Summary
Cells display dynamic and diverse morphologies during development, but the strategies by which differentiated tissues achieve precise shapes and patterns are not well understood. Here we identify a developmental program that generates a highly ordered square cell grid in the Drosophila embryo through sequential and spatially regulated cell alignment, oriented cell division, and apicobasal cell elongation. The basic leucine zipper transcriptional regulator Cnc is necessary and sufficient to produce a square cell grid in the presence of a midline signal provided by the EGF receptor ligand, Spitz. Spitz orients cell divisions through a Pins/LGN-dependent spindle positioning mechanism and controls cell shape and alignment through a transcriptional pathway that requires the Pointed ETS domain protein. These results identify a strategy for producing ordered square cell packing configurations in epithelia and reveal a molecular mechanism by which organized tissue structure is generated through spatiotemporally regulated responses to EGF receptor activation.
Keywords: myosin II, junctional remodeling, planar polarity, EGF receptor, intercalation, oriented cell division
Introduction
Multicellular tissues display highly organized patterns that are essential for tissue function. Cells in the Drosophila wing assemble into a regular honeycomb-like array (Classen et al., 2005; Aigouy et al., 2010), cells at compartmental boundaries adopt rectangular shapes that prevent cell mixing (Landsberg et al., 2009; Monier et al., 2010), single-file columns of chondrocytes facilitate the elongation of bones (Dodds, 1930; Kimmel et al., 1998), and near-crystalline patterns of square-shaped cells are present in mature organs such as the Drosophila heart (Santiago-Martínez et al., 2006), the mouse cochlea (Chacon-Heszele et al., 2012), and the zebrafish retina (Salbreux et al., 2012). The stereotyped architectures of multicellular tissues are generated by a wide range of dynamic and spatially regulated behaviors, including cell proliferation, cell-shape changes, and cell rearrangements. The mechanisms that determine where, when, and in what sequence these cellular processes occur and how they contribute to the final shape and organization of differentiated tissues are not well understood.
Cell topology, or how many neighbors each cell is in contact with, is a central feature of tissue organization that can have a profound effect on intercellular signaling and tissue structure. Topological patterns in epithelia range from ordered hexagonal packing, in which the vast majority of cells are 6-sided, to highly disordered patterns that contain a wide range of polygons, including 4-sided, 5-sided, 7-sided, and 8-sided cells. The dynamic cell behaviors that produce tissue structure during development often increase topological disorder at the cellular level. Cell rearrangements that elongate the Drosophila body axis convert the embryonic epithelium from one in which most of the cells are hexagonal into one that is highly disordered with a wide range of different polygon classes (Zallen and Zallen, 2004). Increased topological disorder can also result from cell proliferation during tissue growth, as cell division alters the topology of both dividing cells and their neighbors (Gibson et al., 2006). Hexagons are the most common polygon class in epithelia, and square cells are rare, as even highly disordered tissues undergoing active cell division or movements contain a characteristically low fraction (<10%) of four-sided cells (Zallen and Zallen, 2004; Classen et al., 2005; Gibson et al., 2006). By contrast, square cells are the predominant topology in many mature organs (Santiago-Martínez et al., 2006; Chacon-Heszele et al., 2012; Salbreux et al., 2012). This striking contrast to the topological patterns present at early stages of development suggests that the disorder produced by cell division and movement must ultimately be reversed to produce the stereotyped patterns of mature tissues. Several strategies have been proposed that can induce square cell packing in theory, including heterogeneous or anisotropic mehanical tension and sequential cell sorting (Salbreux et al., 2012; Bardet et al., 2013). However, the cellular and molecular mechanisms that produce square cell packing configurations in vivo are not known.
Here we show that the midline cells of the developing Drosophila pharynx dynamically reorganize during development to produce a highly ordered square cell grid. This structure forms through sequential and spatially regulated cell alignment, oriented cell division, and apicobasal elongation. We show that the basic leucine zipper transcription factor Cap ‘n’ collar (Cnc) is necessary and sufficient to induce square cell packing in the presence of a short-range signal from the ventral midline. This signal is provided by the EGF receptor (EGFR) ligand Spitz, which directs the planar polarized localization of proteins involved in actomyosin contractility, cell adhesion, and spindle positioning. These results demonstrate that localized EGFR signaling triggers planar polarized cell behaviors that produce square cell packing in epithelia and reveal a developmental program for generating highly ordered structures during development.
Results
Sequential cell behaviors produce a square cell grid in the developing Drosophila pharynx
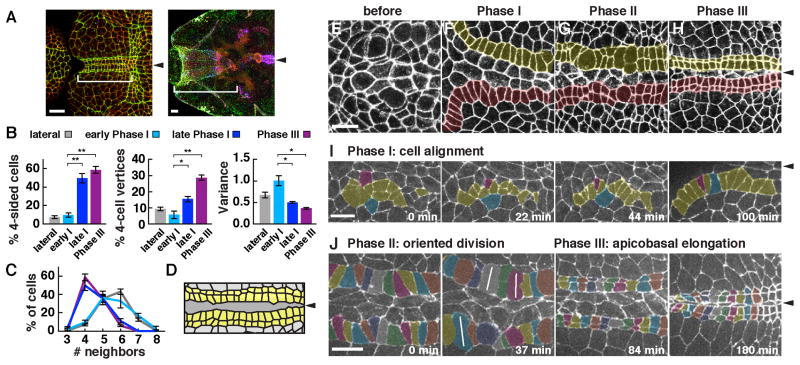
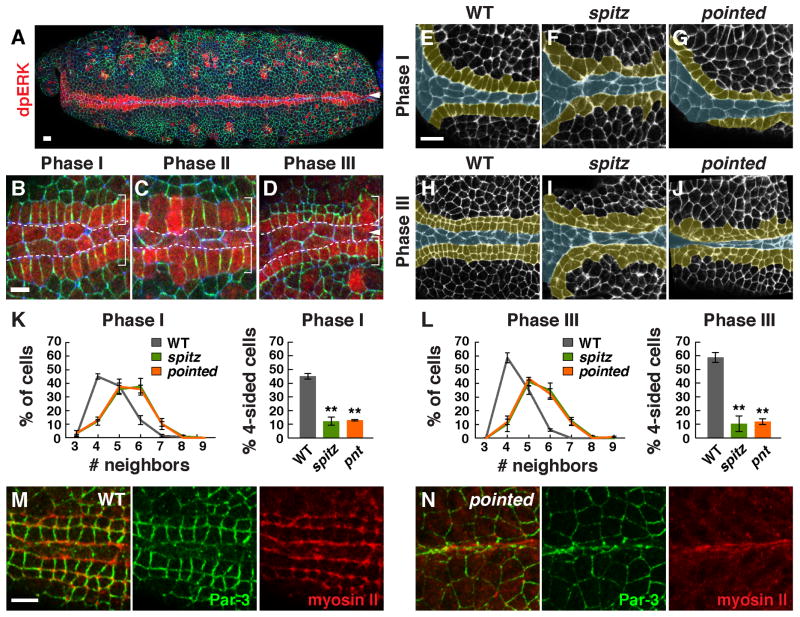
In a quantitative analysis of cell topology in Drosophila, we found that cells in the anterior ventral epithelium of the embryo display a highly ordered square grid pattern (Figure 1A). This square grid will go on to form the midline of the larval pharynx and may act as a force-bearing structure during tissue involution. To visualize the cell behaviors that produce this square grid, we performed time-lapse confocal imaging of embryos expressing β-catenin:GFP (McCartney et al., 2001). We found that these cells transition from an initially topologically disordered organization into four rows of square cells in three phases (Figure 1B–J). In Phase I (stage 11), cells rearrange and change shape on both sides of the embryo to produce two rows of rectangular cells that flank the ventral midline (Figure 1F,I, Movie S1). In Phase II (stage 12), every cell divides once perpendicular to the midline in a wave of oriented and largely synchronous mitoses that converts two rows of rectangular cells into four rows of predominantly square cells (Figure 1G,J, Movie S2). In Phase III (late stage 12 and stage 13), cells elongate along the apical-basal axis and microtubules reorient parallel to this axis, decreasing the total apical area of the grid (Figure 1H,J, Figure 2G–M, Movie S2). Phase III occurs simultaneously with the delamination of the midline cells, joining two rows on either side of the midline to produce a continuous square grid.
