Abstract
Phenotypic variation of important seed traits like seed length, seed breadth, seed thickness, 100 seed weight and seed oil content were recorded in a total of 157 collected accessions of Pongamia. Out of these, fatty acid profiles of 38 accessions selected based on their high and low oil content was analyzed. Fatty acid profile revealed high variability in stearic, oleic and linoleic acid which varied from 0.42 to 10.61 %, 34.34 to 74.58 %, and 7.00 to 31.28 % respectively. Variations in palmitic and linolenic acid were small. Iodine value, saponification number and cetane number (CN) of fatty acid methyl esters (FAME) of seed oil ranges from 186.99 to 201.25, 81.13 to 108.19 and 46.16 to 56.47 respectively. Fatty acid compositions, degree of unsaturation and CN are the important parameters, which are used to determine quality of FAME were used as biodiesel. Some of the Pongamia accessions identified were higher in oil content while some accessions showed higher degree of unsaturation and a few of them had CN values higher than 55. Genetic diversity analysis with six TE-AFLP primers generated a total of 334 bands out of which 174 (52.10 %) were polymorphic. The genetic similarity ranged from 0.11 to 0.47. These findings clearly showed high level of genetic diversity and all economically desirable traits were not present in a single genotype of Pongamia. All these traits could be selected from these CPTs and transfer to a single elite variety through selection and breeding programme and could be utilized for large scale multiplication and plantation to produce high quantity and quality biodiesel in future.
Keywords: Pongamia pinnata, Oil content, Fatty acid methyl esters, Gas chromatography, TE-AFLP, Genetic diversity
Introduction
Energy security is an important issue all over the world because reservoirs of fossil fuels are depleting day by day which also lead to increase in prices of petroleum products. So, we have to think an alternative and renewable source of energy which also emits less carbon to environment. A number of Tree Borne Oilseed species (TBOs), which can produce oil as a source of energy in the form of biodiesel have been identified in India and other countries (Tewari 2003). Pongamia pinnata (L.) Pierre has been found to be one of the most suitable feedstock because its oil can be used as a source of fuel for the biodiesel industry (Karmee and Chadha 2005). Pongamia also have other additional favorable attributes like its hardy nature, capable of fixing atmospheric nitrogen, high yield, high oil recovery and quality of oil. Pongamia tree also has several applications in area of medicine (Elanchezhiyan et al. 1993; Shivanna and Rajakumar 2010) and is also used in insecticides (Pavela 2009). Pongamia is a non-edible oil producing tree legume of India (Lakshmikanthan 1978). The species is found predominantly in Western Ghats of India and Southeast Asia. It is fast growing medium sized tree and starts to yield 7 years after planting (Pavithra et al. 2013). Each mature pod generally encloses 1–2 kidney shaped or elongated brownish red seeds. Oil yield is one of the most important traits determining the commercial viability of Pongamia pinnata as an energy crop. The oil content of Pongamia seed is about 32–42 % (Kaushik et al. 2007), which can be converted into biodiesel (FAME) by transesterification with methanol in the presence of an alkali catalyst (Karmee and Chadha 2005; Naik et al. 2008).
In recent years, significant research efforts have been made to explore plant-based fuels (Martini and Shell 1998). The potentiality of Pongamia oil as an important source of biodiesel is now well recognized (Karmee and Chadha 2005; Azam et al. 2005). The Pongamia seed oil predominantly contains four fatty acids, namely, palmitic, stearic, oleic and linoleic acid. In addition to these major fatty acids, its oil also contains very less amounts of linolenic, behenic and eicosenoic acid. Oleic acid is the major fatty acid among pongamia trees and generally constitute about half of the oil with a range from approximately one to two thirds (Arpiwi et al. 2012; Sunil et al. 2009). The biodiesel properties of seed oil and FAME are: saponification number (SN), which specifies the relative fatty acid chain length of the FAMEs; the iodine value (IV), measures number of double bonds in respective fatty acids and cetane number (CN), which measures indication of ignition quality of fuel. In the US biodiesel standard (BS 2002), the minimum CN value is 47, where the Pongamia accessions analyzed in this study exhibit CN values from 50 to 59 with a mean of 54.4 (Mukta et al. 2009).
The major bottleneck in harnessing the biofuel potential of this tree is the unavailability of any improved and characterized planting stock. The variation in the pattern of fruiting and flowering was observed in different individuals of this species, also few trees bear high fruit numbers as expected. The high phenotypic diversity was observed in this species, thus providing an opportunity for its genetic improvement (Kaushik et al. 2007). It is important to characterize and select candidate plus trees (CPTs) of Pongamia for the improvement of this species. A plus tree is an individual tree of a species possessing superior morphological and reproductive characters than other individuals of the same species (Kesari and Rangan 2010).
Recently, initiatives have been taken towards molecular characterization and identification of superior genotypes of Pongamia. In the previous work, we attempted to analyze changes in oil and fatty acid profile at various developmental stages of seed in Pongamia (Sharma et al. 2015). The present study was carried out to assess variability and divergence of oil, fatty acids profile and important biodiesel traits for selection of CPTs with more efficient biodiesel yielder and their large scale commercial plantation for its development as an economically viable tree for biodiesel production across India and other tropical countries.
Materials and methods
Plant material
Germplasm collection surveys were conducted during the months of April–June, 2007–2011 from different regions of NCT of Delhi. Some accessions were also collected from other states of India (Assam, Karnataka and Andhra Pradesh) and a total of 157 accessions were collected. Passport data including site of collection, number of pods per bunch, number of seeds per pod and important seed traits were recorded. Global Positioning System (Garmin eTrex Vista Cx) used to mark the location of collected germplasm. Explored area lies between 28°32′8″–28°41′22″N and 77°23′28″–77°07′52″E covering parts of East Delhi, Central Delhi, West Delhi, East Delhi and Noida region of NCT of Delhi, India. Representative samples consisting of 1–2 kg mature pods were collected in cloth bags and seeds were separated from pods by manual dehulling. The mature trees which attained age of 6–10 years were selected. Seeds were dried in an oven at 50 °C till no weight loss was seen on further drying.
Oil content analysis
For oil content analysis, several seeds were pooled to represent an accession. Seed oil content was analyzed by solvent extraction using n-hexane as solvent in a Soxhlet apparatus (SOCS PLUS, Pelican Equipments, Chennai). Seed powder (1 g) and 60 mL of n-hexane were used per batch. Extraction was carried out at 80 °C for 1 h after which fast solvent recovery was done at 180 °C for 30 min. Two replicates of each accession were used and means were taken. The oil content for each accession was calculated and expressed as percentage (w/w) of dry seed using following equation
where, Wfb is final beaker weight, Wib is initial beaker weight and Ws is the sample weight.
