Abstract
While the core subunits of Polycomb group (PcG) complexes are well characterized, little is known about the dynamics of these protein complexes during cellular differentiation. We used quantitative interaction proteomics and genome-wide profiling to study PcG proteins in mouse embryonic stem cells (ESCs) and neural progenitor cells (NPCs). We found the stoichiometry and genome-wide binding of PRC1 and PRC2 to be highly dynamic during neural differentiation. Intriguingly, we observed a downregulation and loss of PRC2 from H3K27me3-marked chromatin during differentiation, whereas PRC1 was retained at these sites. Additionally, we found PRC1 at enhancer and promoter regions independent of PRC2 binding and H3K27me3. Finally, overexpression of NPC-specific PRC1 interactors in ESCs led to increased Ring1b binding to and decreased expression of NPC-enriched Ring1b target genes. In summary, our integrative analyses have uncovered dynamic PcG subcomplexes and widespread co-localization with active chromatin marks during differentiation.
Keywords: label-free quantitative proteomics, PR-DUB, cross-linking mass spectrometry, interactomes
Polycomb group proteins are an evolutionary conserved family of protein complexes that are important for cellular differentiation and development1. They play a key role in regulating the expression of Hox gene clusters, which control body segmentation patterns2. Polycomb proteins were first discovered and characterized in Drosophila and were later identified in higher eukaryotes3. Polycomb proteins are known to assemble into three major protein complexes: Polycomb repressive complexes 1 and 2 (PRC1 and PRC2), and the Polycomb repressive deubiquitinase (PR-DUB)4. PRC1 contains a catalytic subunit, Ring1a or b, which are ubiquitin ligases for histone H2A lysine 1195,6. The PRC2 subunits Ezh1 and 2 mono-, di-, and trimethylates histone H3 lysine 277,8. PR-DUB contains a deubiquitinase (BAP1, known as Calypso in Drosophila) for H2A lysine 119 (118 in Drosophila)9. Each of these three protein complexes contain additional subunits and several paralogous subunits define distinct subcomplexes10. Furthermore, a number of presumably substoichiometric interactors have been described11,12. In general, Polycomb proteins are associated with gene repression, although recent reports have also hinted towards a role in gene activation13–16.
Polycomb protein complexes are known to functionally cooperate to transcriptionally silence genes. For example, the histone H3K27me3 modification that is catalyzed by PRC2 can be recognized by the Cbx subunits of PRC117,18. Furthermore, histone H2AK119Ub can serve as a binding scaffold for PRC2 in vivo19 and stimulates the catalytic activity of PRC2 on nucleosomes in vitro20. However, recent evidence suggests that H2AK119Ub is not strictly required for transcriptional repression by PRC1, at least during certain stages of development21,22. Nevertheless, it is believed that the enzymatic activities and cross-talk between PRC1 and PRC2 result in the generation of large domains of H3K27me3- and H2AK119Ub-marked transcriptionally silent chromatin.
During the last decade, genetic, biochemical, and mass spectrometry-based approaches from a number of laboratories have elucidated the subunits and interactors of Polycomb complexes in a variety of species. However, systematic, quantitative, and integrative studies are required to understand the dynamics and function of Polycomb protein complexes during cellular differentiation. Here, we set out to characterize the subunit composition, stoichiometry, and architecture of Polycomb complexes in mouse embryonic stem cells and neural progenitor cells using label-free quantitative mass spectrometry-based proteomics. We complemented this work by profiling genome-wide binding of PRC1 and PRC2 in the same cell types. Our work revealed striking dynamics of Polycomb complexes during differentiation, both at the proteomic and genomic level. For PRC2, we identified both stem cell and neural specific interactors and we observed a global downregulation of PRC2 during differentiation despite maintenance of global H3K27me3 levels. We also observed a switch between variant PRC1 and canonical PRC1 during differentiation, which is accompanied by a drastic change in genome-wide Ring1b binding. In addition to ‘classical’ H3K27me3-marked repressed Polycomb targets, we also observe PRC1 binding at active promoters and enhancers which are enriched for neural functions. Strikingly, these genes do not contain PRC2, thus revealing a partial decoupling between PRC1 and PRC2 during cellular differentiation. Finally, overexpression of NPC-specific Ring1b interactors in ESCs affected the genomic localization of Ring1b and expression of Ring1b target genes, indicating that the observed subunit dynamics at least partially drive Polycomb target gene binding.
Results
Identification of dynamically incorporated PRC2 subunits
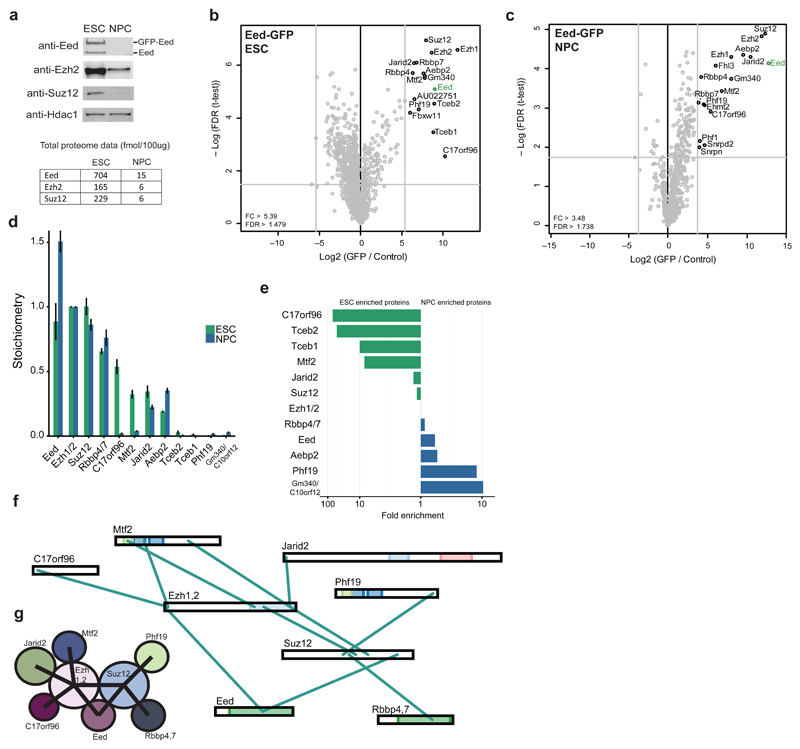
To explore the subunit composition and dynamics of PcG complexes during mouse embryonic stem cell differentiation, we utilized stably integrated bacterial artificial chromosome (BAC) transgenes containing the endogenous promoter of the gene of interest as well as a C-terminal GFP tag as described previously23. For PRC2, we generated an Eed BAC line in mouse embryonic stem cells (ESCs), which was then differentiated into neural progenitor cells (NPCs). Western blotting revealed nearly endogenous expression levels of GFP-Eed in both cell types (Fig. 1a). We also observed extreme downregulation of core PRC2 members during differentiation to NPCs, both at the protein and transcript level (Fig. 1a and Supplementary Fig. 1a). The Western blot data is in agreement with previously obtained total proteome data24, which also shows a strong reduction in PRC2 core subunit abundance levels (Fig. 1a).
Figure 1. PRC2 interactors and architecture during stem cell differentiation.