Figure 1. Spatiotemporally regulated cell alignment, division, and cell-shape change during square cell grid formation.
(A) Cells form a square cell grid (bracket) at the midline of the presumptive pharynx (left, stage 13), which ultimately forms the midline of the pharyngeal tube after involution (right, stage 15). Phosphotyrosine (green), β-catenin (red), and Par-3 (blue). (B,C) The percentage of 4-sided cells increased from 10±2% (mean±sem) in early Phase I to 59±4% in Phase III. The average number of neighbors decreased from 5.6±0.09 in early Phase I to 4.5±0.09 in late Phase I and 4.5±0.04 in Phase III. The percentage of 4-cell vertices significantly increased and the variance of the topological distribution significantly decreased in Phase III compared to early Phase I (p < 0.001, 4-cell vertices, p = 0.01, variance) or lateral cells in Phase III (p < 0.001, 4-cell vertices, p = 0.0002, variance) (unpaired t test). (D) Schematic of the square grid (yellow). (E–H) Wild-type embryos at stages 10 (E), 11 (F), 12 (G), or 13 (H) (β-catenin, white). (I,J) Stills from time-lapse movies of wild-type cells expressing β-catenin:GFP. (I) In Phase I (cell alignment, stage 11), rectangular cells align into rows on either side of the midline. (J) In Phase II (oriented division, stage 12), cells divide largely synchronously perpendicular to the midline. White lines, cells in anaphase or telophase. In Phase III (apicobasal cell elongation, stage 13), cells elongate along the apical-basal cell axis and midline cells delaminate from the epithelium, producing a compact square grid. t = 0 is the onset of Phase I (I) or the onset of the first division in Phase II (J). (n = 299–751 vertices in 86–398 cells in 4–7 embryos/stage) (*, p = 0.005 – 0.03, ** p < 0.005). Ventral views, anterior left. Arrowheads, ventral midline. Bars, 10 μm. See also Supplemental Movies 1 and 2.
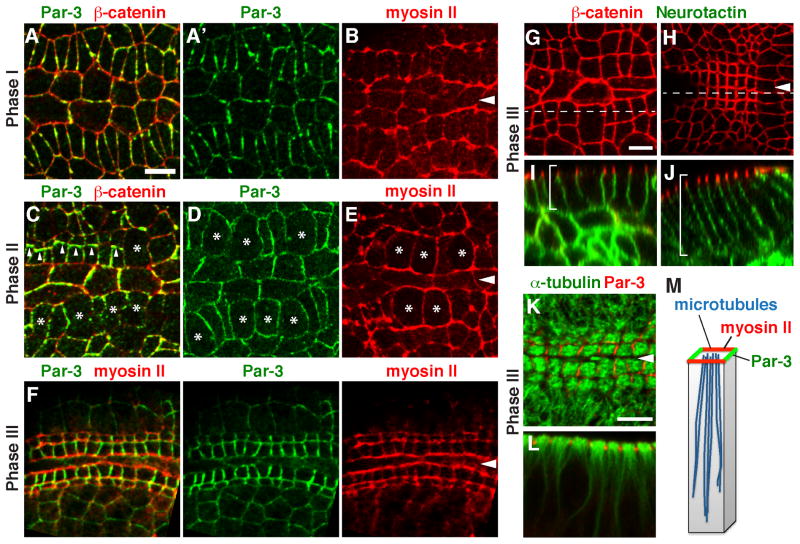
Figure 2. Square grid cells display planar polarized distributions of myosin II and Par-3.
(A–F) Localization of Par-3, β-catenin, and myosin II (Myosin:GFP) in wild-type embryos in Phase I (stage 11) (A, B), Phase II (stage 12) (C–E), or Phase III (stage 13) (F). Myosin II (red) is parallel to the midline in all three phases (B, E, and F). Par-3 (green) is perpendicular to the midline in Phases I (A) and III (F). Par-3 levels were transiently reduced at the cortex of dividing cells (asterisks) and transiently localized to new interfaces between daughter cells (arrowheads) in Phase II (C,D). (G–J) Cells undergo apicobasal elongation in Phase III (early Phase III, left; late Phase III, right). Brackets indicate the lateral cell membrane. (K and L) Microtubules align with the apical-basal axis in Phase III. (M) Schematic. Arrowheads, ventral midline. Bars, 5 μm. See also Figure S1.
Using quantitative measures of cell topology (Zallen and Zallen, 2004), we found that the fraction of 4-sided cells increased from 10% of cells in early Phase I to more than half of cells (59%) in Phase III (Figure 1B,C). This transition was accompanied by an increase in the fraction of vertices where four cells meet and a decrease in the variance of the topological distribution, consistent with a square cell packing configuration (Figure 1B). Cell divisions in Phase II had no effect on the topological distribution (Figure 1C), in contrast to the effects of cell proliferation in other tissues (Gibson et al., 2006), suggesting that square cell packing is actively maintained throughout division. These results demonstrate that the anterior ventral epithelium of the Drosophila embryo converts from a disordered epithelium into a highly organized square cell grid through cell alignment, cell division, and apicobasal cell elongation.
Square grid cells display planar polarity
The spatially regulated cell behaviors in the developing pharynx indicate that square grid cells are polarized in the plane of the tissue. To identify the molecular basis of this polarity, we analyzed the localization of proteins involved in cell adhesion and actomyosin contractility, which are planar polarized in other contexts (Zallen and Wieschaus, 2004). The nonmuscle myosin II motor protein localized to cell interfaces that were oriented perpendicular to the midline during cell alignment in Phase I (Figure S1). As cells reorganized into two rows in late Phase I, myosin redistributed to cell interfaces that were oriented parallel to the midline (Figure 2B). This localization was maintained throughout cell division and apicobasal elongation in Phase II and III (Figure 2E,F).
The adherens junction regulator Par-3 was also dynamically localized during square grid formation. Par-3 localized to cell interfaces oriented perpendicular to the midline in late Phase I, complementary to myosin (Figure 2A). Par-3 was then downregulated at the cortex during cell division in Phase II and transiently accumulated at new interfaces between daughter cells after division (Figure 2C,D). By the end of square grid formation in Phase III, Par-3 relocalized to cell interfaces oriented perpendicular to the midline, complementary to myosin II and rotated by 90° compared to its distribution in the rest of the embryo (Figure 2F) (Zallen and Wieschaus, 2004). These results suggest the presence of signals in the square cell grid that actively reorient planar polarity relative to the ventral midline.
Pins/LGN is required for oriented cell divisions during square grid formation
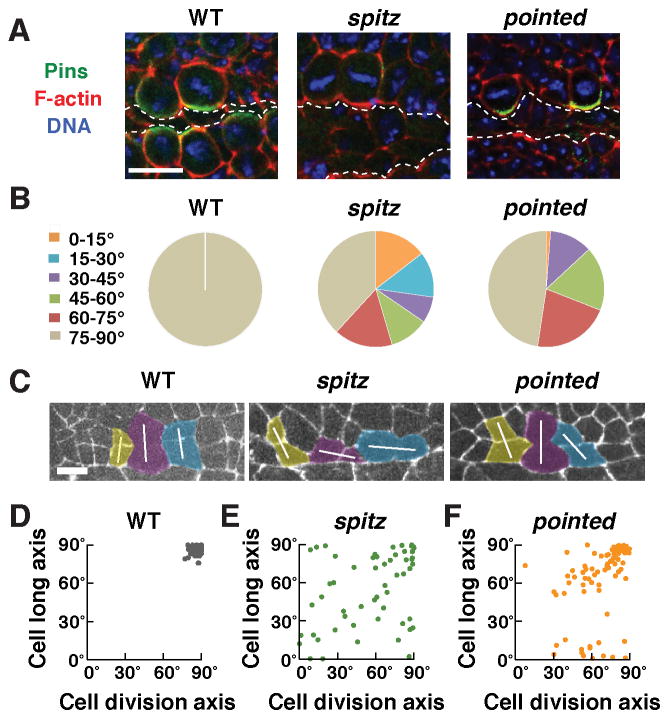
Every cell in the developing grid divides once in Phase II to convert two rows of rectangular cells into four rows of square cells. We analyzed the spatiotemporal pattern of these cell divisions in time-lapse movies. Cells in the square cell grid divided perpendicular to the ventral midline with striking precision (100% of cells divided at 75–90° relative to the midline) (Figure 3A,B). Cell division initiated near the anterior end of the grid and proceeded toward the posterior and extreme anterior ends of the grid in a wave-like fashion (Figure S2B; Movie S2). In contrast to their striking alignment at anaphase, mitotic spindles initially formed at a wide range of orientations in prophase (Figure 3C,G). A majority of spindles rotated within the epithelial plane during prometaphase and metaphase, such that most spindles were perpendicular to the midline at anaphase (Figure 3G). Mitotic spindles in the square cell grid thus rotate by up to 90° to divide perpendicular to the ventral midline.
