoot buds were used to see the effect of different concentration of hygromycin (0, 5, 10, 15, 20, 25, 30 and 35 mg/L). SIM media supplemented with cefotaxime (200 mg/L), with different concentrations of hygromycin was prepared. Regeneration of attached cotyledons and proliferation of multiple shoot buds were observed after 20 days by culturing them on SIM. The experiment was performed in two replicates of 8 attached cotyledons and 4 multiple shoots buds for each treatment.
Preparation of Agrobacterium suspension
The binary vector pCambia1301 was used to transform four A. tumefaciens chemical competent strains (AGL1, EHA101, GV3101, and LBA4404) by freeze and thaw method (Holsters et al. 1978). Transformed colonies of all four strains were selected on Luria agar plate containing proper antibiotics (Supplementary Table 1). Single colony was used to perform colony PCR using primers specific to hpt gene to confirm the presence of plasmid. The confirmed single Agrobacterium colony was inoculated in 5 ml LB medium with their respective antibiotics (Supplementary Table 1) as primary culture. 5 ml of primary culture was used to inoculated 50 ml of LB and grown to an OD600 of 1.2–1.4 at 28 °C with constant shaking at 200 rpm. The culture was centrifuged to form pellet at 5000 rpm for 10 mins at room temperature and resuspended in MSL media.
A. tumefaciens mediated transformation and co-cultivation
The attached cotyledons, leaves and multiple shoot buds were used for transient transformation via A. tumefaciens. These explants were dipped into MSL medium containing Agrobacterium (pH 5.6) and 100 μM acetosyringone. Agrobacterium infection was carried out for 45 min at room temperature under continuous shaking with 90 rpm. The explants were blot dried on sterilized Whatman filter paper to remove excess bacterial suspension, then transferred to SIM for co-cultivation and the plates were kept in dark at 23 °C. After co-cultivation, the explants were rinsed 3–4 times with autoclaved water supplemented with 200 mg/L cefotaxime. Then the explants were blot dried and then kept on SIM(H) containing cefotaxime.
Optimization of different factors to increase transient transformation efficiency
Different factors such as type of bacterial strain (AGL1, EHA101, GV3101, LBA4404), OD600 of bacterial strain (0.5, 1.0, 1.5), co-cultivation duration (1, 2, 4 days) at 23 °C, concentration of acetosyringone (100 μM, 200 μM, 400 μM) and explants type (cotyledon, leaf and multiple shoot buds) were optimized. The efficiency of transformation was calculated on the basis of transient expression of uidA gene by observing the number of blue spots per ten explants during GUS histochemical enzyme assay (Jefferson et al. 1987).
Selection of transformants and hardening
The optimized factors during transient transformation were used for stable transformation. The medium which gave highest multiple shoots and roots was used for stable transformation with hygromycin as selectable marker. SIM(H) and RIM(H) was supplemented with cefotaxime (200 mg/L) and hygromycin (15 mg/L) for proper selection of transgenic plants. Multiple shoot buds were dissected and sub-cultured at an interval of 2 weeks onto fresh selection medium. The proliferated shoot buds were maintained for two months on SIM(H) and then transferred to RIM(H). The rooted plantlet were transferred to soilrite containing pots and covered with transparent plastic bag for proper acclimatization.
GUS histochemical assay
The transient and stable expression of uidA transgene was analysed using GUS histochemical assay. The X-Gluc solution was freshely prepared as suggested by Jefferson et al. 1987. Tissues were incubated within X-Gluc solution at 37 °C for 24 h in dark and washed with water and finally dipped in 70 % ethanol to remove all chlorophyll. The uidA gene in pCambia1301 is interrupted by intron, therefore all blue spots in GUS histochemical analysis were a result of plant protein synthesis machinery.