(a) Western blot (top) of core PRC2 members on nuclear extracts from ESCs and NPCs. Hdac1 is used as a loading control. Uncropped blots appear in Supplementary Data Set 1. The bottom table lists the absolute abundance of Eed, Ezh2 and Suz12 in nuclear extracts from each cell type24. (b,c) Volcano plots from label-free GFP pulldowns on Eed-GFP ESC (b) and NPC (c) nuclear extracts. Statistically enriched proteins in the Eed-GFP pulldown are identified by a permutation-based FDR-corrected t-test. The label-free quantification (LFQ) intensity of the GFP pulldown relative to the control [fold change (FC), x-axis] is plotted against the -log10-transformed P-value of the t-test (y-axis). Dotted grey lines represent statistical cutoffs. The proteins in the upper right corner represent the bait (Eed, green) and its interactors. Snrpn, Snrpd2, and Fhl3 are known GFP contaminants. (d) Stoichiometry of Eed-GFP interactors in ESCs and NPCs. The iBAQ value of each protein group is divided by the iBAQ value of the core PRC2 subunits Ezh1 and 2, then graphed with Ezh1 and 2 set to 1. Data are shown as mean ± s.d. (n = 3 pulldowns). (e) Logarithmic plot of the ratio of ESC enrichment (left) or NPC enrichment (right) for Eed-GFP interacting proteins. (f) Visualization of cross-links identified from single affinity purified Eed-GFP in ESCs. Ambiguous cross-links between paralogous subunits (Rbbp4 and 7, Ezh1 and 2) are combined in this visualization. (g) Summary of PRC2 architecture in mESCs based on cross-links from (f).
Next, we performed label-free GFP pulldowns followed by liquid chromatography tandem mass spectrometry (LC MS-MS) on nuclear extracts from ESCs and NPCs to look at the subunit composition of PRC2 in both cell types. GFP-Eed pulldowns identified all known core PRC2 subunits (Ezh1 or 2, Suz12, Rbbp4 or 7, Mtf2, Jarid2, and Aebp2) in both ESCs and NPCs (Fig. 1b,c and Supplementary Fig. 1b). PRC2 is stable in our pulldown conditions, as Suz12 successfully immunoprecipitated Jarid2 (Supplementary Fig. 2b). C17orf96, a recently identified PRC2 interactor in HeLa cells12,25, is also found as an Eed interactor in ESCs and NPCs (Fig. 1b,c and Supplementary Fig. 1b). Additionally, the uncharacterized Gm340 protein (also known as C10orf1212,25) was found in both ESC and NPC pulldowns. Two PRC2-associated proteins were found only in the ESC pulldowns, the F-box protein Fbxw11 and the uncharacterized protein AU022751, which was recently reported as a PRC2 interactor in ESCs26. Several NPC-specific interactors were identified, including the known PRC2-associated proteins Phf1 and Ehmt2.
We then made use of the iBAQ (intensity based absolute quantification) algorithm27 to calculate the abundance (stoichiometry) of each interactor relative to Ezh1 and 2. Whereas the stoichiometry of the core subunits remains stable during differentiation, the abundance of substoichiometric interactors varies greatly between the two cell types (Fig. 1d). C17orf96, Mtf2, and Tceb 1 and 2 are highly enriched (~10-fold or higher) as Eed-GFP interactors in ESCs relative to NPCs (Fig. 1e). In contrast, Gm340 and Phf19 are nearly 10-fold enriched in the NPC complex. We also purified Eed-GFP from the ESC nuclear pellet (NP) fraction remaining after nuclear extraction (NE) (Supplementary Fig. 1c). These experiments revealed that the core PRC2 complex looks similar in the NE vs NP pulldown, whereas certain substoichiometric interactors are slightly more abundant, either in the NE or NP fraction (Supplementary Fig. 1d). In summary, these results reveal a global downregulation of PRC2 abundance during stem cell differentiation towards a neural lineage. Furthermore, whereas the core of PRC2 is stable during differentiation, substoichiometric interactors are highly dynamic.
Cross-linking MS reveals mESC PRC2 architecture
To determine the architecture of PRC2 in ESCs, we performed single affinity purification cross-linking MS experiments on the endogenous complex following GFP-Eed purification. GFP pulldowns from GFP-Eed ESC nuclear extracts were incubated on-bead with BS3, an amine-to-amine cross-linker, and cross-linked peptides were identified using pLink28 (Fig. 1f). Our data agrees with a PRC2 architecture where Ezh1 or 2, Suz12, and Eed represent a well-connected core complex and auxiliary subunits bind at the periphery29 (Fig. 1g). In addition, we observed cross-links between multiple partially overlapping regions on Mtf2, Jarid2, or C17orf96 and Ezh1 or 2. Mtf2 and Jarid2 utilize overlapping binding surfaces in the Ezh1 and 2 SET domain, while Mtf2 and C17orf96 bind to a similar N-terminal region of Ezh1 or 2. This suggests that these proteins may bind PRC2 in a mutually exclusive manner as previously hypothesized30. Altogether, these findings extend our knowledge of PRC2 architecture, particularly for ESC-enriched substoichiometric interactors.
Most PRC2 is lost from chromatin during differentiation
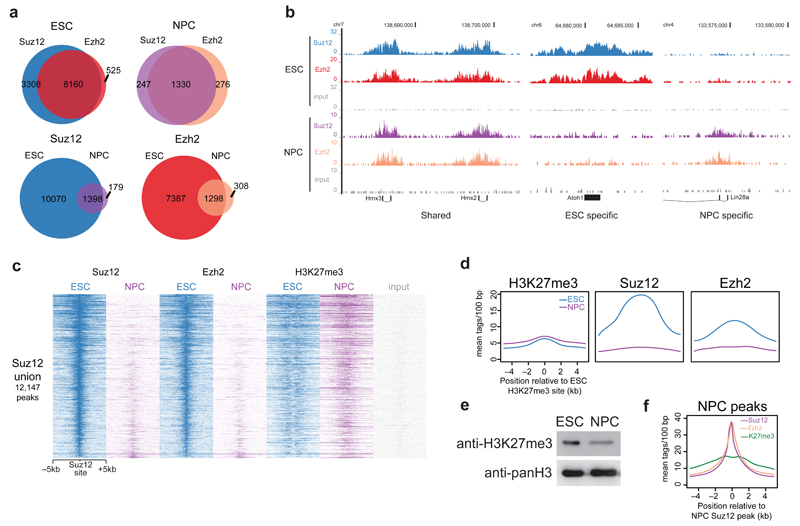
To determine the functional consequences of PRC2 downregulation during differentiation, we performed chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-seq) experiments on core PRC2 complex members in ESCs and NPCs. We used endogenous antibodies raised against the Suz12 and Ezh2 subunits. >80% of Ezh2 peaks overlapped with Suz12 in both ESCs and NPCs (Fig. 2a). More Suz12 peaks were identified than for Ezh2, which could be due to the presence of PRC2 complexes containing Ezh1. GO terms enriched amongst PRC2 bound genes are related to developmental processes or cell-type specification (Supplementary Fig. 2a). Surprisingly, over 85% of Suz12 and Ezh2 peaks are lost during differentiation to NPCs, with very few NPC-specific peaks appearing (Fig. 2a,b and Supplementary Fig. 2b,c). This suggests that lower PRC2 protein levels in NPCs results in a drastic loss of PRC2 binding to chromatin during differentiation. Heat maps sorted on ESC Suz12 peaks confirm the loss of Suz12 and Ezh2 binding in NPCs (Fig. 2c).
Figure 2. ChIP-sequencing of core PRC2 subunits during stem cell differentiation.
(a) Venn diagrams summarizing the number of peaks called from Suz12 and Ezh2 ChIP-seq in ESCs and NPCs. (b) UCSC genome browser screenshots of binding profiles at 3 classes of genes: Cell type independent, ESC specific, or NPC specific binding. (c) Suz12, Ezh2, and H3K27me3 occupancy (ChIP-seq read density) in ESCs and NPCs centered on Suz12 ESC peaks. (d) Average binding profile of Suz12 and Ezh2 in ESCs and NPCs at H3K27me3 peaks in ESCs. (e) Western blot analysis of acid extracted histones from ESCs and NPCs using the indicated antibodies. Uncropped blots appear in Supplementary Data Set 1. (f) Average binding profile of Suz12, Ezh2, and H3K27me3 in NPCs at Suz12 NPC peaks.
