Abstract
Segmentation of the vertebrate head emerges out of earlier processes that establish the anterior-posterior (A-P) axis. Recent genetic studies and comparisons across species have led to a better understanding of the links between A-P patterning and segmentation. These point to similar signals acting on both head and trunk, such as retinoic acid and fibroblast growth factors. These form interacting networks of diffusible morphogen gradients that pattern both hindbrain rhombomeres and mesodermal somites. New computational models, particularly for retinoic acid, have revealed how morphogen gradients are established and made robust to changes in signaling levels. However, the orientations of these gradients, as well as how they interact to generate segments, differ remarkably between germ layers and body regions. Thus, the vertebrate head is, in part, built through modifications of the same processes that link A-P patterning and segmentation in the trunk, but fundamental differences in how these processes are deployed lend further doubt to the notion that head and trunk segments are homologous.
Introduction
The idea that the vertebrate head is fundamentally segmented dates back to the early 19th century with the vertebral theories of Goethe (1820) and others (de Beer 1937; Northcutt 2008). Comparative embryologists emphasized the segmentation of mesoderm and its relationship to the mesodermal somites of the trunk and tail (Balfour 1878; Goodrich 1930). However, the head contains several segmental structures derived largely, or entirely, from ectoderm, namely the rhombomeres of the hindbrain and arches of the pharyngeal apparatus, which are difficult to align with any mesodermal organization. One feature that is shared in common with all of these tissues is an earlier dependence on graded signals that pattern the anterior-posterior (A-P) axis. In this article, I discuss new evidence showing how networks of morphogen gradients are established and deployed in distinct ways in the cranial region.
One main reason for a renewed interest in head segmentation has come from studies on the embryonic hindbrain (Lumsden and Keynes 1989; Fraser et al. 1990; Trevarrow et al. 1990; reviewed by Moens and Prince 2002; Lumsden 2004). These have provided clear evidence for a rhombomeric organization underlying the pattern of cranial nerves, which correlates with the migratory pathways of cranial neural-crest cells that contribute to the pharyngeal arches. In contrast, the spinal cord does not display an obvious, intrinsic, early segmental patterning. Rather, spinal nerve segmentation is dictated by surrounding somites (Keynes and Stern 1984; Eisen and Pike 1991; Ensini et al. 1998). Molecular developmental biology has shown segmental patterns of gene expression in rhombomeres and arches, and requirements for Hox genes in both sets of structures, but these have failed to resolve the issue of the basic segmental plan underlying the organization of the vertebrate head (reviewed by Schilling and Knight 2001). At least two fundamental questions remain: (1) how are A-P patterning and segmentation linked and (2) how do these mechanisms differ between germ layers (i.e., ectoderm and mesoderm) and body regions (i.e., head and trunk)?
A-P patterning of head segments
In vertebrates, regional induction of cell fates along the A-P axis is perhaps best understood for the neural plate. Tissue derived from the dorsal mesoderm of Spemann's organizer in amphibians induces neural identity and also regionally specifies neural tissue as future brain or spinal cord (Mangold 1933). The first signs of segmentation in the brain emerge morphologically soon after gastrulation as a series of bulges along the A-P axis of the neural tube, including 7-8 hindbrain rhombomeres (Fig. 1). Rhombomeres are serially reiterated, each containing the same basic cell types (e.g., commissural interneurons, branchiomotor neurons), but also have distinct identities and contribute to different cranial nerves (Lumsden 2004). In the embryo, each rhombomere also forms a compartment, within which cell lineages are confined and separated by distinct boundary regions (Fraser et al. 1990).
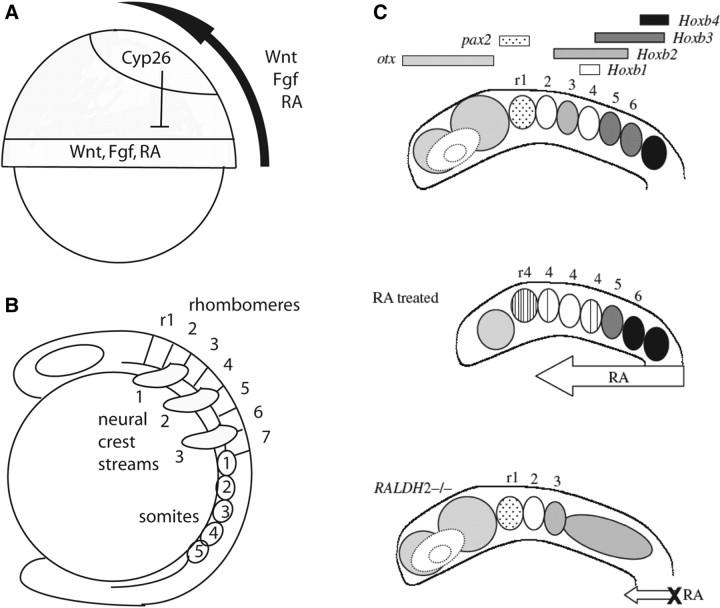
Fig. 1.
A-P patterning signals and hindbrain segmentation. (A) Diagrams of gastrula-stage zebrafish embryos in lateral view, dorsal to the right, illustrating the proposed distribution of RA, Fgf, and Wnt signaling in the neural ectoderm (Kudoh et al. 2002). All three signals are made at the margin of the gastrula, the future posterior, and are thought to diffuse anteriorly to promote posterior development. (B) Segmentally organized structures in vertebrates, including rhombomeres of the hindbrain, streams of migrating cranial neural-crest cells, and mesodermal somites in the trunk, are schematized in a zebrafish at the five-somite stage. (C) Role of RA in A-P patterning of the hindbrain and in Hox regulation (Schilling and Knight 2001). Diagrams illustrate embryonic morphology and patterns of gene expression in the embryonic zebrafish head, lateral views, anterior to the left. Major regions of the brain are indicated, including forebrain, midbrain and rhombomeres (r) 1–7 of the hindbrain. Shaded bars above each diagram indicate the A-P extent of mRNA expression for each gene. Open arrows indicate RA treatments (10−7 M all trans RA) or disruption of RA synthesis (fish mutants or mouse mutants lacking a functional Aldh1a2).