Fatty acid analysis
For fatty acid analysis using gas chromatography (GC), 38 Pongamia accessions were selected from both the tails of the oil content distribution of 157 accessions representing the top as well as the bottom of the distribution table. Analysis was done in two replicates using the method described by Thies (1971) with minor modifications. Dried seed samples of individual accessions were taken and crushed to powder with the help of mortar and pestle. Seed powder (25 mg) was incubated with 750 µL of sodium methylate for 20 min. Then iso-octane (300 µL) was added and incubated for additional 20 min. The clear upper phase was taken in a GC vial and 400 µL of iso-octane was added. Fatty acid profiles were analyzed using a GC (Shimadzu, Kyoto, Japan) fitted with a flame ionization detector (FID). The oven, injector and detector blocks temperature were maintained at 210, 230 and 250 °C respectively and nitrogen gas was used as the carrier. The proportion of FAME was expressed as percentage of total fatty acids excluding the minor constituents.
Biodiesel properties like SN, IV of oil were calculated using FAMEs composition of oil with the help of following equations (Kalayasiri et al. 1996):
where, Ai is the percentage, D is the number of double bonds and MWi is the molecular weight of each component.
Whereas, Cetane Number of FAMEs was calculated by the following equation (Krisnangkura 1986):
Descriptive statistics on oil content and fatty acid data was analyzed. Correlation coefficients between various seed and oil traits of Pongamia genotypes were calculated using SPSS software (Version 17.0). Pongamia accessions were classified into three categories, namely, low (< − 1s), medium ( − 1s to + 1s) and high (> + 1s) based on seed oil and biodiesel traits where and s are the mean and standard deviation respectively (Mukta et al. 2009; Sarkar and Deb 1984).
Three endonuclease (TE)-AFLP assay
The protocol for TE-AFLP marker analysis was based on van der Wurff et al. (2000) with minor modifications. In current study, Eco RI, Pst I and Mse I were used instead of Bam HI, Xba I and Rsa I that were used in the original protocol. The detailed protocol of TE-AFLP has already published by our group (Sharma et al. 2011).
Genotyping and evaluation of molecular marker attributes
A total of 38 accessions were genotyped using six TE-AFLP primers combinations. The amplicons were in the size range of 80–450 bps. Amplified fragments were scored manually for their presence (denoted as ‘1’) or absence (denoted as ‘0’) for each primer combination. For analysis clear, distinct and polymorphic bands were recorded.
The binary matrix was used to estimate genetic dissimilarities using Jaccard’s coefficient [GDij = (b + c)/a + (b + c); (Jaccard 1908)], where GD is the genetic dissimilarity measures between individuals i and j, a is the number of polymorphic bands that are shared by i and j, b is the number of bands present in i and absent in j, c is the number of bands present in j and absent in i. The dissimilarity matrix was subjected to neighbor joining clustering in order to construct the phenetic dendrogram DARWin software version 5.0.158 (Perrier and Jacquemoud-Collet 2006). The robustness and reliability of the phenograms were tested by bootstrap analysis for 1000 bootstraps for computing probabilities in terms of percentage for each node of the tree using the DARWin software (Perrier and Jacquemoud-Collet 2006). Different marker attributes like polymorphic information content (PIC), resolving power (RP), marker index (MI) and degree of polymorphism were analyzed (Sharma et al. 2011).
Results and discussion
Seed and oil traits
The mean performance of seed traits like seed width, seed length, seed thickness, seed weight and total seed oil content of 157 Pongamia accessions revealed significant differences (Table 1). Maximum value for seed length (25.40 mm) was observed in P276 and minimum (16.40 mm) in P002 accession. Seed thickness also varied significantly among all the accessions. It has been observed that maximum 100-seed weight (250 g) was recorded from P276 accession. In this context of seed trait, Pongamia accession P276 from Dwarka region of Delhi has maximum seed length and 100-seed weight. A study on seed and oil traits across twenty four CPTs from Jharkhand revealed that, a genotype CPT-19 recorded maximum values for seed length (27.93 mm), 100-seed weight (202.89 g) and total oil content (44.33 %) (Divakara et al. 2010) which are more similar to results of current study.