Next, we asked what happens to H3K27me3 levels during differentiation in light of the PRC2 downregulation. Strikingly, we observed that H3K27me3-marked nucleosomes in ESCs retain the mark during differentiation to NPCs (Fig. 2c,d), whereas Suz12 and Ezh2 are lost at these sites. A Western blot on bulk H3K27me3 levels showed that the abundance of the histone mark also decreases during differentiation, but not nearly as drastically as PRC2 subunit protein levels (Fig. 2e). H3K27me3 is still deposited at Suz12 and Ezh2 NPC peaks (Fig. 2f), indicating that the loss of PRC2 at ESC-marked sites is not due to loss of a functional complex in NPCs. Loss of PRC2 during differentiation did not lead to global changes in expression of PRC2-target genes, and a large proportion of genes are actually downregulated upon loss of PRC2 (Supplementary Fig. 2d-h). Altogether, these findings suggest that downregulation of PRC2 protein levels during differentiation leads to a loss of binding at most Polycomb target genes during differentiation, while surprisingly, H3K27me3 deposition at most of these sites is maintained.
Identification of dynamically incorporated PR-DUB subunits
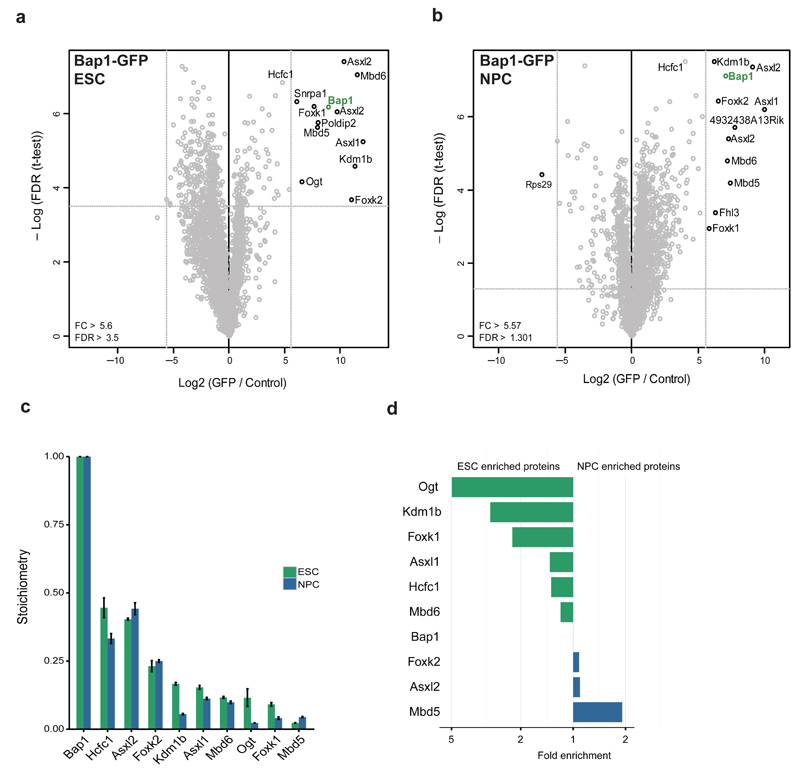
To identify interactors of the PR-DUB complex, we used a BAC line containing the core subunit Bap1 followed by the same mass spectrometry workflow used for PRC2. We identified the core of the complex (Bap1, Asxl1 and 2, Foxk1 and 2, and Kdm1b) in both cell types (Fig. 3,b). Bap1 and the Asxl proteins are present at a ratio of ~2:1, indicating that Bap1 is dimeric within the PR-DUB complex. As previously reported in HeLa cells31, the methyl binding domain proteins Mbd5 and Mbd6 were identified as PR-DUB interactors in both ESCs and NPCs. Using the iBAQ algorithm, we determined that the stoichiometry of the core complex changes very little during differentiation (Fig. 3c). Similar to PRC2, some of the substoichiometric interactors appear to be cell-type specific (Fig. 3d). The glycosylase Ogt and histone demethylase Kdm1b are enriched in ESC PR-DUB, whereas Mbd5 is NPC enriched, consistent with a role for this protein in brain function32. We also purified Asxl2-GFP from ESCs, which revealed that this protein exclusively assembles in a PR-DUB like complex (Supplementary Fig. 3a,b). Furthermore, this analysis showed that Asxl1 and Asxl2 define mutually exclusive PR-DUB subcomplexes. Since the core of PR-DUB remains relatively stable during differentiation, our results suggest a conserved function for this complex in different cell types.
Figure 3. PR-DUB interactors during stem cell differentiation.
(a,b) Volcano plots from label-free GFP pulldowns on Bap1-GFP ESC (a) and NPC (b) nuclear extracts graphed as in Fig. 1b. Snrpa1 and Fhl3 are known GFP contaminants. (c) Stoichiometry of Bap1-GFP interactors in mESCs and NPCs. The iBAQ value of each protein group is divided by the iBAQ value of Bap1, then graphed with Bap1 set to 1. Data are shown as mean ± s.d. (n = 3 pulldowns). (d) Plot of the ratio of ESC enrichment (left) or NPC enrichment (right) for Bap1-GFP interacting proteins.