Rhombomere identities are determined, at least in part, by a combinatorial code of trancription factors such as the Hox genes (Fig. 1; reviewed by Moens and Prince 2002; Lumsden 2004). Stripes of rhombomere-specific Hox gene expression domains appear in the neural ectoderm by the end of gastrulation and these are positioned by early signals along the A-P axis (Fig. 1A). Based primarily on studies in fish and amphibians, three signals in particular form an A-P patterning network: Wnt signals initially repress expression of anterior genes, after which Wnts, Fibroblast growth factors (Fgfs) and the vitamin A derivative, retinoic acid (RA), activate posterior genes in a concentration-dependent manner (Cho and De Robertis 1990; Sive et al. 1990; Holowacz and Sokol 1999; Domingos et al. 2001; Kudoh et al. 2002). Fgf and RA, in particular, are good candidates for the molecular link between A-P patterning and segmentation, potentially acting as graded “morphogens” that establish boundaries of segments at distinct concentration thresholds.
Rhombomeric segmentation is also closely tied to the segmental formation and migration of cranial neural-crest cells that contribute to the pharyngeal arches (Fig. 1B). These migrate in three major streams, each with a distinct rhombomeric origin (stream 1, r2–3; stream 2, r4–5; stream 3, r6–7). Transplantations of rhombomeres along the A-P axis in avian embryos, grafted together with adjacent neural-crest cells bound for one of the three streams, lead to arch transformations in which the segmental fates of surrounding tissues are reorganized according to the identities of the transplanted/donor cells (Noden 1983; reviewed by Noden and Trainor, 2005). This has led to the notion that, unlike the trunk in which primary segmentation occurs in the mesodermal somites, head segments are established first in the ectoderm (e.g., hindbrain and neural crest) and secondarily imposed on surrounding mesoderm. Both require RA and Hox genes for proper segmentation and this is thought to coordinate the cranial nerves derived from individual rhombomeres with structures that they innervate within the arches. Because reducing RA signaling leads to expansion of anterior rhombomeres and arches and to loss of posterior ones (Fig. 1C; Niederreither 2000; Begemann et al. 2001; Dupe and Lumsden 2001), the simplest interpretation of these results is that an early gradient of RA coordinates head segments through its roles in patterning the early neural ectoderm.
RA and the morphogen model
Recently, however, the role of RA as a graded morphogen in this system has been under intense debate, particularly in relation to the hindbrain. RA has long been known to specify posterior rhombomere identities, and to regulate many segment-specific target genes directly, including Hox genes. RA is not produced in the neuroepithelium, but is generated by Raldh2 in paraxial mesoderm in the trunk, and degraded by the cytochrome p450 enzyme Cyp26 (retinoic acid-4-hydroxylase) in the anterior neural plate (Fig. 2A) (Niederreither et al. 1997; Swindell et al. 1999). This has led to a model in which RA forms a gradient, declining from its posterior source in mesoderm to a degradation sink anteriorly, thereby patterning rhombomere identities.
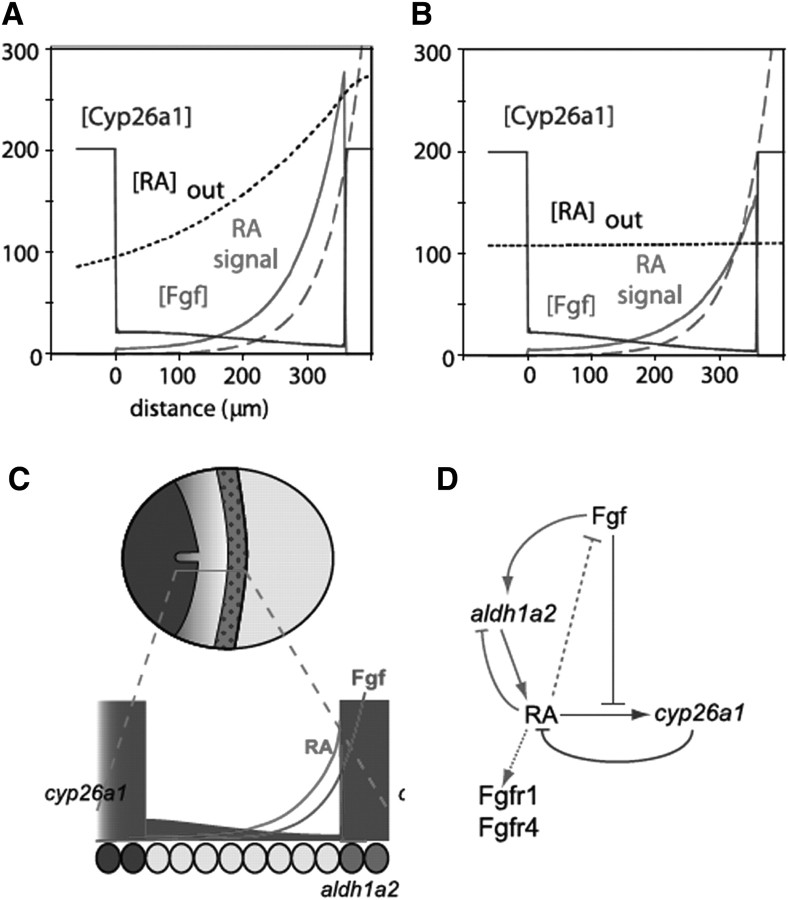
Fig. 2.