Table 1.
Mean values for important seed traits in Pongamia accessions
| Sr. no. | Accession ID | Seed length (mm) | Seed breadth (mm) | Seed thickness (mm) | 100-seed weight (g) | Oil content (%) |
|---|---|---|---|---|---|---|
| 1 | P001 | 22.1 | 16.2 | 5.6 | 115 | 37.80 |
| 2 | P002 | 16.4 | 12.9 | 7.2 | 91 | 29.50 |
| 3 | P002 | 18.0 | 13.5 | 6.2 | 98 | 34.20 |
| 4 | P003 | 21.2 | 13.0 | 6.8 | 132 | 37.93 |
| 5 | P004 | 16.8 | 12.2 | 5.0 | 76 | 33.34 |
| 6 | P007 | 17.2 | 11.3 | 5.8 | 120 | 20.20 |
| 7 | P008 | 21.2 | 12.0 | 8.2 | 100 | 27.83 |
| 8 | P009 | 17.4 | 13.5 | 5.2 | 92 | 33.86 |
| 9 | P010 | 21.2 | 17.4 | 7.2 | 118 | 34.66 |
| 10 | P012 | 18.8 | 13.2 | 5.8 | 76 | 31.81 |
| 11 | P013 | 22.2 | 14.9 | 7.1 | 109 | 38.88 |
| 12 | P015 | 23.4 | 15.0 | 8.4 | 117 | 36.83 |
| 13 | P016 | 21.8 | 12.2 | 7.1 | 112 | 33.13 |
| 14 | P017 | 20.8 | 12.2 | 6.8 | 64 | 27.81 |
| 15 | P018 | 18.2 | 14.4 | 6.2 | 63 | 30.26 |
| 16 | P019 | 17.8 | 16.2 | 6.8 | 115 | 33.53 |
| 17 | P020 | 22.2 | 14.2 | 7.8 | 126 | 30.56 |
| 18 | P021 | 23.4 | 17.2 | 8.1 | 165 | 28.71 |
| 19 | P022 | 21.2 | 11.2 | 7.2 | 103 | 33.60 |
| 20 | P024 | 18.2 | 12.4 | 7.2 | 99 | 30.80 |
| 21 | P028 | 20.2 | 16.5 | 7.2 | 120 | 30.60 |
| 22 | P031 | 17.2 | 12.8 | 6.8 | 76 | 36.69 |
| 23 | P033 | 21.1 | 11.4 | 7.3 | 67 | 25.34 |
| 24 | P037 | 18.2 | 16.8 | 7.2 | 117 | 32.43 |
| 25 | P039 | 17.4 | 16.2 | 6.2 | 112 | 34.58 |
| 26 | P042 | 17.7 | 12.6 | 6.2 | 64 | 35.53 |
| 27 | P051 | 21.2 | 12.0 | 6.9 | 111 | 24.75 |
| 28 | P053 | 19.3 | 12.3 | 6.5 | 91 | 31.85 |
| 29 | P054 | 23.2 | 11.2 | 7.1 | 98 | 30.21 |
| 30 | P056 | 17.3 | 13.6 | 5.3 | 69 | 22.60 |
| 31 | P057 | 22.2 | 12.6 | 7.3 | 76 | 32.47 |
| 32 | P058 | 17.8 | 14.2 | 5.8 | 120 | 33.98 |
| 33 | P059 | 19.4 | 12.6 | 6.7 | 100 | 35.19 |
| 34 | P060 | 23.4 | 11.3 | 7.8 | 92 | 31.54 |
| 35 | P061 | 18.2 | 15.8 | 7.2 | 118 | 31.40 |
| 36 | P062 | 20.8 | 12.0 | 6.5 | 76 | 32.40 |
| 37 | P065 | 19.2 | 13.2 | 6.4 | 67 | 32.74 |
| 38 | P070 | 23.8 | 16.7 | 8.0 | 117 | 35.06 |
| 39 | P071 | 19.2 | 12.2 | 8.2 | 112 | 23.02 |
| 40 | P075 | 18.2 | 12.8 | 6.6 | 60 | 28.77 |
| 41 | P076 | 19.1 | 14.2 | 5.2 | 63 | 26.30 |
| 42 | P077 | 17.5 | 16.2 | 6.2 | 113 | 30.60 |
| 43 | P078 | 19.2 | 14.5 | 7.4 | 126 | 27.20 |
| 44 | P079 | 23.1 | 13.8 | 8.4 | 165 | 31.20 |
| 45 | P082 | 22.2 | 15.2 | 6.2 | 103 | 31.87 |
| 46 | P083 | 18.3 | 13.3 | 6.6 | 99 | 24.95 |
| 47 | P084 | 22.4 | 16.2 | 6.1 | 120 | 36.35 |
| 48 | P085 | 19.2 | 14.4 | 7.8 | 70 | 29.88 |
| 49 | P086 | 23.3 | 11.4 | 6.8 | 90 | 32.80 |
| 50 | P089 | 22.2 | 12.2 | 7.1 | 91 | 29.20 |
| 51 | P094 | 21.3 | 11.8 | 7.5 | 83 | 29.40 |
| 52 | P099 | 20.2 | 13.8 | 8.7 | 133 | 33.20 |
| 53 | P101 | 22.3 | 12.4 | 7.8 | 81 | 31.74 |
| 54 | P103 | 23.2 | 13.5 | 7.4 | 123 | 33.00 |
| 55 | P104 | 21.1 | 12.2 | 7.5 | 105 | 34.51 |
| 56 | P105 | 22.9 | 12.2 | 7.3 | 70 | 37.99 |
| 57 | P106 | 23.3 | 11.4 | 7.4 | 96 | 32.41 |
| 58 | P107 | 22.3 | 12.4 | 7.5 | 91 | 28.60 |
| 59 | P108 | 21.8 | 11.2 | 7.2 | 90 | 35.22 |
| 60 | P110 | 21.6 | 13.2 | 8.2 | 127 | 34.40 |
| 61 | P111 | 21.2 | 11.2 | 8.4 | 113 | 31.40 |
| 62 | P124 | 21.3 | 11.3 | 7.8 | 102 | 28.00 |
| 63 | P126 | 23.7 | 11.8 | 8.2 | 92 | 30.20 |
| 64 | P127 | 20.2 | 14.2 | 7.8 | 128 | 29.75 |
| 65 | P130 | 22.8 | 14.4 | 7.2 | 126 | 33.75 |
| 66 | P133 | 22.0 | 12.3 | 7.2 | 70 | 29.60 |
| 67 | P134 | 23.1 | 11.5 | 7.2 | 96 | 32.00 |
| 68 | P135 | 23.1 | 11.3 | 6.5 | 91 | 30.80 |
| 69 | P137 | 22.8 | 12.4 | 7.2 | 90 | 30.00 |
| 70 | P139 | 22.8 | 13.4 | 6.2 | 94 | 30.20 |
| 71 | P140 | 20.2 | 14.1 | 6.2 | 80 | 28.00 |
| 72 | P147 | 21.5 | 12.8 | 6.8 | 94 | 29.40 |
| 73 | P148 | 22.2 | 13.3 | 8.2 | 87 | 29.00 |
| 74 | P157 | 19.2 | 13.4 | 7.8 | 139 | 34.73 |
| 75 | P158 | 22.2 | 13.8 | 8.8 | 155 | 27.82 |
| 76 | P159 | 20.2 | 14.1 | 6.3 | 86 | 23.66 |
| 77 | P160 | 21.4 | 12.3 | 7.8 | 113 | 28.06 |
| 78 | P161 | 23.1 | 11.4 | 7.1 | 88 | 37.97 |
| 79 | P162 | 21.1 | 14.2 | 7.2 | 99 | 36.47 |
| 80 | P164 | 19.2 | 13.2 | 8.2 | 109 | 34.19 |
| 81 | P165 | 20.1 | 13.3 | 5.8 | 85 | 23.31 |
| 82 | P166 | 17.2 | 12.1 | 6.5 | 51 | 23.47 |
| 83 | P167 | 23.1 | 11.8 | 6.4 | 92 | 29.00 |
| 84 | P168 | 23.3 | 11.4 | 6.5 | 85 | 34.32 |
| 85 | P169 | 22.2 | 15.0 | 7.4 | 154 | 31.87 |
| 86 | P170 | 22.8 | 12.3 | 6.8 | 94 | 34.19 |
| 87 | P171 | 23.3 | 13.2 | 5.5 | 80 | 29.87 |
| 88 | P172 | 23.2 | 12.8 | 6.2 | 90 | 30.49 |
| 89 | P173 | 22.2 | 12.6 | 7.2 | 121 | 34.49 |
| 90 | P174 | 21.8 | 12.2 | 8.0 | 109 | 31.42 |
| 91 | P175 | 22.1 | 10.2 | 7.7 | 114 | 33.77 |