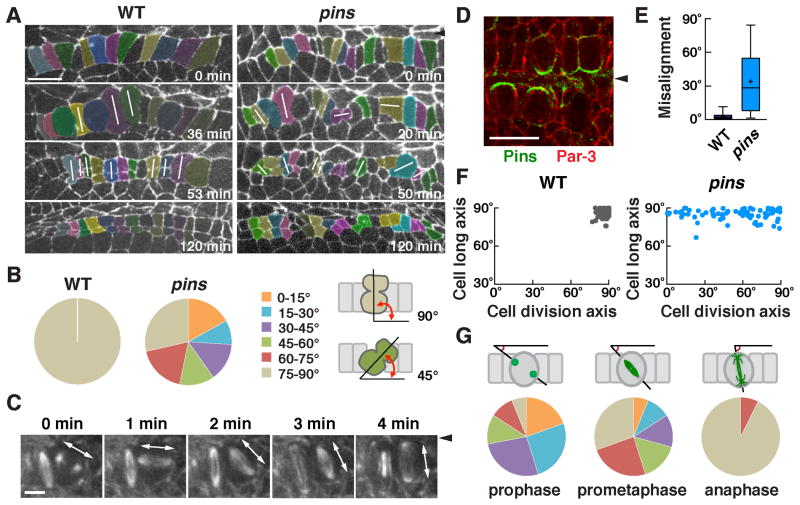
Figure 3. Pins/LGN is asymmetrically localized and is required to orient cell division during square grid formation.
(A) Stills from time-lapse movies of wild-type and pins mutant embryos expressing β-catenin:GFP. White lines indicate cells in anaphase or telophase. (B) Quantification of the cell division axis (0° is parallel to the ventral midline). All wild-type cells divided at 75–90° relative to the midline. Cell divisions were frequently misoriented in pins mutants (p < 0.001) (Chi square test). (C) Stills from a time-lapse movie of wild-type cells expressing Jupiter:mCherry. One spindle rotates 90° in the epithelial plane. (D) Pins/LGN accumulates at cell interfaces contacting midline cells in Phase II (stage 12). (E) The gap between the cell division axis and the cell long axis was significantly larger in pins mutants (p < 0.001) (unpaired t test). Boxes, 25th to 75th percentile; whiskers, 2.5th to 97.5th percentile; horizontal line, median; +, mean. Plot shows the distribution of average values across embryos. (F) The cell division axis and the cell long axis were both close to perpendicular to the ventral midline in wild type. The orientation of the cell long axis in pins mutants (85°±0.5°) was similar to wild type (86°±0.4°), but cell division occurred at a much wider range of orientations. (G) Angular distributions of the aligned centrosomes at prophase and spindles at prometaphase or anaphase. n = 73–77 cells in 3 embryos/genotype in B, E, and F and 82 cells in 5 embryos in G. Arrowheads, ventral midline. Bars, 10 μm in A,D, 5 μm in C. See also Figure S2.
It has been proposed that cell divisions invariably follow cell shape anisotropy so that cells divide along their long axis, a property known as Hertwig’s rule (Thery et al., 2007; Vogel et al., 2007; Minc et al., 2011). Consistent with this, 100% of cell divisions in wild type were aligned with the long axis of the cell (Figure 3A,B). However, it is not known if spindle alignment is the result of mechanical constraints of cell shape, or if the cell division axis is actively controlled by cortical molecular cues that determine spindle position. To distinguish between these models, we analyzed the localization and function of Pins/LGN, a conserved protein that regulates spindle positioning by recruiting the Dynein-dynactin complex (Kotak and Gonczy, 2013; McNally, 2013). We found that Pins localized asymmetrically to the cortical domain in contact with the ventral midline in dividing cells (Figure 3D). The Pins binding partner Mud/NuMA partially colocalized with Pins in this domain (Figure S2A). To test if Pins activity is required for spindle orientation, we analyzed cell division orientation in embryos lacking maternal and zygotic Pins, referred to as pins mutants. Although cell shapes in pins mutants were normal at the onset of cell division, cells divided at a much wider range of orientations than in wild type, such that a majority of divisions (71%) failed to align with the cell long axis (Figure 3A,B,E,F). Notably, 17% of cells divided parallel to the short axis of the cell, which never occurred in wild type (Figure 3B,F). In addition, 17% of cells divided out of the epithelial plane, which occurred in only 3% of wild-type cells. As a consequence, cells in pins mutants failed to organize into rows in Phase III. These results demonstrate that oriented cell divisions in the square cell grid do not simply result from geometric constraints due to spindle alignment with the long axis of the cell. Instead, oriented cell divisions are actively and precisely spatially regulated by the Pins/LGN cortical protein.
The transcriptional activator Cap ‘n’ collar is necessary and sufficient for square grid formation
We next sought to identify the upstream spatial cues that induce square cell packing, planar polarity, and Pins asymmetry in the developing square grid. Pins asymmetry and square grid formation do not require Inscuteable or core components of the PCP pathway, which regulate Pins localization in other contexts (Figure S3 and data not shown) (Schaefer et al., 2000; Yu et al., 2000; Bellaiche et al., 2004), prompting us to search for alternative regulators that control square grid formation. The basic leucine zipper transcription factor Cap ‘n’ collar (Cnc) is expressed in the anterior ventral region of the embryo and is necessary for pharynx development (McGinnis et al., 1998). In addition, ubiquitous Cnc expression transforms the abdominal ventral cuticle into a structure resembling the pharynx (Veraksa et al., 2000). We analyzed the cellular basis of the defects in cnc mutants and found that all three phases of square grid formation were strongly defective (Figure 4B,C). These defects were accompanied by a disruption of several molecular features of square grid formation, including a loss of Par-3 planar polarity, a failure of apicobasal microtubule reorientation, and inconsistent Pins asymmetry (Figure 4C). These results demonstrate that Cnc activity is essential for square grid formation.