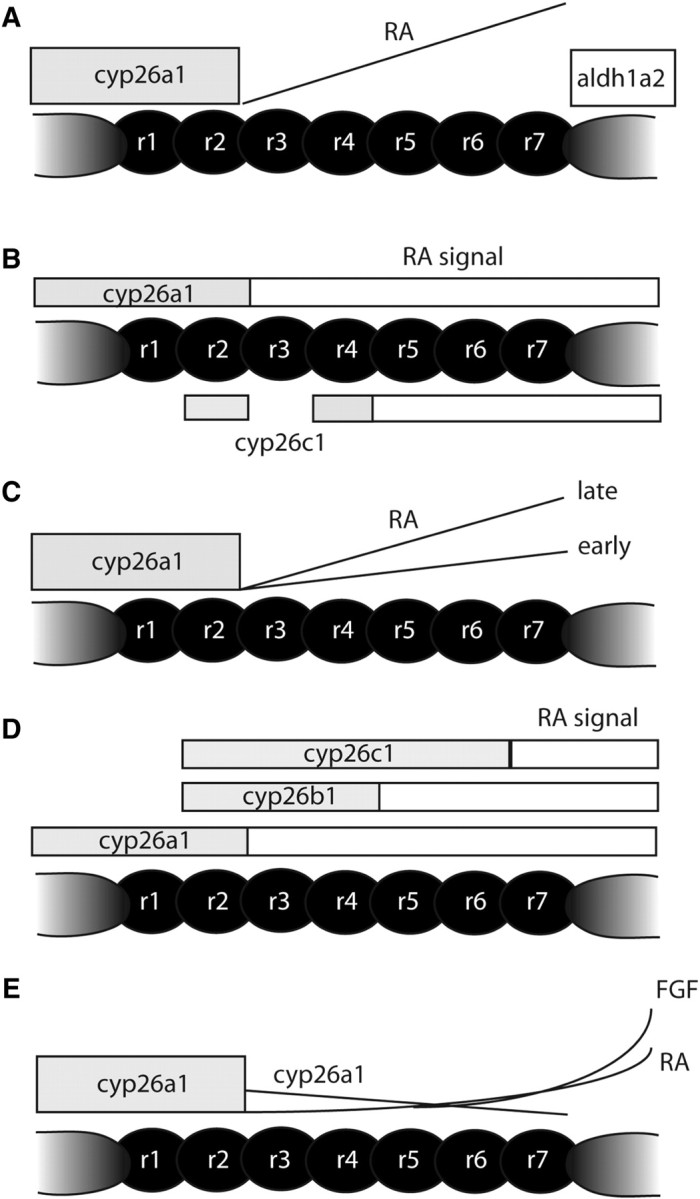
Models for morphogen gradients in the hindbrain. Diagrams illustrating models emerging from recent studies. (A) The classic morphogen model. RA is synthesized posteriorly by Aldh1a2 and degraded anteriorly by Cyp26a1. Diffusion across the presumptive hindbrain creates a gradient that activates distinct target genes in a concentration-dependent manner. (B) A shifting boundaries model (Sirbu et al. 2005). Shifting domains of Cyp26-mediated degradation control the length of exposure to RA. In this model r5–7 are exposed longer than r3/4 because of cyp26c1 expression in the anterior hindbrain after gastrulation. (C) An increasing gradient model (Maves and Kimmel 2005). RA synthesis increases with time, activating posterior genes at higher concentrations and at later stages. (D) A modified shifting boundaries model (Hernandez et al. 2007). In this model, there are three distinct boundaries rather than two. (E) A modified morphogen model (White et al. 2007). Local degradation by Cyp26a1, under the positive control of RA and negative regulation by Fgf, shapes the RA signaling gradient. As the embryo grows, the RA gradient grows without increasing synthesis, integrating both spatial and temporal aspects of hindbrain patterning.
Support for this model came first from treatments of amphibian and avian embryos with exogenous RA, which causes posterior rhombomeres to expand, apparently at the expense of anterior segments, in a concentration-dependent manner (Durston et al. 1989; Sive et al. 1990; Marshall et al. 1992; Godsave et al. 1998). Conversely, reductions in RA signaling cause anterior rhombomeres to expand and replace posterior segments in a concentration-dependent manner (Dupe and Lumsden 2001; Begemann et al. 2004; Maves and Kimmel 2005).
Arguments against the morphogen model have centered around the time-dependent aspects of the formation of rhombomeres. Rhombomeres, like somites, form more or less in an anterior to posterior sequence. Length of exposure to the RA signal may be just as important as its concentration. Timing could be regulated by precise patterns of synthesis and degradation. Consistent with this idea, three Cyp26 enzymes (Cyp26a1, Cyp26b1, and Cyp26c1) are dynamically expressed in the prospective hindbrain and are required for segmentation of the hindbrain (Swindell et al. 1999; Abu-Abed et al. 2001; Sakai et al. 2001; Emoto et al. 2005; Sirbu et al. 2005; Hernandez et al. 2007). Recent studies have argued that “shifting boundaries” of degradation, rather than a gradient of RA, determine the pattern of rhombomeres (Fig. 2B and D). In these models, RA signaling initially extends up to the rhombomere 2/3 (r2/3) boundary, but later becomes restricted posteriorly to the r4/5 boundary due to degradation by Cyp26b1 and Cyp26c1. Posterior rhombomeres are specified by a longer exposure to RA than are more anterior segments, rather than by the concentration of RA.
None of these studies, however, can rule out a concentration-dependent role for RA. Concentration dependence is still the best explanation for the graded expansion or contraction of rhombomeres along the A-P axis with disruptions of RA signaling and for the responses of certain RA-dependent genes (i.e., hoxb1 and vHNF1; Sirbu et al. 2005). Zebrafish or mouse embryos lacking Cyp26c1 also do not show hindbrain defects, arguing against the later steps of degradation in these models (Uehara et al. 2006; Hernandez et al. 2007).
Simple timing of exposure to the RA signal also seems unlikely to play a major role in segmentation of the hindbrain. While Hox genes expressed in the posterior hindbrain are normally expressed later than are more anteriorly-expressed genes, their expression can be induced at earlier stages in zebrafish embryos with high concentrations of RA (Maves and Kimmel 2005). Thus, the thresholds needed to activate posteriorly-expressed genes may only be achieved at later stages, as the RA gradient grows with time, giving the appearance of a time-dependence where it does not exist (Fig. 2C).
If the spatial and temporal activities of RA are controlled by shifting boundaries of Cyp26-mediated degradation, some other A-P patterning information must control the expression of Cyp26 (Sirbu et al. 2005; Hernandez et al. 2007). Hernandez et al. (2007) argued that RA acts permissively, rather than instructively, in hindbrain segmentation and against a gradient model. They base this in part on the phenotypes of Cyp26-deficient zebrafish. Depletion of Cyp26b1 and Cyp26c1 in a Cyp26a1−/−-mutant embryo leads to much more severe posteriorization than does loss of Cyp26a1 alone. However, the argument is also based on the curious fact that exogenous RA, applied uniformly to embryos (fish, frog, bird, or mammal) in an unlocalized manner, can rescue hindbrain defects in an RA-deficient animal. This argues strongly for a permissive role.
Two aspects of the RA signaling pathway, however, have not been taken into account in these models: (1) positive and negative feedback by various RA-induced components within the pathway and (2) their regulation by other signals, such as Fgf and Wnt. We have recently incorporated experiments addressing both of these components into a more computational, systems-level approach to RA signaling in the hindbrain of the zebrafish (Fig. 2E; White et al. 2007). This has led to yet another version of the gradient model, but one that we think can reconcile many features of the models discussed above.