Identification of dynamically incorporated PRC1 subunits
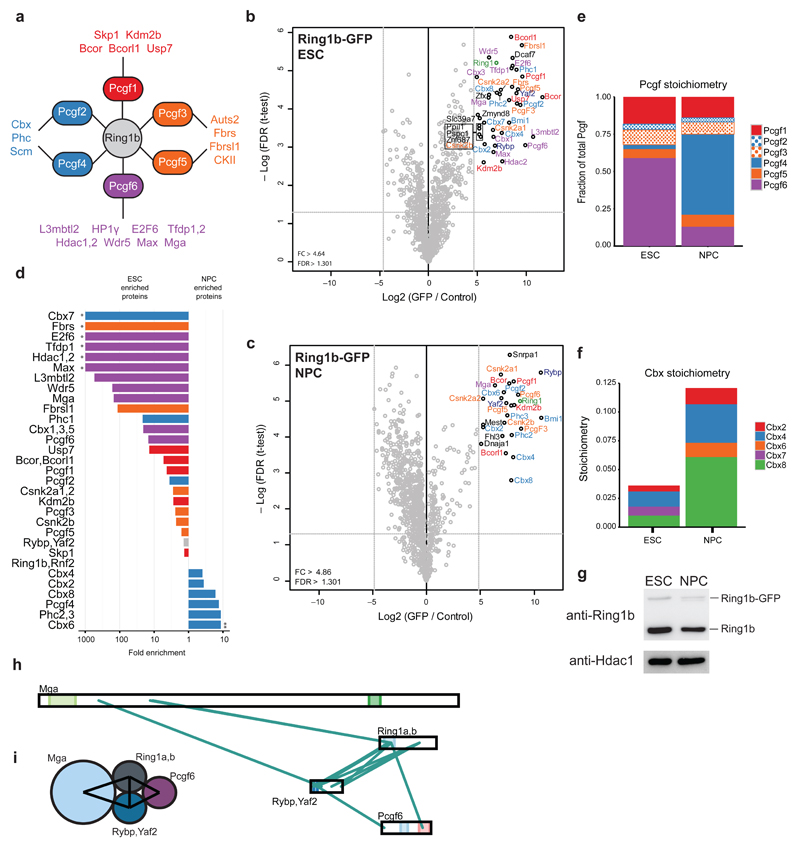
A similar label-free proteomics strategy was used for PRC1. PRC1 interacts with 6 different Pcgf proteins in a mutually exclusive manner11 (Fig. 4a). Each of these Pcgf proteins has a shared (Pcgf2 and 4, Pcgf3 and 5) or unique set of interactors. Nearly the same set of core subunits and substoichiometric interactors was identified in both ESCs (Fig. 4b) and NPCs (Fig. 4c). However, a dramatic switch occurs with respect to the binding of Pcgf and Cbx family members during differentiation (Fig. 4d-f). iBAQ-derived stoichiometry values revealed that Pcgf6 occupies 60% of Ring1b complexes in ESCs (Supplementary Fig. 4a and Fig. 4e). Pcgf6 also remains the predominant Ring1b interactor in the nuclear pellet (NP) fraction (Supplementary Fig. 4c,d) Differentiation to NPCs mainly results in an exchange of Pcgf6 for Pcgf4 (Bmi1)-containing PRC1 (54% of NPC complexes, Fig. 4e and Supplementary Fig. 4a,b). A similar switch is observed for the Cbx proteins. Notably, total Cbx occupancy in Ring1b complexes increases during differentiation (3.5% to 12%), but remains substoichiometric. This is not an overexpression artifact, as Ring1b-GFP levels remain lower than those of the endogenous Ring1b in both cell types (Fig. 4g). Purification of Flag-tagged Pcgf2 from ESCs also revealed substoichiometric Cbx proteins (Supplementary Fig. 4e-f). Cbx7 is only found associated with Ring1b in ESCs, as previously reported33,34, and is replaced by Cbx6 in NPCs (Fig. 4f and Supplementary Fig. 4a,b). However, Cbx4 and Cbx8 have the highest stoichiometry values in both ESCs and NPCs. In general, Pcgf6-specific interactors showed the most dynamic changes between ESCs and NPCs due to the loss of Pcgf6 from the complex during differentiation to a neural lineage (Fig. 4d). Conversely, Pcgf2 and 4 interactors are more abundant relative to Ring1b in NPCs, most likely due to the strong upregulation of Pcgf4 in NPC cells. Curiously, Phc1, a Pcgf2 and 4 interactor, is more abundant in ESCs compared to NPCs. From these results, we conclude that the composition of the core PRC1 complex is highly dynamic during differentiation.
Figure 4. PRC1 interactors and architecture during stem cell differentiation.
(a) Schematic of the PRC1 complex showing the core catalytic subunit Ring1b as well as all 6 Pcgf proteins and a summary of their known interactors. (b,c) Volcano plot from label-free GFP pulldowns on Ring1b-GFP ESC (b) or NPC (c) nuclear extracts graphed as in Fig. 1b. Snrpa1 and Fhl3 are known GFP contaminants. (d) Logarithmic plot of the ratio of ESC enrichment (left) or NPC enrichment (right) for Ring1b-GFP interacting proteins. (*) only detected in ESC pulldown (**) only detected in NPC pulldown (e) Pcgf protein stoichiometry values in ESCs and NPCs. Results are presented as the fraction of total Pcgf bound to the complex. (f) Cbx protein stoichiometry values from ESCs and NPCs. (g) Western blot of GFP-tagged and endogenous Ring1b on nuclear extracts from ESCs and NPCs with Hdac1 used as a loading control. Uncropped blots appear in Supplementary Data Set 1. (h) Visualization of cross-links identified from single affinity purified Ring1b-GFP from ESCs. Ambiguous cross-links between paralogous subunits (Ring1a and Ring1b, Rybp and Yaf2) are combined in this visualization. (i) Summary of PRC1 architecture in ESCs based on cross-links shown in (h).
Cross-linking MS reveals mESC PRC1 architecture
We also performed cross-linking MS following Ring1b-GFP affinity purification from ESC nuclear lysates. Our data suggest Ring1a or Ring1b and Rybp or Yaf2 form a stable core for PRC1 (Fig. 4h). The Pcgf6 interactor Mga cross-linked directly with Ring1a or Ring1b and Rybp or Yaf2, suggesting it stably interacts with a subset of PRC1 complexes. We also identified cross-links between Pcgf6 and Rybp or Yaf2 and between Pcgf6 and Ring1a or Ring1b. This hints that Pcgf6 binds to Ring1a or Ring1b complexes through a Rybp or Yaf2 and Ring1a- or Ring1b-mediated interaction surface (Fig. 4i). We did not identify any cross-links between Ring1b and other Pcgf proteins, likely due to the fact that Pcgf6 is the predominant Pcgf complex in ESCs.
PRC1 is retained at H3K27me3-marked sites in ESCs and NPCs
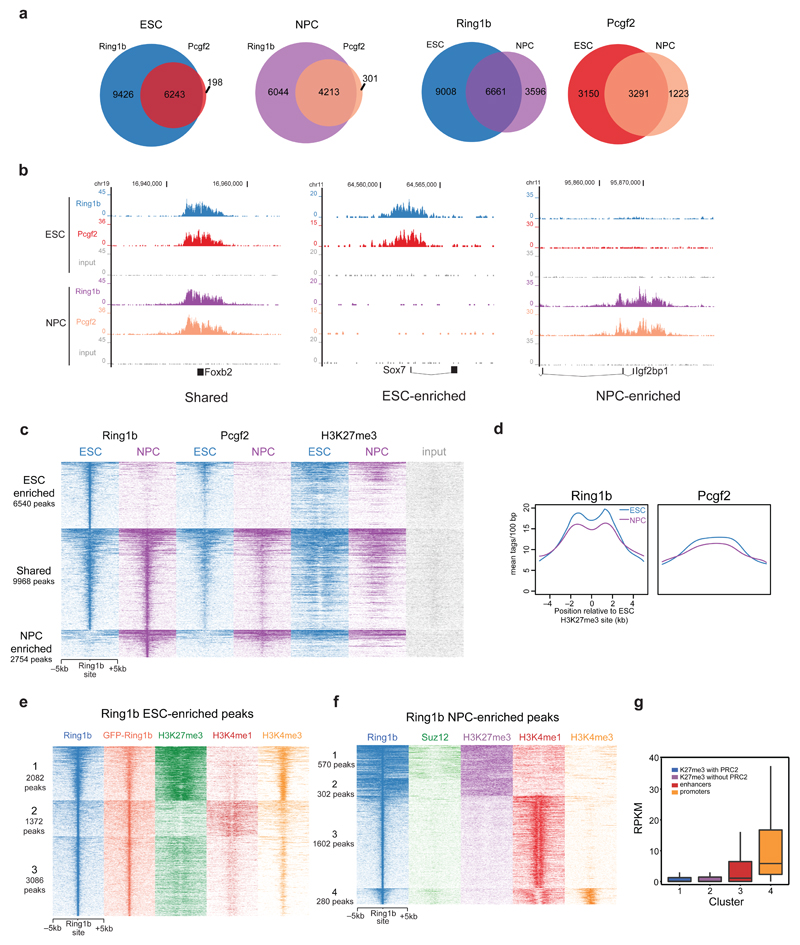
Next, we determined the genomic localization of the PRC1 complex in both cell types using endogenous antibodies raised against Ring1b and Pcgf2. Nearly all Pcgf2 peaks overlap with Ring1b peaks in both ESCs and NPCs, indicating that Pcgf2 binds to chromatin only within the context of PRC1 (Fig. 5a). However, we observed many Ring1b peaks independent of Pcgf2 in both cell types which is likely due to the presence of other Pcgf proteins bound to Ring1b. In contrast to PRC2, we observed that ~50% of Ring1b peaks (n = 9968 peaks) are shared between ESCs and NPCs. There are also large groups of peaks that are >3-fold enriched only in ESCs (n = 6540 peaks) or NPCs (n = 2754 peaks) (Fig. 5b,c and Supplementary Fig. 5a-c). GO-term analysis of PRC1-associated genes revealed that ESC-enriched targets are enriched for developmental and morphogenic functions (Supplementary Fig. 5a). These are also the top functions in the shared gene set and the NPC-enriched set, but many more biosynthetic and metabolic processes are found at the genes shared between cell types, suggesting a conserved role for PRC1 at housekeeping genes. Mining RNA-seq data revealed no global up or downregulation in the three classes of PRC1 bound genes (Supplementary Fig. 5d). However, individual transcripts do show up or downregulation upon gain or loss of PRC1 (Supplementary Fig. 5e-g). Heat maps of the different peak classes revealed good overlap between the Ring1b and Pcgf2 data sets in both ESCs and NPCs (Fig. 5c). We also looked at PRC1 binding at H3K27me3 sites in ESCs and NPCs. To our surprise, both Ring1b and Pcgf2 are retained at H3K27me3 sites during differentiation to NPCs, whereas PRC2 was not (Fig. 5c,d and Fig. 2d). Indeed, clustering of NPC-enriched Ring1b sites identified a cluster of Ring1b that does not overlap with Suz12 in NPCs, but still co-localizes with H3K27me3 (Fig. 5f cluster 2). These results suggest that PRC1 can bind to H3K27me3-marked chromatin independent of PRC2.