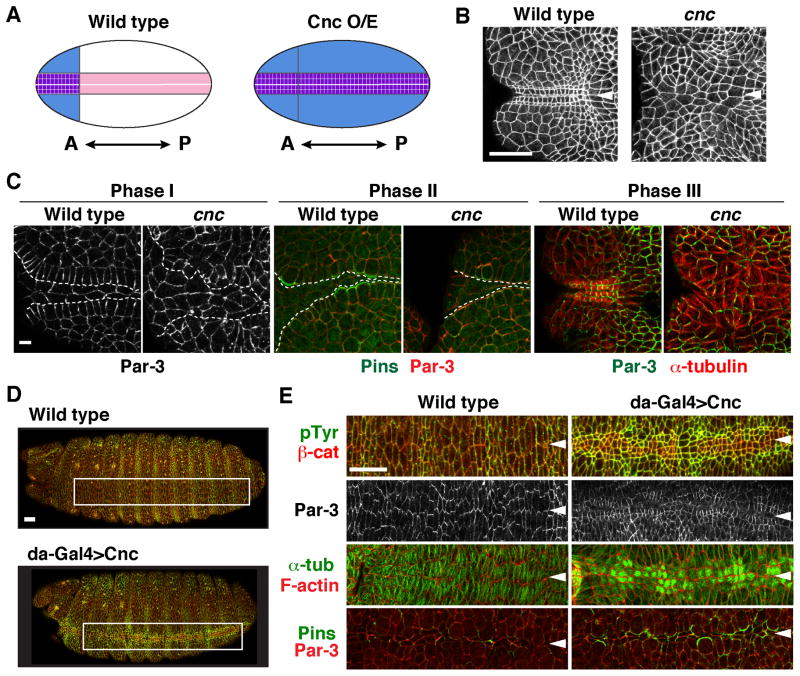
Figure 4. The Cap ‘n’ collar transcription factor is necessary and instructive for square grid formation.
(A) Schematic of results. Cnc (blue) is expressed in the anterior ventral embryo during square grid formation (left). The wild-type grid forms within this domain. Ubiquitous Cnc expression (Cnc O/E) induces a square grid (purple) along the entire ventral midline (right). (B) The square grid fails to form in cnc mutants (stage 13). (C) The cnc mutants displayed defects in multiple processes required for square grid formation, including cell alignment in Phase I (left), Pins asymmetry in Phase II (middle), and apicobasal microtubule reorganization in Phase III (right). Dashed lines, boundaries between midline cells and the square grid. (D) Ubiquitous Cnc expression with the da-Gal4 driver induced square cells along the ventral midline (stage 14, boxed regions shown at higher magnification in E). (E) These cells displayed several features of the wild-type grid, including square cell packing (stage 14) (top panels), Par-3 localization perpendicular to the midline (stage 13), microtubule reorganization (stage 13), and Pins asymmetry (stage 11). Arrowheads, ventral midline. Bars, 5 μm in C, 20 μm in B, D, and E.
To test if Cnc activity is sufficient to induce square cell packing, we analyzed cell morphology in embryos that misexpress Cnc ubiquitously throughout the embryo. In contrast to wild type, in which square cells are restricted to the anterior ventral region of the embryo, ubiquitous Cnc expression induced an ectopic square grid along the entire ventral midline (Figure 4A,D). Ectopic square cells displayed several molecular signatures of the wild-type grid, including square cell shape, apicobasal elongation, Pins asymmetry, microtubule reorganization, and the reorientation of Par-3 planar polarity (Figure 4E). These results demonstrate that the precise positioning of square grid structures requires the spatially restricted expression of the Cnc transcriptional regulator.
A midline signal is required for square grid formation
Although Cnc was expressed throughout the embryo in these misexpression experiments, uniform Cnc only produced an ectopic square grid in cells closest to the ventral midline. Moreover, Cnc is normally expressed throughout the anterior ventral epithelium, but only the cells closest to the midline produce a square grid. These results suggest that other signals in addition to Cnc are necessary for square grid formation. To identify the source of these signals, we analyzed embryos mutant for single-minded (sim), which specifies midline cells (Mayer and Nusslein-Volhard, 1988). The square grid did not form in sim mutants (Figure S4), indicating that the ventral midline is necessary for square grid formation.
Several extracellular ligands are secreted at the ventral midline, including the EGFR ligand Spitz. Phospho-ERK, a readout of Spitz-EGFR signaling, is upregulated in cells near the ventral midline (Gabay et al., 1997). We analyzed the spatial pattern of phospho-ERK activity relative to the square cell grid and found that the high phospho-ERK signal was specific to ventral midline cells and the first row of square grid cells in contact with the midline (Figure 5A,B). Phospho-ERK signal rapidly decreased in daughter cells that were no longer in contact with the midline after division (Figure 5C,D; Figure S5A,D). These results demonstrate that EGFR signaling is specifically upregulated in square grid cells that contact the ventral midline.
Figure 5. EGF receptor signaling is required for square grid formation.
(A–D) MAP kinase activity (indicated by dpERK signal) was increased in the first row of square grid cells adjacent to the midline in phase I (A and B). DpERK signal decreased in lateral daughter cells after division (C,D). DpERK (red), Par-3 (green), E-cadherin (blue). (E–J) Cell alignment and apicobasal cell elongation were disrupted in spitz (F,I) and pointed (G,J) mutants compared to wild type (E,H) (F-actin, white; square grid region, yellow). (K, L) The percentage of four-sided cells was significantly lower in spitz (p = 0.0001 in late Phase I, 0.0002 in Phase III) and pointed mutants (p ≤ 0.0001 in late Phase I and Phase III) compared to wild type (unpaired t test, n = 70–145 cells in 4 embryos/genotype in Phase I and 145–398 cells in 4–7 embryos/genotype in Phase III). (M,N) Myosin II localization parallel to the midline did not occur between adjacent rows of square grid cells in pointed mutants, but myosin accumulation at interfaces contacting the midline occurred normally. Bars, 10 μm (A, E–J), 5 μm (B–D,M,N).
EGF receptor signaling is required for planar polarized cell behavior in the square cell grid
To test if EGFR signaling is required for square grid formation, we anlayzed mutants that lack the EGFR ligand Spitz or its downstream effector Pointed, a conserved ETS domain transcription factor (Brunner et al., 1994; O’Neill et al., 1994). In spitz mutant embryos, cells failed to align into two rows of cells in Phase I and very few square cells were present at the end of Phase III (Figure 5E–L). In addition, spitz mutants displayed a strong reduction in myosin II and Par-3 planar polarity and microtubules failed to reorient parallel to the apical-basal axis (Figure S5F,G and data not shown). Moreover, whereas cell divisions in wild type were consistently oriented perpendicular to the midline, more than half of cells divided at nonoptimal orientations in spitz and pointed mutants (Figure 6B). These results demonstrate that EGFR signaling is necessary for multiple aspects of square grid formation.
Figure 6. The EGF receptor ligand Spitz is required for Pins asymmetry and oriented cell division.
(A) Pins asymmetry was absent in spitz mutants (0/19 cells in 6 embryos), but occurred normally in pointed mutants (25/27 cells in 7 embryos), similar to wild type (33/35 cells in 5 embryos). (B) Quantification of the cell division axis (0° is parallel to the ventral midline). All wild-type cells divided at 75–90° relative to the midline. Cell divisions were frequently misoriented in spitz (62%) and pointed (52%) mutants (55–84 cells in 3–4 embryos/genotype). (C) Stills from time-lapse movies of wild-type, spitz, and pointed mutant embryos expressing β-catenin:GFP. White lines indicate the division axis. (D) The cell division axis correlates with the long axis of the cell in wild type. (E,F) Cells divided at a wider range of orientations and the cell division axis was not well correlated with the cell long axis in spitz (linear correlation coefficient R2 = 0.14) and pointed (R2 = 0.18) mutants. The orientation of the cell long axis in spitz mutants (55°±4°) was significantly different from wild type (86°±0.4°), but was less strongly affected in pointed mutants (64°±3°) (p = 0.06, spitz vs. pointed) (unpaired t test). Bars, 10 μm in A, 5 μm in C.
We next tested whether the EGFR transcriptional effector Pointed mediates some or all of the effects of Spitz on square grid formation, as some outputs of EGFR signaling, such as phospho-ERK activation, are retained in pointed mutants (Figure S5C). We found that square cell shape, microtubule reorientation, and myosin II and Par-3 planar polarity were all strongly defective in pointed mutants (Figure 5G,J–N; Figure S5F,H). However, in contrast to spitz mutants, in which Pins was no longer detected at the cortex of dividing cells, the asymmetric accumulation of Pins at the cortex of dividing cells occurred normally, and cell divisions were less strongly misoriented in pointed mutants (Figure 6). Only 1% of cells in pointed mutants divided at an orientation that was rotated by more than 60° from the correct orientation, compared to 27% of cells in spitz mutants and no wild-type cells (Figure 6B,C). The cell division axis was insensitive to the cell long axis in spitz mutants, consistent with the idea that the mitotic spindle is unable to sense cell shape in the absence of cortical Pins asymmetry (Figure 6D,E). By contrast, Pins localization was uncoupled from cell shape in pointed mutants. The division axis largely followed the long axis of the cell when these two inputs agreed and was more variable when these two signals conflicted in cells whose long axis was parallel to the midline (Figure 6F). Together, these results demonstrate that Spitz regulates cell shape and Par-3 and myosin II planar polarity through Pointed-dependent mechanisms but controls Pins asymmetry independently of Pointed.