Polymerase chain reaction (PCR)
Genomic DNA was isolated from 12 putative transgenic shoots (T0) of A. pallens by CTAB method (Devi et al. 2013). The quality of the genomic DNA was checked on 0.8 % agarose gel and quantification done by measuring absorbance (260/280) using a spectrophotometer. PCR was performed using total DNA as template using PCR Master Mix (Bangalore Genei) and primers specific to uidA and hpt genes to confirm the transgene integration. Additionally, primers specific to virG gene was also used to ensure that none of bacteria was adhered to transgenic plants (Table 2). The PCR cycle condition was: 94 °C for 5 min, 94 °C for 40 s, 56 °C for 30 s, and 72 °C for 60 s with a final extension at 72 °C for 5 min. The PCR product was electrophoresed onto 0.8 % agarose gel, detected by staining in ethidium bromide solution.
Table 2.
Primers and sequences
| Primers name | Sequence of nucleotides | Amplicon size |
|---|---|---|
|
UidA_For: UidA_Rev: |
5’TGGAGCATCAGGGCGGCTATAC3’ 5’GTCGGGTCGAGTTTACGCGTTGC3’ |
1000 bp |
|
Hpt_For: Hpt_Rev: |
5’ATGAAAAAGCCTGAACTCACCGC3’ 5’CCAGAAGAAGATGTTGGCGACC3’ |
700 bp |
|
VirG_For: VirG_Rev: |
5′CTGGCGGCAAAGTCTGAT3’ 5′TGTCGTAAACCTCCTCGT3’ |
450 bp |
Results
Effect of different media composition
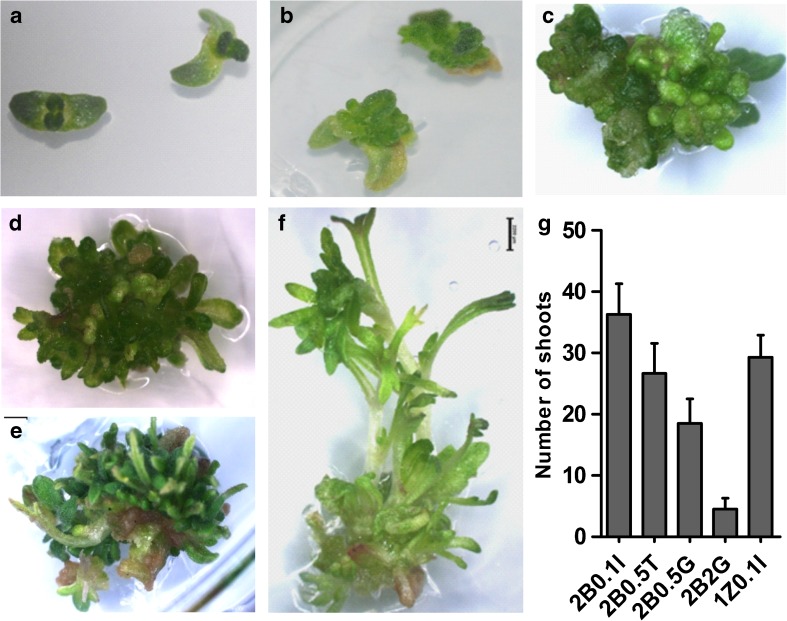
The seeds of A. pallens showed germination in 3 days on sterile moist filter paper whereas, on plain MS media germination happened after 7 days. The germination rate of seeds was similar at 22 °C and 26 °C temperatures. The attached cotyledons showed better response than single cotyledon on different media combination; therefore, in all the cases we used attached cotyledons. The explants showed swelling within 15 days (Fig. 1a, b) of initial culture and multiple shoot buds (MSB) were initiated after 30 days (Fig. 1c). The MSB were continuously proliferated after dissecting it into 2 or 4 parts according to their size (Fig. 1d, e). The average number of shoots, formed after two month of initiation on different media, was recorded (Fig. 1g). The maximum no. of shoots (average 36 shoots per explant) produced via organogenesis was observed in 2B0.1 N medium, whereas the lowest was in 2B2G (average 4.5 shoots per explant) (Fig. 1g). The individual elongated shoot having at least 2 cm stem were dissected and transferred to rooting medium (Fig. 1f). Callus like structures appeared in the lower part of stem, when shoots were transferred to 1 mg/L IBA (Fig. 2a). The rooting started after 7–10 days on half MS media, and the best root induction was observed on half MS media (Fig 2b). Delayed rooting was observed in case of MS and 1 mg/L NAA along with MS.