| 92 | P176 | 23.4 | 11.2 | 6.8 | 91 | 30.20 |
| 93 | P178 | 22.3 | 12.2 | 6.5 | 86 | 34.03 |
| 94 | P179 | 18.2 | 13.2 | 8.2 | 107 | 33.13 |
| 95 | P185 | 20.2 | 14.4 | 6.2 | 124 | 25.46 |
| 96 | P186 | 22.2 | 15.1 | 6.5 | 105 | 32.02 |
| 97 | P187 | 23.4 | 11.3 | 6.5 | 91 | 30.51 |
| 98 | P188 | 21.4 | 13.0 | 8.4 | 134 | 33.93 |
| 99 | P189 | 22.4 | 12.4 | 7.2 | 132 | 37.98 |
| 100 | P190 | 21.4 | 14.2 | 7.2 | 125 | 31.20 |
| 101 | P191 | 21.2 | 12.3 | 7.6 | 110 | 33.80 |
| 102 | P192 | 20.4 | 14.2 | 6.2 | 84 | 36.60 |
| 103 | P193 | 22.2 | 12.5 | 6.5 | 92 | 34.80 |
| 104 | P194 | 21.2 | 13.2 | 7.2 | 88 | 33.40 |
| 105 | P195 | 22.4 | 12.8 | 6.8 | 178 | 34.40 |
| 106 | P196 | 22.2 | 14.2 | 9.8 | 44 | 29.80 |
| 107 | P197 | 20.3 | 13.4 | 7.2 | 114 | 30.00 |
| 108 | P198 | 21.2 | 14.2 | 6.8 | 118 | 37.00 |
| 109 | P200 | 22.2 | 12.1 | 8.3 | 132 | 30.20 |
| 110 | P201 | 19.5 | 12.8 | 6.6 | 96 | 28.40 |
| 111 | P202 | 22.2 | 12.2 | 6.2 | 91 | 31.80 |
| 112 | P203 | 22.4 | 12.2 | 6.6 | 90 | 35.60 |
| 113 | P204 | 21.2 | 14.2 | 7.4 | 127 | 30.80 |
| 114 | P205 | 21.1 | 11.4 | 7.5 | 113 | 32.20 |
| 115 | P206 | 22.2 | 15.2 | 6.4 | 102 | 25.40 |
| 116 | P207 | 21.2 | 12.4 | 6.2 | 92 | 24.00 |
| 117 | P208 | 21.2 | 15.2 | 7.1 | 128 | 31.60 |
| 118 | P209 | 21.2 | 14.2 | 7.3 | 126 | 37.20 |
| 119 | P210 | 21.1 | 14.2 | 7.8 | 117 | 35.80 |
| 120 | P211 | 19.3 | 13.5 | 6.5 | 94 | 33.20 |
| 121 | P212 | 21.2 | 14.4 | 7.2 | 102 | 30.20 |
| 122 | P213 | 22.1 | 15.2 | 7.2 | 163 | 36.00 |
| 123 | P214 | 18.8 | 13.2 | 5.8 | 77 | 28.37 |
| 124 | P215 | 20.2 | 14.2 | 8.2 | 133 | 32.87 |
| 125 | P216 | 23.8 | 14.2 | 10.1 | 224 | 33.60 |
| 126 | P217 | 19.5 | 14.3 | 7.4 | 144 | 34.72 |
| 127 | P218 | 22.2 | 13.4 | 6.8 | 145 | 33.80 |
| 128 | P219 | 21.2 | 14.2 | 7.0 | 168 | 29.60 |
| 129 | P220 | 23.2 | 16.3 | 8.2 | 181 | 33.40 |
| 130 | P221 | 22.6 | 12.4 | 7.7 | 136 | 29.00 |
| 131 | P222 | 19.2 | 12.3 | 6.8 | 101 | 32.00 |
| 132 | P223 | 21.3 | 16.2 | 6.3 | 118 | 33.80 |
| 133 | P224 | 22.2 | 14.2 | 7.2 | 148 | 34.60 |
| 134 | P225 | 19.8 | 13.6 | 6.8 | 93 | 33.80 |
| 135 | P226 | 21.3 | 12.8 | 7.2 | 84 | 31.00 |
| 136 | P227 | 21.4 | 14.2 | 9.2 | 155 | 36.85 |
| 137 | P228 | 21.0 | 13.8 | 8.2 | 135 | 40.28 |
| 138 | P229 | 22.2 | 14.4 | 9.2 | 159 | 32.00 |
| 139 | P230 | 21.8 | 13.2 | 7.6 | 109 | 37.80 |
| 140 | P232 | 20.2 | 14.2 | 7.2 | 120 | 31.42 |
| 141 | P233 | 20.2 | 14.4 | 7.4 | 133 | 35.00 |
| 142 | P234 | 24.4 | 16.3 | 8.0 | 182 | 32.34 |
| 143 | P235 | 21.0 | 13.5 | 9.0 | 135 | 33.80 |
| 144 | P236 | 22.2 | 14.8 | 8.2 | 151 | 36.00 |
| 145 | P237 | 21.2 | 15.1 | 7.8 | 162 | 34.20 |
| 146 | P273 | 21.0 | 14.2 | 6.3 | 80 | 31.20 |
| 147 | P274 | 22.8 | 15.2 | 9.6 | 180 | 31.80 |
| 148 | P275 | 23.3 | 15.4 | 8.5 | 175 | 34.00 |
| 149 | P276 | 25.4 | 16.4 | 9.2 | 250 | 37.20 |
| 150 | P277 | 23.2 | 16.2 | 8.8 | 192 | 32.00 |
| 151 | Assam-1 | 22.2 | 14.8 | 8.2 | 155 | 28.20 |
| 152 | Assam-2 | 22.4 | 12.8 | 8.2 | 182 | 29.00 |
| 153 | Assam-3 | NA | NA | NA | 104 | 28.60 |
| 154 | Assam-4 | NA | NA | NA | 122 | 28.40 |
| 155 | Bangalore-1 | 23.0 | 12.2 | 7.5 | 133 | 28.00 |
| 156 | Eluru-1 | NA | NA | NA | 102 | 26.00 |
| 157 | Eluru-2 | 21.2 | 13.4 | 7.2 | 113 | 23.50 |
| Mean | 12.14 | 13.45 | 7.21 | 111.45 | 31.70 | |
| SD | 1.84 | 1.51 | 0.93 | 32.60 | 3.71 |
Based on seed oil content, 157 Pongamia accessions were categorized in five groups (Table 2). Oil content of accessions varied from 20.20 to 40.28 % with a mean of 31.7 %. About 55 % of the accessions had oil content between 30 and 35 %. Ten accessions had oil content less than 25 % whereas one accession had oil content more than 40 %. Other studies also showed oil content was highly variable trait among trees. Across 75 germplasm accessions from Andhra Pradesh oil content ranges from 9.5 to 46 % (Mukta et al. 2009) and across 123 accessions from Andhra Pradesh and Orissa oil content ranges from 15 to 47 % (Sunil et al. 2009).
Table 2.
Distribution of 157 Pongamia pinnata accessions for seed oil content
| Phenotypic trait | Range (%) | Number of accession |
|---|---|---|
| Seed oil content (%) | >40 | 1 |
| 35–40 | 25 | |
| 30–35 | 86 | |
| 25–30 | 35 | |
| <25 | 10 |
There was a positive correlation (all at P = 0.01) between seed weight and seed length (r = 0.35), seed breadth, (r = 0.46) and seed thickness (r = 0.56). There was no significant correlation these seed traits and oil content (Table 3). The results obtained in this study correlated with the earlier study by Sunil et al. (2009) and Arpiwi et al. (2012).
Table 3.