Ectopic EGFR signaling is sufficient to induce square cell packing in epithelia
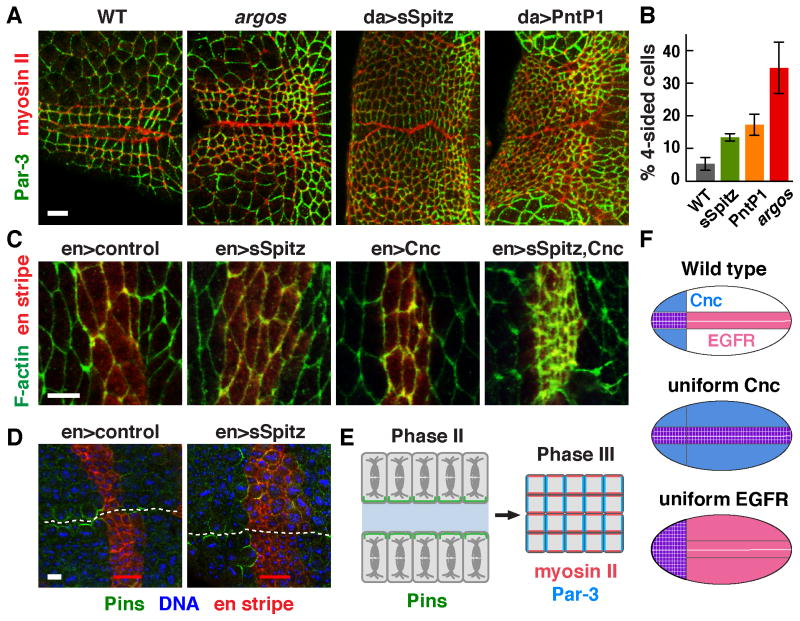
As EGFR signaling is necessary for square grid formation, we next asked if EGFR signaling is sufficient for square cell packing. To test this possibility, we analyzed two types of embryos with altered EGFR signaling. We first analyzed embryos mutant for the inhibitory EGFR ligand Argos, which have a broader but still graded pattern of EGFR signaling (Figure S5E) (Gabay et al., 1997). Second, we ubiquitously expressed a secreted form of Spitz (sSpitz) or a constitutively active form of Pointed (PntP1) using the da-Gal4 driver to induce EGFR signaling in cells where this pathway is not normally active. We found that the square grid was expanded by 3–4 rows of cells in argos mutants and by 14–16 rows of cells in embryos expressing sSpitz or PntP1 (Figures 7A,B). In addition, microtubule reorientation, apical myosin recruitment, and apicobasal cell elongation occurred in a broader domain (Figure 7A; Figure S6A,S6B). Similar changes in cell morphology were induced when both Cnc and sSpitz were expressed together in the main body of the embryo, but not when either one was expressed alone (Figure 7C). These results indicate that Cnc and EGFR signaling act combinatorially to induce multiple features of square grid formation (Figure 7F).
Figure 7. Ectopic EGF receptor signaling induces square cell packing and Pins asymmetry.
(A) Expanded EGFR signaling induces a broader region of square-like cells in argos mutants and in embryos that ubiquitously express secreted Spitz (sSpitz) or Pointed P1 (PntP1) with the da-Gal4 driver. (B) Increased percentage of four-sided cells in the lateral region (excluding cells in the two most ventral rows on both sides of the midline) of argos (p = 0.02), da>sSpitz (p = 0.02) and da>PntP1 (p = 0.03) embryos compared to wild type (unpaired t test, n = 193–397 cells in 3 embryos/genotype). (C) Using the en-Gal4 driver to produce ectopic expression in stripes outside of the Cnc domain, expression of sSpitz and Cnc together, but not either one alone, induced apicobasal cell elongation characteristic of the square cell grid. (D) Expression of sSpitz with the en-Gal4 driver induced Pins asymmetry in dividing cells adjacent to this domain (0/24 dividing cells in control en>mCherry:Moesin embryos compared to 39/45 dividing cells in en>sSpitz, mCherry:Moesin embryos). These effects were restricted to the anteriormost en stripe located within the Cnc domain. En stripes were visualized by coexpression of mCherry:Moesin (red). (E) Patterned EGF receptor signaling and Cnc activity activate cell alignment, oriented cell division, and apicobasal cell elongation to produce a square cell grid at the midline of the developing pharynx. (F) Uniform Cnc expression or EGFR activity produce an expanded square grid in the region where both signals overlap. Bars, 5 μm.
To ask if localized EGFR signaling plays an instructive role in establishing planar polarity, we misexpressed sSpitz in stripes of cells that are oriented perpendicular to the ventral midline using the en-Gal4 driver, which ectopically activated EGFR signaling in 3–4 rows of cells parallel to the stripe (Figure S6D). A striped source of sSpitz induced F-actin and myosin apical relocalization in cells anterior to the stripe, reminiscent of square grid cells (Figure S6E,F). We found that striped sSpitz expression induced Pins asymmetry in cells anterior to the first en stripe, which overlaps with cells that endogenously express Cnc (Figure 7D). Notably, Pins asymmetry was primarily induced in dividing cells anterior to the first en stripe within the Cnc domain (39/45 cells), whereas Pins asymmetry was much less frequent in dividing cells posterior to the first en stripe, which are just outside of the Cnc domain (2/21 cells). Conversely, uniform sSpitz expression disrupted Pins asymmetry (Figure S6C). These results support a model in which a Spitz midline signal orients planar polarity in the square cell grid by activating spatially regulated EGFR activity (Figure 7E).
Discussion
Cells display dynamic and diverse morphologies during development, but the strategies by which differentiated tissues achieve precise shapes and patterns are not well understood. Here we identify a developmental program that generates a highly ordered square cell grid in the Drosophila embryo through spatiotemporally regulated cell alignment, cell division, and apicobasal cell elongation. EGFR signaling activated by the midline ligand Spitz is necessary for all aspects of planar polarity in the square cell grid. Spitz provides an instructive signal that induces and orients Pins/LGN asymmetry, and Pins in turn is required to orient mitotic spindles perpendicular to the midline. The square grid is positioned at the anterior ventral midline of the embryo at the intersection of EGFR activation and Cnc expression, and ectopic Cnc expression or EGFR activation produces an expanded domain of square cell packing in the region where both signals overlap. These results indicate that Cnc transcriptional targets modulate the cellular response to the Spitz ligand to allow EGFR signaling to stimulate cells to adopt a square packing configuration. Unique functional outputs produced at the intersection of EGFR signaling with transcriptional patterning systems may provide a general mechanism that refines the response to EGFR signaling to generate diverse tissue architectures during development.
The organization of the square cell grid deviates substantially from a low-energy hexagonal cell packing configuration (Classen et al., 2005; Farhadifar et al., 2007), providing a new challenge for determining how highly ordered and apparently unstable square cell configurations are generated. Rectangular arrays are present in mature organs in many organisms, including pillar cells in the mouse cochlea, which are required for hearing, and in the zebrafish retina, which is essential for vision (Mueller et al., 2002; Salbreux, et al., 2012), as well as during development in other organisms (Gerberding et al., 2002). The square grid in the Drosophila pharynx displays three distinctive features. First, cells meet at predominantly four-cell vertices, which are established prior to and maintained throughout dramatic cell-shape changes during cell division. Vertices where more than three cells meet are extremely transient in remodeling epithelia (Zallen and Zallen, 2004; Blankenship et al., 2006). This suggests that specific mechanisms are required to actively stabilize higher-order interactions between cells in the square cell grid, perhaps making them resistant to further rearrangement. Second, cell boundaries meet precisely at right angles to produce a square cell shape. This shape is not recapitulated by current models of differential tension or adhesion (Farhadifar et al., 2007; Salbreux et al., 2012), suggesting that additional physical properties are required to achieve this regular cell geometry. Third, square grid cells elongate substantially along the apical-basal axis to achieve a localized, highly columnar morphology. These three features are predicted to thicken and stabilize this structure and allow it to withstand the strong mechanical forces generated by involution to produce the final three-dimensional organization of the pharynx.