Figure 5. ChIP-sequencing of core PRC1 subunits during stem cell differentiation.
(a) Venn diagrams summarizing the number of peaks called from Ring1b and Pcgf2 ChIP-seq in ESCs and NPCs. (b) UCSC genome browser screenshots of binding profiles at 3 classes of genes: Shared, ESC enriched, or NPC enriched binding. (c) Ring1b, Pcgf2, and H3K27me3 occupancy (ChIP-seq read density) in ESCs and NPCs at 3 clusters of genes centered on the union of all Ring1b peaks. Enriched peaks have >3-fold more reads at the Ring1b binding site relative to the other cell type. (d) Average binding profile of Ring1b and Pcgf2 in ESCs and NPCs at H3K27me3 peaks in ESCs. (e) k-means clustering of Ring1b ESC enriched peaks on Ring1b, GFP-Ring1b, H3K27me3, H3K4me1, and H3K4me3 ChIP-sequencing results. (f) k-means clustering of Ring1b NPC enriched peaks on Ring1b, Suz12, H3K27me3, H3K4me1, and H3K4me3 ChIP-sequencing results. (g) Read per kilobase per million mapped reads (RPKM) values for genes located nearby Ring1b peaks from each of the four clusters identified in (f). For all box plots: midline, median; box limits, 25th percentile (first quartile) and 75th percentile (third quartile); upper whisker, min(max(x)), third quartile + 1.5× interquartile range (IQR; third-quartile minus first-quartile values); lower whisker, max(min(x)), first quartile − 1.5× IQR.
Ring1b binds enhancers independent of H3K27me3
K-means clustering revealed three discrete classes of ESC enriched Ring1b binding sites (Fig. 5e). The first cluster represents H3K4me3- and H3K27me3-marked bivalent genes enriched for developmental GO terms (Fig. 5e, Supplementary Fig. 5h). Cluster two represents H3K4me1-marked enhancers, which are also enriched for developmental GO terms (Fig. 5e, Supplementary Fig. 5i). A third cluster contains low amounts of H3K27me3, H3K4me1 and H3K4me3. We also performed ChIP-seq in the GFP-Ring1b BAC line using a GFP antibody, which shows a very good overlap with the endogenous Ring1b ChIP-seq reads (Fig. 5e). NPC enriched Ring1b sites also co-localize with histone marks for enhancers (H3K4me1, Fig. 5f cluster 3, 58% of peaks) or active genes (H3K4me3, Fig. 5f cluster 4, 10% of peaks). RNA-seq analysis (Fig. 5g) of genes from the H3K27me3-marked clusters shows these loci are repressed independent of PRC2 binding (Cluster 1 vs Cluster 2). There is an appreciable amount of transcription from genes nearby the H3K4me1-marked chromatin, and transcript levels from the H3K4me3-marked chromatin are high as expected. Interestingly, genes nearby H3K4me1-marked chromatin are almost exclusively involved in neural function (Supplementary Fig. 5j), suggesting that these putative enhancer regions are required for NPC propagation or terminal differentiation.
Altering PRC1 composition affects genomic PRC1 binding
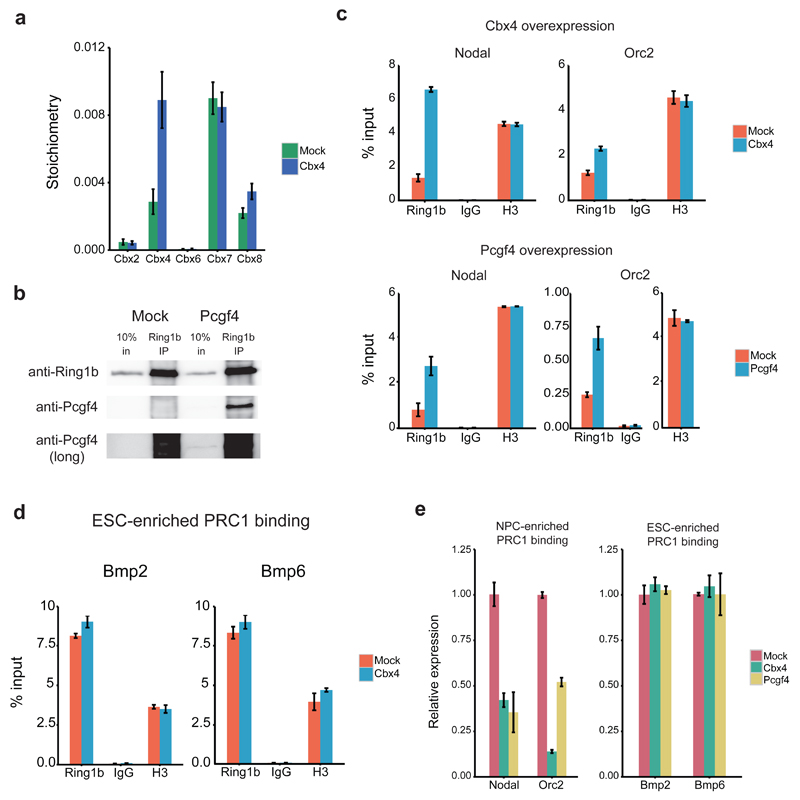
Finally, we wanted to determine if the subunit switching we observed for PRC1 during differentiation affects genomic localization of Ring1b and, consequently, gene expression. To address this, we overexpressed two NPC-enriched PRC1 subunits, Cbx4 and Pcgf4, in ESCs. Cbx4 overexpression in the Ring1b BAC-GFP line followed by label-free GFP-Ring1b purification resulted in a nearly 3-fold increase in Cbx4 stoichiometry relative to Ring1b when compared to a mock-transfected control (Fig. 6a). No change was observed in the Cbx7 stoichiometry value, suggesting that Cbx4 overexpression only increases the pool of Ring1b containing Cbx4, rather than replacing Cbx7 within the complex. Furthermore, co-immunoprecipitation analysis revealed that the transfected Pcgf4 interacts with Ring1b (Fig. 6b). Next, we performed ChIP-qPCR to determine if Cbx4 and Pcgf4 overexpression results in increased Ring1b binding at genomic sites which are predominantly bound in NPCs. We chose two target genes for this analysis (Nodal and Orc2) that are highly expressed in ESCs relative to NPCs (see Supplementary Fig. 5g) and which gain Ring1b binding during differentiation (Supplementary Table 1). Cbx4 and Pcgf4 overexpression in ESCs led to increased Ring1b binding at these NPC-enriched sites relative to the mock-transfected control (Fig. 6c). ChIP-qPCR at ESC-enriched loci (Fig. 6d) revealed no changes in Ring1b recruitment upon Cbx4 overexpression. Increased Ring1b binding to Nodal and Orc2 correlated with decreased expression of these genes, whereas the ESC-enriched loci were not affected upon Cbx4 and Pcgf4 overexpression (Fig. 6e). These experiments serve as a proof-of-principle example that perturbation of Ring1b interactors can affect the genomic localization of the PRC1 complex and expression of PRC1 target genes.