Fig. 1.
Different stages of organogenesis a 4 day old explants, b swelled explants, c and d multiple shoot buds (MSB), e newly emerged shoots, f elongated shoot and g average no. of shoots in different media
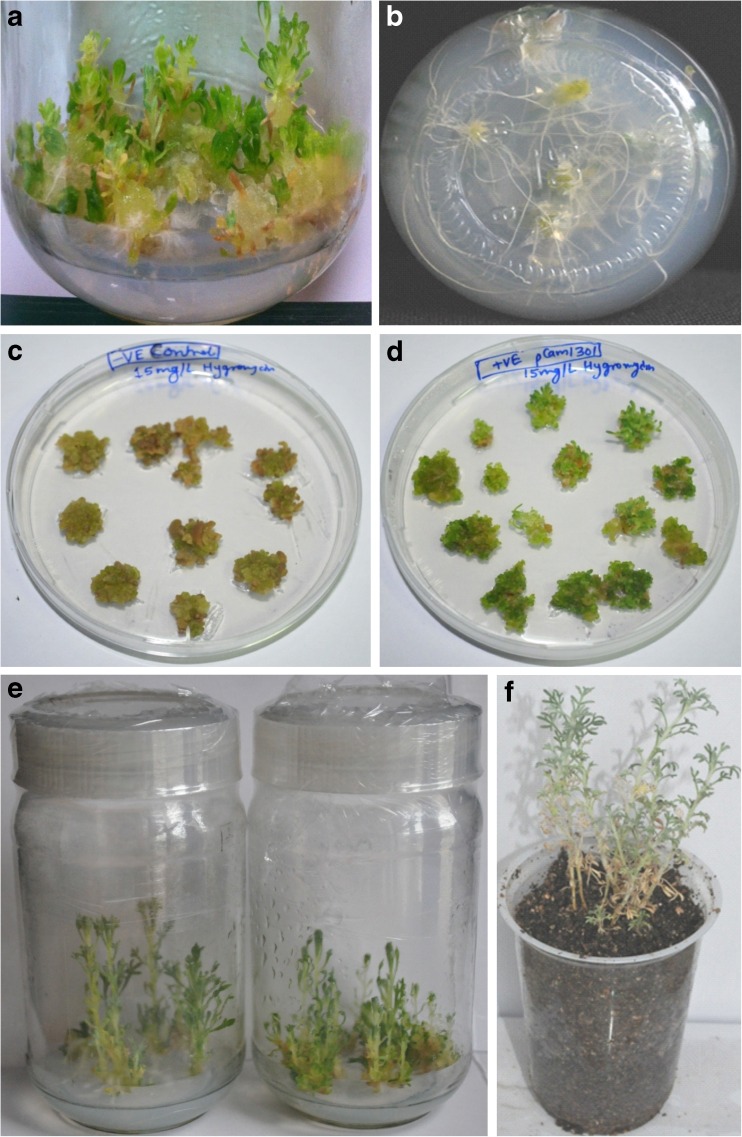
Fig. 2.