Several fundamental mechanisms that control cell shape are spatially and temporally regulated to produce the distinctive structure of the square cell grid. Square grid cells display a planar polarized distribution of proteins involved in contraction and adhesion, which could provide a molecular basis for physical parameters that produce square-like packing in theoretical models (Salbreux et al., 2012; Bardet et al., 2013). Differences in EGFR signaling have been shown to activate localized myosin contractility, which drives cell-shape changes and cell rearrangements in other contexts (Nishimura et al., 2007; Saxena et al., 2014). Alternatively, square cell assemblies could be induced by selective cell adhesion, as planar polarized Par-3 localization regulates differential adhesion in Drosophila (Simoes et al., 2010), and heterotypic interactions between transmembrane nectin proteins organizes cells into a checkerboard pattern in the mouse cochlea (Togashi et al., 2011). EGFR signaling promotes trapezoidal cell shapes in Drosophila wing vein cells (O’Keefe et al., 2012), reminiscent of cell behaviors in the square cell grid. The reorganization of contractile and adhesive proteins by EGFR signaling could play a general role in inducing square cell packing configurations in epithelia.
Oriented cell divisions are required to maintain the square grid structure. We show that these divisions require the asymmetrically localized Pins/LGN protein, which systematically aligns mitotic spindles perpendicular to the midline. Pins aligns cell divisions with the apical-basal axis in Drosophila neuroblasts (Parmentier et al., 2000; Schaefer et al., 2000; Yu et al., 2000) and mouse skin (Lechler and Fuchs, 2005) and maintains the axis of cell division within the plane of the tissue in Drosophila sensory organ precursors (David et al., 2005). In addition, uniform Pins at the lateral cell cortex allows cells to divide within the plane of the tissue, but not along any particular axis (Hao et al., 2010; Bergstralh et al., 2013). Here we demonstrate that Pins regulates the precise orientation of cell division in the plane of the tissue during square grid formation. Pins planar polarity in this system does not require core PCP pathway components or Inscuteable, which control Pins localization in other cell types (Schaefer et al., 2000; Yu et al., 2000; Bellaiche et al., 2004). Instead, we show that Pins planar polarity in the square cell grid requires the spatially localized EGFR ligand Spitz. Spitz activity is necessary for all aspects of square grid formation, and an ectopic source of Spitz is sufficient to induce and orient Pins asymmetry in adjacent dividing cells. These results define a novel mechanism by which spatially regulated EGFR signaling regulates tissue organization by directing Pins localization and oriented cell division. Notably, cells fail to divide along their long axis in the absence of Pins, demonstrating that a localized cortical signal, rather than geometric constraints of cell shape, is the critical signal that regulates spindle positioning in this system. However, Pins asymmetry is not sufficient to achieve consistently oriented cell divisions in pointed mutants, suggesting that cell shape may be a parallel input that reinforces the ability of cortical spindle positioning cues to faithfully orient cell division.
EGFR signaling is required for epithelial organization in an extraordinarily diverse range of tissues, including the Drosophila eye, wing, embryo, and oocyte (Nilson and Schupbach, 1999; Shilo, 2003) and the mammalian skin, lung, pancreas, gastrointestinal tract, and mammary gland (Wieduwilt and Moasser, 2008). Crosstalk with other developmental pathways allows EGFR signaling to specify diverse cell fates (Shilo, 2014). In addition, deregulated EGFR signaling can lead to cancer (Scaltriti and Baselga, 2006; da Cunha Santos et al., 2011). The mammalian homolog of Cnc, Nrf2, is highly expressed in non-small-cell lung cancer cells and promotes cell proliferation in response to EGFR activation (Yamadori et al., 2012), suggesting that the functional interaction between Cnc/Nrf2 and EGFR signaling is conserved. Although transcriptional activation is the ultimate outcome of EGFR signaling in many contexts, the effector mechanisms by which EGFR signaling carries out its diverse functions are not well understood. We show that cell alignment and apicobasal cell elongation in the square cell grid require the major transcriptional effector of the EGFR pathway, Pointed, similar to the effects of EGFR signaling on actomyosin contractility and cell adhesion in the Drosophila eye and trachea (Gaengel and Mlodzik, 2003; Brown et al., 2006; Nishimura et al., 2007; Robertson et al., 2012). The transcriptional targets of Pointed that regulate actomyosin contractility, and how these targets influence the organization of the contractile and junctional machinery within cells, remain to be identified. By contrast, Pins asymmetry in the square cell grid is independent of Pointed and requires alternative and possibly nontranscriptional effectors of EGFR signaling. EGFR signaling directs many biological processes independently of Pointed, including cell survival (Bergmann et al., 2002), tissue growth (Cabernard and Affolter, 2005), cell adhesion (Cela and Llimargas, 2006), and cell migration (Bianco et al., 2007). Identification of the effectors of EGFR signaling that regulate diverse properties of cell and tissue organization will help to elucidate the cell biological mechanisms that underlie the important roles of EGFR signaling in development and disease.
Experimental procedures
Fly stocks and genetics
Oregon-R was the wild-type control unless otherwise specified. Embryos were raised at 21–23°C. Alleles were cnc03921 (McGinnis et al., 1998), spi1 (Mayer and Nüsslein-Volhard, 1988), pntΔ88 (Scholz et al., 1993), sim2 (Thomas et al., 1988), pinsp62 (Yu et al., 2000), argosΔ7 (Freeman et al., 1992), dsh1 (Krasnow et al., 1995), and vangA3 (Taylor et al., 1998). Germline clones were generated with the FLP-DFS system (Chou and Perrimon, 1996). Larvae of the following genotypes were heat-shocked and crossed to mutant males that contain twist-Gal4, UAS-GFP balancers:
hs-FLP; pinsp62 FRT82B/ovoD1 FRT82B
hs-FLP; β-catenin:GFP/+; pinsp62 FRT82B/ovoD1 FRT82B
hs-FLP; vangA3 FRT42D/ovoD1 FRT42D
Homozygous mutant embryos were identified by the absence of GFP from the balancers. β-catenin:GFP (McCartney et al., 2001), Myosin:GFP (GFP fused to the myosin regulatory light chain Sqh) (Royou et al., 2004), and Jupiter:mCherry (Conduit et al., 2014) were expressed from endogenous promoters and UAS-mCherry:Moesin (Millard and Martin, 2008) was expressed from the en-Gal4 driver. For overexpression of Cnc, secreted Spitz, and Pointed P1, UAS-CncB (Veraksa et al., 2000), UAS-sSpitz (Schweitzer, et al., 1995) and UAS-pntP1 (Klaes et al., 1994) males were crossed to females of the following genotypes and the progeny were analyzed: (1) da-Gal4, (2) Myosin:GFP; da-Gal4,(3) en-Gal4, UAS-mCherry:Moesin.