Correlation between important seed and oil traits of Pongamia genotypes
| Seed length | Seed breadth | Seed thickness | 100-seed weight | Oil content | |
|---|---|---|---|---|---|
| Seed length | 1.00 | ||||
| Seed breadth | −0.05 | 1.00 | |||
| Seed thickness | 0.410** | 0.12 | 1.00 | ||
| 100-seed weight | 0.359** | 0.465** | 0.568** | 1.00 | |
| Oil content | 0.201** | 0.192** | 0.166* | 0.239** | 1.00 |
** Correlation is significant at the 0.01 level
* Correlation is significant at the 0.05 level
Fatty acid profile analysis
The fatty acid composition of oil governs its chemical properties and its efficacy as biodiesel. Gas chromatography analysis of the selected accessions exhibited very high variability in palmitic, stearic, oleic, linoleic and linolenic acid content (Table 4). Accessions P001, P003, P189 and P198 had high (>67.75 %) oleic acid content whereas accessions P017, P056, P105, P140, P158 and P161 had high (>22.57 %) linoleic acid content. The oleic acid content observed in these accessions is slightly higher than the previous report (Mukta et al. 2009). Accessions P056, P105, P140, P161 and P166 had high (>12.49 %) palmitic acid content whereas accessions P070, P105, P140, P161 and P165 had high (>7.82 %) stearic acid content (Table 5). Total unsaturated fatty acid content in the accessions varied from 68.7 to 87.6 %. Considering high genetic and phenotypic variability observed in Pongamia from India, effect of genetic factors on seed oil composition is very likely (Sharma et al. 2011; Kaushik et al. 2007). Bala and coworkers (Bala et al. 2011) reported the presence of significant levels of erucic acid in Pongamia trees from Western India. The absence of erucic acid in the present accessions may be due to genetic as well as environmental factors. Sharma et al. (2015) reported the significant variation in seed oil and fatty acid composition during seed development in Pongamia.
Table 4.
Phenotypic variations in seed oil content, fatty acid profile and biodiesel properties of 38 of Pongamia accessions
| Accession | Oil content (%) | Palmitic | Stearic | Oleic | Linoleic | Linolenic | Oleic/linoleic ratio | Saponification number | Iodine value | Cetane number |
|---|---|---|---|---|---|---|---|---|---|---|
| P001 | 37.80 | 8.66 | 5.01 | 67.98 | 13.06 | 1.07 | 5.21 | 191.77 | 87.72 | 55.02 |
| P003 | 37.93 | 9.69 | 0.52 | 70.29 | 15.61 | 1.67 | 4.50 | 196.07 | 96.05 | 52.52 |
| P007 | 20.20 | 11.02 | 5.94 | 62.33 | 16.59 | 1.49 | 3.76 | 195.45 | 90.18 | 53.94 |
| P008 | 27.83 | 12.36 | 7.44 | 63.58 | 10.07 | 2.09 | 6.31 | 191.99 | 81.13 | 56.47 |
| P013 | 38.88 | 9.02 | 4.39 | 63.79 | 16.02 | 3.45 | 3.98 | 193.73 | 95.82 | 52.91 |
| P015 | 36.83 | 10.16 | 4.18 | 61.67 | 17.55 | 1.98 | 3.51 | 191.70 | 92.67 | 53.92 |
| P017 | 27.81 | 11.38 | 3.74 | 52.63 | 27.92 | 3.76 | 1.89 | 199.87 | 108.19 | 49.26 |
| P031 | 36.69 | 9.04 | 4.56 | 60.98 | 17.94 | 2.77 | 3.40 | 191.00 | 94.91 | 53.52 |
| P033 | 25.34 | 7.98 | 7.35 | 60.71 | 17 | 1.61 | 3.57 | 189.43 | 89.79 | 54.91 |
| P042 | 35.53 | 10.45 | 6.43 | 56.05 | 19.53 | 2.53 | 2.87 | 190.68 | 92.70 | 54.07 |
| P051 | 24.75 | 10.18 | 4.55 | 61.68 | 17.17 | 2.11 | 3.59 | 192.00 | 92.34 | 53.95 |
| P056 | 22.60 | 13.72 | 1.46 | 50.85 | 31.28 | 0.68 | 1.63 | 197.48 | 104.25 | 50.48 |
| P059 | 35.19 | 9.96 | 5.49 | 60.75 | 16.66 | 2.99 | 3.65 | 192.27 | 92.99 | 53.76 |
| P070 | 35.06 | 10.09 | 8.04 | 58.81 | 15.87 | 2.09 | 3.71 | 190.34 | 87.35 | 55.32 |
| P071 | 23.02 | 10.84 | 6.52 | 62.49 | 16.7 | 0.32 | 3.74 | 194.38 | 87.32 | 54.73 |
| P076 | 26.30 | 9.94 | 7.46 | 58.08 | 19.24 | 3.19 | 3.02 | 196.36 | 95.81 | 52.54 |
| P078 | 27.20 | 10.46 | 4.73 | 58.34 | 21.11 | 3.05 | 2.76 | 196.10 | 99.05 | 51.85 |
| P083 | 24.95 | 12.07 | 4.91 | 63.72 | 15.98 | 3.02 | 3.99 | 200.33 | 94.51 | 52.28 |
| P105 | 37.99 | 15.49 | 8.05 | 45.35 | 24.96 | 5.1 | 1.82 | 199.67 | 99.95 | 51.15 |
| P108 | 35.22 | 8.06 | 6.44 | 65.6 | 14.4 | 1.08 | 4.56 | 191.25 | 88.03 | 55.03 |
| P124 | 28.00 | 9.99 | 6.46 | 57.11 | 18.25 | 4.28 | 3.13 | 192.80 | 96.13 | 52.98 |
| P140 | 28.00 | 14.41 | 9.83 | 34.34 | 27.13 | 7.27 | 1.27 | 187.68 | 99.92 | 52.90 |
| P158 | 27.82 | 10.65 | 5.47 | 51.01 | 25.93 | 2.69 | 1.97 | 192.34 | 100.20 | 52.13 |
| P161 | 37.97 | 13.33 | 10.02 | 43.8 | 24.67 | 3.86 | 1.78 | 192.68 | 94.64 | 53.33 |
| P162 | 36.47 | 9.33 | 5.85 | 64.24 | 16.07 | 2.13 | 4.00 | 195.62 | 92.71 | 53.34 |
| P165 | 23.31 | 8.06 | 10.61 | 64.1 | 14.14 | 0.18 | 4.53 | 194.15 | 83.75 | 55.57 |
| P166 | 23.47 | 14.18 | 6.1 | 63.62 | 14.22 | 1.87 | 4.47 | 201.25 | 88.09 | 53.60 |
| P189 | 37.98 | 8.9 | 5.55 | 74.58 | 10.8 | 0.16 | 6.91 | 200.10 | 87.07 | 53.99 |
| P198 | 37.00 | 7.82 | 5.51 | 71.42 | 11.61 | 0.57 | 6.15 | 193.84 | 86.81 | 54.92 |
| P203 | 35.60 | 9.79 | 6.71 | 60.52 | 14.17 | 2.84 | 4.27 | 188.57 | 87.86 | 55.47 |
| P206 | 25.40 | 12.18 | 4.93 | 62.13 | 14.07 | 3.72 | 4.42 | 195.05 | 91.54 | 53.69 |
| P209 | 37.20 | 10.58 | 5.05 | 59.93 | 17.18 | 2.26 | 3.49 | 190.71 | 91.20 | 54.40 |
| P210 | 35.80 | 10.18 | 4.79 | 62.07 | 16.58 | 3.14 | 3.74 | 194.14 | 94.44 | 53.16 |
| P213 | 36.00 | 10.5 | 5.34 | 59.73 | 15.54 | 2.04 | 3.84 | 186.99 | 87.44 | 55.81 |
| P228 | 40.28 | 11.78 | 4.74 | 54.75 | 21.38 | 3.85 | 2.56 | 194.03 | 98.50 | 52.27 |
| P230 | 37.80 | 12.48 | 0.42 | 65.29 | 16.67 | 1.39 | 3.92 | 193.61 | 92.71 | 53.63 |
| P236 | 36.00 | 9.53 | 5.59 | 66.42 | 12.66 | 2.78 | 5.25 | 194.36 | 90.27 | 54.07 |
| P276 | 37.20 | 8.99 | 5.18 | 63.58 | 18.97 | 1.23 | 3.35 | 196.23 | 94.90 | 52.76 |
| Mean | 32.06 | 10.61 | 5.67 | 60.11 | 17.76 | 2.43 | 3.70 | 193.84 | 92.87 | 53.57 |
| SD | 6.06 | 1.87 | 2.15 | 7.63 | 4.82 | 1.43 | 1.27 | 3.49 | 5.58 | 1.49 |
| Range | 20.20–38.88 | 7.82–15.49 | 0.42–10.61 | 34.34–74.58 | 10.07–31.28 | 0.16–7.27 | 1.27–6.91 | 186.99–201.25 | 81.13–108.19 | 46.16–56.47 |
Table 5.