Selection and rooting a unusual callus like swelling occurs at base of stem in media containing 1 mg/L IBA, b roots formed in half MSA c browning and death of non transgenics MSB, d selection of putative transgenic MSB, e rooted plants at half MSA, and f in-vitro raised plantlet after acclimatization and hardening
Effective dose of cefotaxime and hygromycin
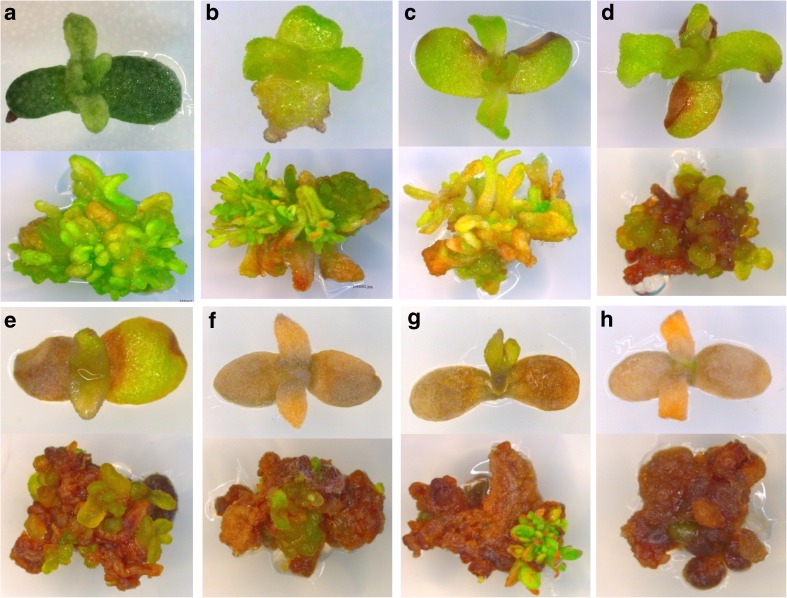
The effect of different concentration of the cefotaxime was tested against all four bacterial strains by disc-diffusion assay. The result showed that, cefotaxime (>200 mg/L) was more effective in eliminating all the bacterial strains. The effect of different concentration of hygromycin on attached cotyledons and multiple shoot buds were tested (Fig. 3). The attached cotyledons and MSB in the media without hygromycin remained green and swelled (Fig. 3a); whereas all explants on media having different concentration of hygromycin became brown and necrosis occured according to their dose (Fig. 3b to h). The optimal dose of hygromycin is 15 mg/L, which is suitable for selection of transgenics and mortality of non transgenic (Fig. 3d). With increasing concentration of hygromycin in media the explants became browner and finally the explants died. The data are based on two replicates of 10 attached cotyledon and 4 multiple shoot bud as explants for each treatment.
Fig. 3.
Effect of hygromycin on explants after 20 days a at 0 mg/L hygromycin, b at 5 mg/L hygromycin, c at 10 mg/L hygromycin, d at 15 mg/L hygromycin, e at 20 mg/L hygromycin, f at 25 mg/L hygromycin, g at 30 mg/L hygromycin and h at 35 mg/L hygromycin
Colony PCR and transient transformation
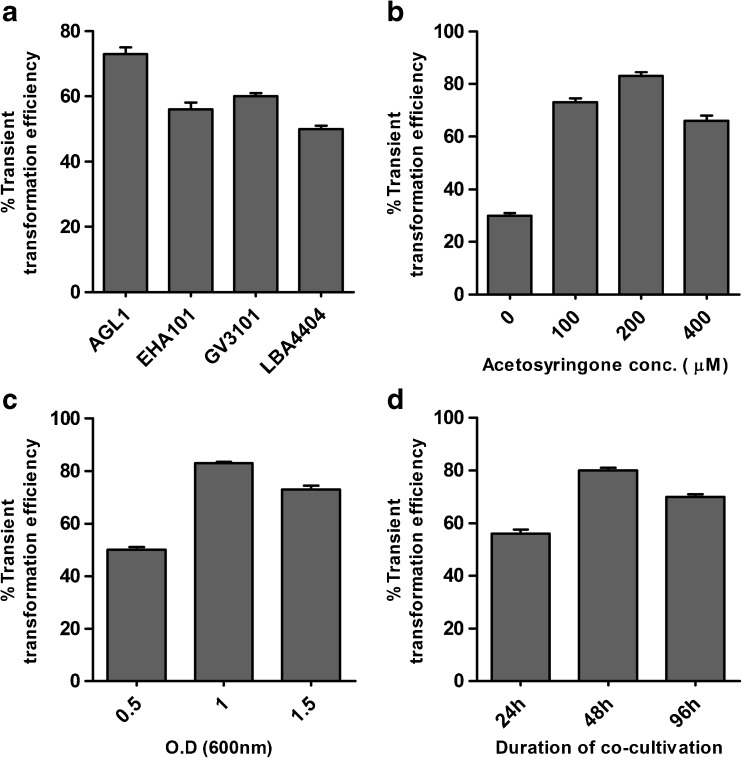
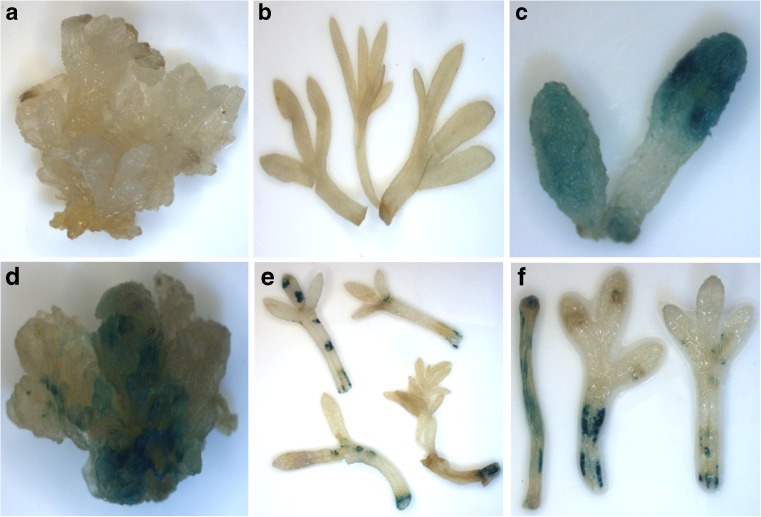