Immunohistochemistry
Antibodies were mouse Arm/β-catenin (1:25, Developmental Studies Hybridoma Bank, DSHB), rat DE-cadherin (1:25, DSHB), mouse Neurotactin (1:100, DSHB), guinea pig Bazooka/Par-3 (1:1000) (Blankenship et al., 2006), mouse phosphotyrosine (1:250, 4G10 Millipore), rabbit GFP (1:100, Torrey Pines), mouse GFP (1:25, Roche), mouse α-tubulin (1:400, Sigma), and rabbit phospho-ERK1/2 (Thr202/Tyr204) (dpERK) (1:100, D13.14.4E, Cell Signaling). Rabbit Pins (1:50) and rabbit Mud (1:50) were gifts from F. Matsuzaki (Izumi et al., 2006). Embryos were fixed 9 min in 1:1 37% formaldehyde:heptane (Sigma) and manually devitellinized. For Neurotactin and phosphotyrosine antibodies, embryos were boiled 10 s in 0.03% Triton X-100/0.4% NaCl, cooled on ice, and devitellinized in heptane/methanol. Secondary antibodies conjugated to Alexa-488, Alexa-568, or Alexa-647 and phalloidin (Molecular Probes) were used at 1:500. Embryos were mounted in Prolong Gold with or without DAPI (Molecular Probes) and imaged on a Zeiss LSM700 META confocal microscope with a PlanNeo 40×/1.3NA objective. 1.0 μm z slices were acquired at 0.5 μm steps. Maximum intensity projections of 1.5–3 μm in the apical junctional domain were analyzed for cell division, cell topology, myosin II, Par-3, α-tubulin, F-actin, and dpERK localization, and 1.5–2 μm more basal slices were projected to analyze Pins and Mud localization.
Quantitative image analysis
To quantify cell topology, we analyzed one row of cells on each side of the ventral midline in Phase I and two rows of cells on each side of the ventral midline in Phase III. Embryos were staged by salivary gland morphology. The cell division axis was analyzed in time-lapse movies of embryos expressing β-catenin:GFP. The cell division axis was measured relative to the ventral midline at the onset of cytokinesis. The cell long axis was the orientation of the longest span of the cell in interphase before cells rounded up to initiate division.
Time-lapse imaging
Embryos were dechorionated 1 min in 50% bleach, washed in water, mounted in halocarbon oil 27 (Sigma), and imaged with a Perkin Elmer Ultraview RS5 spinning disk confocal using Metamorph or Volocity software and a PlanApo 63x/1.4NA oil-immersion objective (Zeiss). For β-catenin:GFP, maximum intensity projections of a 1–4 μm region containing the junctional signal were analyzed. For Jupiter:mCherry, all slices containing microtubule signal were projected.
Supplementary Material
Acknowledgments
We thank Fumio Matsuzaki, William McGinnis, Dirk Bohmann, and the Bloomington Drosophila Stock Center at Indiana University for fly stocks and antibodies and WIll Razzell for the en-Gal4, mCherry:Moesin recombinant. We thank the members of the Zallen lab and Richard Zallen for comments on the manuscript. This work was supported by NIH/NIGMS R01 grant GM079340 and a Burroughs Wellcome Fund Career Award in the Biomedical Sciences to JAZ. JAZ is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aigouy B, Farhadifar R, Staple DB, Sagner A, Roper JC, Julicher F, Eaton S. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophlia. Cell. 2010;142:772–786. doi: 10.1016/j.cell.2010.07.042. [DOI] [PubMed] [Google Scholar]
- Bardet PL, Guirao B, Paoletti C, Serman F, Leopold V, Bosveld F, Goya Y, Mirouse V, Graner F, Bellaiche Y. PTEN controls junction lengthening and stability during cell rearrangement in epithelial tissue. Dev Cell. 2013;25:534–546. doi: 10.1016/j.devcel.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Beaudoin-Massiani O, Stuttem I, Schweisguth F. The planar cell polarity protein Strabismus promotes Pins anterior localization during asymmetric division of sensory organ precursor cells in Drosophila. Development. 2004;131:469–478. doi: 10.1242/dev.00928. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Tugentman M, Shilo BZ, Steller H. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev Cell. 2002;2:159–70. doi: 10.1016/s1534-5807(02)00116-8. [DOI] [PubMed] [Google Scholar]
- Bergstralh DT, Lovegrove HE, St Johnston D. Discs large links spindle orientation to apical-basal polarity in Drosophila epithelia. Curr Biol. 2013;23:1707–1712. doi: 10.1016/j.cub.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rørth P. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–5. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell. 2006;11:459–470. doi: 10.1016/j.devcel.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Brown KE, Baonza A, Freeman M. Epithelial cell adhesion in the developing Drosophila retina is regulated by Atonal and the EGF receptor pathway. Dev Biol. 2006;300:710–21. doi: 10.1016/j.ydbio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- Cabernard C, Affolter M. Distinct roles for two receptor tyrosine kinases in epithelial branching morphogenesis in Drosophila. Dev Cell. 2005;9:831–42. doi: 10.1016/j.devcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Cela C, Llimargas M. Egfr is essential for maintaining epithelial integrity during tracheal remodelling in Drosophila. Development. 2006;133:3115–25. doi: 10.1242/dev.02482. [DOI] [PubMed] [Google Scholar]
- Chacon-Heszele MF, Ren D, Reynolds AB, Chi F, Chen P. Regulation of cochlear convergent extension by the vertebrate planar cell polarity pathway is dependent on p120-catenin. Development. 2012;139:968–978. doi: 10.1242/dev.065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–9. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen AK, Anderson KI, Marois E, Eaton S. Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Dev Cell. 2005;9:805–817. doi: 10.1016/j.devcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Conduit PT, Richens JH, Wainman A, Holder J, Vicente CC, Pratt MB, Dix CI, Novak ZA, Dobbie IM, Schermelleh L, et al. A molecular mechanism of mitotic centrosome assembly in Drosophila. Elife. 2014;3:e03399. doi: 10.7554/eLife.03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- David NB, Martin CA, Segalen M, Rosenfeld F, Schweisguth F, Bellaïche Y. Drosophila Ric-8 regulates Galphai cortical localization to promote Galphai-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat Cell Biol. 2005;7:1083–1090. doi: 10.1038/ncb1319. [DOI] [PubMed] [Google Scholar]
- Dodds GS. Row formation and other types of arrangement of cartilage cells in endochondral ossification. Anat Rec. 1930;46:385–399. [Google Scholar]
- Farhadifar R, Röper JC, Aigouy B, Eaton S, Jülicher F. The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr Biol. 2007;17:2095–104. doi: 10.1016/j.cub.2007.11.049. [DOI] [PubMed] [Google Scholar]
- Freeman M, Klambt C, Goodman CS, Rubin GM. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992;69:963–975. doi: 10.1016/0092-8674(92)90615-j. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Gaengel K, Mlodzik M. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development. 2003;130:5413–5423. doi: 10.1242/dev.00759. [DOI] [PubMed] [Google Scholar]
- Gerberding M, Browne WE, Patel NH. Cell lineage analysis of the amphipod crustacean Parhyale hawaiensis reveals an early restriction of cell fates. Development. 2002;129:5789–5801. doi: 10.1242/dev.00155. [DOI] [PubMed] [Google Scholar]
- Gibson MC, Patel AB, Nagpal R, Perrimon N. The emergence of geometric order in proliferating metazoan epithelia. Nature. 2006;442:1038–1041. doi: 10.1038/nature05014. [DOI] [PubMed] [Google Scholar]
- Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr Biol. 2010;20:1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Kruze G, Ullmann B, BreMiller RA, Larison KD, Snyder HC. The shaping of pharyngeal cartilages during early development of the zebrafish. Dev Biol. 1998;15:245–63. doi: 10.1006/dbio.1998.9016. [DOI] [PubMed] [Google Scholar]
- Klaes A, Menne T, Stollewerk A, Scholz H, Klambt C. The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell. 1994;78:149–160. doi: 10.1016/0092-8674(94)90581-9. [DOI] [PubMed] [Google Scholar]
- Kotak S, Gönczy P. Mechanisms of spindle positioning: cortical force generators in the limelight. Curr Opin Cell Biol. 2013;25:741–748. doi: 10.1016/j.ceb.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Krasnow RE, Wong LL, Adler PN. Dishevelled is a component of the frizzled signaling pathway in Drosophila. Development. 1995;121:4095–4102. doi: 10.1242/dev.121.12.4095. [DOI] [PubMed] [Google Scholar]