Distribution of Pongamia pinnata accessions based on seed oil content, fatty acid profile and biodiesel properties
| Trait | Low (< − 1s) | Medium ( − 1s to + 1s) | High (> + 1s) |
|---|---|---|---|
| Seed oil content (%) | 007, 033, 051, 056, 071, 083, 165, 166, 206 | 001, 003, 008, 015, 017, 031, 042, 049, 070, 076, 078, 105, 108, 124, 140, 158, 161, 162, 189, 203, 209, 210, 213, 230, 236, 276 | 013, 228 |
| Fatty acid content (%) | |||
| Palmitic | 001, 033, 108, 165, 198 | 003, 007, 008, 013, 015, 017, 031, 042, 051, 059, 070, 071, 076, 078, 083, 124, 158, 162, 189, 203, 206, 209, 210, 213, 218, 230, 236,276 | 056, 105, 140,161, 166 |
| Stearic | 003, 056, 230 | 001, 007, 008, 013, 015, 017, 031, 033, 042, 051, 059, 071, 076, 078, 083, 108, 124, 158, 162, 166, 189, 198, 203, 206, 209, 210, 213, 228, 236, 276 | 070, 105, 140, 161, 165 |
| Oleic | 056, 105, 140, 158, 161 | 007,008, 013, 015, 017, 031, 033, 042, 051, 059, 070, 071, 076, 078, 083, 108, 124, 162, 165, 166, 203, 206, 209, 210, 213, 228, 230, 236, 276 | 001, 003, 189, 198 |
| Linoleic | 008, 189, 198, 236 | 001, 003, 007, 013, 015, 031, 033, 042, 059, 070, 071, 076, 078, 083, 108, 124, 162, 165, 166, 203, 206, 209, 210, 213, 228, 230, 276 | 017, 056, 105, 140, 158,161 |
| Linolenic | 056, 071, 165, 189, 198 | 001, 003, 007, 008, 013, 015, 017, 031, 033, 042, 051, 059, 070, 076, 078, 083, 108, 158, 162, 166, 203, 206, 209, 210, 213, 228, 230, 236, 276 | 105, 124, 140, 161 |
| Oleic/linoleic ratio | 017, 056, 105, 140, 158, 161 | 003, 007, 013, 015, 031, 033, 042, 051, 059, 070, 071, 076, 078, 083, 108, 124, 162, 165, 166, 203, 206, 209, 210, 213, 228, 230, 276 | 001, 008, 189,198, 236 |
| Saponification number | 033, 070, 140, 203, 213 | 001, 003, 007, 008, 013, 015, 031, 042, 051, 059, 071, 076, 078, 108, 124, 158, 161, 162, 165, 198, 206, 209, 210, 228, 230, 236, 276 | 017, 056, 083, 105, 166, 189 |
| Iodine value | 008, 165, 189, 198 | 001, 003, 007, 013, 015, 031, 033, 042, 051, 059, 070, 071, 076, 083, 108, 124, 161, 162, 166, 203, 206, 209, 210, 213, 230, 236, 276 | 017, 056, 078, 105, 140, 158, 228 |
| Cetane number | 017, 056, 078, 105 | 001, 003, 007, 013, 015, 031, 033, 042, 051, 059, 071, 076, 083, 108, 124, 140, 158, 161, 162, 166, 189, 198, 206, 209, 210, 228, 230, 236, 276 | 008, 070, 165, 203, 213 |
Assessment of genetic diversity
In the present study, a total of 38 accessions were genotyped using six TE-AFLP primer combinations (Table 6), which were standardized previously for their polymorphism content (Sharma et al. 2011). The representative TE-AFLP gel profile obtained with primer combination E-AG × P-C is shown in (Fig. 1). Six primer combinations produced a total of 334 bands of which 174 (52.10 %) were polymorphic (Table 6). The maximum genetic similarity (Jc = 0.11) was between accessions P070 and P071, both from South Delhi. In contrast, the minimum genetic similarity (Jc = 0.47) was found between accessions P078 (from Eastern NCR) and P209 (from Kerala). The mean genetic distance among accessions was 0.32, which indicates that high level of genetic diversity exists in the analyzed accessions.
Table 6.
Summary of the number of TE-AFLP fragments scored for different primer pairs used in selective amplification
| Sl no. | Primer combination | Total bands | Polymorphic bands | Polymorphism (%) | PIC | MI | RP |
|---|---|---|---|---|---|---|---|
| 1 | E-AG × P-A | 63 | 36 | 57.14 | 0.33 | 11.99 | 17.69 |
| 2 | E-AG × P-C | 61 | 37 | 60.66 | 0.29 | 10.82 | 16.00 |
| 3 | E-AG × P-G | 39 | 21 | 53.85 | 0.31 | 5.58 | 10.00 |
| 4 | E-AT × P-A | 73 | 31 | 42.47 | 0.34 | 10.41 | 15.34 |
| 5 | E-AC × P-C | 45 | 18 | 40.00 | 0.35 | 6.36 | 9.78 |
| 6 | E-AC × P-A | 53 | 31 | 58.49 | 0.26 | 8.21 | 11.21 |
| Total | 334 | 174 | 52.10 | 0.31 | 8.90 | 13.34 |
PIC polymorphic information content, MI marker index, RP resolving power
Fig. 1.