Colony PCR was performed using primers specific to uidA gene and electrophoresed on 1 % agarose gel. Positive colonies were used further to perform transient and stable transformation. The T-DNA of pCambia1301 contains uidA gene interrupted by a modified castor bean catalase intron and driven under a constitutive CaMV35S promoter. Transient transformation efficiency (T.T.E) was calculated based on the presence of GUS blue spots on leaves. Different strains of A. tumefaciens were examined for A. pallens transformation. Average T.T.E of A. pallens was 73 % using AGL1, followed by GV3101 (60 %), EHA101 (56 %), and LBA4404 (50 %), when leaf was used as explant (Fig. 4a). The O.D of bacterial culture in co-cultivation media affected the transformation efficiency. MSL having bacterial O.D 1 at 600 nm resulted in higher transformation efficiency (83 %) as compared to that with O.D 0.5 and 1.5 (Fig. 4c). Initially, the transformation efficiency was examined keeping the concentration of acetosyringone (100 μM) constant. After this, different concentrations of acetosyringone were also tried. The highest T.T.E (83 %) was observed at a concentration of 200 μM acetosyringone (Fig. 4b). Co cultivation for two days resulted in maximum T.T.E as compared to one and four days (Fig. 4d). We have also analysed T.T.E in different explants and found that the attached cotyledons and MSB were showing uniform blue color where as leaf showed less blue color (Fig. 5c, d, e, and f). Gus staining of non-transformed explants showed no blue color (Fig. 5a, b).
Fig. 4.
Transient transformation efficiency with different parameter a effect of bacterial strains, b effect of different concentration of acetosyringone c, effect of different O.D at 600 nm and d effect of co-cultivation duration
Fig. 5.