We found that, in contrast to many previous studies, zebrafish cyp26a1 is expressed at low levels within the prospective hindbrain itself during gastrulation, rather than being restricted to presumptive forebrain and midbrain (Fig. 3; White et al. 2007). In addition, we showed that this low-level expression in the hindbrain is RA-dependent. RA induces Cyp26a1 expression, forming a negative feedback loop through inducible degradation that could play major roles in shaping the RA-signaling gradient. Thus, Cyp26 enzymes may not simply act as RA sinks wherever they are expressed, but rather act dynamically to modulate signaling gradients locally and compensate for fluctuations in RA levels. This makes sense for a signal derived from a dietary precursor (vitamin A) that may vary dramatically in its availability to the embryo.
Fig. 3.
The modified morphogen model. A one-dimensional mathematical model of the gastrula-stage zebrafish embryo (White et al. 2007). The model incorporates RA synthesis, diffusion, cell permeability, degradation and signaling, as well as a stable Fgf gradient, and expression of cyp26a1 under the control of RA and Fgf signals. (A and B) Zero on the abscissa denotes the posterior boundary of high cyp26a1 expression near the r1/2 border. Values on the ordinate are arbitrary. Fgf concentration is shown in green, cyp26a1 in blue, extracellular RA ([RA]out) in black, and RA signaling, which is a function of intracellular RA, in red. (A) Typical patterns generated by the model. Low-level cyp26a1 expression declines from anterior to posterior. (B) RA supplied in a delocalized manner can produce a relatively normal morphogen gradient. (C) A diagram illustrating a dorsal view of a gastrulating zebrafish embryo and corresponding gradients, anterior to the left. RA is produced by Aldh1a2 in somitic mesoderm (red) and diffuses through the neural ectoderm. Cyp26a1 (blue) degrades RA at differing rates to produce a gradient that specifies hindbrain fates. (D) Diagram of interactions between RA, Fgf, and Cyp26a1. Only those shown in blue are included in the mathematical model; dotted lines are extrapolated from results in other systems.
In contrast, Cyp26a1 expression (and induction by RA) is inhibited by other posteriorizing signals, such as Fgfs (Kudoh et al. 2002). These form A-P gradients that parallel that of RA and yet appear to have the opposite effects on Cyp26a1. In our model, we propose that this creates an integrated signaling network that is extremely robust—i.e., able to compensate for changes in RA levels (Fig. 3; White et al. 2007). The model helps resolve questions surrounding both spatial/temporal and permissive/instructive aspects of RA signaling.
For instance, previous studies suggested that the RA gradient grows with time, possibly through a gradual increase in RA synthesis (Maves and Kimmel 2005). However, our model points instead to growth of the gradient through a gradual reduction in degradation as inhibition of Cyp26a1 by Fgf decreases during embryonic growth (Fig. 3A). This also seems preferable to a system dependent on precise control of RA synthesis from varying levels of dietary vitamin A. Instead, the RA gradient is tightly coupled to the less environmentally labile Fgf gradient. Our model also provides a very interesting resolution to the dilemma of why exogenous RA, applied uniformly, rescues an RA-deficient embryo (Hernandez et al. 2007). The key is regulated degradation. By having RA induce and Fgf inhibit Cyp26a1 expression, uniform RA is rapidly converted into an intracellular concentration gradient (Fig. 3B).
It is important, however, to note that so far our model has only considered interrelationships between RA and Fgf signaling at early stages of gastrulation, when the synthesis of RA in trunk mesoderm and degradation by Cyp26a1 are only separated by a short distance (100–150 micrometers in zebrafish). Wnt signaling also inhibits Cyp26a1 expression at these early stages and may act upstream to regulate competence to respond to RA and Fgf (Kudoh et al. 2002). In addition, a complex hierarchy of inter-rhombomeric interactions exists in the hindbrain by the end of gastrulation. For example, Fgfs (e.g., Fgf8) are expressed at the midbrain-hindbrain boundary and in r4, where in zebrafish they are required for patterning neighboring segments (Maves et al. 2002). Cyp26b1 and Cyp26c1 are expressed in different subsets of rhombomeres, in species-specific patterns, where they very likely protect cells from excess RA (Swindell et al. 1999; Sirbu et al. 2005; Hernandez et al. 2007). These may have important roles in modulating the later influences of RA on neurogenesis in the hindbrain, and this is an important topic for future research.
A reversal of the orientation of morphogen gradients in the head and trunk
While the hindbrain subdivides into rhombomeres, the spinal cord remains unsegmented. However, both are patterned along the A-P axis by Hox genes. Fgf and RA also influence Hox gene expression and neurogenesis in the prospective spinal cord, as well as segmentation of the paraxial mesoderm into somites, suggesting that common signals control A-P patterning in head and trunk. Unlike the hindbrain, however, the gradients of RA and Fgf oppose one another in the trunk after gastrulation, with RA levels highest in the most anterior somites and Fgfs highest posteriorly (Fig. 4). This difference in the spatial deployment of at least one morphogen, RA, suggests a fundamental difference in the mechanisms by which these signals establish segmentation in the head and trunk.
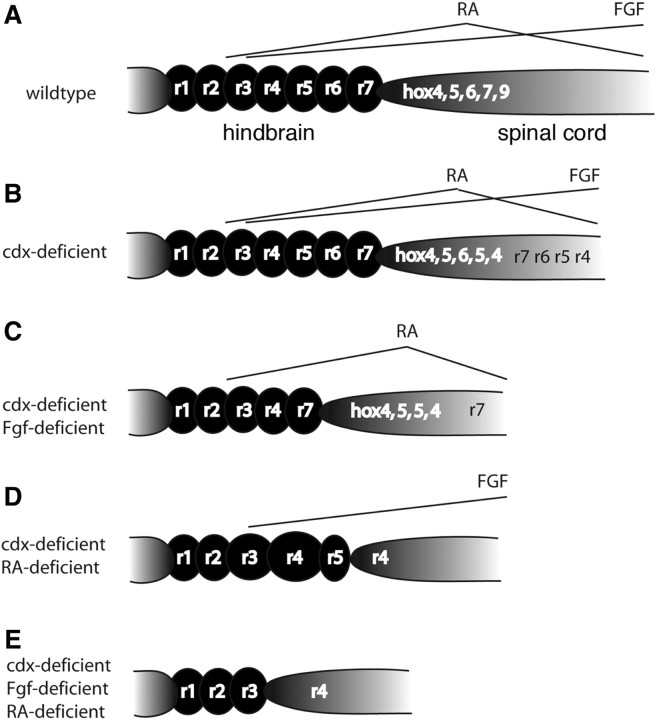
Fig. 4.