Representative gel profiles of Pongamia using TE-AFLP primer E-AG × P-C. Lane numbers are the same as the serial numbers of the accessions mentioned in Table 4
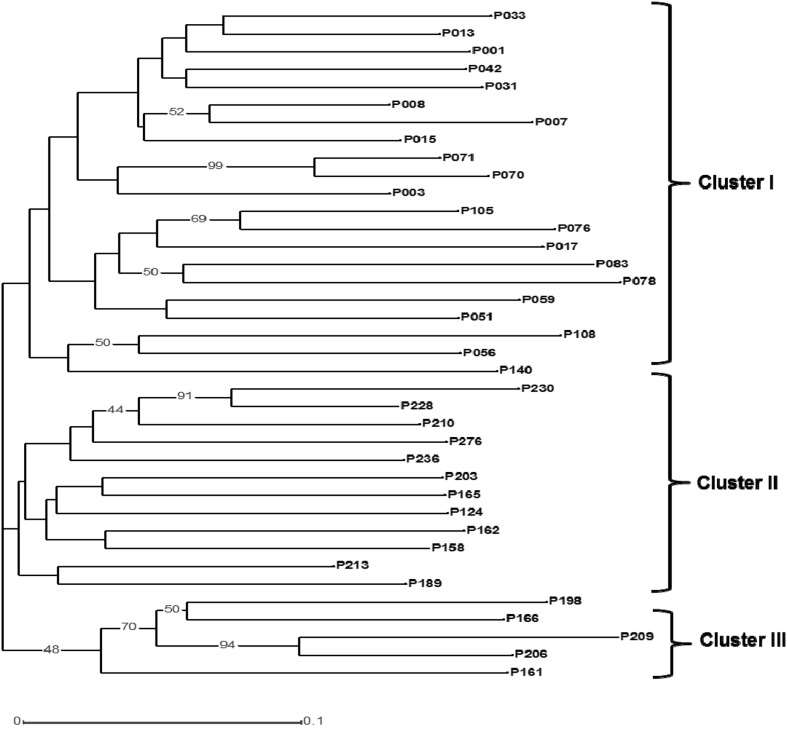
For cluster analysis neighbor joining method was used and all accessions were grouped into three major clusters (Fig. 2). The cluster I grouped all accessions belongs to South Delhi and Eastern NCR, cluster II grouped accessions from East and West Delhi region whereas cluster III includes accessions from both clusters. This indicates that these accessions had significant similarity with accessions within Delhi and NCR used in this study. No significant grouping were observed based on oil content and fatty acid profile. The bootstrap values of all major nodes were higher than 40 % and a majority of them were in the range of 50–99 % indicating robustness of data and clustering results.
Fig. 2.
Dendrogram showing neighbor joining clustering of Pongamia. The numbers on nodes indicate % bootstrap values
Marker attributes
Discriminatory power of each TE-AFLP primers combination was studied by calculated PIC, MI and RP with polymorphic bands only. PIC value ranged from 0.26 (E-AC × P-A) to 0.35 (E-AC × P-C) and MI value from 5.58 (E-AG × P-G) to 11.99 (E-AG × P-A) while RP varied from 9.78 (E-AC × P-C) to 17.69 (E-AG × P-A) (Table 6).
In current years, studies have been directed to assess molecular diversity in P. pinnata using molecular markers such as RAPD, ISSR, AFLP and TE-AFLP (Kesari et al. 2010; Sahoo et al. 2010; Thudi et al. 2010; Sharma et al. 2011, 2014). Both TE-AFLP and AFLP indicated a high level of genetic diversity of P. pinnata collected from different locations of NCT of Delhi, India (Sharma et al. 2011) whilst ISSR indicated narrow genetic diversity within the trees from several regions of Orissa, India (Sahoo et al. 2010). AFLP detected higher levels of genetic diversity (100 %) in natural populations of P. pinnata on contrary the RAPD and ISSR showed lesser genetic diversity (aprox. 10 %) (Kesari et al. 2010). Another study on genetic diversity of Pongamia populations using AFLP markers in 33 CPT was undertaken in five agro-ecological zones of Southern Peninsular India (Pavithra et al. 2014). This study revealed relatively higher levels of gene diversity and high number of unique bands from eastern dry zone of Karnataka and southern dry and transition zone of Karnataka.
In the recent study a total of 38 Pongamia individuals from different locality of NCT of Delhi genotyped using six TE-AFLP primer combinations. Pongamia exhibited very high genetic diversity even within the accessions from Delhi and surrounding regions. All accessions grouped into three major clusters and highest genetic similarity was found in the accessions from South Delhi. It may be noted that this area of National Capital harbours the largest number of Pongamia trees. High diversity in pod and fruit traits in Pongamia from a neighboring area of NCT of Delhi has been reported earlier (Kaushik et al. 2007). A high level of genetic diversity in Pongamia is expected because of its presumed Indian origin. Scott et al. (2008) also suggested that P. pinnata were probably introduced from India to Australia early in human history.
Conclusions
Currently, no elite variety of Pongamia is available. Therefore, this species has high scope of improvement to make it suitable feedstock for biodiesel production through selection of economically important traits such as high seed yield, high oil content and desirable fatty acid composition. The current study showed that Pongamia accession P276 is high in 100 seed weight and seed yield, the accessions P013 and P228 high in oil content, the accessions P001, P003, P189 and P198 high in unsaturated fatty acids and likewise, accessions P008, P070, P165, P203 and P213 have CN values higher than 55. These findings clearly showed that all economically desirable traits are not present in a single genotype of Pongamia. Further, high level of genetic diversity was observed in collected germplasm of Pongamia and therefore there is a scope for its genetic improvement. In future, these identified CPTs could be used as parental material to combine all desirable traits in a single elite variety though selection and tree breeding programme in this species and further can be utilized for large scale multiplication and plantation to produce high quantity and quality biodiesel production.
Acknowledgments
The authors sincerely acknowledge the funding received from the Department of Science and Technology, Government of India, for the project (Grant Number SR/SO/PS-08/2006). Mr. Shyam Sundar Sharma thanks to Council of Scientific and Industrial Research, Government of India, for providing fellowship for this work. We thank Dr. Y. S. Sodhi, The Centre for Genetic Manipulation of crop plants, university of Delhi for their assistance in fatty acid profiling.