Gus histochemical assay of different explants a non-transgenic MSB, b non-transgenic leaf, c transformed attached cotyledon, d transformed MSB, e and f transformed leaf after GUS staining
Selection and screening of T0 transgenic plants
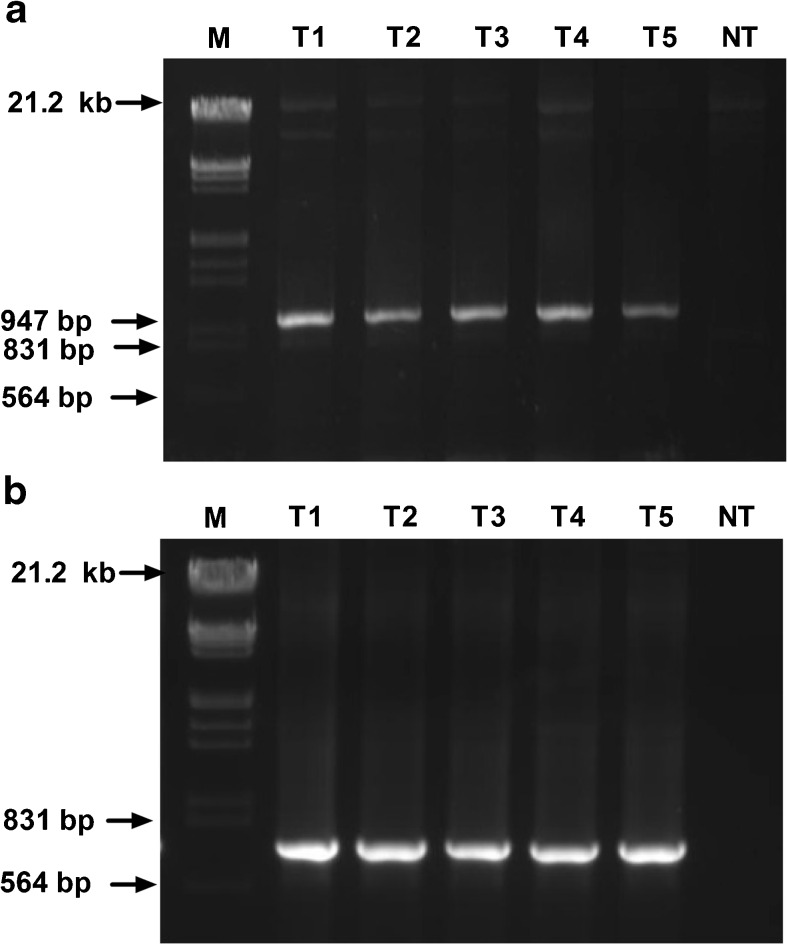
The attached cotyledons and MSB were used for stable transformation by keeping all optimized parameter constant. The co-cultivated cotyledons and MSB were washed with 200 mg/L cefataxime and kept for 10 days on 2B0.1 N(C) without hygromycin. After 10 days explants were transferred to selected SIM(H), and at every subculture steps explants or multiple shots buds were dissected according to their size and the dead brown tissue were excised off and removed. After 30 days of incubation non transgenic MSB became brown and dead (Fig. 2c) whereas transgenics survived at a concentration of 15 mg/L hygromycin (Fig. 2d). The selected shoots were transferred to rooting media containing 20 mg/L hygromycin (Fig. 2e). The putative transgenics were transferred into plastic pot containing soilrite for hardening (Fig. 2f). Screening of 12 putative T0 transgenics resulted in five positive plants by PCR. Agarose gel elctrophoresis gave a amplification of 700 bp and 1000 bp corresponding to hpt and uidA gene respectively in transgenic plantlets whereas no amplification was observed in control plants (Fig. 6a, b). PCR using virG gene primers showed no band amplified in either transgenic or control plants.
Fig. 6.
Gel image of PCR positive T0 transgenics lines a Amplification of uidA gene (1000 bp), M-Lamda marker, T1 to T2- different transgenic lines and NT- nontransgenic line, b Amplification of hpt (700 bp), M-Lamda marker, T1 to T2- different transgenic lines and NT- nontransgenic line
Discussion
In this present study, we tried shoot organogenesis of A. pallens from individual cotyledons, attached cotyledons, leaf blade and found best response in attached cotyledons. It might be due to fact that attached cotyledons has meristematic tissue which remains intact compared to dissected individual cotyledons. However, shoot organogenesis in different species of Artesmisia was achieved by using leaf, petiole, lamina, node, hypocotyl segment and internode as explants (Han et al. 2005; Sujatha and Ranjitha Kumari 2007; Nathar and Yatoo 2014; Sharafi et al. 2014a). On supplementing with both BAP and TDZ in media, we found average of 26.6 shoots per explants where as some explants gave rise to green callus like mass. Highest numbers of shoots (Average 36.3 shoots) were produced when 2 mg/L BAP and 0.1 mg/L NAA was used (Fig. 1g). This is in agreement of the result reported from A. aucheri (Sharafi et al. 2014a). Nathar and Yatoo (2014) reported callus induction and regeneration of A. pallens. However, they found maximum shoots (14.25) when they used 3 mg/L kinetin and 10 shoots using 2 mg/L BAP. The rooting was best when it was subjected onto half MS media, whereas in case of IBA and IAA lower part of stem swelled and aerial roots were formed (Fig. 2a). Whereas in another report the rooting of A. pallens was found best on IBA and NAA (Nathar and Yatoo 2014).