- Kraut R, Campos-Ortega JA. Inscuteable, a neural precursor gene of Drosophila, encodes a candidate for a cytoskeleton adaptor protein. Dev Biol. 1996;174:65–81. doi: 10.1006/dbio.1996.0052. [DOI] [PubMed] [Google Scholar]
- Landsberg KP, Farhadifar R, Ranft J, Umetsu D, Widmann TJ, Bittig T, Said A, Julicher F, Dahmann C. Increased cell bond tension governs cell sorting at the Drosophila anteroposterior compartment boundary. Curr Biol. 2009;19:1950–1955. doi: 10.1016/j.cub.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Nusslein-Volhard C. A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev. 1988;2:1496–1511. doi: 10.1101/gad.2.11.1496. [DOI] [PubMed] [Google Scholar]
- McCartney BM, McEwen DG, Grevengoed E, Maddox P, Bejsovec A, Peifer M. Drosophila APC2 and Armadillo participate in tethering mitotic spindles to cortical actin. Nat Cell Biol. 2001;3:933–938. doi: 10.1038/ncb1001-933. [DOI] [PubMed] [Google Scholar]
- McGinnis N, Ragnhildstveit E, Veraksa A, McGinnis W. A cap ‘n’ collar protein isoform contains a selective Hox repressor function. Development. 1998;125:4553–4564. doi: 10.1242/dev.125.22.4553. [DOI] [PubMed] [Google Scholar]
- McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;21:131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard TH, Martin P. Dynamic analysis of filopodial interactions during the zippering phase of Drosophila dorsal closure. Development. 2008;135:621–626. doi: 10.1242/dev.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc N, Burgess D, Chang F. Influence of cell geometry on division-plane positioning. Cell. 2011;4:414–426. doi: 10.1016/j.cell.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier B, Pelissier-Monier A, Brand AH, Sanson B. An actomyosin-based barrier inhibits cell mixing at compartmental boundaries in Drosophila embryos. Nat Cell Biol. 2010;12:60–65. doi: 10.1038/ncb2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KL, Jacques BE, Kelley MW. Fibroblast growth factor signaling regulates pillar cell development in the organ of Corti. J Neurosci. 2002;22:9368–9377. doi: 10.1523/JNEUROSCI.22-21-09368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson LA, Schupbach T. EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol. 1999;44:203–243. doi: 10.1016/s0070-2153(08)60471-8. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Inoue Y, Hayashi S. A wave of EGFR signaling determines cell alignment and intercalation in the Drosophila tracheal placode. Development. 2007;134:4273–4282. doi: 10.1242/dev.010397. [DOI] [PubMed] [Google Scholar]
- O’Keefe DD, Gonzalez-Niño E, Edgar BA, Curtiss J. Discontinuities in Rap1 activity determine epithelial cell morphology within the developing wing of Drosophila. Dev Biol. 2012;15:223–234. doi: 10.1016/j.ydbio.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Parmentier ML, Woods D, Greig S, Phan PG, Radovic A, Bryant P, O’Kane CJ. Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J Neurosci. 2000;20:RC84. doi: 10.1523/JNEUROSCI.20-14-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberston F, Pinal N, Fichelson P, Pichaud F. Atonal and EGFR signalling orchestrate rok- and Drak-dependent adherens junction remodelling during ommatidia morphogenesis. Development. 2012;139:3432–3431. doi: 10.1242/dev.080762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A, Field C, Sisson JC, Sullivan W, Karess R. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol Biol Cell. 2004;15:838–850. doi: 10.1091/mbc.E03-06-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salbreux G, Barthel LK, Raymond PA, Lubensky DK. Coupling mechanical deformations and planar cell polarity to create regular patterns in the zebrafish retina. PLoS Comput Biol. 2012;8:e1002618. doi: 10.1371/journal.pcbi.1002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Martinez E, Soplop NH, Kramer SG. Lateral positioning at the dorsal midline: Slit and Roundabout receptors guide Drosophila heart cell migration. Proc Natl Acad Sci USA. 2006;103:12441–12446. doi: 10.1073/pnas.0605284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A, Denholm B, Bunt S, Bischoff M, VijayRaghavan K, Skaer H. Epidermal growth factor signalling controls myosin II planar polarity to orchestrate convergent extension movements during Drosophila tubulogenesis. PLoS Biol. 2014;12:e1002013. doi: 10.1371/journal.pbio.1002013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted theraphy. Clin Cancer Res. 2006;12:5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Shevchenko A, Knoblich JA. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Scholz H, Deatrick J, Klaes A, Klambt C. Genetic dissection of pointed, a Drosophila gene encoding two ETS-related proteins. Genetics. 1993;135:455–468. doi: 10.1093/genetics/135.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R, Shaharabany M, Seger R, Shilo BZ. Secreted spitz triggers the DER signalling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev. 1995;9:1518–1529. doi: 10.1101/gad.9.12.1518. [DOI] [PubMed] [Google Scholar]
- Shilo BZ. Signaling by the Drosophila epidermal growth factor receptor pathway during development. Exp Cell Res. 2003;284:140–149. doi: 10.1016/s0014-4827(02)00094-0. [DOI] [PubMed] [Google Scholar]
- Shilo BZ. The regulation and functions of MAPK pathways in Drosophila. Methods. 2014;68:151–159. doi: 10.1016/j.ymeth.2014.01.020. [DOI] [PubMed] [Google Scholar]
- Simoes SM, Blankenship JT, Weitz O, Farrell DL, Tamada M, Fernandez-Gonzalez R, Zallen JA. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev Cell. 2010;19:377–388. doi: 10.1016/j.devcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Abramova N, Charlton J, Adler PN. Van gogh. A new Drosophila tissue polarity gene. Genetics. 1998;150:199–210. doi: 10.1093/genetics/150.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery M, Jimenez-Dalmaroni A, Racine V, Bornens M, Julicher F. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–496. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- Thomas JB, Crews ST, Goodman CS. Molecular genetics of the single-minded locus: a gene involved in the development of the Drosophila nervous system. Cell. 1988;52:133–141. doi: 10.1016/0092-8674(88)90537-5. [DOI] [PubMed] [Google Scholar]
- Togashi H, Kominami K, Waseda M, Komura H, Miyoshi J, Takeichi M, Takai Y. Nectins establish a checkerboard-like cellular pattern in the auditory epithelium. Science. 2011;26:1144–1147. doi: 10.1126/science.1208467. [DOI] [PubMed] [Google Scholar]
- Veraksa A, McGinnis N, Li X, Mohler J, McGinnis W. Cap ‘n’ collar B cooperates with a small Maf subunit to specify pharyngeal development and suppress deformed homeotic function in the Drosophila head. Development. 2000;127:4023–4037. doi: 10.1242/dev.127.18.4023. [DOI] [PubMed] [Google Scholar]
- Vogel SK, Raabe I, Dereli A, Maghelli N, Toic-Norrelykke I. Interphase microtubules determine the initial alignment of the mitotic spindle. Curr Biol. 2007;17:438–444. doi: 10.1016/j.cub.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Wieduwilt MJ, Moasser MM. The epidermal growth factor receptor family: Biology driving targeted therapeutics. Cell Mol Life Sci. 2008;65:1566–1584. doi: 10.1007/s00018-008-7440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamadori T, Ishii Y, Homma S, Morishima Y, Kurishima K, Itoh K, Yamamoto M, Minami Y, Noguchi M, Hizawa N. Molecular mechanisms for the regulation of Nrf2-mediated cell proliferation in non-small-cell lung cancers. Oncogene. 2012;31:4768–4777. doi: 10.1038/onc.2011.628. [DOI] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- Zallen JA, Wieschaus E. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev Cell. 2004;6:343–355. doi: 10.1016/s1534-5807(04)00060-7. [DOI] [PubMed] [Google Scholar]
- Zallen JA, Zallen R. Cell-pattern disordering during convergent extension in Drosophila. J Physics: Condensed Matter. 2004;16:S5073–S5080. doi: 10.1088/0953-8984/16/44/005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.