References
- Arpiwi NL, Yan G, Barbour EL, Plummer JA. Genetic diversity, seed traits and salinity tolerance of Millettia pinnata (L.) Panigrahi, a biodiesel tree. Genet Res Crop Evol. 2012 [Google Scholar]
- Azam MM, Waris A, Nahar NM. Prospects and potential of fatty acid methyl esters of some non-traditional seed oils for use as biodiesel in India. Biomass Bioenergy. 2005;29:293–302. doi: 10.1016/j.biombioe.2005.05.001. [DOI] [Google Scholar]
- Bala M, Nag TN, Kumar S, Vyas M, Kumar A, Bhogal NS. Proximate composition and fatty acid profile of Pongamia pinnata, a potential biodiesel crop. J Am Oil Chem Soc. 2011;88:559–562. doi: 10.1007/s11746-010-1699-2. [DOI] [Google Scholar]
- BS (2002) Biodiesel Standard ASTM D6751, USA
- Divakara BN, Alur AS, Tripati S. Genetic variability and relationship of pod and seed traits in Pongamia Pinnata (L.) Pierre., a potential agroforestry tree. Int J Plant Prod. 2010;4:131–141. [Google Scholar]
- Elanchezhiyan M, Rajarajan S, Rajendran P, Subramaniyan S, Thyagarajan SP. Antiviral properties of the seed extracts of an Indian medicinal plant, Pongamia pinnata, Linn., against herpes simplex virus: in vitro studies on Vero cells. J Med Microbial. 1993;38:262–264. doi: 10.1099/00222615-38-4-262. [DOI] [PubMed] [Google Scholar]
- Jaccard P. Nouvelles rescherches sur la distribution florale. Bulletin de la Société vaudoise des sciences naturelles. 1908;44:223–270. [Google Scholar]
- Kalayasiri P, Jayashke N, Krisnangkura K. Survey of seed oils for use as diesel fuels. J Am Oil Chem Soc. 1996;73:471–474. doi: 10.1007/BF02523921. [DOI] [Google Scholar]
- Karmee SK, Chadha A. Preparation of biodiesel from crude oil of Pongamia pinnata. Bioresour Technol. 2005;96:1425–1429. doi: 10.1016/j.biortech.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Kaushik N, Kumar S, Kumar K, Beniwal R, Kaushik N, Roy S. Genetic variability and association studies in pod and seed traits of Pongamia pinnata (L.) Pierre in Haryana, India. Genet Res Crop Evol. 2007;54:1827–1832. doi: 10.1007/s10722-006-9204-3. [DOI] [Google Scholar]
- Kesari V, Rangan L. Development of Pongamia pinnata as an alternative biofuel crop—current status and scope of plantations in India. J Crop Sci Biotechnol. 2010;13:127–137. doi: 10.1007/s12892-010-0064-1. [DOI] [Google Scholar]
- Kesari V, Madurai V, Sathyanarayana Parida A, Rangan L. Molecular marker-based characterization in candidate plus trees of Pongamia pinnata, a potential biodiesel legume. AoB Plants. 2010 doi: 10.1093/aobpla/plq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisnangkura K. A simple method for estimation of cetane index of vegetable oil methyl esters. J Am Oil Chem Soc. 1986;63:552–553. doi: 10.1007/BF02645752. [DOI] [Google Scholar]
- Lakshmikanthan V. Tree borne oil seeds. Mumbai: Directorate of non edible oils and soap industries, Khadi and Village Industries Commission; 1978. p. 10. [Google Scholar]
- Martini N, Shell JS, editors. Plant oils as fuels—present state of science and future development. Berlin: Springer; 1998. [Google Scholar]
- Mukta N, Murthy IYLN, Sripal P. Variability assessment in Pongamia pinnata (L.) Pierre germplasm for biodiesel traits. Ind Crops Prod. 2009;29:536–540. doi: 10.1016/j.indcrop.2008.10.002. [DOI] [Google Scholar]
- Naik M, Meher LC, Naik SN, Das LM. Production of biodiesel from high free fatty acid Karanja (Pongamia pinnata) oil. Biomass Bioenergy. 2008;32:354–357. doi: 10.1016/j.biombioe.2007.10.006. [DOI] [Google Scholar]
- Pavela R. Effectiveness of some botanical insecticides against Spodoptera littoralis boisduvala (Lepidoptera: Noctudiae), Myzus persicae Sulzer (Hemiptera: Aphididae) and Tetranychus urticae Koch (Acari: Tetranychidae) Plant Prot Sci. 2009;45:161–167. [Google Scholar]
- Pavithra HR, Gowda B, Prasann KT, Shivanna MV. Pod and seed traits in candidate plus trees of Pongamia pinnata (L.) Pierre from southern peninsular India in relation to provenance variation and genetic variability. J Crop Sci Biotechnol. 2013;16:131–142. doi: 10.1007/s12892-012-0052-8. [DOI] [Google Scholar]
- Pavithra HR, Shivanna MB, Chandrika K, Prasanna KT, Gowda B. Genetic analysis of Pongamia pinnata (L.) Pierre populations using AFLP markers. Tree Genet Genomes. 2014;10:173–188. doi: 10.1007/s11295-013-0674-0. [DOI] [Google Scholar]
- Perrier X, Jacquemoud-Collet JP (2006) DARwin software, Version 5.0.158. http://darwin.cirad.fr/darwin
- Sahoo D, Aparajita S, Rout G. Inter and intra-population variability of Pongamia pinnata: a bioenergy legume tree. Plant Syst Evol. 2010;285:121–125. doi: 10.1007/s00606-009-0254-9. [DOI] [Google Scholar]
- Sarkar AK, Deb DL. Micronutrient cation extracting ability of some improved rice cultivars. J Ind Soc Soil Sci. 1984;32:115–119. [Google Scholar]
- Scott PT, Pregelj L, Chen N, Hadler JS, Djordjevic MA, Gresshoff PM. Pongamia pinnata: an untapped resource for the biofuels industry of the future. Bioenergy Res. 2008;1:2–11. doi: 10.1007/s12155-008-9003-0. [DOI] [Google Scholar]
- Sharma SS, Negi MS, Sinha P, Kumar K, Tripathi SB. Assessment of genetic diversity of biodiesel species Pongamia pinnata accessions using AFLP and three endonuclease-AFLP. Plant Mol Biol Rep. 2011;29:12–18. doi: 10.1007/s11105-010-0204-2. [DOI] [Google Scholar]
- Sharma SS, Aadil K, Negi MS, TripathI SB. Efficacy of two dominant marker systems, ISSR and TE-AFLP for assessment of genetic diversity in biodiesel species Pongamia pinnata. Curr Sci. 2014;106:1576–1580. [Google Scholar]
- Sharma SS, Islam MA, Negi MS, Tripathi SB. Changes in oil content and fatty acid profiles during seed development in Pongamia pinnata (L.) Pierre. Ind J Plant Physiol. 2015 [Google Scholar]
- Shivanna MB, Rajakumar N. Ethno-medico-botanical knowledge of rural folk in Bhadravathi taluk of Shimoga district, Karanataka. Ind J Tradit Knowl. 2010;9:158–162. [Google Scholar]
- Sunil N, Kumar V, Sivaraj N, Lavanya C, Prasad RBN, Rao BVSK, Varaprasad KS. Variability and divergence in Pongamia pinnata (L.) Pierre germplasm—a candidate tree for biodiesel. GCB Bioenergy. 2009;1:382–391. doi: 10.1111/j.1757-1707.2009.01030.x. [DOI] [Google Scholar]
- Tewari DN. Report of the committee on development of Bio-fuel. New Delhi: Planning Commission, Government of India; 2003. [Google Scholar]
- Thies W. Rapid and easy analysis of fatty acid composition in individual rapeseed cotyledons. Z Pflanzenzhchtg. 1971;65:181–202. [Google Scholar]
- Thudi M, Revathi M, Wani SP, Tatikonda L, Hoisington DA, Varshney RK. Analysis of genetic diversity in Pongamia [Pongamia pinnata (L.) Pierre] using AFLP markers. J Plant Biochem Biotechnol. 2010;19:209–216. doi: 10.1007/BF03263342. [DOI] [Google Scholar]
- van der Wurff AWG, Chan YL, van Straalen NM, Schouten J. TE-AFLP: combining rapidity and robustness in DNA fingerprinting. Nucleic Acids Res. 2000;28:e105. doi: 10.1093/nar/28.24.e105. [DOI] [PMC free article] [PubMed] [Google Scholar]