Figure 6. Overexpression of NPC-enriched PRC1 subunits can affect genomic localization of Ring1b.
(a) Stoichiometry values for Cbx proteins bound to Ring1b in mock or Cbx4-transfected Ring1b-GFP BAC ESCs. Data are shown as mean ± s.d. (n = 3 pulldowns) (b) Western blots of Ring1b co-immunoprecipitations in mock or Pcgf4-transfected ESCs. Uncropped blots appear in Supplementary Data Set 1. (c-d) ChIP-qPCR of Ring1b, IgG, and histone H3 at 2 NPC-enriched loci (c) and at 2 ESC-enriched loci (d) in mock, Cbx4 or Pcgf4-transfected ESCs. Data shown are from one representative ChIP (n = 3 ChIPs) and graphed as mean ± s.d. (n = 3 technical replicates). (e) qRT-PCR analysis of gene expression levels in mock, Cbx4 or Pcgf4-transfected ESCs. Data shown are from one representative transfection (n = 3 transfections) and graphed as mean ± s.d. (n = 3 technical replicates).
Discussion
We have provided both proteomic and genomic characterization of Polycomb group protein complexes during embryonic stem cell differentiation to a neural lineage. This work revealed several key observations regarding Polycomb biology. First, we found that the interactomes of PRC1 and PRC2 are highly cell-type specific, whereas the subunits of PR-DUB are largely cell-type invariant. The total abundance of Ring1b-containing complexes is mildly downregulated during differentiation, whereas PRC2 is drastically downregulated (at least 30-fold) in neural progenitor cells relative to ESCs. Genome-wide analyses confirm a loss of PRC2 from chromatin, even though bulk H3K27me3 levels are largely maintained. Strikingly, only a fraction (32%) of genome-wide Ring1b sites in neural progenitor cells represent ‘canonical’ Polycomb targets which are characterized by broad H3K27me3 domains. The remaining 68% of Ring1b sites in NPCs are at enhancers and active promoters. Finally, we integrated our proteomic and genomic data to show that expression of cell type-enriched Polycomb interactors can affect target gene specificity and expression.
Our quantitative mass spectrometry-based proteomics workflows allowed us to identify known and novel components of the Polycomb machinery during stem cell differentiation. Furthermore, the application of the iBAQ algorithm to our data adds an additional layer of information, stoichiometry, which was essential for the identification of dynamic interactors. For example, C17orf96 and Mtf2 were identified as highly confident Eed interactors in ESCs and NPCs, but only after calculation of the iBAQ values were we able to determine that both of these proteins are highly enriched in the ESC PRC2 complex. We envision that the workflow presented here will be a powerful tool to characterize dynamic protein-protein interactions in a variety of cellular contexts and upon cellular perturbations (cell cycle stages, growth factor stimulation, DNA damage, etc.)35.
Our data revealed a number of substoichiometric PRC2 interactors including Mtf2, C17orf96, Gm340, and Tceb1 and 2. Based on our data, we cannot conclude whether these proteins assemble together in a subset of PRC2 complexes or whether all of these proteins interact stoichiometrically with PRC2 in cells and are partially lost during affinity purification. Alternatively, some of the substoichiometric PRC2 interactors may define mutually exclusive PRC2 subcomplexes with distinct functions. Our cross-linking data supports the latter hypothesis since we find that some of the substoichiometric interactors share a binding surface on Ezh1 or 2. Clearly, the tagging, purification and cross-linking of substoichiometric PRC2 interactors is needed to substantiate these findings.
One intriguing observation is the near complete loss of PRC2 from chromatin in NPCs despite the continued presence of H3K27me3. Previously, it was shown that PRC2 binds to and co-localizes with H3K27me3 in the G1 phase of the cell cycle and remains at sites of DNA replication in S phase, thus ensuring propagation of the modification during cell division36. The cell cycle duration of NPCs is nearly 3 times longer than that of ESCs (30hrs vs ~10hrs), which results in more time spent in S-phase per cell division37,38. We hypothesize that a longer S phase requires less PRC2 to propagate the mark during cell division. Alternatively, once cells are lineage committed, less ‘de novo’ H3K27me3 and, consequently, less PRC2 is required to maintain H3K27me3 domains.
Finally, we identified thousands of Ring1b binding sites in the NPC genome which are devoid of H3K27me3. These binding sites are typically very narrow (<1kb), especially when compared to the classical Polycomb domains. These binding sites may represent examples of permissive chromatin ‘sampling’ by PRC139. However, the functional relevance of Ring1b binding at these sites remains unclear. Interestingly, several recent studies have also reported a link between PRC1 and gene activation13–16. Clearly, further work is required to determine whether distinct PRC1 subcomplexes also play a role in gene activation during neural lineage commitment.
Online Methods
Cell culture and BAC line generation
R1 mESCs were obtained from the ATCC and cultured as in Spruijt et al24. NPCs were differentiated and propagated following the protocol from Conti et al38. All cell lines have been tested for mycoplasma contamination. BACs were tagged according to the protocol from Poser et al23. Tagged BAC lines were prepped on NucleoBond BAC 100 columns (Macherey-Nagel) and transfected into ESCs using Lipofectamine LTX Plus (Invitrogen), followed by G418 selection for 10-12 days. Individual colonies were picked, expanded, and screened for GFP expression.
Nuclear extracts and nuclear pellet solubilization
Nuclear extracts were prepared essentially according to Dignam et al40. Cells were harvested with trypsin, washed twice with PBS, and centrifuged at 400xg for 5 min at 4°C. Cells were swelled for 10 min at 4°C in five volumes of Buffer A (10 mM Hepes-KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl), and then pelleted at 400xg for 5 min at 4°C. Cells were resuspended in two volumes of Buffer A plus protease inhibitors and 0.15% NP-40 and transferred to a Dounce homogenizer. After 30-40 strokes with a Type B pestle, the lysates were spun at 3200xg for 15 min at 4°C. The nuclear pellet was washed once with PBS, and spun at 3200xg for 5 min at 4°C. The nuclear pellet was resuspended in 2 volumes Buffer C (420 mm NaCl, 20 mm Hepes-KOH, pH 7.9, 20% v/v glycerol, 2 mm MgCl2, 0.2 mm EDTA) with 0.1% NP-40, protease inhibitors, and 0.5 mm dithiothreitol. The suspension was incubated with rotation for 1 h at 4°C, and then spun at 18000xg for 15 min at 4°C. The supernatant was aliquoted and stored at -80°C until further use.
The nuclear pellets remaining after nuclear extraction were solubilized by resuspension in four volumes of RIPA buffer (150 mM NaCl, 50 mM Tris pH 8.0, 1% NP-40, 5 mM MgCl2, 10% glycerol) plus benzonase (Millipore) at 1000U per 100ul nuclear pellet. Samples were incubated at 37°C with shaking until solubilized, then spun at 18000xg for 15 min at 4°C. The supernatant was aliquoted and stored at -80°C until further use.