Distinct gradients in the head and trunk and the roles of Cdx genes. Diagrams illustrating neural patterning in Cdx-deficient embryos in the presence or absence of Fgf and RA signaling (Shimizu et al. 2006; Skromne et al. 2007). Anterior to the left, Rhombomere-specific domains (r1-7) are indicated. The A-P order of hox gene expression in the spinal cord is also indicated. (A) Wild-type. (B) Cdx-deficient. (C) Cdx-deficient and Fgf-deficient. (D) Cdx-deficient and RA-deficient. (E) Cd-deficient, Fgf-deficient, and RA-deficient.
Are apparent differences in A-P patterning between hindbrain and spinal cord due to some intrinsic difference in their ability to respond to the same signals? Two recent studies in zebrafish have shown that the spinal cord is competent to acquire a hindbrain identity.
Competence is under the control of the Cdx genes, homeobox-containing members of the ParaHox clusters, closely related to Drosophila caudal. Cdx genes suppress anterior development and promote posterior development both in vertebrates and invertebrates (Lohnes 2003; Deschamps and van Nes 2005). In mice, Cdx genes help integrate RA, Fgf and Wnt signals to regulate patterns of Hox expression in the paraxial mesoderm. Cdx proteins bind to cis-regulatory elements of posterior hox genes and directly activate their transcription (Subramanian et al. 1995; Pownall et al. 1996; Charite et al. 1998; Isaacs et al. 1998). cdx4-/--mutant zebrafish have reductions in posterior hox gene expression (Davidson et al. 2003; Hammerschmidt et al. 1996) and combined depletion of cdx4 and cdx1a causes loss of hoxb7a and hoxb9a expression and severe posterior truncations (Davidson and Zon 2006; Shimizu et al. 2005).
Surprisingly, loss of Cdx function in these experiments on zebrafish also leads to ectopic hindbrain tissue in the spinal cord (Fig. 4; Shimizu et al. 2006; Skromne et al. 2007). Numerous hindbrain-specific hox genes, as well as differentiated branchiomotor neurons, are detected in the trunks and tails of Cdx-deficient embryos. Furthermore, genes marking different rhombomeres are spatially localized within this ectopic hindbrain tissue in a reversed orientation; anterior markers such as hoxb4 are expressed further posteriorly than is hoxb6 (Fig. 4B). The authors interpret this to mean that the Cdx genes control the responsiveness of spinal-cord cells to RA, and that RA gradients are reversed between the hindbrain and spinal cord, with the pivot point occurring somewhere around the level of the third somite. These studies also demonstrate that, at least within the neural ectoderm, cells are competent to acquire hindbrain fates and possibly to become organized into rhombomeres all along the A-P axis.
Consistent with the hypothesis that Cdx genes modulate the responses of cells to posteriorizing signals, ectopic hindbrain tissue in Cdx-deficient embryos is suppressed by inhibiting RA or Fgf (Shimizu et al. 2006; Skromne et al. 2007) (Fig. 4C–E). Embryos deficient for both Cdx1a/4 and Fgf signaling still form mirror-image duplicates of hindbrain tissue in the spinal cord, but these duplicates only retain markers of more posterior rhombomeres and appear to lack r4-6 (Fig. 4C). Likewise, Cdx-deficient embryos with severely reduced RA signaling only show duplications of r4 and r5 (Fig. 4D). Without either RA or Fgf signaling, Cdx-deficient embryos fail to form any duplicate tissue and lack posterior rhombomeres altogether (Fig. 4E).
Cdx-Hox functions also permit both RA and Fgf signaling pathways to operate. They are required autonomously (within the cells that express them) for Hox expression, and restoration of Hox expression in Cdx-deficient zebrafish embryos can at least partially rescue Krox20 expression (Shimizu et al. 2006). Like Fgfs, Cdx proteins are also negative regulators of Cyp26a1 (Wingert et al. 2007). In addition, Cdx transcription factors regulate, to some extent, the formation of the mesodermal sources of RA and Fgfs. For example, Raldh2 expression in the paraxial mesoderm shifts posteriorly in Cdx-deficient embryos, possibly altering the spatial relationships between RA and Fgf gradients such that both are high posteriorly.
Mesodermal segmentation
Strikingly, the same mirror-image reversal in the orientation of the RA gradient between the head and trunk is also a crucial feature of recent models of somite formation and segmentation of the paraxial mesoderm in the trunk and tail (Fig. 5). Somites and their derivatives (e.g., vertebrae, axial muscles) are the most clearly segmented portions of the vertebrate body. Somite formation depends on a “clock and wavefront” mechanism involving oscillating patterns of gene expression that move progressively toward the posterior end. An oscillating signal combines with a moving wavefront from anterior to posterior that sets the boundaries of somites within the presomitic mesoderm. A working model at the molecular level suggests that the clock involves Notch signaling, the wavefront involves RA and Fgf signaling, and the two are interconnected through cycles of Wnt signaling (Palmeirim et al. 1997; Dequeante et al. 2006; reviewed by Andrade et al. 2005; Dequeante and Pourquie 2008). The discovery of this precise control of the timing of gene expression has revolutionized our understanding of the mechanisms underlying mesodermal segmentation. Can similar rules be applied to head segments?