The transformation efficiency varies among different cultivars of Artemisia therefore; we optimized different parameters which affect T-DNA transfer. There are various parameter which affects transformation efficiency as explored in A. annua (Elfahmi and Chahyadi, 2014). The selection of strain plays a crucial for Agrobacterium mediated transformation. The transformation frequency of AGL1 was reported to be 70.91 % in A. annua when leaves were used as explants (Elfahmi and Chahyadi, 2014). However, our study shows highest transient transformation efficiency (83 %) with AGL1 strain using 200 μM acetosyringone in media (Fig. 4b). This may be due to high virulence of AGL1 strain compared to nopaline strain (GV3101) and octopine strain (LBA4404). The optical density of Agrobacterium solution is also an important parameter. In case of A. pallens the O.D.600 1.0 gave maximum T.T.E (83 %) while an O.D of 1.5 resulted in 73 % T.T.E. Higher bacterial density in the medium may leads to severe necrosis of cotyledon cells (Li et al. 2009). Co-cultivation is an important parameter for genetic transformation of plants because in this step, T-DNA is incorporated into plant genomic DNA (Han et al. 2005). Our study showed that, the T.T.E was high (80 %) at 48 h while it decreased at 96 h (70 %). Co-cultivation from 48 to 60 h was shown to be acceptable in A. annua for better transformation while the longer for co-cultivation does not help to increase transformation efficiency (Vergauwe et al. 1998). It is reported that, secretion of acetosyringone during wounding plant cells induces the transcription of the virulence genes of Agrobacterium. Addition of Acetosyringone can improve transformation efficiency, in various monocots and some dicots plants which fail to secrete acetosyringone (Han et al. 2005). However in our study, addition of 200 μM acetosyringone increased efficiency to 83 % while at 400 μM T.T.E decreased to 66 % (Fig. 4b). Transient expression of any gene depends upon the number of T-DNA copies transferred to the nucleus of plant cell but not yet integrated into plant genome. Therefore, transient transformation might give higher expression with respect to stable transformation. To the best of our knowledge, this is the first report of Agrobacterium mediated transformation of A. pallens which could pave way for functional genomics studies to increase Artemisinin production.
Electronic supplementary material
(DOCX 11 kb)
Acknowledgment
The authors express their gratitude for providing research facilities to School of Biotechnology and Bioinformatics, D. Y. Patil University, Navi Mumbai, India.
Author contribution
Conceived and designed the experiments: AA and ND. Performed the experiments: AA, VS, ZP, SS and JK. Analyzed the data: AA, VS and ND. Wrote the paper: AA, JK and ND.