Label-free pulldowns and cross-linking MS
Label-free GFP pulldowns were performed in triplicate as previously described12 with the following modifications. For GFP pulldowns, 2mg of nuclear extract was incubated with 7.5 ul GFP Nano trap beads (Chromotek) and 50 ug/mL ethidium bromide in Buffer C (300 mm NaCl, 20 mm Hepes-KOH, pH 7.9, 20% v/v glycerol, 2 mm MgCl2, 0.2 mm EDTA) with 0.1% NP-40, protease inhibitors, and 0.5 mm dithiothreitol in a total volume of 400 ul. 6 washes were performed: 2 with Buffer C and 0.5% NP-40, 2 with PBS and 0.5% NP-40, and 2 with PBS. Flag pulldowns were performed as above but with 10 ul anti-Flag M2 affinity gel (Sigma). Affinity purified proteins were subject to on-bead trypsin digestion as previously described31. Tryptic peptides were acidified and desalted using StageTips41 prior to mass spec analyses.
Single affinity cross-linking MS was performed according to Makowski et al42. Briefly, 30 uL of GFP-Trap bead slurry (Chromotek) and 1 mL of nuclear extracts were used per pulldown (~7 mg/mL). On-bead cross-linking was performed by immediately resuspending beads in 50 mM borate-buffered saline containing 1 mM BS3 (Thermo Scientific) following GFP affinity purification. Cross-linking reactions were performed for one hour at room temperature with shaking at 1000 rpm. Reactions were quenched by adding 100 mM ammonium bicarbonate and incubating for ten minutes at room temperature with shaking at 1000 rpm. On-bead digest and StageTipping were performed as above.
Label-free quantitation (LFQ) LC MS-MS analysis
Tryptic peptides were separated with an Easy-nLC 1000 (Thermo Scientific). Buffer A was 0.1% formic acid and Buffer B was 80% acetonitrile and 0.1% formic acid. Eed-GFP LFQ samples were separated using a 120-min gradient from 7% until 32% Buffer B followed by step-wise increases up to 95% Buffer B. Mass spectra were recorded on a LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific), selecting the 15 most intense precursor ions of every full scan for fragmentation. The tryptic peptides from Ring1b-GFP ESC and NPC pulldowns, Bap1-GFP ESC pulldowns, and Flag-Pcgf2 pulldowns were separated using a 94-min gradient from 9-32% Buffer B followed by washes at 50% then 95% Buffer B for 120 min of total data collection time. Mass spectra were recorded on an LTQ-Orbitrap QExactive mass spectrometer (Thermo Fisher Scientific), selecting the top 10 most intense precursor ions for fragmentation. The tryptic peptides from Bap1-GFP NPC pulldowns, Asxl2-GFP ESC pulldowns, and all nuclear pellet pulldowns were measured by developing a gradient from 9-32% Buffer B for 114 minutes before washes at 50% then 95% Buffer B, for 140 minutes of total data collection time. Mass spectra were recorded on an LTQ-Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific). Scans were collected in data-dependent top speed mode with dynamic exclusion set at 60 seconds.
LFQ peptide identification and analysis
Thermo RAW files from LFQ AP MS-MS were analyzed with MaxQuant version 1.5.1.0 using default settings and searching against the UniProt mouse proteome, release 2014_09. Additional options for Match between runs, LFQ, and iBAQ were selected. Stoichiometry calculations and volcano plots were produced essentially as described12 using Perseus version 1.4.0.8 and in-house R scripts. Proteins used to generate the volcano plots are listed in Supplementary Table 2. Statistical cutoffs were chosen such that no proteins were present as outliers on the control, non-GFP side of the volcano plot.
Single affinity cross-linking LC MS-MS and data analysis
Samples were purified by GFP affinity purification from nuclear extracts using high stringency washing conditions as described above. Cross-linked and digested peptides were measured on an LTQ-Orbitrap QExactive as described previously42. Thermo RAW files were converted to mgf format using MSConvert43 and analyzed using pLink28. Identifications were accepted with a 0.05 FDR. To increase confidence in reported identifications, all spectral matches were additionally filtered to include only matches of peptide length >=5 and <=40, a precursor error of <10ppm, and an e-value of <0.01. Filtered cross-links are listed in Supplementary Table 3. Cross-link maps were produced with xiNET44.
Chromatin preparation
Attached mESCs and NPCs were cross-linked with 1% formaldehyde for 10 min at room temperature with gentle shaking. Cross-linking was quenched with the addition of 1/10 volume 1.25 M glycine. Cells were washed with PBS, then harvested by scraping in Buffer B (20 mM Hepes, 0.25% Triton X-100, 10 mM EDTA, and 0.5 mM EGTA). Cells were pelleted by spinning at 600xg for 5 min at 4°C. Cell pellet was resuspended in Buffer C (150 mM NaCl, 50 mM Hepes, 1 mM EDTA, and 0.5 mM EGTA) and rotated for 10 min at 4°C. Cells were pelleted by spinning at 600xg for 5 min at 4°C. The cell pellet is then resuspended in 1x incubation buffer (0.15% SDS, 1% Triton X-100, 150 mM NaCl, 1mM EDTA, 0.5mM EGTA, and 20mM Hepes) at 15 million cells/mL. Cells were sheared in a Bioruptor Pico sonicator (Diagenode) at 4°C using 5 or 7 cycles of 30 sec ON, 30 sec OFF for mESCs and NPCs, respectively. Sonicated material was spun at 18000xg for 10 min at 4°C, then aliquoted and stored at -80°C.
Chromatin immunoprecipitation
10 million cells were used as input for library prep, and 5 million cells were used as input for ChIP-qPCR experiments. Chromatin is incubated overnight at 4°C in 1x incubation buffer (0.15% SDS, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and 20 mM Hepes) supplemented with protease inhibitors and 0.1% BSA. Antibody amounts, catalog numbers, and validation information are listed in Supplementary Table 4. A 50/50 mix of Protein A and G Dynabeads (Invitrogen) are added the following day followed by a 90 minute incubation. The beads are washed 2x with Wash Buffer 1 (0.1% SDS, 0.1% sodium deoxycholate, 1% Triton, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and 20mM Hepes), 1x with Wash Buffer 2 (Wash Buffer 1 with 500 mM NaCl), 1x with Wash Buffer 3 (250 mM LiCl, 0.5% sodium deoxycholate, 0.5% NP-50, 1 mM EDTA, 0.5 mM EGTA, and 20 mM Hepes), and 2x with Wash Buffer 4 (1 mM EDTA, 0.5 mM EGTA, and 20 mM Hepes). After washing, beads are rotated for 20 min at room temperature in Elution Buffer (1% SDS, 0.1 M NaHCO3). The supernatant is decrosslinked with 200mM NaCl and 100 ug/mL Proteinase K for 4 hours at 65°C. Decrosslinked DNA is purified using MinElute PCR Purification columns (Qiagen). DNA amount is quantitated using Qubit fluorometric quantitation (ThermoFisher Scientific). qPCR analysis of ChIP DNA was performed using iQ SYBR Green Supermix (Bio-Rad) on a CFX96 Real-Time System C1000 Thermal Cycler (Bio-Rad). Primers used for qPCR analysis are listed in Supplementary Table 4.