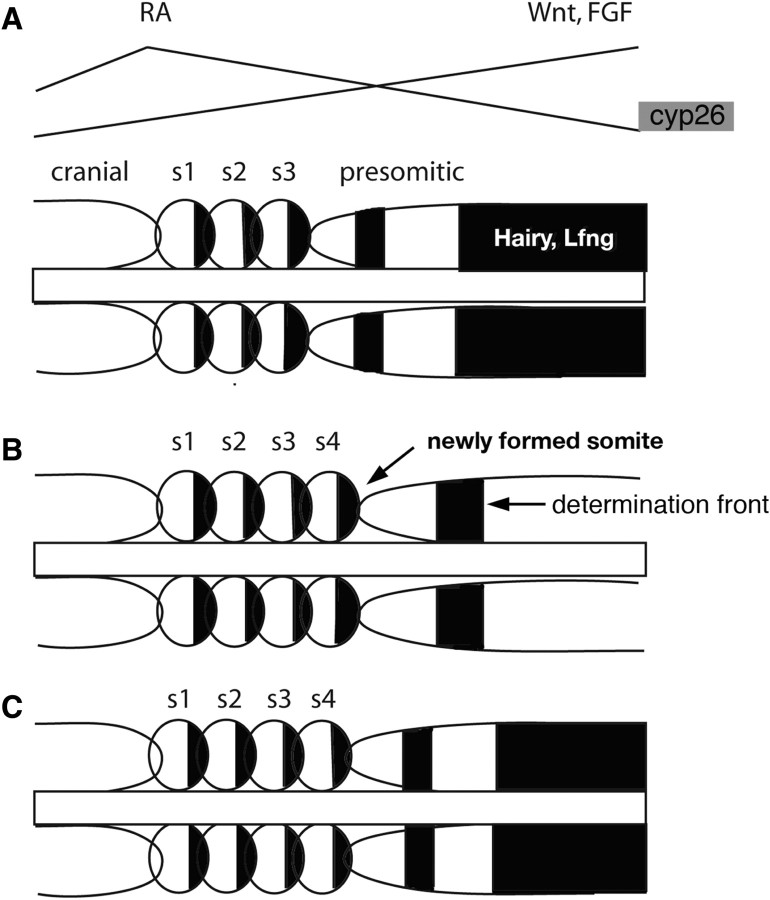
Fig. 5.
Gene oscillations and mesodermal segmentation. Diagrams illustrating gene oscillations involved in mesodermal patterning and in the clock and wavefront model for somite formation. (B) Cranial mesoderm shows no such gene oscillations or molecular domains corresponding to early head segments, but becomes secondarily segmented within the pharyngeal arches through influences from surrounding neural crest and endoderm.
At first glance, many of the signals involved are the same. However, unlike the hindbrain, RA and Fgf in the presomitic mesoderm are thought to form opposing gradients, with RA highest anteriorly and Fgf posteriorly (Fig. 5; Goldbeter et al. 2007). This parallels their orientations in the spinal cord, as revealed by the studies of Cdx genes (Fig. 4), and reflects the fact that after gastrulation most of the RA in the embryo is synthesized in anterior somites near the head-trunk boundary. An elegant computational model incorporating these components of the wavefront argues that a delicate balance between levels of these signals, where their morphogen gradients intersect, determines the boundaries of somites (Fig. 5; Goldbeter et al. 2007). Mutually inhibitory gradients, in this model, generate sharp morphogen thresholds through bistability at two steady states, and in this way synchronize activation of segmentation genes along a determination front. Interestingly, a second domain of Cyp26a1 expression at the posterior end of the embryo forms a mirror-image sink to complement the anterior source of RA in the somites, and future studies are needed to determine the roles of degradation in the context of this system.
All of this new information on the mechanisms of mesodermal segmentation in the trunk also has important implications for understanding the relationship between head and trunk segmentation. Comparative embryologists have long wondered if the equivalent of mesodermal somites and their segmental derivatives extend into the cranial region (Goethe 1820; Balfour 1876; Kuratani and Schilling 2008). We can now ask if similar cycles of gene expression extend into the cranial mesoderm. For the best-studied elements of this system, such as Hairy, Lunatic fringe, and other components of Notch signaling, the answer appears to be “no” (Fig. 5). If any cycling occurs in presumptive head mesoderm, it occurs during gastrulation, prior to formation of the first somite. Only two pulses of Hairy or Lfng expression have been observed in avian embryos at these early stages, which are insufficient to account for the full set of head segments (Jouve et al. 2002). Head segments may form independently of the segmentation clock or a similar clock may act through a distinct set of cycling genes.
Cranial mesoderm also shows no obvious morphological signs of segmentation. As discussed extensively at our symposium, a lack of any molecular evidence for reiterated patterning within this tissue has led most researchers to the conclusion that cranial “somitomeres” do not exist (Kuratani and Schilling 2008). Instead, cranial mesoderm appears to become secondarily segmented within the pharyngeal arches through influences from surrounding neural crest and endoderm (Noden and Trainor 2005). Thus, consistent with the “new head” hypothesis (Northcutt 2008), the cranial mesoderm does not appear to be built simply through modifications of the same processes that pattern segments in the trunk (i.e., somites).
Conclusions and perspectives—evolution of systems of A-P patterning
Given all of this new information, how do we envisage the link between A-P patterning and head segmentation and how do these mechanisms differ between germ layers and body regions? One major take-home message of this work, and of the entire Head Segmentation symposium at the SICB Meeting in January 2008, is that it seems unlikely that a single global patterning system controls all types of segments. Ectoderm differs in fundamental ways from mesoderm, and both germ layers become segmented quite differently in the head, compared with more posterior body regions. Part of this difference appears to lie in the links between early A-P patterning and the formation of segments.
In the cranial region, most of the evidence points to a primary role for the ectoderm, including the hindbrain and neural crest, in establishing segments. These seem to impart patterning on an otherwise unsegmented mesoderm during formation of the pharyngeal arches. Pharyngeal endoderm also segments into a series of pouches, which form the boundaries between adjacent arches, and less is known about the mechanisms that underlie pouch formation. Pouches do form in the absence of cranial neural crest (Veitch et al. 1999) but also require interactions with adjacent mesoderm and with both RA and Fgf signaling. Cdx genes also specify a posterior domain within the endoderm that alters its responsiveness to RA signaling, similar to their roles in the nervous system (Kinkel et al. 2008). Future studies are needed to determine how segmentation in the endodermal germ layer is coordinated with that of other germ layers, and whether or not the endoderm is subjected to similar morphogen gradients during its early development.
Much of the distinction between mechanisms of A-P patterning in the head and trunk centers around the role of RA. This is interesting from a comparative perspective, since RA seems to have acquired roles in A-P patterning in deuterostomes (Schilling and Knight 2001). Protostomes (e.g., nematodes and arthropods) lack both RARs and the enzymes that synthesize RA (Marletaz et al. 2006). In contrast, Retinoid X receptors (RXRs) and cytochrome p450 enzymes similar to Cyp26s are found throughout the animal kingdom (Niwa et al. 2004; Szanto et al. 2004; Bogwitz et al. 2005; King-Jones et al. 2006). Our model for regulation of the degradation of RA in the hindbrain suggests that an RA gradient system could have evolved simply by bringing one of these degrading enzymes under the control of an ancestral A-P patterning system, so that its expression in embryos became graded from posterior to anterior. This would automatically form a gradient of any substrate creating a primitive RA-like gradient system that could later be refined by localizing the storage and synthesis of precursors. Gradients of nuclear hormone–receptor ligands, together with the feedback loops discussed here, may have distinct advantages for forming sharp boundaries such as those between hindbrain rhombomeres or mesodermal somites (Kerszberg 1996). Likewise, the ability to evolve gradients readily through changes in localized degradation could help explain many of the perceived differences between the head and trunk segments of vertebrates.