References
- Bhalothia P, Alok A, Mehrotra S, Mehrotra R. AACA element negatively regulates expression of protein phosphatase 2C (PP2C) like promoter in Arabidopsis thaliana. Am J Plant Sci. 2013;04:549–554. doi: 10.4236/ajps.2013.43071. [DOI] [Google Scholar]
- Devi KD, Punyarani K, Singh NS, Devi HS. An efficient protocol for total DNA extraction from the members of order Zingiberales- suitable for diverse PCR based downstream applications. Springerplus. 2013;2:669. doi: 10.1186/2193-1801-2-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfahmi SS, Chahyadi A. Optimization of genetic transformation of Artemisia annua L. Using Agrobacterium for artemisinin production. Pharmacogn Mag. 2014;10:S176–S180. doi: 10.4103/0973-1296.127372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider SZ, Mohan M, Andola HC. Constituents of Artemisia indica Willd. From Uttarakhand Himalaya: a source of davanone. Pharmacognosy Res. 2014;6:257–259. doi: 10.4103/0974-8490.132607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Wang H, Ye H, Li G. High efficiency of genetic transformation and regeneration of Artemisia annua L. Via Agrobacterium tumefaciens-mediated procedure. Plant Sci. 2005;168:73–80. doi: 10.1016/j.plantsci.2004.07.020. [DOI] [Google Scholar]
- Holsters M, de Waele D, Depicker A, Messens E, van Montagu M, Schell J. Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet MGG. 1978;163:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-F, Park E, von Arnim AG, Nebenführ A. The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods. 2009;5:6. doi: 10.1186/1746-4811-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu GR, Kulkarni RN, Baskaran K, Rao L, Ramesh S. Influence of plant growth stage on the essential oil content and composition in Davana (Artemisia pallens Wall.) J Agric Food Chem. 1999;47:254–258. doi: 10.1021/jf980624c. [DOI] [PubMed] [Google Scholar]
- Nathar VN, Yatoo GM (2014) Micropropagation of an antidiabetic medicinal plant. Artemisia pallens Turk J Botany 38:491–498
- Rech EL, Vianna GR, Aragao FJL. High-efficiency transformation by biolistics of soybean, common bean and cotton transgenic plants. Nat Protoc. 2008;3:410–418. doi: 10.1038/nprot.2008.9. [DOI] [PubMed] [Google Scholar]
- Ruikar AD, Khatiwora E, Ghayal NA, Misar AV, Mujumdar AM, Puranik VG, Deshpande NR. Studies on aerial parts of Artemisia pallens wall for phenol, flavonoid and evaluation of antioxidant activity. J Pharm Bioallied Sci. 2011;3:302–305. doi: 10.4103/0975-7406.80768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharafi A, Sohi H, Mirzaee H, Azadi P. In vitro regeneration and Agrobacterium mediated genetic transformation of Artemisia aucheri Boiss. Physiol Mol Biol Plants. 2014;20:487–494. doi: 10.1007/s12298-014-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharafi A, Sohi H, Sharafi AA, Azadi P, Mausavi A. Tissue culture and regeneration of an antima larial plant, Artemisia sieberi Besser. Res J Pharmacogn. 2014;1:15–20. [Google Scholar]
- Shukla V, Pala Z, Alok A, Desai N. Screening of different Artemisia spp. from Western Ghats of Maharashtra for an anti-malarial compound-artemisinin. Am J Plant Sci. 2015;06:1619–1632. doi: 10.4236/ajps.2015.69162. [DOI] [Google Scholar]
- Subramoniam A, Pushpangadan P, Rajasekharan S, Evans DA, Latha PG, Valsaraj R. Effects of Artemisia pallens Wall. On blood glucose levels in normal and alloxan-induced diabetic rats. J Ethnopharmacol. 1996;50:13–17. doi: 10.1016/0378-8741(95)01329-6. [DOI] [PubMed] [Google Scholar]
- Sujatha G, Ranjitha Kumari BD. High-frequency shoot multiplication in Artemisia vulgaris L. Using thidiazuron. Plant Biotechnol Rep. 2007;1:149–154. doi: 10.1007/s11816-007-0028-1. [DOI] [Google Scholar]
- Sujatha G, Zdravković-Korać S, Ćalić D, Flamini G, Kumari Ranjita BD. High-efficiency Agrobacterium rhizogenes-mediated genetic transformation in Artemisia vulgaris: hairy root production and essential oil analysis. Ind Crop Prod. 2013;44:643–652. doi: 10.1016/j.indcrop.2012.09.007. [DOI] [Google Scholar]
- Tiwari S, Tuli R. Optimization of factors for efficient recovery of transgenic peanut (Arachis hypogaea L.). Plant cell. Tissue Organ Cult. 2012;109:111–121. doi: 10.1007/s11240-011-0079-4. [DOI] [Google Scholar]
- Upadhyay SK, Kumar J, Alok A, Tuli R. RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 2013;3:2233–2238. doi: 10.1534/g3.113.008847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergauwe A, Van Geldre E, Inzé D, Van Montagu M, Van den Eeckhout E. Factors influencing Agrobacterium tumefaciens-mediated transformation of Artemisia annua L. Plant Cell Rep. 1998;18:105–110. doi: 10.1007/s002990050540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 11 kb)