Illumina high-throughput sequencing and data analysis
Libraries were prepared using the Kapa Hyper Prep Kit for Illumina sequencing (Kapa Biosystems) according to the manufacturer’s protocol with the following modifications. 5ng DNA was used as input, with NEXTflex adapters (Bioo Scientific) and 10 cycles of PCR amplification. Post-amplification cleanup was performed with QIAquick MinElute columns (Qiagen) and size selection was done with an E-gel (300bp fragments) (ThermoFisher Scientific). Size-selected samples were analyzed for purity using a High Sensitivity DNA Chip on a Bioanalyzer 2100 system (Agilent). Samples were sequenced on an Illumina HiSeq2000 or NextSeq genome analyzer. The 43bp tags were mapped to the reference mouse genome mm9 (NCBI build 37) using the Burrows-Wheeler Alignment tool (BWA) allowing one mismatch. Only uniquely mapped reads were used for data analysis and visualization. External files used for analysis are listed in Supplementary Table 445–48. Mapped reads were filtered for quality and duplicates were removed. Peak-calling was performed with the MACS 2.0 tool against a reference input sample from the same cell line with a q-value of 0.001 and with ‘—broad’ option enabled for the H3K27me3 ESC sample. Heat maps and k-means clustering were performed using a Python package available at http://simonvh.github.io/fluff/. Average binding profiles were generated using an in-house Perl package. All average profiles are obtained by counting tags per 100bp window. ChIP-seq datasets used for generating heat maps and average profiles were normalized for total number of uniquely mapped reads. For gene expression analysis, RPKM values were calculated using RNA-seq data from Guttman et al49. GREAT50 was used for GO term analysis, and P-values were computed using a hypergeometric distribution with FDR correction. R was used to generate most of the graphs.
Quantitative reverse transcriptase PCR (qRT-PCR)
RNA was isolated using the RNeasy Mini Kit (Qiagen) and 1ug of RNA was used for cDNA synthesis with the iScript cDNA Synthesis Kit (Bio-Rad). qRT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad) on a CFX96 Real-Time System C1000 Thermal Cycler (Bio-Rad). Primers used for qRT-PCR analysis are listed in Supplementary Table 4. Gapdh was used as the reference gene.
List of Supplementary Information
Supplementary Figure 1: Extended analysis of PRC2 pulldown data
Supplementary Figure 2: Extended analysis of PRC2 ChIP-seq
Supplementary Figure 3: Asxl2-GFP pulldowns and stoichiometry
Supplementary Figure 4: Extended analysis of PRC1 pulldown data
Supplementary Figure 5: Extended analysis of PRC1 ChIP-seq
Supplementary Table 1: List of genes within 5kb of Suz12 and Ring1b peaks
Supplementary Table 2: Filtered MaxQuant protein group files used to generate volcano plots
Supplementary Table 3: Filtered list of cross-linked peptides
Supplementary Table 4: List of antibodies, primers, and ChIP-seq tracks used in this study
Supplementary Data Set 1: Uncropped Western blots
Acknowledgements
We are grateful to I. Poser (MPI-CBG Dresden) for contributing the BAC lines and to D. Reinberg (NYU), J. Gil (MRC Clinical Sciences), and M. van Lohuizen (NKI) for contributing antibodies and plasmids. We thank the Sequencing and Bioinformatics teams of the RIMLS Mol(Dev)Bio departments for ChIP-seq support. We thank N. Hubner and H. Stunnenberg (RIMLS) for providing access to the QExactive mass spectrometer. The Vermeulen and Di Croce groups are supported by the EU FP7 framework program 277899, 4DCellFate. The Vermeulen group is supported by the EU FP7 ITN 607142, DevCom, and an ERC Starting Grant 309384.
Footnotes
Accession codes
ChIP-seq reads and peak files have been deposited in the GEO repository under GSE74330. Proteomics data have been submitted to the ProteomeXchange via the PRIDE partner repository under identifier PXD003758.
Author contributions
S.L.K performed most of the experiments and analyzed the data. M.M.M. performed and analyzed the cross-linking MS experiments. H.I.B. purified PR-DUB from ESCs and NPCs. L.V.V. purified PRC1 from ESCs. I.D.K. purified PRC1 and PRC2 from the nuclear pellet fraction. P.W.T.C.J. assisted with the mass spectrometry experiments. A.S. performed the co-IP for PRC2. L.D.C. supervised A.S. S.L.K. and M.V. wrote the paper. M.V. supervised the project.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 2014;14:735–51. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Montavon T, Duboule D. Chromatin organization and global regulation of Hox gene clusters. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120367. doi: 10.1098/rstb.2012.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–24. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–55. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 6.de Napoles M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 8.Muller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 9.Scheuermann JC, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–7. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14:853–64. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 11.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–56. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013;41:e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Z, et al. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature. 2014;516:349–54. doi: 10.1038/nature13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morey L, et al. Polycomb Regulates Mesoderm Cell Fate-Specification in Embryonic Stem Cells through Activation and Repression Mechanisms. Cell Stem Cell. 2015;17:300–15. doi: 10.1016/j.stem.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–64. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Boom V, et al. Non-canonical PRC1.1 Targets Active Genes Independent of H3K27me3 and Is Essential for Leukemogenesis. Cell Rep. 2016;14:332–46. doi: 10.1016/j.celrep.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 17.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–81. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–8. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–59. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalb R, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol. 2014;21:569–71. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 21.Illingworth RS, et al. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev. 2015;29:1897–902. doi: 10.1101/gad.268151.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pengelly AR, Kalb R, Finkl K, Muller J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 2015;29:1487–92. doi: 10.1101/gad.265439.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poser I, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–15. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–59. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2488–93. doi: 10.1073/pnas.1400648111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier VK, et al. Functional Proteomic Analysis of Repressive Histone Methyltransferase Complexes Reveals ZNF518B as a G9A Regulator. Mol Cell Proteomics. 2015;14:1435–46. doi: 10.1074/mcp.M114.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 28.Yang B, et al. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–6. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 29.Ciferri C, et al. Molecular architecture of human polycomb repressive complex 2. Elife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen X, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–14. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baymaz HI, et al. MBD5 and MBD6 interact with the human PR-DUB complex through their methyl-CpG-binding domain. Proteomics. 2014;14:2179–89. doi: 10.1002/pmic.201400013. [DOI] [PubMed] [Google Scholar]
- 32.Maussion G, et al. Investigation of genes important in neurodevelopment disorders in adult human brain. Hum Genet. 2015;134:1037–53. doi: 10.1007/s00439-015-1584-z. [DOI] [PubMed] [Google Scholar]
- 33.Morey L, et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 34.O'Loghlen A, et al. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell. 2012;10:33–46. doi: 10.1016/j.stem.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smits AH, Vermeulen M. Characterizing Protein-Protein Interactions Using Mass Spectrometry: Challenges and Opportunities. Trends Biotechnol. 2016 doi: 10.1016/j.tibtech.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 37.Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9:809–18. [PubMed] [Google Scholar]
- 38.Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. Chromatin sampling--an emerging perspective on targeting polycomb repressor proteins. PLoS Genet. 2013;9:e1003717. doi: 10.1371/journal.pgen.1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 42.Makowski MM, Willems E, Jansen PW, Vermeulen M. Cross-linking immunoprecipitation-MS (xIP-MS): Topological Analysis of Chromatin-associated Protein Complexes Using Single Affinity Purification. Mol Cell Proteomics. 2016;15:854–65. doi: 10.1074/mcp.M115.053082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–20. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Combe CW, Fischer L, Rappsilber J. xiNET: cross-link network maps with residue resolution. Mol Cell Proteomics. 2015;14:1137–47. doi: 10.1074/mcp.O114.042259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiwari VK, et al. A chromatin-modifying function of JNK during stem cell differentiation. Nat Genet. 2012;44:94–100. doi: 10.1038/ng.1036. [DOI] [PubMed] [Google Scholar]
- 47.Riising EM, et al. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell. 2014;55:347–60. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–10. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.