References
- Abu-Abed S, Dolle P, Metzger D, Beckett B, Chambon P, Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15:226–40. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade RP, Pascoal S, Palmeirim I. Thinking clockwise. Brain Res Rev. 2005;49:114–19. doi: 10.1016/j.brainresrev.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Balfour FM. The development of the elasmobranchial fishes. J Anat Physiol. 1878;11:405–706. [Google Scholar]
- Begemann G, Marx M, Mebus K, Meyer A, Bastmeyer M. Beyond the neckless phenotype: influence of reduced retinoic acid signaling on motor neuron development in the zebrafish hindbrain. Dev Biol. 2004;271:119–29. doi: 10.1016/j.ydbio.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–94. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Bogwitz MR, Chung H, Magoc L, Rigby S, Wong W, O'Keefe M, Batterham P, Daborn PJ. Cyp12a4 confers lufenuron resistance in a natural population of Drosophila melanogaster. Proc Natl Acad Sci USA. 2005;102:12807–12. doi: 10.1073/pnas.0503709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charite J, de Graaff W, Rossant J, Deschamps J, Beck F. Transducing positional information to the Hox genes: critical interaction of Cdx gene products with position-sensitive regulatory elements. Development. 1998;125:4349–58. doi: 10.1242/dev.125.22.4349. [DOI] [PubMed] [Google Scholar]
- Cho KW, De Robertis EM. Differential activation of Xenopus homeobox genes by mesoderm-inducing factors and retinoic acid. Genes Dev. 1990;4:1910–6. doi: 10.1101/gad.4.11.1910. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Ernst P, Wang Y, Dekens MP, Kingsley PD, Palis J, Korsmeyer SJ, Daley GQ, Zon LI. Cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425:300–6. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Zon LI. The caudal-related homeobox genes cdx1a and cdx4 act redundantly to regulate hox expression and the formation of putative hematopoietic stem cells during zebrafish embryogenesis. Dev Biol. 2006;292:506–18. doi: 10.1016/j.ydbio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- de Beer GR. The development of the vertebrate skull. London: Oxford University Press; 1937. [Google Scholar]
- Dequeant M-L, Glynn E, Gaudenz K, Wahl M, Chen J, Mushegian A, Pourquie O. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–8. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- Dequeant M-L, Pourquie O. Segmental patterning of the vertebrate embryonic axis. Nat Rev Genet. 2008;9:370–82. doi: 10.1038/nrg2320. [DOI] [PubMed] [Google Scholar]
- Domingos PM, Itasaki N, Jones CM, Mercurio S, Sargent MG, Smith JC, Krumlauf R. The Wnt/beta-catenin pathway posteriorizes neural tissue in Xenopus by an indirect mechanism requiring Fgf signaling. Dev Biol. 2001;239:148–60. doi: 10.1006/dbio.2001.0431. [DOI] [PubMed] [Google Scholar]
- Dupe V, Lumsden A. Hindbrain patterning involves graded response to retinoic acid signaling. Development. 2001;128:2199–208. doi: 10.1242/dev.128.12.2199. [DOI] [PubMed] [Google Scholar]
- Durston AJ, Timmermans JP, Hage WJ, Hendriks HF, de Vries NJ, Hiedeveld M, Niewkoop PD. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature. 1989;340:140–4. doi: 10.1038/340140a0. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Pike SH. The spt-1 mutation alters segmental arrangement and axonal development of identified neurons in the spinal cord of the embryonic zebrafish. Neuron. 1991;6:767–76. doi: 10.1016/0896-6273(91)90173-w. [DOI] [PubMed] [Google Scholar]
- Emoto Y, Wada H, Okamoto H, Kudo A, Imai Y. Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish. Dev Biol. 2005;278:415–27. doi: 10.1016/j.ydbio.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Ensini M, Tsuchida TN, Belting HG, Jessell TM. The control of rostrocaudal pattern in the developing spinal cord: specification of motor neuron subtype identity is initiated by signals from paraxial mesoderm. Development. 1998;125:969–82. doi: 10.1242/dev.125.6.969. [DOI] [PubMed] [Google Scholar]
- Fraser S, Keynes R, Lumsden A. Segmentation in the chick embryo hindbrain is defined by cell lineage restrictions. Nature. 1990;344:431–5. doi: 10.1038/344431a0. [DOI] [PubMed] [Google Scholar]
- Furlong RF, Younger R, Kasahara M, Reinhardt R, Thorndyke M, Holland PW. A degenerate ParaHox gene cluster in a degenerate vertebrate. Mol Biol Evol. 2007;24:2681. doi: 10.1093/molbev/msm194. [DOI] [PubMed] [Google Scholar]
- Godsave SF, Koster CH, Getahun A, Mathu M, Hooiveld M, van der Wees J, Hendriks J, Durston AJ. Graded retinoid responses in the developing hindbrain. Dev Dyn. 1998;213:39–49. doi: 10.1002/(SICI)1097-0177(199809)213:1<39::AID-AJA4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Goethe JW. Stuttgart and Tubingen (Germany) 1820. Zur Naturwissenschaften uberhaupt, besonders zur Morphologie. I, J.G. Cotta. [Google Scholar]
- Goldbeter A, Gonze D, Pourquie O. Sharp developmental thresholds defined through bistability by antagonistic gradients of retinoic acid and FGF signaling. Dev Dyn. 2007;236:1495–508. doi: 10.1002/dvdy.21193. [DOI] [PubMed] [Google Scholar]
- Goodrich ES. Studies on the structure and development of vertebrates. London: MacMillan; 1930. [Google Scholar]
- Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;1134:177–87. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowacz T, Sokol S. FGF is required for posterior neural patterning but not for neural induction. Dev Biol. 1999;205:296–308. doi: 10.1006/dbio.1998.9108. [DOI] [PubMed] [Google Scholar]
- Isaacs HV, Pownall ME, Slack JM. Regulation of hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J. 1998;17:3413–27. doi: 10.1093/emboj/17.12.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouve C, Iimura T, Pourquie O. Onset of the segmentation clock in the chick embryo: evidence for oscillations in the somite precursors in the primitive streak. Development. 2002;129:1107–17. doi: 10.1242/dev.129.5.1107. [DOI] [PubMed] [Google Scholar]
- Keenan ID, Sharrard RM, Isaacs HV. FGF signal transduction and the regulation of Cdx gene expression. Dev Biol. 2006;299:478–88. doi: 10.1016/j.ydbio.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Kerszberg M. Accurate reading of morphogen concentrations by nuclear receptors: a formal model of complex transduction pathways. J Theor Biol. 1996;183:95–104. doi: 10.1006/jtbi.1996.0205. [DOI] [PubMed] [Google Scholar]
- Keynes RJ, Stern CD. Segmentation in the vertebrate nervous system. Nature. 1984;310:786–9. doi: 10.1038/310786a0. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Kinkel MD, Eames SC, Alonzo MR, Prince VE. Cdx4 is required in the endoderm to localize the pancreas and limit beta-cell number. Development. 2008;135:919–29. doi: 10.1242/dev.010660. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–46. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- Kuratani S, Schilling TF. Head segmentation in vertebrates. Integ Comp Biol. 2008 doi: 10.1093/icb/icn036. [Epub ahead of print; doi: 10.1093/icb/icn036] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden A. Segmentation and compartition in the early avian hindbrain. Mech Dev. 2004;121:1081–8. doi: 10.1016/j.mod.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Keynes R. Segmental patterns of neuronal development in the chick hindbrain. Nature. 1989;337:424–8. doi: 10.1038/337424a0. [DOI] [PubMed] [Google Scholar]
- Lumsden A. Segmentation and compartition in the early avian hindbrain. Mech Dev. 2004;121:1081–8. doi: 10.1016/j.mod.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Mangold O. Uber die Induktionsfahigkeit der verschiedenen Bezirke der Neurula von Urodelen. Naturwissenschaften. 1933;21:761–6. [Google Scholar]
- Marletaz F, Holland LZ, Laudet V, Schubert M. Retinoic acid signaling and the evolution of chordates. Int J Biol Sci. 2006;2:38–47. doi: 10.7150/ijbs.2.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall H, Nonchev S, Sham MH, Muchamore I, Lumsden A, Krumlauf R. Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature. 1992;360:737–41. doi: 10.1038/360737a0. [DOI] [PubMed] [Google Scholar]
- Maves L, Jackman W, Kimmel CB. Fgf3 and Fgf8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–37. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- Maves L, Kimmel CB. Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev Biol. 2005;285:593–605. doi: 10.1016/j.ydbio.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Moens CB, Prince VE. Constructing the hindbrain: insights from the zebrafish. Dev Dyn. 2002;224:1–17. doi: 10.1002/dvdy.10086. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dolle P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127:75–85. doi: 10.1242/dev.127.1.75. [DOI] [PubMed] [Google Scholar]
- Niwa R, Matsuda T, Yoshiyama T, Namiki T, Mita K, Fujimoto Y, Kataoka H. CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J Biol Chem. 2004;279:35942–9. doi: 10.1074/jbc.M404514200. [DOI] [PubMed] [Google Scholar]
- Noden DM. The role of neural crest in patterning of avian cranial skeletal, connective and muscle tissues. Dev Biol. 1983;96:144–65. doi: 10.1016/0012-1606(83)90318-4. [DOI] [PubMed] [Google Scholar]
- Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt RG. Historical hypotheses regarding segmentation of the vertebrate head. Integ Comp Biol. 2008 doi: 10.1093/icb/icn065. [Epub ahead of print; doi: 10.1093/icb/icn065] [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O. Avian hair gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–48. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Tucker AS, Slack JM, Isaacs HV. eFGF, Xcad3 and HOx genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development. 1996;122:3881–92. doi: 10.1242/dev.122.12.3881. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the antero-posterior axis within the mouse embryo. Genes Dev. 2001;115:213–25. doi: 10.1101/gad.851501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, Knight RD. Origins of anteroposterior patterning and Hox gene regulation during chordate evolution. Phil Trans R Soc Lond B Biol Sci. 2001;356:1599–613. doi: 10.1098/rstb.2001.0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Hibi M. Cdx-Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue. Development. 2006;133:4709–19. doi: 10.1242/dev.02660. [DOI] [PubMed] [Google Scholar]
- Sirbu IO, Gresh L, Barra J, Duester G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development. 2005;131:2653–67. doi: 10.1242/dev.01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Draper BW, Harland RM, Weintraub H. Identification of a retinoic acid-sensitive period during primary axis formation in Xenopus laevis. Genes Dev. 1990;4:932–42. doi: 10.1101/gad.4.6.932. [DOI] [PubMed] [Google Scholar]
- Skromne I, Thorsen D, Hale M, Prince VE, Ho RK. Repression of the hindbrain developmental program by Cdx factors is required for the specification of the vertebrate spinal cord. Development. 2007;134:2147–58. doi: 10.1242/dev.002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V, Meyer BI, Gruss P. Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell. 1995;83:641–53. doi: 10.1016/0092-8674(95)90104-3. [DOI] [PubMed] [Google Scholar]
- Swindell EC, Thaller C, Sockanathan S, Petkovich M, Jessell TM, Eichele G. Complementary domains of retinoic acid production and degradation in the early chick embryo. Dev Biol. 1999;216:282–96. doi: 10.1006/dbio.1999.9487. [DOI] [PubMed] [Google Scholar]
- Szanto A, Narkar V, Shen Q, Uray IP, Davies PJA, Nagy L. Retinoid X receptors: X-ploring their (patho)physiological functions. Cell Death Differ. 2004;11(Suppl 2):S126–43. doi: 10.1038/sj.cdd.4401533. [DOI] [PubMed] [Google Scholar]
- Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–79. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- Uehara M, Yashiro K, Mamiya S, Nishino J, Chambon P, Dolle P, Sakai Y. CYP26A1 and CYP26C1 cooperatively regulate anterior-posterior patterning of the developing brain and the production of migratory cranial neural crest cells in the mouse. Dev Biol. 2006;302:399–411. doi: 10.1016/j.ydbio.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Veitch E, Begbie J, Schilling TF, Smith MM, Graham A. Pharyngeal arch patterning in the absence of neural crest. Curr Biol. 1999;9:1481–4. doi: 10.1016/s0960-9822(00)80118-9. [DOI] [PubMed] [Google Scholar]
- White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, et al. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3:1922–38. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]